It has been demonstrated that porcine circovirus 2 (PCV2) attaches to cells via heparan sulfate (HS) and chondroitin sulfate B (CSB) glycosaminoglycans; however, the underlying structural mechanism describing the HS/CSB recognition by PCV2 remains to be explored. We used cryo-electron microscopy with single-particle analysis, symmetry expansion, and focused classification to visualize the interaction between the PCV2 capsid and heparin, an analog of heparan sulfate, to better than 3.6-Å resolution. We observed that the interaction between PCV2 and heparin does not adhere to the icosahedral symmetry of the capsid. To the best of our knowledge, this is the first example where the interaction between heparin and an icosahedral capsid does not follow the symmetry elements of the capsid. Our findings also suggest that anionic polymers, such as dextran sulfate, may act to inhibit PCV2 infection.
KEYWORDS: circovirus, cellular attachment, chondroitin sulfate B, electron microscopy, focused classification, glycosaminoglycan, heparan sulfate, heparin, symmetry expansion
ABSTRACT
Porcine circovirus 2 (PCV2) is the smallest pathogenic virus capable of autonomous replication within its host. Infections result in immunosuppression and subsequent death of the host and are initiated via the attachment of the PCV2 icosahedral capsid to heparan sulfate (HS) and chondroitin sulfate B (CSB) glycosaminoglycans on the cell surface. However, the underlying mechanism of structural recognition remains to be explored. Using heparin, a routinely used analog of heparan sulfate, we demonstrate that increasing lengths of heparin exhibit a greater affinity toward PCV2. Our competition assays indicate that dextran sulfate (8 kDa) has a higher affinity for PCV2 than heparin (12 kDa), chondroitin sulfate B (41 kDa), hyaluronic acid (1.6 MDa), and dextran (6 kDa). This suggests that polymers high in sulfate content are capable of competing with the PCV2-heparan sulfate interaction and, thus, have the potential to inhibit PCV2 infection. Finally, we visualized the interaction between heparin and the PCV2 capsid using cryo-electron microscopy single-particle analysis, symmetry expansion, and focused classification. The image reconstructions provide the first example of an asymmetric distribution of heparin on the surface of an icosahedral virus capsid. We demonstrate that each of the 60 capsid subunits that generate the T=1 capsid can bind heparin via one of five binding sites. However, not all of the binding sites were occupied by heparin, and only one-third to two-thirds of the binding sites were occupied. The binding sites are defined by arginine, lysine, and polar amino acids. Mutating the arginine, lysine, and polar amino acids to alanine diminished the binding capacity of PCV2 to heparin.
IMPORTANCE It has been demonstrated that porcine circovirus 2 (PCV2) attaches to cells via heparan sulfate (HS) and chondroitin sulfate B (CSB) glycosaminoglycans; however, the underlying structural mechanism describing the HS/CSB recognition by PCV2 remains to be explored. We used cryo-electron microscopy with single-particle analysis, symmetry expansion, and focused classification to visualize the interaction between the PCV2 capsid and heparin, an analog of heparan sulfate, to better than 3.6-Å resolution. We observed that the interaction between PCV2 and heparin does not adhere to the icosahedral symmetry of the capsid. To the best of our knowledge, this is the first example where the interaction between heparin and an icosahedral capsid does not follow the symmetry elements of the capsid. Our findings also suggest that anionic polymers, such as dextran sulfate, may act to inhibit PCV2 infection.
INTRODUCTION
Porcine circovirus (PCV) belongs to the Circovirus genus of the Circoviridae family (1, 2). Members of this family are nonenveloped viruses and possess a circular single-stranded DNA (ssDNA) genome. Circoviruses are widely distributed in nature and infect terrestrial, avian, and aquatic members of the animal kingdom (3, 4). Three genotypes of PCV have been identified: PCV1, PCV2, and PCV3. PCV1 (1,759 nucleotides [nt]) was first detected in porcine kidney (PK-15) cell lines and later found to be a nonpathogenic virus (5, 6). PCV2 (1,767 nt to 1,768 nt) is morphologically similar to but genetically and antigenically distinct from PCV1 and was isolated from pigs with postweaning multisystemic wasting syndrome (PMWS) (3, 7–10). PMWS, later named porcine circovirus-associated disease (PCVAD) or porcine circovirus disease (PCVD), culminates in the immunosuppression of the host and death from secondary infection (11–14). Autopsy of infected pigs identifies PCV2 in nearly every tissue, indicating that it has a broad tissue tropism (1, 2, 11, 15). The promiscuous nature of PCV2 is further exhibited by its ability to infect and induce its pathogenic phenotype in rodents and bovines living in the vicinity of infected farms and BALB/c mice and human cells in the laboratory (3, 4, 16–19). PCV3 (2,000 nt) was recently identified and shown to be associated with porcine dermatitis, reproductive failure, and nephropathy syndrome (5, 6, 20).
PCV2 is the smallest pathogenic virus capable of replicating in cells without the need for additional viruses (3, 7–10, 14). Its ∼1.7-kilonucleotide ambisense genome encodes a replicase (ORF1) responsible for the rolling circle replication of the genome, a capsid protein (ORF2) responsible for forming the capsid and enclosing the genome, and ORF3 and ORF4, which may be responsible for causing cellular apoptosis and the pathogenic nature of PCV2 (5, 11–14, 21–24). PCV2 has been shown to initiate cellular infection via attachment to the glycosaminoglycans (GAGs) heparan sulfate (HS) and chondroitin sulfate B (CSB) (25). HS and CSB are ubiquitously expressed on mammalian cells and act as attachment factors for a variety of macromolecules, such as proteases, chemokines, receptors, and pathogens (26, 27). Heparan sulfate is a 30- to 70-kDa linear polysaccharide (40 to 300 sugar residues and approximately 20 to 150 nm long) composed of alternating sulfated (NS) domains and unsulfated (NA) domains (26, 28). The NS domains are composed of three to eight repeating disaccharides of l-iduronic acid (IdoA) and d-glucosamine (GlcN) (see Fig. S1 in the supplemental material). An NS disaccharide can possess two to three sulfates. The NA domains are composed of 2 to 12 repeating disaccharides of N-acetyl-d-glucosamine (GlcNAc) and d-glucuronic acid (GlcA) (Fig. S1) (29). Thus, a single chain of HS is composed of multiple NS and NA domains. The structure of the NS domain is similar to the heparin structure, and heparin is routinely used as a reagent to study the interaction between the NS domains and their interacting partners (28). Heparin is not a cellular attachment factor but is stored in the granules of mast cells and released into the vascular system as an anti-inflammatory agent (30). CSB, also a linear polysaccharide, is composed of the repeating disaccharide N-acetylgalactosamine (GalNAc) and IdoA. A CSB disaccharide has one to two sulfates (27).
PCV2 has been shown to internalize via two distinct pathways: clathrin-mediated endocytosis in monocytic (31) and dendritic (1) cells and caveola-, clathrin-, and dynamin-independent small GTPase-regulated pathways in epithelial cell lines of porcine kidney (PK-15), swine kidney (SK), and swine testicles (ST) (3). The PCV2 nucleocapsid has been shown to gain cellular entry by escaping the acidified endosome-lysosome of 3D4/31 cells (31). A bipartite nuclear localization signal (NLS) at the N terminus of the capsid protein has been implicated in the guiding of the nucleocapsid into the nucleus for genome replication (32). The PCV2 replicase (ORF1) recruits the cellular machinery to initiate rolling circle replication of the PCV2 genome in the nucleus (33). The newly synthesized PCV2 capsid protein is transported into the nucleus, via its NLS, for genome encapsidation and assembly of infectious virions. The virions then egress from the cell to initiate another cycle of infection (3, 8, 10).
