Abstract
Objectives: To investigate the specific role of Toll-like receptor 4 (TLR4) in the regulation of the intestinal mucosa-associated microbiota by vitamin A (VA).
Methods: Both TLR4-/- (knockout, KO) and wild-type (WT) female mice were randomly fed a VA normal (VAN) or VA deficient (VAD) diet for 4 weeks to establish the following four mouse model groups: TLR4-/- mice fed a VAN diet (KO VAN), TLR4-/- mice fed a VAD diet (KO VAD), WT mice fed a VAN diet (WT VAN), and WT mice fed a VAD diet (WT VAD). Then, the mice from each experimental group were mated with male mice with the same genetic background. The pups in the KO VAD and WT VAD groups were subsequently fed the VAD diet after weaning, while the pups in the KO VAN and WT VAN groups were fed the VAN diet continuously after weaning. The serum retinol levels of 7-week-old offspring were determined using high-performance liquid chromatography, and colons were collected from mice in each group and analyzed via 16S rRNA gene sequencing using an Illumina MiSeq platform to characterize the overall microbiota of the samples.
Results: The abundance and evenness of the colon mucosa-associated microbiota were unaffected by dietary VA and TLR4 KO. VAD decreased the abundance of Anaerotruncus (Firmicutes), Oscillibacter (Firmicutes), Lachnospiraceae _NK4A136 _group (Firmicutes) and Mucispirillum (Deferribacteres) and increased the abundance of Parasutterella (Proteobacteria). TLR4 KO decreased the abundance of Bacteroides (Bacteroidetes) and Alloprevotella (Bacteroidetes). However, the abundance of Allobaculum (Firmicutes), Ruminiclostridium_9 (Firmicutes), Alistipes (Bacteroidetes), and Rikenellaceae_RC9 (Bacteroidetes) impacted the interaction between VA and TLR4.
Conclusion: TLR4 may play a pivotal role in regulation of the intestinal mucosa-associated microbiota by VA to maintain the intestinal microecology.
Keywords: toll-like receptor 4 (TLR4), vitamin A normal (VAN), vitamin A deficiency (VAD), intestinal mucosa-associated microbiota, intestinal innate immunity
Introduction
Vitamin A is an essential fat-soluble vitamin that maintains normal growth and development, participates in the immune response, promotes reproduction, and maintains visual function (Stephensen, 2001; Clagett-Dame and DeLuca, 2002). VAD remains a significant public health concern in many regions of the world (World Health Organization [WHO], 1995). Children experiencing VAD are especially prone to gastrointestinal (GI) tract infections (Thornton et al., 2014). Studies by us and others have shown that VAD can decrease gut integrity and impact the immune response of the GI tract in humans and animals (Quadro et al., 2000; Liu et al., 2014). The diversity and balance of the gut microbiota is important for maintaining the normal biological barrier function of the intestine (Topping and Clifton, 2001; Round et al., 2010). Recent studies have shown that the VA nutritional status can affect the total amount of bacteria in the GI tract and alter the intestinal microflora (Amit-Romach et al., 2009). Our preliminary study found that VAD impacts the structural segregation of the gut microbiota in children with persistent diarrhea (Lv et al., 2016).
Toll-like receptors (TLRs) are membrane-anchored proteins that are expressed on immune cells and enterocytes (Takeda et al., 2003). TLRs act as pathogen recognition receptors (PRRs), identifying microbe-associated molecular patterns (MAMPs) to activate specific signaling pathways (Frosali et al., 2015). A total of 10 TLRs are expressed in humans, and TLR4 is the best characterized PRR. Recognition of MAMPs by TLR4 is involved in protective innate immune response mechanisms against bacterial invasion (Furuta et al., 2006). In addition, TLR4-/- mice exhibited a striking reduction in acute inflammatory cells, impaired epithelial cell proliferation and marked bacterial translocation during injury compared with WT mice (Fukata et al., 2005). Moreover, mouse epithelial cells overexpressing the TLR4 signaling pathway exhibited increased bacterial density in the colonic mucosa and increased bacterial translocation (Dheer et al., 2016). In our previous study, we confirmed that RARβ enhanced ZO-2 expression by regulating TLR4 to improve intestinal epithelial barrier function both in vivo and in vitro (Li et al., 2017). However, the role of TLR4 in regulation of the gut microbiota by VA is unclear.
Therefore, the purpose of this study was to determine the effect of TLR4 on the intestinal mucosal microbiota associated with VA nutritional levels. In the present study, TLR4-/- and WT mice were acquired to establish both VAN and VAD mouse models. 16S rRNA deep sequencing was used to examine the distribution and structural characteristics of the intestinal mucosa-associated microbiota.
Materials and Methods
Animals, Diets and Sample Collection
This study was approved by the Animal Experimentation Ethics Committee of Chongqing Medical University (Chongqing, China) and was conducted in accordance with the guidelines of the Animal Care Committee of Chongqing Medical University. TLR4-/- (knockout, KO) and WT mice obtained from Jackson laboratories (Maine, United States) were purchased from the Model Animal Research Center of Nanjing University (MARC). The TLR4-/- mouse strain was C57BL/10ScNJNju, which is based on the C57BL/10JNju mouse strain (WT). The mice were housed in the same room with a constant airflow system, controlled temperature (22–24°C), and a 12-h light/dark cycle. The VAN and VAD animal models were constructed according to methods described previously (Liu et al., 2014). Half of the female KO and WT mice (3 weeks of age), which were randomly selected, were fed a VAD-inducing diet comprising 400 IU/kg VA for 4 weeks to establish a TLR4-/- mouse model with VAD (KO VAD) and a WT mouse model with VAD (WT VAD), and the other half received a VAN diet containing 6,500 IU/kg VA for 4 weeks to establish a VAN TLR4-/- mouse model (KO VAN) and a VAN WT mouse model (WT VAN). Then, the (female) mice from each experimental group were mated with the corresponding male mice with the same genetic background. Pregnant mice were fed either the VAD or VAN diet during both gestation and lactation to maintain stable serum retinol levels. Once the pups had weaned, their mothers were sacrificed, and blood was collected from the eyeballs. The serum retinol levels of the maternal VAN mice increased to 1.05 μmol/L, and those of the maternal VAD mice decreased to 0.7 μmol/L. The offspring were used for subsequent experiments. The pups in the KO VAD and WT VAD groups were subsequently fed the VAD diet continuously for 4 weeks, while the pups in the KO VAN and WT VAN groups were fed the VAN diet continuously for the same time period. Next, the mice were sacrificed, and blood was immediately harvested from the eyeball. The colons were extracted from the mice in each group, and after cleaning with 0.01 M PBS, the colons were stored at -80°C until further study.
Serum Retinol Detection
The serum retinol levels in the collected mouse blood were determined using HPLC. VA standard curve preparation and testing methods were modified slightly following methods described previously (Li et al., 2017), and VA standard compound was purchased from Sigma (R7632, United States). Briefly, 200 μL of serum was deproteinized with the same volume of anhydrous ethanol. Then, 1000 μL of hexane was used to extract the retinol from the serum, and the hexane was evaporated using nitrogen gas. The retinol residue was dissolved in 100 μL of the mobile phase mixture (methanol:water = 97:3). Finally, the prepared sample was measured using an HPLC apparatus (DGU-20As, Shimadzu Corporation, Japan). The retinoids were separated by chromatography on an analytical column (Hypersil phenyl 120 A 5 mm, 250 mm × 4.6 mm, Phenomenex, United States) via gradient elution of the mobile phase in a liquid chromatograph equipped with a 315-nm ultraviolet photodiode array detector.
