Abstract
BACKGROUND
Long QT syndrome (LQTS) genetic test reports commonly exclude potentially proarrhythmic common variants such as p.Asp85Asn-KCNE1.
OBJECTIVE
The purpose of this study was to determine whether a discernible phenotype is associated with p.Asp85Asn-KCNE1 and whether relatively common KCNE1 variants underlie transient QT prolongation pedigrees with negative commercial LQTS genetic tests.
METHODS
Retrospective review was used to compare demographics, symptomatology, and QT parameters of individuals with p.Asp85Asn-KCNE1 in the absence of other rare/ultra-rare variants in LQTS-susceptibility genes and those who underwent comprehensive LQTS genetic testing.
RESULTS
Compared to the Genome Aggregation Database, p.Asp85Asn-KCNE1 was more prevalent in individuals undergoing LQTS genetic testing (33/1248 [2.6%] vs 1552/126,652 [1.2%]; P = .0001). In 19 of 33 patients (58%), only p.Asp85Asn-KCNE1 was observed. These patients were predominantly female (90% vs 62%; P = .01) and were less likely to experience syncope (0% vs 34%; P = .0007), receive β-blockers (53% vs 85%; P = .001), or require an implantable cardioverter-defibrillator (5.3% vs 33%; P = .01). However, they exhibited a similar degree of QT prolongation (QTc 460 ms vs 467 ms; P = NS). Whole exome sequencing of 2 commercially genotype-negative pedigrees revealed that p.Asp85Asn-KCNE1 and p.Arg36His-KCNE1 traced with a transient QT prolongation phenotype. Functional characterization of p.Arg36His-KCNE1 demonstrated loss of function, with a 47% reduction in peak IKs current density in the heterozygous state.
CONCLUSION
We provide further evidence that relatively common variants in KCNE1 may result in a mild QT phenotype designated as “LQT5-Lite” to distinguish such potentially proarrhythmic common variants (ie, functional risk alleles) from rare pathogenic variants that truly confer monogenic disease susceptibility, albeit with incomplete penetrance.
Keywords: Arrhythmia, Genetics, Long QT syndrome, LQT5-Lite, Sudden death
Introduction
Long QT syndrome (LQTS) is a heritable disorder of cardiac repolarization characterized clinically by QT-interval prolongation on electrocardiogram (ECG) and an increased risk of syncope, seizure, and sudden death.1 Over the past 2 decades, 17 LQTS-susceptibility genes have been published.2 Coupled with sequencing advances, our increased understanding of the molecular basis of LQTS has allowed LQTS genetic testing to transition from an experimental exercise to the current slate of clinically relevant and commercially available genetic tests.3
However, large-scale sequencing efforts have revealed that monogenic disease-causative genes, including those responsible for LQTS, tolerate a substantial amount of non-synonymous genetic variation.4–6 As a result, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) released a set of revamped variant interpretation and reporting guidelines in 2015, to help standardize the classification and reporting of genetic variants.7 Although the current ACMG/AMP guidelines adequately address rare genetic variants, they provide little guidance regarding the classification and reporting of common genetic variants that occur too frequently in the general population to be designated as disease-causative for any reasonably penetrant monogenetic disease but nevertheless may modify or result in circumstance-dependent expression of a Mendelian disease phenotype.8 Consequently, most relatively common genetic variants, including those with substantive functional and epidemiologic evidence to support a potential contribution in disease pathogenesis, have been designated as benign and excluded from commercial genetic testing reports.8 As a result, potentially clinically relevant information is being withheld from both patients and providers. In turn, this may result in clinical LQTS genetic tests that fail to communicate the full extent of an individual’s genetic risk.
Previous epidemiologic and functional studies have shown that the common p.Asp85Asn-KCNE1 variant can reduce repolarization reserve and may result in a clinical phenotype in certain situations.9,10 As a result, we hypothesized that (1) p.Asp85Asn-KCNE1 in isolation may be overrepresented in our large cohort of patients referred for evaluation of congenital LQTS; (2) such p.Asp85Asn-KCNE1-positive individuals may exhibit a distinct, albeit milder, clinical phenotype; and (3) p.Asp85Asn-KCNE1, or other relatively common KCNE1 variants, may underlie transient QT prolongation pedigrees who remain genetically elusive after commercial LQTS genetic testing.