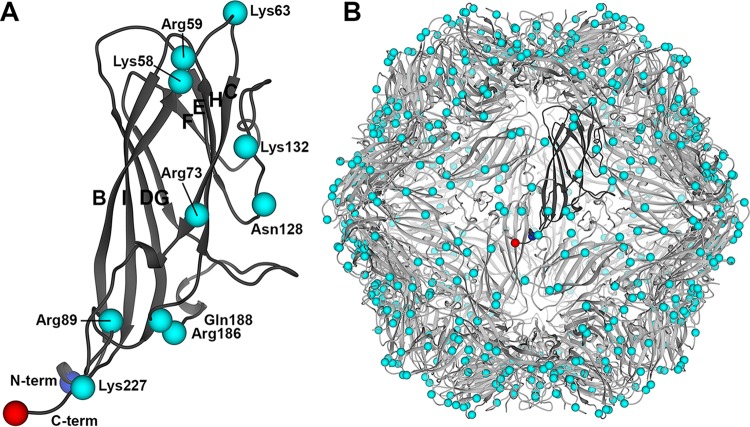
The PCV capsids are 20 nm in diameter, possess icosahedral (I1) symmetry, and are composed of 60 copies (T=1) of a capsid protein (34, 35). From here forth we use the terminology “capsid subunit” to refer to the capsid protein in the context of the assembled capsid. The crystal structure of the PCV2 virus-like particle (VLP) visualized the capsid subunit fold to be that of the canonical viral jelly roll, first observed for the capsid subunit of tobacco bushy stunt virus (35, 36). The subunit can be described as a β-sandwich, or β-barrel, with a right-hand twist (Fig. 1A). The β-sandwich is composed of two β-sheets. One β-sheet is made of the four antiparallel β-strands BIDG, and the second β-sheet is composed of the four antiparallel β-strands CHEF (Fig. 1A). The interface between the two β-sheets is the hydrophobic core of the protein. Loops connecting the strands generate the surface topography of the VLP. Strands BC, DE, FG, and HI are short and surround the 5-fold vertex of the VLP (Fig. 1B). Loops connecting strands CD, EF, and GH are longer. Loops C and D from neighboring subunits contact one another across the icosahedral 2-fold axes of symmetry, and loops G and H from neighboring subunits contact one another across the icosahedral 3-fold axes of symmetry (Fig. 1B).
FIG 1.
Cartoon depiction of the PCV2 subunit and VLP atomic coordinates. (A) Ribbon diagram of the capsid subunit. The β-strands (BCDEFGHI) are labeled according to the sequence in which they occur. The N and C termini are shown as blue and red spheres, respectively. The C-α atoms for basic and polar residues identified as heparin binding residues in this study are depicted as cyan spheres. (B) Ribbon diagram of the VLP showing the molecular organization of the subunits. A single subunit is shown in dark gray. Images were produced with the UCSF Chimera program (65).
Because the structural mechanism of cellular recognition is not understood, we used a combination of cryo-electron microcopy (cryo-EM), biochemistry, and site-directed mutagenesis to describe this interaction. We used heparin as an analog for studying the interaction between heparan sulfate and PCV2. We asked, does the length of heparin affect its affinity for PCV2, what chemical moieties of GAGs interact with PCV2, where are the heparin binding sites on PCV2, does the binding of heparin follow the icosahedral symmetry of the capsid, and does PCV2 undergo a conformational change upon binding of heparin? Our biochemical results demonstrate that longer oligosaccharides of heparin bind with a greater affinity to the capsid and that the interaction is predominantly driven by the sulfates of the polysaccharide. Our structural results visualize heparin to bind one of five binding sites per capsid subunit and the interaction to not adhere to the capsid’s icosahedral symmetry. Finally, binding of heparin does not induce the C-α backbone to undergo a conformational change. To our knowledge, this is the first example of a nonsymmetric distribution of heparin on an icosahedral virus. Furthermore, our studies provide the first structural study of a member of the Circovirus genus interacting with the cellular attachment factor of a cell to initiate infection. The knowledge gained in this study can pave the path for developing molecules to interfere with this interaction and inhibit PCV2 infection.
(This article was submitted to an online preprint archive [37].)
RESULTS
The interaction between PCV2 and heparin is reversible, dependent on the size of heparin, and primarily dictated by sulfates.
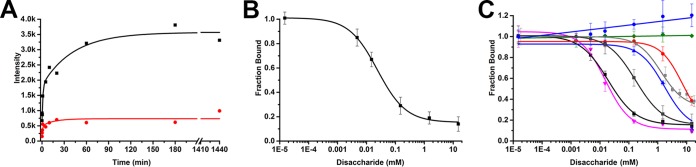
To study the interaction between heparan sulfate and PCV2, we used an in vitro binding assay that involves interacting PCV2 virus-like particles (VLPs) with chromatography sorbent conjugated to 15-kDa porcine intestinal mucosa heparin. Heparin is routinely used as an analog of heparan sulfate (HS) for studying the interaction between macromolecules and HS (26, 29). This is because the structure of heparin is similar to that of the NS domain of HS (see Fig. S1 in the supplemental material). We first determined the concentration of baculovirus-expressed PCV2 VLPs (GenBank accession number ACA51584.1) and the time necessary to interact with the sorbent to achieve a robust readout (Fig. 2A). Two concentrations of VLPs were used (370 nM and 92 nM). Maximum binding occurred within 30 min for the lower concentration of VLPs and within 3 h for the higher concentration of VLPs. We chose to use the higher concentration of VLPs (370 nM) for our subsequent studies because of the improved signal in our binding assays (Fig. S2A). We then examined if the interaction was reversible by incubating the PCV2-bound sorbent with increasing concentrations of 12-kDa porcine intestinal mucosa heparin (Fig. 2B and S2B). Elution of PCV2 from the sorbent demonstrated that the interaction is reversible.
FIG 2.
Interaction between PCV2 and heparin-conjugated chromatography sorbent. (A) Binding kinetics between PCV2 VLPs and the chromatography sorbent with 370 nM (black) and 92 nM (red) concentrations of VLP. (B) Elution of PCV2 from the chromatography sorbent in the presence of increasing concentration of heparin. (C) Competition assays testing the ability of various GAGs and polysaccharides to displace PCV2 from the chromatography sorbent. Red, dp6 heparin (6-hexose); blue, dp10 heparin (10-hexose); dark gray, dp20 heparin (20-hexose); black, dp36 heparin (36-hexose); magenta, 8-kDa dextran sulfate; brown, 6-kDa dextran sulfate; light gray, 41-kDa chondroitin sulfate B; green, hyaluronic acid. Error bars represent the standard deviation for three measurements. Images were made with Origin 2016 software (OriginLab).
To determine the chemical moieties of GAGs that PCV2 recognizes for cellular attachment, we employed a competition assay to identify the effectiveness of different ligands to compete with the above-mentioned chromatography sorbent for binding PCV2 (Fig. 2C). We examined the length of heparin, the pyranose structure, sulfate content, and branched polysaccharide chains. The results demonstrate that while the efficiency with which the degree of polymerization (dp) 6 (6-hexose) (Fig. 2C and S2C), dp10 (10-hexose) (Fig. 2C and S2D), and dp20 (20-hexose) (Fig. 2C and S2E) heparins inhibit 15-kDa heparin from binding to PCV2 increases, they are not nearly as effective as dp36 (36-hexoses, 12-kDa) heparin (Fig. 2C and S2F). A plausible explanation could be that there are multiple weak binding sites on the PCV2 capsid and that longer oligosaccharides have a higher affinity to PCV2 because they can bind to multiple sites concurrently. It has been shown that heparin fragments below dp18 are helical, extended, and rigid but become more bent with increasing size (38). Thus, the dp6 and dp10 heparins may bind to a few binding sites on a PCV2 facet, while the dp20 and dp36 heparins may bend and bind to multiple facets on PCV2.
To address if a polysaccharide with a high sulfate content can interact with PCV2, we examined the ability of dextran sulfate (8 kDa) to inhibit the interaction between PCV2 and the chromatography sorbent (Fig. S2G). The dextran sulfate (8 kDa) used in our studies is a highly branched polysaccharide with glucose repeating units connected via 1 → 4 linkages and branching points at 1 → 6. There are 2.8 sulfates per glucose monomer for this product (personal communication with the manufacturer, Alfa Aesar). Dextran sulfate is the ligand most efficient at inhibiting the interaction between PCV2 and 15-kDa heparin (Fig. 2C). However, dextran, which has the same branched structure as dextran sulfate, appears to slightly enhance the affinity of the VLP for the sorbent (Fig. S2H). This may be due to dextran acting as a crowding agent (39). The ability of dextran sulfate to inhibit the PCV2-heparin interaction while dextran slightly enhances the interaction suggests that the sulfates are sufficient for binding to PCV2 and that the backbone polysaccharide common to dextran and dextran sulfate does not appear to interact with PCV2.
To address if longer GAGs with a lower sulfate content exhibit a reduced affinity for PCV2, we examined the ability of the 1.6-MDa hyaluronic acid (HA) and 41-kDa chondroitin sulfate B (CSB) glycosaminoglycans to inhibit the interaction between PCV2 and 15-kDa heparin. HA demonstrates no affinity for PCV2 (Fig. 2C and S2I). HA possesses a single carboxylate on each of its repeating disaccharide units. The hydrogen bonds formed within and between its repeating disaccharides produce stiff helical polysaccharide chains that entangle at very low concentrations (40). Thus, the carboxylate, stiff, and entangled properties of HA may not provide the appropriate interface for PCV2 to bind. The CSB used in our studies is longer and more flexible (41) than the 12-kDa heparin that we used; however, CSB demonstrates a much lower affinity for PCV2 than heparin. This may be because of the lower sulfate content of CSB (an average of 1.1 sulfates per repeating disaccharide) than heparin (an average of 2.4 sulfates per repeating disaccharide) (Fig. 2C and S2J).