DNA Extraction and PCR Amplification
Microbial DNA was extracted from colon samples using an OMEGA DNA Kit (Omega Bio-Tek, United States) according to the manufacturer’s protocol. The final concentration of the purified DNA was determined with a NanoDrop 2000 UV-vis spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States), and DNA quality was checked via 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Xu et al., 2016) using a PCR thermocycler system (GeneAmp 9700, ABI, United States). The PCRs were conducted using the following program: 3 min of denaturation at 95°C; 27 cycles of 30 s at 95°C, 30 s for annealing at 55°C, and 45 s for elongation at 72°C; and a final extension at 72°C for 10 min. The PCRs were performed in triplicate in 20-μL mixtures containing 4 μL of 5 × FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu polymerase and 10 ng of template DNA. The resulting PCR products were extracted from a 2% agarose gel, further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using QuantiFluorTM-ST (Promega, United States) according to the manufacturer’s protocol.
Illumina MiSeq Sequencing
Colon samples were collected from 40 mice from 4 groups: KO VAN group, KO VAD group, WT VAN group, and WT VAD group, with 10 samples per group. After DNA extraction and PCR amplification, the target band size and concentration of the samples to be sequenced were correct. Purified amplicons were pooled in equimolar amounts, and paired-end (2 × 300) sequencing was performed on an Illumina MiSeq platform (TruSeqTM DNA Sample Prep Kit, United States) according to the standard protocols recommended by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP: 158355).
Processing of Sequencing Data
Raw fastq files were demultiplexed, quality-filtered with Trimmomatic and merged using FLASH with the following criteria: (i) The reads were truncated at any site that received an average quality score <20 over a 50-bp sliding window. (ii) Primers were exactly matched, allowing 2-nucleotide mismatches, and reads containing ambiguous bases were removed. (iii) Sequences with overlaps longer than 10 bp were merged at the overlap sequence.
Operational taxonomic units were clustered with a 97% similarity cutoff using UPARSE (version 7.11 ), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier algorithm2 compared against the Silva (SSU128) 16S rRNA database using a confidence threshold of 70%.
Microbial Analysis
Sets of sequences with 97% identity were clustered into OTUs using USEARCH (version 7.03). OTUs that reached 97% similarity levels were used for community richness (Chao, ACE), community diversity (Shannon, Simpson), and rarefaction curve analyses. The β-diversity was estimated by computing unweighted UniFrac distances and visualized with principal coordinate analysis. To effectively distinguish between the four groups, a partial least squares discriminant analysis (PLS-DA) was conducted. In addition, linear discriminant analysis (LDA) effect size (LEfSe) was determined using LEfSe software to determine the community or species that most influenced the group division. After features that were significantly different at various bacterial taxonomic levels were identified by LEfSe, the nonparametric factorial Kruskal-Wallis (KW) sum-rank test and LDA were performed to determine whether these features were consistent with the expected behaviors of the different bacterial taxonomic levels; genera with LDA scores greater than three were defined as having a significant impact on the group.
Statistical Analyses
All data were obtained from ten biological replicates and are presented as the mean ± SEM. Significant differences were calculated via two-way analysis of variance (ANOVA) with a Bonferroni post hoc test using the GraphPad Prism version 5.0 software package. The interaction between the effect of the different VA nutrition levels and the effect of TLR4 deletion was investigated with a Bonferroni post hoc test. When there was a statistically significant interaction, all the experimental groups were compared using a Bonferroni post hoc test. However, when no interaction was observed, the effect of the different VA nutrition levels or TLR4 deletion was assessed using Student’s t-test. Only the relevant comparisons of the combined groups are presented in the Results section. Significance was accepted at P < 0.05.
Results
The VAD Diet Decreased Serum Retinol Levels in Mice
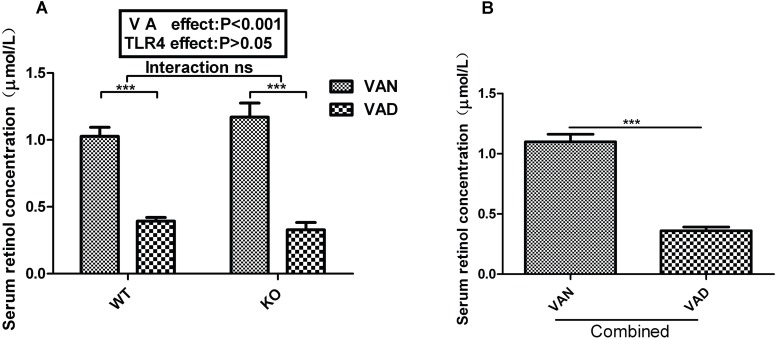
To explore whether TLR4 participates in regulation of intestinal microbial homeostasis by VA, we established VAN and VAD mouse models in both TLR4-/- (KO VAN and KO VAD) and WT mice (WT VAN and WT VAD). As shown in Figure 1A, the serum retinol levels of offspring in WT mice fed the VAD diet (0.393 ± 0.027 μmol/L) were significantly lower than those in WT mice fed the VAN diet (1.027 ± 0.067 μmol/L) (P < 0.001). Similar results were observed for the TLR4-/- mice, and the serum retinol levels were significantly repressed in the KO VAD group (0.328 ± 0.054 μmol/L) compared with those in the KO VAN group (1.171 ± 0.104 μmol/L). However, there was no significant interaction between the effects of VA and TLR4, determined using two-way ANOVA with a Bonferroni post hoc test, but differences in VA nutritional levels had an effect on the serum retinol levels (P < 0.001, Figure 1A). After combining the data for the WT and KO groups, we found that the serum retinol levels in the combined VAD group were markedly lower than those in the combined VAN group (P < 0.001) (Figure 1B). The above data demonstrate that the VAD diet was an important factor associated with the decreased serum retinol levels in mice, providing us with a solid foundation for subsequent studies.
FIGURE 1.
Effects of different VA nutritional levels and TLR4 deletion on the serum retinol levels of seven-week-old offspring mice. (A) Changes in the serum retinol levels of the WT and TLR4-/- mice fed VAN or VAD diets (n = 10 for each group). (B) The main effect of VA, independent of TLR4-/- challenge, on the differences in serum retinol levels between the combined VAN and VAD groups (n = 20). The values are the means ± SEMs; “Interaction” indicates an effect of the different VA nutritional levels in the TLR4 knockout vs. WT mice; ∗∗∗P < 0.001. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
Rarefaction Curves and Alpha Diversity Index
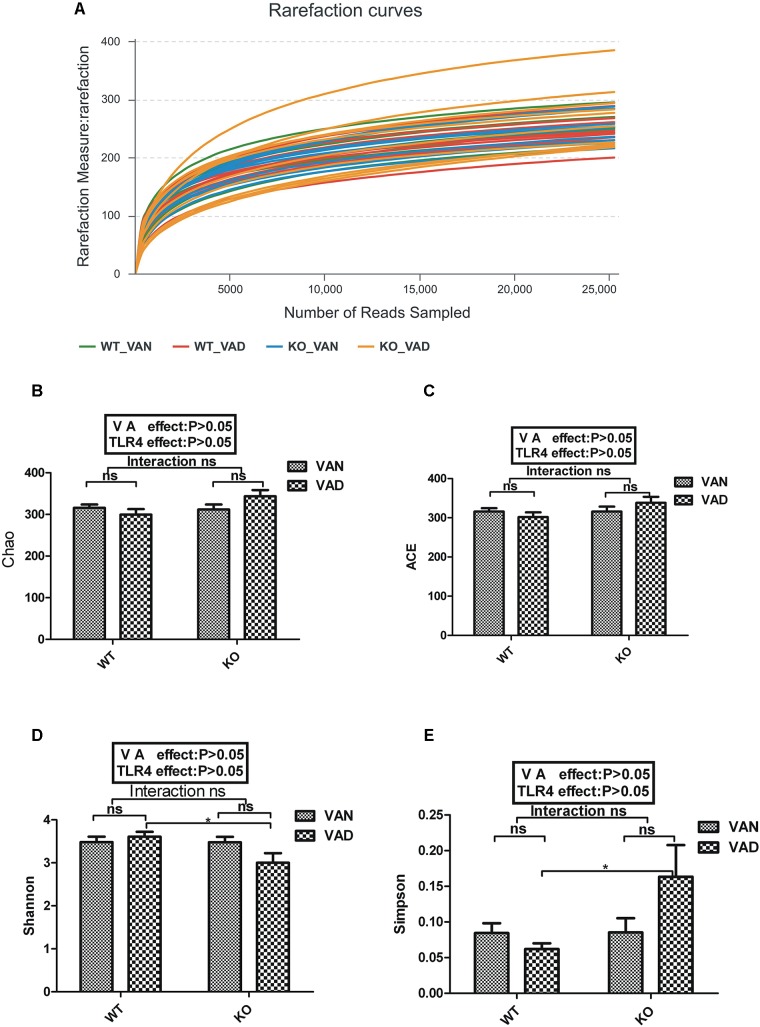
After optimization, a total of 1,487,466 high-quality sequences were obtained from 40 samples, and there were 37,187 high-quality sequences per sample on average according to MiSeq sequencing. We acquired numerous OTUs from valid sequences that exhibited 97% similarity for further statistical analyses. Along with an increase in the number of reads, the rarefaction curves for all the samples shown in Figure 2A exhibited smooth increasing trends and approached saturation plateaus, demonstrating that the sequencing data volume acquired was suitable for the present study.