Methods
Study population
In this Institutional Review Board–approved study, a retrospective analysis of the 3041 patients seen in the Genetic Heart Rhythm Clinic from January 2000 to April 2017 was used to identify genotype-negative patients with p.Asp85Asn-KCNE1. Genotype-negative status was defined as absence of ultra-rare and rare variants in all LQTS-susceptibility genes tested (at a minimum: KCNQ1, KCNH2, and SCN5A). Demographics, clinical parameters (including ECG parameters as outlined in the Supplementary Materials), therapy history, and history of cardiac events were sourced from the patients’ electronic medical records.
Genetic analyses
Identification of p.Asp85Asn-KCNE1 was accomplished through either commercial genetic testing, largely undertaken before the release of the 2015 ACMG/AMP guidelines, or use of laboratory-based genetic testing. For the 2 commercially genotype-negative transient QT prolongation pedigrees, written informed consent was obtained, genomic DNA extracted from whole blood, and whole exome sequencing (WES) performed at the Mayo Clinic Medical Genome Facility (Rochester, Minnesota), as described previously.11 Variant filtering was restricted to all variants, regardless of minor allele frequency (MAF), in the 17 published LQTS-susceptibility genes (AKAP9, ANK2, CACNA1C, CALM1, CALM2, CALM3, CAV3, KCNE1, KCNE2, KCNH2, KCNJ2, KCNJ5, KCNQ1, SCN4B, SCN5A, SNTA1, and TRDN) and assessed under an autosomal-dominant model using the Ingenuity Variant Analysis platform (Qiagen, Hilden, Germany).
p.Arg36His-KCNE1 functional characterization
Details regarding the construction of KCNQ1 and KCNE1 mammalian expression vectors, cell culture/transfection, and whole-cell patch-clamp technique are given in the Supplementary Materials.
Statistical analysis
Categorical data are given as frequency and percentage, and were compared using the Pearson χ2 test or Fisher’s exact, as appropriate. Continuous variables are expressed as mean ± SD if normally distributed, or median and interquartile range if not normally distributed. They were compared using the Student t test if normally distributed or the Wilcoxon rank sum if not normally distributed. Normality was checked using the Shapiro-Wilk W test. For all tests, P <.05 was considered significant. Statistical tests were performed using JMP software, version 13.0 (SAS Institute, Cary, NC). Heterologous expression data are given as mean ± SEM, and 1-way analysis of variance was performed to determine statistical significance among multiple groups.
Results
Frequency of p.Asp85Asn-KCNE1 in an LQTS referral cohort
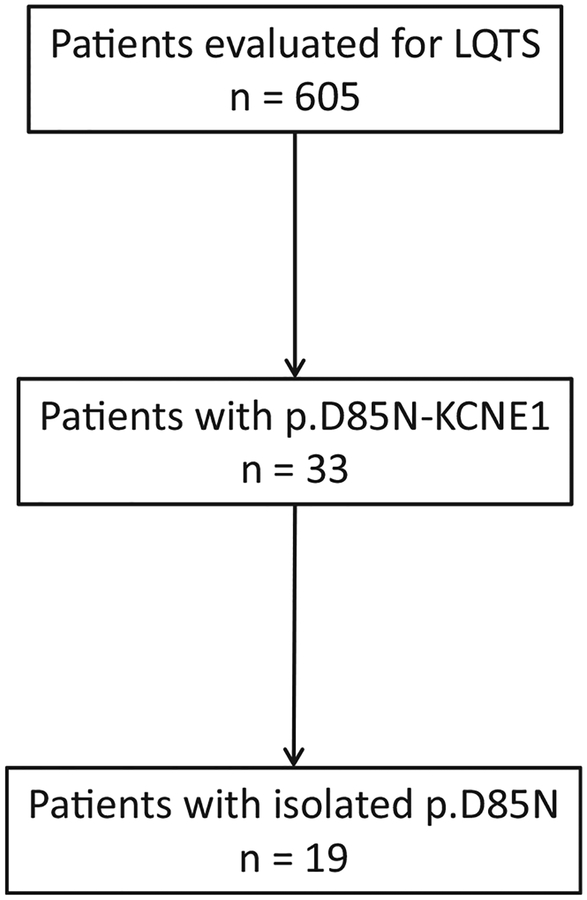
Overall, 624 patients who underwent full LQTS genetic evaluation (ie, forms of diagnostic LQTS genetic testing that allowed for ascertainment of the presence/absence of common genetic variants) and had a 12-lead ECG available for review were included (Figure 1). Patients with only confirmatory genetic testing were excluded.