Collectively, our data indicate that the PCV2-heparin interaction is reversible, that the strength of the interaction increases with longer lengths of heparin, that oligosaccharides high in sulfate content bind with a greater affinity, that long oligosaccharides with a low sulfate content bind weakly, and that the interaction between PCV2 and the polysaccharide is predominantly driven by sulfates on the polysaccharide.
A capsid subunit can bind heparin via multiple sites.
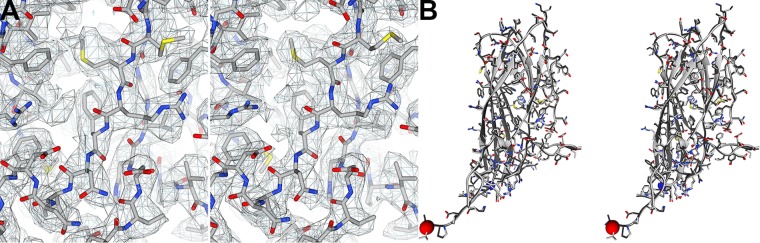
To identify the binding sites of heparin on PCV2, we determined the cryo-EM image reconstruction of PCV2 in the absence and presence of 12-kDa (dp36) porcine intestinal mucosa heparin at pH 7. The purpose of the unliganded image reconstruction was to identify if a conformational change occurred in the subunit upon binding to heparin. We generated icosahedral image reconstruction of the unliganded and heparin-liganded PCV2 to 3.3-Å and 2.8-Å resolutions, respectively. The image reconstructions were of suitable quality to allow us to confidently model the atomic coordinates of PCV2 (Table 1; Fig. 3A). The refined atomic models for the two image reconstructions overlay with a C-α root mean square deviation (RMSD) of 0.27 Å and an all-atom RMSD of 0.98 Å (42). These differences are small and can be a result of modeling error at these resolutions (Fig. 3B) (43, 44). The symmetrized heparin-liganded image reconstruction does not allow us to confidently identify the molecular envelopes for heparin.
TABLE 1.
Statistics for cryo-EM image reconstructions and refined atomic coordinates
| Parameter | Value(s) for: |
|
|---|---|---|
| Unliganded PCV2 | PCV2 + heparin | |
| Image reconstruction | ||
| No. of micrographs | 1,149 | 3,725 |
| Defocus range (μm) | −0.28 to −3.2 | −0.25 to −3.6 |
| Total dose (e− Å−2) | 35 | 32 |
| Dose rate (e− Å−2 s−1) | 7.0 | 6.4 |
| No. of frames | 50 | 25 |
| No. of particles extracted | 54,409 | 166,369 |
| No. of particles used for reconstruction | 28,938 | 93,725 |
| FSC resolution (Å)a | 3.3 | 2.8 |
| B-factor sharpening (Å2)b | −152.2 | −122.92 |
| Atomic modeling | ||
| Correlation coefficient (CC) (%) | 82 | 83 |
| B-factor average (Å2) | 35.5 | 38.1 |
| RMSDc | ||
| Bonds | 0.012 | 0.016 |
| Angles | 1.278 | 1.271 |
| Molprobityd | 1.76 | 1.62 |
| EMRinger | 4.89 | 5.02 |
| Ramachandran plot (%) | ||
| Favored | 76 | 87 |
| Allowed | 24 | 13 |
| EMDB accession no. | 8939 | 8973, 8975, 8974, 8969, 8970, 8971, 8972 |
| PDB accession no. | 6DZU | 6E32, 6E39, 6E34, 6E2R, 6E2X, 6E2Z, 6E30 |
FSC, the Fourier shell correlation, reported by Relion (version 2.0) using the gold standard method at a CC of 0.143.
The B-factor sharpening reported by Relion (version 2.0) during the postrefinement process.
The root mean standard deviation for bonds and angles reported by the phenix.real_space_refine tool.
Molprobity overall score.
FIG 3.
Symmetrized cryo-EM image reconstructions of unliganded and heparin-liganded PCV2. (A) Full-atom models of unliganded (left) and heparin-liganded (right) PCV2 subunits shown as stick models. Overlaid onto the stick models are mesh representations of the corresponding 3.3-Å (left) and 2.8-Å (right) symmetrized image reconstructions. The unliganded image reconstruction is rendered at 1.5 sigma, and the liganded image reconstruction is rendered at 2.6 sigma. The rendering thresholds correspond to 100% mass content (see the Materials and Methods section). (B) Cross-eyed stereo representation comparing the atomic models derived from the unliganded (light gray) and liganded (dark gray) image reconstructions. The backbone atoms are depicted as a ribbon diagram, and the side chains are shown as stick models. Blue and red spheres represent the N and C termini, respectively. Images were generated with the UCSF Chimera program (65).
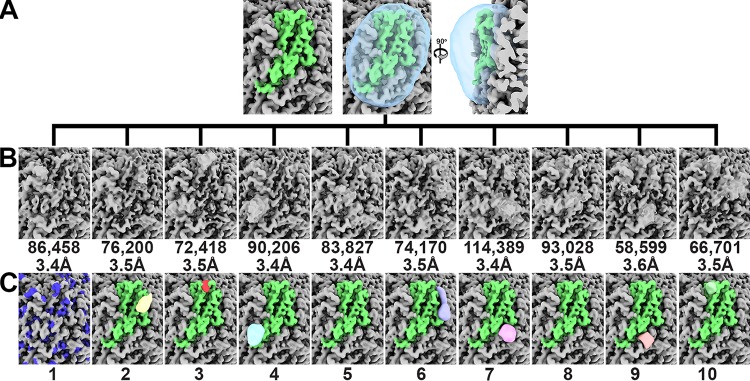
We postulated that if each subunit possessed multiple binding sites for heparin but only one site could be occupied per subunit, then each capsid might be composed of a unique combination of subunits liganded to heparin; thus, an icosahedral averaging strategy could eliminate the molecular envelope of heparin. To test this possibility, we employed the symmetry expand capability of the Relion program on the alignment parameters from the image reconstruction where icosahedral (I1) symmetry was imposed (45). Briefly, the symmetry expansion protocol generates a copy of each particle for each symmetry operator used during the symmetrized image reconstruction. In our case, 60 equivalent particles were generated for each imaged VLP because 60 matrices describe the I1 symmetry used by Relion. The orientation parameters of these particles were determined by modifying the orientation parameters of the parent particle by the 60 symmetry operators that describe the I1 symmetry. Consequently, a single imaged VLP that was assigned an orientation parameter for I1 symmetry was replaced with 60 copies of the VLP assigned orientation parameters for asymmetry (C1). These 60 orientation parameters were related to one another via I1 symmetry. The consequence of this strategy is to align the projections of all the subunits from the VLP to one another (46). We then used the Frealign program to perform focused classification of a subunit (47, 48). Seven of the 10 classes exhibited molecular envelopes distinct from the symmetrized image reconstruction (Fig. 4). To generate a difference map for each class average, we subtracted from each class average an image reconstruction generated using the same number of particles as the class average. These particles were randomly selected from the symmetry-expanded data set. Calculation of the local resolution for each of the 10 classes with the MonoRes program suggested that the resolution of the peaks varies from 7 Å to 15 Å (Fig. S3) (49). The difference peaks were low-pass filtered to 7-Å resolution and rendered at 27 sigma (Fig. 4C). We used 27 sigma as the criterion to remove small and spurious density.
FIG 4.
Focused classification of the PCV2 VLPs liganded to heparin image reconstruction. Symmetry expansion of 13,600 particles generated 816,000 capsid subunits for analysis. The image reconstructions are shown at 2.6 sigma. (A) The molecular envelope for a subunit is shown in green. The mask used for the classification is shown as a transparent cyan blob. (B) Ten classes were requested. The number of particles and the resolution reported by the Frealign program are provided below each class. (C) Difference peaks for each class average shown in panel B. There was no difference peak for class 1. The blue patches represent the molecular envelope of the Arg/Lys side chains. The class numbers referred to in the text are presented at the bottom. Images were made with the UCSF ChimeraX program (70).
The difference peaks may correspond to segments of the 12-kDa heparin molecule incubated with PCV2. We note that the amorphous shapes of these peaks indicate that heparin may use multiple binding modes/poses to bind to the capsid. Indeed, heparin is known to use multiple binding modes/poses to interact with proteins (28). We modeled segments of heparin as rigid bodies into the difference peaks to see if the models were chemically reasonable. We then examined if mutation of the basic and polar amino acids that interact with the sulfates in our models diminished the mutant VLPs’ affinity for heparin. The VLPs for these studies were generated using an in vitro assembly protocol of capsid protein expressed in Escherichia coli (see Materials and Methods).
