The genetic basis of many spastic ataxias remains unknown. Minnerop et al. identify intronic mutations in POLR3A as a frequent cause of recessive spastic ataxia. Surprisingly, >80% of carriers have the same c.1909+22G>A mutation. The mutation causes a homogeneous phenotype without overt leukodystrophy, previously considered the unifying feature of POLR3A-disease.
Keywords: spastic ataxia, hereditary spastic paraplegia, cerebellar ataxia, POLR3A, leukodystrophy
Abstract
Despite extensive efforts, half of patients with rare movement disorders such as hereditary spastic paraplegias and cerebellar ataxias remain genetically unexplained, implicating novel genes and unrecognized mutations in known genes. Non-coding DNA variants are suspected to account for a substantial part of undiscovered causes of rare diseases. Here we identified mutations located deep in introns of POLR3A to be a frequent cause of hereditary spastic paraplegia and cerebellar ataxia. First, whole-exome sequencing findings in a recessive spastic ataxia family turned our attention to intronic variants in POLR3A, a gene previously associated with hypomyelinating leukodystrophy type 7. Next, we screened a cohort of hereditary spastic paraplegia and cerebellar ataxia cases (n = 618) for mutations in POLR3A and identified compound heterozygous POLR3A mutations in ∼3.1% of index cases. Interestingly, >80% of POLR3A mutation carriers presented the same deep-intronic mutation (c.1909+22G>A), which activates a cryptic splice site in a tissue and stage of development-specific manner and leads to a novel distinct and uniform phenotype. The phenotype is characterized by adolescent-onset progressive spastic ataxia with frequent occurrence of tremor, involvement of the central sensory tracts and dental problems (hypodontia, early onset of severe and aggressive periodontal disease). Instead of the typical hypomyelination magnetic resonance imaging pattern associated with classical POLR3A mutations, cases carrying c.1909+22G>A demonstrated hyperintensities along the superior cerebellar peduncles. These hyperintensities may represent the structural correlate to the cerebellar symptoms observed in these patients. The associated c.1909+22G>A variant was significantly enriched in 1139 cases with spastic ataxia-related phenotypes as compared to unrelated neurological and non-neurological phenotypes and healthy controls (P = 1.3 × 10−4). In this study we demonstrate that (i) autosomal-recessive mutations in POLR3A are a frequent cause of hereditary spastic ataxias, accounting for about 3% of hitherto genetically unclassified autosomal recessive and sporadic cases; and (ii) hypomyelination is frequently absent in POLR3A-related syndromes, especially when intronic mutations are present, and thus can no longer be considered as the unifying feature of POLR3A disease. Furthermore, our results demonstrate that substantial progress in revealing the causes of Mendelian diseases can be made by exploring the non-coding sequences of the human genome.
Introduction
Novel sequencing technologies have been essential for expediting our understanding of the genetics of rare diseases. They have contributed to elucidating genetically unsolved cases and unmasking unusual overlap between disorders previously regarded clinically as different disease entities, such as hereditary spastic paraplegias and cerebellar ataxias. However, in both diseases, despite extensive efforts, 40–50% of patients remain genetically unexplained, implicating novel genes and unrecognized mutations in known genes (Schule et al., 2016).
Strategies to search for genes and mutations have so far mainly focused on coding variations. Attention is now increasingly turned toward the non-coding space as variants in non-coding regions located beyond the highly conserved splice sites at the intron–exon boundary are likely candidates to explain at least part of the ‘diagnostic gap’ in rare Mendelian disorders like hereditary spastic paraplegias and cerebellar ataxias. Unfortunately, although whole-exome sequencing (WES) covers at least some non-coding regions, particularly those adjacent to known exons and untranslated regions, variants within these non-coding regions are often excluded from analysis due to sparse annotation and challenging functional validation of these variants.
In this study, WES findings in a large recessive spastic ataxia family turned our attention to intronic variants in POLR3A, a gene previously associated with hypomyelinating leukodystrophy type 7 (Bernard et al., 2011; Saitsu et al., 2011; Daoud et al., 2013). Consequently, we screened a cohort of hereditary spastic paraplegia and cerebellar ataxia cases (n = 618) for mutations in POLR3A and identified compound heterozygous POLR3A mutations in ∼3.1% of index cases. Interestingly, >80% of carriers presented with an identical splice variant caused by a nucleotide change more than 20 base pairs apart from the canonical splice site (c.1909+22G>A). This variant was clinically associated with a distinct and uniform novel phenotype characterized by adolescent onset of progressive spastic ataxia with frequent occurrence of tremor, involvement of the central sensory tracts and dental abnormalities. Interestingly, these patients did not present any overt signs of hypomyelinating leukodystrophy, previously considered a unifying feature of POLR3A-related syndromes. Our results demonstrate that substantial insights into genetic causes of Mendelian diseases can be obtained by focusing on non-coding DNA sequences. Specifically, our data identified a major genetic cause for hereditary spastic ataxias.
Material and methods
Patients and controls
A large non-consanguineous German family (Family F1) with two non-affected (a third sibling had died in a traffic accident) and four affected children and with apparently recessive spastic ataxia (Fig. 1A) underwent detailed clinical and genetic studies. An extensive genetic workup was performed to identify the causative gene, with negative results for spastic paraplegia genes SPG4, SPG7, SPG11, SPG15, SPG20, SPG21, SPG35, SPG39, SPG44, spinocerebellar ataxia loci SCA1, SCA2, SCA3, SCA6, SCA7, SCA17, Friedreich ataxia, ataxia with oculomotor apraxia type 2, autosomal recessive spastic ataxia of Charlevoix-Saguenay type, and adrenomyeloneuropathy. Additionally, a cohort of 618 index cases with sporadic or recessive hereditary spastic paraplegia, spastic ataxia or cerebellar ataxia was analysed for mutations in POLR3A based on available next generation sequencing data (Supplementary Table 1).
Figure 1.
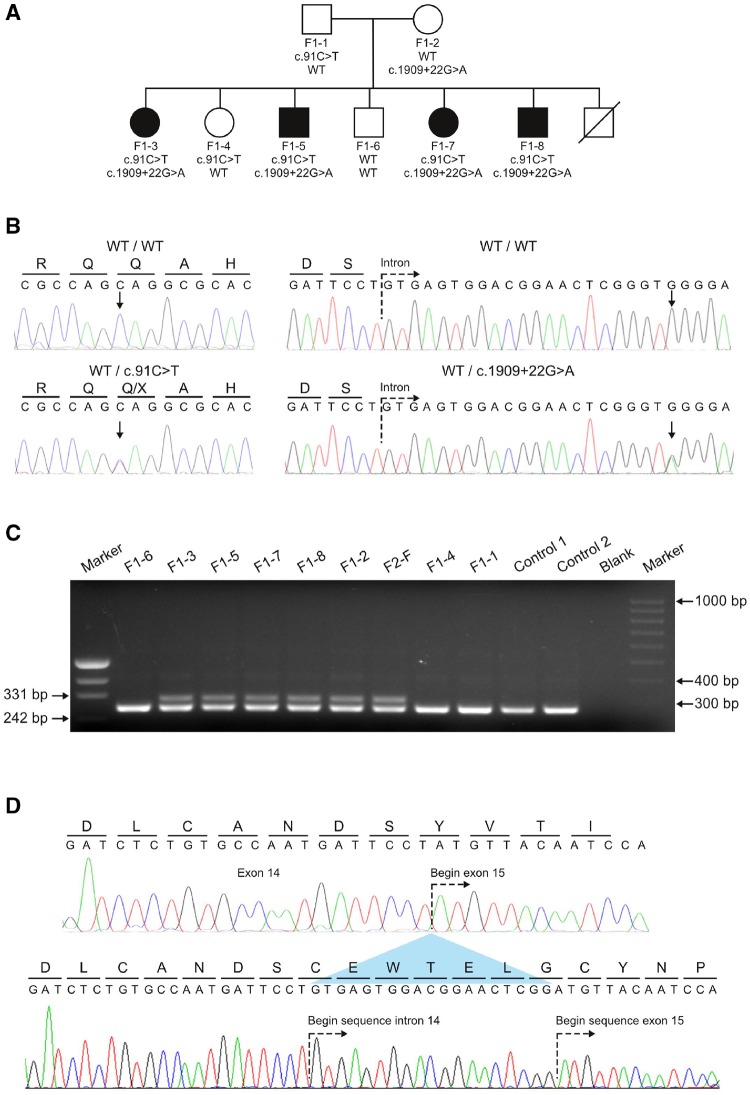
Genetic screening of a non-consanguineous German family. (A) Pedigree of Family F1 affected by the novel spastic ataxia syndrome. Pedigree symbols: circle, female; square, male; filled, affected; unfilled, unaffected; strike through, deceased. The compound heterozygous mutations found in POLR3A are shown below each symbol. Both mutations segregated with the disease phenotype in the family. (B) Sequencing traces showing the wild-type alleles and the mutant alleles c.91C>T and c.1909+22G>A. Arrows denote the nucleotides undergoing a change for the respective mutations. All affected members in Family F1 are compound heterozygous carriers for both mutations. (C) Agarose gel showing PCR product obtained from the amplification of cDNA template derived from patients, healthy family members, and two controls to show the effect of c.1909+22G>A on the splicing of exon 14. A primer pair flanking exon 14 was used to amplify leucocyte-derived cDNA template from six carriers of the c.1909+22G>A variant and three additional family members of Family F1 that carry the wild-type allele (Patients F1-1, F1-4 and F1-6). An additional aberrant larger fragment (298 bp) in carriers of c.1909+22G>A is not present in the five non-carriers of the splice site mutation. (D) Sequencing analysis of the aberrant amplicon. The upper sequence trace shows the wild-type cDNA sequence obtained for exon 14 and 15. The predicted amino acid translation of this sequence is shown above the nucleotides. The lower sequence trace represents direct sequencing of the aberrant fragment obtained in carriers of the c.1909+22G>A allele. An out-of-frame inclusion of the first 19 nucleotides of intron 14 (caused by the activation of a cryptic splice site) causes a shift of the reading frame.
Cases selected for WES and gene panel sequencing were recruited by the Universities of Bonn, Essen, Miami, Tübingen, or Newcastle. All subjects included in the study gave their written informed consent according to protocols approved by the respective institutional human ethics review boards. Genomic DNA was extracted from EDTA blood samples according to standard protocols.
Clinical characterization
A detailed neurological examination by a movement disorder and/or neuromuscular specialist was performed in all affected individuals as well as in some unaffected family members. Additional exams in subsets including neurophysiological and neuropsychological testing, detailed dental and oro-facial examination and MRI of the brain were performed as indicated in Table 1.
