Abstract
(Quantitative) structure–activity relationship or (Q)SAR predictions of DNA-reactive mutagenicity are important to support both the design of new chemicals and the assessment of impurities, degradants, metabolites, extractables and leachables, as well as existing chemicals. Aromatic N-oxides represent a class of compounds that are often considered alerting for mutagenicity yet the scientific rationale of this structural alert is not clear and has been questioned. Because aromatic N-oxide-containing compounds may be encountered as impurities, degradants and metabolites, it is important to accurately predict mutagenicity of this chemical class. This article analysed a series of publicly available aromatic N-oxide data in search of supporting information. The article also used a previously developed structure–activity relationship (SAR) fingerprint methodology where a series of aromatic N-oxide substructures was generated and matched against public and proprietary databases, including pharmaceutical data. An assessment of the number of mutagenic and non-mutagenic compounds matching each substructure across all sources was used to understand whether the general class or any specific subclasses appear to lead to mutagenicity. This analysis resulted in a downgrade of the general aromatic N-oxide alert. However, it was determined there were enough public and proprietary data to assign the quindioxin and related chemicals as well as benzo[c][1,2,5]oxadiazole 1-oxide subclasses as alerts. The overall results of this analysis were incorporated into Leadscope’s expert-rule-based model to enhance its predictive accuracy.
Introduction
The use of computational or (quantitative) structure–activity relationship [(Q)SAR] methodologies to predict DNA-reactive mutagenicity is increasing due in part to the International Council for Harmonisation (ICH) M7 guideline (Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk) (1). This guideline recommends the use of two complementary (Q)SAR methodologies to predict the results of the reverse bacterial mutagenicity assay [Organisation for Economic Co-operation and Development (OECD) 1997] when no or inadequate experimental mutagenicity data are available for the impurity.
One of these (Q)SAR methodologies is referred to as ‘expert rule-based’ (1,2). This is a computational system that uses a series of structural rules or alerts to predict mutagenicity. When no alerts are present in the test chemical, it is possible to conclude that the compound is non-mutagenic if other factors are satisfied, such as the test chemical being within the model’s applicability domain (3–5) of the alert system, i.e. the region of chemical space where a model makes reliable predictions. Such structural alerts should ideally be based on an understanding of known mechanism(s) of mutagenicity. In addition, it is essential as part of the development of such systems that the alerts are validated based on a sufficient number of relevant chemicals to conclude that the class has an increased likelihood of being mutagenic. As part of this validation process, mutagenic compounds containing the alert may need to be removed from consideration if their mutagenicity can be explained by other structural elements. There should also be a sufficient number of clear examples (defined as at least four clear examples) because anecdotal mutagenic examples may be explained by experimental artefacts or other reasons (6).
The second recommended methodology is referred to as ‘statistical-based’ (1,2). This is based on a mathematical model calculated from a training set containing both mutagenic and non-mutagenic examples. Fragment-based descriptors (e.g. the presence or absence of specific functional groups) are often used to describe the training set examples. Models that use knowledge of DNA-reactive chemical moieties (e.g. structural alerts for mutagenicity from the literature) enhance predictivity and are easier to explain; therefore, it is important to consider such knowledge in the selection of these structural descriptors (6).
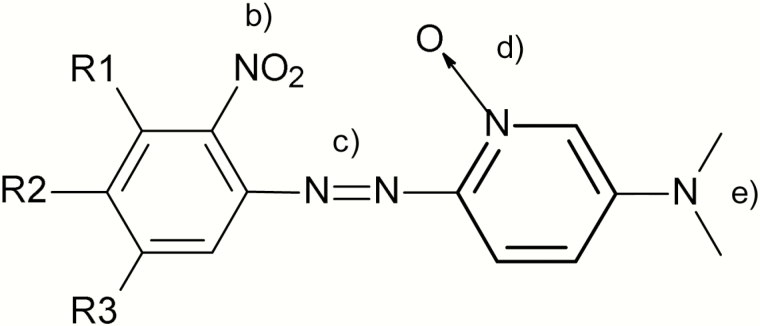
One of the first series of structural alerts for mutagenicity, known as the Ashby Tennant alerts, was proposed in the late 1980s (7,8). These expert-rule-based alerts were identified based on an analysis of mutagenic compounds and an understanding of electrophilic theory (9). Ashby and Tennant proposed 18 structural alerts, which were illustrated as part of a large supermolecule in their original publication. Figure 1 shows a portion of this supermolecule to highlight the inclusion of ‘(d) aromatic ring N-oxides’ as one of the proposed alerts. These alerts are based on the potential for entering into an electrophilic reaction with DNA. However, Ashby and Tennant (7) do not discuss any specific mechanistic basis for this alert nor provide any examples of mutagenic aromatic N-oxide compounds.
Fig. 1.
Portion of the Ashby Tenant supermolecular listing mutagenicity alerts.
Major structures units ... which led to a chemical being classified as structure-activity positive ... (b) aromatic nitro groups; (c) aromatic azo group ...; (d) aromatic ring N-oxides; (e) aromatic mono- and dialkylamino groups ... (emphasis [bold] added)
Snodin and McCrossen (10) reviewed available information supporting the use of aromatic N-oxides as a DNA-reactive mutagenicity alert and concluded that there is a lack of adequate information to support their use as alerts. However, many current expert-rule-based computational systems use the Ashby–Tennant alerts as a starting point in their development. For example, the aromatic N-oxide alert has also been incorporated into the collection of structural alerts described by Benigni et al. (11); however, the article indicates that no clear mechanism for the aromatic ring N-oxide alert has been identified [Plošnik et al. (12) reviews aromatic N-oxides and describes the mechanism as redox cycling (p. 176) and assigns the class under the SnAr alert category (p. 177); however, the references they cite have no discussion on aromatic N-oxides.]. The article also provides N′-nitrosonornicotine-1-N-oxide (CAS 78246-24-9) as an example; however, this is inappropriate because the chemical also contains the well-known DNA reactive alert N-nitroso. Separately, the OECD report (13) states that no definitive mechanism can be identified for the aromatic ring N-oxide alert. In addition, Galloway et al. (14) discusses the link between the aromatic N-oxide from the Ashby & Tennant publications (7,8) to the Derek alert ‘Aromatic N-oxide or N-hydroxy Tautomer’ as part of a comparison of the Ashby alerts to the Derek categories.
An assessment of impurities, metabolites, extractable and leachables showed that many contain aromatic N-oxides (15). For example, aromatic N-oxide-containing compounds are used as reagents (16). Pharmaceutical degradation products and metabolites may also contain aromatic N-oxides (17). In addition, there are examples where aromatic N-oxides are functional groups in active ingredients (18). Furthermore, a retrospective analysis of 1440 ICH M7-compliant (Q)SAR assessments conducted on drug impurities by Food and Drug Administration (FDA)/Center for Drug Evaluation and Research (CDER) found that 24 contained aromatic N-oxides (FDA/CDER, unpublished results). It is therefore important to know whether an aromatic N-oxide or any specific subclass demonstrates a correlation with observed mutagenicity and can serve as an alert for predicting DNA-reactive mutagenicity. As recommended by the ICH M7 guideline, if a compound is predicted to be mutagenic by at least one of the (Q)SAR methodologies, a follow-on reverse bacterial mutagenicity test may be performed or it may be necessary to control the amount of the impurity in the final product to levels defined by the guideline. This additional work can be both time consuming and costly as alternative synthetic routes may need to be explored. In the case of metabolites, difficult to synthesise, or unstable compounds, it may not be feasible to obtain enough material to test. The ICH M7 guideline also states that, if warranted, an expert review of the (Q)SAR results may be conducted. Such a review may refute any positive, out-of-domain or equivocal results with sufficient supporting evidence (19–21). In addition, such expert review may be supported by publications, such as the current publication, that have analysed the evidence for any DNA-reactive structural alerts.
The following study analyses available public and proprietary databases to determine whether the aromatic N-oxide or any of its subclasses are structural alerts for predicting DNA-reactive mutagenicity.
Methods
Analysis of public data
An initial analysis of public data was performed to understand whether aromatic N-oxides or any subclasses are alerts for predicting mutagenicity. This can be broken down into two questions:
(i) Are there any Ames-positive compounds that can be definitely attributed to the presence of a general aromatic N-oxide alert (i.e. there are no other structural alerts most likely responsible for the positive mutagenic experimental call)?
(ii) Are there experimentally Ames-negative examples of aromatic N-oxides?
The reference database from the Leadscope Genetox Expert Alerts version 4 (22), PharmaPendium® (trademark of Reed Elsevier Properties SA, used under license) (23) and PubMed (24) were searched as part of this analysis. The Leadscope reference database includes over 10 600 structured chemicals with detailed bacterial mutagenicity experimental data (6,22). The Leadscope Genetox Expert Alerts version 4 (System: Leadscope Model Applier v2.2.1.1) was used to understand whether any additional alerts for mutagenicity are also present in aromatic N-oxide compounds that may explain their mutagenicity. This system is suitable for this task because there are currently 224 alerts for mutagenicity in the Leadscope Genetox Expert Alerts version 4, derived from the literature and from analysis of public and proprietary databases (6,25) and the alerts have been validated using multiple public and proprietary databases covering over 35 000 chemicals. The alerts are annotated with information on the biological mechanisms and, wherever possible, indicate the bacterial tester strains that are sensitive for each alert (26).