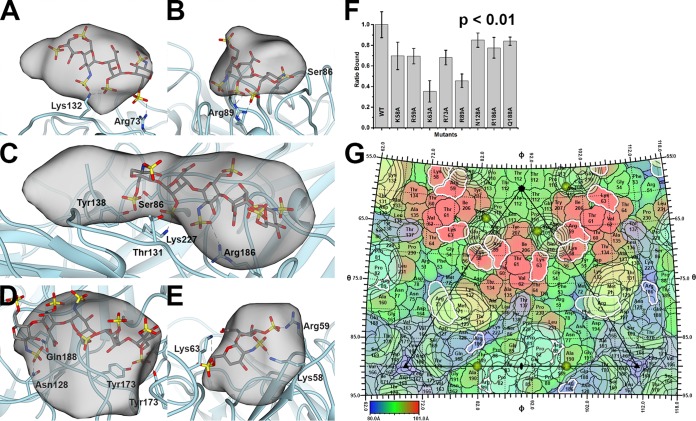
Class 1 (11% of subunits) did not possess a difference peak and thus represents an unliganded subunit. The difference peak for class 2 (9% of subunits) was located near the Arg73 and Lys132 of a subunit. A four-hexose segment of heparin could be modeled into this difference peak (Fig. 5A). Mutation of Arg73 to Ala diminished the capsid-heparin affinity to 68% of that of the wild type, whereas mutation of Lys132 to Ala abolished capsid assembly (Fig. 5F). The difference peak for class 3 (9% of subunits) was located near Lys58 and Arg59 but was too small to accommodate heparin. The difference peak for class 4 (11% of subunits) was near Arg89 and next to the icosahedral 2-fold axis of symmetry. A three-hexose segment of heparin could be modeled into this density (Fig. 5B). Mutation of Arg89 to Ala diminished the capsid-heparin affinity to 46% of that of the wild type (Fig. 5F). The difference peak for class 5 (10% of subunits) was extremely weak. The difference peak for class 6 (9% of subunits) ran between adjacent facets. It was located near Arg186 and Lys227 of the adjacent subunit. A five-hexose segment of heparin could be modeled into this difference peak. A number of hydrogen bonds between the sulfates of heparin and the side chains of Thr131, Tyr138, and Ser86 were also possible (Fig. 5C). Mutation of Arg186 to Ala diminished the capsid-heparin affinity to 78% of that of the wild type, whereas mutation of Lys227 to Ala abolished capsid assembly (Fig. 5F). The difference peak for class 7 (14% of subunits) was distal to Arg/Lys, but a five-hexose segment of heparin modeled into the difference peak suggested that hydrogen bonds could form between the sulfates of heparin and the side chains of Asn128, Gln188, and the three Tyr173 at the icosahedral 3-fold axis (Fig. 5D). Mutation of Asn128 or Gln188 to Ala diminished the capsid-heparin affinity to 85% and 84% of that of the wild type, respectively (Fig. 5F). The difference peak for class 8 (12% of subunits) was also very weak. The difference peak for class 9 (7% of subunits) was located near Arg73 and Lys132 of the neighboring subunit and was equivalent to the peak of class 2. This class was identified by our procedure because the mask used for classification includes regions of neighboring subunits that are related to one another via the icosahedral symmetry of the capsid. Given that this is a redundant class, we excluded it from further analysis. The difference peak for class 10 (8% of subunits) was located near the icosahedral 5-fold axis and surrounded by Lys58 and Arg59 of one subunit and Lys63 of a neighboring subunit. A sulfate was observed at this location in the crystal structure of PCV2 (35). A two-hexose segment of heparin could be modeled into this density (Fig. 5E). Mutation of either Lys58 or Arg59 to Ala diminished the PCV2-heparin interaction to 69%, while mutation of Lys63 diminished the affinity to 38% (Fig. 5F).
FIG 5.
PCV2-heparin interaction. The PCV2 capsid is shown as a cyan ribbon cartoon. The difference peak (at 27 sigma) is shown as a gray transparent blob low-pass filtered to 7 Å. A modeled heparin is shown as a gray stick model. The dashed lines represent hydrogen bonds and/or salt bridges to the sulfates (red and yellow sticks) of heparin. (A) Class 2 binding site. (B) Class 4 binding site. (C) Class 6 binding site. (D) Class 7 binding site. (E) Class 10 binding site. Images were made with the UCSF Chimera program (65). (F) Bar graph representing the efficiency of 370 nM mutant PCV2 VLPs for binding to heparin, with the value for wild-type (WT) VLPs having been normalized to 1. The error bars represent the errors from five independent measurements. The image was made with the Origin 2016 program (OriginLab). The P value was calculated using the ranksum (Wilcoxon rank-sum test) routine of MATLAB 2018a (MathWorks). (G) The surface amino acids of PCV2 are plotted on a stereographic projection and colored from blue (80 Å) to red (101 Å) according to the distance from the center of the particle (see the color bar). Overlaid are contour plots at 27 sigma (black outline) for the six difference peaks. The plots are colored as described in the legend to Fig. 4. Residues with a white outline were mutated to Ala in this study, assembled into capsids, and tested for their ability to bind to heparin. Note that Lys137 and Lys227 are not colored because they did not assemble into capsids and a binding assay could not be performed. The sulfates observed in the crystal structure of PCV2 are shown as gold spheres (35). The icosahedral 5-fold, 3-fold, and 2-fold symmetry elements are depicted as a black pentagon, triangle, and ellipse, respectively. The figure was generated with the RIVEM program (50).
To compare the difference peaks on the surface of the PCV2 capsid, we generated a stereographic projection of the PCV2 surface with the program RIVEM (Fig. 5G) (50). Overlaid onto this image were contour plots of the symmetrized difference peaks, the sulfates observed in the PCV2 VLP crystal structure, and the 532 icosahedral symmetry elements (35). The difference peaks were symmetrized to visualize all the binding sites available on the surface of the capsid. The sulfates observed in the crystal structure were located near the difference peaks from classes 4 and 10. The stereographic projection demonstrated that the difference peaks from classes 6 and 7 partially overlap and share a number of amino acids that include Asn128 and Gln188.
The resolution of the image reconstructions from each class was of sufficient quality to refine the atomic models. Refining the atomic model generated from the image reconstruction with imposed icosahedral symmetry into the subunit from each of the seven classes did not identify significant movements in the main-chain atoms that could be associated with a conformational change upon binding to heparin. The largest C-α RMSD between subunits from two classes was 0.313 Å.
The occupied heparin binding sites do not follow the icosahedral symmetry of the capsid.
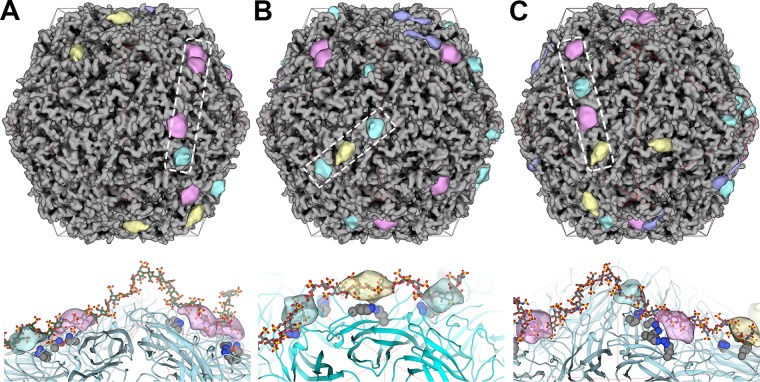
The result of focused classification was to assign each of the 60 subunits belonging to an imaged VLP to 1 of 10 classes. We used this information along with the inverse of the Relion I1 matrices to position the difference peaks onto the originating VLPs (Fig. 6). We refer to these models as composite VLPs, as they are composed of a symmetrized PCV2 image reconstruction and the positioned difference peaks. The number of difference peaks present per VLP followed a Gaussian distribution, with the VLPs possessing a minimum of 18 peaks (Fig. 6A and S5), an average of 31 peaks (Fig. 6B and S5), and maximum of 43 peaks (Fig. 6C and S5). The dp36 heparin used to generate these data prompted us to look for occupied binding sites that could accommodate its physicochemical properties (Fig. 6). Indeed, we could manually model the dp24 and dp36 heparins (PDB accession numbers 3IRJ and 3IRL, respectively) as rigid bodies into these composite VLPs (Fig. 6A to C). The bend in the dp36 heparin allows it to bind binding sites distributed across two adjacent facets of the capsid.
