Abstract
Type IV P-type ATPases (P4 ATPases) are lipid flippases that catalyze phospholipid transport from the exoplasmic to the cytoplasmic leaflet of cellular membranes, but the mechanism by which they recognize and transport phospholipids through the lipid bilayer remains unknown. In the present study, we succeeded in purifying recombinant aminophospholipid ATPase 2 (ALA2), a member of the P4 ATPase subfamily in Arabidopsis thaliana, in complex with the ALA-interacting subunit 5 (ALIS5). The ATP hydrolytic activity of the ALA2–ALIS5 complex was stimulated in a highly specific manner by phosphatidylserine. Small changes in the stereochemistry or the functional groups of the phosphatidylserine head group affected enzymatic activity, whereas alteration in the length and composition of the acyl chains only had minor effects. Likewise, the enzymatic activity of the ALA2–ALIS5 complex was stimulated by both mono- and di-acyl phosphatidylserines. Taken together, the results identify the lipid head group as the key structural element for substrate recognition by the P4 ATPase.
Keywords: Arabidopsis thaliana, FLAG-tag purification, lipid flippase, lysophospholipid, membrane protein complex
Introduction
P-type ATPases constitute a large family of active membrane pumps that are driven by ATP-dependent autophosphorylation of a conserved aspartate residue during the catalytic cycle, hence the designation P-type [1]. Based on sequence similarity, the P-type ATPase family is divided into five subfamilies with different transport specificities, among which P4 ATPases translocate phospholipids from the exoplasmic to the cytosolic side of cellular membranes. P4 ATPases are unique to eukaryotes and are found in every eukaryotic genome that has been sequenced, whereas they are absent from eubacteria and archaea. Most family members function in a heterodimer with a β-subunit of the Cdc50p/Lem3p protein family. Their activity is responsible for the generation and maintenance of a non-random distribution of phospholipids across cellular membranes, with the aminophospholipids phosphatidylserine (PS) and phosphatidylethanolamine (PE) restricted to the cytosolic leaflet. This asymmetric lipid arrangement plays a crucial role in multiple cellular processes related to the function of secretory and endocytic pathways [2].
Eukaryotes express several P4 ATPases that display different subcellular localizations and substrate specificities (recently reviewed in [3,4]). With regard to substrate specificity, the P4 ATPases can be divided into three categories: enzymes that preferentially flip PS and to a lesser extent PE, enzymes that preferentially flip phosphatidylcholine (PC) and PE, and enzymes with broad substrate specificity. Yeast Saccharomyces cerevisiae, for example, expresses five members of this family: Neo1p in the endosomal membranes, Drs2p and Dnf3p present in the trans-Golgi network, and Dnf1p and Dnf2p at the plasma membrane [5–8]. Dnf1p, Dnf2p, and Dnf3p have been identified as PE and PC flippases using cell-based assays [8,9], while Drs2p transports PS and PE [9,10]. Neo1 has been implicated in the transport of PE and PS, but the lipid substrate for this P4 ATPase remains to be determined [11]. Several of the 14 P4 ATPases identified in humans have been linked to phospholipid translocation and have been localized to the plasma membrane, the Golgi, and early endosomes (reviewed in [3]). Similarly, the Arabidopsis thaliana genome encodes 12 P4 ATPases, which have been termed ALA1 through ALA12 (for aminophospholipid ATPase). ALA2 promotes flipping of PS in the endosomal system, whereas other ALAs that have been characterized show a broader transport specificity and also transport PE, PC, or sphingomyelin (SM) [12–15].
Understanding the structural basis for substrate recognition by P4 ATPases is important for understanding their role in membrane biology and physiology, and to explain how P4 ATPases can transport such giant substrates compared with the much smaller ions transported by related P-type ATPase such as the Ca2+-ATPases, Na+/K+-ATPases, and plasma membrane H+-ATPases. Herein, we describe the purification and biochemical characterization of the recombinant plant P4 ATPase ALA2 in association with the Cdc50p homolog ALIS5 (ALA-interacting subunit 5). We tested the specificity of ALA2 in order to determine the minimal requirements the substrate must fulfill to be transported by the P4 ATPase. We demonstrate that ALA2 displays a strong preference for the PS head group and is relatively insensitive to the composition of the acyl groups bound to the glycerol unit.
Materials and methods
Materials
N-dodecyl-β-maltoside (DDM) was obtained from GLYCON Biochemicals (Luckenwalde, Germany). All other detergents were obtained from Affymetrix (Maumee, Ohio, U.S.A.). [γ-32P]ATP (3000 Ci/mmol) was purchased from PerkinElmer. PageRuler™ prestained ladder from Thermo Scientific was used for size determination in SDS–PAGE gels. 1-Palmitoyl-2-oleoyl-sn-phosphatidyl-hydroxy-propionate (PP), 1-palmitoyl-2-oleoyl-sn-phosphatidylserine methyl ester (PS-O-Me), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-d-serine (P-d-S), 1-palmitoyl-2-oleoyl-sn-phosphatidylhomoserine (PhS), and 1-NBD-dodecanoyl-2-hydroxy-sn-glycero-3-phospho-serine (NBD-lyso-PS) were synthesized by headgroup exchange using phospholipase D [16]; other lipids were purchased from Avanti Polar Lipids (Alabaster, AL, U.S.A.). All synthetic lipids were characterized by thin layer silica gel chromatography, 1H-NMR, mass spectrometry, and circular dichroism for structural and stereochemical purity. The purity of all synthetic lipids was higher than 95%. All other chemicals were obtained from Sigma–Aldrich (Taufkirchen, Germany) unless otherwise indicated.
Strains, media and growth conditions
Escherichia coli TOP10 (Invitrogen) was used for all plasmid amplifications and isolations according to standard protocols. Yeast strains were grown at 30°C in yeast extract–peptone medium with glucose (YPD) or galactose (YPG), or in selective synthetic dextrose (SD) or galactose (SG) medium supplemented with appropriate auxotrophic components [17]. For solid media, 2% agar was added. The protease-deficient strain MP3848, deleted in PEP4, a gene encoding a key hydrolytic enzyme, was created from w303-1b/Gal4 (MATα, leu2, his3, trp1::TRP1-GAL10-GAL4, ura3, ade2-1, canr, cir+; [18]) using a loxP–HIS3–loxP cassette as described [19]. Lipid translocation assays were carried out employing the S. cerevisiae mutant strain ZHY709 (MATα his3 leu2 ura3 met15 dnf1Δ dnf2Δ drs2::LEU2 [6]. Yeast cells were transformed by the lithium acetate method [20].