Structure–activity relationship fingerprint analysis of public and proprietary data
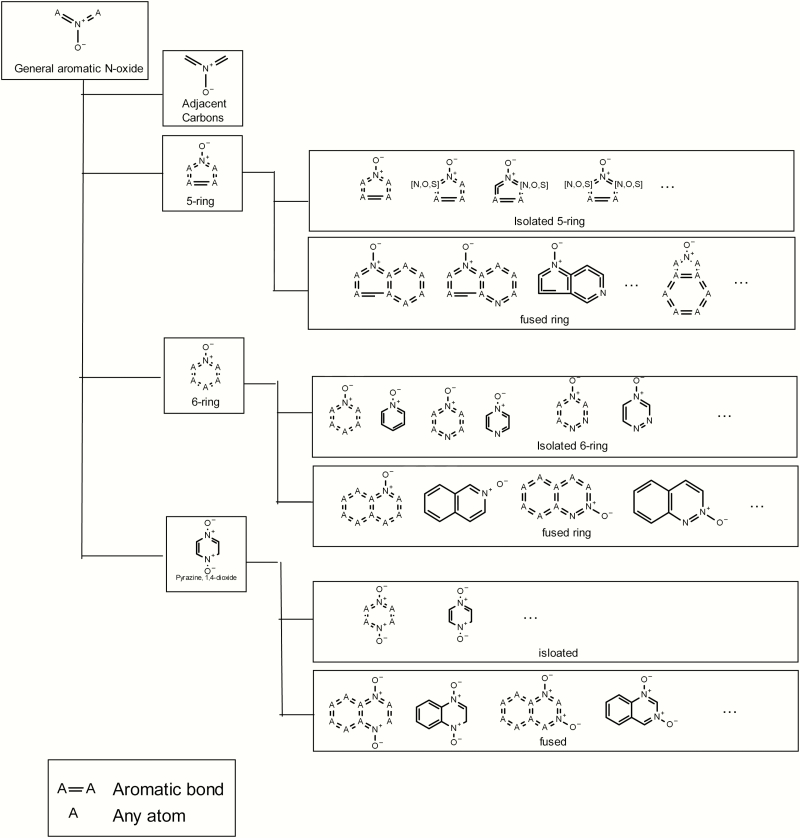
An analysis of public and proprietary data was performed to see if there was any further evidence, particularly from proprietary data collections, to determine whether aromatic N-oxides or any subclasses correlate with observed mutagenicity and hence would serve as alerts for predicting mutagenicity. This analysis used a series of predefined aromatic N-oxide substructure searches, as illustrated in Figure 2. This analysis is referred to as structure–activity relationship (SAR) fingerprints (6). These 101 substructure searches are organised hierarchically and are specifically designed to explore the chemical space of aromatic N-oxide compounds in order to understand any SARs. A general aromatic N-oxide substructure was included to help understand the significance of the general class. A series of isolated 5- and 6-membered rings were included (e.g. pyridinium, 1-oxide-; pyridazinium, 1-oxide-; pyrimidinium, 1-oxide- and pyrazinium, 1-oxide-). These substructures helped to understand the significance of single unfused 5- and 6-membered rings containing an aromatic N-oxide, and if the position of any heteroatoms relative to the N-oxide was significant. A series of fused aromatic substructures containing an N-oxide were included (e.g. quinolinium, 1-oxide-; isoquinolinium, 2-oxide-; quinoxalinium, 1-oxide-; quinazolinium, 3-oxide- and quinazolinium, 1-oxide-). Dioxin-related substructures were also included (e.g. dioxin, quindioxin, quinazolium and quidioxin).
Fig. 2.
Hierarchical organisation of selected substructures incorporated into the fingerprint.
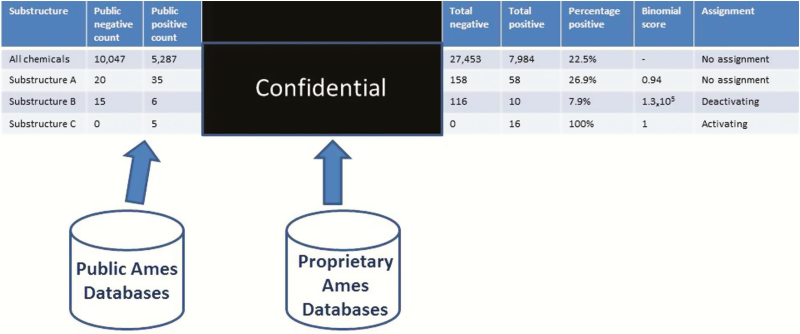
The SAR fingerprint analysis was used to assess the significance of the aromatic N-oxide alert and any subclasses (6). As part of this analysis, all 101 substructures (a subset is illustrated in Figure 2) were matched against a series of public and proprietary databases containing chemical structures with bacterial mutagenicity experimental data. This included the Leadscope Genetox Expert Alerts version 4 reference set, the National Institute of Health Sciences (Tokyo, Japan) databases (27) and a series of proprietary databases maintained by GlaxoSmithKline, Sanofi, Pfizer, Bristol-Myers Squibb and Bayer AG, Pharmaceuticals Division. These tests included both full GLP Ames tests as well as limited strain tests (28). The number of mutagenic (positive) and non-mutagenic (negative) examples from each database matching these substructures was identified using the Leadscope software (29). The number of positive and negative examples across all sources was then summed, and a binomial score (calculated using Microsoft™ Excel) was generated to help understand the statistical significance of the individual chemical classes (6). This analysis uses the percentage of positives across all compounds as a baseline. Subsets containing a significantly greater percentage of positive compounds were designated as activating and those with significantly lower percentage of positive compounds were designated as deactivating. This process is shown conceptually in Figure 3. As part of this exercise, no proprietary information about the chemical structures or the corresponding data were exchanged. Only counts of the number of positive and negative examples per substructure were exchanged.
Fig. 3.
Searching the list of substructures over multiple databases (it should be noted that substructure A, substructure B and substructure C are used to illustrate different predefined substructure queries that are incorporated into the SAR fingerprint).
Results
Analysis of public data
Forty-one relevant mutagenic chemicals were identified from public sources, as shown in Figures 4–12. The chemicals were analysed using the Leadscope Genetox Expert Alerts version 4 and all contained an alert for mutagenicity. Table 1 lists all the data corresponding to these 41 compounds.
Fig. 4.
Mutagenic isolated and fused examples containing both an aromatic N-oxide (highlighted in bold) and one or more structural alerts (highlighted in grey).
Table 1.
Summary of data and matching alerts for the 35 public mutagenic aromatic N-oxide chemicals
| Compound ID | Leadscope ID | Name | Positive tester strains | Reference |
|---|---|---|---|---|
| 1 | LS-131487 | 2,3-Dimethyl-4-nitropyridine 1-oxide | Positive tester strains not reported (conducted according to the OECD TG471 guideline) | (31) |
| 2 | LS-131865 | 4-Nitropyridine N-oxide | TA102 ± S9;TA100 ± S9; TA1538 + S9 | (32–35) |
| 3 | LS-1376 | 4-Nitroquinoline N-oxide | WP2uvra/pKM101 ± S9; TA102 ± S9;TA100 ± S9; TA98 ± S9;TA1535 − s9;TA1537 − s9 | (36,37) |
| 4 | LS-1278 | 4-(Hydroxyamino)quinoline 1-oxide | WP2uvra ± S9; TA100 ± S9;TA1537 ± S9 | (34,35, 37–39) |
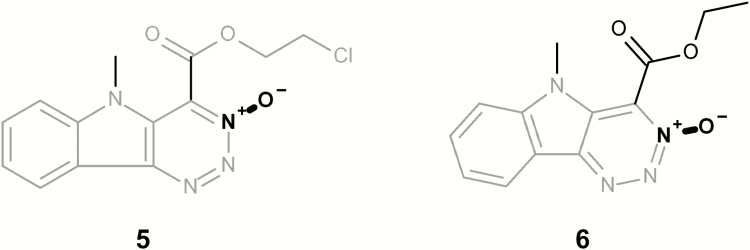
| 5 | LS-189132 | 2-Chloroethyl 1,2,3-triazino[5,4-b]indole-4-carboxylate N(3)-oxide | TA97 ± S9; TA98 ± S9 | (40) |
| 6 | LS-189651 | Ethyl 1,2,3-triazino[5,4-b]indole-4-carboxylate N(3)-oxide | TA102 − S9; TA100 + S9; TA98 ± S9 | (40) |
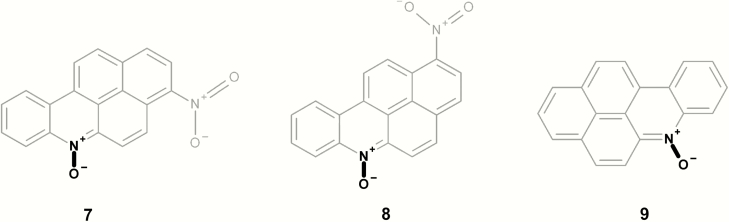
| 7 | LS-94153 | 3-Nitro-6-azabenzo[a]pyrene N-oxide | TA98 − S9 | (41) |
| 8 | LS-94152 | 1-Nitro-6-azabenzo[a]pyrene N-oxide | TA98 − S9 | (41) |
| 9 | LS-212651 | 6-Azabenzo[a]pyrene N-oxide | TA100 − S9; TA98 − S9 | (40) |
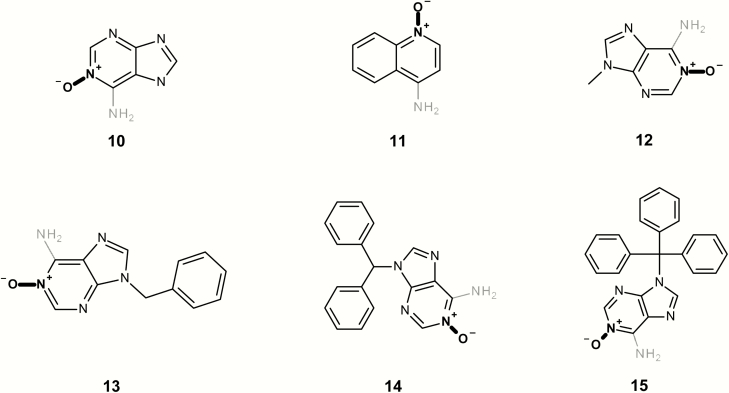
| 10 | LS-15080 | Adenine 1-N-oxide | TA98 + S9 | (42) |
| 11 | LS-141320 | 4-Aminoquinoline 1-oxide | Positive tester strains not reported | (43) |
