Abstract
Existing approaches to managing genetic and genomic test results from external laboratories typically include filing of text reports within the electronic health record, making them unavailable in many cases for clinical decision support. Even when structured computable results are available, the lack of adopted standards requires considerations for processing the results into actionable knowledge, in addition to storage and management of the data. Here, we describe the design and implementation of an ancillary genomics system used to receive and process heterogeneous results from external laboratories, which returns a descriptive phenotype to the electronic health record in support of pharmacogenetic clinical decision support.
Keywords: electronic health record, genetic testing, pharmacogenomics
BACKGROUND AND SIGNIFICANCE
Many institutions have adopted genetic testing as part of clinical care, with some exploring how computable representations of genetic and genomic results in the electronic health record (EHR) can facilitate clinical decision support (CDS).1–8 However, despite established standards for the computable representation of these results,9 this level of integration has not seen wide adoption outside of the research setting.10,11 Genetic test results are generally returned as a plain-text representation of the interpretation report and transmitted using the Health Level 7 (HL7) v2 standard, or more commonly as PDFs that must be scanned from a fax or downloaded from an external laboratory portal.
Several national initiatives, including the electronic Medical Records and Genomics (eMERGE) network,12,13 the Clinical Sequencing Exploratory Research consortium,14 the Implementing Genomics in Practice network,15 and the Displaying and Integrating Genetic Information Through the EHR Action Collaborative16 have proposed models, as well as demonstrated successful strategies, for better integration of results. The eMERGE network proposed the concept of an omics ancillary system, which receives structured, computable results from a laboratory, stores the results in an optimized manner for specialized processing (similar to a picture archiving and communications system for imaging), and returns actionable information to the EHR.17
Additionally, individual institutions have developed tools to manage genetic data5,18; however these are typically tied a single laboratory source, and do not facilitate transmission of structured genetic results to the EHR itself. As many healthcare institutions work with several different third-party laboratories to provide genetic testing services, the ability to manage structured results from different external sources is important, yet not well addressed.
In this article, we present the design and implementation of an ancillary genomics system (AGS) to manage genetic test results from 2 external laboratories and return them to the EHR in a computable manner to facilitate CDS.
MATERIALS AND METHODS
This work was done as part of the eMERGE Pharmacogenomics (eMERGE-PGx) project,12 and was approved by the Northwestern University Institutional Review Board. We consented patients from Northwestern Medicine’s internal medicine practice who provided blood samples that were genotyped at 1 of 2 CLIA (Clinical Laboratory Improvement Amendments)-certified laboratories (Mount Sinai Genetic Testing Laboratory and the Johns Hopkins DNA Diagnostic Laboratory). Both patients and their primary care providers received genotype results for 4 genes (CYP2C19, SLCO1B1, CYP2C9, and VKORC1) and genotype-guided prescribing information for 3 drugs (clopidogrel, simvastatin, and warfarin).
The development of the AGS to manage the processing and return of results was conducted by a multidisciplinary team and included physicians, genetic counselors, informaticians, health information technology leadership, EHR analysts, and software developers. During biweekly meetings, the team discussed design objectives and provided feedback to the software development team during the development process.
RESULTS
Design criteria
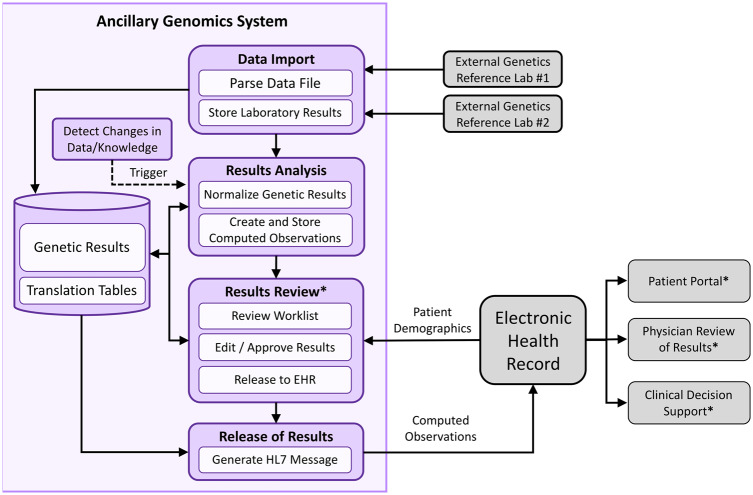
When approaching the design of the AGS, we sought to meet several objectives identified in the literature19: (1) anticipate receipt and processing of heterogeneous data (eg, pharmacogenetic star alleles, single nucleotide polymorphisms [SNPs]), (2) support the reinterpretation of results given new data or knowledge, (3) convert genetic and genomic test results into a more clinically relevant and intuitive form, and (4) design the system in a modular fashion to support future expansion. The overall system architecture is shown in Figure 1.