Figure 1.
Patient selection flow chart. LQTS = long QT syndrome.
In comparison to an ethnically similar cohort in the Genome Aggregation Database (gnomAD), p.Asp85Asn-KCNE1 was statistically more common in the LQTS referral cohort (MAF 33/1248 [2.6%] vs 1552/126,652 [1.2%]; P = .0001]). Of the 33 p.Asp85Asn-KCNE1-positive patients, 19 (58%) were negative for pathogenic, likely pathogenic, or variants of uncertain significance (VUS) in all analyzed LQTS-susceptibility genes. Given the overrepresentation of p.Asp85Asn-KCNE1 in our referral LQTS cohort and previously mentioned epidemiologic/functional data, we next sought to determine whether individuals with only p.Asp85Asn-KCNE1 manifest a discernible shared clinical phenotype.
Phenotype of patients with p.Asp85Asn-KCNE1 in the absence of rare/ultra-rare variants in LQTS-susceptibility genes
The clinical characteristics of the 19 patients with p.Asp85Asn-KCNE1 and no additional rare/ultra-rare variants in LQTS-susceptibility genes were compared to the remaining 605 patients in our referral LQTS cohort are summarized in Table 1. Overall, a female predominance was observed in these p.Asp85Asn-KCNE1-positive individuals (90% vs 62%; P = .01) (Table 1). Furthermore, p.Asp85Asn-KCNE1 patients had a lower symptom burden (syncope/seizure 0% vs 34%; P = .0007) and lacked a family history of LQTS (5.3% vs 51%; P <.0001) (Table 1). However, the rate of sudden cardiac arrest was not statistically different (5.3% vs 11%; P = .7) (Table 1). Interestingly, the average QTc and magnitude of QTc variation were no different than those of the unfiltered cohort (Table 1). Lastly, p.Asp85Asn-KCNE1-positive patients were less likely to be placed on beta-blockers (53% vs 85%; P = .001) and to require an implantable cardioverter-defibrillator (5.3% vs 33%; P = .01). Individual p.Asp85Asn-KCNE1-positive case descriptions are detailed in Supplemental Table 1.
Table 1.
Clinical characteristics of Mayo Clinic LQTS cohort
| Demographic | p.Asp85Asn-KCNE1 only (n = 19) | LQTS genetic testing (n = 605) | P value |
|---|---|---|---|
| Female | 17 (90) | 376 (62) | .01 |
| Age at ECG (years) | 23.2 | 23.4 | 1 |
| Proband status | |||
| Index | 15 (79) | 375 (62) | .2 |
| Family | 4(21) | 230 (38) | .2 |
| Family history | 1 (5.3) | 311 (51) | <.0001 |
| Shortest QTc (ms) | 436.6 | 439.5 | .8 |
| Longest QTc (ms) | 486.1 | 497.9 | .8 |
| Average QTc (ms) | 460 | 466.6 | .5 |
| ΔQTc (ms) | 49.5 | 58.5 | .5 |
| QTc >99th percentile for sex | 12 (63) | 381 (63) | 1 |
| Syncope/seizure | 0 | 204 (34) | .0007 |
| Sudden cardiac arrest | 1 (5.3) | 68 (11) | .7 |
| Beta-blocker | 10 (53) | 516 (85) | .001 |
| ICD | 1 (5.3) | 198 (33) | .01 |
| LCSD | 0 | 93 (15) | .09 |
| AADs | 0 | 65 (11) | .2 |
Values are n (%) or mean.