FIG 6.
Composite VLPs, where each model represents an imaged VLP-heparin complex. The coloring scheme used for the difference peaks is identical to that described in the legend to Fig. 4. (Top) The dashed box outlines the occupied binding sites that a heparin oligosaccharide is modeled into (bottom). (Bottom) A heparin oligosaccharide (gray stick model) occupying the difference peaks (transparent blobs) identified by the dashed box (top). The binding site residues are shown as gray space-filling models, and the PCV2 capsid is shown as a cyan ribbon model. A red icosahedral cage distinguishes the facets. (A) A VLP with 18 binding sites occupied by heparin. A dp36 heparin has been modeled into the outlined sites. (B) A VLP with 31 binding sites occupied by heparin. A dp24 heparin has been modeled into the outlined sites. (C) A VLP with 40 binding sites occupied by heparin. A dp36 heparin has been modeled into the outlined sites. Images were made with the UCSF Chimera program (65).
DISCUSSION
The cellular attachment factors responsible for initiating infection of porcine monocytic 3D4/31 and porcine kidney epithelial PK-15 cells by PCV2 were demonstrated to be heparan sulfate (HS) and chondroitin sulfate B (CSB), using competition assays, enzymatic removal of HS and CSB, and mutant CHO cells deficient in HS (pgsD-677) or HS and CSB (pgsA-745) (25). Both HS and CSB are long linear polysaccharides known as glycosaminoglycans (GAGs). HS is composed of two alternating domains: a variably sulfated and rigid NS domain and a nonsulfated and flexible NA domain (26, 29, 51). Three to 8 repeating disaccharides generate the NS domain, and 2 to 12 repeating disaccharides generate the NA domain. The NS domain has a chemical structure similar to that of heparin with a length of dp6 (6-hexoses) to dp16 (16-hexoses) (see Fig. S1 in the supplemental material). The NS domains bind to a plethora of ligands that include but that are not limited to cytokines, enzymes and their inhibitors, receptors, chemokines, and pathogens (26). A number of nonenveloped icosahedral viruses are known to use HS for cellular attachment. Examples include serotypes of adenovirus (52), serotypes of adeno-associated virus (53), foot-and-mouth disease virus (54), and human papillomavirus (55). Here we note that while heparin is routinely used as an analogue of HS (29), it is not a cellular attachment factor. Heparin is stored in the secretory granules of mast cells and released into the vascular system as an anti-inflammatory agent (30). An extensive review by Xu and Esko describes that a large portion of the binding energy between heparin and proteins can be attributed to the ionic interaction between the Arg/Lys amino acids of proteins and the sulfates of heparin (28). However, polar amino acids, such as Asn, Gln, and His, can also make hydrogen bonds with the sulfates of heparin and can contribute 20% to 70% of the binding energy. Analysis of structures deposited into the Protein Data Bank (PDB) indicates that heparin binding sites vary in size (200 Å2 to 360 Å2), the number of lysine and arginine residues (0 to 7), and the number of polar amino acids (0 to 5). Interestingly, the size of the interface and the number of basic residues in the binding site do not predict the affinity of the interaction. The protein-heparin binding strength also varies from 0.3 nM to 7 μM (Kd [dissociation constant]). The majority of the heparin binding amino acids are present in the loops of the proteins (28).
We have used an in vitro competition assay to demonstrate that PCV2 virus-like particles (VLPs) bind to increasing lengths of heparin fragments with a greater affinity. The binding trend suggests that the PCV2 VLPs may possess multiple weak binding sites such that longer fragments of heparin are able to bind additional sites for producing greater affinity, a phenomenon referred to as “avidity” (56). Our assays also demonstrate that PCV2 has a greater affinity toward dp36 heparin (12 kDa) than toward the much longer 41-kDa CSB. This is in agreement with the results of cellular competition assays that demonstrate that the incubation of PCV2 with heparin reduces infection greater than incubation of PCV2 with CSB (25). Moreover, infection of the Chinese hamster ovary (CHO) mutant cell line pgsA-677 (lacking HS) was shown to be reduced to ∼28% of that of wild-type cells, while infection of the mutant cell line pgsA-745 (lacking HS and CSB) was reduced to ∼10% of that of wild-type cells, suggesting that HS may be the preferred attachment factor on the surface of the cell.
Our in vitro competition assays also demonstrated that PCV2 VLPs exhibit a greater affinity toward sulfated polysaccharides. In particular, the observation that PCV2 VLPs bind to 8-kDa dextran sulfate (DS) but not to 6-kDa dextran suggests that PCV2 recognizes and interacts with the sulfates of the polysaccharide. Indeed, our competition experiments demonstrate that DS is the most potent of the competitors tested in our assays, suggesting that DS may be used as an inhibitor of PCV2 infection.
Our structural data identified six class averages where additional molecular envelopes modify the molecular envelope of the capsid subunits. Subtracting the molecular envelope of an unmodified capsid from these class averages generates six difference peaks. The difference peaks are large enough to accommodate heparin oligosaccharides. Heparin oligosaccharides can be modeled into the difference peaks such that their sulfates are in close proximity to amino acids Lys58, Arg59, Lys63, Arg73, Arg89, Asn128, Lys132, Arg186, Gln188, and Lys227. A single point substitution of these amino acids to Ala diminished the capsid’s affinity for heparin, which thus provides biochemical support for the suggestion that the difference peaks represent bound heparin. A single mutation is not sufficient to tremendously diminish the binding of heparin; therefore, we anticipate that multiple mutations may be necessary to eliminate the interaction. Comparison of the difference peaks suggests that the peaks belonging to two class averages are equivalent. Thus, heparin can bind to one of five binding sites per PCV2 capsid subunit, such that a PCV2 VLP can possess a maximum of 60 sites occupied by heparin. Given that each difference peak originates from a subunit at a specific position in a VLP, the difference peak can be mapped to that position in the VLP to generate a heparin-liganded molecular envelope for each imaged VLP. These molecular envelopes demonstrate that heparin asymmetrically decorates rather than symmetrically decorates the surface of the capsid, the latter of which has been demonstrated for a number of icosahedral viruses. This may be because prior structural studies of icosahedral viruses in complex with heparin utilized averaging techniques that may have eliminated the asymmetric distribution of heparin on the capsid surface (53, 54, 57–60). The packing of capsids into a crystal for X-ray crystallography inherently imposes the averaging of structures (61). Similarly, cryo-EM image reconstructions with imposed icosahedral symmetry average nonicosahedral entities to symmetric structures with reduced occupancy or amorphous blobs (e.g., the molecular envelope of viral genomes). However, the use of symmetry expansion and focused classification forgoes symmetry averaging to provide a more detailed analysis.
Our modeling suggests that the reported solution structures of dp24 and dp36 heparins (38) can bind to adjacent facets of the PCV2 capsid. This is primarily because of the bent structure of these oligosaccharides (38). Consequently, it is anticipated that shorter heparin lacking such a bend is unable to interact with adjacent facets. The longest NS domain of HS is reported to be eight disaccharides (dp16) (28) and, thus, unlikely to bend and bind to adjacent facets. This creates the likelihood that an NS domain binds to a single facet and the neighboring flexible NA domain allows the HS chain to bend for the subsequent NS domain to bind to an additional facet on the surface of PCV2. Heparan sulfate possesses 40 to 300 sugar molecules (20 to 150 nm long) and is composed of multiple alternating NS domains (dp6 to dp16) and NA domains (dp4 to dp24) (26, 28, 51).
In conclusion, we demonstrate that PCV2 capsids bind increasing lengths of heparin with a greater affinity and that this interaction is predominantly driven by the affinity for the sulfates rather than the pyranose. Our structural studies visualize heparin to bind one of five binding sites per subunit and the binding of heparin to the capsid to not adhere to the icosahedral symmetry of the capsid. Binding of heparin does not induce a conformational change in the capsid. We also show that longer heparin oligosaccharides (dp24 and dp36) can be modeled to bind adjacent icosahedral facets. Our studies strongly imply that polymers of sufficient length and sulfate content, including 12-kDa heparin and 8-kDa dextran sulfate, may act as inhibitors of PCV2 infection and that such inhibitors may act as therapeutics in swine farms where PCV2 infection is present. Future studies include assessing if capsid dynamics are altered upon binding of heparin, if certain binding sites are dispensable for binding to heparin and heparan sulfate, and if the asymmetric distribution of heparin on the capsid surface is important for the life cycle of the virus.
MATERIALS AND METHODS
PCV2 VLP expression and purification.