Plasmid constructions
Primers and plasmids used are listed in Supplementary Tables S1 and S2. All PCRs were carried out using Phusion® High-Fidelity DNA Polymerase (New England Biolabs) according to the manufacturer's instructions. For co-expression ALA2 and ALIS5, a DNA fragment corresponding to FLAG–ALIS5 was excised from pMP3836 [21] using AgeI and SacI and inserted into plasmid pMP3395 [14] cut with the same restriction enzymes, yielding plasmid pMP3681. The new multicopy (2 µm) plasmid containing His10-tagged ALA2 and FLAG-tagged ALIS5 on each side of a bidirectional galactose-inducible GAL1-10 promoter was named pMP3681. HA-tagged ala2E159Q was generated by overlapping PCR, cloned into pENTR-D/TOPO (Invitrogen), and then transferred to a modified version of yeast plasmid pRS423-GAL [13] using the Gateway technology, rendering plasmid pMP3464. Cloning of HA-tagged ala2D381A into pRS423-GAL (pMP2766) has been described elsewhere [13]. An N-terminal FLAG-tagged version of ala2E159Q was generated by overlapping PCR and cloned in frame with the FLAG tag present in plasmid pMP3074 [21], using BamHI and SacI digestion. The final plasmid containing FLAG-tagged ala2E159Q was named pMP4131.
Purification and solubilization of the ALA2–ALIS5 complex
All steps were performed at 4°C unless otherwise stated. Protease-deficient yeast strain MP3848 was transformed with plasmid pMP3681 (for expression of ALA2 and ALIS5) or pMP4131 (for purification of ala2E159Q in the absence of ALIS5). After selection, protein expression was induced in YPG medium. Unless otherwise indicated, cells were collected 8 h after induction, re-suspended in 10 ml of lysis buffer (100 mM NaCl, 50 mM Tris–HCl, pH 7.4, 1 mM EGTA, 20% glycerol) containing protease inhibitors (1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 5 µg/ml antipain, and 157 µg/ml benzamidine, 1 mM phenylmethanesulfonyl fluoride), and lysed by vortexing 16 times for 30 s with acid-washed glass beads (10 g; 0.5 mm). The lysate was clarified by centrifugation (2000×g; 5 min). For solubilization tests, membranes were collected from the precleared lysates by centrifugation (100 000×g; 1 h) and detergent-solubilized for 75 min with end-over-end rotation at a protein concentration of 3 mg/ml in S-buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 2% glycerol) supplemented with protease inhibitors and 0.5% (w/v) detergent. Insoluble material was removed by centrifugation (78 000×g; 1 h) and supernatant and pellet were analyzed by Western blot (WB). For purification, 0.8% (w/v) DDM was used during solubilization and the supernatant was incubated with 12 µl/ml anti-FLAG-M2 affinity resin for 16 h with end-over-end rotation. The resin was washed three times for 10 min with S-buffer containing 0.05% (w/v) DDM. Proteins were eluted using S-buffer containing 0.05% (w/v) DDM and 400 µg/ml FLAG peptide. The protein was finally concentrated on Vivaspin™ columns (GE Healthcare) with a molecular mass cutoff of 10 kDa.
Protein analysis
ALA2 concentration was quantified by densitometry analysis of Coomassie-stained SDS–PAGE gels including known amounts of the A. thaliana H+-ATPase isoform 2 (AHA2) as standard [22]. Gels were scanned using a Gel Doc 2000 (Bio-Rad) and the bands were quantified using ImageJ (National Institutes of Health). Analysis by mass spectrometry was performed using previously described methods [23,24]. FLAG-tagged ALIS5 was immunodetected using monoclonal mouse antibodies anti-FLAG-M2 (Qiagen, Hilden, Germany), and wild-type and mutant ALA2 with a polyclonal antibody raised against its C terminus [21]. Dpm1p was immunodetected using an anti-dolichol phosphate mannose synthase antibody (Molecular Probes), and Pma1p with a polyclonal antibody raised against its C terminus [25]. Secondary antibodies were polyclonal goat-anti-rabbit-AP (alkaline phosphatase) and polyclonal rabbit-anti-mouse-AP (Dako, Glostrup, Denmark). All antibodies were used at 1 : 5000 dilution. Blots were developed using BCIP/NBT Color Development Substrate (Promega, Madison) according to the manufacturer's instructions. Peptide-N-glycosidase F (PNGAse F) treatment was carried out at room temperature essentially as described [26]. Discontinuous sucrose-gradient fractionation of plasma membrane-enriched membrane fractions was performed as previously described [13].
Phosphoenzyme and ATPase activity assays
Purified protein (100 ng) in S-buffer (usually at pH 7.4) containing 0.05% DDM, in the absence or presence of exogenous lipids (0.32 mg/ml), was incubated for 20 min at room temperature for protein activation and then assayed for phosphorylation and ATPase activity using 1 mM ATP and 2 µCi of [γ-32P]ATP as described previously [27,28]. Exogenous lipids were prepared in round-bottom glass tubes. To ensure complete relipidation for all samples, the lipid of interest was mixed with PC 16 : 0/18 : 1 in chloroform at a molar ratio of 30 : 70. The solvent was removed under a stream of nitrogen, and the lipids were solubilized in S-buffer containing 0.05% DDM to a concentration of 0.32 mg/ml by vortexing with 5-mm glass beads. Inhibition was assayed in mixtures supplemented with 1 mM orthovanadate unless otherwise stated. Curves of ATPase activity versus lipid concentration were fitted to v = VmaxL/(L + Ka), where L is the lipid concentration. For analysis in the pH range 5.5–8.5, pH was adjusted using varying proportions of low pH buffer (35 mM HEPES, 40 mM MES, 150 mM NaCl, pH 5.5) and high pH buffer (35 mM HEPES, 40 mM MES, 150 mM NaCl, pH 8.5). For pH 4, a 100 mM Tris-buffered succinic acid solution was used. ATPase activity versus orthovanadate curves were fit to the equation , where y is the ATPase activity; x is the orthovanadate concentration; IC50 is the half maximal inhibitory concentration; max and min are approximated by the program SigmaPlot 11.0 (Systat Software, Inc., U.S.A.) automatically during the calculation.
