Abstract
Human skin is morphologically and physiologically different from the skin of other primates. However, the genetic causes underlying human-specific skin characteristics remain unclear. Here, we quantitatively demonstrate that the epidermis and dermis of human skin are significantly thicker than those of three Old World monkey species. In addition, we indicate that the topography of the epidermal basement membrane zone shows a rete ridge in humans but is flat in the Old World monkey species examined. Subsequently, we comprehensively compared gene expression levels between human and nonhuman great ape skin using next-generation cDNA sequencing (RNA-Seq). We identified four structural protein genes associated with the epidermal basement membrane zone or elastic fibers in the dermis (COL18A1, LAMB2, CD151, and BGN) that were expressed significantly greater in humans than in nonhuman great apes, suggesting that these differences may be related to the rete ridge and rich elastic fibers present in human skin. The rete ridge may enhance the strength of adhesion between the epidermis and dermis in skin. This ridge, along with a thick epidermis and rich elastic fibers might contribute to the physical strength of human skin with a low amount of hair. To estimate transcriptional regulatory regions for COL18A1, LAMB2, CD151, and BGN, we examined conserved noncoding regions with histone modifications that can activate transcription in skin cells. Human-specific substitutions in these regions, especially those located in binding sites of transcription factors which function in skin, may alter the gene expression patterns and give rise to the human-specific adaptive skin characteristics.
Keywords: primates, epidermal basement membrane zone, rete ridge, elastic fibers, transcriptional regulatory region
Introduction
Skin is an important organ that is constantly exposed to external environments. It protects the inside of the body from external stresses, such as physical, chemical, and microbial insults. It is likely that skin phenotypes evolved to protect the inside of the body in species adapted to external environments such as terrestrial amniotes, including humans.
Human skin is morphologically and physiologically different from the skin of other primates. The reduced amount of hair (Dávid-Barrett and Dunbar 2016) and the high number of sweat glands (Folk and Semken 1991) are examples of human-specific skin characteristics that are not found in other primates such as chimpanzees, gorillas, and orangutans. It has been proposed that these human-specific characteristics allowed for efficient thermoregulation and adaptation to the savannah environment after our human ancestors abandoned the forest (Folk and Semken 1991).
Skin is generally composed of three layers: the epidermis, dermis, and subcutaneous tissue (Smoller 2009). The epidermal basement membrane (BM) zone forms adhesion between the epidermal underside and the dermal upside through anchoring structures (Has and Nystrom 2015). Until the early 1980s, the histological differences between human and other primate skin were solely reported from a qualitative perspective. For example, the epidermis of human skin is thicker than that of other primates (Montagna 1982, 1985). In addition, the epidermal underside and dermal upside (the position of the epidermal BM zone) in the furred skin of most nonhuman primates are flat (Montagna 1982). On the other hand, those in human skin, including those in the hairy skin of the scalp, are strongly sculptured and penetrate each other (Montagna 1982, 1985), resulting in undulating topography of the epidermal BM zone known as a rete ridge. In the furred skin of chimpanzees and gorillas, the epidermal underside has been reported with inconsistent descriptions; a degree of sculpturing in chimpanzees and gorillas (Montagna 1982), discrete and moderate sculpturing in chimpanzees (Montagna and Yun 1963), and a nearly flat topography in gorillas (Ellis and Montagna 1962). Another striking difference between human and other primate skin is the amount of elastic fibers, which give skin elasticity (Kielty et al. 2002). Elastic fibers are rich in the human dermis but those in most other primates are not as numerous as humans (Montagna 1982, 1985). Also in this case, the amount of elastic fibers in the furred skin of chimpanzees and gorillas has not been reported consistently; although it was reported to be similar to the content in humans (Montagna 1982, 1985), the elastic fibers in the chimpanzee dermis were also described as nowhere numerous (Montagna and Yun 1963). According to these qualitative studies, a thick epidermis, extensive rete ridge formation, and an abundance of elastic fibers seem to be human-specific skin characteristics. However, to the best of our knowledge, no recent study has quantitatively compared the characteristics of human and other nonhuman primate skin.
It is widely accepted that most of the phenotypic differences observed between closely related species are a result of different quantitative and spatiotemporal expression patterns in functionally relevant genes, rather than amino acid differences in the protein-coding regions of those genes (King and Wilson 1975; Wray 2007; Carroll 2008). Certain noncoding regions within the genome (e.g., promoters and enhancers) are important for the regulation of gene expression (Lindblad-Toh et al. 2011). Such transcriptional regulatory regions generally harbor a multitude of binding sites for sequence-specific transcription factors (TFs) that modulate gene expression (Carroll et al. 2013). From an adaptive standpoint, these regions tend to evolve under functional constraint and are thus more conserved between species than the surrounding nonfunctional noncoding regions (Pennacchio et al. 2006; He et al. 2011). Transcriptional regulatory regions of a certain gene of interest can separately reside at various positions: immediately 5′ of the transcription start site, in the adjacent intergenic regions, in the introns of the gene itself or neighboring genes, or/and even in the noncoding regions at considerable distances from the gene (Kleinjan and van Heyningen 2005). Mutations in transcriptional regulatory regions can change the expression level of the target gene by altering TF-binding affinities (Wittkopp and Kalay 2012), which plays important roles in phenotypic diversity for morphology, physiology, and behavior between species (Wray 2007).
Transcription is also regulated by histone modification. In eukaryotic cells, genomic DNA is folded into chromatin, which is composed of nucleosomes and linker DNA (Luger et al. 2012). Histones comprise core histones (H2A, H2B, H3, and H4) and are wrapped by ∼147 bp of DNA in nucleosomes (Luger et al. 1997). Core histones are subjected to posttranslational modifications on various amino acid residues, mostly in their N-terminal tails that extrude from the nucleosomes (Kimura 2013). Histone modifications, including methylation and acetylation, regulate the structure of chromatin, resulting in changing accessibility of TFs to potential transcriptional binding sites (Kouzarides 2007). Chromatin state is different between diverse cell types in a multicellular organism and contributes to cell type-specific gene expression patterns in the presence of an essentially identical genome in each cell (Heintzman et al. 2009). The active promoters and enhancers of transcribed genes generally possess some specific modifications: mono-, di-, and tri-methylation of histone H3 lysine 4 (represented by H3K4m1, H3K4m2, and H3K4m3, respectively) and acetylation of histone H3 lysine 9 and histone H3 lysine 27 (H3K9ac and H3K27ac, respectively) (Barski et al. 2007; Wang et al. 2008; Karmodiya et al. 2012; Kimura 2013). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is used to determine the genome-wide distribution of each histone modification in multiple cell types (Zhou et al. 2011; Consortium 2012). The output from ChIP-seq allows us to estimate transcriptional regulatory regions in the genome, and ChIP-seq data are widely available in public databases (e.g., Kent et al. 2002).
The genetic causes that underlie human-specific characteristics, including those in skin, remain poorly understood, although a few cases are known (Enard et al. 2002; Stedman et al. 2004; Prabhakar et al. 2008). In this study, we quantitatively distinguished histological skin differences between humans and other primates to investigate human-specific characteristics in skin structure. We then comprehensively compared gene expression levels between human and nonhuman great ape (chimpanzee, gorilla, and orangutan) skin using next-generation cDNA sequencing (RNA-Seq). We identified genes with human-specific expression patterns that may be related to human-specific characteristics in skin structure. Finally, we identified possible transcriptional regulatory regions and DNA sequence substitutions likely responsible for the human-specific expression patterns of the genes. Taken together, these analyses may provide insight into the evolution of adaptive human-specific skin characteristics.
Materials and Methods
Skin Specimens
The skin tissue specimens from Pan troglodytes verus, Gorilla gorilla gorilla, and Pongo pygmaeus (n = 3 for each species) (supplementary table S1, Supplementary Material online) were collected by the Primate Research Institute, Kyoto University via the Great Ape Information Network (GAIN) from zoos and the Kumamoto sanctuary, Wildlife Research Institute, Kyoto University. The use of human skin tissues was authorized by the Ethics Committee of the University of the Ryukyus for Medical and Health Research Involving Human Subjects (#18-1295). The research using Old World monkey (anubis baboons [Papio anubis], Sykes’ monkeys [Cercopithecus albogularis], vervet monkeys [Chlorocebus pygerythrus]) skin tissues was approved by the Institutional Review Committee of the Institute of Primate Research, National Museum of Kenya (No. IRC/05/14). The dorsal skin tissues were collected by A.M.-O. under research permission (No. NCST/RRI/12/1/BS/240), and transferred to the University of the Ryukyus under the regulation of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES: No. 0830732).