FIGURE 2.
Rarefaction curves and α diversity analysis for WT and TLR4-/- mice fed VAN or VAD diets. (A) Rarefaction curves were calculated for OTUs with 97% identity in the gut microbiota in the WT VAN, WT VAD, KO VAN, and KO VAD groups (n = 10). The green curves represent the WT VAN group; red curves represent the WT VAD group; blue curves represent the KO VAN group; yellow curves represent the KO VAD group (n = 10). Comparison of (B) Chao index, (C) ACE index, (D) Shannon index and (E) Simpson index among the four groups (n = 10). Mean ± SEM; ns. = not significant. “Interaction” indicates an effect of the different VA nutritional levels in the TLR4 knockout vs. WT mice; ∗P < 0.05. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
In general, Chao and ACE diversity indexes reflect the richness of the microbiota, while Shannon and Simpson diversity indexes are considered to be indicators of colony richness and evenness. After two-way ANOVA with a post hoc test, no significant interaction was found between the effects of VAD and TLR4 KO on the Chao, ACE, Shannon and Simpson diversity indexes (Figure 2B–E). Furthermore, these four indexes were not affected by either VAD or TLR4 KO, even though the Simpson index of the KO VAD group was slightly higher than that of the WT VAD group (P < 0.05, Figure 2E) and the Shannon index of the KO VAD group was significantly decreased when compared with that of the WT VAD group (P < 0.05, Figure 2D). The above data demonstrate that the abundance and evenness of the colonic mucosa-associated microflora were unaffected by dietary VA and TLR4 KO.
Different VA Levels in the Diet May Affect the Microbial Community Structure of the Colonic Mucosa in Both the TLR4 KO and WT Mice
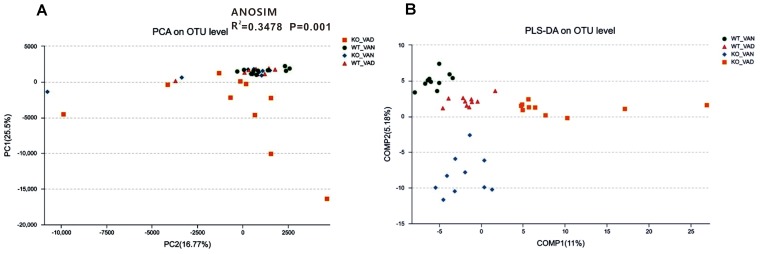
To further evaluate structural differences in the microbial communities among the four groups, an unweighted UniFrac distance matrix was calculated based on the OTUs of each sample. Figure 3A shows that samples from the WT VAN, WT VAD, and KO VAN groups were relatively concentrated compared with the KO VAD group samples. Principle component analysis (PCA) revealed a separation of the TLR4-/- and WT mice fed VAN or VAD diets based on the first two principal component (PC) scores, accounting for 25.5 and 16.77% of explained variances (Figure 3A). Meanwhile, ANOSIM analysis showed that the difference among the four groups was significantly greater than the difference within the group (R2 = 0.3478, P = 0.001, Figure 3A), indicating that our grouping was meaningful. The results of the subsequent PLS-DA are shown in Figure 3B, and the variance among the KO VAD group samples was greater than that among samples from the other three groups. These data suggest that there may be differences in the distribution of the colonic mucosal microbiota due to the different levels of VA in the diet and due to TLR4 deletion.
FIGURE 3.
PCA and PLS-DA of samples from the TLR4-/- and WT mice fed VAN or VAD diets. (A) PCA scores were plotted based on the relative abundance of the OTUs (n = 10). (B) PLS-DA was plotted based on the unweighted UniFrac distance metrics (n = 10). The green circles represent the WT VAN group; red triangles represent the WT VAD group; blue diamonds represent the KO VAN group; orange squares represent the KO VAD group (n = 10). VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
Both VA and TLR4 Are Involved in the Community Abundance of the Colonic Mucosal Microbiota at the Phylum Level
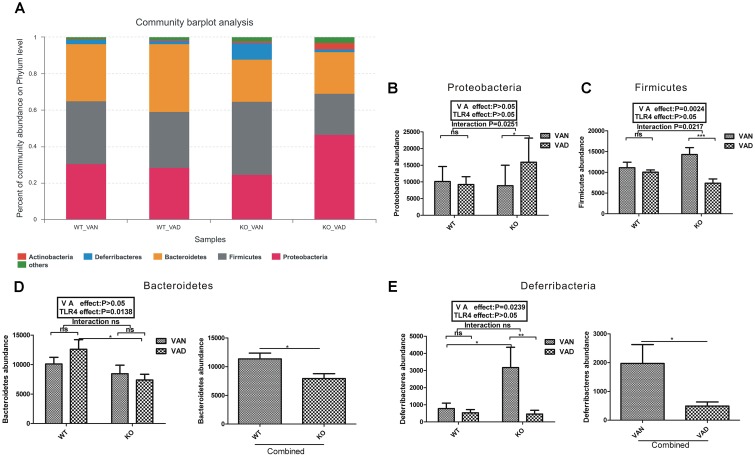
Figure 4A shows the composition of the dominant microflora with a relative abundance of more than 1% at the phylum level in the TLR4 KO and WT mice fed the VAN or VAD diet. In the WT VAN group, the predominant phyla were Firmicutes (34.47%), Bacteroidetes (31.34%), Proteobacteria (30.62%), and Deferribacteres (2.372%); however, the predominant phyla in the WT VAD group were Bacteroidetes (37.33%), Firmicutes (30.91%), Proteobacteria (28.38%), and Deferribacteres (1.718%). The most abundant phyla in the KO VAN group, in decreasing order, were Firmicutes (40%), Proteobacteria (24.66%), Bacteroidetes (23.42%), and Deferribacteres (8.928%). Notably, the percentage of Actinobacteria was highest in the KO VAD group compared with that in the other three groups, and the most abundant phyla, in decreasing order, in the KO VAD group were Proteobacteria (46.58%), Bacteroidetes (22.92%), Firmicutes (22.69%), Actinobacteria (3.644%), and Deferribacteres (1.357%).
FIGURE 4.