AAD = antiarrhythmic drug; ECG = electrocardiogram; ICD = implantable cardioverter-defibrillator; LQTS = long QT syndrome; QTc = Bazett corrected QT interval; ΔQTc = difference in QTc; LCSD = left cardiac sympathetic denervation.
Genotype-negative transient QT prolongation pedigrees
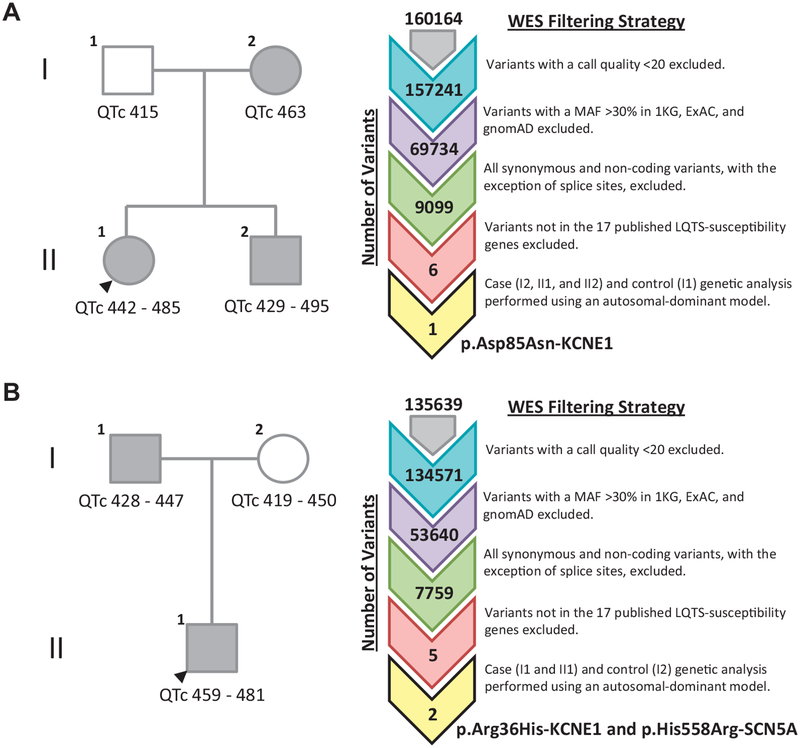
Given the “LQT5-Lite” phenotype detailed earlier, we elected to re-examine 2 transient QT prolongation pedigrees of European descent previously found to be negative for pathogenic, likely pathogenic, or VUS after commercial LQTS genetic testing. Because attempts to elucidate a novel genetic basis for the shared transient QT prolongation phenotype using research-based WES were unsuccessful, all variants, regardless of MAF, in the 17 published LQTS-susceptibility genes were reassessed in order to determine whether a single functional common variant, a combination of multiple common functional variants (so-called oligogenic/polygenic LQTS), or a limited evidence gene not covered by commercial genetic testing was responsible (Figure 2).
Figure 2.
Research-based whole exome sequencing analysis suggests relatively common variants in the KCNE1-encoded minK β-subunit may underlie genetically elusive familial transient QT prolongation. A: Two-generation transient QT prolongation pedigree found to harbor p.Asp85Asn-KCNE1 after pedigree-based WES analysis. B: Two-generation transient QT prolongation pedigree found to harbor p.Arg36His-KCNE1 and p.His558Arg-SCN5A after pedigree-based WES analysis. Affected individuals are indicated by gray circles/squares. Unaffected individuals are indicated by white circles/squares. Heart rate-corrected QT interval ranges (QTc) are displayed beneath each individual. WES variant filtering strategy is displayed to the right of each respective transient QT prolongation pedigree. LQTS = long QT syndrome; MAF = minor allele frequency; WES = whole exome sequencing.