Lipid uptake assay and microscopic visualization of lipid translocation
Uptake of NBD lipids was analyzed as described previously [13]. For microscopic visualization of lipid translocation, yeast cells were added 4 ng/µl DAPI (4′,6-diamidino-2-phenylindole) immediately before the addition of fluorescent lipid analogs. After incubation with lipids, cells were washed twice in SG medium without galactose but containing 2% sorbitol, 20 mM NaN3, and 4% (w/v) bovine serum albumin to extract NBD lipids from the cell surface. At this point, 5-µl aliquots of the washed cell samples were collected and mounted on microscopic slides for visualization, using a Leica SP5-X confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany). DAPI and NBD lipids were excited at 355 and 478 nm, respectively. Emission spectra were recorded between 430 and 475 nm for DAPI and between 509 and 594 nm for NBD. A 63×/1.40 numerical aperture oil-immersion objective was used. For flow cytometry analysis, aliquots of 50 µl of cells were labeled with 1 µl of 1 mg/ml propidium iodide for staining of non-viable cells. Flow cytometry was performed on a Becton Dickinson FACS Calibur system (San Jose, CA) equipped with a 488 nm argon laser for excitation as described [29]. For NBD and propidium iodide detection, fluorescence emission was measured with a 530/30 nm band-pass filter and a 670 nm long-pass filter, respectively. Twenty thousand cells were analyzed without gating during the acquisition. Data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR) as previously described [30]. Briefly, live yeast cells were selected based on forward/side-scatter gating and propidium iodide exclusion. The NBD fluorescence of living cells was plotted on a histogram and the geometrical mean of the NBD fluorescence intensity of the living cell population was used for quantification.
Results
Expression, solubilization, and purification of the plant ALA2–ALIS5 complex
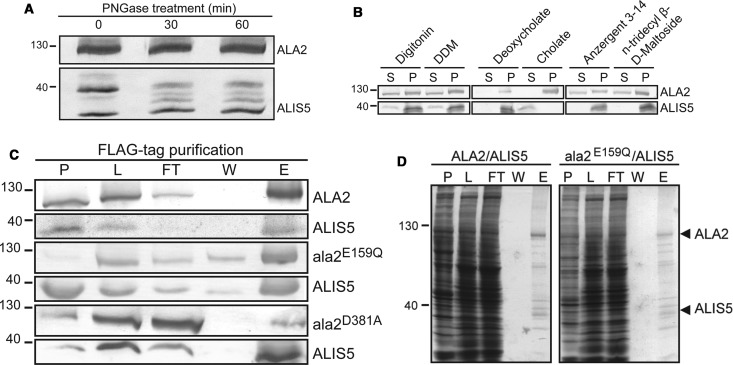
To facilitate purification, we co-expressed N-terminally His10-tagged ALA2 and N-terminally FLAG-tagged ALIS5 under the control of an inducible bidirectional galactose promoter in a protease-deficient strain of the yeast S. cerevisiae. Upon galactose induction, the expression of both proteins could be detected by immunoblotting of total membrane preparations as a prominent band at ∼120 kDa for ALA2 and three weaker bands between 33 and 40 kDa for ALIS5 (Figure 1A). Deglycosylation experiments using PNGase F demonstrated that the multiple bands observed for ALIS5 are due to protein glycosylation. To determine optimal solubilization conditions, membrane preparations from galactose-induced cells were exposed to a range of detergents (Figure 1B). Of the tested detergents, only digitonin and DDM showed sufficient solubilization of both proteins. DDM has been successfully used for solubilization and biochemical characterization of the yeast P4 ATPase Drs2p [26] and was chosen for further purification of ALA2 by anti-FLAG affinity chromatography. This resulted in efficient co-purification of ALA2 and ALIS5 (Figure 1C). The purity of the ALA2–ALIS5 complex was assessed by SDS–PAGE followed by Coomassie blue staining (Figure 1D). A band of the expected molecular mass for HIS–ALA2 (≍120 kDa), a band at the expected molecular mass for FLAG–ALIS5 (≍37 kDa), and several contaminating proteins were detected. Identities of proteins were further confirmed by mass spectrometric analysis (Supplementary Table S3).
Figure 1. Expression and purification of the ALA2–ALIS5 complex.
(A) Yeast membranes expressing N-terminally His10-tagged ALA2 and FLAG-tagged ALIS5 were incubated with PNGase F for the indicated times and analyzed by immunoblotting. (B) Solubilization efficiency by non-ionic and zwitterionic detergents at a concentration of 0.5% (w/v). S, supernatant; P, insoluble material. (C) Western blot analysis of fractions from FLAG-tag affinity purification for wild-type and two non-functional ALA2 mutants. (D) Coomassie blue-stained SDS–PAGE gels representative of at least two purifications of the ALA2–ALIS5 and ala2E159Q–ALIS5 complexes. Arrows indicate the positions of purified ALA2 and its glycosylated ALIS5 subunit. Abbreviations: P, insoluble material; L, solubilized material, loaded onto affinity beads; FT, flow through; W, pooled washes; E, eluate. The positions of molecular mass protein markers are given in kDa.
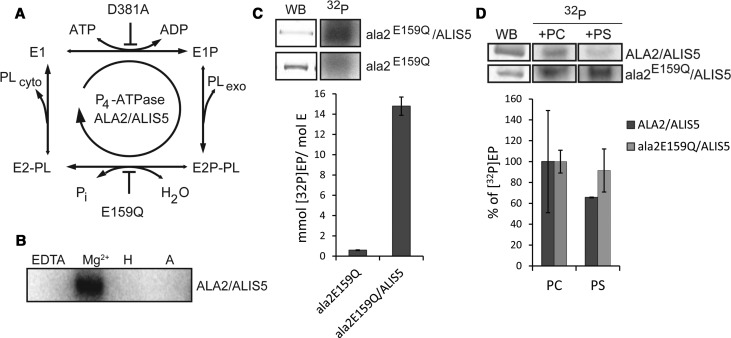
For further biochemical analysis of the ALA2–ALIS5 complex, two non-functional ALA2 mutants were generated that are blocked at different transition steps during the reaction cycle (Figure 2A). The mutant ala2D381A lacks the aspartic acid residue in the conserved DKTG motif that is usually phosphorylated at the beginning of the cycle and can therefore not be phosphorylated. The mutant ala2E159Q lacks the glutamate in the conserved DGE motif, which is the key amino acid required for dephosphorylation of D381, and consequently is blocked in the turnover of the phosphorylated protein. Both mutants were co-expressed with ALIS5, purified and analyzed in the same manner as wild-type ALA2. While the E159Q mutant is efficiently co-purified with FLAG–ALIS5, recovery of the D381A mutant with FLAG–ALIS5 was not sufficient to allow for further studies (Figure 1C).
Figure 2. Purified ALA2 is active and the catalytic cycle includes a phosphorylated intermediate.