Measurement of Skin Thickness
Digital photographs of dissected skins of anubis baboons (n = 6), Sykes’ monkeys (n = 6), vervet monkeys (n = 6), and humans (n = 4) were provided by the Department of Dermatology, Graduate School of Medicine, University of the Ryukyus. Epidermis and dermis thickness was measured at ten sites on dissected skin photographs for each individual using iViewer version 5.5.7 (http://www.pathimaging.jp; last accessed February 8, 2019). Measurements that were unreliable due to skin condition were discarded. The average values of the ten measurements for epidermis and dermis thickness for each individual were used to calculate the average thickness in the species. We compared the average thickness between humans and each of the three Old World monkey species using a t-test with Bonferroni correction.
RNA Extraction and Sequencing
Total RNA was extracted from skin tissue samples of Pan troglodytes verus, Gorilla gorilla gorilla, and Pongo pygmaeus using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). Human skin total RNA of five individuals was obtained from commercial sources, and these individuals were not the same as ones used for measurement of skin thickness (total RNA: BioChain, Newark, CA; MVP total RNA, human skin: Agilent Technologies, Santa Clara, CA; supplementary table S1, Supplementary Material online). Skin total RNA was used to construct libraries for high-throughput RNA sequencing using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA). Short cDNA sequences were determined from the libraries using the Illumina HiSeq2000 (paired-end, 100 bp) or HiSeq2500 (paired-end, 125 bp) platform.
Comparison of RNA Expression in Skin
The procedure to compare skin RNA expression patterns between humans and nonhuman great apes is shown in supplementary figure S1, Supplementary Material online. Sequenced reads from all libraries were mapped to each of the four reference genome sequences of human, chimpanzee, gorilla, and orangutan (supplementary fig. S1 and supplementary table S2, Supplementary Material online). In each of four mapping results, the expression values, Reads Per Kilobase of an exon model per Million mapped reads (RPKM) values, were calculated for each gene in each sample. We focused on genes with average RPKM values for humans or nonhuman great apes ≥1 in each mapping result. We normalized the expression values by Quantile normalization (Bolstad et al. 2003). The normalized expression data were checked by boxplot. The normalized expression values of five human individuals were compared with those of nine nonhuman great apes by Baggerley’s test (Baggerly et al. 2003). The genes showing statistically significant differences (P < 0.05, with false discovery rate (FDR) P value correction) in their average normalized RPKM values between humans (n = 5) and nonhuman great apes (n = 9) were extracted in each of four mapping results. The mapping and comparison of normalized RPKM values were conducted using CLC Genomics Workbench (https://www.qiagenbioinformatics.com/; last accessed February 8, 2019). Then, the genes that were common to each of the extracted results were selected as differentially expressed genes between humans and nonhuman great apes.
Inference of Substitutions Responsible for the Human-Specific Expression Patterns
Noncoding regions that were conserved in nonhuman lineages (fig. 1, gray lines) were identified to estimate transcriptional regulatory regions for the four focused genes. The analyzed genomic regions were set to include noncoding regions at both sides of the genes of interest and were 372, 100, 100, and 78 kb in length for the COL18A1, LAMB2, CD151, and BGN genes, respectively, in the human genome (GRCh38). Each of the four genes of interest was located in the center of their respective regions. The genomic sequence alignments of human, chimpanzee, gorilla, orangutan, and rhesus macaque were obtained from Ensembl (https://asia.ensembl.org/index.html; last accessed February 8, 2019). Alignment sites that showed one or more gaps in at least one of the five species were removed.
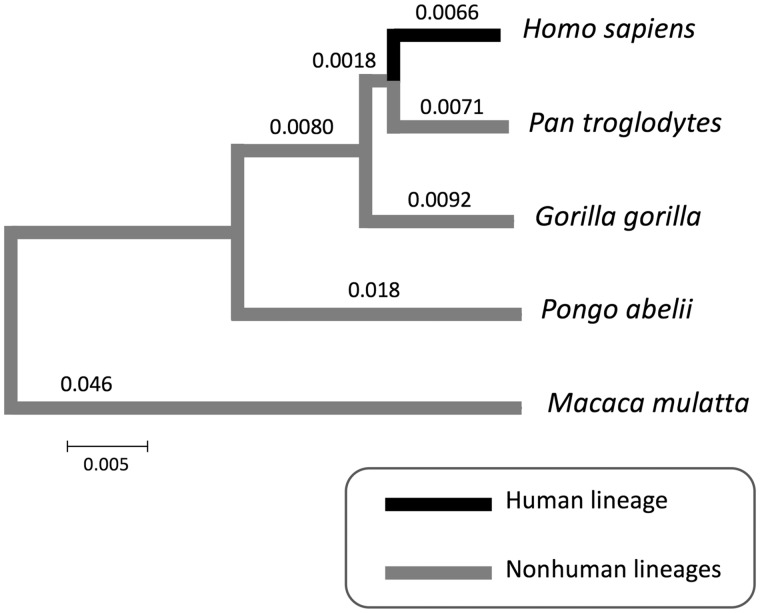
Fig. 1.
—Phylogenetic relationships between human, nonhuman great apes, and rhesus macaque. A phylogenetic tree was constructed using the Neighbor-Joining method and the pair-wise nucleotide divergence of whole genome sequences (Scally et al. 2012). The scale bar represents 0.005 substitutions per site. The distance on each branch was calculated by the Fitch–Margoliash algorithm (Fitch and Margoliash 1967) using the pair-wise nucleotide divergence.
To identify conserved domains throughout the analyzed genomic regions, a sliding-window analysis was performed using a 120-bp window size and a 4-bp step size (supplementary fig. S2, Supplementary Material online). For each window, pair-wise nucleotide differences between the sequences of the species were estimated using the Jukes–Cantor model implemented in DnaSP 5.0 (Rozas et al. 2003). Then, the numbers of substitutions in nonhuman lineages (fig. 1, gray lines) for each window were calculated using the Fitch–Margoliash algorithm (Fitch and Margoliash 1967). For each analyzed genomic sequence alignment, the pair-wise nucleotide divergences between species excluding exonic and unaligned regions and their standard errors were calculated with the Jukes–Cantor model and a bootstrap method (1,000 replicates), respectively, using MEGA 7 (Kumar et al. 2016). Using the same algorithm, the average expected number of substitutions in nonhuman lineages for a 120-bp region in each analyzed genomic region was calculated using these pair-wise nucleotide divergence values for noncoding sequences. The 120-bp regions with the significantly smaller numbers of substitutions in nonhuman lineages than expected under a Poisson distribution (P < 0.05) were identified as the conserved regions. When multiple conserved regions were continuous, these regions were concatenated into a single conserved region (supplementary fig. S2, Supplementary Material online).
Subsequently, conserved regions completely overlapping with exonic regions were eliminated by referring to exonic positions in the human genome (University of California–Santa Cruz Genome Browser [https://genome.ucsc.edu; last accessed February 8, 2019], Human GRCh38/hg38). We then extracted conserved regions harboring human-specific substitutions in noncoding regions.
Next, we selected regions with histone modifications (H3K4m1, H3K4m2, H3K4m3, H3K9ac, and H3K27ac) for active transcription from the conserved regions with human-specific substitutions identified above. ChIP-seq data for two skin cell strains, the normal human epidermal keratinocytes (NHEK) and normal human dermal fibroblasts from adult skin (NHDF-Ad), in the University of California–Santa Cruz Genome Browser was used. Each gene of interest is expressed in either or both of these skin cell strains (Iivanainen et al. 1995; Saarela, et al. 1998; Li et al. 2013; Has and Nystrom 2015). The histone modifications around the transcription start sites of neighboring genes are expected to regulate the transcription of those genes, but not the genes of interest. Therefore, conserved regions with such histone modifications were not selected. The human-specific substitutions in the selected regions were estimated to be the candidate substitutions responsible for the human-specific expression patterns in the four genes of interest. The ancestral allele frequencies at the candidate substitution loci in human populations were investigated using the 1000 Genomes Project data (phase 3) in Ensembl (https://asia.ensembl.org/index.html; last accessed February 8, 2019).
Evolutionary Analyses and TF-Binding Site Search for the Candidate Substitutions
We assumed that the candidate substitutions most likely to change the gene expression levels of the genes of interest would be 1) in highly conserved 120-bp regions or 2) in conserved 120-bp regions with the larger numbers of substitutions in the human lineage (fig. 1, black line). Among the most conserved 120-bp regions for each candidate substitution, regions with the significantly smaller numbers of substitutions in nonhuman lineages than expected (P < 0.01, Poisson distribution) were regarded as matches to condition (1), above. Next, we focused on the conserved 120-bp regions with the largest numbers of substitutions in the human lineage for each candidate substitution. The expected numbers of substitutions in the human lineage in each region were calculated from the numbers of substitutions in nonhuman lineages in the same 120-bp regions, according to the ratio of the numbers of substitutions in nonhuman and the human lineage(s) in each analyzed genomic region. The 120-bp regions with the significantly larger numbers of substitutions in the human lineage than expected (P < 0.05, Poisson distribution) were regarded as matches to condition (2), above.