| 12 | LS-188416 | 9-Methyladenine 1-N-oxide | TA98 + S9 | (42) |
| 13 | LS-188320 | 9-Benzyladenine 1-N-oxide | TA98 + S9 | (42) |
| 14 | LS-189066 | 9-Benzhydryladenine 1-N-oxide | TA98 + S9 | (42) |
| 15 | LS-189065 | 9-Trityladenine 1-N-oxide | TA98 + S9 | (42) |
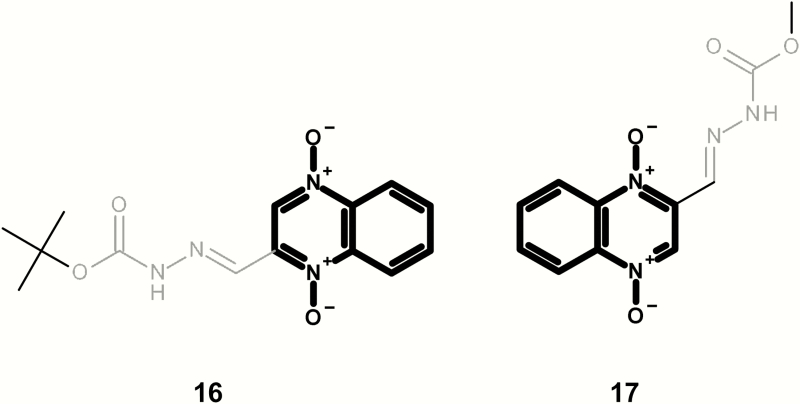
| 16 | LS-76683 | tert-Butyl-3-(2-quinoxalinylmethylene)carbazate 1,4-dioxide | WP2uvra/pKM101 ± S9; TA100 ± S9; TA98 ± S9 | (44) |
| 17 | LS-51665 | Carbadox | WP2uvra/pKM101 ± S9;TA98 ± S9 | (44) |
| 18 | LS-12821 | 1-Acetyl-2-(2-quinoxalinylmethylene)hydrazine 1,4-dioxide | WP2uvra/pKM101 ± S9; TA100 ± S9; TA98 ± S9 | (44) |
| 19 | LS-143005 | 2-Quinoxalinecarboxylic acid 1,4-dioxide | WP2uvra/pKM101 ± S9; TA100 ± S9; TA98 ± S9 | (44) |
| 20 | LS-142996 | Bayonox; N-(2-hydroxyethyl)-3-methyl-4-oxido- 1-oxoquinoxalin-1-ium-2-carboxamide | WP2uvra/pKM101 ± S9; TA100 ± S9; TA98 ± S9 | (44) |
| 21 | LS-110641 | 4-(4-Formylpiperazinyl)-7-nitrobenzofuroxan | Positive tester strains not reported | (37) |
| 22 | LS-35380 | Benzofurazan, 4-nitro-, 1-oxide | Positive tester strains not reported | (37) |
| 23 | LS-35377 | Benzofurazan, 7-(4-methyl-1-piperazinyl)- 4-nitro-, 1-oxide | Positive tester strains not reported | (37) |
| 24 | LS-35368 | 4-(4-(4-Methoxyphenyl)-1-piperazinyl)-7-nitrobenzofurazan 3-oxide | Positive tester strains not reported | (37) |
| 25 | LS-35367 | 4-(4-(2-Methoxyphenyl)-1-piperazinyl)-7-nitrobenzofurazan 3-oxide | Positive tester strains not reported | (37) |
| 26 | LS-35364 | 7-(4-(2-Hydroxyethyl)-1-piperazinyl)-4-nitrobenzofurazan 1-oxide | Positive tester strains not reported | (37) |
| 27 | LS-35355 | Benzofuran, 4-nitro-7-(4-(phenylmethyl)-1-piperazinyl)-, 1-oxide | Positive tester strains not reported | (37) |
| 28 | LS-35386 | Benzofuran, 4-nitro-7-(4-propyl-1-piperazinyl)-, 1-oxide | Positive tester strains not reported | (37) |
| 29 | LS-35383 | 4-Nitro-7-(4-phenyl-1-piperazinyl)benzofurazan 1-oxide | Positive tester strains not reported | (37) |
| 30 | LS-201311 | 7-(4-(Ethoxycarbonyl)piperazin-1-yl)- 4-nitrobenzo[c][1,2,5]oxadiazole 1-oxide | Positive tester strains not reported | (43) |
| 31 | LS-201438 | 7-(Bis(2-hydroxyethyl)amino)-4-nitrobenzo[c] [1,2,5]oxadiazole 1-oxide | Positive tester strains not reported | (43) |
| 32 | LS-35354 | 4-Ethylenimino-7-nitrobenzofuroxan | Positive tester strains not reported | (37) |
| 33 | LS-201105 | Benzo[c][1,2,5]oxadiazole 1-oxide | Positive tester strains not reported | (45) |
| 34 | LS-460268 | (E)-6-(2-(Phenylsulfonyl)vinyl)benzo[c][1,2,5] oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 35 | LS-460269 | (Z)-6-(2-(Phenylsulfonyl)vinyl)benzo[c][1,2,5] oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 36 | LS-460270 | 6-((2-(5-Methyl-1H-imidazole-4-carbonyl) hydrazineylidene)methyl)benzo[c][1,2,5] oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 37 | LS-417996 | (Z)-6-(2-(Benzo[d][1,3]dioxol-5-yl)vinyl) benzo[c][1,2,5]oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 38 | LS-460271 | (E)-6-(2-(Phenylsulfinyl)vinyl)benzo[c][1,2,5] oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 39 | LS-460272 | 6-((2-Nicotinoylhydrazineylidene)methyl) benzo[c][1,2,5]oxadiazole 1-oxide | TA98 ± S9 | (46) |
| 40 | LS-189357 | Pyrido[3,4-d]pyridazine 3-oxide | TA98 ± S9 | (47) |
| 41 | LS-189356 | Pyrido[3,4-d]pyridazine 6-oxide | TA100 + S9; TA98 ± S9 | (47) |
Figure 5 shows two mutagenic aromatic N-oxide chemicals; however, both contain at least one additional structural alert. Compounds 5 and 6 contain a dibenzofuran alert and compound 5 also contains a strong activating alkyl methyl halide. Similarly, Figure 6 presents three mutagenic aromatic N-oxide chemicals; however, they all contain a polycyclic aromatic system and compounds 7 and 8 also contain an aromatic nitro.
Fig. 5.
Mutagenic examples containing both an aromatic N-oxide (highlighted in bold) and one or more structural alerts (highlighted in grey).
Fig. 6.
Mutagenic examples containing both an aromatic N-oxide (highlighted in bold) and a polycyclic aromatic system (highlighted in grey).
Figure 7 contains a series of mutagenic N-oxides; however, all contain a primary aromatic amine that is alerting in the Leadscope expert alerts. The Leadscope alerts for primary aromatic amines take into consideration many factors that may activate and deactivate aromatic amine mutagenicity, including the position of heteroatoms and the position of the amine proximate to any fused ring (6).
Fig. 7.
Mutagenic examples containing both aromatic N-oxides (highlighted in bold) and an alerting primary aromatic amine (highlighted in grey).
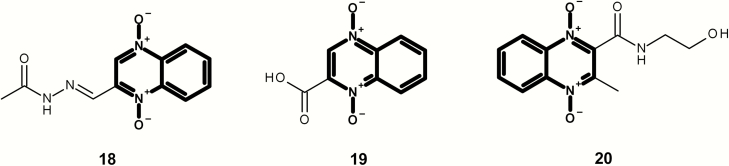
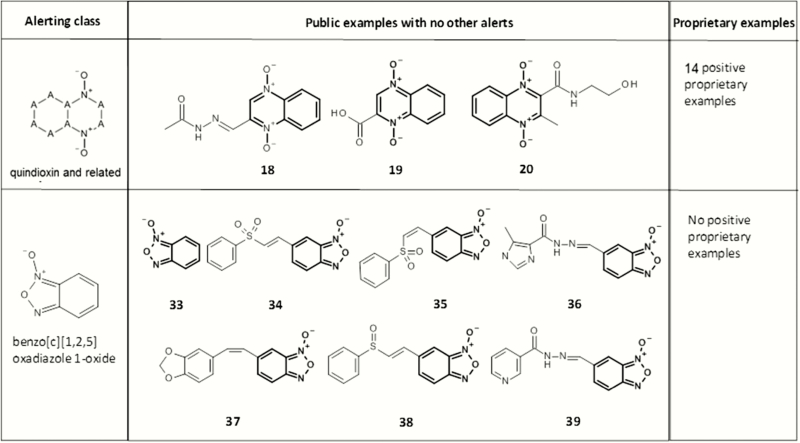
Figures 8 and 9 present a series of mutagenic quindioxins [this class is often referred to as quindioxins; however, the article will refer to it as quindioxin to be consistent with its citation in Enoch and Cronin (30)]. It should be noted that quindioxin is an alert within the Leadscope expert alerts v4; however, the examples shown in Figure 8 contain an additional alert (aryl methylene semicarbazones). Figure 9 shows three additional examples with no additional structural alert (based on the Leadscope expert alerts v4).
Fig. 8.
Mutagenic examples containing a quindioxin (highlighted in bold) and an alerting aryl methylene semicarbazones (highlighted in grey).
Fig. 9.
Mutagenic examples containing a quindioxin (highlighted in bold) and no additional structural alerts.