(A) Schematic representation of the Post-Albers catalytic cycle for P4 ATPases. ALA2 mutations blocking different steps of the cycle, D381A and E159Q, are indicated. Abbreviations: Exo, exoplasmic side; cyto, cytosolic side; Pi, inorganic phosphate; PL, phospholipid. (B) Radiogram showing the phosphoenzyme state of ALA2 in the presence ALIS5. The phosphoenzyme state was tested in the presence of 20 mM EDTA (EDTA), without inhibitors (Mg2+), upon treatment with 20 mM hydroxylamine (H) after the reaction, or pre-incubation with 2 mM AMP-PCP (A). (C) Radiograms showing phosphoenzyme levels of ala2E159Q in the presence or absence of ALIS5. The amount of protein present in the reactions was compared by Western blot (WB) analysis. Excised bands in the radiograms are from the same gels ensuring equal exposure. (D) Dephosphorylation analysis of phosphointermediates for ALA2 and ala2E159Q in the presence of ALIS5 and PC 16:0/18:1 (set to 100%) as compared with PS 16:0/18:1. The phosphoenzyme state of ALA2 was monitored 50 s after addition of cold ATP (40 µM). Excised bands in the radiograms are from the same gels ensuring equal exposure. WB analysis served as loading control. Data represent the mean of triplicate determinations and are representative of at least two independent experiments. Error bars show SD.
Phosphoenzyme formation and PS-stimulated dephosphorylation
To verify that the purified protein complex was an active P-type ATPase, we subjected it to phosphorylation and dephosphorylation assays (Figure 2). Incubation of the purified complex with [γ-32P]ATP resulted in the Mg2+-dependent formation of an acid-stable phosphorylated ALA2 (Figure 2B). This phosphorylation was found to be strongly dependent on association with ALIS5. Thus, ala2E159Q purified in the absence of ALIS5 failed to become phosphorylated (Figure 2C), demonstrating that interaction with the β-subunit is a key element of P4 ATPase activity. Importantly, dephosphorylation of the wild-type protein was PS-dependent, demonstrating that full ATP turnover requires the presence of the substrate (Figure 2D). The ala2E159Q mutant was not dephosphorylated, which confirms that the DGE motif is essential for dephosphorylation during catalysis (Figure 2D). Furthermore, phosphorylation levels for ALIS5-bound ala2E159Q were consistently higher than those found for the wild-type protein (Figure 2D).
Lipid-stimulated ATPase activity of the ALA2–ALIS5 complex
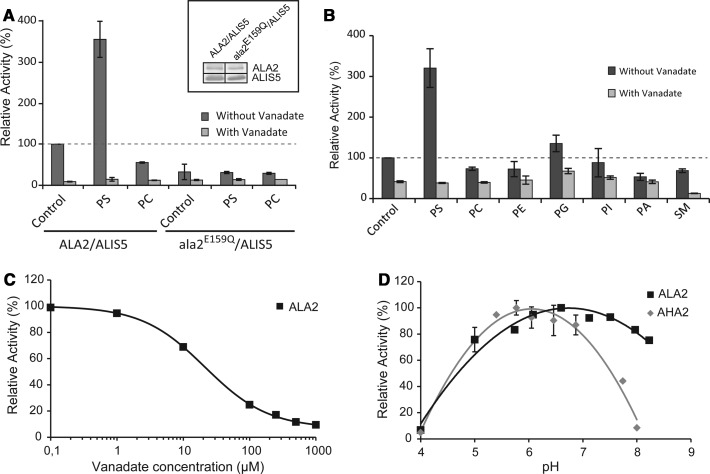
We next analyzed lipid-stimulated ATPase activity by assaying ATP hydrolytic activities of wild-type ALA2–ALIS5 and ala2E159Q–ALIS5 (Figure 3). ATPase activity for the purified, detergent-solubilized complex varied considerably among preparations with a basal activity between 80 and 150 nmol of ATP/min/mg protein, probably due to the presence of cellular lipids carried over in the purification steps. The wild-type protein complex was specifically activated by the presence of PS (3- to 9-fold increase, depending on the preparation), while ala2E159Q–ALIS5 was inactive under the same conditions (Figure 3A), confirming that the observed PS-stimulated ATPase activity is due to ALA2–ALIS5. Weak activation (<1.3-fold) was also observed in the presence of phosphatidylglycerol (PG 18 : 1/18 : 1) but not with other lipids, including PC (16 : 0/18 : 1) alone, PE (16 : 0/18 : 1), phosphatidic acid (PA 18 : 1/18 : 1), phosphatidylinositol (PI 16 : 0/18 : 1), and SM (16 : 0) (Figure 3B). Orthovanadate inhibited the purified ALA2–ALIS5 complex in a concentration-dependent manner, with a calculated IC50 of ∼22 µM (Figure 3C). This value is comparable to that reported for the yeast P4 ATPase Drs2p (Table 1). ALA2–ALIS5 was active in a broad pH range (Figure 3D), with an optimum at pH 7, similar to other purified P4 ATPases (Table 1).
Figure 3. ALA2–ALIS5 is a PS-stimulated ATPase.
The effect of various phospholipids (30 mol%) in mixture with PC 16:0/18:1 (70 mol%) on the ATPase activity of purified ALA2–ALIS5 was assayed in the absence and presence of 1 mM vanadate. (A) ATPase activity of purified ALA2–ALIS5 and ala2E159Q–ALIS5. Inset: Western blot analysis to verify the amount of protein present in the reactions. (B) Effect of changes in the lipid head group. (C) Orthovanadate sensitivity of ALA2–ALIS5 activity in the presence of PS/PC 16:0–18:1 (molar ratio, 30:70). The line corresponds to the best fit of the data as described in Material and methods. (D) pH dependence of ALA2–ALIS5 ATPase activity in comparison with the plant proton pump AHA2. Values are normalized with respect to the values at pH 6.5 (ALA2) or pH 6.0 (AHA2). The lines correspond to a second-order polynomial regression. All data are representative of at least two independent experiments. Error bars show SD. Abbreviations: PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidyl-l-serine; SM, sphingomyelin.