The 51-bp sequence regions in which each candidate substitution locus was located at the center were screened for TF-binding sites using the JASPAR 2016 database (Mathelier et al. 2016). We screened two sequences that differed at one base pair in the candidate substitution locus: 1) the human sequence with the human-specific allele and 2) the human sequence with the ancestral allele. Relative scores in the JASPAR database were used to show the similarity with the consensus sequences of TF-binding sites.
Results
Histological Differences in Skin Structure between Humans and Three Old World Monkey Species
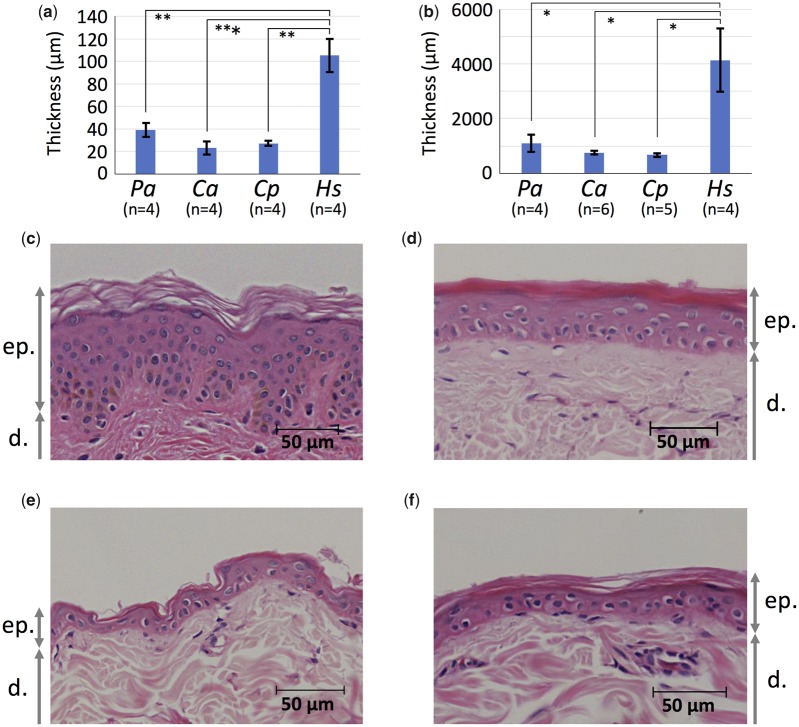
To clarify the human-specific characteristics in skin structure, we measured the thickness of the epidermis and dermis in humans and three Old World monkey species, anubis baboons, Sykes’ monkeys, and vervet monkeys. The epidermis and dermis were significantly thicker in humans than in the three Old World monkey species (P < 0.05, t-test with Bonferroni correction) (fig. 2a and b). We also observed that the epidermal BM zone topography in human skin was undulating (i.e., showed a rete ridge) (fig. 2c), whereas that in the three Old World monkey species was flat (fig. 2d–f).
Fig. 2.
—The thickness and epidermal BM zone topography of skin in humans and three Old World monkey species. The comparison of thickness of the epidermis (a) and dermis (b). Pa, Ca, Cp, and Hs indicate Papio anubis, Cercopithecus albogularis, Chlorocebus pygerythrus, and Homo sapiens, respectively. The numbers of individuals used for measurements are shown under each species name abbreviation. Photographs of hematoxylin–eosin stained histologic skin sections are from H. sapiens (c), P. anubis (d), C. albogularis (e), and Ch. pygerythrus (f). Scale bars are shown in each panel. ep. and d. indicate the epidermis and dermis, respectively. *P < 0.05, **P < 0.01, and ***P < 0.001, t-test with Bonferroni correction.
Differentially Expressed Genes between Human and Nonhuman Great Ape Skin
To investigate genes associated with human-specific skin characteristics, we identified differentially expressed genes between human and nonhuman great ape skin using RNA-Seq (supplementary fig. S1, Supplementary Material online). We used total RNA from the skin of five human and nine nonhuman great ape individuals (three individuals each from chimpanzees, gorillas, and orangutans) and sequenced their cDNA transcripts using the Illumina HiSeq platforms. The 25–45 million reads from each sample were mapped to the human reference genome. To avoid a mapping bias caused by genetic divergences between the human reference genome and the mapped reads, we also mapped reads from each sample to the chimpanzee, gorilla, and orangutan reference genomes. Details of mapped read depth for each sample were shown in supplementary table S3, Supplementary Material online. For each of the four mapping results, the expression values (i.e., RPKM values) were calculated for each gene of each sample and subsequently normalized (supplementary fig. S3, Supplementary Material online). The average normalized RPKM values of five humans were compared with those of nine nonhuman great apes for each gene. The genes that showed statistically significant differences in their average normalized RPKM values (P < 0.05, Baggerley’s test with FDR P value correction) were extracted for each of the four mapping results. Finally, we selected the genes that were common to each of the extracted results as differentially expressed genes.
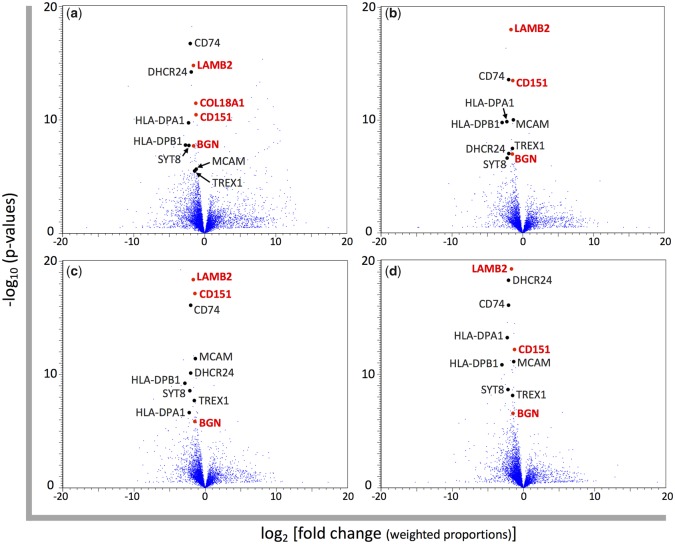
As a result, we extracted 487, 126, 165, and 166 genes (including unannotated genes and pseudogenes) with differential expression from the mapping results using human, chimpanzee, gorilla, and orangutan reference genomes, respectively. Among these genes, 30 genes were common to all mapping results (fig. 3a–d). Differential expression of COL18A1 was only detected in mapping to the human reference genome (fig. 3a); however, the annotations of COL18A1 in the three nonhuman great ape reference genomes were incomplete, and so the majority of reads could not be mapped to these genomes. Therefore, we selected COL18A1 as a differentially expressed gene between humans and nonhuman great apes. In total, 31 genes were assigned as differentially expressed genes (table 1). Twenty-five and six genes showed higher and lower expression in humans than nonhuman great apes, respectively. In this study, we focused on structural differences between human and other primate skin, therefore we further analyzed structural protein genes in our differential expression results, namely, BGN, COL18A1, CD151, and LAMB2.
Fig. 3.
—Volcano plots for the gene expression differences between humans and nonhuman great apes. Each blue dot represents the gene expressed in the skin. The normalized RPKM values are based on the mapping to the reference genome of (a) human, (b) chimpanzee, (c) gorilla, and (d) orangutan. The log 2-fold changes of average normalized RPKM values of nonhuman great apes compared with those of humans and the −log 10 P values resulting from Baggerley’s test comparing average normalized RPKM values between humans and nonhuman great apes for each gene are plotted on the x and y axis, respectively. Differentially expressed genes between humans and nonhuman great apes selected in this study are shown by black and red dots. The gene names colored in red indicate structural protein genes.
Table 1.