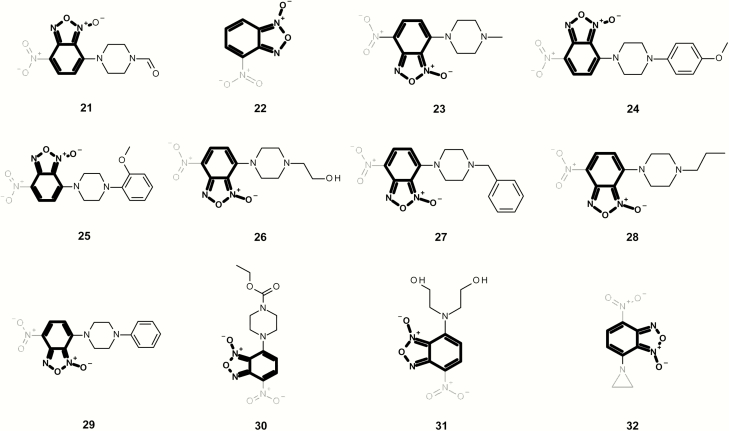
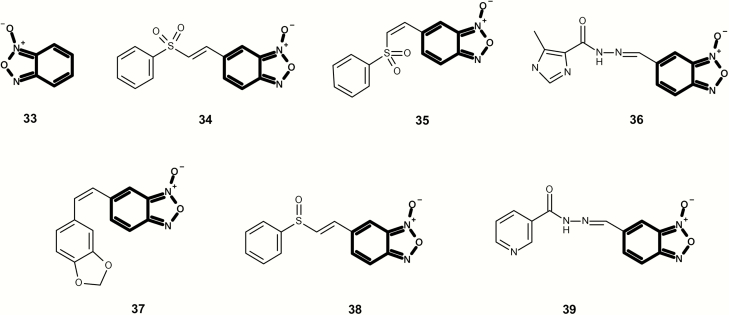
Figures 10 and 11 contain a series of mutagenic benzo[c][1,2,5]oxadiazole 1-oxides, which are alerting in the current Leadscope expert alerts. In Figure 10, all examples contain an aromatic nitro alert with chemical 32 also containing an aziridine. Although chemicals 34 and 35 are considered Michael acceptors, no chemicals in Figure 11 contain other alerts based on the Leadscope Expert Alerts system v4.
Fig. 10.
Mutagenic examples containing a benzo[c][1,2,5]oxadiazole 1-oxide (highlighted in bold) that also contain one or more additional structural alerts (highlighted in grey).
Fig. 11.
Mutagenic examples containing a benzo[c][1,2,5]oxadiazole 1-oxide (highlighted in bold) with no additional structural alerts.
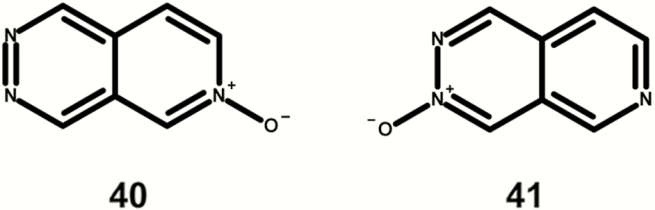
Figure 12 shows two mutagenic examples containing 2,7-napthyridine 2-oxides, which are alerting in the current Leadscope expert alerts v4 but marked as indeterminate due to the limited number of examples.
Fig. 12.
Mutagenic examples containing a 2,7-napthyridine 2-oxide (highlighted in bold).
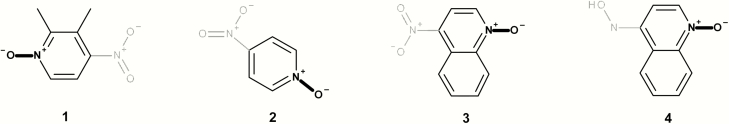
Figure 4 shows a series of isolated and fused mutagenic chemicals containing an aromatic N-oxide that also contain a separate mutagenic structural alert. Compounds 1, 2 and 3 contain an aromatic nitro and compound 4 contains a N-hydroxy aromatic amine.
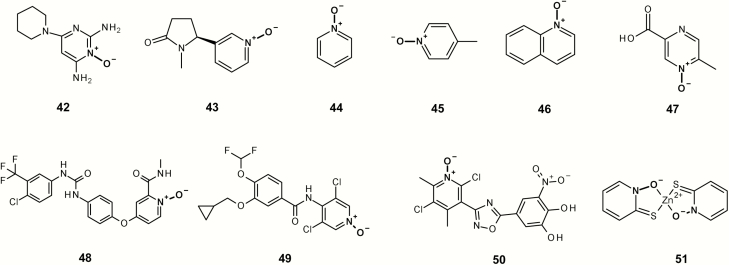
A number of diverse non-mutagenic chemicals (Figure 13 and Table 2) that contain an aromatic N-oxide were identified. Many of these chemicals were tested in four or more of the standard bacterial tester strains with and without metabolic activation or were part of a submission to a regulatory authority. In addition, all chemicals were predicted to be non-mutagenic using the Leadscope Genetox Expert Alerts version 4 except 50 (although the aromatic nitro alert was deactivated because of the adjacent hydroxyl groups). Minoxidil 42 is a vasodilator and over-the-counter treatment for hair loss in products such as Rogaine® from Johnson and Johnson with negative Ames and rodent carcinogenicity data (48). In addition, N-oxides are formed as metabolites of the parent substances. Cotinine N-oxide 43 is a metabolite of nicotine in human urine (49). Pyridine N-oxide 44 is a significant mammalian metabolite of pyridine, which is detected at around one-third of the urinary radioactivity in human volunteers administered [14C]pyridine (50). Olbetam 46 is a lipid-lowering agent that is reported as negative for bacterial mutagenicity (51). Both sorafenib and roflumilast form N-oxide metabolites (48,49) in patients (52,53). Opicapone 50 is a catechol-o-methyltransferase inhibitor for patients with Parkinson’s disease with negative Ames and rodent carcinogenicity data (54), and pyrithione zinc 51 is the active ingredient in shampoos such as Head & Shoulders® and is Ames negative.
Fig. 13.
Non-mutagenic aromatic N-oxides examples.
Table 2.
Negative aromatic N-oxides
| Chemical id | Leadscope id | Name | Negative tester strains | Reference |
|---|---|---|---|---|
| 42 | LS-135040 | Minoxidil | Negative tester strains not reported | (55) |
| 43 | LS-138926 | Cotinine N-oxide | TA98 ± S9; TA100 ± S9; TA1535 ± S9; TA1537 ± S9; TA1538 ± S9 | (56) |
| 44 | LS-131876 | Pyridine N-oxide | TA98; TA100 | (45) |
| 45 | LS-131819 | 4-Picoline N-oxide | TA98; TA100 | (45) |
| 46 | LS-142087 | Quinoline N-oxide | TA98; TA100 | (45) |
| 47 | LS-127587 | Olbetam | Negative tester strains not reported | (51) |
| 48 | LS-456979 | Sorafenib N-oxide | Negative tester strains not reported | (52) |
| 49 | LS-442852 | Roflumilast N-oxide | Negative tester strains not reported | (53) |
| 50 | LS-453011 | Opicapone | Negative tester strains not reported | (54) |
| 51 | LS-784 | Pyrithione zinc | TA98 ± S9, TA1535 ± S9, TA1537 ± S9, TA100 ± S9, TA1538 ± S9 | (57–59) |
Analysis of public and proprietary data using the SAR fingerprint
Table 3 summarises results from the SAR fingerprint exercise analysing public and proprietary databases. Thirty five thousand four hundred and thirty-seven compounds were analysed with 22.5% testing positive in the bacterial reverse mutation assay. For the general class of aromatic N-oxides (shown in row 2), there were 242 examples and 27.7% were mutagenic. Specific subclasses are included in this table where they included four or more examples or where they represent a general subclass of aromatic N-oxides. An analysis is included in the comments field including cases where positive data are from public sources.
Table 3.
Summary of results from the SAR fingerprints
| Row id | Substructure name | Substructure query | Number of negative | Number of positive | Total | Percentage of positives (%) | Comments |
|---|---|---|---|---|---|---|---|
| 1 | All compounds | 27 453 | 7 984 | 35 437 | 22.5 | ||
| 2 | Aromatic N-oxide |
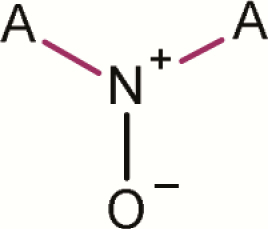
|
175 | 67 | 242 | 27.7 | 35 of the positives are attributable to subclasses shown in row ids 5, 11, 13 |
| 3 | Aromatic N-oxide- (5-ring isolated; any atom in ring) |
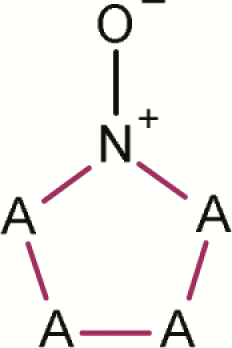
|
4 | 0 | 4 | 0.0 | No isolated 5-membered rings containing an aromatic N-oxide are mutagenic. It should be noted that these examples are from a single proprietary source |
| 4 | 5,6 fused rings N-Oxide (any atom in ring) |
|
17 | 16 | 33 | 48.5 | 13 of the 16 positive examples are from row id 5 |
| 5 | 1H-[2,3-[aza|oxo|thia]] indole 1-oxide |
|
3 | 13 | 16 | 81.3 | Many positive examples are from the public domain and contain other alerts. The negative proprietary examples are not members of the class benzo[c][1,2,5]oxadiazole 1-oxide |
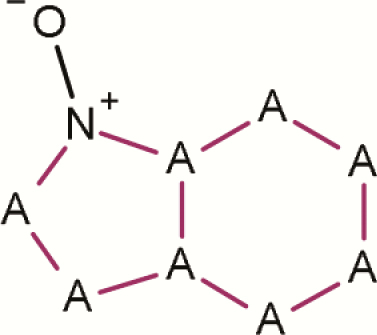
| 6 | Aromatic N-oxide- (6-ring isolated; any atom in ring) |
|
120 | 10 | 130 | 7.7 | Strong evidence that isolated 6-membered aromatic rings with an aromatic N-oxide attached are not mutagenic (P-value < 0.001) |
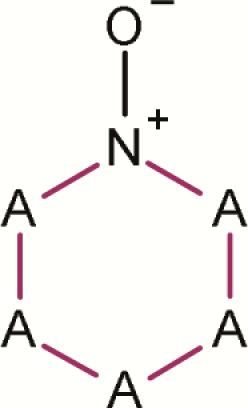