Table 1. Comparison of biochemical properties of ALA2 with other purified P4 ATPases.
| ATPase | Specificity | Vmax for PS activation (µmol min−1 mg−1) | K0.5 for activation by PS (μM) | K0.5 for inhibition by vanadate (μM) | pH range | References | |
|---|---|---|---|---|---|---|---|
| Head group | Fatty acid | ||||||
| ALA2 | PS | Di-acyl | 0.8 | 3.5 | 22 | 5–8 | The present study |
| Mono-acyl | 0.35 | 20 | – | – | |||
| Drs2p | PS, PE | Di-acyl | 0.45 | – | 5–10 | 6–8 | [10,26] |
| ATP8A1 (bovine) | PS, PE | Di-acyl | 3–9 | – | – | – | [41] |
| ATP8A1 (murine) | PS (PE) | Di-acyl | 14.6 | 32 | – | – | [42] |
| ATP8A1 (human) | PS* | Di-acyl | 38.7 | 59.1 | – | – | [39] |
| ATP8A2 | PS, PE | Di-acyl | 45–107 | 38–98 | 2.7 | 7–9 | [37,38] |
| ATP11A | PS, PE | Di-acyl | 15.1 | 87 | – | – | [40] |
| ATP11B | PS, PE | Di-acyl | 17.3 | 178 | – | – | [40] |
| ATP11C | PS, PE | Di-acyl | 34.9; 9.5 | 69; 0.8 | – | – | [40,43] |
PE not analyzed.
Effect of modification of the head and tail group of phosphatidylserine
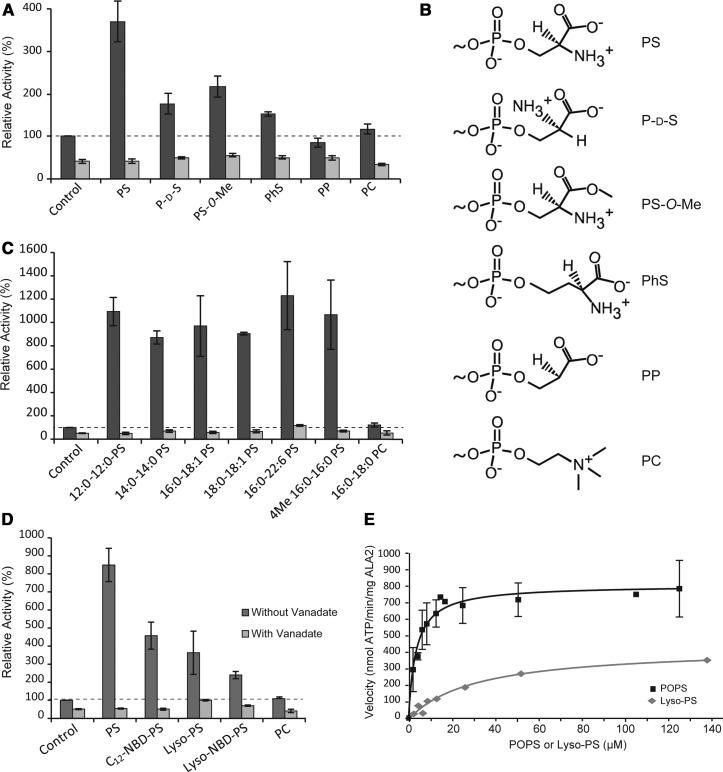
To provide further insights into the structural specificity of the observed PS effect, we explored the ability of PS stereoisomers and structural derivatives to stimulate ATPase activity (Figure 4A,B). Maximal activation occurred in the presence of the naturally occurring phosphatidyl-l-serine (PS). The stereoisomer phosphatidyl-d-serine (P-d-S) was less than half as effective at stimulating ATPase activity compared with PS. Similarly, modification of the size of the PS head group (phosphatidylhomoserine, PhS), or esterification of the carboxyl group (phosphatidylserine methyl ester, PS-O-Me) halved the activity. Notably, removal of the amine group (PP) completely prevented ATPase stimulation. Taken together, this is evident that ALA2 recognizes the phospholipid head group with high specificity.
Figure 4. Activation of ALA2–ALIS5 by PS and lyso-PS.
The effect of various PS phospholipids (30 mol%) in mixture with PC 16:0/18:1 (70 mol%) on the ATPase activity of purified ALA2–ALIS5 was assayed in the absence and presence of 1 mM vanadate. Values were normalized to the rate of ATP hydrolysis by the purified ALA2–ALIS5 without lipid addition (control). (A) Effect of changes in the PS head group chemistry. (B) Polar head group structures of the lipid derivatives employed in (A). (C,D) Effect of PS acyl chain composition. (E) ATPase activity was measured as a function of increasing PS 16:0/18:1 and lyso-PS 16:0 concentrations. Lines correspond to the best fit of the data as described in Materials and methods. The data are representative of at least two independent experiments. Error bars show SD. Abbreviations: PC, phosphatidylcholine; PS, phosphatidyl-l-serine; P-d-S, phosphatidyl-d-serine; PS-O-Me, phosphatidylserine methyl ester; PhS, phosphatidylhomoserine; PP, phosphatidyl-hydroxy-propionate; C12-NBD-PS, palmitoyl-(NBD-dodecanoyl)-PS; Lyso-PS, 1-hexadecanoyl-sn-glycero-3-phospho-l-serine; Lyso-NBD-PS, 1-NBD-dodecanoyl-sn-glycero-3-phospho-l-serine.
Finally, we analyzed the ATPase activity of ALA2–ALIS5 in the presence of PS with various acyl compositions (Figure 4C). ALA2–ALIS5 showed equally high stimulated ATPase activity by PS containing various combinations of saturated (lauric acid 12:0, myristic acid 14 : 0, palmitic acid 16 : 0, and stearic acid 18 : 0), unsaturated (oleic acid 18 : 1, and docosahexaenoic acid 22 : 6) or two-branched fatty acids (phytanoyl chains, 4Me 16 : 0) at the sn-1 and sn-2 positions. Remarkably, even 1–16 : 0-lysophosphatidyl-serine (lyso-PS), fluorescent PS and lyso-PS analogs carrying a bulky NBD group were able to stimulate the ATPase activity of the complex, albeit to a lesser extent than di-acyl PS (Figure 4D). The Vmax and Ka values for lyso-PS were 450 ± 40 nmol ATP/min/mg and 34 ± 7 µM, respectively, as compared with 814 ± 38 nmol ATP/min/mg and Ka of 3.48 ± 0.5 µM for 16 : 0–18 : 1 PS (Figure 4E). Collectively, these findings indicate that the pump complex tolerates drastic changes in both the length and structure of the acyl chains, which is evidence that the phospholipid tail is hardly recognized by ALA2.