Table 1.
Clinical characteristics of POLR3A patients
| Study ID | F1-3 | F1-5 | F1-7 | F1-8 | F2-1 | F3-1 | F4-1 | F5-1 | F6-1 | F7-1 | F8-1 | F9-1 | F10-1 | F11-1 | F12-1 | F13-1 | F14-1 | F15-1 | F16-1 | F16-2 | F17-1 | F18-1 | F19-1 | F20-1 | F21-3 | F22-6 | F22-7 | F23-3 | F23-5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.1909 + 22G>A | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Het | Hom | − | − | − | − | − | − |
| Second mutation | Q31* | Q31* | Q31* | Q31* | D372N | F431 Sfs*26 | E12 61K | L454F | S825 Qfs*18 | G904* | A515V | G854 Afs*5 | splice | Q511* | V1315fs*7 | K71 3Kfs*3 | C109S | G350 Gfs*27 | R873* | R873* | M85 2fs*7 | L356P | − | V1033A (hom) | c.1771-7C>G (hom) | ||||
| Mode of inheritance | AR | AR | AR | AR | AR | S | S | S | S | AR | S | S | S | S | S | S | S | AR | AR | AR | S | S | S | ARa | S | AR | AR | AR | AR |
| Gender | F | M | F | M | F | M | M | M | M | F | M | M | F | M | M | F | F | F | F | F | M | M | F | M | M | F | F | M | F |
| Race | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU | CAU |
| Origin | GER | GER | GER | GER | GER | GER | GER | GER | GER | USA | UK | GER | GER | GER | GER | GER | GER | GER | UK | UK | UK | UK | USA | BEL | GER | TR | TR | IL | IL |
| Age at onset (y) | 51 | 15 | 20 | 14 | 20 | 15 | 20 | 28 | 13 | 17 | 18 | 12 | 26 | 12 | 18 | 6 | 12 | 31 | 21 | 23 | 12 | 20 | 11 | 2 | 16 | 0b | 0b | 2 | 2 |
| Age at exam (y) | 57 | 53 | 50 | 48 | 47 | 68 | 56 | 38 | 27 | 30 | 31 | 42 | 56 | 66 | 41 | 50 | 45 | 45 | 30 | 35 | 25 | 42 | 18 | 20 | 55 | 29 | 21c | 12 | 19 |
| Loss of independent walkingd | N/A | 23 | 8 | 18 | 26 | 29 | 27 | N/A | N/A | N/A | 13 | 27 | 29 | 26 | N/A | 37 | 31 | N/A | 7 | 7 | 8 | 10 | 12 | 32 | N/A | 14 | 0e | N/A | 16 |
| Cognitive deficits | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | +f | − | − | + | + | + |
| Depression | + | + | + | + | − | − | − | + | − | − | − | − | + | − | − | + | + | − | − | − | − | + | − | + | − | + | + | − | − |
| Pyramidal and peripheral motor system | |||||||||||||||||||||||||||||
| UL/LL spasticity | −/− | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | +/+ | −/− | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/− | −/− | −/+ | −/+ | −/+ | −/+ | −/+ | −/− | −/− | −/+ | −/− | −/− | −/− | −/− |
| UL/LL tendon reflexes | ↓/↓ | ⌋/⌋ | ⌋/↑ | −/⌋ | ↓/↓ | −/− | ⌋/↓ | ⌋/↓ | ⌋/↓ | ⌋/↑ | −/↓ | ⌋/↑ | ↑/⌋ | ↑/↑ | ⌋/↓ | −/⌋ | −/↓ | ⌋/⌋ | −/− | −/− | −/⌋ | −/⌋ | −/− | ⌋/⌋ | ⌋/⌋ | ⌋/↓ | ⌋/↓ | ⌋/⌋ | ⌋/⌋ |
| Extensor plantar response | − | + | + | + | + | − | + | + | + | + | + | + | − | + | + | + | + | + | − | − | − | − | − | − | − | − | − | + | + |
| UL/LL weakness | −/− | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/− | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | −/+ | +/+ | +/+ | −/+ | −/− | −/+ | −/+ | −/+ | −/− | −/+ |
| Muscle atrophy | − | LL | − | − | − | − | − | − | LL | − | − | − | − | UL/LL | UL/LL | − | − | − | − | − | LL | UL/LL | − | − | LL | n.d. | LL | − | − |
| Cerebellar system | |||||||||||||||||||||||||||||
| Sacc. pursuit | + | + | − | + | − | + | + | + | − | + | − | − | − | + | − | + | + | − | + | + | + | + | − | + | + | + | + | + | + |
| Dysarthria/dysphagia | −/− | +/− | +/+ | +/− | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | +/+ | −/− | −/− | +/− | −/− | +/− | +/− | −/− | +/− | −/− | +/− | +/− | +/+ | +/+ | +/+ | +/+ |
| UL/LL intention tremor | +/− | +/− | −/− | +/− | +/− | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− | +/− | −/+ | +/− | +/− | −/+ | +/− | −/− | +/+ | −/− | −/− | +/+ | +/+ | −/− | −/− | −/− | −/− |
| Ataxia | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Vibration/surface sens. (LL) | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/↓ | ↓/⌋ | ↓/↓ | ↓/⌋ | ↓/⌋ | ↓/⌋ | ↓/↓ | ↓/↓ | ↓/⌋ | ↓/⌋ | ↓/↓ | n.d. | ↓/↓ | n.d. | ↓/⌋ | ⌋/⌋ | ↓/⌋ | ↓/⌋ | ↓/↓ | n.d. | n.d. |
| Urinary/faecal urgency | −/− | +/− | +/− | +/− | −/− | −/− | +/+ | +/− | −/− | +/− | −/− | +/− | +/n.d | +/− | +/− | +/− | +/− | −/− | −/+ | −/+ | −/− | −/− | −/− | −/− | +/− | −/− | +/+ | −/− | −/− |
| Dentition abnormalities | + | + | + | + | + | + | + | n.d. | + | n.d. | n.d. | + | n.d. | − | − | + | + | n.d. | − | − | − | − | n.d. | n.d. | n.d. | n.d. | − | − | − |
| Others | RLS, sleep disturbance | Facial hemispasm, sleep disturbance | Sleep disturbance | RLS, sleep disturbance | Sleep disturbance | RLS, sleep disturbance | − | Aggressive behaviour | − | Tremor (head, voice, UL) | − | − | Tremor (head, voice) | Severe dystonic tremor (head, voice) | Mild dystonic tremor (head) | Tremor (head), RLS, vitiligo | Pes cavus | − | − | − | − | − | Pes cavus | Autism | Rigor | Pes cavus | Infantile seizures dystonia | Rigor, hypokinesia | Rigor, hypokinesia |
| Neurophysiology | |||||||||||||||||||||||||||||
| Abnormal nerve conduction studies | − | + | − | + | − | + | − | − | − | n.d. | − | − | − | + | + | − | − | − | n.d. | n.d. | n.d. | n.d. | + | − | n.d. | n.d. | − | n.d. | n.d. |
| Abnormal MEPs | + | + | + | + | + | + | + | + | + | n.d. | n.d. | + | + | n.d. | + | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. | n.d. |
| Abnormal SEPs | + | + | + | + | + | + | + | + | n.d. | n.d. | + | + | + | n.d. | n.d. | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. | n.d. |
| Abnormal VEPs | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | + | + | n.d. | n.d. | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. | − | n.d. | n.d. | n.d. | n.d. | n.d. | + | n.d. | n.d. | n.d. | n.d. | n.d. |
| MRI | |||||||||||||||||||||||||||||
| Spinal cord atrophy | + | + | + | + | + | n.d. | + | + | n.d. | n.d. | + | n.d. | + | + | + | + | + | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d.g | n.d. | n.d. | − | n.d. | n.d. |
| Cerebellar atrophy | − | − | − | − | − | n.d. | − | − | − | n.d. | − | n.d. | − | − | − | − | − | − | − | − | − | − | n.d. | + | n.d. | n.d. | − | − | n.d. |
| TCC | + | + | + | + | + | n.d. | − | − | − | N/E | − | n.d. | N/E | − | − | + | − | N/E | N/E | N/E | N/E | N/E | N/E | + | n.d. | n.d. | − | − | n.d. |
| SCP hyperintensity (FLAIR) | + | + | + | + | + | n.d. | + | + | + | N/E | − | n.d. | N/E | + | n.d. | + | + | N/E | N/E | N/E | N/E | N/E | N/E | N/E | n.d. | n.d. | − | − | n.d. |
| Hypomyelinaton | − | − | − | − | − | n.d. | − | − | − | n.d. | − | n.d. | − | − | n.d. | − | − | − | − | − | − | − | n.d. | n.d. | n.d. | n.d. | − | − | n.d. |
AR = autosomal recessive; BEL = Belgium; CAU = Caucasian; F = female; gen = generalized; GER = Germany; IL = Israel; LL = lower limb; M = male; MEP = motor evoked potential; n.d. = not done; N/E = not evaluated; RLS = restless legs syndrome; S = sporadic; SCP = superior cerebellar peduncle; SEP = cortical latencies of somatosensory evoked potentials; TCC = thin corpus callosum; TR = Turkey; UL = upper limb; VEP = cortical latencies of visual evoked potentials. + = clinical sign is present; − = clinical sign is absent; ↑ = increase of clinical sign; ↓ = decrease of clinical sign; ↔ = normal/unchanged findings.
aChild from incestuous relationship between brother and sister.
bStart at ∼2 months of age.
cDeath at age 24 years.
dIn years after disease onset.
eNever able to walk independently.
fSevere cognitive decline at the age of 39 years.
gAtrophy of brainstem and pons.
Linkage analysis in Family F1
Samples were genotyped using the Illumina HumanOmniExpressExome-8v1-2 array following the manufacturer’s instructions (Illumina) at the Department of Genomics, Life and Brain Center, University of Bonn, Germany. Samples were processed using the Illumina’s proprietary GenomeStudio software in version V2011 and the genotyping module in version 1.9.4 (Illumina). Samples were annotated and clustered using the latest manifest and cluster files provided by Illumina. All samples were checked for genotyping quality (call rate >98%) and gender discrepancies.