The codon sequence for the PCV2 capsid protein (GenBank accession number ACN59889.1) was optimized by GenScript for use in a baculovirus expression system. GenScript also generated the baculovirus stock used in these studies. Trichoplusia ni insect cells were propagated in ESF921 medium (Expression Systems) and infected with the baculovirus at a multiplicity of infection (MOI) of 0.1. Infected cells were grown for 5 days (50% viability, according to trypan blue staining) and harvested via centrifugation at 3,000 × g for 20 min.
PCV2 VLPs were purified from the nucleus of the infected cells. Cells from 500 ml of medium were gently centrifuged at 800 × g for 10 min at 4°C. The cell pellet was suspended and lysed in 20 mM HEPES, pH 7.5, 250 mM sodium chloride (NaCl), 2 mM magnesium chloride (MgCl2), 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 1% (vol/vol) NP-40 and placed on a revolver at 4°C for 30 min. The lysate was centrifuged for 30 min at 720 × g and 4°C to pellet the nuclei. The nucleus pellet was suspended in nuclear lysis buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 2 mM MgCl2, 0.2mM TCEP, 2 mM deoxycholate, 1 μl salt-activated nuclease [catalog number SRE0015-5KU; Sigma-Aldrich]) and lysed using a sonicator. The lysate was centrifuged at 28,000 × g for 30 min at 4°C. The supernatant was overlaid on a 30% sucrose cushion and centrifuged at 184,000 × g and 11°C for 2.5 h using a 45Ti Beckman Coulter rotor. The pellet was suspended in 2 ml of nucleus lysis buffer and processed on a discontinuous 20% to 70% sucrose gradient. The gradient was centrifuged, using a SW40 Beckman Coulter rotor, at 160,000 × g and 4°C for 18 h. The fraction containing the VLPs was visually identified using light scattering and SDS-PAGE. The fraction containing VLPs was dialyzed overnight in 20 mM HEPES, pH 7.0, 250 mM NaCl, 0.2 mM TCEP, and 2 mM EDTA. Samples were concentrated using a 100-kDa-molecular-weight-cutoff (MWCO) ultrafiltration device (Pall Corporation) and stored at 4°C. The sample concentration was estimated to be 370 nM using SDS-PAGE with Coomassie staining against a standard of a similar molecular weight with a known concentration.
Binding studies.
Heparin HyperD M affinity chromatography sorbent conjugated to 15-kDa porcine intestinal mucosa heparin was purchased from Pall Corporation. The HyperD M sorbent was washed and equilibrated with 20 mM HEPES, pH 7.0, 250 mM NaCl, 0.1 mM TCEP, 2 mM EDTA and allowed to settle to the bottom of the tube. Each reaction mixture contained 2 μl of settled HyperD M sorbent and the amount of VLPs indicated below. Samples were incubated at 20°C for the amount of time indicated below [see “(i) Kinetic binding assays” below] on a revolver to allow PCV2 to bind to the sorbent. The HyperD M sorbent was gently pelleted at 100 × g for 30 s, washed two times using the equilibration buffer, suspended in water, and processed using SDS-PAGE. The polyacrylamide gel was stained using InstantBlue from Expedeon, and band intensities were quantitated using a Li-Cor Odyssey blot imager equipped with Image Studio software (version 5.0). The reported fraction bound is the quotient of the intensity for each band (see Fig. S2 in the supplemental material) and the intensity of the band for the lowest concentration of polysaccharide tested. The washes did not remove VLPs from the sorbent, as exhibited by SDS-PAGE (data not shown).
(i) Kinetic binding assays. Four microliters of VLP at concentrations of 370 nM and 92.5 nM were incubated with 2 μl of the HyperD M sorbent for 5 s, 10 s, 30 s, 1 min, 5 min, 10 min, 20 min, 1 h, 3 h, and 24 h on a revolver at 20°C. The sorbent was processed as described above.
(ii) Elution assays. VLPs bound to the HyperD M sorbent (as described above) were incubated with increasing concentrations of porcine intestinal mucosa heparin [see “(iii) Competition assays” below] for 3 h at 20°C. The bound VLPs were quantitated as described above.
(iii) Competition assays. PCV2 was incubated with the GAG reagents at 20°C for 3 h prior to incubation with 2 μl of settled HyperD M sorbent for 3 h. The sorbent was processed as described above for SDS-PAGE.
Glycosaminoglycans and polysaccharide source.
Heparin oligosaccharides (catalog numbers HO06, HO10, and HO20) and chondroitin sulfate B (catalog number DS01) were purchased from Galen Laboratories and dissolved in the VLP dialysis buffer. Porcine intestinal mucosa heparin (average molecular weight, 12 kDa) was purchased from J. T. Baker, hyaluronic acid and 8-kDa dextran sulfate were purchased from Alfa Aesar, and dextran 6000 was purchased Sigma-Aldrich. All reagents were used within 48 h after being dissolved in the VLP dialysis buffer. Concentrations were calculated based on the disaccharide molecular weight of 659 g/mol for heparin, 551 g/mol for chondroitin sulfate B, 389 g/mol for hyaluronic acid, and 728 g/mol for 8-kDa dextran sulfate. Samples were stored at 4°C.
Electron microscopy.
Ultrathin carbon film on Lacey carbon support film (catalog number 01824; TedPella) was used for cryo-electron microscopy. The grids were glow discharged using a Pelco easiGlow system for 30 s. Unliganded and heparin-liganded VLPs (4 μl) were adsorbed to the grid for 15 s, blotted with a force of 2 for 6 s, and plunge frozen into liquid ethane using an FEI Mark IV Vitrobot sample vitrification robot. Data for the unliganded PCV2 were collected using a side entry FEI Titan Halo transmission electron microscope with Leginon software (62). Images were collected at a calibrated pixel size of 1.09 Å on a Gatan K2 camera operating in counting mode. Cryo-EM grids for the PCV2 liganded to heparin were prepared by incubating 20 μl of the VLPs with 7 μl of 60 mg/ml 12-kDa heparin for 3 h. The mixture was then incubated with 2 μl of HyperD M sorbent for 3 h at 20°C on a revolver to remove VLPs that were not bound to heparin. Cryo-EM grids were prepared as described above for the unliganded PCV2. Data for the heparin-liganded PCV2 were collected on a Titan Krios cryo-transmission electron microscope with Leginon software. Images were collected at a calibrated pixel size of 1.09 Å on a Gatan K2 camera that follows a GIF quantum energy filter with a slit width of 15 eV. The data collection parameters for both data sets are reported in Table 1.
Image reconstruction.
Motion correction was performed using the MotionCor2 package (63). The first frame of each movie was discarded during the alignment. With the exception of a patch 5 option, the default options were used for the alignment and dose weighting. Particles were automatically detected using the Gautomatch program (version 0.53), and per-particle contrast transfer function (CTF) estimation was performed on the aligned micrographs using the Gctf program (version 0.50) (64). Particles were extracted from the dose-weighted micrographs using the coordinates identified by Gautomatch. Particles from the unliganded data set were extracted into boxes of 256 by 256 pixels, and particles from the heparin-liganded data set were extracted into boxes of 300 by 300 pixels. Three iterations of the reference free two-dimensional classification with a diameter of 250 Å, 128 classes, and default Relion (version 2.0) parameters were used to discard bad particles between each iteration (45). The PCV2 crystal structure (PDB accession number 3R0R) was used to generate a 60-Å initial model using the molmap function of the UCSF Chimera program (65). The three-dimensional (3D) classification was carried out with the default Relion (version 2.0) parameters using C1 symmetry. A total of 39,048 particles for the unliganded data set and a total of 118,853 particles for the heparin-liganded data set were classified using the 3D classification. A total of 4 classes were requested for the unliganded data set, and a total of 10 classes were requested for the liganded data set. From each data set, particles from the 3D class with the highest-resolution image reconstruction were selected for further full-resolution refinement using I1 symmetry. The default parameters of the Refine 3D program in Relion (version 2.0) were used to achieve the highest resolution possible. For each data set, binary masks for postprocessing were generated using the relion_mask_create program of Relion (version 2.0) and the following options: (i) a low-pass-filtered final image reconstruction at a 15-Å resolution, (ii) a threshold value just high enough to eliminate the noise exterior to the PCV2 capsid, (iii) mask extension by 7 pixels, and (iv) an additional 2 soft edge pixel expansion. Masks were manually inspected with the UCSF Chimera program to ensure that no cavities existed in the interior of the masks. Postprocessing was performed with the default parameters of Relion (version 2.0) (45).