The relative abundances of bacterial phyla in the colonic mucosa of the TLR4-/- and WT mice fed the VAN or VAD diet. (A) The dominant bacterial phyla with relative abundances greater than 1% in the four groups (n = 10). The combined effects of the different VA nutritional levels and the TLR4 deletion on the relative abundance of (B) Proteobacteria and (C) Firmicutes determined by two-way analysis of variance with a Bonferroni post hoc test (n = 10). (D) The main effect of TLR4, independent of VA nutritional level, on the relative abundance of Bacteroidetes in the four groups (n = 10). (E) The main effect of VA, independent of TLR4 deletion, on the relative abundance of Deferribacteres in the four groups (n = 10). Mean ± SEM; ns. = not significant. “Interaction” indicates an effect of the different VA nutritional levels in the TLR4 knockout vs. WT mice; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
As shown in Figure 4B, although VA and TLR4 had no effect on the abundance of Proteobacteria (P > 0.05), the P-value of the interaction between the VA and TLR4-/- challenges was 0.0251 after two-way ANOVA with a Bonferroni post hoc test. The Firmicutes abundance in the VAD group was lower than that in the VAN group in the TLR4-/- mice (Figure 4C). The VA nutritional level had significant effects on the Firmicutes abundance (P = 0.0024), and the P-value of the interaction between the VA and TLR4-/- challenges for the Firmicutes abundance was 0.0217 according to the post hoc test (Figure 4C). However, for the abundance of Bacteroidetes and Deferribacteres, no significant interaction was observed between the VA and TLR4-/- challenges, determined using two-way ANOVA (Figure 4D,E). After combining the VAN and VAD groups, the Bacteroidetes abundance in the combined KO group was significantly lower than that in the combined WT group (P < 0.05, Figure 4D). The Deferribacteres abundance in the combined VAD group was significantly lower than that in the combined VAN group (P < 0.05, Figure 4E). Based on these data, VA and TLR4 have interactive effects on the abundance of Proteobacteria and Firmicutes; the Bacteroides abundance was affected by TLR4 and that of Deferribacteres was affected by VA.
VA and TLR4 Altered the Community Structure of the Colonic Microbiota at the Genus Level
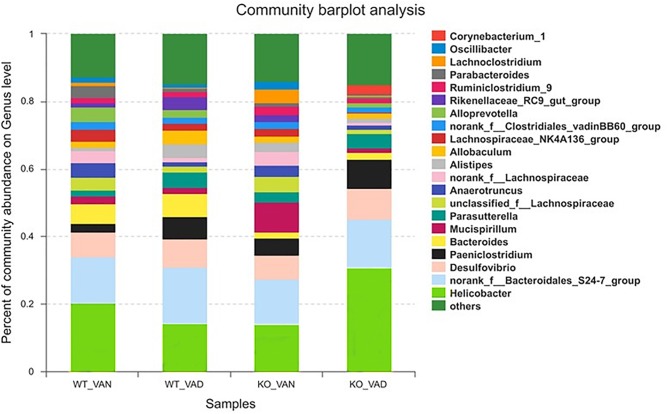
To further understand the effect of VA and TLR4 on colonic bacteria, we analyzed the community structure at the genus level in the four groups. Figure 5 shows the composition of the dominant microflora with relative abundances greater than 2% at the genus level in the TLR4-/- and WT mice fed the VAN or VAD diet. We noticed that the percentage of Helicobacter in the KO VAD group was highest compared with the percentage in the other three groups.
FIGURE 5.
The dominant bacterial genera with relative abundances greater than 2% in the colonic mucosa of TLR4-/- and WT mice fed the VAN or VAD diet (n = 10). VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
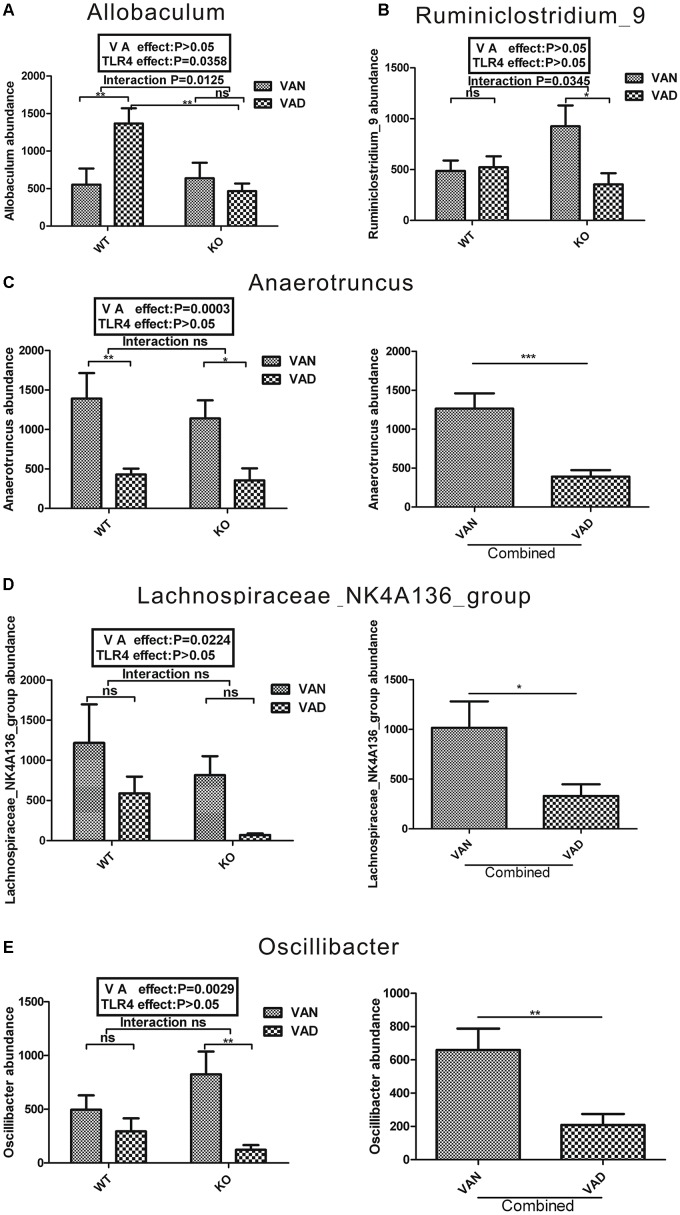
Two-way ANOVA was conducted for each strain, as shown in Figure 6, 7. There were five genera of the phylum Firmicutes that were affected by VA or both VA and TLR4. A significant interaction was observed between the effects of VA and TLR4 on the abundance of Allobaculum and Ruminiclostridium_9 according to a Bonferroni post hoc analysis, and the P-values were 0.0125 and 0.0345, respectively (Figure 6A,B). On the other hand, the main effects of VA, independent of TLR4, were on the relative abundance of Anaerotruncus (P = 0.0003), Lachnospiraceae _NK4A136 _group (P = 0.0224), and Oscillibacter (P = 0.0029) in the four groups (Figure 6C–E). After combining the WT and KO groups, the abundance of Anaerotruncus (P < 0.001), Lachnospiraceae_NK4A136_group (P < 0.05), and Oscillibacter (P < 0.01) was markedly decreased in the combined VAD group compared with that in the combined VAN group (Figure 6C–E).
FIGURE 6.
Two-way ANOVA of the five genera with relative abundances greater than 2% from the phylum Firmicutes in the colonic mucosa of TLR4-/- and WT mice fed the VAN or VAD diet. The combined effects of the different VA nutritional levels and the TLR4 deletion on the relative abundance of (A) Proteobacteria and (B) Ruminiclostridium_9 (n = 10). The main effect of VA, independent of TLR4 deletion, on the relative abundance of (C) Anaerotruncus, (D) Lachnospiraceae_NK4A136_group and (E) Oscillibacter in the four groups (n = 10). Mean ± SEM; ns. = not significant. “Interaction” indicates an effect of the different VA nutritional levels in the TLR4 knockout vs. WT mice; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
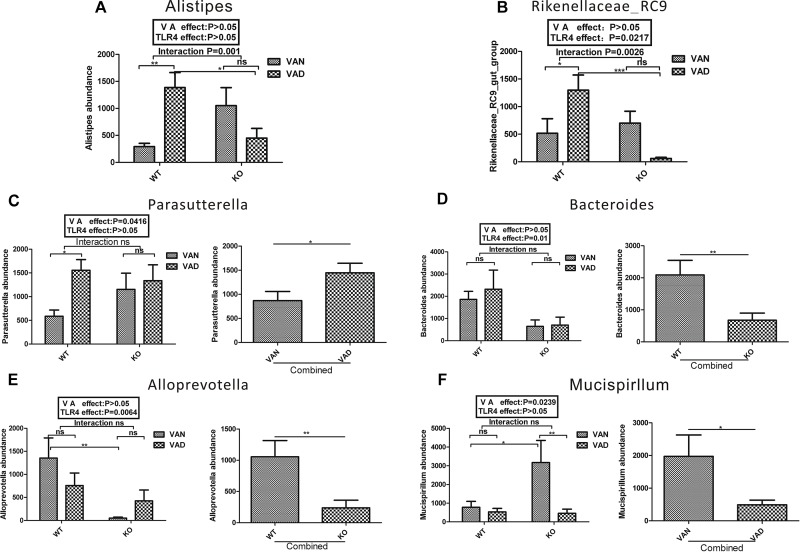
FIGURE 7.