The statistical package EasyLinkage Plus v5.08 designed to perform automated linkage analyses using large-scale single nucleotide polymorphism (SNP) data was used to perform all analyses (http://nephrologie.uniklinikum-leipzig.de/nephrologie.site,postext,easylinkage,a_id,797.html) (Hoffmann and Lindner, 2005). PedCheck (v1.1) was used to search for Mendelian genotyping inconsistencies, which were removed from the analyses. Allegro v1.2c software (incorporated in the EasyLinkage Plus v5.08 package) was used to perform fully automated two-point and multipoint linkage analysis (Gudbjartsson et al., 2000). LOD [logarithm (base 10) of odds] scores in Family F1 were obtained using a recessive model of inheritance, with a penetrance of 100% and a disease allele frequency of 0.0001. Allele frequencies of all genotyped SNPs (derived from Caucasian control individuals), map order, and genetic inter-SNP distances were taken from the Illumina website.
Initial whole genome linkage analysis was performed including SNPs with a predefined intermarker distance of 1 cM. In chromosomal regions showing suggestive signals, fine mapping was performed by including additional SNPs with an intermarker distance of 0.1 cM and the results were used to construct haplotypes. Graphical visualization of haplotypes was performed with HaploPainter v 1.043 (Thiele and Nurnberg, 2005).
Whole-exome sequencing
For WES performed in Family F1, the SeqCap EZ Human Exome Library version 2.0 kit (Roche NimbleGen) was used for enrichment of exonic and adjacent intronic sequences comprising a total of 44 Mb of target sequence. Enriched DNA was then sequenced using a HiSeq 2000 Sequencer (Illumina). Mean coverage was 87-fold (±21), and 85% (±5) of the target sequence was covered at least 30-fold. The Varbank pipeline version 2.1 was used for data analysis (Kawalia et al., 2015) and filtering.
For the patients recruited at the University of Newcastle, WES sequencing was performed using Illumina TruSeqTM 62MB (Illumina) and the HiSeq 2000 instrument (Illumina). Data were analysed as described previously in Pyle et al. (2015). For the remaining cases, WES sequencing was performed at the University of Miami using the Agilent SureSelect Human All Exon 50MB capture (Agilent) and the HiSeq2000 and HiSeq2500 instruments (Illumina). Data were analysed on the Genesis/GEM.app platform (Gonzalez et al., 2015).
Panel sequencing
Genes associated with hereditary spastic paraplegia (n = 98) or cerebellar ataxia (n > 120) were enriched using Agilent in-solution (Agilent) or Haloplex technology (Agilent) and then sequenced on an Illumina MiSeq device. Genes included in the panel approach are provided on request.
Sanger sequencing
All reported POLR3A variants were confirmed by Sanger sequencing and the allelic phase and segregation were analysed by sequencing additional family members whenever possible. Mutation numbering was assigned using the cDNA sequence of the GenBank entry NM_007055.3.
Quantitative real-time PCR
To analyse the effect of the POLR3A mutation on mRNA expression, blood samples were collected into PAXgene™ Blood RNA System tubes (PreAnalytiX, Qiagen) for total mRNA isolation from leucocytes using PAXgene™ reagents according to the manufacturer’s protocol. For fibroblasts, total RNA was prepared by using the RNeasy® protect kit (Qiagen) following the manufacturer’s instructions. RNA concentration and purity was determined using the NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific). Total RNA (500 ng) was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. Gene expression using the cDNA template was quantified by real-time PCR using cDNA specific TaqMan® Gene Expression Assays on a StepOnePlus™ Real-Time PCR System (Applied Biosystems) or alternatively on the Real-Time PCR System on a LightCycler 480 device (Roche Applied Science). For specific quantification of the aberrant splice variant produced by c.1909+22G>A, specific primers were designed directed to the retained intronic sequence. Power SYBR® Green PCR Master Mix was used to quantify the transcript on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). A melting curve was generated for each SYBR® assay to check for specificity of the designed primers. Primer sequences are available upon request.
All PCR experiments were performed with three technical replicates from three independent cDNA preparations. For TaqMan® assays, the endogenous expression levels of POLR3A were quantified using the Hs01086939_m1 (exon boundery 8-9) TaqMan® Gene Expression Assay (Thermo Fisher Scientific). The expression levels of POLR3A were normalized using two reference genes, YWHAZ (Hs03044281_g1, exon boundary 4–5, Hs01122445_g1, exon boundary 3-4, Thermo Fisher Scientific) and SDHA (Hs00417200_m1, exon boundary 4–5, Thermo Fisher Scientific). Both reference genes showed the most stable expression across samples as determined with the geNorm stability M-value (Hellemans et al., 2007). For the quantification analysis, raw Ct (cycle threshold) values for POLR3A, SDHA and YWHAZ were imported into the qbase+ software (Biogazelle). The qbase+ algorithm enables normalization to more than one reference gene and also takes gene amplification efficiencies into account, as well as the errors on all measured parameters along the entire calculation track (Hellemans et al., 2007). The effect of c.1771-7C>G on POLR3A levels was quantified using a LightCycler 480 device. The LightCycler assay for POLR3A was quantified in relation to three reference genes: RNF10, RNF111 and RPLP0. For quantification, the advanced relative quantification module of the LightCycler software was used. The linearity and efficiency of each assay was tested over three 2-fold serial dilutions of input cDNA. All data were expressed as calibrated normalized relative quantities.
Protein quantification
Leucocytes from EDTA blood were isolated by Biocoll (Biochrom) gradient centrifugation for Patients F1‐3, F1‐5 to F1‐8, F2-1 and F3-1. Protein extracts were prepared by resuspending the cells in lysis buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM MgCl2, 0.5% (v/v) Triton™ X-100, 0.1% (v/v) Benzonase (Millipore), 1 × protein inhibitor mix Complete Mini (Roche Diagnostics)]. After centrifugation, 50 µg of protein per lane was separated by SDS-PAGE, and detection was performed with the ECL western blotting detection system (GE Healthcare) using secondary antibodies conjugated with horseradish peroxidase. Antibodies were used at the following dilutions: anti-POLR3A (D5Y2D; 1:1000; #12825, Cell Signaling) and anti-rabbit immunoglobulin G (1:2000; #7074, Cell Signaling).
Cell culture, induced pluripotent stem cell generation and neural differentiation
After giving informed consent, skin biopsies from two affected adult members of Family F1 (Patients F1‐3 and F1‐7), were taken to establish fibroblast lines. These fibroblast lines were cultured in Dulbecco’s modified eagle medium (DMEM) (Thermo Fisher Scientific) and supplemented with 10% (v/v) foetal calf serum (FCS) (Thermo Fisher Scientific) and 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific). All cells were maintained at 37°C under saturated humidity atmosphere containing 5% CO2.
For induced pluripotent stem cell (iPSC) generation, 80000 fibroblasts were plated in one well of a 24-well plate in fibroblast medium containing 10% (v/v) FCS. The next day, cells were infected twice for 30 min at 1500 rcf (relative centrifugal force) and 32°C with 2% (v/v) of a CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific) in a total volume of 250 µl Advanced DMEM containing 5% (v/v) FCS. The cells were incubated for another 24 h. For the following 5 days the medium was changed daily with Advanced DMEM containing 5% (v/v) FCS. At Day 6 cultures were passaged with trypsin and plated onto one 10 cm dish coated with Geltrex® (Thermo Fisher Scientific). The next day, medium was changed to E7 medium prepared in-house (Chen et al., 2011). E7 medium was changed every day and switched to E8 medium around Day 18. Primary iPSC colonies were picked around Day 20 and expanded in E8 medium on Geltrex® as described below.
Human iPSC lines were cultured on plates coated with Geltrex® 1:100 dilution in KnockOut DMEM (Thermo Fisher Scientific) in self-made E8 medium (Chen et al., 2011). Human iPSC lines were passaged twice a week using an EDTA dissociation solution (0.5 M EDTA and 1.8 mg/ml NaCl in phosphate buffered saline; pH 8.0) according to Beers et al. (2012). For neural differentiation, human iPSCs were singularized by Accutase® treatment and 2.5 × 106 cells were seeded in one well of a six-well plate coated with Matrigel®, 1:30 dilution (BD Biosciences) in E8 medium. The following day cells were able to form a confluent layer and the medium was then switched to neural induction medium [50% (v/v) DMEM/F12, 50% (v/v) Neurobasal® medium, 1:200 N2 supplement, 1:100 B27 (Thermo Fisher Scientific), 15 µM SB431542 and 500 nM LDN-193189 (Axon)]. Beginning on Day 6 the medium was additionally supplemented with 10 ng/ml fibroblast growth factor-2 (R&D systems). The medium was changed daily until Day 10.
Nonsense-mediated mRNA decay
Primary fibroblast cell lines derived from Patients F1‐3 and F1‐7 were cultured in 10 cm dishes (Sarstedt) up to a confluence of 70–80%. Fibroblast cultures were incubated with cycloheximide (100 µg/ml, Sigma), a known nonsense-mediated mRNA decay inhibitor, for 4 h. DMSO (Sigma) was used a negative control. Cells were then washed twice in PBS buffer and harvested. Total RNA was prepared using the RNeasy® protect kit (Qiagen) according to the manufacturer’s instructions. RNA concentration and purity was determined using the NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific). Total RNA (500 ng) was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions.
Sequence alignment and protein modelling
Sequence retrieval of the entire protein family was performed using the program PsiBlast (Altschul et al., 1997) on the UNIPROT database (http://www.uniprot.org/). The family of curated sequences was then used to build a multiple sequence alignment using the PROMALS program (Pei et al., 2007). The alignment was analysed with the HHPred program (Soding et al., 2005) for template search. A multi-template approach was used to build the model. To investigate the putative functional consequences of the substitutions a three-step strategy was followed: (i) the structure of the largest subunit of human RNA polymerase III (RPC1) was modelled using homology modelling techniques. The template chosen was RNA polymerase II from Schizosaccharomyces pombe (PDB accession code: 3H0G), which shares 38% sequence identity with RPC1; (ii) the mutants were introduced and the putative gain/loss of interactions were analysed; and (iii) interaction partners of RPC1 were modelled, including RPAB5, RPB2 (Martinez-Calvillo et al., 2007).
Association study of the variant rs191875469 within the POLR3A gene
To test association between rs191875469 (c.1909+22G>A) and specific phenotypic features, SNP genotype data were retrieved from three different cohorts: (i) cases with features of the reported POLR3A disease spectrum (ataxia, spastic paraplegia, peripheral neuropathy; ‘POLR3A-spectrum cohort’; n = 1139) (Gonzalez et al., 2015; Pyle et al., 2015); (ii) a cohort of cases with neurological disorders and features not related to the POLR3A-spectrum (e.g. Parkinson’s disease, Alzheimer’s disease, cardiomyopathy, retinopathy, deafness and others; ‘disease controls’, n = 1448) (Gonzalez et al., 2015); and (iii) a control cohort of healthy elderly subjects (‘healthy controls’, n = 853) (Supplementary Table 2). Cohort 1 was selected from WES and whole-genome sequencing datasets available in the GEM.app/Genesis and in the University of Newcastle databases (Gonzalez et al., 2015; Pyle et al., 2015).