The particle orientation parameters attained from the full-resolution refinement described above were expanded from I1 to C1 symmetry using the relion_particle_symmetry_expand program of Relion (version 2.0) for 13,600 heparin-liganded particles. The consequence of this was to generate 60 × 13,600 (816,000) subunits that were aligned to one another. The molecular envelope for a PCV2 subunit that was refined (see below) into the I1 image reconstruction was calculated to a 25-Å resolution using the molmap function of UCSF Chimera, threshold masked, extended by 7 pixels, and extended by 2 soft edge pixels using the relion_mask_create program of Relion (version 2.0) (45, 65). The mask was manually moved radially outward to include the area peripheral to the capsid in case heparin may be present on the surface of the capsid. The alignment parameters and images were converted to a format for the Frealign program using the relion_preprocess function of Relion and the conversion script provided by the Frealign authors. Twenty rounds of masked classification (10 classes), no alignment, were performed with Frealign (47, 66). Difference maps were calculated using the vop function of UCSF Chimera (65). The image reconstruction from each class was subtracted from an image reconstruction generated using the same number of particles randomly selected from the data set. This was done so that both image reconstructions had comparable signal-to-noise ratios. The image reconstructions from the randomly selected particles were visually indistinguishable from the icosahedrally averaged image reconstruction.
The image reconstructions were rendered at 100% mass content of the capsid shell (60 × 27,853 Da, or 1.67 MDa). The threshold level to render the images was calculated using the volume command from EMAN software with the pixel size of the image reconstruction (67).
Structure refinement.
The reported crystal structure of PCV2 (35) was manually docked into the symmetrized image reconstructions using UCSF Chimera. The resulting coordinates were iteratively refined using phenix.real_space_refine from the Phenix software package with noncrystallographic symmetry (NCS) constraints applied and manual fitting with Coot software (68, 69). Heparin molecules (PDB accession numbers 3IRI, 3IRJ, 3IRK, and 3IRL) were treated as rigid bodies and manually fitted into the difference peaks of the five classes described above. The capsid subunits pertaining to these molecular envelopes were further refined using manual fitting with Coot and phenix.real_space_refine and without NCS.
Generating composite VLPs and representative models of individual VLPs.
The focused classification identifies 1 of 10 class averages that represents each subunit of a symmetry-expanded VLP, and the difference peaks represent each of these 10 classes. Consequently, the class averages can be combined with the symmetry elements used in the symmetry expansion process to generate a model of each imaged VLP. The icosahedral image reconstructions were generated using the I1 symmetry of Relion, and symmetry expansion was performed using the I1 symmetry matrices. Consequently, inverse matrices for the Relion I1 matrices should position the class averages or their corresponding difference peaks onto the VLP from which they originated. Inverse matrices were calculated for the I1 symmetry matrices using the MATLAB R2018a program (MathWorks). These symmetry operators were then applied to difference peaks representing each subunit of a VLP with UCSF Chimera (65).
Mutagenesis and binding efficiency.
Single point mutants of the PCV2 capsid were made by mutating the consensus sequence construct described by Khayat et al. (35). Briefly, this construct possesses a hexahistidine and a thrombin cleavage site at the N terminus in lieu of the 42 amino acids in the sequence with GenBank accession number ACN59889.1. The remaining amino acid sequence is identical. Mutations were made using a New England Biolabs Inc. site-directed mutagenesis kit. Sequences were confirmed using the services of Genewiz. Proteins were purified as previously described (35). Virus-like particles (VLPs) were assembled by mixing equal volumes of the capsid protein at a 37 μM concentration with 12% polyethylene glycol 3350, 5% isopropanol, and 0.6 M ammonium citrate, pH 5.0, at 4°C overnight. VLPs form crystals in this solution. The crystals were pelleted via centrifugation at 14,000 × g for 10 min at 4°C. The pellet was suspended in 0.3 M HEPPS, pH 9.0, 0.6 M NaCl, and 5 mM β-mercaptoethanol (β-Me) and processed through a HiPrep 16/60 Sephacryl S-500 HR column (GE Healthcare Life Sciences) equilibrated with 20 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (HEPPS) pH 9.0, 0.6 M NaCl, and 5 mM β-Me. The eluted peak was concentrated using a 100-kDa-MWCO ultrafiltration device (Pall Corporation) and assessed by negative-stained electron microscopy. Briefly, 3 μl of the sample was applied to a glow-discharged carbon-coated 400-mesh copper grid (catalog number 01814-F; TedPella) and stained with 2% uranyl formate. Images were collected using a JEOL JEM-1230 microscope that was operated at 120 kV and that was equipped with a Gatan US-4000 charge-coupled device.
Binding studies were performed as described above. The interaction between the sorbent and a 370 nM concentration of VLPs was measured five times to generate error bars. Statistical analysis was performed using the MATLAB R2018a program to generate the P values via the Wilcoxon rank sum test (MathWorks).
Accession number(s).
The image reconstructions and atomic coordinates have been deposited into the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB). Accession numbers are as follows: EMDB accession number 8939 and PDB accession number 6DZU for the unliganded image reconstruction, EMDB accession number 8969 and PDB accession number 6E2R for the symmetrized liganded image reconstruction, EMDB accession number 8970 and PDB accession number 6E2X for class 2, EMDB accession number 8971 and PDB accession number 6E2Z for class 4, EMDB accession number 8972 and PDB accession number 6E30 for class 6, EMDB accession number 8973 and PDB accession number 6E32 for class 7, EMDB accession number 8974 and PDB accession number 6E34 for class 9, and EMDB accession number 8975 and PDB accession number 6E39 for class 10.
Supplementary Material
ACKNOWLEDGMENTS
S.D. performed electron microscopy experiments and helped write the manuscript, B.A. performed electron microscopy experiments, S.F. performed sample preparation, and R.K. designed and performed experiments, analyzed data, and wrote the manuscript.
Funds supporting these studies were provided by the NIH National Institute of General Medical Sciences and National Institute of Allergy and Infectious Diseases (5SC1AI114843) and by grant 5G12MD007603-30 from the National Institute on Minority Health and Health Disparities. Data collection for the PCV2 heparin-liganded sample was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy, located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310).
Data collection for the unliganded PCV2 sample was performed at the Imaging Facility of the City University of New York (CUNY) Advanced Science Research Center (ASRC). We acknowledge the scientific and technical assistance from the NYSBC and CUNY ASRC.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02222-18.