Differentially Expressed Genes between Human and Nonhuman Great Ape Skin
| Average Normalized RPKMa |
FDc | |||
|---|---|---|---|---|
| Humans | NH Great Apesb | |||
| Higher expression in humans | ||||
| DHCR24 | 24-Dehydrocholesterol reductase | 137.0 | 32.1 | 4.3*** |
| BGN | Biglycan | 138.3 | 41.8 | 3.3** |
| CDHR1 | Cadherin related family member 1 | 20.9 | 0.7 | 30.8*** |
| CD151 | CD151 molecule | 61.3 | 27.3 | 2.2*** |
| CD207 | CD207 molecule | 29.3 | 1.9 | 15.5*** |
| CD74 | CD74 molecule | 547.6 | 112.8 | 4.9*** |
| COL18A1 | Collagen type XVIII alpha 1 chain | 41.7 | 18.3 | 2.3*** |
| CFD | Complement factor D | 986.0 | 130.0 | 7.6*** |
| FAM57A | Family with sequence similarity 57 member A | 34.3 | 12.5 | 2.7*** |
| GEMIN4 | Gem nuclear organelle associated protein 4 | 7.0 | 2.8 | 2.5* |
| GRINA | Glutamate ionotropic receptor NMDA type subunit associated protein 1 | 42.8 | 19.1 | 2.2** |
| LAMB2 | Laminin subunit beta 2 | 83.1 | 27.3 | 3.0*** |
| LCE2A | Late cornified envelope 2A | 75.2 | 2.0 | 36.9* |
| LCE6A | Late cornified envelope 6A | 56.2 | 6.4 | 8.8* |
| HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 30.8 | 7.0 | 4.4*** |
| HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | 64.8 | 10.4 | 6.2*** |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 20.5 | 9.3 | 2.2* |
| HLA-DQB2 | Major histocompatibility complex, class II, DQ beta 2 | 40.1 | 2.4 | 17.0*** |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | 361.7 | 103.5 | 3.5*** |
| NFE2L1 | Nuclear factor, erythroid 2 like 1 | 80.0 | 42.1 | 1.9*** |
| SCRN2 | Secernin 2 | 12.0 | 5.0 | 2.4** |
| SYT8 | Synaptotagmin 8 | 14.3 | 3.2 | 4.4*** |
| TREX1 | Three prime repair exonuclease 1 | 14.0 | 6.4 | 2.2* |
| TSR3 | TSR3, acp transferase ribosome maturation factor | 47.2 | 23.3 | 2.0** |
| WFDC5 | WAP four-disulfide core domain 5 | 118.6 | 45.5 | 2.6** |
| Lower expression in humans | ||||
| BNIP3 | BCL2 interacting protein 3 | 14.7 | 39.3 | 2.7* |
| CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | 4.8 | 22.9 | 4.7** |
| HMGB2 | High mobility group box 2 | 19.6 | 45.6 | 2.3* |
| ID3 | Inhibitor of DNA binding 3, HLH protein | 41.1 | 74.2 | 1.8** |
| NPM3 | Nucleophosmin/nucleoplasmin 3 | 19.7 | 47.0 | 2.4*** |
| SULT1C4 | Sulfotransferase family 1C member 4 | 0.4 | 7.7 | 18.6*** |
Note.—Bold letters: structural protein genes.
In mapping to the human reference genome.
NH great apes: nonhuman great apes.
FD: fold difference.
P < 0.05,
P < 0.01, and
P < 0.001, Baggerley’s test with FDR P value correction.
Both COL18A1 and BGN colocalize with other collagen proteins; therefore, we also analyzed the expression of the other collagen genes using the mapping result in the human reference genome. COL18A1 forms collagen XVIII (Marneros and Olsen 2005), which is a structural component of epidermal BM (Has and Nystrom 2015). Epidermal BM and its associated anchoring structures include collagens IV, VII, and XVII as well as collagen XVIII in the epidermal BM zone (Has and Nystrom 2015). All five relatively highly expressed genes (COL4A1, COL4A2, COL7A1, COL17A1, and COL18A1) encoding proteins that form collagens in the epidermal BM zone (average normalized RPKM values for humans or nonhuman great apes ≥10) showed higher expression in humans than in nonhuman great apes (table 2). Among them, the expression differences in the two genes, COL17A1 and COL18A1, were statistically significant (P < 0.05, t-test with Bonferroni correction).
Table 2.
Collagen Genes with Relatively High Expression in Skin
| Average Normalized RPKMa |
Fold Difference | |||
|---|---|---|---|---|
| Humans | NH Great Apesb | |||
| (a) Genes encoding proteins that form collagens in the epidermal BM zone | ||||
| COL4A1 | Collagen type IV alpha 1 chain | 54.6 | 21.5 | 2.5 |
| COL4A2 | Collagen type IV alpha 2 chain | 77.5 | 32.9 | 2.4 |
| COL7A1 | Collagen type VII alpha 1 chain | 32.9 | 16.8 | 2.0 |
| COL17A1 | Collagen type XVII alpha 1 chain | 181.7 | 76.4 | 2.4* |
| COL18A1 | Collagen type XVIII alpha 1 chain | 41.7 | 18.3 | 2.3*** |
| (b) Genes encoding proteins that form BGN-binding collagens | ||||
| COL1A1 | Collagen type I alpha 1 chain | 444.4 | 198.5 | 2.2 |
| COL1A2 | Collagen type I alpha 2 chain | 311.3 | 134.9 | 2.3 |
| COL3A1 | Collagen type III alpha 1 chain | 404.7 | 148.9 | 2.7 |
| COL6A1 | Collagen type VI alpha 1 chain | 276.3 | 106.2 | 2.6 |
| COL6A2 | Collagen type VI alpha 2 chain | 459.3 | 185.1 | 2.5** |
| COL6A3 | Collagen type VI alpha 3 chain | 45.6 | 27.3 | 1.7 |
In mapping to the human reference genome.
NH great apes: nonhuman great apes.
P < 0.05,
P < 0.01, and
P < 0.001, t-test with Bonferroni correction.
BGN binds to collagens I, II, III, VI, and IX (Chen and Birk 2013) and regulates collagen fibrillogenesis in skin (Halper 2014). All six relatively highly expressed genes (COL1A1, COL1A2, COL3A1, COL6A1, COL6A2, and COL6A3) encoding proteins that form BGN-binding collagens (average normalized RPKM values for humans or nonhuman great apes ≥10) showed higher expression in humans than in nonhuman great apes (table 2). Among them, the expression difference in the gene COL6A2 was statistically significant (P < 0.05, t-test with Bonferroni correction).
In the same manner as above, we identified differentially expressed genes in each of nonhuman great ape species with results summarized in supplementary table S4, Supplementary Material online.
Inference of Substitutions Responsible for the Human-Specific Expression Patterns
We inferred substitutions in transcriptional regulatory regions responsible for the expression differences between humans and nonhuman great apes in the four structural protein genes of interest (i.e., COL18A1, LAMB2, CD151, and BGN). Transcriptional regulatory regions are expected to be conserved noncoding regions due to functional constraint (Pennacchio et al. 2006; He et al. 2011). Substitutions responsible for human-specific gene expression patterns are expected to be human-specific among the four primate species. Therefore, we identified regions that 1) were noncoding and conserved in nonhuman lineages (fig. 1, gray lines) and 2) harbored human-specific substitutions.
The genomic sequence alignments of human, chimpanzee, gorilla, and orangutan were used for this analysis. We hypothesized that the expression patterns of the four genes of interest in the skin of one Old World monkey species, the rhesus macaque (Macaca mulatta), would be similar to those of the three nonhuman great ape species, and included the genomic sequence of Macaca mulatta in the multiple primate sequence alignments to improve detection of conserved regions. We intended to infer transcriptional regulatory regions located at short distance from each gene of interest. In general, transcriptional regulatory regions located at long distance from a target gene of interest are difficult to infer accurately due to the increased possibility that the inferred region is part of the regulatory network of a nontarget neighboring gene. The analyzed genomic regions were set to include intergenic regions adjacent to the genes of interest and to locate target genes in the center of the analyzed region. When adjacent genes were close to the genes of interest (CD151 and LAMB2), we set the analyzed regions as 100 kb to increase the lengths of regions under analysis. As a result, the size of our analyzed genomic regions was 372, 100, 100, and 78 kb in length for COL18A1, LAMB2, CD151, and BGN, respectively.
In our comparative analysis, genetic distances of noncoding regions within the analyzed genomic regions between species were similar to the average divergence based on whole genome sequences (Scally et al. 2012) (supplementary table S5, Supplementary Material online). We designated regions as conserved if they showed the significantly smaller numbers of substitutions compared with the divergence of the analyzed genomic region (P < 0.05, Poisson distribution). We identified such conserved regions with a 120-bp sliding-window analysis throughout the analyzed genomic regions. Subsequently, we eliminated conserved regions completely overlapping with exonic regions from the analysis. We then extracted regions harboring human-specific substitutions in noncoding regions from the conserved regions. As a result, the numbers of extracted regions finally obtained were 49, 39, 10, and 32 for COL18A1, LAMB2, CD151, and BGN, respectively (supplementary fig. S4, Supplementary Material online, black and orange vertical lines).