The POLR3A-spectrum cohort included 1139 mostly Caucasian index cases of US and European ancestry with a diagnosis of hereditary ataxia (n = 213), hereditary spastic paraplegia (HSP; n = 482) or Charcot-Marie-Tooth disease (n = 444). WES was performed for 975 index cases at the University of Miami and at the University of Newcastle using the protocol described above. The remaining cases were analysed using disease-specific next generation sequencing-based gene panels targeting HSP (n = 114) or ataxia relevant genes (n = 50), respectively.
The disease controls group (n = 1448) were extracted from the Genesis/GEM.app database by selecting index cases with disease features not related to the clinical spectrum published for POLR3A or observed in our study. Only genotypes with coverage of at least 20 reads were included in the analysis.
German controls (n = 853) were obtained from the longitudinal cohort AgeCoDe (Jessen et al., 2011; Luck et al., 2012). The AgeCoDe cohort is a German multicentre prospective cohort study. The samples were randomly selected from general practice registries in six German cities, and the selected individuals were contacted by mail. The main inclusion criteria were age 75 years or above and the absence of dementia according to the clinical judgement of the respective general practitioner. A total of 3327 AgeCoDe participants were recruited between January 2002 and November 2003. Each participant was assessed at baseline and at 18 month intervals thereafter. All assessments are performed at the participant’s home by a trained study psychologist or physician. For the current study, healthy elderly individuals from the AgeCoDe cohort, in whom cognitive impairment had been excluded at the last follow-up visit, were genotyped. To genotype rs191875469 in the AgeCoDe sample, a custom TaqMan® SNP genotyping assay was designed using the available Applied Biosystems online assay design tool. Oligonucleotide primers were ordered from Applied Biosystems (Thermo Fisher Scientific) and performed according to the manufacturer’s instructions in a StepOnePlus™ Real-Time PCR Systems (ThermoFisher Scientific).
Statistical analysis
Association between case-control status and variant carrier/non-carrier status was calculated using Fisher’s exact test for 2 × 2 contingency tables. The statistical analyses were performed using the implementation in R (The R Project for Statistical Computing. www.r-project.org). The alpha-level for statistical significance was set at ≤0.05. The genotyping data for controls were inspected to detect deviations from Hardy-Weinberg equilibrium. Descriptive analysis for censored data (walking aid dependency, wheelchair dependency) was done using the Kaplan Meier Method (Jmp v11 for Mac).
Results
Pathogenic variants in POLR3A cause familial recessive spastic ataxia
In a large non-consanguineous German family (Family F1) with apparently recessive spastic ataxia (Fig. 1A and Table 1) we performed linkage analysis (Supplementary Table 3) followed by WES in two affected (Patients F1‐3 and F1‐8) and one unaffected family member (Patient F1‐6). Three genes with compound heterozygous rare variants co-localized within suggestive linkage regions: HMCN1, POLR3A, ADCY4 (Supplementary Table 4). However, the variants in HMCN1 and ADCY4 were intronic and predicted in silico to have either no or only marginal effect on splicing. The POLR3A gene carried a rare heterozygous nonsense mutation in exon 2 (c.91C>T, p.Q31*). The second variant in POLR3A was located 22 base pairs downstream of the donor site of intron 14 and predicted in silico to activate a cryptic splice site (c.1909+22G>A, rs191875469, www.fruitfly.org) (Supplementary Fig. 1A). Sanger sequencing confirmed the compound-heterozygosity of both POLR3A variants and segregation of the variants with the phenotype in Family F1 (Fig. 1A and B). Amplification of cDNA from whole blood (PAXgene™) using forward and reverse primers located in exons 13 and 15, respectively, revealed an aberrant larger (298 bp) fragment in all carriers of c.1909+22G>A (Fig. 1C). Specific amplification (Supplementary Fig. 2) and sequencing of this aberrant band demonstrated the inclusion of 19 nucleotides of intron 14 sequence leading to a frameshift and a premature stop codon following 13 extraneous amino acids (p.Y637Cfs*14, Fig. 1D). POLR3A encodes the largest subunit of human RNA polymerase III (RPC1) (Dumay-Odelot et al., 2010) and previous studies have linked the gene to the phenotypic spectrum of hypomyelinating leukodystrophies (Bernard et al., 2011; Saitsu et al., 2011; Daoud et al., 2013).
Pathogenic variants in POLR3A are a frequent cause of recessive and sporadic spastic ataxia
The findings in Family F1 prompted us to search for additional rare variants in POLR3A in spastic ataxia related phenotypes. A total of 618 sporadic or recessive index cases with hereditary spastic paraplegia (n = 355) or cerebellar ataxia (n = 263), were screened with either next generation or Sanger sequencing to identify rare homozygous or compound heterozygous POLR3A variants. In 3.1% of the index cases mutations were identified (n = 19, Patients F2–F20) (Tables 1 and 2). Surprisingly, all but one of the patients (Patient F20) carried the intronic variant c.1909+22G>A on at least one allele (Table 1). This observation led us to hypothesize that c.1909+22G>A could be a founder mutation. However, patients carrying the 1909+22G>A variant do not share a common haplotype at the POLR3A locus (Supplementary Fig. 3). Moreover, this intronic variant is present in nearly all populations included in the ExAC browser at similar allele frequencies, including the European [minor allele frequency (MAF) 0.0019], South Asian (MAF 0.0008), Latino (MAF 0.0009) and African (MAF 0.0004) population. A common founder for the c.1909+22G>A variant is thus unlikely.
Table 2.
POLR3A variants identified in this screening
| Family | cDNA | Exon/intron position | Protein effect | EVS6500 alleles | ExAC alleles | PhastCons | GERP | CADD score v1.3 | Patient | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| F1 | c.91C>T | Exon 2 | p.Q31* | Unknown | A = 1/C=2/G = 121251 | 1.0 | 5.1 | 36 | F1‐3 | |
| F14 | c.325T>A | Exon 4 | p.C109S | Unknown | Unknown | 1.0 | 5.67 | 26 | F14-1 | |
| F15 | c.1048+5G>T | Intron 7 | p.G350Gfs*27 | Unknown | Unknown | 0.104 | 2.98 | 17.86 | F15-1 | |
| F18 | c.1067T>C | Exon 8 | p.L356P | Unknown | Unknown | 1.0 | 5.58 | 31 | F18-1 | |
| F2 | c.1114G>A | Exon 8 | p.D372N | T = 1/C = 13005 | T = 1/C = 121187 | 1.0 | 5.58 | 34 | F2-1 | rs267608673, (Bernard et al., 2011) |
| F3 | c.1290-2A>G | Intron 9 | p.F431Sfs*26 | Unknown | Unknown | 0.875 | 5.90 | 25.2 | F3-1 | |
| F5 | c.1360C>T | Exon 10 | p.L454F | Unknown | Unknown | 1.0 | 5.90 | 27.5 | F5-1 | |
| F11 | c.1531C>T | Exon 11 | p.Q511* | Unknown | Unknown | 1.0 | 5.46 | 40 | F11‐1 | |
| F8 | c.1544C>T | Exon 11 | p.A515V | Unknown | Unknown | 1.0 | 5.69 | 33 | F8-1 | |
| F21, F22, and F23 | c.1771‐7C>G | Intron 13 | p.G548_Y637del / p.P591Mfs*9 | Unknown | C = 18/G = 121394 | 0.071 | 2.87 | 8.949 | F21‐3 - F23-3 | |
| F1 to F19 | c.1909+22G>A | Intron 14 | p.Y637Cfs*14 | T = 36/C = 12970 | T = 161/C = 121249 | 0.007 | 3.81 | 7.377 | F1‐3 - F19-1 | rs191875469 |
| F13 | c.2247+2T>G | Intron 16 | p.K713Kfs*3 | Unknown | Unknown | 1.0 | 5.95 | 25.2 | F13-1 | |
| F6 | c.2472_2472delC | Exon 18 | p.S825Qfs*18 | Unknown | Unknown | 1.0 | 1.93 | 34 | F6-1 | |
| F17 | c.2554_2554delA | Exon 19 | p.M852Wfs*7 | Unknown | Unknown | 1.0 | 6.17 | 35 | F17-1 | |
| F9 | c.2561_2561delG | Exon 19 | p.G854Afs*5 | Unknown | Unknown | 0.998 | 6.17 | 36 | F9-1 | |
| F10 | c.2617-1G>A | Intron 19 | Splice | Unknown | T = 2/C = 120060 | 1.0 | 5.87 | 28.1 | F10-1 | RCV000024142 (Bernard et al., 2011), rs181087667 |
| F16 | c.2617C>T | Exon 20 | p.R873* | A=1/G = 13005 | A = 4/G = 120128 | 1.0 | 3.42 | 39 | F16-1 | |
| F7 | c.2710G>T | Exon 20 | p.G904* | Unknown | Unknown | 1.0 | 5.87 | 44 | F7-1 | |
| F20 | c.3098T>C | Exon 24 | p.V1033A | Unknown | Unknown | 0.762 | 5.95 | 27.9 | F20-1 | |
| F4 | c.3781G>A | Exon 29 | p.E1261K | T = 1/C = 13005 | T = 1/C = 121399 | 0.996 | 5.37 | 35 | F4-1 | (Daoud et al., 2013), rs371703979 |
| F12 | c.3944_3945delTG | Exon 30 | p.V1315Afs*7 | Unknown | Unknown | 1.0 | 5.84 | 36 | F12-1 |
All cDNA positions refer to transcript NM_007055; CADD scores are PHRED-scaled.