REFERENCES
- 1.Vincent IE, Carrasco CP, Guzylack-Piriou L, Herrmann B, McNeilly F, Allan GM, Summerfield A, McCullough KC. 2005. Subset-dependent modulation of dendritic cell activity by circovirus type 2. Immunology 115:388–398. doi: 10.1111/j.1365-2567.2005.02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol 29:373–394. doi: 10.1080/030794500750047126. [DOI] [PubMed] [Google Scholar]
- 3.Nauwynck HJ, Sanchez R, Meerts P, Lefebvre DJ, Saha D, Huang L, Misinzo G. 2012. Cell tropism and entry of porcine circovirus 2. Virus Res 164:43–45. doi: 10.1016/j.virusres.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Lorincz M, Csagola A, Farkas SL, Szekely C, Tuboly T. 2011. First detection and analysis of a fish circovirus. J Gen Virol 92:1817–1821. doi: 10.1099/vir.0.031344-0. [DOI] [PubMed] [Google Scholar]
- 5.Tischer I, Gelderblom H, Vettermann W, Koch MA. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 6.Tischer I, Rasch R, Tochtermann G. 1974. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralb Bakteriol Orig A 226:153–167. [PubMed] [Google Scholar]
- 7.Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J 39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 8.Finsterbusch T, Mankertz A. 2009. Porcine circoviruses—small but powerful. Virus Res 143:177–183. doi: 10.1016/j.virusres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Morozov I, Sirinarumitr T, Sorden SD, Halbur PG, Morgan MK, Yoon KJ, Paul PS. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol 36:2535–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Cariñg C, Segalés J. 2009. Ultrastructural findings in lymph nodes from pigs suffering from naturally occurring postweaning multisystemic wasting syndrome. Vet Pathol 46:729–735. doi: 10.1354/vp.08-VP-0141-R-FL. [DOI] [PubMed] [Google Scholar]
- 11.Opriessnig T, Meng X-J, Halbur PG. 2007. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest 19:591–615. doi: 10.1177/104063870701900601. [DOI] [PubMed] [Google Scholar]
- 12.Segalés J. 2012. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res 164:10–19. doi: 10.1016/j.virusres.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Mankertz A. 2012. Molecular interactions of porcine circoviruses type 1 and type 2 with its host. Virus Res 164:54–60. doi: 10.1016/j.virusres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Ramamoorthy S, Meng X-J. 2009. Porcine circoviruses: a minuscule yet mammoth paradox. Anim Health Res Rev 10:1–20. doi: 10.1017/S1466252308001461. [DOI] [PubMed] [Google Scholar]
- 15.Meng X-J. 2013. Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 1:43–64. doi: 10.1146/annurev-animal-031412-103720. [DOI] [PubMed] [Google Scholar]
- 16.Hattermann K, Roedner C, Schmitt C, Finsterbusch T, Steinfeldt T, Mankertz A. 2004. Infection studies on human cell lines with porcine circovirus type 1 and porcine circovirus type 2. Xenotransplantation 11:284–294. doi: 10.1111/j.1399-3089.2004.00134.x. [DOI] [PubMed] [Google Scholar]
- 17.Pinheiro ALBC, Bulos LHS, Onofre TS, de Paula Gabardo M, de Carvalho OV, Fausto MC, Guedes RMC, de Almeida MR, Silva Júnior A. 2013. Verification of natural infection of peridomestic rodents by PCV2 on commercial swine farms. Res Vet Sci 94:764–768. doi: 10.1016/j.rvsc.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Kiupel M, Stevenson GW, Choi J, Latimer KS, Kanitz CL, Mittal SK. 2001. Viral replication and lesions in BALB/c mice experimentally inoculated with porcine circovirus isolated from a pig with postweaning multisystemic wasting disease. Vet Pathol 38:74–82. doi: 10.1354/vp.38-1-74. [DOI] [PubMed] [Google Scholar]
- 19.Halami MY, Freick M, Shehata AA, Müller H, Vahlenkamp TW. 2014. Susceptibility of calves to porcine circovirus-2 (PCV2). Vet Microbiol 173:125–131. doi: 10.1016/j.vetmic.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. 2017. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol 91:e01879-16. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Chen I, Kwang J. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol 79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Wei Y, Zhang C, Lu Y, Kong X. 2007. Construction and characterization of porcine circovirus type 2 carrying a genetic marker strain. Virus Res 127:95–99. doi: 10.1016/j.virusres.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 23.Chaiyakul M, Hsu K, Dardari R, Marshall F, Czub M. 2010. Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus. J Virol 84:11440–11447. doi: 10.1128/JVI.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv Q, Guo K, Wang T, Zhang C, Zhang Y. 2015. Porcine circovirus type 2 ORF4 protein binds heavy chain ferritin. J Biosci 40:477–485. doi: 10.1007/s12038-015-9551-3. [DOI] [PubMed] [Google Scholar]
- 25.Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ. 2006. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J Virol 80:3487–3494. doi: 10.1128/JVI.80.7.3487-3494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarrazin S, Lamanna WC, Esko JD. 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizumoto S, Yamada S, Sugahara K. 2015. Molecular interactions between chondroitin-dermatan sulfate and growth factors/receptors/matrix proteins. Curr Opin Struct Biol 34:35–42. doi: 10.1016/j.sbi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Xu D, Esko JD. 2014. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem 83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabenstein DL. 2002. Heparin and heparan sulfate: structure and function. Nat Prod Rep 19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 30.Zehnder JL, Galli SJ. 1999. Mast-cell heparin demystified. Nature 400:714–715. doi: 10.1038/23360. [DOI] [PubMed] [Google Scholar]
- 31.Misinzo G, Meerts P, Bublot M, Mast J, Weingartl HM, Nauwynck HJ. 2005. Binding and entry characteristics of porcine circovirus 2 in cells of the porcine monocytic line 3D4/31. J Gen Virol 86:2057–2068. doi: 10.1099/vir.0.80652-0. [DOI] [PubMed] [Google Scholar]
- 32.Shuai J, Wei W, Li X, Chen N, Zhang Z, Chen X, Fang W. 2007. Genetic characterization of porcine circovirus type 2 (PCV2) from pigs in high-seroprevalence areas in southeastern China. Virus Genes 35:619–627. doi: 10.1007/s11262-007-0121-0. [DOI] [PubMed] [Google Scholar]
- 33.Mankertz A, Mankertz J, Wolf K, Buhk HJ. 1998. Identification of a protein essential for replication of porcine circovirus. J Gen Virol 79:381–384. doi: 10.1099/0022-1317-79-2-381. [DOI] [PubMed] [Google Scholar]
- 34.Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D. 2003. Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus. J Virol 77:13036–13041. doi: 10.1128/JVI.77.24.13036-13041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, Johnson JE. 2011. The 2.3-angstrom structure of porcine circovirus 2. J Virol 85:7856–7862. doi: 10.1128/JVI.00737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. 1978. Tomato bushy stunt virus at 2.9 Å resolution. Nature 276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 37.Dhindwal S, Avila B, Feng S, Khayat R. 2019. Porcine circovirus 2 uses a multitude of weak binding sites to interact with heparan sulfate, and the interactions do not follow the symmetry of the capsid. bioRxiv 10.1101/370411. [DOI] [PMC free article] [PubMed]
- 38.Khan S, Gor J, Mulloy B, Perkins SJ. 2010. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes. J Mol Biol 395:504–521. doi: 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 39.Laurent TC. 1995. An early look at macromolecular crowding. Biophys Chem 57:7–14. doi: 10.1016/0301-4622(95)00048-3. [DOI] [PubMed] [Google Scholar]
- 40.Necas J, Bartosikova L, Brauner P, Kolar J. 2008. Hyaluronic acid (hyaluronan): a review. Vet Med 53:397–411. doi: 10.17221/1930-VETMED. [DOI] [Google Scholar]
- 41.Thelin MA, Bartolini B, Axelsson J, Gustafsson R, Tykesson E, Pera E, Oldberg Å, Maccarana M, Malmstrom A. 2013. Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J 280:2431–2446. doi: 10.1111/febs.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. 2004. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burra PV, Zhang Y, Godzik A, Stec B. 2009. Global distribution of conformational states derived from redundant models in the PDB points to non-uniqueness of the protein structure. Proc Natl Acad Sci U S A 106:10505–10510. doi: 10.1073/pnas.0812152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kufareva I, Abagyan R. 2012. Methods of protein structure comparison. Methods Mol Biol 857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilca SL, Kotecha A, Sun X, Poranen MM, Stuart DI, Huiskonen JT. 2015. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat Commun 6:8843. doi: 10.1038/ncomms9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. 2013. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol 183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigorieff N. 2016. Frealign: an exploratory tool for single-particle cryo-EM methods. Methods Enzymol 579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilas JL, Gómez-Blanco J, Conesa P, Melero R, la Rosa-Trevín de JM, Otón J, Cuenca J, Marabini R, Carazo JM, Vargas J, Sorzano COS. 2018. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26:337–344.e4. doi: 10.1016/j.str.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 50.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mobli M, Nilsson M, Almond A. 2008. The structural plasticity of heparan sulfate NA-domains and hence their role in mediating multivalent interactions is confirmed by high-accuracy 15N-NMR relaxation studies. Glycoconj J 25:401–414. doi: 10.1007/s10719-007-9081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dechecchi MC, Tamanini A, Bonizzato A, Cabrini G. 2000. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology 268:382–390. doi: 10.1006/viro.1999.0171. [DOI] [PubMed] [Google Scholar]
- 53.O'Donnell J, Taylor KA, Chapman MS. 2009. Adeno-associated virus-2 and its primary cellular receptor—cryo-EM structure of a heparin complex. Virology 385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fry EE, Lea SM, Jackson T, Newman JW, Ellard FM, Blakemore WE, Abu-Ghazaleh R, Samuel A, King AM, Stuart DI. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J 18:543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vauquelin G, Charlton SJ. 2013. Exploring avidity: understanding the potential gains in functional affinity and target residence time of bivalent and heterobivalent ligands. Br J Pharmacol 168:1771–1785. doi: 10.1111/bph.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spillmann D. 2001. Heparan sulfate: anchor for viral intruders? Biochimie 83:811–817. doi: 10.1016/S0300-9084(01)01290-1. [DOI] [PubMed] [Google Scholar]
- 58.Xie Q, Lerch TF, Meyer NL, Chapman MS. 2011. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6). Virology 420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Smith DL. 1993. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure. Protein Sci 2:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guan J, Bywaters SM, Brendle SA, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2017. Cryoelectron microscopy maps of human papillomavirus 16 reveal L2 densities and heparin binding site. Structure 25:253–263. doi: 10.1016/j.str.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Johson JE, Rueckert RR. 1997. Chapter 10. Packaging and release of the viral genome In Chiu W, Burnett RM, Garcea RL (ed), Structural biology of viruses, p 269–287. Oxford University Press, New York, NY. [Google Scholar]
- 62.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Grigorieff N. 2007. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol 157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 68.Afonine PV, Headd JJ, Adams TCTPD. 2013. New tool: phenix.real_space_refine.
- 69.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2017. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.