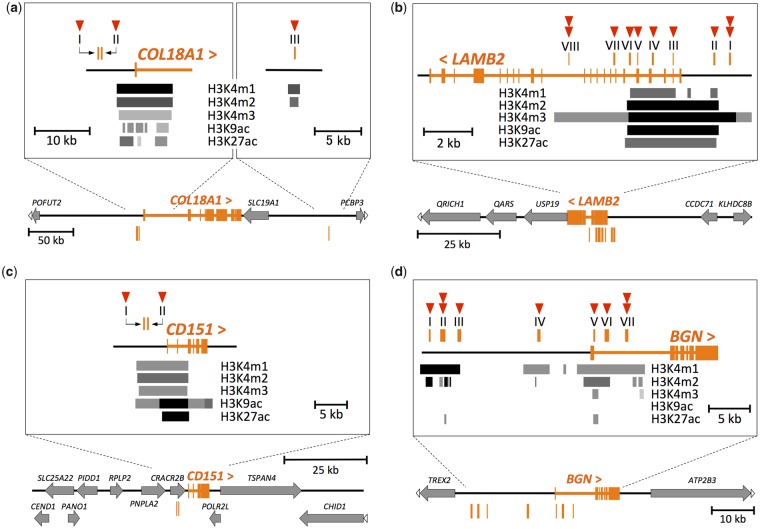
The activity of a transcriptional regulatory region differs among diverse cell types in a multicellular organism (Heintzman et al. 2009). Active promoters and enhancers generally possess specific histone modifications (H3K4m1, H3K4m2, H3K4m3, H3K9ac, and H3K27ac). From our conserved regions with human-specific substitutions, we selected regions with these histone modifications in human skin cells. As a result, the numbers of selected regions were one, six, two, and seven for COL18A1, LAMB2, CD151, and BGN, respectively (fig. 4 and supplementary fig. S4, Supplementary Material online, orange vertical lines). In addition to these regions, we also selected two regions each for COL18A1 and LAMB2. Regions I and II for COL18A1 were near (∼2-kb proximity) the histone modifications around the transcription start site of this target gene, making it likely that they regulate the expression of COL18A1 (fig. 4a). Moreover, these regions showed dimethylation of histone H3 lysine 79 (H3K79m2), which is known to correlate with multiple roles including active transcription (Farooq et al. 2016). Regions VII and VIII for LAMB2 were near (∼2-kb proximity) four histone modifications (H3K4m1, H3K4m2, H3K9ac, and H3K27ac), and in a weak histone modification (H3K4m3) (fig. 4b). These two regions possessed monomethylation of histone H3 lysine 9 (H3K9m1), which may be associated with active transcription (Barski et al. 2007), as well as H3K79m2. The selected regions were assumed to be putative transcriptional regulatory regions for each of the genes of interest. Human-specific substitutions in these regions were regarded as candidate substitutions that would result in human-specific gene expression patterns. In total, the numbers of the candidate substitutions were three, ten, two, and nine for COL18A1, LAMB2, CD151, and BGN, respectively (fig. 4).
Fig. 4.
—The positions of the putative transcriptional regulatory regions with the candidate substitutions. The exons of (a) COL18A1, (b) LAMB2, (c) CD151, and (d) BGN genes are shown in orange in each panel. The symbols “<” and “>” indicate the direction of the genes. All genes (except genes of interest) are shown by gray arrows. White arrowheads at the end of horizontal lines indicate that the genes are continuous beyond the schematic representation. Orange vertical lines indicate the position of the putative transcriptional regulatory regions numbered by Roman numerals. The zoomed-in view around these regions are shown in the rectangles of each panel. H3K4m1, H3K4m2, H3K4m3, H3K9ac, and H3K27ac indicate monomethylation of histone H3 lysine 4, dimethylation of histone H3 lysine 4, trimethylation of histone H3 lysine 4, acetylation of histone H3 lysine 9, and acetylation of histone H3 lysine 27, respectively. The positions and density of the gray scale bars indicate the positions and intensity of histone modifications shown at the side of the bars, respectively. The skin cell strains referred for the histone modifications were as below: a: NHEK, b: NHDF-Ad, c: NHEK, and d: NHDF-Ad. For LAMB2 and BGN, we also referred to the NHEK strain, in which the genes are also expressed (supplementary fig. S4b and d, Supplementary Material online). The red arrowheads indicate the numbers of the candidate substitutions located in each putative transcriptional regulatory region. A substitution in COL18A1 region I was later removed from the candidate substitutions because of the high ancestral allele frequency in the locus.
Among the 24 candidate substitution loci for the genes of interest, the ancestral alleles were found at one and two loci for CD151 and LAMB2, respectively, with low frequencies (ancestral allele frequencies: 0.0126 for CD151; 0.0002 and 0.0016 for LAMB2) in human populations, based on data from the 1000 Genomes Project (supplementary table S6, Supplementary Material online). The human-specific alleles at these loci are not fixed completely. However, they could still have a possibility to be responsible for the observed human-specific expression patterns in the genes because the ancestral alleles could remain in low frequencies when the human-specific alleles make a small contribution to the human-specific expression patterns. Therefore, we included these almost-fixed mutations in the candidate substitutions putatively responsible for the human-specific expression patterns. On the other hand, an ancestral allele was found at the candidate substitution locus in COL18A1 region I with high frequency (ancestral allele frequency: 0.683). This mutation was thus removed from the list of candidate substitutions putatively responsible for the human-specific gene expression patterns. The expression changes of the genes of interest in the human lineage may be attributable to the independent or combined effects of these candidate substitutions. The positions of the putative transcriptional regulatory regions and the candidate substitutions within those regions in the human genome are shown in supplementary table S6, Supplementary Material online.
We further explored the possibility that these candidate substitutions change the gene expression levels by using two independent evolutionary analyses. We focused on conserved 120-bp regions (P < 0.05, Poisson distribution) where each candidate substitution was located. First, we assumed that regions conserved more in nonhuman lineages (P < 0.01, Poisson distribution) than the other regions (0.01 ≤ P < 0.05, Poisson distribution) were likely to have a role in gene expression regulation. Among the most conserved 120-bp regions for each candidate substitution, three and two regions for BGN and LAMB2, respectively, matched with this condition. They contained four and two candidate substitutions for BGN and LAMB2, respectively (table 3, bold letters). Second, we assumed that the conserved 120-bp regions with the significantly larger numbers of substitutions in the human lineage than expected from the numbers of substitutions in nonhuman lineages in the same 120-bp regions (P < 0.05, Poisson distribution) are likely to change their functions in gene expression regulation. Among the conserved 120-bp regions with the largest numbers of substitutions in the human lineage for each candidate substitution, two regions for each of BGN and CD151 and three regions for LAMB2 had the significantly larger numbers of substitutions (two or three substitutions) in the human lineage. They contained four, two, and four candidate substitutions for BGN, CD151, and LAMB2, respectively (table 3, bold letters). We hypothesize that the candidate substitutions in the conserved 120-bp regions indicated by these two evolutionary analyses have a high likelihood of changing target gene expression.
Table 3.
The Numbers of Substitutions in Conserved 120-bp Regions with Candidate Substitutions
| Region | Candidate Substitutiona | Substitutionsb in Nonhuman Lineages | Substitutionsc in the Human Lineage |
|---|---|---|---|
| BGN | |||
| I | S1 | 3.79* | 1 |
| II | S1 | 1.01d,** | 2d,** |
| S2 | |||
| III | S1 | 2.52** | 1 |
| IV | S1 | 2.02** | 1 |
| V | S1 | 4.05* | 1 |
| VI | S1 | 4.05* | 1 |
| VII | S1 | 4.06d,* | 2d,* |
| S2 | |||
| COL18A1 | |||
| II | S1 | 3.03* | 1 |
| III | S1 | 3.80* | 1 |
| CD151 | |||
| I | S1 | 3.03* | 2* |
| II | S1 | 3.03* | 2* |
| LAMB2 | |||
| I | S1 | 4.09* | 1 |
| S2 | 4.06* | 1 | |
| II | S1 | 3.04* | 1 |
| III | S1 | 3.04* | 1 |
| IV | S1 | 3.03* | 2* |
| V | S1 | 4.06* | 1 |
| VI | S1 | 1.00** | 2* |
| VII | S1 | 2.02** | 1 |
| VIII | S1 | 4.25d,* | 3d,** |
| S2 | |||
Note.—Bold letters: significantly highly conserved regions in nonhuman lineages (P < 0.01, Poisson distribution) or conserved regions with the significantly larger numbers of substitutions in the human lineage (P < 0.05, Poisson distribution).
S1 and S2 represent substitutions 1 and 2 in one region, respectively.
The 120-bp regions with the smallest numbers of substitutions were selected for each candidate substitution.
The 120-bp regions with the largest numbers of substitutions were selected for each candidate substitution.
Two substitutions (S1 and S2) were located in the same 120-bp region.
P < 0.05 and **P < 0.01, Poisson distribution.
In addition to the evolutionary analyses above, we searched for TF-binding sites that contain each candidate substitution locus. Candidate substitutions in TF-binding sites have a high likelihood of changing target gene expression levels. Searching for TF-binding sites showed that all of the candidate substitutions were located in TF-binding sites (supplementary table S7, Supplementary Material online). Especially, one, six, two, and five candidate substitutions for COL18A1, LAMB2, CD151, and BGN, respectively, were in binding sites of TFs that are reported to function in skin and were also expressed based on our RNA-Seq analyses (average normalized RPKM values for humans and nonhuman great apes ≥1 in the mapping result of the human reference genome) (table 4). The candidate substitutions from the ancestral alleles to the human-specific alleles changed the similarity with the consensus sequences of binding sites for such TFs associated with skin function (table 4). However, without further experimentation, it remains difficult to infer whether these differences in similarity values would affect the TF-binding affinities.