The alternatively spliced larger transcript observed in Family F1 could be amplified in all additional carriers of c.1909+22G>A for whom cDNA from whole blood (PAXgene™) could be obtained (Fig. 1C and Supplementary Fig. 4). However, the aberrant transcript appeared to be of low abundance, suggesting either incomplete activation of the cryptic splice site or nonsense-mediated mRNA decay of the aberrant splice variant. To answer this question, we inhibited nonsense-mediated mRNA decay with cycloheximide in fibroblast cultures of two affected members of Family F1. Inhibition of nonsense-mediated mRNA decay led to almost a 5-fold increase of the amount of aberrantly spliced transcript compared to untreated cells (Fig. 2A and Supplementary Fig. 5A), demonstrating that the aberrantly spliced transcript is subjected to nonsense-mediated mRNA decay. Along these lines, nonsense-mediated mRNA decay inhibition also allowed detection of the aberrant transcript alongside the wild-type transcript by Sanger sequencing (Supplementary Fig. 5B). In contrast, c.91C>T is observed at equal amounts as the wild-type suggesting that this allele escapes nonsense-mediated mRNA decay (Supplementary Fig. 5C). The escape of nonsense-mediated mRNA decay has been previously described in nonsense-mediated mRNA decay-resistant beta-globin transcripts carrying truncating mutation close to the amino-terminal (Neu-Yilik et al., 2011). The molecular mechanism is still poorly understood but it involves the use of an alternative methionine localized downstream of the stop codon introduced by the nonsense mutations.
Figure 2.
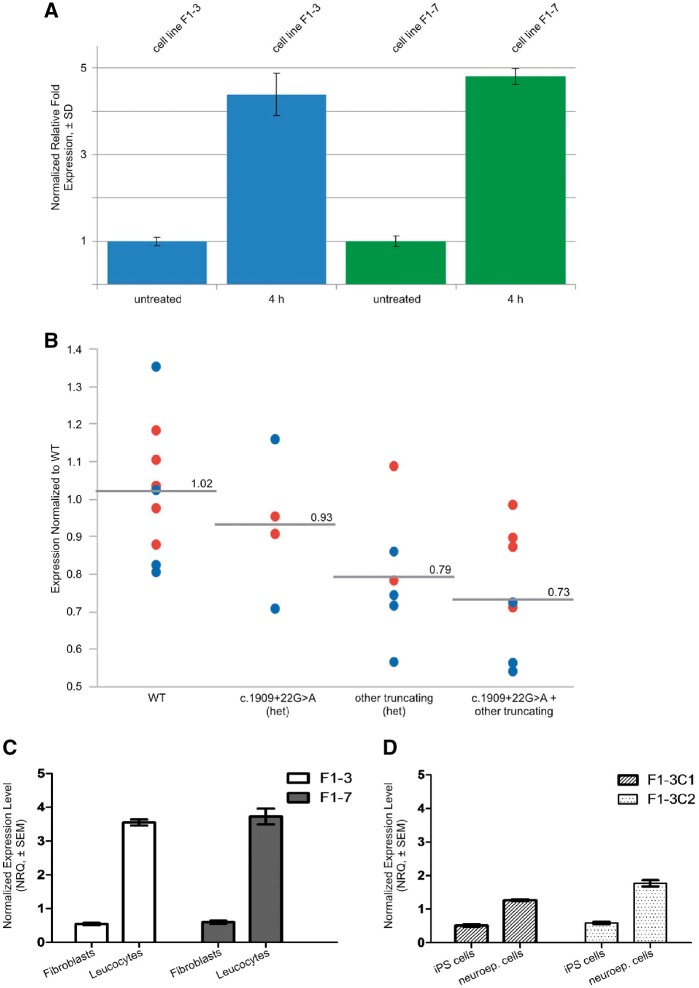
Expression analysis of POLR3A. (A) Specific quantitative-PCR amplification of aberrant splice variant in fibroblast cells from two affected members of Family F1. Expression of POLR3A is normalized with YWHAZ and SDHA as reference genes. Expression levels are shown as relative fold expression levels of cells treated with cycloheximide (100 ng/ml) for 4 h compared to untreated cells. Inhibition of nonsense-mediated mRNA decay led to almost a 5-fold increase of the amount of aberrantly spliced transcript compared to untreated cells. (B) Figure demonstrating interindividual variability in POLR3A expression in controls (wild-type, WT), healthy heterozygous carriers of c.1909+22G>A, healthy heterozygous carriers of other truncating mutations, and affected individuals with compound heterozygous POLR3A mutations (i.e. c.1909+22G>A plus an additional truncating POLR3A mutation). Blue circles are Light Cycler (LC) data from Tübingen (normalized to controls that were available in Tübingen), red circles are TaqMan® data from Bonn (normalized to controls available in Tübingen). Group-wise comparison showed the strongest reduction of total POLR3A levels in the group of affected individuals with compound heterozygous POLR3A mutations of ∼39% of that observed in wild-type. (C) Specific expression levels of the aberrant transcript of POLR3A in adult fibroblasts and leukocytes from Patients F1‐3 and F1‐7 shown relative to average expression level. (D) Specific expression levels of the aberrant transcript of POLR3A in different developmental stages. Fibroblasts from Patients F1‐3 and F1‐7 were reprogrammed to establish iPSC lines. Two iPSC clones from Patient F1‐3 (F1‐3C1 and F1‐3C2), were differentiated to neuroepithelial cells (neuroep. cells). The plot shows expression of cDNA template derived from iPSC lines and iPSC-derived neuroepithelial cells. The level of cryptic splice site activation is lowest in the iPSCs reprogrammed from patient fibroblasts.
To explore further the effect of c.1909+22G>A and other truncating variants on POLR3A expression, we quantified the gene expression of POLR3A in cDNA derived from whole blood (PAXgene™). For the analysis, we decided to cluster individuals in four different groups, namely healthy controls, healthy heterozygous carriers of c.1909+22G>A, healthy heterozygous carriers of other truncating mutations, and affected individuals with compound heterozygous POLR3A mutations (i.e. c.1909+22G>A plus an additional variable POLR3A mutation). We first observed that healthy controls displayed considerable variability in POLR3A expression in blood (Fig. 2B). This variability, however, seems to be a true biological variability and not a technical artefact for two reasons. First, we carefully tested our real-time assay for specificity by performing a melting curve analysis prior to quantification. Second, POLR3A expression levels were found to be highly reproducible in repeated experiments using the same blood sample drawn from an individual, but separate cDNA synthesis and amplification. Despite this large variability among healthy controls, we detected reduction of POLR3A expression in both healthy heterozygous carrier groups the c.1909+22G>A group and the carriers of other truncating mutations. Moreover, we observed the strongest reduction of total POLR3A levels in the group of patients with compound heterozygous POLR3A mutations of ∼39% of that observed in wild-type. Importantly, this ‘moderate’ reduction corresponds very well to the reduction of POLR3A protein levels of ∼60–80% shown in Supplementary Fig. 6. A similar level of reduction in POLR3A expression has been reported for patients with homozygous c.1771-6C>G mutation in POLR3A (Azmanov et al., 2016).
Next, to gain additional insights into the splicing activation produced by c.1909+22G>A, we explored expression of the 298 bp band in cDNA from fibroblasts and leucocytes as well as in iPSC and IPSC-derived neuroepithelial cells derived from patients carrying the c.1909+22G>A mutation. This analysis revealed that activation of the cryptic splice site depends on the tissue type (Fig. 2C) and developmental stage (Fig. 2D) with higher activation of the cryptic splice site in more mature differentiated neuroepithelial cells compared to iPSCs.
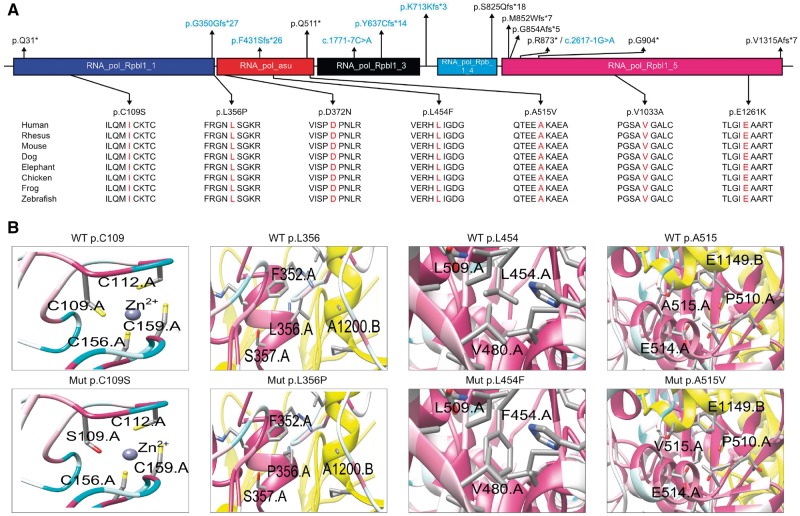
Among the 20 distinct variants we identified in our screening, only three have been previously associated with the POLR3A spectrum of disease (Table 2) (Bernard et al., 2011; Daoud et al., 2013). In 13 of these mutations, including c.1909+22G>A, a protein truncating effect was predicted (Fig. 3A), including four nonsense variants, four deletions leading to a shift of the reading frame and five splice variants (including c.1909+22G>A). For all novel splice variants, analysis of cDNA derived from patient leucocytes revealed the presence of aberrantly spliced transcripts, which were further characterized by Sanger sequencing (Supplementary Fig. 7).
Figure 3.
Schematic representation of RPC1 and 3D structure of amino acid exchanges. (A) Schematic representation of the RPC1 protein. Exonic and intronic mutations that result in missense, nonsense, or frameshift mutations are mapped to the respective domains. Intronic variants that affect splicing are labelled in blue. The amino acid conservation across species was determined for all missense variants from the UCSC Genome Browser (genome.ucsc.edu). (B) Protein 3D representations of POLR3A missense mutations p.C109S, p.L356P, p.L454F and p.A515V. 3D representations of POLR3A missense mutations p.D372N, p.E1261K and p.V1033A are shown in Supplementary Fig. 8A. In the panels, the colour of the main chain of the protein indicates highly conserved (pink) to poorly conserved (white) amino acid residues (based on the entire phylogenetic tree, see ‘Materials and methods’ section). The interacting subunits (chain B: RPB2, chain E: RPABC1 and chain K: RPABC5) are indicated in yellow, green and red, respectively. For a model of the full-length RPC1 and the localization of each of the mutants in relation with the accessory proteins with the same colour code, see Supplementary Fig. 8B.
Among the remaining seven missense variants, five had never previously been associated with disease. Public database searches (EVS6500 and ExAC) revealed that these five missense variants (Table 2) were unreported and affected highly conserved amino acid residues. The PHRED-scaled CADD (combined annotation dependent depletion) scores for these mutations were all above 25, indicating a more deleterious effect than >99% of SNPs annotated in the GRCh37/hg19 reference (Kircher et al., 2014). Moreover, 3D models of RPC1 with these mutations suggest that all these variants may affect either polymerase activity or interaction with other proteins (Fig. 3B, Supplementary Fig. 8A and B, and Supplementary material).