Two-way ANOVA of genera with relative abundances greater than 2% from the phyla Proteobacteria, Bacteroidetes, and Deferribacteres in the colonic mucosa of the TLR4-/- and WT mice fed the VAN or VAD diet. The combined effects of the different VA nutritional levels and TLR4 deletion on the relative abundance of (A) Alistipes and (B) Rikenellaceae_RC9 (n = 10). The main effect of VA, independent of TLR4 deletion, on the relative abundance of (C) Parasutterella and (F) Mucispirillum in the four groups (n = 10). The main effect of TLR4, independent of VA levels, on the relative abundance of (D) Bacteroides and (E) Alloprevotella among the four groups (n = 10). Mean ± SEM; ns. = not significant. “Interaction” indicates an effect of the different VA nutritional levels in the TLR4 knockout vs. WT mice; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
Vitamin A and TLR4 also impacted the community structure at the genus level within the phyla Bacteroidetes, Proteobacteria, and Deferribacteres. According to two-way ANOVA, there was a significant interaction between the effects of VA and TLR4 on the abundance of Alistipes and Rikenellaceae_RC9 in the phylum Bacteroidetes, and the P-values were 0.001 and 0.0026, respectively (Figure 7A,B). However, no significant interaction was observed between the effects of VA and TLR4 on the abundance of Bacteroides (Figure 7D) and Alloprevotella (Figure 7E). After combining the VAN and VAD groups, the abundance of Bacteroides and Alloprevotella in the combined KO group was markedly lower than that in the combined WT group. TLR4 seems to be the main factor affecting the abundance of Bacteroides and Alloprevotella at the Bacteroidetes level. However, the Parasutterella abundance (Proteobacteria) was significantly increased in the combined VAD group compared with that in the combined VAN group, and the abundance of Mucispirillum (Deferribacteres) was significantly reduced in the combined VAD group compared with that in the combined VAN group.
Based on these data, both VA and TLR4 affected the abundance of Allobaculum (Firmicutes), Ruminiclostridium_9 (Firmicutes), Alistipes (Bacteroidetes), and Rikenellaceae_RC9 (Bacteroidetes), while the abundance of Anaerotruncus (Firmicutes), Lachnospiraceae_NK4A136_group (Firmicutes), Oscillibacter (Firmicutes), Parasutterella (Proteobacteria), and Mucispirillum (Deferribacteres) was mainly affected by VA, and that of Bacteroides (Bacteroidetes) and Alloprevotella (Bacteroidetes) was primarily affected by TLR4.
Key Phylotypes in the TLR4-/- and WT Mice With Different VA Levels
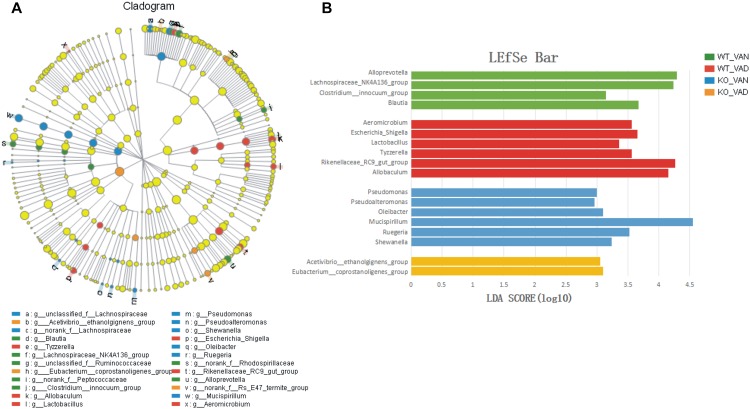
A metagenomic analysis approach (LEfSe) was used to identify the key phylotypes responsible for the differences among the TLR4 KO and WT mice fed the VAN or VAD diet. Figure 8 shows a comparison of the bacterial populations in the four groups at the genus level. The results indicated that the key genera in the WT VAN group were Alloprevotella (LDA = 4.34, P = 0.003751), Lachnospiraceae_NK4A136_group (LDA = 4.14, P = 0.000845), Clostridium_innocuum_group (LDA = 3.45, P = 0.000844), and Blautia (LDA = 3.68, P = 0.000227). In the WT VAD group, the key genera were Aeromicrobium (LDA = 3.81, P = 0.004725), Escherichia_Shigella (LDA = 3.63, P = 0.000193), Lactobacillus (LDA = 3.29, P = 0.007436), Tyzzerella (LDA = 3.58, P = 0.005026), Rikenellaceae_RC9_gut_group (LDA = 4.26, P = 0.000131), and Allobaculum (LDA = 4.13, P = 0.010607). Oleibacter (LDA = 3.19, P < 0.003641), Pseudomonas (LDA = 3.00, P = 0.000502), Mucispirillum (LDA = 4.54, P = 0.012631), Ruegeria (LDA = 3.51, P = 0.000837), Shewanella (LDA = 3.52, P = 0.023659), and Pseudoalteromonas (LDA = 3.07, P < 0.0001) were the key genera in the KO VAN group. However, only two genera played key roles in the KO VAD group: Acetivibrio_ethanolgignens_group (LDA = 3.07, P = 0.004929) and Eubacterium_coprostanoligenes_group (LDA = 3.15, P = 0.022025).
FIGURE 8.
LEfSe analysis of the different structures of the colonic mucosal microbiota in the TLR4 KO and WT mice fed the VAN or VAD diet. (A) A cladogram of the statistical and biological differences in the colonic microbiota among the WT VAN, WT VAD, KO VAN, and KO VAD groups, which are shown by the color of the most abundant phylotypes (n = 10). (B) A histogram of the LDA scores for the most abundant phylotypes (n = 10). Mean ± SEM. VAN, vitamin A normal; VAD, vitamin A deficiency. WT VAN refers to WT mice fed a VAN diet; WT VAD refers to WT mice fed a VAD diet; KO VAN refers to TLR4-/- mice fed a VAN diet; KO VAD refers to TLR4-/- mice fed a VAD diet.
Discussion
The GI tract harbors a complex community of bacteria in the mucosa, lumen and feces. Feces may primarily contain bacteria that are not adherent (Zoetendal et al., 2002), and the luminal microbiota is more variable than the mucosal microbiota (Li et al., 2015). Therefore, in our study, the mucosal bacteria may better reflect the actual intestinal microbiota (Amit-Romach et al., 2009). Some ecologists have noted that the Chao and ACE diversity indexes reflect the richness of the microbiota. The Shannon and Simpson diversity indexes are considered indicators of colony richness and evenness. The Simpson index is sensitive to dominant species, and the Shannon index is sensitive to rare species (Magurran, 1988). In the present study, TLR4 KO reduced the Shannon index and upregulated the Simpson index in the VAD group. TLR4 KO may decrease the abundance of rare species and increase the dominant intestinal mucosa-associated microbiota in VAD rats. However, VA had no effect on the Shannon and Simpson indexes in WT mice; therefore, these two indexes were not found to be affected by either VA or TLR4 after two-way ANOVA with a post hoc test. Although VA and TLR4 had no effect on the abundance and evenness of the colonic mucosa-associated microflora, the results of PCA and PLS-DA showed that VA and TLR4 affected the distribution and composition of the colonic mucosa-associated bacteria.