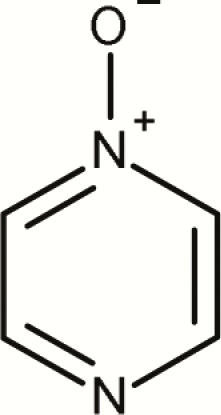
| 7 | Pyrazinium, 1-oxide- |
|
4 | 0 | 4 | 0.0 | No evidence that pyrazinium, 1-oxide- subclass is a mutagenic alert |
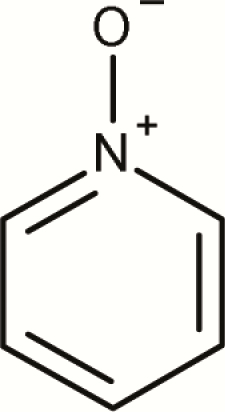
| 8 | Pyridinium, 1-oxide- |
|
114 | 10 | 124 | 8.1% | No evidence that pyridinium, 1-oxide-subclass is a mutagenic alert |
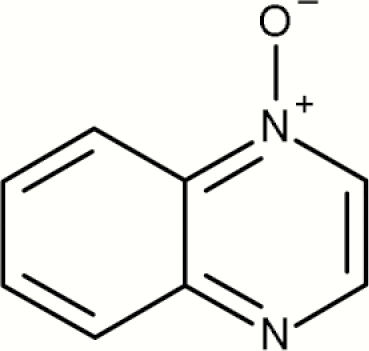
| 9 | Quinoxalinium, 1-oxide- |
|
4 | 1 | 5 | 20.0 | No evidence that quinoxalinium, 1-oxide- is a mutagenic alert |
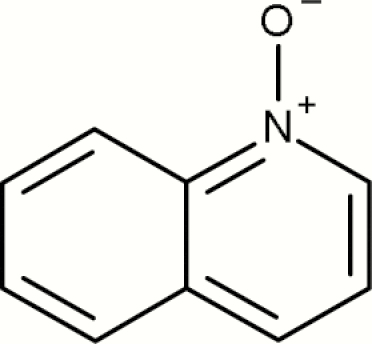
| 10 | Quinolinium, 1-oxide- |
|
9 | 6 | 15 | 40.0 | The three positive public examples (structure ids: 3, 4, 11) contain other alerts; two proprietary positive examples also contained other alerting structures |
| 11 | Quindioxin and related |
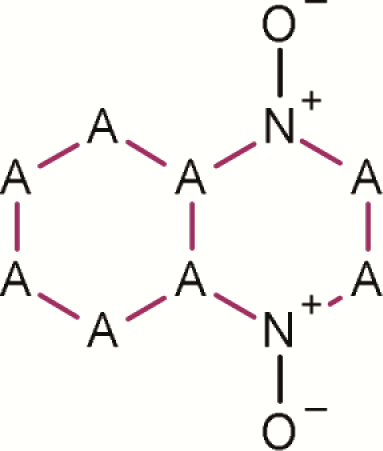
|
1 | 20 | 21 | 95.2 | Strong evidence that quindioxin and related chemicals are mutagenic (P-value < 0.001) |
| 12 | Isoquinolinium, 2-oxide- |
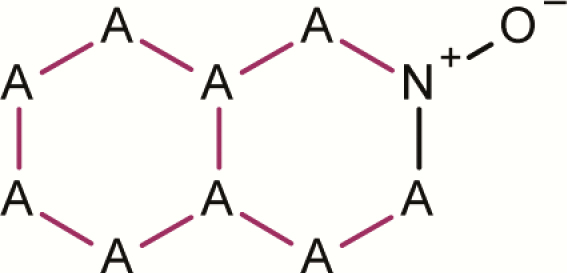
|
7 | 3 | 10 | 30.0 | Two positive examples contain the substructure from row id 13 |
| 13 | 2,7-[1,3,4,4a,5,6,8,8a- any]naphthyridine 2-oxide |
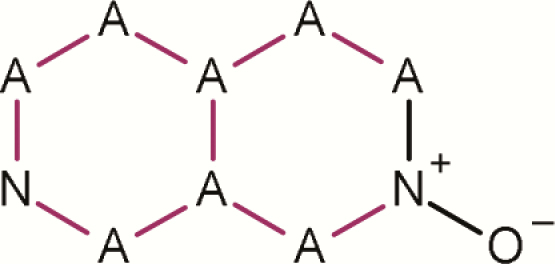
|
0 | 2 | 2 | 100.0 | There are only two public mutagenic examples(40,41) |
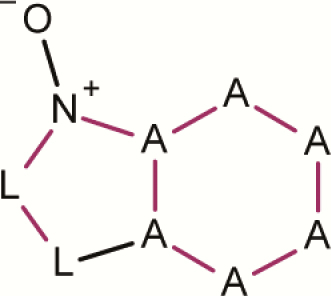
| 14 | 6,5 Fused ring N-oxide |
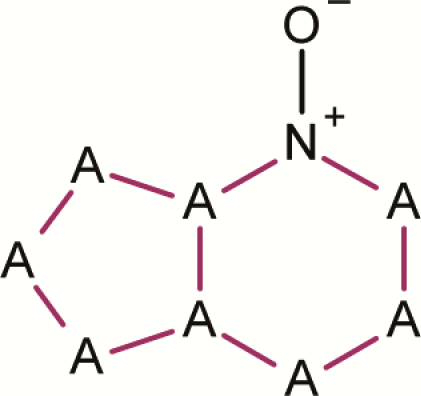
|
3 | 0 | 3 | 0.0 | No evidence for assignment as a mutagenic alert |
| 15 | 6,5 Fused ring N-oxide |
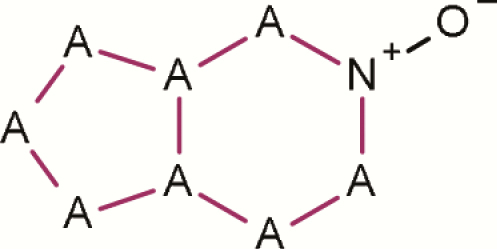
|
1 | 7 | 8 | 87.50 | All 7 positive examples (structure ids: 5, 6, 10, 12, 13, 14, 15) contain other alerts |
A is any atom; L is N or S or O; A-A, A-L, L-L, A-N and A-N+ are aromatic bonds; all P-values are calculated based on the binomial score.
Discussion
Are aromatic N-oxides, in general, associated with DNA-reactive mutagenicity?
The initial analysis of available public data found no evidence that the general class of aromatic N-oxides is a structural alert for DNA-reactive mutagenicity. All mutagenic examples contained another alert most likely responsible for the mutagenicity (e.g. presence of an aromatic nitro alert in compound 1 in Figure 2). There are also structurally diverse examples of non-mutagenic compounds containing aromatic N-oxides, and for many the data have been accepted by regulatory authorities. Moreover, a review of the published literature did not identify a plausible mechanism of mutagenicity for simple aromatic N-oxides.
An additional retrospective analysis of 24 ICH-M7-compliant (Q)SAR evaluations of impurities containing aromatic N-oxides conducted by FDA/CDER with the application of expert knowledge found that 6 were predicted to be mutagenic and 18 predicted to be non-mutagenic (not shown in the article). Those predicted to be positive were found to contain other confounding structural alerts that could explain their mutagenic activity. The negatives consisted of simple, monocyclic N-oxides, which appeared to be accurately predicted by the models. Although not experimentally verified, these observations are supportive of the hypothesis that aromatic N-oxides are not generally an alerting class and consistent with the data from Table 3.
The combined analysis of public and proprietary data confirmed this finding. In fact, if the 4 negative and 33 positive examples are removed from consideration based on the two classes shown in Figure 14, the number of non-mutagenic aromatic N-oxide examples (covering monocyclic and fused aromatic N-oxides) is 171 and the number of mutagenic aromatic N-oxide examples is 34. The proportion of mutagenic examples is 16.6%, which is statistically significantly lower (P-value = 0.023) than the baseline computed across all compounds of 22.5%. This is comparable with other functional groups that are generally considered to be non-mutagenic. For example, the non-mutagenic carboxylate functional group was included in this SAR fingerprinting exercise and the proportion of mutagenic chemicals for these 5000 examples was 16.7%. The remaining positive rate can be explained by other factors responsible for the positive test results, such as additional mutagenic functional groups, impurities of the tested compound and unstable compounds resulting in mutagenic cleavage products.
Fig. 14.
Summary of two validated alerts with mutagenic examples.
Using the information in Table 3, the SAR fingerprint analysis provides additional affirmative support, because an isolated 6-membered ring with an N-oxide is assigned as deactivating based on the 7.7% positive rate from the 130 examples (P-value < 0.001). There was also no evidence that five-membered isolated aromatic rings (e.g. furan, pyrrole) containing an N-oxide are mutagenic.
As discussed earlier, a structural alert must be based on a sufficient number of examples to conclude that chemicals in a class have an increased likelihood of being active (e.g. mutagenic). Strege et al. (17) identified two chemicals containing aromatic N-oxides as being mutagenic. However, when combined with the other data reported in this article, it is not sufficient to support assigning aromatic N-oxides as alerts.
Are subclasses of aromatic N-oxide alerts DNA-reactive mutagens?
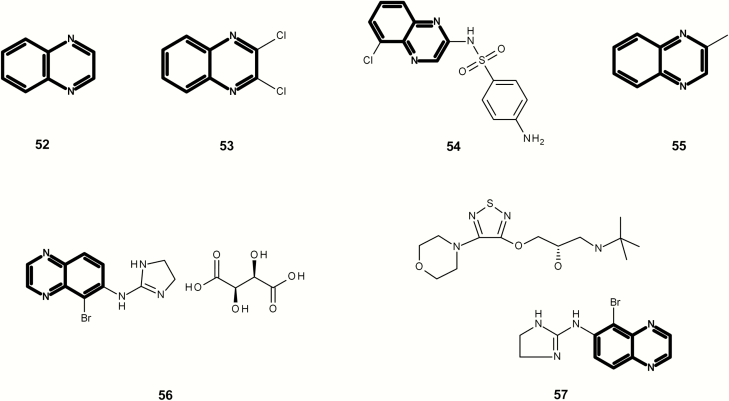
On the basis of the analysis of public and proprietary data, there is sufficient evidence to conclude and define a DNA-reactive mutagenic alert for quindioxin and related compounds (see Figure 14 and Table 4). Out of the 21 identified examples, 20 were positive and the association between chemicals containing this class and positive mutagenicity data was determined to be statistically significant (P-value < 0.001 based on the binomial score as discussed previously). There are 17 positive examples (public and proprietary) with no additional alerts (shown in Figure 14). The public compounds had similar positive tester strains suggesting a common mechanism. This quindioxin class was also identified as a structural alert in Enoch and Cronin (30).