ALA2–ALIS5 flips both mono- and di-acyl phosphatidylserine
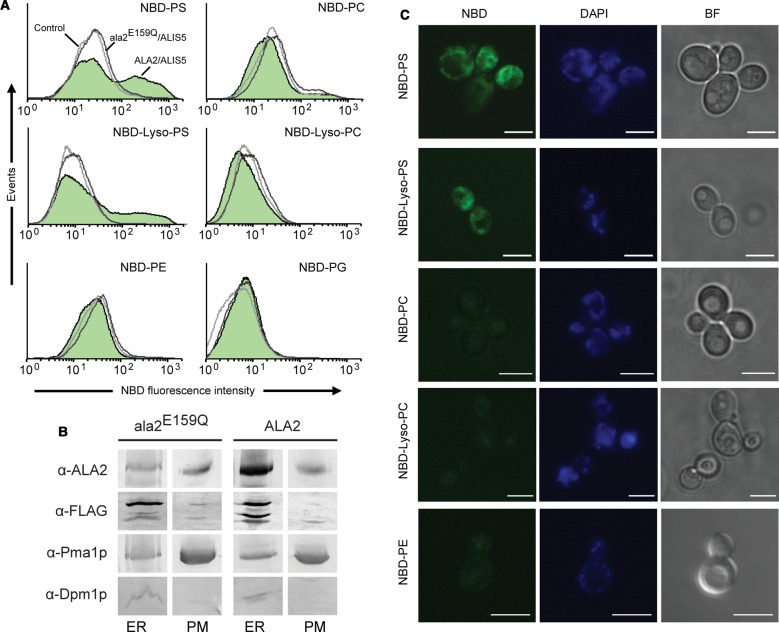
To confirm that the observed PS-induced stimulation of ATPase activity actually reflects lipid transport by ALA2–ALIS5, we analyzed the in vivo uptake of two fluorescent PS analogs, NBD-PS and NBD-lyso-PS into Δdrs2Δdnf1Δdnf2 yeast cells expressing or lacking ALA2–ALIS5. This triple yeast mutant strain lacks three endogenous P4 ATPases and thus presents a low background of lipid uptake through the plasma membrane. Co-expression of ALA2 with ALIS5 in the Δdrs2Δdnf1Δdnf2 yeast strain resulted in a population of cells with increased internalization of both NBD-PS and lyso-NBD-PS, but not of NBD-PC, -PE, and -PG (Figure 5A). The catalytically-‘dead’ mutant ala2E159Q in combination with ALIS5 was capable of reaching the plasma membrane but unable to support NBD-lipid translocation (Figure 5A,B), confirming that P4 ATPase activity is a requirement for lipid internalization at the plasma membrane. Fluorescence microscopy performed on the same ALA2–ALIS5-expressing cells revealed intense labeling of intracellular membranes in the presence of C6-NBD-PS and lyso-NBD-PS but not for the other derivatives (Figure 5C). Based on these findings, we conclude that stimulation of ATP hydrolytic activity of ALA2–ALIS5 by PS and lyso-PS is coupled to the transport of these lipids.
Figure 5. Activation of ALA2–ALIS5 by PS and lyso-PS is coupled with their transport in vivo.
Yeast Δdrs2Δdnf1Δdnf2 cells expressing ALIS5 together with wild-type ALA2 or ala2E159Q were labeled with DAPI and the indicated NBD-lipids. (A) Analysis by flow cytometry. NBD fluorescence of living cells was plotted on an overlay histogram. Graphs represent cells transformed with an empty vector (control, light gray line), ala2E159Q–ALIS5 (dark gray line) or ALA2–ALIS5 (green shadow). Results show representative histograms of at least three independent experiments. Results show representative histograms of at least two independent experiments. (B) Density-gradient fractionation of plasma membrane-enriched membrane preparations. Fractions corresponding to 30 and 48% sucrose, enriched, respectively, in endomembranes (ER) and plasma membranes (PM), were analyzed. Western blots were probed using the following antibodies: anti-Pma1p, plasma membrane; anti-Dpm1p, ER; anti-FLAG, ALIS5, and anti-ALA2. (C) Analysis by fluorescence (NBD, DAPI) and bright-field (BF) microscopy. Scale bar, 5 μm. The figure shows representative images of cell populations with NBD-lipid uptake from at least two independent experiments.
Discussion
A striking characteristic of the ALA2 protein is its high selectivity for the PS head group. The structural basis for the different substrate specificities between P4 ATPases has not been elucidated. Recent mutational studies suggest two opposing pathways by which P4 ATPases could transport their substrate [15,31,32]: (i) the classical pathway with the lipid transported through the interior of P4 ATPases in analogy with the cation transport mechanism of well-characterized P2-ATPases and (ii) a non-classical pathway in which transport occurs at the protein–membrane interface. In both cases, the hydrophilic phospholipid head group makes direct contact with the P4 ATPase protein during transport, while the acyl chains remain within the surrounding lipid environment of the membrane at all stages. Yet, the structure of the lipid tail may influence the correct positioning of the glycerol backbone and thereby affect the recognition of mono- and di-acyl phospholipids. Results presented here define the phosphoryl head group of the lipid as the key element for substrate selection by ALA2–ALIS5. Thus, P4 ATPases must at least provide a sizeable hydrophilic pathway for the polar head group to pass through the membrane.
Modification of the carboxyl group (POPS-O-Me) or serine stereochemistry (P-d-S) resulted in a reduction in ATPase stimulation, while PE and PP were incapable of activating the enzyme. In line with these results, expression of ALA2–ALIS5 supported the uptake of fluorescent PS analogs but not of PE and PC in Δdrs2Δdnf1Δdnf2 yeast cells (Figure 5, [13]). Thus, the presence of both an amine and a carboxyl group in the head group of the lipid substrate are necessary for maximal transport. On the other hand, and in contrast with other aminophospholipid-specific P4 ATPases (Table 1; [33]), the protein tolerated drastic changes of the fatty acid chains attached to the glycerol group. Removing one of the two acyl chains (lyso-PS), changing the length and structure of the acyl chains or even adding a bulky fluorescent NBD group at the sn-1 or sn-2 chain (lyso-NBD-PS, C6/C12-NBD-PS) of the PS molecules still allowed recognition as a transport substrate, albeit with different affinity. These data are consistent with a broader degree of substrate specificity for P4 ATPases than has been previously recognized. Notably, some P4 ATPases, such as yeast Dnf1p and Dnf2p, are capable of transporting synthetic alkylphospholipids lacking a glycerol backbone [34].