The POLR3A variant c.1909+22G>A is specifically enriched in spastic ataxia-related phenotypes
The c.1909+22G>A variant occurs at low frequency in the general population (MAF EVS6500 0.28%; ExAC 0.13%). To gain further evidence for the phenotype-specific enrichment of this variant we therefore compared the frequency of its occurrence in a cohort of cases with the phenotypic spectrum in the above reported biallelic POLR3A mutation carriers to other disease phenotypes and healthy controls. Indeed, the minor adenine allele of c.1909+22G>A was significantly enriched (P = 1.3 × 10−4, odds ratio = 3.11, 95% confidence interval = 1.69–5.75) in cases with hereditary spastic paraplegia, cerebellar ataxia, and/or neuropathy (n = 1139, MAF = 1.14%) compared to other disease phenotypes (n = 1448, MAF = 0.38%), and to healthy controls (n = 853, MAF = 0.35%) (Supplementary Table 2).
Distinct and homogeneous phenotype in compound heterozygous carriers of c.1909+22G>A
We investigated clinical data from 29 affected POLR3A mutation carriers belonging to 23 different families (additional clinical description is provided in the Supplementary material). Age at onset varied between 0 and 51 with a median of 15 years [interquartile range (IQR) 11.5–20 years]. The median disease duration at examination was 22 years (IQR 13–33.5 years).
Interestingly, we noticed that the phenotype of individuals carrying the c.1909+22G>A variant at heterozygous state in combination with a second POLR3A mutation (Patients F1–F18) was surprisingly homogenous (Tables 1 and 2). In these patients, the median age of onset was 18 years (IQR 12.75–21.5), whereby only five patients manifested after the second decade of life. The progressive disease course led to walking aid dependency in 15/22 patients after a median disease duration of 27 years (95% CI 23–30 years; Kaplan-Meier analysis). Without exception, all 22 affected carriers of a heterozygous c.1909+22G>A mutation (Patient F1‐18) displayed a combination of ataxia and pyramidal involvement. The latter was clinically manifest in all but the mildest affected individual (Patient F1‐3; disease duration 6 years), in whom pyramidal involvement was subclinical and only demonstrated by prolonged central motor latencies in motor evoked potentials (Table 1). Ataxia, mostly a combination of limb and gait ataxia, was manifest in all, with additional saccadic pursuit in 14/22 (64%). Ten of 22 reported dysarthria (45%) and 2/22 dysphagia (9%). Intention tremor of upper (11/22, 50%) or lower (3/22, 14%) limbs was commonly present. Vibration sense was reduced at the lower limbs in almost all cases (not assessed in two patients), some additionally had reduction of surface sensation (12/20, 60%). Other commonly observed features included a sometimes severe action tremor of head and voice (5/22, 23%).
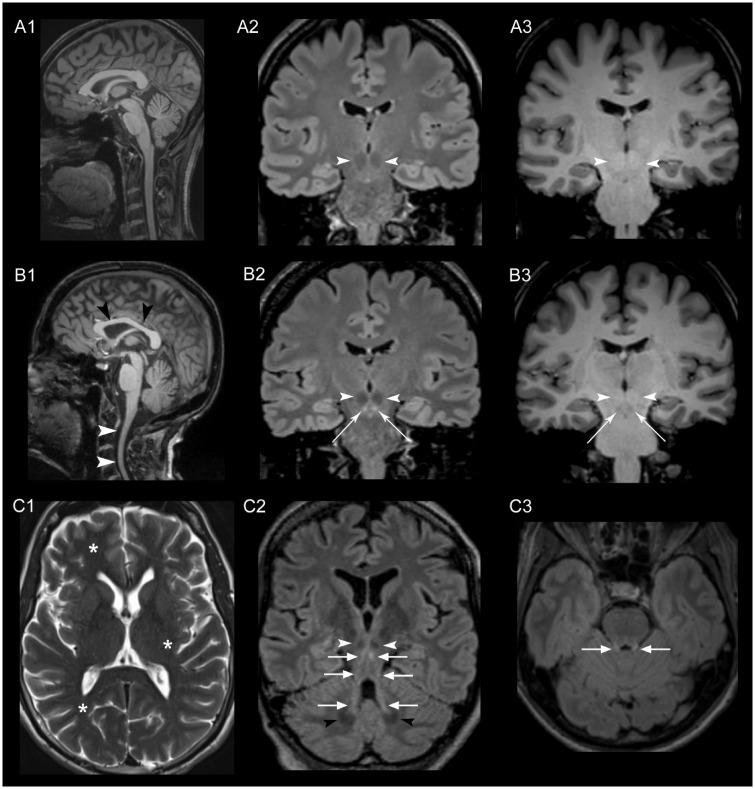
Next, we searched specifically for clinical features within the spectrum of symptoms described for hypomyelinating leukodystrophy due to mutations in POLR3A (Bernard et al., 2011; Saitsu et al., 2011; Daoud et al., 2013; Wolf et al., 2014). Herein, none of the cases spontaneously reported ‘dental abnormalities’. However, when asked specifically, 11 of 17 cases (65%) reported dental problems, including hypodontia and early loss of teeth due to severe and aggressive periodontal disease in young age. There were neither clinically overt signs of gonadal dysfunction nor cognitive abnormalities. The brain imaging demonstrated on 3D FLAIR (fluid attenuated inversion recovery) MRI images a striking bilateral hyperintensity along the superior cerebellar peduncles ranging from the dentate nucleus up to the midbrain just below the red nucleus (11/12). This hyperintensity had a hypointense correlate in the T1-weighted images. In addition, most cases showed cervical cord atrophy and some slight hypoplasia of the corpus callosum (Fig. 4).
Figure 4.
Brain MRI. MRI images of a healthy control (A1–3) and Patients F1‐5 (B1), F13-1 (B2, B3), F1‐5 (C1), F1‐7 (C2) and F5-1 (C3), obtained using a 3 T Scanner (MAGNETOM Skyra, Siemens Healthcare). White arrowheads in B1 indicate the atrophy of the cervical spinal cord and the comparatively hypoplastic corpus callosum (black arrowheads) in a sagittal T1-weighted image compared to a healthy control in A1. In B2, C2 and C3 a standard coronal (B2), standard axial (C3) and a paracoronal view (C2)—the latter angulated along the course of the superior cerebellar peduncle—were reformatted from the 3D FLAIR sequence dataset. Striking bilateral hyperintensity (bright signal) along the entire superior cerebellar peduncles (arrows in B2, C2 and C3) ranging from the cerebellar dentate nucleus (black arrowheads in C2) to the midbrain just below the red nucleus (white arrowheads in B2 and C2) was found. The hyperintensity in the FLAIR images had a hypointense correlate in T1-weighted images (B3) indicating secondary myelin degradation rather than hypomyelination (Schiffmann and van der Knaap, 2009). No signs of generalized hypomyelination in T2 images as described in hypomyelinating leukodystrophy were observed (asterisks in C1, axial T2-weighted image indicating a normal myelination in subcortical white matter and internal capsule exemplarily shown in Patient F1‐5).
The clinical presentation differed considerably in the patient carrying the homozygous c.1909+22G>A mutation (Patient F19-1) and the patient with a classical homozygous POLR3A mutation (p.V1033A, F20-1). In Patient F19-1, childhood-onset axonal neuropathy predominated. Marked gait ataxia was noted, the degree of which seemed disproportionate to the only mild peripheral sensory involvement. In Patient F20-1, the phenotype was characterized by cerebellar ataxia with intention tremor, and electrophysiological signs of pyramidal and somatosensory involvement. Additionally, autism and cognitive decline became evident at the age of 39 years.
Splicing disturbances in POLR3A are a main pathogenic mechanism underlying variant-specific phenotypes
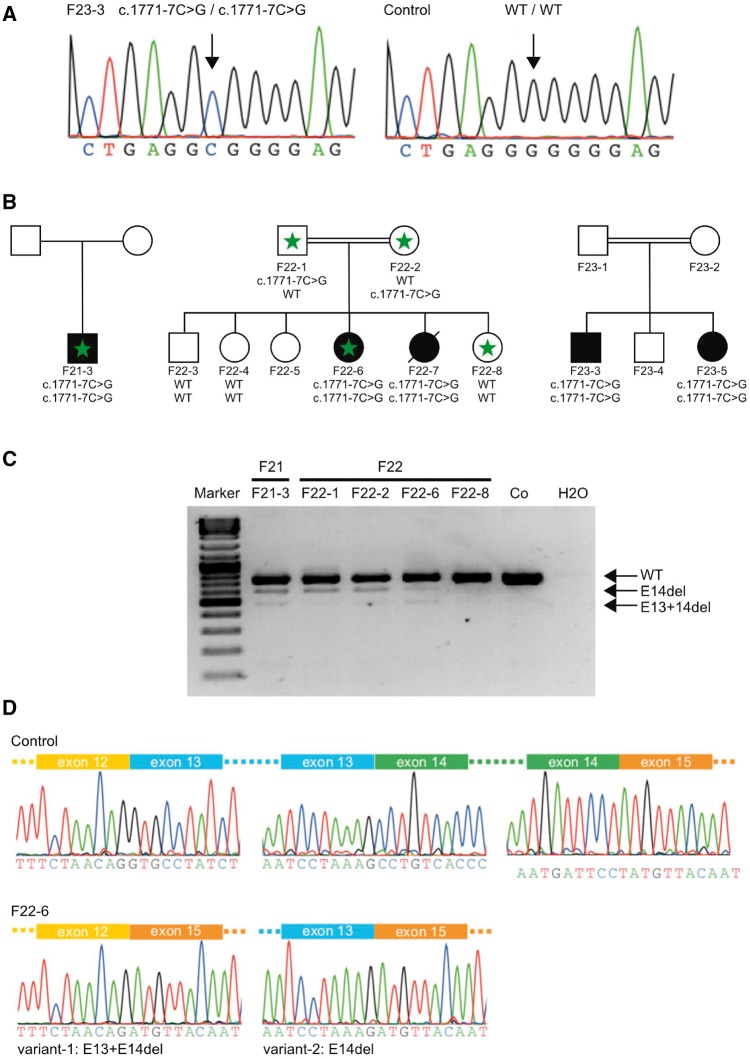
We hypothesized that the remaining levels of wild-type protein function due to the incomplete inactivation of the canonical splice site by the c.1909+22G>A variant might drive the phenotypic shift from hypomyelinating leukodystrophy towards the spastic ataxia phenotype observed here. In support of this hypothesis, the screening of POLR3A in additional exome cohorts with hereditary spastic paraplegia and/or cerebellar ataxia revealed a homozygous intronic variant in POLR3A (c.1771-7C>G, Fig. 5A and B) in three independent families with spastic ataxia complicated by extrapyramidal features. The mutant G allele of c.1771-7C>G was predicted to reduce the splicing score of the native acceptor site of exon 14 (Supplementary Fig. 1). Furthermore, the G allele creates a binding site for the splicing enhancer protein SC35 that overlaps with a native enhancer sequence for SRp40 (Azmanov et al., 2016). This overlap may lead to competition for this binding position, additionally affecting the native acceptor splice site. Indeed, PCR amplification of leucocyte-derived cDNA in two families yielded two additional bands in mutation carriers, which were absent in healthy family members and unrelated controls (Fig. 5C). Direct sequencing of the extra bands revealed two aberrant transcripts lacking either exons 13 and 14 (variant 1) or only exon 14 (variant 2) (Fig. 5D). Hence, both c.1909+22G>A and c.1771-7C>G mutations activate a leaky splice site producing both wild-type and aberrant transcripts.