Table 4.
Candidate Substitutions in Binding Sites of TFs with a Function in Skin
| Region | Candidate Substitutiona | TF | Binding Siteb | Relative Scorec |
|
|---|---|---|---|---|---|
| Ancestral Allele | Human-Specific Allele | ||||
| BGN | |||||
| II | S2 | HOXB2 | 22–31 (f) | 0.819 | 0.713 |
| III | S1 | FLI1 | 22–31 (r) | 0.701 | 0.827 |
| VI | S1 | KLF4 | 20–29 (f) | 0.808 | 0.719 |
| VII | S1, S2d | SPDEF | 20–30 (f) | 0.595 | 0.803 |
| FLI1 | 21–30 (f) | 0.614 | 0.825 | ||
| COL18A1 | |||||
| II | S1 | HOXB2 | 23–32 (f) | 0.755 | 0.821 |
| MSX2 | 24–31 (f) | 0.686 | 0.810 | ||
| PRRX2 | 24–31 (f) | 0.757 | 0.858 | ||
| CD151 | |||||
| I | S1 | FOSL2 | 26–36 (f) | 0.859 | 0.841 |
| SNAI2 | 23–31 (f) | 0.763 | 0.902 | ||
| II | S1 | KLF4 | 21–30 (f) | 0.809 | 0.814 |
| LAMB2 | |||||
| I | S1 | SNAI2 | 20–28 (r) | 0.971 | 0.876 |
| II | S1 | KLF4 | 25–34 (f) | 0.818 | 0.727 |
| SPDEF | 19–29 (r) | 0.842 | 0.731 | ||
| III | S1 | FLI1 | 20–29 (r) | 0.736 | 0.866 |
| V | S1 | SNAI2 | 20–28 (f) | 0.779 | 0.929 |
| SNAI2 | 24–32 (f) | 0.937 | 0.842 | ||
| VI | S1 | SNAI2 | 22–30 (r) | 0.840 | 0.785 |
| MSX2 | 21–28 (f) | 0.713 | 0.810 | ||
| VII | S1 | HOXB2 | 21–30 (r) | 0.854 | 0.748 |
S1 and S2 represent substitutions 1 and 2 in one region, respectively.
The numbers represent the positions within the 51-bp sequences retrieved for the TF search. Each candidate substitution is located at position 26. “f” and “r” indicate TF-binding site on forward and reverse strands, respectively.
This score shows the similarity with the consensus sequence of TF-binding site in the JASPAR database. The score changes by the candidate substitutions from the ancestral alleles to the human-specific alleles are shown.
These two substitutions (S1 and S2) are located next to each other. The 25-bp sequences on both sides of these substitutions were retrieved for the TF search (candidate substitutions: positions 26 and 27).
Discussion
The Identification of Human-Specific Gene Expression Patterns That May Be Related to Human-Specific Skin Characteristics
More than three decades ago, histological differences between human and other primate skin were qualitatively described (Montagna 1982, 1985). However, to the best of our knowledge, there is no report that quantifies the differences in skin structure between humans and other primates. In this study, we quantified two of the primary skin differences between humans and three Old World monkey species. As for these Old World monkey species, anubis baboons are larger than both Sykes’ monkeys and vervet monkeys (Strasser 1992; Smith and Jungers 1997; Sandel 2013) and are the largest primates in the savannah ecosystem where the early hominin species with a large body size had evolved (McHenry 1994). Because the effect of body mass on immune system evolution was reported (Semple et al. 2002), the skin tissue samples of the three Old World monkey species were originally collected for a different study related to body mass. In the present study, we used these skin samples to examine skin differences between humans and nonhuman primates. One of the primary skin differences is that the epidermis and dermis in human skin are significantly thicker than those of the three Old World monkey species investigated. The other difference is that the epidermal BM zone topography shows a rete ridge in humans but is flat in the three Old World monkey species.
To definitively clarify the human-specific characteristics in skin structure, it would be logical to compare human skin with closely related species. However, we could not investigate the histological traits in nonhuman great ape skin due to the absence of dissected skin digital photographs. Previous studies report that the epidermis in humans is thicker than that in other primates including nonhuman great apes, although without quantitative analysis (Montagna 1982, 1985). In most nonhuman primates, the epidermal underside (the position of the epidermal BM zone) of the furred skin is described as flat (Montagna 1982). Those in chimpanzees and gorillas are reported with inconsistent descriptions; a degree of sculpturing (Montagna 1982), discrete and moderate sculpturing (Montagna and Yun 1963), and a nearly flat topography (Ellis and Montagna 1962). Thus, it is assumed that the thicker epidermis and strongly sculptured epidermal underside (i.e., rete ridge) may be human-specific skin characteristics. To fully clarify these points, additional quantitative comparisons between human and nonhuman great ape skin are required.
Based on our RNA-Seq analyses, expression levels of 25 and 6 genes in skin were found to be significantly higher and lower in humans than in nonhuman great apes, respectively. Four of them (COL18A1, LAMB2, CD151, and BGN) encode structural proteins and showed higher expression in humans, suggesting the possibility that the expression changes of these genes influence human skin structure.
COL18A1, LAMB2, and CD151 are genes that encode proteins structurally associated with the epidermal BM zone. Epidermal BM and its associated anchoring structures contain collagens IV, VII, XVII, and XVIII as their structural components (Has and Nystrom 2015), and COL18A1 forms collagen XVIII (Marneros and Olsen 2005). The gene encoding protein that form collagen XVII (COL17A1) also showed significantly higher expression levels in humans. Mice lacking COL18A1 have a broadened epidermal BM (Utriainen et al. 2004), and a patient with a known mutant COL17A1 gene exhibits junctional epidermolysis bullosa (McGrath et al. 1996). Both of these observations indicate a structural role of these genes in epidermal BM zone integrity. LAMB2 is a component of the network structure of laminins in the epidermal BM (Has and Nystrom 2015). Therefore, it is plausible that the higher expression of COL17A1, COL18A1, and LAMB2 may make structure in the epidermal BM zone in human skin different from that in nonhuman great ape skin, perhaps leading to an undulating epidermal BM zone and the high number of anchoring structures corresponding to the increased adhesive area of rete ridge in human skin.
CD151 functions as an adhesion protein between the epidermis and epidermal BM (Has and Nystrom 2015). Thus, the higher expression of this gene may contribute to the strong adhesion between the epidermis and epidermal BM in human skin. If the rete ridge is specific to human skin, the higher expression of CD151 could correlate with the increased adhesive area.
BGN is localized to both the epidermis and dermis (Li et al. 2013). In the epidermis, BGN is on the cell surface of differentiating keratinocytes of the prickle cell layer (Bianco et al. 1990). Although the function of this protein in epidermis is unknown, the higher expression of BGN may somehow correlate with the human-specific thicker epidermis.
BGN is a component of the extracellular matrix in the dermis (Li et al. 2013). This protein interacts with collagens and regulates collagen fibrillogenesis to make the tensile strength of skin (Halper 2014). Because BGN interacts with collagens I, II, III, VI, and IX (Chen and Birk 2013), it was predicted that the expression of the COL genes encoding proteins that form the BGN-binding collagens would also be higher in human skin. The expression of the gene (COL6A2) encoding a protein that forms collagen VI was significantly higher in humans. Thus, the increased expression of BGN and COL6A2 may produce a stronger tensile strength in human skin. Elastic fibers are another component of the extracellular matrix in the dermis (Smoller 2009) and give skin elasticity (Kielty et al. 2002). Elastin is one of the components of elastic fibers (Kielty et al. 2002), and BGN regulates elastin formation (Reinboth et al. 2002). It is known that the human dermis is more enriched with elastic fibers compared with most other primates (Montagna 1982, 1985). The amount of elastic fibers in the furred skin of chimpanzees and gorillas is reported with inconsistent descriptions; similar to the content in humans (Montagna 1982, 1985) and nowhere numerous (Montagna and Yun 1963). Although the amount of elastic fibers in nonhuman great apes is ambiguous, it is possible that the higher expression of BGN might contribute to the richness of elastic fibers in human skin.
Humans have a low amount of hair on their body compared with other primates, which gives humans a high level of thermoregulation (Folk and Semken 1991). However, it is believed that human skin has lost the ability to protect the internal tissues from external physical stresses by hair. Rete ridge increases the area where the epidermis and dermis connect compared with flat topography of the epidermal BM zone, which may make strong adhesion between these two layers. The rete ridge, thick epidermis, and rich elastic fibers in skin might contribute to the physical strength of human skin. Actually, it has been proposed that human skin has developed adaptive structural changes that give it strength and resilience (Montagna 1982). Although additional quantitative comparison between human and nonhuman great ape skin is required to examine the human-specific skin characteristics, the human-specific expression patterns found in this study may contribute to adaptive skin characteristics specific to humans with a low amount of hair on their body.