Amit-Romach showed that VAD led to a reduction in the proportion of Lactobacillus spp. and resulted in the appearance of pathogenic Escherichia coli strains among the mucosa-associated bacteria (Amit-Romach et al., 2009). Matthew C. found that acute VAD had an effect on bacterial community structure, leading to an increase in the abundance of Bacteroides vulgatus (Hibberd et al., 2017). Studies on changes in gut microbial diversity have shown varying results. However, in the present study, at the genus level, VAD decreased the abundance of Anaerotruncus, Oscillibacter, Lachnospiraceae_NK4A136_group, and Mucispirillum and increased the abundance of Parasutterella.
Anaerotruncus is a newly described bacterial genus isolated from human stool (Lawson et al., 2004). Currently, Anaerotruncus colihominis and Anaerotruncus massiliensis are the identified species of this genus (Lau et al., 2006). A. H. Togo isolated Anaerotruncus massiliensis from an obese patient after bariatric surgery (Togo et al., 2016). Although Anaerotruncus is not well known, studies have shown that Anaerotruncus species might be optimal probiotic strains because these species express enzymes that favor the production of butyrate (Polansky et al., 2015). Butyrates are important nutrients for cells lining the mammalian colon. As critical short-chain fatty acid derivatives that regulate colon homeostasis, butyrates participate in colon inflammation (Donohoe et al., 2011). In a study of human gut microflora, Oscillibacter valericigenes was identified in a significantly greater number of samples from healthy controls than from patients with Crohn’s disease (Man et al., 2011; Mondot et al., 2011). Anaerotruncus and Oscillibacter appear to play a positive role in maintaining intestinal immune homeostasis. Lachnospiraceae species are also associated with maintenance of gut health, and members of this family may protect against colon cancer in humans by producing butyric acid (Tap et al., 2009; Meehan and Beiko, 2014; Liu et al., 2017). Mice administered retinoic acid (RA) orally and then subjected to partial hepatectomy had higher levels of Lachnospiraceae than mice in the control group, which were not treated with RA (Liu et al., 2016). In the present study, VAD reduced the abundance of Anaerotruncus and Oscillibacter in the colonic mucosa, and the abundance of Lachnospiraceae _NK4A136 _ group was also lower in the combined VAD group than in the VAN group, while VAD upregulated the abundance of Parasutterella. Previously, the Parasutterella abundance was found to be increased and the Lachnospiraceae abundance was decreased in the submucosal tissues of patients with Crohn’s disease (Chiodini et al., 2015). In addition, the Parasutterella abundance was increased significantly in rats with hypertriglyceridemia-related acute necrotizing pancreatitis (Huang et al., 2017). These results suggest that VA may be involved in regulation of the intestinal mucosa-associated microbiota.
Mucispirillum is a core member of the laboratory mouse microbiota and can colonize the intestinal tract from the stomach to the colon; this genus is represented by a single species, namely, Mucispirillum schaedleri (Robertson et al., 2005). As part of the phylum Deferribacteres, Mucispirillum has been shown to be associated with both inflammatory markers (El Aidy et al., 2014) and active colitis in a T-bet-/- Rag2-/- mouse model (Berry et al., 2012; Rooks et al., 2014) and was even associated with Citrobacter rodentium infection (Hoffmann et al., 2009). A recent study showed that M. schaedleri possesses specialized systems to handle oxidative stress during inflammation (Loy et al., 2017). Interestingly, our data showed that VAD downregulated the abundance of Mucispirillum. We speculate that this downregulation may be a self-regulatory effect of the intestinal mucosa-associated microbiota.
Activation of TLRs by commensal microflora is critical for protection against gut injury (Rakoff-Nahoum et al., 2004; Fukata et al., 2005; Fukata et al., 2006). A large amount of research has indicated that TLR4 signaling affects the intestinal microbiota (Anitha et al., 2012). However, the effects of TLR4 on the intestinal mucosa-associated microbiota are complex and remain unclear. Our results suggest that TLR4 KO decreased the abundance of Bacteroides and Alloprevotella. Bacteroides, a commensal bacterium that colonizes the lower digestive tract, can strongly affect the host immune system (Swidsinski et al., 2009). A study by Erin B showed that Bacteroides species produce a capsular polysaccharide, polysaccharide A (PSA), to repress proinflammatory cytokines (Troy and Kasper, 2010). Many studies have implicated decreased levels of Bacteroides in the development of IBD (Zhou and Zhi, 2016). In our study, TLR4-/- mice exhibited a defective immune response, which may be associated with the marked reduction in Bacteroides abundance. Alloprevotella is a genus of Prevotellaceae, and the clinical significance of this genus remains unclear. Decreased inflammatory cytokine expression in the mouse intestine following interferon tau (IFNT) supplementation led to increased Alloprevotella abundance in the colon (Ren et al., 2016), indicating that Alloprevotella has positive effects on the intestinal mucosa. Intriguingly, the abundance of Alloprevotella and Bacteroides was distinctly reduced in the TLR4 KO group, indicating a complex relationship among TLR4, the microbiota, and intestinal immunity.
Notably, at the genus level, VA and TLR4 had a combined effect on the abundance of Allobaculum (Firmicutes), Ruminiclostridium_9 (Firmicutes), Alistipes (Bacteroidetes), and Rikenellaceae_RC9 (Bacteroidetes). Allobaculum has been shown to prevent dextran sulfate sodium (DSS)-induced inflammation (Wang et al., 2015), and the abundance of this genus was seen to be positively correlated with markers of ileal immunity (Cox et al., 2014). However, in the present study, the abundance of Allobaculum in the WT VAD group was significantly higher than that in the WT VAN group, but there was no difference in the abundance of this genus between the VAN and VAD groups of TLR4 KO mice, suggesting that VA might regulate Allobaculum via TLR4. Ruminiclostridium_9 belongs to the Ruminococcaceae family and is associated with the release of inflammatory and cytotoxic factors from the gut for maintenance of a stable intestinal microecology (Cheng et al., 2017; Ma et al., 2017; Wu et al., 2017). Although there were no differences among the WT VAN, WT VAD, and KO VAD groups in terms of the abundance of Ruminiclostridium_9, we found that the Ruminiclostridium_9 abundance in the KO VAN group was significantly higher than that in the other three groups. These results suggest that TLR4 may be involved in the regulation of intestinal microbiota by VA, but the specific mechanism of regulation and the clinical significance of this regulation need to be further explored. A cross-study analysis showed that Alistipes species are associated with healthy subjects rather than with individuals with gut disease (Mancabelli et al., 2017). In a murine model of DSS-induced colitis, Alistipes finegoldii (Alistipes) was seen to be protective against colitis (Dziarski et al., 2016). However, the Alistipes genus was found to be associated with colorectal cancer, and the abundance of this genus exhibited a negative correlation with the consumption of fruits and vegetables (Feng et al., 2015; Dai et al., 2018). Rikenellaceae_RC9, similar to Alistipes, belongs to the Rikenellaceae family. The abundance of both these genera was affected by VA and TLR4. However, the specific regulatory mechanisms remain unclear.