Figure 1.
The Northwestern Medicine ancillary genomics system. Overall flow of data from external laboratories, which are imported into the ancillary genomics system, analyzed by the system to create computed observations (results), manually reviewed by a genetic counselor, and then released to the electronic health record (EHR). Steps denoted with an asterisk involve human interaction or decision making; all other steps are automated. HL7: Health Level 7.
Computed observations
To address design objective 3, we introduced the concept of the “computed observation.” The computed observation is a synthesis of the genetic test results and describes the predicted phenotype as opposed to requiring a provider to mentally translate this from the genotype. For example, instead of reporting the specific diplotypes for a gene (eg, CYP2C19 *2/*2), the computed observation describes the predicted metabolizer category for a medication (eg, “Clopidogrel Poor Metabolizer”). This not only makes CDS easier to implement, but also is intended to be more intuitively understandable by practicing clinicians.
Laboratory data format
As part of the eMERGE-PGx study, each laboratory agreed to provide results as Microsoft Excel documents containing structured results and interpretive text. Each laboratory used unique formats for returning results: one provided results as pharmacogenetic star alleles for each of the 4 genes of interest (CYP2C19, CYP2C9, SLCO1B1, VKORC1), while the other provided 19 SNPs across 6 genes (CYP2C19, CYP2C9, SLCO1B1, VKORC1, TPMT, CYP3A5). A list of fields provided by each laboratory is provided in Supplementary Appendix A.
Laboratory data import
Given the heterogeneity of results, we developed a separate importer for each laboratory that extracted participants’ results from the Excel document. Results were stored in a relational database using the entity-attribute-value format. Attributes were represented as a hierarchy with parent-child relationships. An example of this is the top of the tree representing a generic attribute for results in the CYP2C19 gene, and more specific representations (eg, CYP2C19 SNP) are represented as descendant attributes (additional details in the Supplementary Material). The hierarchical nature simplified querying related concepts (eg, “return all CYP2C19 results”). The attribute hierarchy used was developed manually for the project in response to the types of results provided by each laboratory and is not derived from any standard ontology.
Result analysis
Data analysis modules converted laboratory results into computed observations. A separate analysis module was developed for each of the 3 genotype-guided drugs, using the C# language. The processing flow is illustrated in Figure 2 and generally follows the sequence outlined in Table 1. During these processing steps, the AGS tracked all data transformation and lookup steps applied to the data. Processing included several steps to normalize the results (described further in Supplymentary Appendix C). A lookup table mapped the normalized results to an actionable interpretation, based on guidelines developed by the Clinical Pharmacogenetics Implementation Consortium.20 If no entry was found, the record was noted as having an “Unknown” interpretation, which flagged it for further review.
Figure 2.
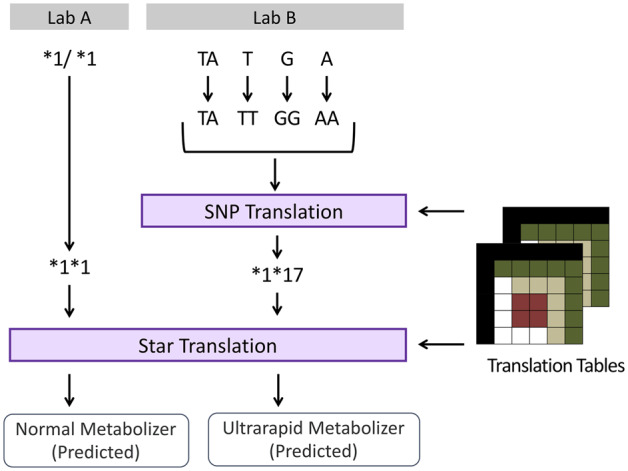
Translation and normalization steps from heterogeneous laboratory results to computed observations in the ancillary genomics system. For Lab A, the star variant results are converted to a normalized string representation and then translated to a computed observation. For Lab B, the single nucleotide polymorphism (SNP) results are normalized, translated to a star variant representation, and then translated to a computed observation.