As for the differentially expressed genes other than structural protein genes, the comparison of nonstructural traits between human and other primate skin could reveal the correlation between the differential expression of these genes and human-specific characteristics. Among those, late cornified envelope (LCE) genes may function practically in skin. The LCE gene family consists of 18 members subdivided into 6 subgroups, LCE1 to LCE6, based on similarities of amino acid sequences, genomic organization, and patterns of expression (Jackson et al. 2005; Bergboer et al. 2011). Among the differentially expressed genes, LCE2A and LCE6A genes, which showed higher expression in humans, are the members of this gene family. LCE6A sequences were intact in humans and nonhuman great apes (human: NM_001128600.1, chimpanzee: XM_003308479.1, gorilla: XM_004026698.1, and orangutan: XM_002810185.1), whereas LCE2A sequence was intact in humans (NM_178428.3) but those were independently pseudogenized by a premature stop codon in chimpanzees (LOC736270), by a frameshift in gorillas (LOC109029453), and by a large deletion in the coding region in orangutans (LOC103891408). The functions of LCE2A and LCE6A proteins were unknown so far.
In humans, LCE2A is expressed in normal healthy skin (Bergboer et al. 2011), as observed in our RNA-Seq analyses. Upregulation of this gene was induced by high concentration of extracellular calcium, ultraviolet irradiation (Jackson et al. 2005), and Th17 cytokine stimulation (Niehues et al. 2017), indicating functional roles of this gene in skin. Thus, only humans retain the functional LCE2A, which might be related to human-specific skin characteristics.
In the LCE gene family, the function of a few members was reported. A deletion of LCE3B and LCE3C is strongly associated with psoriasis (De Cid et al. 2009), and the deletion is traced back to the ancient Homo lineage (Lin et al. 2015; Pajic et al. 2016). Recently, the antimicrobial activity against a variety of bacterial taxa was shown in LCE3A, LCE3B, and LCE3C proteins (Niehues et al. 2017). The LCE2A and LCE6A proteins might also have antimicrobial defensive roles predominant in human skin.
Because humans have a much less hair than nonhuman great apes, it was predicted that the expression of the genes associated with the components of hair would be lower in humans. However, no such gene showed significantly lower expression in humans than in nonhuman great apes based on the present RNA-Seq analyses. Actually, the average normalized RPKM values of hair keratin genes were tens to thousands of fold higher in nonhuman great apes than in humans. However, several nonhuman great ape individuals showed the RPKM values similar to human individuals in those genes, resulting in insignificant differences in expression levels between humans and nonhuman great apes (examples are shown in supplementary table S8, Supplementary Material online). This variation in RPKM values did not depend on differences in sex, age, or body part between nonhuman great ape individuals.
It is conceivable that the genes with human-specific expression patterns found in this study cooperate with other differentially expressed genes to change human skin structure. For example, although significant differences in gene expression were detected only in a few genes, the expression of all the relatively highly expressed genes encoding proteins that form collagens associated with the epidermal BM zone and BGN was higher in humans than in nonhuman great apes. These genes might also contribute to the rete ridge formation and stronger tensile strength in human skin. In this study, we used skin specimens from individuals of different sex and age and from different body parts for RNA-Seq analyses. Therefore, the identified genes were consistently differentially expressed in human skin compared with nonhuman great ape skin, regardless of these differences. In the future, increasing the number of skin specimens and comparing gene expression levels between humans and nonhuman great apes using the skin specimens from individuals of the same condition (e.g., the same sex, age, and body part) would identify other differentially expressed genes specific to that condition. This approach would allow us to further reveal the genetic causes of human-specific skin characteristics.
Inference of Substitutions Possibly Related to Human-Specific Skin Characteristics
Substitutions in transcriptional regulatory regions can change the expression of their target genes (Wittkopp and Kalay 2012). We hypothesized that the substitutions responsible for the human-specific expression patterns in the four structural protein genes of interest (COL18A1, LAMB2, CD151, and BGN) were 1) in noncoding regions that were conserved in nonhuman lineages and 2) specific to humans. Conserved regions in nonhuman great ape and rhesus macaque lineages are difficult to find through sequence alignment, as the species are closely related and the sequences are largely identical. Therefore, we utilized a sliding-window analysis to identify regions that exhibited the significantly smaller numbers of substitutions than expected from the divergences of the each analyzed genomic region between the species.
The conserved noncoding regions with human-specific substitutions we identified were taken for the next analysis. We suggested that the human-specific substitutions in those regions with histone modifications for active transcription in skin cells could be the candidate substitutions responsible for the human-specific expression patterns in the genes of interest.
Substitutions changing the expression of target genes in transcriptional regulatory regions generally alter the binding affinities for TFs that modulate the gene expression (Wittkopp and Kalay 2012). According to the TF-binding site searches, all of the candidate substitution loci were expected to be located in TF-binding sites, suggesting a possibility that the regions containing the candidate substitution loci are associated with gene expression regulation. Expected TFs listed in table 4 are reported to function in skin, and therefore they are likely to regulate the expression of the genes of interest. Comparison in skin injury repair between MSX2 null mutant and wild type mice suggested that MSX2 regulates the cellular competence of keratinocytes and fibroblasts (Yeh et al. 2009). Deletion of the PRRX2 gene in mice reduced fetal fibroblast proliferation during wound healing (White et al. 2003). FOSL2 mutant mice caused skin barrier defects due to reduced expression of epidermal differentiation genes, and ectopic expression of FOSL2 induced expression of those genes (Wurm et al. 2015). Microarray analysis showed that HOXB2 and SPDEF genes were highly expressed in the regenerating skin during tissue expansion (Yang et al. 2011). The other three TFs function in skin and are somewhat associated with the genes of interest. Homozygous KLF4 deletion mutant mice lose the skin barrier function (Segre et al. 1999), and KLF4 accelerated epidermal barrier acquisition (Patel et al. 2006). KLF4 regulates the expression of some members of the laminin family (Ghaleb and Yang 2017). The reepithelialization component was reduced during wound healing in SNAI2 null mice (Hudson et al. 2009), and SNAI2 is intrinsically linked to CD151 (Yin et al. 2014). The homozygous deletion of the C-terminal transcriptional activation domain of the Fli1 gene upregulates expression levels of the genes encoding collagens I and III components in mouse skin (Asano et al. 2009). These collagens are known to bind to BGN (Chen and Birk 2013). Their predicted binding sites suggest that KLF4, SNAI2, and FLI1 are likely to regulate the expression of LAMB2, CD151, and BGN, respectively. The candidate substitutions from the ancestral alleles to the human-specific alleles changed the similarity with the consensus sequences of binding sites for the TFs associated with skin function. This result suggests that these substitutions may change the binding affinities for the predicted TFs and may change the expression levels in the genes of interest. In addition, it is likely that the candidate substitutions located in highly conserved regions in nonhuman lineages and in conserved regions with the larger numbers of substitutions in the human lineage than expected would change the expression levels in the genes of interest.
In this study, we suggested that the candidate substitutions in the putative transcriptional regulatory regions may cause the human-specific gene expression patterns that possibly lead to the human-specific characteristics in skin structure. In the future, to examine whether these candidate substitutions are responsible for the expression differences between humans and nonhuman great apes, we will conduct a promoter assay in skin cells using the putative transcriptional regulatory regions with the ancestral alleles and the human-specific alleles located at the candidate substitution loci. Identifying substitutions that may give humans adaptive skin characteristics through human-specific gene expression patterns will contribute to the understanding of how human-specific characteristics have been genetically acquired.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by an internal SOKENDAI grant to Y.T. and the Cooperative Research Program of Primate Research Institute, Kyoto University. This work was supported in part by SOKENDAI and the GAIN. This work was supported by JSPS KAKENHI (grant number JP26650172) to A.M.-O. We thank the following facilities for providing nonhuman great ape skin samples: Kumamoto sanctuary, Himeji Central Park, Fukuoka City Zoological Garden, Kamine Zoo, Kobe Oji Zoo, Kyoto City Zoo, Osaka-city Tennoji Zoo, and Sapporo Maruyama Zoo. These samples were provided through the GAIN. We thank the Institute of Primate Research, National Museum of Kenya for help with collection of Old World monkey skin samples.
Author Contributions
N.A. performed measurements of skin thickness, RNA extraction, library preparation for RNA-Seq, gene expression analyses, evolutionary analyses for extraction of conserved regions, extraction of regions with histone modifications, and the search for TF-binding sites, and wrote the manuscript. D.U. provided the digital photographs of dissected skin and helpful discussion. K.T. provided the digital photographs of dissected skin and helpful discussion. A.M.-O. arranged Old World monkey sampling and collected the skin samples. A.N. arranged Old World monkey sampling and helped with collection of the skin samples. D.C.C. arranged Old World monkey sampling and collected the skin samples. N.J. managed Old World monkey sampling and helped with collection of the skin samples. H.I. managed and provided the skin samples of nonhuman great apes and provided helpful discussion. Y.S. developed the concept and plan of evolutionary analyses for extraction of conserved regions and contributed to manuscript writing. Y.T. developed the research concept, planned the research, and performed all experiments and analyses with N.A., and wrote the manuscript.