A large number of studies have shown that the VA nutritional state affects the abundance and composition of the intestinal microbiota (Amit-Romach et al., 2009; Hibberd et al., 2017). There is also considerable variation and discrepancy associated with identification of bacterial markers of VAD among different studies (Chiodini et al., 2015; Liu et al., 2016) and different disease models, such as models of persistent diarrhea and necrotizing enterocolitis (Lv et al., 2016; Xiao et al., 2018). On the other hand, the intestinal microflora was shown to affect the bioavailability of dietary α- and β-carotene and the activity of VA in rats (Grolier et al., 1998). These results indicate that VA levels and the intestinal microbiota are interrelated. Many studies have shown that VAD impairs GI mucosal barrier integrity by altering bacterial populations, the expression of innate immunity-related genes and the number of immune cells (Amit-Romach et al., 2009; Liu et al., 2014; Li et al., 2017). Our study further demonstrated the independent effects of VA and TLR4 on intestinal mucosa-related bacteria. Our study is the first to indicate that TLR4 is involved in regulation of the colonic mucosal microbiota by VA, providing a foundation for elucidating the relationships among VA, the intestinal microecology and intestinal innate immunity. This result further shows that VA regulates TLR4 to improve intestinal barrier function, as described in our previous study (Li et al., 2017). However, elucidation of the specific regulatory mechanism remains challenging and requires further study and the development of novel approaches.
Conclusion
VAD decreased the abundance of Anaerotruncus (Firmicutes), Oscillibacter (Firmicutes), Lachnospiraceae _NK4A136 _group (Firmicutes), and Mucispirillum (Deferribacteres) and increased the abundance of Parasutterella (Proteobacteria). TLR4 KO decreased the abundance of Bacteroides (Bacteroidetes) and Alloprevotella (Bacteroidetes). However, the abundance of Allobaculum (Firmicutes), Ruminiclostridium_9 (Firmicutes), Alistipes (Bacteroidetes), and Rikenellaceae_RC9 (Bacteroidetes) impacted the interaction between VA and TLR4. Therefore, TLR4 may play a pivotal role in the regulation of the intestinal mucosa-associated microbiota and maintenance of the intestinal microecology mediated by VA.
Author Contributions
LX performed the experiments and analyzed the data. BC and DF assisted in completion of the experiments. TY provided technical guidance regarding the use of HPLC. LX and JC wrote the manuscript. TL and JC designed the study. JC provided financial support for the study. All authors read and approved the final manuscript.
Conflict of Interest Statement
Our team bears a patent (Patent No. ZL201010233032.8) on the formula of the vitamin A normal and vitamin A deficiency animal feed in China.
Abbreviations
- HPLC
high-performance liquid chromatography
- IBD
inflammatory bowel disease
- Mean ± SEM
mean ± standard error of mean
- OTU
operational taxonomic units
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RARβ
retinoic acid receptor β
- TLR4-/-
Toll-like receptor 4 knockout
- TLR4
Toll-like receptor 4
- VA
vitamin A
- VAD
vitamin A deficiency
- VAN
vitamin A normal
- WT
wild-type
- ZO-2
zonula-occludens 2
Funding. This work was supported by grants from the National Nature Science Foundation of China (81770526 and 81470799) and from the Science & Technology Project of Yuzhong District of Chongqing (2012) to JC.
References
- Amit-Romach E., Uni Z., Cheled S., Berkovich Z., Reifen R. (2009). Bacterial population and innate immunity-related genes in rat gastrointestinal tract are altered by vitamin A-deficient diet. J. Nutr. Biochem. 20 70–77. 10.1016/j.jnutbio.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Anitha M., Vijay-Kumar M., Sitaraman S. V., Gewirtz A. T., Srinivasan S. (2012). Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143 1006.e4–1016.e4. 10.1053/j.gastro.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D., Schwab C., Milinovich G., Reichert J., Ben Mahfoudh K., Decker T., et al. (2012). Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. ISME J. 6 2091–2106. 10.1038/ismej.2012.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Liang J., Zhang Y., Hu L., Gong P., Cai R., et al. (2017). The bacteriophage EF-P29 efficiently protects against lethal vancomycin-resistant Enterococcus faecalis and alleviates gut microbiota imbalance in a murine bacteremia model. Front. Microbiol. 8:837. 10.3389/fmicb.2017.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini R. J., Dowd S. E., Chamberlin W. M., Galandiuk S., Davis B., Glassing A., et al. (2015). Microbial population differentials between mucosal and submucosal intestinal tissues in advanced Crohn’s disease of the ileum. PLoS One 10:e0134382. 10.1371/journal.pone.0134382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clagett-Dame M., DeLuca H. F. (2002). The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 22 347–381. 10.1146/annurev.nutr.22.010402.102745E [DOI] [PubMed] [Google Scholar]
- Cox L. M., Yamanishi S., Sohn J., Alekseyenko A. V., Leung J. M., Cho I., et al. (2014). Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158 705–721. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Coker O. O., Nakatsu G., Wu W. K. K., Zhao L., Chen Z., et al. (2018). Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 6:70. 10.1186/s40168-018-0451-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheer R., Santaolalla R., Davies J. M., Lang J. K., Phillips M. C., Pastorini C., et al. (2016). Intestinal epithelial Toll-like receptor 4 signaling affects epithelial function and colonic microbiota and promotes a risk for transmissible colitis. Infect. Immun. 84 798–810. 10.1128/IAI.01374-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D. R., Garge N., Zhang X., Sun W., O’Connell T. M., Bunger M. K., et al. (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13 517–526. 10.1016/j.cmet.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R., Park S. Y., Kashyap D. R., Dowd S. E., Gupta D. (2016). Pglyrp- regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PLoS One 11:e0146162. 10.1371/journal.pone.0146162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S., Derrien M., Aardema R., Hooiveld G., Richards S. E., Dane A., et al. (2014). Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef. Microbes 5 67–77. 10.3920/BM2013.0018 [DOI] [PubMed] [Google Scholar]
- Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., et al. (2015). Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 6:6528. 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- Frosali S., Pagliari D., Gambassi G., Landolfi R., Pandolfi F., Cianci R. (2015). How the intricate interaction among Toll-like receptors, microbiota, and intestinal immunity can influence gastrointestinal pathology. J. Immunol. Res. 2015:489821. 10.1155/2015/489821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Chen A., Klepper A., Krishnareddy S., Vamadevan A. S., Thomas L. S., et al. (2006). Cox-2 is regulated by Toll-like receptor-4 (TLR4) signaling: role in proliferation and apoptosis in the intestine. Gastroenterology 131 862–877. 10.1053/j.gastro.2006.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Michelsen K. S., Eri R., Thomas L. S., Hu B., Lukasek K., et al. (2005). Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 288 G1055–G1065. 10.1152/ajpgi.00328.2004 [DOI] [PubMed] [Google Scholar]
- Furuta T., Kikuchi T., Akira S., Watanabe N., Yoshikawa Y. (2006). Roles of the small intestine for induction of toll-like receptor 4-mediated innate resistance in naturally acquired murine toxoplasmosis. Int. Immunol. 18 1655–1662. 10.1093/intimm/dxl099 [DOI] [PubMed] [Google Scholar]
- Grolier P., Borel P., Duszka C., Lory S., Alexandre-Gouabau M. C., Azais-Braesco V., et al. (1998). The bioavailability of α- and β-carotene is affected by gut microflora in the rat. Br. J. Nutr. 80 199–204. [PubMed] [Google Scholar]