PS is a relatively minor constituent of most biological membranes but of high physiological importance. It is found preferentially in the inner leaflet of the plasma membrane and in endocytic membranes and known to regulate the recruitment and activity of numerous enzymes and structural components (reviewed by Leventis and Grinstein [35]). We have localized the ALA2–ALIS5 complex to the endosomal system in plants [13]. It is thus feasible that ALA2–ALIS5 might help generate and maintain lipid asymmetry for PS in the endosomal system. Apart from establishing lipid asymmetry, several members of the P4 ATPase family are required for vesicle-mediated protein transport within the endosomal and secretory pathways, indicating a role for phospholipid translocation in vesicular transport [36]. Similarly, the ALA2–ALIS5 complex might contribute to vesicle biogenesis and membrane trafficking in plants. In line with this notion, overexpression of ALA2 together with ALIS5 in tobacco cells resulted in the formation of an aberrant prevacuolar compartment [13].
P4 ATPases, in complex with their Cdc50p β-subunits, have emerged as key regulators of transmembrane phospholipid transport in eukaryotes, yet key features of their activity remain to be elucidated. To date, only yeast Drs2p and few mammalian P4 ATPases (ATP8A1, ATP8A2, ATP11A, ATP11B, and ATP11C) have been purified and studied under defined conditions [10,37–40]. Here, we describe the first successful purification of a plant P4 ATPase in complex with a Cdc50p homolog. This material provided us with the possibility to get detailed insight into the phospholipid substrate specificity of a P4 ATPase and should also be useful for future structural studies.
Acknowledgements
Imaging data were collected at the Center for Advanced Bioimaging (CAB) Denmark, University of Copenhagen.
Abbreviations
- AP
alkaline phosphatase
- DAPI
4′,6-diamidino-2-phenylindole
- DDM
N-dodecyl-β-maltoside
- NBD
7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PA
phosphatidic acid
- PC
phosphatidylcholine
- P-d-S
phosphatidyl-d-serine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PhS
phosphatidylhomoserine
- PI
phosphatidylinositol
- PP
phosphatidyl-hydroxy-propionate
- PS
phosphatidylserine
- PS-O-Me
phosphatidylserine methyl ester
- SD
synthetic glucose
- SG
synthetic galactose
- SM
sphingomyelin
- YPD
yeast extract–peptone–glucose
- YPG
yeast extract–peptone–galactose
Author Contribution
T.G.P. and R.L.L.-M. designed and supervised the project. R.L.L.-M. and L.T. performed cloning. L.T. and K.F. purified protein and analyzed ATPase activity. D.M.S. performed phosphorylation studies. R.M., G.D. and J.S. performed and analyzed mass spectrometric experiments. L.T. and D.L.D. synthesized lipids. L.T. performed lipid transport experiments. R.L.L.-M. performed confocal microscopy. L.T. and T.G.P. wrote the first version of the manuscript and revised it with the help of K.F., R.L.L.-M., D.L.D., and M.P. All authors commented on the final version.
Funding
This work was supported by the UNIK research initiative of the Danish Ministry of Science, Technology and Innovation through the ‘Center for Synthetic Biology’ at the University of Copenhagen, the Danish National Research Foundation through the PUMPKIN Center of Excellence [DNRF85], the Danish Council for Independent Research/Natural Sciences [FNU, project number 10-083406] (R.L.L.M., T.G.P.), the Deutsche Forschungsgemeinschaft [GU 1133/11-1; T.G.P.] and the Lundbeck Foundation [R34-A3562] (TGP).
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Axelsen K.B. and Palmgren M.G. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 10.1007/PL00006286 [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Marques R.L., Theorin L., Palmgren M.G. and Pomorski T.G. (2014) P4-ATPases: lipid flippases in cell membranes. Pflügers Arch. 466, 1227–1240 10.1007/s00424-013-1363-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen J.P., Vestergaard A.L., Mikkelsen S.A., Mogensen L.S., Chalat M. and Molday R.S. (2016) P4-ATPases as phospholipid flippases—structure, function, and enigmas. Front. Physiol. 7, 275 10.3389/fphys.2016.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roland B.P. and Graham T.R. (2016) Decoding P4-ATPase substrate interactions. Crit. Rev. Biochem. Mol. Biol. 51, 513–527 10.1080/10409238.2016.1237934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C.Y., Ingram M.F., Rosal P.H. and Graham T.R. (1999) Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147, 1223–1236 10.1083/jcb.147.6.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hua Z., Fatheddin P. and Graham T.R. (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13, 3162–3177 10.1091/mbc.e02-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicky S., Schwarz H. and Singer-Krüger B. (2004) Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol. Cell. Biol. 24, 7402–7418 10.1128/MCB.24.17.7402-7418.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomorski T., Lombardi R., Riezman H., Devaux P.F., van Meer G. and Holthuis J.C.M. (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14, 1240–1254 10.1091/mbc.e02-08-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alder-Baerens N., Lisman Q., Luong L., Pomorski T. and Holthuis J.C.M. (2006) Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 17, 1632–1642 10.1091/mbc.e05-10-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X.M. and Graham T.R. (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl Acad. Sci. U.S.A. 106, 16586–16591 10.1073/pnas.0904293106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takar M., Wu Y. and Graham T.R. (2016) The essential Neo1 protein from budding yeast plays a role in establishing aminophospholipid asymmetry of the plasma membrane. J. Biol. Chem. 291, 15727–15739 10.1074/jbc.M115.686253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulsen L.R., López-Marqués R.L., McDowell S.C., Okkeri J., Licht D., Schulz A. et al. (2008) The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20, 658–676 10.1105/tpc.107.054767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Marqués R.L., Poulsen L.R., Hanisch S., Meffert K., Buch-Pedersen M.J., Jakobsen M.K. et al. (2010) Intracellular targeting signals and lipid specificity determinants of the ALA/ALIS P4-ATPase complex reside in the catalytic ALA α-subunit. Mol. Biol. Cell 21, 791–801 10.1091/mbc.e09-08-0656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen L.R., López-Marqués R.L., Pedas P.R., McDowell S.C., Brown E., Kunze R. et al. (2015) A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat. Commun. 6, 7649 10.1038/ncomms8649 [DOI] [PubMed] [Google Scholar]