Table 1.
Typical processing steps performed when analyzing laboratory results
| Step | Description |
|---|---|
| 1. Retrieve all relevant results | Given the relevant high-level attribute identifiers in the system (eg, CYP2C19 Results), retrieve all descendant attribute identifiers, and then pull all result values of those attribute types. |
| 2. Normalize SNP genotypes | Ensure 2 SNP genotypes are present with no additional characters separating them (eg, A/A -> AA or T -> TT). |
| 3. Convert SNP genotypes to pharmacogenetic star alleles | Using a lookup table, translate the SNP genotype(s) to the appropriate star alleles as necessary. |
| 4. Normalize star alleles | Ensure 2 star alleles are present with no additional characters separating them (eg, *1/*1 -> *1*1). |
| 5. Convert genotype to observation | Using a lookup table, translate the star variant or alleles into the appropriate computed observation representation. |
| 6. Generate report from template | Given the computed observation and original genotype results, generate an interpretive report. |
Depending on the type of data present, certain steps did not apply.
SNP: single nucleotide polymorphism.
Results review
The AGS portal provided a web-based interface through which genetic counselors involved with the study could review and release results. The AGS portal contained a worklist for managing the review process and was designed to provide a 2-step review where a genetic counselor would first review the generated result and interpretive text. Users had the ability to modify the generated report if additional supporting text was needed, and the system would confirm that the patient demographics in the AGS matched those in the EHR.
Once reviewed, the result was marked as “Approved.” Because of the large number of results returned in a single batch from the laboratories, and based on feedback from the physicians enrolled in the study, a second step was added to manage releasing approved results to the EHR in smaller quantities (∼10-25) at more consistent intervals (typically once or twice per week).
Release of results
When a result was released, the AGS generated an HL7 v2 observation result, which contained the laboratory test, result and descriptive interpretation report. These messages were sent to our EHR via an HL7 interface and displayed in the laboratory results section of the patient’s chart.
Real-world application
For this project, 746 participants had results processed by the AGS. Each participant had 3 observations generated (1 each for clopidogrel, simvastatin, and warfarin), for 2238 observations total. Observations were reviewed and returned to the EHR between June 6, 2014, and September 14, 2015. As of October 2018, there have been no changes in how results were interpreted that required results to be reprocessed.
The AGS received results for 448 participants from Mount Sinai, and 304 from Johns Hopkins DDL. There were 3 participants with results from both laboratories, with 100% concordance in the AGS interpretation of both sets.
DISCUSSION
In this study, we demonstrated an ancillary genomics system capable of processing heterogeneous genomic results from external laboratories and representing them as computed observations, in accordance with our design objectives. Although not demonstrated in the course of this study, our AGS was designed to reinterpret results as new data or changes in interpretation are made available. The use of a portal to review and release results to the EHR allowed us to manage the number of results a provider received at one time. The format of the computed observations allowed us to also simplify the implementation of CDS rules in the EHR. A full description of our CDS implementation is outside of the scope of this article, but briefly a CDS rule for a particular drug metabolism status could be written as a single condition (eg, “If observation = ‘Clopidogrel Poor Metabolizer’”) as opposed to all possible permutations of laboratory values (eg, “If observation = *2/*2 OR observation = *2/*3…”).