- Hibberd M. C., Wu M., Rodionov D. A., Li X., Cheng J., Griffin N. W., et al. (2017). The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci. Transl. Med. 9:eaal4069. 10.1126/scitranslmed.aal4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C., Hill D. A., Minkah N., Kirn T., Troy A., Artis D., et al. (2009). Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect. Immun. 77 4668–4678. 10.1128/IAI.00493-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Chen J., Wang J., Zhou H., Lu Y., Lou L., et al. (2017). Dysbiosis of intestinal microbiota and decreased antimicrobial peptide level in paneth cells during hypertriglyceridemia-related acute necrotizing Pancreatitis in rats. Front. Microbiol. 8:776. 10.3389/fmicb.2017.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K., Woo P. C., Woo G. K., Fung A. M., Ngan A. H., Song Y., et al. (2006). Bacteraemia caused by Anaerotruncus colihominis and emended description of the species. J. Clin. Pathol. 59 748–752. 10.1136/jcp.2005.031773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson P. A., Song Y., Liu C., Molitoris D. R., Vaisanen M. L., Collins M. D., et al. (2004). Anaerotruncus colihominis gen. nov., sp. nov., from human faeces. Int. J. Syst. Evol. Microbiol. 54(Pt 2), 413–417. 10.1099/ijs.0.02653-0 [DOI] [PubMed] [Google Scholar]
- Li G., Yang M., Zhou K., Zhang L., Tian L., Lv S., et al. (2015). Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J. Microbiol. Biotechnol. 25 1136–1145. 10.4014/jmb.1412.12047 [DOI] [PubMed] [Google Scholar]
- Li Y., Gao Y., Cui T., Yang T., Liu L., Li T., et al. (2017). Retinoic acid facilitates Toll-like receptor 4 expression to improve intestinal barrier function through retinoic acid receptor beta. Cell. Physiol. Biochem. 42 1390–1406. 10.1159/000479203 [DOI] [PubMed] [Google Scholar]
- Liu H. X., Hu Y., Wan Y. J. (2016). Microbiota and bile acid profiles in retinoic acid-primed mice that exhibit accelerated liver regeneration. Oncotarget 7 1096–1106. 10.18632/oncotarget.6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zhao J., Guo P., Lu W., Geng Z., Levesque C. L., et al. (2017). Dietary corn bran fermented by Bacillus subtilis MA139 decreased gut cellulolytic bacteria and microbiota diversity in finishing pigs. Front. Cell. Infect. Microbiol. 7:526. 10.3389/fcimb.2017.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li Y., Wang Y., Wang Q., Li X., Bi Y., et al. (2014). Gestational vitamin A deficiency reduces the intestinal immune response by decreasing the number of immune cells in rat offspring. Nutrition 30 350–357. 10.1016/j.nut.2013.09.008 [DOI] [PubMed] [Google Scholar]
- Loy A., Pfann C., Steinberger M., Hanson B., Herp S., Brugiroux S., et al. (2017). Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2:e00171-16. 10.1128/mSystems.00171-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Wang Y., Yang T., Zhan X., Li Z., Hu H., et al. (2016). Vitamin A deficiency impacts the structural segregation of gut microbiota in children with persistent diarrhea. J. Clin. Biochem. Nutr. 59 113–121. 10.3164/jcbn.15-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Wu Y., Xie F., Du K., Wang Y., Shi L., et al. (2017). Dimethyl fumarate reduces the risk of mycotoxins via improving intestinal barrier and microbiota. Oncotarget 8 44625–44638. 10.18632/oncotarget.17886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran A. E. (1988). Ecological Diversity and Measurement. Princeton, NJ: Princeton University Press; 10.1007/978-94-015-7358-0 [DOI] [Google Scholar]
- Man S. M., Kaakoush N. O., Mitchell H. M. (2011). The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat. Rev. Gastroenterol. Hepatol. 8 152–168. 10.1038/nrgastro.2011.56 [DOI] [PubMed] [Google Scholar]
- Mancabelli L., Milani C., Lugli G. A., Turroni F., Cocconi D., van Sinderen D., et al. (2017). Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 93:fix153. 10.1093/femsec/fix153 [DOI] [PubMed] [Google Scholar]
- Meehan C. J., Beiko R. G. (2014). A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 6 703–713. 10.1093/gbe/evu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S., Kang S., Furet J. P., Aguirre de Carcer D., McSweeney C., Morrison M., et al. (2011). Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm. Bowel Dis. 17 185–192. 10.1002/ibd.21436 [DOI] [PubMed] [Google Scholar]
- Polansky O., Sekelova Z., Faldynova M., Sebkova A., Sisak F., Rychlik I. (2015). Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl. Environ. Microbiol. 82 1569–1576. 10.1128/AEM.03473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadro L., Gamble M. V., Vogel S., Lima A. A., Piantedosi R., Moore S. R., et al. (2000). retinol and retinol-binding protein: gut integrity and circulating immunoglobulins. J. Infect. Dis. 182 S97–S102. 10.1086/315920 [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118 229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Ren W., Chen S., Zhang L., Liu G., Hussain T., Hao X., et al. (2016). Interferon Tau Affects Mouse Intestinal Microbiota and Expression of IL-17. Mediat. Inflamm. 2016:2839232. 10.1155/2016/2839232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson B. R., O’Rourke J. L., Neilan B. A., Vandamme P., On S. L., Fox J. G., et al. (2005). Mucispirillum schaedleri gen. Nov., sp. Nov., a spiral-shaped bacterium colonizing the mucuslayer of the gastrointestinal tract of laboratory rodents. Int. J. Syst. Evol. Microbiol. 55(Pt 3), 1199–1204. 10.1099/ijs.0.63472-0 [DOI] [PubMed] [Google Scholar]
- Rooks M. G., Veiga P., Wardwell-Scott L. H., Tickle T., Segata N., Michaud M., et al. (2014). Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 8 1403–1417. 10.1038/ismej.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L., O’Connell R. M., Mazmanian S. K. (2010). Coordination of tolerogenic immune responses by the commensal microbiota. J. Autoimmun. 34 J220–J225. 10.1016/j.jaut.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen C. B. (2001). Vitamin A, infection, and immune function. Annu. Rev. Nutr. 21 167–192. 10.1146/annurev.nutr.21.1.167 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Herber A. (2009). Mucosal flora in Crohn’s disease and ulcerative colitis - an overview. J. Physiol. Pharmacol. 60(Suppl. 6), 61–71. [PubMed] [Google Scholar]
- Takeda K., Kaisho T., Akira S. (2003). Toll -like receptors. Annu. Rev. Immunol. 21 335–376. 10.1146/annurev.immunol.21.120601.141126 [DOI] [PubMed] [Google Scholar]
- Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J. P., et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11 2574–2584. 10.1111/j.1462-2920.2009.01982.x [DOI] [PubMed] [Google Scholar]
- Thornton K. A., Mora-Plazas M., Marín C., Villamor E. (2014). Vitamin A deficiency is associated with gastrointestinal and respiratory morbidity in school-age children. J. Nutr. 144 496–503. 10.3945/jn.113.185876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo A. H., Valero R., Delerce J., Raoult D., Million M. (2016). “Anaerotruncus massiliensis,” a new species identified from human stool from an obese patient after bariatric surgery. New Microbes New Infect. 14 56–57. 10.1016/j.nmni.2016.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping D. L., Clifton P. M. (2001). Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81 1031–1064. 10.1152/physrev.2001.81.3.1031 [DOI] [PubMed] [Google Scholar]
- Troy E. B., Kasper D. L. (2010). Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front. Biosci. 15 25–34. 10.2741/3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tang H., Zhang C., Zhao Y., Derrien M., Rocher E.et al. (2015). Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 9 1–15. 10.1038/ismej.2014.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (1995). Global Prevalence of Vitamin A Deficiency. Micronutrient Deficiency Information System, Nutrition Unit. Geneva: World Health Organization. [Google Scholar]
- Wu W., Lv L., Shi D., Ye J., Fang D., Guo F., et al. (2017). Protective effect of Akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front. Microbiol. 8:1804. 10.3389/fmicb.2017.01804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Li Q., Hu K., He Y., Ai Q., Hu L., et al. (2018). Vitamin A and retinoic acid exhibit protective effects on necrotizing enterocolitis by regulating intestinal flora and enhancing the intestinal epithelial barrier. Arch. Med. Res. 49 1–9. 10.1016/j.arcmed.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Xu N., Tan G., Wang H., Gai X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74 1–8. 10.1016/j.ejsobi.2016.02.004 [DOI] [Google Scholar]
- Zhou Y., Zhi F. (2016). Lower level of Bacteroides in the gut microbiota is associated with inflammatory bowel disease: a meta-analysis. Biomed Res. Int. 2016:5828959. 10.1155/2016/5828959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E. G., von Wright A., Vilpponen-Salmela T., Ben-Amor K., Akkermans A. D., de Vos W. M. (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68 3401–3407. PMCID:PMC126800 10.1128/AEM.68.7.3401-3407.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]