Table 4.
Study summary for compounds containing the free form of quindioxins
| Compound id | Leadscope id | Name | Study result | Strains tested | Positive tester strain | Reference |
|---|---|---|---|---|---|---|
| 52 | LS-142960 | Quinoxaline | Positive | TA98(2); TA100(2); TA102(2) | TA98 + S9 | (71) |
| 52 | LS-142960 | Quinoxaline | Negative | TA98(3); TA100(3); TA1535(3); TA1537(3); TA1538(3) | (72) | |
| 53 | LS-1019 | 2,3-Dichloroquinoxaline | Negative | TA100(6); TA1535(6); TA1537(6); TA98(6) | (73) | |
| 53 | LS-1019 | 2,3-Dichloroquinoxaline | Negative | TA98(3); TA100(3); TA1535(3); TA1537(3); TA1538(3) | (74) | |
| 53 | LS-1019 | 2,3-Dichloroquinoxaline | Negative | TA100(3); TA1535(3); TA1537(3); TA98(3) | (75) | |
| 54 | LS-31236 | Chlorsulfaquinoxaline | Negative | TA98(3); TA100(3); TA1535(3); TA1537(3); TA1538(3) | (76) | |
| 55 | LS-143043 | 2-Methylquinoxaline | Negative | TA98(2); TA100(2); TA102(2) | (71) | |
| 56 | LS-177484 | Brimonidine tartrate | Negative | TA1535(2); TA1537(2); TA1538(2); TA98(2); TA100(2) | (77) | |
| 56 | LS-177484 | Brimonidine tartrate | Negative | TA98(2); TA1537(2); TA1538(2); TA1535(2); TA100(2) | (78) | |
| 57 | LS-445000 | Brimonidine tartrate and timolol maleate | Negative | TA98(4); TA100(4); TA102(4); TA1535(4); TA1537(4) | (79) |
An understanding of the biological mechanism would provide additional support for the alert. Generally, biological reduction, either bacterial or by S9, is considered to be a necessary step in the generation of the proximate mutagen, which is frequently postulated to be the hydroxyl radical (44,60–62). In view of the likely radical-based mechanism, it is distinctly possible that mutagenic activity detected in vitro may be eliminated or diminished in the presence of radical scavengers/antioxidants such as glutathione (63).
It was also determined that benzo[c][1,2,5]oxadiazole 1-oxide had eight positive public examples (33–39), which is significant enough to warrant assigning it as a DNA-reactive mutagenicity alert (see Figure 14). Gabay et al. (46) investigated the mutagenicity of a number of candidates for pharmaceutical development using Salmonella typhimurium TA98 ± S9 and found that benzo[c][1,2,5]oxadiazole 1-oxide was associated with mutagenicity whereas N-oxides of furoxans lacked mutagenic activity.
The 2,7-napthyridine 2-oxide class had two positive examples (40,41); however, this is not enough to conclude with sufficient confidence that this class should be assigned as a DNA-reactive mutagenicity alert (6).
Aromatic N-oxides and carcinogenicity
The ICH M7 guideline is predicated on the notion that actual or predicted bacterial mutagenicity of a pharmaceutical impurity is an indicator of potential carcinogenicity. Consequently, the validity of a particular conventional structural alert for DNA-reactive mutagenicity is likely to be considerably more secure if there are examples of compounds containing this alert that are convincingly carcinogenic. The literature on aromatic N-oxide carcinogenicity is focused almost entirely on 4-nitroquinoline N-oxide (3,64), whereas no other aromatic N-oxides appear to have been evaluated in lifetime rodent bioassays (apart from carbadox for which the di-N-oxide moiety, absent from desoxycarbadox, is not a critical structural requirement for carcinogenicity). For 4-nitroquinoline N-oxide and its derivatives, whose carcinogenic properties have been investigated over many decades (65), both the nitro and N-oxide substituents are essential structural requirements. The compound is a potent bacterial mutagen that produces positive results in in vitro and in vivo genotoxicity assays and has been shown to form a number of DNA adducts (66). However, the mechanism of action is still not completely elucidated: initial hypotheses on bioreduction of the nitro substituent to the corresponding 4-amino compound have been supplemented by evidence indicating a possible contribution by reactive-oxygen species based on the formation of 8-hydroxydeoxyguanosine in human fibroblasts (67).
Evidence currently available supports an indirect mutagenic mode of action for aromatic N-oxides involving reactive-oxygen species such as the hydroxyl radical. A literature review of the role of hydroxyl radicals in chemically induced carcinogenicity (68) mentions a number of carcinogens including a variety of metals (including Fe, Cu, Cr, As and V) and peroxisome proliferators where this type of mechanism has been proposed. To a broad generalisation, the examples suggest that carcinogenic effects are probably thresholded because high/prolonged exposure is required in order to deplete/overcome endogenous defence mechanisms (68). Hydrogen peroxide, which is a direct-acting mutagen in multiple bacterial strains and a weak local carcinogen in the gastrointestinal tract in the mouse (69), is a well-established source of hydroxyl radicals, yet its cosmetic use as a tooth whitener is considered to be without significant risk and it is not classifiable as to its carcinogenicity in humans (70).
Analysis of the free form of quindioxins
To help verify that the mutagenicity is attributable to the quindioxin or the free form, a separate analysis of compounds without the oxide was performed, shown in Figure 15 and Table 4. All examples were negative, with the exception of quinoxaline (49) where there are conflicting study results for strain TA98 (but concordant negative in TA100 as well as negative results in TA102, TA1535, TA1537 and TA1538). In this case, quinoxaline may be positive in TA98 with metabolic activation; however, there are also conflicting negative data present. Therefore, this analysis did not identify any association with the free form of quindioxin and mutagenicity.
Fig. 15.
Compounds containing the free form of quindioxin (shown in bold).
SAR fingerprint analysis
This article describes how information from public and proprietary sources can be combined to refine alerts for predicting mutagenicity. Through refinement of the set of mutagenicity alerts, it is possible to streamline the assessment of impurities, degradants and other types of chemicals to avoid unnecessary testing and implementation of control strategies. This project made use of proprietary data using a much larger set of compounds than previously available for analysis of aromatic N-oxides to better define the structure–activity (mutagenicity) relationships for this class. It should be noted that it was not necessary to release any proprietary information on individual compounds or data, because only derived aggregated information on chemical classes was used in this analysis. It was, however, possible to ask questions to the owners of the proprietary data to support this analysis. For example, questions were asked for a number of entries to determine whether, for specific classes, they contained other functional groups most likely responsible for DNA-reactive mutagenicity (e.g. Table 3, row 10).
Conclusion
On the basis of the analysis of aromatic N-oxide structure–activity relationships for bacterial mutagenicity presented in this article, we conclude that the general class of aromatic N-oxide is not predictive of DNA-reactive mutagenicity. Two subclasses were determined to be predictive for DNA-reactive mutagenicity: quindioxin and benzo[c][1,2,5]oxadiazole-1-oxide. Currently available evidence suggests Ames-positive aromatic N-oxides may be indirectly DNA-reactive by a mode of action involving hydroxyl radicals. The conclusions from this article may be used to support the development of (Q)SAR methods as well as support an expert review of the (Q)SAR results, mentioned in the ICH M7 guideline, where an aromatic N-oxide is highlighted as responsible for a DNA-reactive mutagenic prediction.
Acknowledgement
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R44ES026909. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We wish to thank Veronique Thybaud from Sanofi for the helpful comments on the manuscript.
Footnotes
FDA CDER disclaimer: The findings and conclusions in this manuscript have not been formally disseminated by the FDA and should not be construed to represent any agency determination or policy. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Conflict of interest statement: It is disclosed that FDA’s Center for Drug Evaluation and Research (CDER) and Leadscope Inc. are parties to a formal Research Collaboration Agreement (RCA). Dr. Naomi Kruhlak is the FDA CDER Principal Investigator for this agreement and Dr. Kevin Cross is the Leadscope Principal Investigator for this agreement.
References
- 1.International Council for Harmonisation. (2017)Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk M7(R1) http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Multidisciplinary/M7/M7_R1_Addendum_Step_4_31Mar2017.pdf (accessed August 14, 2018).