Figure 5.
Functional consequences and co-segregation of c.1771-7C>G. (A) Sequencing of genomic DNA of the proband Patient F23-3 and a healthy control. The proband carries a homozygous intronic variant (c.1771-7C>G). (B) The same intronic variant was found in three different unrelated families. Segregation of the variant with the disease status was demonstrated for two of the families (Families F22 and F23). Genotypes are depicted below the pedigree symbols. Family members with available leucocyte RNA for analysis are marked by a green asterisk. (C) PCR products obtained from the amplification of patient and control cDNA leucocyte samples showing the effect of the c.1771-7C>G variant on the splicing of exon 14. The sequence traces for controls showing the exon–intron boundaries of exons 12 to 15 are depicted. In the probands carrying the c.1771-7C>G variant (the sequence shown is for proband Patient F22-6) two additional splice variants occur (E13+E14del and E14del). (D) Sequence analysis of proband Patient F22-6. In variant-1 exons 13 (128 bp) and 14 (139 bp) are skipped, leading to a product missing 267 bp. The combined loss of exons 13 and 14 does not lead to a shift of the reading frame. However, a total of 90 amino acids are predicted to be missing from the final protein product (aa 548–637) including a part of the Rpb1_3 domain, which likely represents the pore through which nucleotides enter the active site (Severinov et al., 1996). In variant-2, only exon 14 (139 bp) is skipped, resulting in a shift of the reading frame and the introduction of a pre terminal stop codon nine codons after the mutated codon (p.P591Mfs*9).
Clinically all five affected carriers of the homozygous c.1771-7C>G mutation (Patients F21–F23) presented with early onset cerebellar ataxia, which was the most prominent feature with saccadic pursuit (5/5), dysarthria (5/5), dysphagia (4/5) and gait and limb ataxia (5/5) as main manifestations. Pyramidal involvement was much less pronounced than in carriers of c.1909+22G>A (Table 1). Extrapyramidal involvement (hypokinesia, rigor, dystonia) was the most consistent accompanying feature. Disease onset was earlier (median 2 years) and progression more severe than in the c.1909+22G>A mutation carriers.
Discussion
In this study, we demonstrate that autosomal-recessive mutations in POLR3A are a frequent cause of hereditary spastic ataxias, accounting for ∼3% of hitherto genetically unclassified autosomal recessive and sporadic cases. POLR3A encodes the largest subunit of human RNA polymerase III (RPC1) (Dumay-Odelot et al., 2010) and previous studies have linked the gene to the phenotypic spectrum of hypomyelinating leukodystrophies that comprises clinically overlapping disease entities including tremor-ataxia with central hypomyelination (TACH), ataxia, delayed dentition and hypomyelination (ADDH), hypomyelination, hypodontia and hypognadotropic hypogonadism (4H), leukodystrophy with oligodontia, and hypomyelination with cerebellar atrophy and hypoplasia of the corpus callosum (HCAHC) (Bernard et al., 2011; Saitsu et al., 2011; Potic et al., 2012; Terao et al., 2012; Daoud et al., 2013; Wolf et al., 2014). Hypomyelination has so far been considered the unifying feature of POLR3A-related syndromes and patients without the typical hypomyelination pattern within the cerebellar and cerebral white matter in combination with preserved myelination in the anterolateral thalamus, globus pallidus, optic radiation, corticospinal tracts and dentate nucleus were considered unlikely to carry mutations in POLR3A or the related POLR3B gene (Cayami et al., 2015). This notion has to be revised in view of the high frequency of compound heterozygous POLR3A mutations reported here.
The new spastic ataxia phenotype appears to be specifically caused by the intronic allele c.1909+22G>A, which was present in the vast majority of our POLR3A-positive spastic ataxia cases (80%) and co-segregated perfectly with the phenotype in all tested families including the large Family F1.
From a clinical perspective, the typical discriminating features of POLR3A-associated hypomyelinating leukodystrophies, namely the characteristic MRI features of hypomyelination, cerebellar atrophy and corpus callosum hypoplasia, endocrine dysfunction and hypo-/oligodontia were missing or unusually subtle in our cohort. In contrast, cases carrying the c.1909+22G>A variant presented with a surprisingly uniform phenotype characterized by typically adolescent-onset progressive spastic ataxia with frequent occurrence of tremor and involvement of the central sensory tracts, in most cases leading to the diagnosis of complicated hereditary spastic paraplegia. Dental problems (hypodontia, early onset of severe periodontal disease) were frequent (65%); however, subtle in comparison to classical POLR3A phenotypes and discovered only after the genetic diagnosis was made and the patients were specifically re-evaluated for common POLR3A-associated features. Brain imaging in compound heterozygous patients carrying in one allele c.1909+22G>A differed from published MRI findings in POLR3A cases (Steenweg et al., 2010; Takanashi et al., 2014; Wolf et al., 2014; La Piana et al., 2016). However, similar to the uniform clinical presentation, the MRI phenotype was also rather homogeneous. The most consistent finding on FLAIR images was bilateral hyperintensities along the superior cerebellar peduncles ranging from the dentate nucleus up to the midbrain just below the red nucleus and combined with a hypointense correlate in T1-weighted images—indicating secondary myelin degradation rather than hypomyelination (Schiffmann and van der Knaap, 2009). In fact, the MRI sign of degradation of the superior cerebellar peduncle may represent the structural correlate to the cerebellar symptoms developing in the disease course in our patient group. Importantly, although cerebellar abnormalities (atrophy, T2-hyperintense white matter, T2-hypointense dentate nucleus) have been described in POLR3A/B-related hypomyelinating leukodystrophies and may even differ between POLR3A and POLR3B mutation carriers, the cerebellar MRI pattern we observed in our study has not been described before (Steenweg et al., 2010; Takanashi et al., 2014; Wolf et al., 2014; La Piana et al., 2016). In addition, most of our cases showed cervical cord atrophy and some slight hypoplasia of the corpus callosum. Recently, La Piana et al. (2016) reported a small series of cases without the typical POLR3A hypomyelination pattern. In that cohort, the authors described two recurring MRI patterns: (i) selective T2 hyperintensity of the corticospinal tracts most pronounced at the level of the internal capsule; and (ii) cerebellar atrophy without gross signs of hypomyelination. In our sample, neither of these patterns were identified indicating that our MRI findings may indeed be mutation-specific.
The recurring intronic variant c.1909+22G>A leads to inclusion of parts of intron 14 into the mRNA transcript, which hypothetically translates into a truncated RPC1 protein (p.Y637Cfs*14). Interestingly, a known mutation, which is located just four nucleotides upstream (c.1909+18G>A), activates the same cryptic splice site and has been previously associated with hypomyelinating leukodystrophy (Bernard et al., 2011). However, c.1909+18G>A was not observed in any of the samples analysed, suggesting that the enrichment observed for c.1909+22G>A is specific for a spastic ataxia phenotype. In support of the pathogenic role in these cases, the c.1909+22G>A mutation was present at compound heterozygous state with variants previously described in association with hypomyelinating leukodystrophies and novel variants predicting a truncation of RPC1. However, we also identified novel missense mutations with unclear pathogenic significance. Although ultimately functional data confirming the pathogenic role of these variants is needed, 3D modelling supported their potential involvement in key functions of the protein, including binding of Zn2+ ions and interaction with subunits of the RPC1 interactor protein RPB2 (for details see Supplementary material). Strikingly, regardless of these various ‘secondary’ mutations, the homogeneous presentation of the spastic ataxia phenotype led us to hypothesize that c.1909+22G>A mutation seems to be the main phenotypic determinant in this phenotype. Thus, the ‘leaky’ nature of the splice site change produced by c.1909+22G>A causes an incomplete loss of wild-type allele that in turn may modulate the phenotype away from the classical hypomyelination phenotype towards the milder spastic ataxia phenotype. In line with this hypothesis, homozygous presence of c.1909+22G>A, as observed in Patient F19-1, leads to an even milder ataxia-neuropathy phenotype. Further support for this line of reasoning is added by our identification of another intronic mutation (c.1771-7C>G), present at homozygous state in three unrelated families with ataxia complicated by extrapyramidal features that similarly to c.1909+22G>A activates a leaky splice site. A homozygous mutation affecting the neighbouring nucleotide c.1771-6C>G leading to skipping of exon 14 (but not exon 13) was recently described in a Roma family with a combined extrapyramidal-ataxia phenotype (Azmanov et al., 2016) that is similar to the phenotype we observed in c.1771-7C>G mutation carriers.