Footnotes
Data deposition: This project has been deposited at DDBJ Sequenced Read Archive under the accessions DRX121122–DRX121135.
Literature Cited
- Asano Y, et al. 2009. Transcription factor Fli1 regulates collagen fibrillogenesis in mouse skin. Mol Cell Biol. 292:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly KA, Deng L, Morris JS, Aldaz CM.. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 1912:1477–1483. [DOI] [PubMed] [Google Scholar]
- Barski A, et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 1294:823–837. [DOI] [PubMed] [Google Scholar]
- Bergboer JG, et al. 2011. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol. 1784:1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Fisher LW, Young MF, Termine JD, Robey PG.. 1990. Expression and localization of the two small proteoglycans biglycan and decorin in developing human skeletal and non-skeletal tissues. J Histochem Cytochem. 3811:1549–1563. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP.. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 192:185–193. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 1341:25–36. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD.. 2013. From DNA to diversity: molecular genetics and the evolution of animal design. New York:John Wiley & Sons. [Google Scholar]
- Chen S, Birk DE.. 2013. The regulatory roles of small leucine‐rich proteoglycans in extracellular matrix assembly. FEBS J. 28010:2120–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávid-Barrett T, Dunbar RI.. 2016. Bipedality and hair loss in human evolution revisited: the impact of altitude and activity scheduling. J Hum Evol. 94:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cid R, et al. 2009. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 412:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RA, Montagna W.. 1962. The skin of primates. VI. The skin of the gorilla (Gorilla gorilla). Am J Phys Anthropol. 202:79–93. [DOI] [PubMed] [Google Scholar]
- Enard W, et al. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 4186900:869. [DOI] [PubMed] [Google Scholar]
- Farooq Z, Banday S, Pandita TK, Altaf M.. 2016. The many faces of histone H3K79 methylation. Mutat Res Rev Mutat Res. 768:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E.. 1967. Construction of phylogenetic trees. Science 1553760:279–284. [DOI] [PubMed] [Google Scholar]
- Folk GE, Semken A.. 1991. The evolution of sweat glands. Int J Biometeorol. 353:180–186. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Yang VW.. 2017. Krüppel-like factor 4 (KLF4): what we currently know. Gene 611:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper J. 2014. Proteoglycans and diseases of soft tissues In: Progress in heritable soft connective tissue diseases. Berlin:Springer; p. 49–58. [Google Scholar]
- Has C, Nystrom A.. 2015. Epidermal basement membrane in health and disease. Curr Top Membr. 76:117–170. [DOI] [PubMed] [Google Scholar]
- He Q, et al. 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 435:414. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 4597243:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LG, et al. 2009. Cutaneous wound reepithelialization is compromised in mice lacking functional Slug (Snai2). J Dermatol Sci. 561:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iivanainen A, et al. 1995. The human laminin β2 chain (s-laminin): structure, expression in fetal tissues and chromosomal assignment of the LAMB2 gene. Matrix Biol. 146:489–497. [DOI] [PubMed] [Google Scholar]
- Jackson B, et al. 2005. Late cornified envelope family in differentiating epithelia—response to calcium and ultraviolet irradiation. J Invest Dermatol. 1245:1062–1070. [DOI] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L.. 2012. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 131:424.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. 2002. The human Genome Browser at UCSC. Genome Res. 126:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielty CM, Sherratt MJ, Shuttleworth CA.. 2002. Elastic fibres. J Cell Sci. 115(Pt 14):2817–2828. [DOI] [PubMed] [Google Scholar]
- Kimura H. 2013. Histone modifications for human epigenome analysis. J Hum Genet. 587:439.. [DOI] [PubMed] [Google Scholar]
- King M-C, Wilson AC.. 1975. Evolution at two levels in humans and chimpanzees. Science 1884184:107–116. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V.. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 761:8–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 1284:693–705. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. 2013. Age-dependent alterations of decorin glycosaminoglycans in human skin. Sci Rep. 3:2422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Pavlidis P, Karakoc E, Ajay J, Gokcumen O.. 2015. The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol Biol Evol. 324:1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 4787370:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ.. 2012. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 137:436.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ.. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 3896648:251.. [DOI] [PubMed] [Google Scholar]
- Marneros AG, Olsen BR.. 2005. Physiological role of collagen XVIII and endostatin. FASEB J. 197:716–728. [DOI] [PubMed] [Google Scholar]
- Mathelier A, et al. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44(D1):D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, et al. 1996. Compound heterozygosity for a dominant glycine substitution and a recessive internal duplication mutation in the type XVII collagen gene results in junctional epidermolysis bullosa and abnormal dentition. Am J Pathol. 148: 1787. [PMC free article] [PubMed] [Google Scholar]
- McHenry HM. 1994. Behavioral ecological implications of early hominid body size. J Hum Evol. 27(1–3):77–87. [Google Scholar]
- Montagna W, editor. 1982. Advanced Views in Primate Biology. The Evolution of Human Skin, Berlin: Springer; 35–41.
- Montagna W. 1985. The evolution of human skin (?). J Hum Evol. 141:3–22. [Google Scholar]
- Montagna W, Yun JS.. 1963. The skin of primates. XV. The skin of the chimpanzee (Pan satyrus). Am J Phys Anthropol. 212:189–203. [DOI] [PubMed] [Google Scholar]
- Niehues H, et al. 2017. Psoriasis-associated late cornified envelope (LCE) proteins have antibacterial activity. J Invest Dermatol. 13711:2380–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P, Lin Y-L, Xu D, Gokcumen O.. 2016. The psoriasis-associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence. BMC Evol Biol. 161:265.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA.. 2006. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci U S A. 10349:18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 4447118:499. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, et al. 2008. Human-specific gain of function in a developmental enhancer. Science 3215894:1346–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinboth B, Hanssen E, Cleary EG, Gibson MA.. 2002. Molecular interactions of biglycan and decorin with elastic fiber components: biglycan forms a ternary complex with tropoelastin and microfibril-associated glycoprotein 1. J Biol Chem. 2776:3950–3957. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R.. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 1918:2496–2497. [DOI] [PubMed] [Google Scholar]
- Saarela J, Rehn M, Oikarinen A, Autio-Harmainen H, Pihlajaniemi T.. 1998. The short and long forms of type XVIII collagen show clear tissue specificities in their expression and location in basement membrane zones in humans. Am J Pathol. 1532:611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandel AA. 2013. Brief communication: hair density and body mass in mammals and the evolution of human hairlessness. Am J Phys Anthropol. 1521:145–150. [DOI] [PubMed] [Google Scholar]
- Scally A, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 4837388:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E.. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 224:356.. [DOI] [PubMed] [Google Scholar]
- Semple S, Cowlishaw G, Bennett PM.. 2002. Immune system evolution among anthropoid primates: parasites, injuries and predators. Proc R Soc Lond B Biol Sci. 2691495:1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Jungers WL.. 1997. Body mass in comparative primatology. J Hum Evol. 326:523–559. [DOI] [PubMed] [Google Scholar]
- Smoller BR. 2009. Lever’s histopathology of the skin. J Cutan Pathol. 36:605–605. [Google Scholar]
- Stedman HH, et al. 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 4286981:415. [DOI] [PubMed] [Google Scholar]
- Strasser E. 1992. Hindlimb proportions, allometry, and biomechanics in Old World monkeys (Primates, Cercopithecidae). Am J Phys Anthropol. 872:187–213. [DOI] [PubMed] [Google Scholar]
- Utriainen A, et al. 2004. Structurally altered basement membranes and hydrocephalus in a type XVIII collagen deficient mouse line. Hum Mol Genet. 1318:2089–2099. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 407:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P, et al. 2003. Deletion of the homeobox gene PRX-2 affects fetal but not adult fibroblast wound healing responses. J Invest Dermatol. 1201:135–144. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G.. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 131:59.. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 83:206.. [DOI] [PubMed] [Google Scholar]
- Wurm S, et al. 2015. Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 292:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, et al. 2011. A preliminary study of differentially expressed genes in expanded skin and normal skin: implications for adult skin regeneration. Arch Dermatol Res. 3032:125–133. [DOI] [PubMed] [Google Scholar]
- Yeh J, et al. 2009. Accelerated closure of skin wounds in mice deficient in the homeobox gene Msx2. Wound Repair Regen. 175:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, et al. 2014. CD151 represses mammary gland development by maintaining the niches of progenitor cells. Cell Cycle 1317:2707–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE.. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 121:7.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.