Organizations have different needs for managing genetic and genomic results, as is evident when reviewing existing implementations.1–6,18,21,22 With respect to the granularity of results, St. Jude’s Children Hospital successfully integrated diplotypes from an external laboratory in their EHR,1,6 and demonstrated that this level of granularity is acceptable to providers. The NEXT program at the University of Washington represented results at the gene level—flagging if the gene had any known variants—which was found to be too coarse of a level of granularity for CDS.7 The Mayo Clinic has successfully implemented PGx CDS at their institution,3,4 working with their internal laboratory to return data to their EHR, including populating problem and alert lists with phenotype results per gene, as opposed to a specific medication (eg, “Metabolizer CYP2C19 Poor”). Given the myriad approaches, additional research is needed to better understand physicians’ perspectives on the optimal level of granularity. The architecture of our AGS allows us to modify the granularity of results for future studies.
Mount Sinai developed and implemented a system as part of their CLIPMERGE PGx program,5 which receives results from their internal laboratory and acts as their CDS rules engine. The GeneInsight platform developed at Partner’s HealthCare18 supports a laboratory workflow to annotate results, generate interpretive reports that are transmitted to the EHR, and manages re-interpretation of results when the underlying knowledge base is updated.23 GeneInsight offers a clinical portal for the review of results,24–26 and is capable of returning structured results in addition to PDF reports. Each of these implementations follows an ancillary omics model,17 with variations based on institutional preference (eg, the CLIPMERGE decision to use the system to fire CDS rules), and workflow (eg, GeneInsight’s management of laboratory processes). Institutions integrating genomic results into their EHR will have similar considerations to make when implementing a solution.
We note that the lack of an adopted standardized format for results increased the overall programmatic work, requiring us to import and translate results from each laboratory. Ongoing work within HL727,28 and the Global Alliance for Genomics and Health29 are evaluating the use of standards for sequencing results, but broad implementation within laboratory systems will be critical to scale this approach. For the computed observations created by the AGS, while the work presented here used HL7 v2, the computed observations could be represented using the Fast Healthcare Interoperability Resources standard.30 This is a planned area of development, as more EHRs provide inbound Fast Healthcare Interoperability Resources interfaces. Likewise, no computable knowledge bases exist to translate laboratory results into phenotypic representations. A shareable source of knowledge to perform this translation would aid future adoption by other institutions and system developers, and could be based upon translation tables provided by the Clinical Pharmacogenetics Implementation Consortium (eg, simvastatin/SLCO1B131).
Within our study, we note some limitations. The AGS was optimized for genetic counselors to review and release computed observations, as opposed to aiding in the interpretation of rare variants, and may not meet the needs of all institutions. In addition, although the AGS is capable of reprocessing results given new knowledge, the absence of changes during our PGx study precluded us from demonstrating this. This capability was tested during development. Furthermore, while it has been architected to be modular and transportable, to date it has only been implemented at our institution.
Future work currently underway includes better integration with sites such as ClinGen,32 which offer supporting, context-aware information resources to aid in decision making; the inclusion of sequencing results, which include more than PGx findings; and the use of a structured XML format to represent individual sequencing results.
CONCLUSIONS
Given the number of workflows by which genomic and genetic data may enter the healthcare system, we have developed and implemented an AGS to manage results from external laboratories in support of PGx. The AGS furthers the idea that complex and heterogeneous genetic and genomic results may be kept outside of the core EHR but still linked in a way that provides sufficient information to clinicians caring for patients.
FUNDING
This work was funded by the National Human Genome Research Institute through grant U01HG006388.
AUTHOR CONTRIBUTORS
JBS and CC conceptualized the project. LVR and FA led the system architecture and system design, and developed core components of the AGS. All authors were involved in the process of developing, reviewing, and/or refining requirements for the AGS. LVR created the first draft of the manuscript. MES, SDP, LJRT, JAP, TMH, and JBS provided substantial feedback in the refinement of the manuscript. All authors reviewed and approved of the submitted manuscript and have agreed to be accountable for its contents.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge Chris Bethman, Darren Kaiser, Deb Knoner, Steve Lee, Robert Milfajt, and Sameem Samad for their assistance in the implementation of the ancillary genomics system at Northwestern Medicine.
Conflict of interest statement: TMH and JBS founded Ancillary Genomic Systems, LLC, to explore the commercial potential of omic ancillary systems. They have not received any payments for this work and are not currently marketing any products. All other authors have no competing interests to declare.