RPC1, the gene product of POLR3A, contributes to the catalytic activity of Pol III and is part of the active centre of the polymerase. Pol III is responsible for the transcription of untranslated RNAs (Dieci et al., 2007; Dumay-Odelot et al., 2010; Haurie et al., 2010) and dysregulation of tRNA production has been regarded as potentially pivotal for the phenotype caused by mutations in POLR3A due to an extreme vulnerability of myelin homeostasis to tRNA levels (Dittmar et al., 2006; Scheper et al., 2007; Yee et al., 2007; Feinstein et al., 2010; Boespflug-Tanguy et al., 2011). The regulatory nature of the genes transcribed by Pol III, which goes beyond tRNA production solely, may help to understand the range of phenotypes produced by mutations in POLR3A. Thus, ‘moderate’ reduction in levels of Pol III function may first dysregulate functional modules relevant to maintenance of the long corticospinal tract axons that are primarily affected in HSP. In line with this, Azmanov et al. (2016) found that POLR3A expression in homozygous carriers of c.1771-6C>G is only reduced by 37%, yet still led to dysregulation of several non-coding RNAs. Interestingly, patients carrying c.1771-6C>G in homozygous state presented a phenotype similar to that described in our study. As levels of wild-type Pol III are further reduced due to more severe mutations, additional protein pathways and different cell types (e.g. oligodendrocytes, the glia cells producing myelin in the CNS) may also be affected leading to the more severe classical hypomyelinating leukodystrophy phenotypes. An example of this is the leukodystrophy with oligodontia phenotype caused by homozygous c.1909+18G>A wherein the cryptic splice seems to produce more splice variant than c.1909+22G>A leading to this more severe phenotype (Bernard et al., 2011). In addition, missense mutations may contribute to the more severe leukodystrophy phenotype by interfering with normal assembly of Pol III in affected cell types. This loss of Pol III function, together with its broad tissue expression, may also explain involvement of additional tissues in POLR3A phenotypes, such as gonad and teeth. Nevertheless, it is important to note that several pathogenic mechanisms may contribute to this phenotype beyond the simple loss of POLR3A expression, including missense variants and truncated Pol III protein produced by alleles escaping nonsense-mediated mRNA decay (c.91C>T). Furthermore, POLR3A function seems to be extremely relevant for several cellular functions, and cells consequently react with an increase in POLR3A expression if reduction due to any truncating mutation is detected. The importance of POLR3A function is supported by the observation that no single patient has been identified so far carrying mutations completely abolishing POLR3A function.
We cannot completely rule out the possibility that c.1909+22G>A is in linkage disequilibrium with another ‘truly pathogenic’ variant that causes the phenotype in our families. However, several lines of evidence argue in favour of a direct pathogenic contribution of c.1909+22G>A to POLR3A-related spastic ataxia. This includes co-segregation of c.1909+22G>A with the phenotype in the context of numerous independent haplotypes, specific enrichment of c.1909+22G>A in spastic ataxia phenotypes, its effect on mRNA and protein expression, and the functional analogy to the adjacent known pathogenic c.1909+18G>A mutation.
In conclusion, our data clearly show that mutations in POLR3A are a frequent cause of autosomal recessive spastic ataxia. The phenotypic expression appears to be mutation-specific and seems to depend on the amount of wild-type transcript produced from the mutant alleles. While the heterozygous c.1909+22G>A mutation in combination with a null allele is associated with spastic ataxia combined with tremor, involvement of the central sensory tracts and dental problems, the same variant in the homozygous state causes an ataxia-neuropathy phenotype, and the intronic variant c.1771-7C>G leads to ataxia complicated by extrapyramidal features.
From a more general perspective, our data also highlight the pivotal role of intronic variants in disease aetiology, particularly those variants located beyond the highly conserved canonical splice sequences, not captured by WES. As whole-genome sequencing becomes a routine diagnostic tool and prediction strategies for non-coding variants improve this will allow to reliably identify and characterize a comprehensive spectrum of intronic and regulatory variants thus providing a genetic diagnosis for patients with yet unexplained and often rare diseases.
Supplementary Material
Acknowledgements
We want to thank all participating patients and family members from the different studies and cohorts included in our study, as well as all contributing physicians for their good collaboration. We also thank all participants from the AgeCoDe cohort.
Glossary
Abbreviations
- iPSC
induced pluripotent stem cell
- MAF
minor allele frequency
- Pol III
RNA-polymerase III
- RPC1
RNA polymerase III subunit C1
- SNP
single nucleotide polymorphism
- WES
whole-exome sequencing
Funding
This study was supported by the European Union within the 7th European Community Framework Program for Research and Technological Development through funding for the NEUROMICS network (F5-2012-305121 to L.S.), the E-Rare Networks NEUROLIPID (01GM1408B to R.S.) and PREPARE ([Bundesministerium für Bildung und Forschung (BMBF) 01GM1607 to M.S. and TÜBITAK 315S094 to E.L.]), and a Marie Curie International Outgoing Fellowship (grant PIOF-GA-2012-326681 to R.S. and L.S.). This work was further supported by the US National Institutes of Health (NIH) (grants R01NS075764 and U54NS065712 to S.Z:, grant 5R01NS072248 to R.S. and S.Z.), the Australian National Health and Medical Research Council (grant APP104668 to M.L.K and G.A.N.), the German Research Foundation (DFG) (grant SCHO754/5-2 to L.S.), the BMBF (KND grant 01 GI 0422 and KNDD grant 01GI 0711), the German HSP-Selbsthilfegruppe e.V. (grant to R.S. and L.S.), and the Association Belge contre les Maladies Neuromusculaire (ABMM) - Aide à la Recherche ASBL. T.W.R. receives funding from the University of Tübingen, medical faculty, for the Clinician Scientist Program (grant 386-0-0). J.B. is supported by a Senior Clinical Researcher mandate of the Research Fund - Flanders (FWO).
Supplementary material
Supplementary material is available at Brain online.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmanov DN, Siira SJ, Chamova T, Kaprelyan A, Guergueltcheva V, Shearwood AJ. et al. Transcriptome-wide effects of a POLR3A gene mutation in patients with an unusual phenotype of striatal involvement. Hum Mol Genet 2016; 25: 4302–14. [DOI] [PubMed] [Google Scholar]
- Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA. et al. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc 2012; 7: 2029–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard G, Chouery E, Putorti ML, Tetreault M, Takanohashi A, Carosso G. et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet 2011; 89: 415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boespflug-Tanguy O, Aubourg P, Dorboz I, Begou M, Giraud G, Sarret C. et al. Neurodegenerative disorder related to AIMP1/p43 mutation is not a PMLD. Am J Hum Genet 2011; 88: 392–3; author reply 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayami FK, La Piana R, van Spaendonk RM, Nickel M, Bley A, Guerrero K. et al. POLR3A and POLR3B mutations in unclassified hypomyelination. Neuropediatrics 2015; 46: 221–8. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD. et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods 2011; 8: 424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud H, Tetreault M, Gibson W, Guerrero K, Cohen A, Gburek-Augustat J. et al. Mutations in POLR3A and POLR3B are a major cause of hypomyelinating leukodystrophies with or without dental abnormalities and/or hypogonadotropic hypogonadism. J Med Genet 2013; 50: 194–7. [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet 2007; 23: 614–22. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2006; 2: e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 2010; 9: 3687–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein M, Markus B, Noyman I, Shalev H, Flusser H, Shelef I. et al. Pelizaeus-Merzbacher-like disease caused by AIMP1/p43 homozygous mutation. Am J Hum Genet 2010; 87: 820–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Falk M, Gai X, Schule R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat 2015; 36: 950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000; 25: 12–13. [DOI] [PubMed] [Google Scholar]
- Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M. et al. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci USA 2010; 107: 4176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007; 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus–automated linkage analyses using large-scale SNP data. Bioinformatics 2005; 21: 3565–7. [DOI] [PubMed] [Google Scholar]
- Jessen F, Wiese B, Bickel H, Eifflander-Gorfer S, Fuchs A, Kaduszkiewicz H. et al. Prediction of dementia in primary care patients. PloS One 2011; 6: e16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalia A, Motameny S, Wonczak S, Thiele H, Nieroda L, Jabbari K. et al. Leveraging the power of high performance computing for next generation sequencing data analysis: tricks and twists from a high throughput exome workflow. PloS One 2015; 10: e0126321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Natre Genet 2014; 46: 310–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Piana R, Cayami FK, Tran LT, Guerrero K, van Spaendonk R, Ounap K. et al. Diffuse hypomyelination is not obligate for POLR3-related disorders. Neurology 2016; 86: 1622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck T, Luppa M, Wiese B, Maier W, van den Bussche H, Eisele M. et al. Prediction of incident dementia: impact of impairment in instrumental activities of daily living and mild cognitive impairment-results from the German study on ageing, cognition, and dementia in primary care patients. Am J Geriatr Psychiatry 2012; 20: 943–54. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Saxena A, Green A, Leland A, Myler PJ. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int J Parasitol 2007; 37: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik G, Amthor B, Gehring NH, Bahri S, Paidassi H, Hentze MW. et al. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA 2011; 17: 843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim BH, Tang M, Grishin NV. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res 2007; 35: W649–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potic A, Brais B, Choquet K, Schiffmann R, Bernard G. 4H syndrome with late-onset growth hormone deficiency caused by POLR3A mutations. Arch Neurol 2012; 69: 920–3. [DOI] [PubMed] [Google Scholar]
- Pyle A, Smertenko T, Bargiela D, Griffin H, Duff J, Appleton M. et al. Exome sequencing in undiagnosed inherited and sporadic ataxias. Brain 2015; 138 (Pt 2): 276–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Osaka H, Sasaki M, Takanashi J, Hamada K, Yamashita A. et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet 2011; 89: 644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper GC, van der Klok T, van Andel RJ, van Berkel CG, Sissler M, Smet J. et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet 2007; 39: 534–9. [DOI] [PubMed] [Google Scholar]
- Schiffmann R, van der Knaap MS. Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology 2009; 72: 750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S. et al. Hereditary spastic paraplegia -clinico-genetic lessons from 608 patients. Ann Neurol 2016; 79: 646–58. [DOI] [PubMed] [Google Scholar]
- Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst SA. et al. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the beta and beta’ subunits of Escherichia coli RNA polymerase. J Biol Chem 1996; 271: 27969–74. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005; 33: W244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenweg ME, Vanderver A, Blaser S, Bizzi A, de Koning TJ, Mancini GM. et al. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain 2010; 133: 2971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi J, Osaka H, Saitsu H, Sasaki M, Mori H, Shibayama H. et al. Different patterns of cerebellar abnormality and hypomyelination between POLR3A and POLR3B mutations. Brain Dev 2014; 36: 259–63. [DOI] [PubMed] [Google Scholar]
- Terao Y, Saitsu H, Segawa M, Kondo Y, Sakamoto K, Matsumoto N. et al. Diffuse central hypomyelination presenting as 4H syndrome caused by compound heterozygous mutations in POLR3A encoding the catalytic subunit of polymerase III. J Neurol Sci 2012; 320: 102–5. [DOI] [PubMed] [Google Scholar]
- Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 2005; 21: 1730–2. [DOI] [PubMed] [Google Scholar]
- Wolf NI, Vanderver A, van Spaendonk RM, Schiffmann R, Brais B, Bugiani M. et al. Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology 2014; 83: 1898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee NS, Gong W, Huang Y, Lorent K, Dolan AC, Maraia RJ. et al. Mutation of RNA Pol III subunit rpc2/polr3b Leads to deficiency of subunit Rpc11 and disrupts zebrafish digestive development. PLoS Biol 2007; 5: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.