- 2. Myatt G. J., Beilke L. D. and Cross K. P (2016)In silico tools and their application. In Reedijk, J (ed.) Reference Module in Chemistry, Molecular Sciences and Chemical Engineering. Elsevier, Oxford. doi: 10.1016/B978-0-12-409547-2.12379-0 [DOI] [Google Scholar]
- 3. Netzeva T. I., Worth A., Aldenberg T., et al. (2005)Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships. The report and recommendations of ECVAM Workshop 52. Altern. Lab. Anim., 33, 155–173. [DOI] [PubMed] [Google Scholar]
- 4. Carrió P. Pinto M. Ecker G. Sanz F. and Pastor M (2014)Applicability Domain ANalysis (ADAN): a robust method for assessing the reliability of drug property predictions. J. Chem. Inf. Model., 54, 1500–1511. [DOI] [PubMed] [Google Scholar]
- 5. Patlewicz G. Worth A. P. and Ball N (2016)Validation of computational methods. Adv. Exp. Med. Biol., 856, 165–187. [DOI] [PubMed] [Google Scholar]
- 6. Ahlberg E., Amberg A., Beilke L. D., et al. (2016)Extending (Q)SARs to incorporate proprietary knowledge for regulatory purposes: a case study using aromatic amine mutagenicity. Regul. Toxicol. Pharmacol., 77, 1–12. [DOI] [PubMed] [Google Scholar]
- 7. Ashby J. and Tennant R.W (1988)Chemical structure, Salmonella mutagenicity and extent of carcinogenicity as indicators of genotoxic carcinogenesis among 222 chemicals tested in rodents by the U.S. NCI/NTP. Mutat. Res., 204,17–115. doi: 10.1016/0165-1218(88)90114-0. [DOI] [PubMed] [Google Scholar]
- 8. Ashby J. and Tennant R. W (1991)Definitive relationships among chemical structure, carcinogenicity and mutagenicity for 301 chemicals tested by the U.S. NTP. Mutat. Res., 257, 229–306. [DOI] [PubMed] [Google Scholar]
- 9. Miller J. A. and Miller E. C (1977)Ultimate chemical carcinogen as reactive mutagenic electrophiles. In Hiatt H. H., Watson J. D. and Winsten, J. A (eds), Origin of Human Cancers. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 605–627. [Google Scholar]
- 10. Snodin D. J. and McCrossen S. D (2013)Mutagenic impurities in pharmaceuticals: a critique of the derivation of the cancer TTC (Threshold of Toxicological Concern) and recommendations for structural-class-based limits. Regul. Toxicol. Pharmacol., 67, 299–316. [DOI] [PubMed] [Google Scholar]
- 11. Benigni R., Bossa C., Jeliazkova N. G., Netzeva T. I. and Worth A. P (2008)The Benigni/Bossa Rulebase for Mutagenicity and Carcinogenicity - A Module of Toxtree. Technical Report EUR 23241 EN, European Commission - Joint Research Centre, Brussels, Belgium. [Google Scholar]
- 12. Plošnik A., Vračko M. and Dolenc M. S (2016)Mutagenic and carcinogenic structural alerts and their mechanisms of action. Arh. Hig. Rada. Toksikol., 67, 169–182. doi: 10.1515/aiht-2016-67-2801. [DOI] [PubMed] [Google Scholar]
- 13.Organisation for Economic Co-operation and Development. (2010) OECD Environment, Health and Safety Publications Series on Testing and Assessment: Report of the expert consultation on scientific and regulatory evaluation of organic chemistry mechanism-based structural alerts for the identification of DNA binding chemicals - series on testing and assessment number 120 ENV/JM/MONO(2010)8/PART1. http://www.oecd.org/env/ehs/risk-assessment/45401393.pdf (accessed August 14, 2018).
- 14. Galloway S. M. Vijayaraj Reddy M. McGettigan K. Gealy R. and Bercu J (2013)Potentially mutagenic impurities: analysis of structural classes and carcinogenic potencies of chemical intermediates in pharmaceutical syntheses supports alternative methods to the default TTC for calculating safe levels of impurities. Regul. Toxicol. Pharmacol., 66, 326–335. [DOI] [PubMed] [Google Scholar]
- 15. Yang R. S., Beard A., Sheng H., Zhang L. K. and Helmy R (2015)Applications of TiCl3 as a diagnostic reagent for the detection of nitro- and N-oxide-containing compounds as potentially mutagenic impurities using ultrahigh-performance liquid chromatography coupled with high-resolution mass spectrometry. Org. Process Res. Dev., 20, 59–64. doi: 10.1021/acs.oprd.5b00312. [DOI] [Google Scholar]
- 16. Chandrasekhar S., Reddy C. R., Rao R. J. and Rao J. M (2002)Efficient and chemoselective deoxygenation of amine N-Oxides using polymethylhydrosiloxane. Synlett, 2002, 0349–0351. doi:10.1055/s-2002– 19751. [Google Scholar]
- 17. Strege M. A., Osborne L. M., Hetrick E. M., et al. (2015)Assessing the risk of formation of potential genotoxic degradation products in a small-molecule kinase inhibitor drug substance and drug product. Org. Process Res. Dev., 19, 1458–1464. doi: 10.1021/acs.oprd.5b00112. [DOI] [Google Scholar]
- 18. Mfuh A. M. and Larionov O. V (2015)Heterocyclic N-Oxides - an emerging class of therapeutic agents. Curr. Med. Chem., 22, 2819–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Powley M. W. (2015)(Q)SAR assessments of potentially mutagenic impurities: a regulatory perspective on the utility of expert knowledge and data submission. Regul. Toxicol. Pharmacol., 71, 295–300. [DOI] [PubMed] [Google Scholar]
- 20. Barber C., Amberg A., Custer L., et al. (2015)Establishing best practise in the application of expert review of mutagenicity under ICH M7. Regul. Toxicol. Pharmacol., 73, 367–377. [DOI] [PubMed] [Google Scholar]
- 21. Amberg A., Beilke L., Bercu J., et al. (2016)Principles and procedures for implementation of ICH M7 recommended (Q)SAR analyses. Regul. Toxicol. Pharmacol., 77, 13–24. [DOI] [PubMed] [Google Scholar]
- 22. Bower D., Cross K. P., Esche S., Myatt G. J. and Quigley D. P.(2017)In silico toxicology: an overview of toxicity databases, prediction methodologies, and expert review. In Richardson R. J. and Johnson, D. E (eds), Computational Systems Pharmacology and Toxicology. Royal Society of Chemistry, Cambridge. doi: 10.1039/9781782623731-00209. [DOI] [Google Scholar]
- 23.Reed Elsevier Prop. SA. (2013)PharmaPendium 2017: Expanded Access to Drug Safety Information https://www.elsevier.com/solutions/pharmapendium-clinical-data/drug-safety (accessed August 14, 2018).
- 24. Wheeler D. L., Barrett T., Benson D. A., et al. (2005)Database resources of the national center for biotechnology information. Nucleic Acids Res., 33, D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leadscope Genetox Expert Alerts Suite (2018) Leadscope. http://www.leadscope.com/genetox_expert_alerts/ (accessed August 14, 2018).
- 26. Cross K. P., Hasselgren C. and Myatt G. J (2016)Mitigation of Bacterial Mutation (Q)SAR Alerts Using Limited‐Ames and Tester Strain Data 2016 SOT poster 3738. https://www.toxicology.org/pubs/docs/Tox/2016ToxSup.pdf (accessed August 14, 2018).
- 27. Honma M. (2018)AMES/QSAR International Collaborative Study. Division of Genetics and Mutagenesis, National Institute of Health Scienceshttp://www.nihs.go.jp/dgm/amesqsar.html (accessed August 14, 2018).
- 28. OECD Guidelines for the Testing of Chemicals, Section 4 - Test No. 471: Bacterial Reverse Mutation Test (1997)OECDiLibrary http://www.oecd- ilibrary.org/environment/test-no-471-bacterial-reverse-mutation- test_9789264071247-en (accessed August 14, 2018).
- 29. Leadscope Applications (2018)Leadscope http://www.leadscope.com/data_management/ (accessed August 14, 2018).
- 30. Enoch S. J. and Cronin M. T (2012)Development of new structural alerts suitable for chemical category formation for assigning covalent and non-covalent mechanisms relevant to DNA binding. Mutat. Res., 743, 10–19. [DOI] [PubMed] [Google Scholar]
- 31.NIHS: AMES/QSAR International Collaborative Study. (2016)Strong Positive (Class A) in Ames Assay in Phase I Trial (183 Chemicals) http://www.nihs.go.jp/dgm/ClassA183.pdf (accessed August 14, 2018).
- 32.NIHS: AMES/QSAR International Collaborative Study. (2016)Strong Positive (Class A) in Ames Assay in Phase II Trial (253 Chemicals)http://www.nihs.go.jp/dgm/ClassA253.pdf (accessed August 14, 2018).
- 33. Blahová M. Sokolík J. Lahitová N. and Murín A (1994)[Mutagenic activity of copper phenoxyacetate complexes]. Ceska Slov. Farm., 43, 240–242. [PubMed] [Google Scholar]
- 34. Rinkus S. J. and Legator M. S (1979)Chemical characterization of 465 known or suspected carcinogens and their correlation with mutagenic activity in the Salmonella typhimurium system. Cancer Res., 39, 3289–3318. [PubMed] [Google Scholar]
- 35. Sugimura T., Sato S., Nagao M., Yahagi T., Matsushima T., Seino Y., Takeuchi M. and Kawac Hi T (1976)Overlapping of carcinogens and mutagens. In Magee P. N., Takayama S., Sugimura T. and Matsushima, T (eds), Fundamentals in Cancer Prevention. University Park Press, Baltimore, MD, pp. 191–215. [Google Scholar]
- 36. Brusick D. J. Simmon V. F. Rosenkranz H. S. Ray V. A. and Stafford R. S (1980)An evaluation of the Escherichia coli WP2 and WP2 uvrA reverse mutation assay. Mutat. Res., 76, 169–190. [DOI] [PubMed] [Google Scholar]
- 37. Kier L. D., Brusick D. J., Auletta A. E., et al. (1986)The Salmonella typhimurium/mammalian microsomal assay. A report of the U.S. Environmental Protection Agency Gene-Tox Program. Mutat. Res., 168, 69–240. [DOI] [PubMed] [Google Scholar]
- 38. Dunkel V. C. Zeiger E. Brusick D. McCoy E. McGregor D. Mortelmans K. Rosenkranz H. S. and Simmon V. F (1984)Reproducibility of microbial mutagenicity assays: I. Tests with Salmonella typhimurium and Escherichia coli using a standardized protocol. Environ. Mutagen., 6Suppl 2, 1–251. [DOI] [PubMed] [Google Scholar]
- 39. Mccann J. and Ames B.N (1976)Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals: discussion. Proceedings of the National Academy of Sciences, 73, 950–954. doi: 10.1073/pnas.73.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopez-De-Cerain A., Garcia E., Gullón A., Recalde J. I. and Monge A. A (1990)Mutagenic evaluation of some triazino indoles using the Salmonella/mammalian microsome assay. Mutagenesis, 5, 307–312. doi: 10.1093/mutage/5.4.307. [DOI] [PubMed] [Google Scholar]
- 41. Fukuhara K. Hakura A. Sera N. Tokiwa H. and Miyata N (1992)1- and 3-nitro-6-azabenzo[a]pyrenes and their N-oxides: highly mutagenic nitrated azaarenes. Chem. Res. Toxicol., 5, 149–153. [DOI] [PubMed] [Google Scholar]
- 42. Gorrod J. W. Ioannides C. Lam S. P. and Neville S (1993)Mutagenicity testing of 9-N-substituted adenines and their N-oxidation products. Environ. Health Perspect., 101Suppl 3, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stavitskaya L., Minnier R., Benz D. and Kruhlak N, (2013)Development of Improved Salmonella Mutagenicity QSAR Models Using Structural Fingerprints of Known Toxicophores, FDA Center for Drug Evaluation and Research, Poster presented at: Society of Toxicology (SOT) 52nd Annual Meeting. http://www.leadscope.com/media/SOT%202013%20Stavitskaya%20CDER%20poster.pdf (accessed August 14, 2018). [Google Scholar]
- 44. Nunoshiba T. and Nishioka H (1989)Genotoxicity of quinoxaline 1,4-dioxide derivatives in Escherichia coli and Salmonella typhimurium. Mutat. Res., 217, 203–209. [DOI] [PubMed] [Google Scholar]
- 45. Voogd C. E. Van der Stel J. J. and Jacobs J. J (1980)On the mutagenic action of some enzyme immunoassay substrates. J. Immunol. Methods, 36, 55–61. [DOI] [PubMed] [Google Scholar]
- 46. Gabay M. Cabrera M. Maio R. D. Paez J. A. Campillo N. Lavaggi M. L. Cerecetto H. and González M (2014)Mutagenicity of N-oxide containing heterocycles and related compounds: experimental and theoretical studies. Curr. Top. Med. Chem., 14, 1374–1387. [DOI] [PubMed] [Google Scholar]
- 47. Morita T. Naitou H. and Oishi E (1995)Mutagenicity of condensed pyridazines with different substituents. Biol. Pharm. Bull., 18, 363–367. [DOI] [PubMed] [Google Scholar]
- 48. Drug label LONITEN- minoxidil tablet Approved Drug Label (2015)https://dailymed.nlm.nih.gov/dailymed/archives/fdaDrugInfo.cfm? archiveid=166610 (accessed August 14, 2018).