REFERENCES
- 1. Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM.. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014; 166c (1): 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beyan T, Aydin Son Y.. Incorporation of personal single nucleotide polymorphism (SNP) data into a national level electronic health record for disease risk assessment, part 2: the incorporation of SNP into the national health information system of Turkey. JMIR Med Inform 2014; 2 (2): e17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014; 89 (1): 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caraballo PJ, Bielinski SJ, St Sauver JL, Weinshilboum RM.. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin Pharmacol Ther 2017; 102 (2): 254–64. [DOI] [PubMed] [Google Scholar]
- 5. Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013; 94 (2): 214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther 2012; 92 (5): 563–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishimura AA, Shirts BH, Dorschner MO, et al. Development of clinical decision support alerts for pharmacogenomic incidental findings from exome sequencing. Genet Med 2015; 17 (11): 939–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 2014; 166C (1): 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bosca D, Marco L, Burriel V, et al. Genetic testing information standardization in HL7 CDA and ISO13606. Stud Health Technol Inform 2013; 192: 338–42. [PubMed] [Google Scholar]
- 10. Hoffman MA. The genome-enabled electronic medical record. J Biomed Inform 2007; 40 (1): 44–6. [DOI] [PubMed] [Google Scholar]
- 11. Kho AN, Rasmussen LV, Connolly JJ, et al. Practical challenges in integrating genomic data into the electronic health record. Genet Med 2013; 15 (10): 772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther 2014; 96 (4): 482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013; 15 (10): 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green RC, Goddard KA, Jarvik GP, et al. Clinical Sequencing Exploratory Research Consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet 2016; 98 (6): 1051–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weitzel KW, Alexander M, Bernhardt BA, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 2016; 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Academy of Sciences. DIGITizE: Displaying and Integrating Genetic Information Through the EHR. 2018. http://www.nationalacademies.org/hmd/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/DIGITizE.aspx. Accessed February 6, 2019.
- 17. Starren J, Williams MS, Bottinger EP.. Crossing the omic chasm: a time for omic ancillary systems. JAMA 2013; 309 (12): 1237–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat 2011; 32 (5): 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform 2012; 45 (3): 419–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Relling MV, Klein TE.. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther 2011; 89 (3): 464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang L, Fernandes H, Zia H, et al. The cancer precision medicine knowledge base for structured clinical-grade mutations and interpretations. J Am Med Informat Assoc 2017; 24 (3): 513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lærum H, Bremer S, Bergan S, Grünfeld T.. A taste of individualized medicine: physicians' reactions to automated genetic interpretations. J Am Med Inform Assoc 2014; 21 (e1): e143–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet Med 2012; 14 (8): 713–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilcox AR, Neri PM, Volk LA, et al. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc 2014; 21 (e1): e117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neri PM, Pollard SE, Volk LA, et al. Usability of a novel clinician interface for genetic results. J Biomed Inform 2012; 45 (5): 950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klinkenberg-Ramirez S, Neri PM, Volk LA, et al. Evaluation: a qualitative pilot study of novel information technology infrastructure to communicate genetic variant updates. Appl Clin Inform 2016; 7 (2): 461–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Health Level 7. Genomics - FHIR v3.0.1. 2018. https://www.hl7.org/fhir/genomics.html.
- 28.Health Level 7. Clinical Genomics. 2018. http://www.hl7.org/Special/committees/clingenomics/.
- 29.Global Alliance for Genomics and Health. 2017. https://www.ga4gh.org/.
- 30.Health Level 7. Index - FHIR v3.0.1. 2017. https://www.hl7.org/fhir/index.html.
- 31.Clinical Pharmacogenetics Implementation Consortium. 2014. SLCO1B1 translation table. 2014. https://cpicpgx.org/content/guideline/publication/simvastatin/2014/24918167.xlsx.
- 32. Overby CL, Heale B, Aronson S, et al. Providing access to genomic variant knowledge in a healthcare setting: a vision for the ClinGen Electronic Health Records Workgroup. Clin Pharmacol Ther 2016; 99 (2): 157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.