- 15.Jensen M.S., Costa S.R., Duelli A.S., Andersen P.A., Poulsen L.R., Stanchev L.D. et al. (2017) Phospholipid flipping involves a central cavity in P4 ATPases. Sci. Rep. 7, 17621 10.1038/s41598-017-17742-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juneja L.R., Kazuoka T., Goto N., Yamane T. and Shimizu S. (1989) Conversion of phosphatidylcholine to phosphatidylserine by various phospholipases D in the presence of l- or d-serine. Biochim. Biophys. Acta 1003, 277–283 10.1016/0005-2760(89)90233-6 [DOI] [Google Scholar]
- 17.Rose A.B. and Broach J.R. (1990) Propagation and expression of cloned genes in yeast: 2-μm circle-based vectors. Methods Enzymol. 185, 234–279 10.1016/0076-6879(90)85024-I [DOI] [PubMed] [Google Scholar]
- 18.Lenoir G., Menguy T., Corre F., Montigny C., Pedersen P.A., Thinès D. et al. (2002) Overproduction in yeast and rapid and efficient purification of the rabbit SERCA1a Ca2+-ATPase. Biochim. Biophys. Acta 1560, 67–83 10.1016/S0005-2736(01)00458-8 [DOI] [PubMed] [Google Scholar]
- 19.Sauer B. (1987) Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 2087–2096 10.1128/MCB.7.6.2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz R.D. and Woods R.A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 10.1016/S0076-6879(02)50957-5 [DOI] [PubMed] [Google Scholar]
- 21.Costa S.R., Marek M., Axelsen K.B., Theorin L., Pomorski T.G. and Lopez-Marques R.L. (2016) Role of post-translational modifications at the β-subunit ectodomain in complex association with a promiscuous plant P4-ATPase. Biochem. J. 473, 1605–1615 10.1042/BCJ20160207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahn T., Dietrich J., Andersen B., Leidvik B., Otter C., Briving C. et al. (2001) Large scale expression, purification and 2D crystallization of recombinant plant plasma membrane H+-ATPase. J. Mol. Biol. 309, 465–476 10.1006/jmbi.2001.4688 [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A., Tomas H., Havlis J., Olsen J.V. and Mann M. (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 24.de Godoy L.M.F., Olsen J.V., Cox J., Nielsen M.L., Hubner N.C., Fröhlich F. et al. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 10.1038/nature07341 [DOI] [PubMed] [Google Scholar]
- 25.Monk B.C., Montesinos C., Ferguson C., Leonard K. and Serrano R. (1991) Immunological approaches to the transmembrane topology and conformational changes of the carboxyl-terminal regulatory domain of yeast plasma membrane H+-ATPase. J. Biol. Chem. 266, 18097–18103 PMID: [PubMed] [Google Scholar]
- 26.Jacquot A., Montigny C., Hennrich H., Barry R., le Maire M., Jaxel C. et al. (2012) Phosphatidylserine stimulation of Drs2p-Cdc50p lipid translocase dephosphorylation is controlled by phosphatidylinositol-4-phosphate. J. Biol. Chem. 287, 13249–13261 10.1074/jbc.M111.313916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marek M., Milles S., Schreiber G., Daleke D.L., Dittmar G., Herrmann A. et al. (2011) The yeast plasma membrane ATP binding cassette (ABC) transporter Aus1: purification, characterization, and the effect of lipids on its activity. J. Biol. Chem. 286, 21835–21843 10.1074/jbc.M111.244525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørensen D.M., Møller A.B., Jakobsen M.K., Jensen M.K., Vangheluwe P., Buch-Pedersen M.J. et al. (2012) Ca2+ induces spontaneous dephosphorylation of a novel P5A-type ATPase. J. Biol. Chem. 287, 28336–28348 10.1074/jbc.M112.387191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen M.S., Costa S., Günther-Pomorski T. and López-Marqués R.L. (2016) Cell-based lipid flippase assay employing fluorescent lipid derivatives. Methods Mol. Biol. 1377, 371–382 10.1007/978-1-4939-3179-8_33 [DOI] [PubMed] [Google Scholar]
- 30.Jensen M.S., Costa S.R., Theorin L., Christensen J.P., Pomorski T.G. and López-Marqués R.L. (2016) Application of image cytometry to characterize heterologous lipid flippases in yeast. Cytometry A 89, 673–680 10.1002/cyto.a.22886 [DOI] [PubMed] [Google Scholar]
- 31.Baldridge R.D. and Graham T.R. (2013) Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl Acad. Sci. U.S.A. 110, E358–E367 10.1073/pnas.1216948110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vestergaard A.L., Coleman J.A., Lemmin T., Mikkelsen S.A., Molday L.L., Vilsen B. et al. (2014) Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl Acad. Sci. U.S.A. 111, E1334–E1343 10.1073/pnas.1321165111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smriti T., Nemergut E.C. and and Daleke D.L. (2007) ATP-dependent transport of phosphatidylserine analogues in human erythrocytes. Biochemistry 46, 2249–2259 10.1021/bi061333x [DOI] [PubMed] [Google Scholar]
- 34.Hanson P.K., Malone L., Birchmore J.L. and Nichols J.W. (2003) Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 278, 36041–36050 10.1074/jbc.M305263200 [DOI] [PubMed] [Google Scholar]
- 35.Leventis P.A. and Grinstein S. (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 10.1146/annurev.biophys.093008.131234 [DOI] [PubMed] [Google Scholar]
- 36.Muthusamy B.P., Natarajan P., Zhou X.M. and Graham T.R. (2009) Linking phospholipid flippases to vesicle-mediated protein transport. Biochim. Biophys. Acta 1791, 612–619 10.1016/j.bbalip.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman J.A., Kwok M.C.M. and Molday R.S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 10.1074/jbc.M109.047415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coleman J.A. and Molday R.S. (2011) Critical role of the β-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 10.1074/jbc.M111.229419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S., Uchida Y., Wang J., Matsudaira T., Nakagawa T., Kishimoto T., et al. (2015) Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34, 669–688 10.15252/embj.201489703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Molday L.L., Hii T., Coleman J.A., Wen T., Andersen J.P. et al. (2018) Proteomic analysis and functional characterization of P4-ATPase phospholipid flippases from murine tissues. Sci. Rep. 8, 10795 10.1038/s41598-018-29108-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding J., Wu Z., Crider B.P., Ma Y., Li X., Slaughter C. et al. (2000) Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase: effect of isoform variation on the ATPase activity and phospholipid specificity. J. Biol. Chem. 275, 23378–23386 10.1074/jbc.M910319199 [DOI] [PubMed] [Google Scholar]
- 42.Paterson J.K., Renkema K., Burden L., Halleck M.S., Schlegel R.A., Williamson P. et al. (2006) Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II). Biochemistry 45, 5367–5376 10.1021/bi052359b [DOI] [PubMed] [Google Scholar]
- 43.Segawa K., Kurata S. and Nagata S. (2018) The CDC50A extracellular domain is required for forming a functional complex with and chaperoning phospholipid flippases to the plasma membrane. J. Biol. Chem. 293, 2172–2182 10.1074/jbc.RA117.000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.