- 49. Benowitz N. L., Hukkanen J. and Jacob P (2009)Nicotine chemistry, metabolism, kinetics and biomarkers. Handbook of Experimental Pharmacology Nicotine Psychopharmacology, 192, 29–60. doi:10.1007/ 978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. D’Souza J. Caldwell J. and Smith R. L (1980)Species variations in the N-methylation and quaternization of [14C]pyridine. Xenobiotica., 10, 151–157. [DOI] [PubMed] [Google Scholar]
- 51. Data Sheet Olbetam®(2014): http://www.medsafe.govt.nz/Profs/Datasheet/o/Olbetamcap.pdf (accessed August 14, 2018).
- 52. Nexavar (Sorafenib Tosylate) Review and Evaluation of Pharmacology/Toxicology Data. (2005)Center for Drug Evaluation and Research; Application Number: 21–923. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2005/021923_s000_Nexavar_PharmR.pdf (accessed August 14, 2018). [Google Scholar]
- 53. Roflumilast Review and Evaluation of Pharmacology/Toxicology Data. (2011)Center for Drug Evaluation and Research; Application Number: 22–522. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2011/022522Orig1s000PharmR.pdf (accessed August 14, 2018). [Google Scholar]
- 54. EMEA: Assessment report Ongentys. (2016)http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/002790/WC500209538.pdf (accessed August 14, 2018).
- 55. Center for Drug Evaluation and Research Application Number: NDA 20–834. Pharmacological Review(s) 1997. https://www.accessdata.fda.gov/drugsatfda_docs/nda/97/20834_ROGAINE%20EXTRA%20STRENGTH%20FOR%20MEN%205%25_PHARMR.PDF#page=27 (accessed August 14, 2018).
- 56. Doolittle D. J. Winegar R. Lee C. K. Caldwell W. S. Hayes A. W. and de Bethizy J. D (1995)The genotoxic potential of nicotine and its major metabolites. Mutat. Res., 344, 95–102. [DOI] [PubMed] [Google Scholar]
- 57. Skoulis N. P. Barbee S. J. Jacobson-Kram D. Putman D. L. and San R. H (1993)Evaluation of the genotoxic potential of zinc pyrithione in the Salmonella mutagenicity (Ames) assay, CHO/HGPRT gene mutation assay and mouse micronucleus assay. J. Appl. Toxicol., 13, 283–289. [DOI] [PubMed] [Google Scholar]
- 58.National Toxicology Program. Study Information (1982)https://tools.niehs.nih.gov/cebs3/ntpViews/?activeTab=detail&studyNumber=053040 (accessed August 14, 2018).
- 59.National Toxicology Program. Study Information (1982)https://tools.niehs.nih.gov/cebs3/ntpViews/?activeTab=detail&studyNumber=941167 (accessed August 14, 2018).
- 60. Chowdhury G. Sarkar U. Pullen S. Wilson W. R. Rajapakse A. Fuchs-Knotts T. and Gates K. S (2012)DNA strand cleavage by the phenazine di-N-oxide natural product myxin under both aerobic and anaerobic conditions. Chem. Res. Toxicol., 25, 197–206. [DOI] [PubMed] [Google Scholar]
- 61. Brown J. M. and Wilson W. R (2004)Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer, 4, 437–447. [DOI] [PubMed] [Google Scholar]
- 62. Cheng G. Sa W. Cao C. Guo L. Hao H. Liu Z. Wang X. and Yuan Z (2016)Quinoxaline 1,4-di-N-Oxides: biological activities and mechanisms of actions. Front. Pharmacol., 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Słoczyńska K., Powroźnik B., Pękala E. Å. and Waszkielewicz A. M (2014)Antimutagenic compounds and their possible mechanisms of action. J. Appl. Genet., 55, 273–285. doi: 10.1007/s13353-014-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Endo H., Ono T. and Sugimura T.(1971)Chemistry and Biological Actions of 4-Nitroquinoline 1-Oxide. Springer, Berlin. [Google Scholar]
- 65. Nakahara W. (1961)Critique of carcinogenic mechanism. Prog. Exp. Tumor Res., 2, 158–202. [DOI] [PubMed] [Google Scholar]
- 66. Kirkland D. Kasper P. Martus H. J. Müller L. van Benthem J. Madia F. and Corvi R (2016)Updated recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 795, 7–30. [DOI] [PubMed] [Google Scholar]
- 67. Arima Y. Nishigori C. Takeuchi T. Oka S. Morimoto K. Utani A. and Miyachi Y (2006)4-Nitroquinoline 1-oxide forms 8-hydroxydeoxyguanosine in human fibroblasts through reactive oxygen species. Toxicol. Sci., 91, 382–392. [DOI] [PubMed] [Google Scholar]
- 68. Kanno T. Nakamura K. Ikai H. Kikuchi K. Sasaki K. and Niwano Y (2012)Literature review of the role of hydroxyl radicals in chemically-induced mutagenicity and carcinogenicity for the risk assessment of a disinfection system utilizing photolysis of hydrogen peroxide. J. Clin. Biochem. Nutr., 51, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. HSDB hydrogen peroxide (2012) Toxicology Data Network. https://toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+547 (accessed August 14, 2018).
- 70. Public Health England (2009)Hydrogen Peroxide: Toxicological Overview. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/337708/Hydrogen_Peroxide_Toxicological_Overview_phe_v1.pdf (accessed August 14, 2018). [Google Scholar]
- 71. Aeschbacher H. U. Wolleb U. Löliger J. Spadone J. C. and Liardon R (1989)Contribution of coffee aroma constituents to the mutagenicity of coffee. Food Chem. Toxicol., 27, 227–232. [DOI] [PubMed] [Google Scholar]
- 72. NCI: Short-term test program sponsored by the division of cancer biology (91-19-0), (1995)National Cancer Institute, Ms. Ellen Zaika, Assistant Project Officer, p. Y95; https://toxnet.nlm.nih.gov/cgi-bin/sis/search2/r?dbs+ccris:@term+@rn+91-19-0 (accessed August 14, 2018). [Google Scholar]
- 73.National Toxicology Program. (1982)Study Information http://tools.niehs.nih.gov/cebs3/ntpViews/?activeTab=detail&studyNumber=267151 (accessed August 14, 2018).
- 74. NCI: Short-term test program sponsored by the division of cancer biology (2213-63-0), (1995)National Cancer Institute, Ms. Ellen Zaika, Assistant Project Officer, p. Y95; https://toxnet.nlm.nih.gov/cgi-bin/sis/search2/r?dbs+ccris:@term+@rn+2213-63-0 (accessed August 14, 2018). [Google Scholar]
- 75. Zeiger E., Anderson B., Haworth S., Lawlor T., Mortelmans K. and Speck W (1987)Salmonella Mutagenicity tests: III. Results from the testing of 255 chemicals. Environ. Mutagen., 9, 61–109. doi:10.1002/em.2860090603; https://toxnet.nlm.nih.gov/cgi-bin/sis/search2/r?dbs+ccris:@term+@rn+2213-63-0 (accessed August 14, 2018). [PubMed] [Google Scholar]
- 76. NCI: Short-term test program sponsored by the division of cancer biology (1986)National Cancer Institute, Ms. Ellen Zaika, Assistant Project Officer, p. Y86; https://toxnet.nlm.nih.gov/cgi-bin/sis/search2/r?dbs+ccris:@term+@rn+97919-22-7 (accessed August 14, 2018). [Google Scholar]
- 77. NDA: Alphagan (Brimonidine Tartrate) Ophthalmic Solution Drug Approval Package (2001)http://www.accessdata.fda.gov/drugsatfda_docs/nda/2001/20-490s007_Alphagan.cfm (accessed August 14, 2018).
- 78. Brimonidine Tartrate Review and Evaluation of Pharmacology/Toxicology Data. (2000)Center for Drug Evaluation and Research; Application Number: 21–262. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2001/21-262_Alphagan%20P%20Ophthalmic_pharmr.pdf (accessed August 14, 2018). [Google Scholar]
- 79. Combidan™ (2006)Review and Evaluation of Pharmacology/Toxicology Data. Center for Drug Evaluation and Research. Application Number: 21-398. http://www.accessdata.fda.gov/drugsatfda_docs/nda/2007/ 021398s000_PharmR.pdf#page=14 (accessed August 14, 2018).