The first transient QT prolongation pedigree was identified after the proband (II1) experienced a presyncopal episode during soccer practice (Figure 2A). Serial ECGs revealed transient QT prolongation (QTc 442 ms to 485 ms). Screening of first-degree relatives revealed a similar phenotype in the index case’s brother (QTc 429 ms to 495 ms) and mother (QTc 463 ms). The biological father had a normal QTc (415 ms). Research-based WES revealed that the affected proband, brother, and mother were positive for p.Asp85Asn-KCNE1, whereas the unaffected father was negative for this functional common variant (Figure 2A). No other rare or common variants in the 17 published LQTS-susceptibility genes were shared between affected individuals.
The second transient QT prolongation pedigree was identified after a screening ECG obtained on the asymptomatic proband (II1) revealed QT prolongation (QTc 459 ms to 481 ms) (Figure 2B). Screening of first-degree relatives was initiated and revealed the patient’s father had marked QT prolongation during exercise recovery (QTc 5 500 ms at 1 minute of recovery). His mother’s evaluation was normal. Research-based WES revealed that the proband and his affected father shared a relatively common variant in KCNE1, p.Arg36His-KCNE1, observed in 21 of 63,310 individuals (0.03%) in gnomAD and the common p.His558Arg-SCN5A polymorphism (Figure 2B). Both variants were absent in the unaffected mother. In order to determine whether p.Arg36His-KCNE1, like p.Asp85Asn-KCNE1, could be contributing to the proband’s transient QT prolongation phenotype and to provide additional evidence needed to elevate this variant from VUS status, we proceeded to characterize this variant.
Functional characterization of the transient QT prolongation-associated p.Arg36His-KCNE1 genetic variant
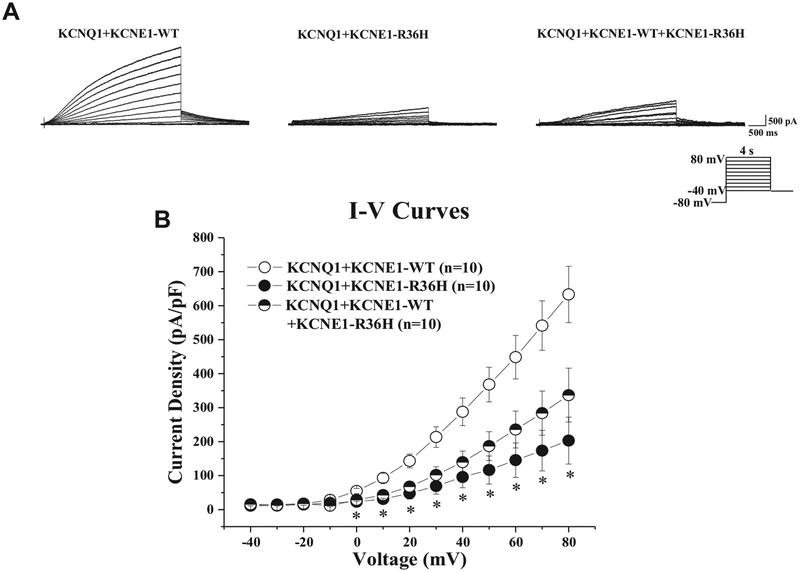
Typical IKs tracings of voltage-dependent activation from wild-type KCNQ1/Kv7.1 plus wild-type KCNE1, wild-type KCNQ1/Kv7.1 plus homozygous p.Arg36His-KCNE1 overexpression, and wild-type KCNQ1/Kv7.1 plus heterozygous p.Arg36His-KCNE1 overexpression with holding potential at −80 mV to various depolarization potentials are shown in Figure 3A. Analysis of the current-voltage relationship revealed that IKv7.1 (IKs) current densities were significantly reduced by both the homozygous and heterozygous coexpression of p.Arg36His-KCNE1 across the voltage from 0 mV to +80 mV compared with wild-type (P <.05), indicating an IKs loss of function. At +80 mV, the IKs current density was significantly reduced by 67.9% and 46.8%, respectively, from 633.1 ± 82.8 pA/pF (wild-type, n = 10) to 203.2 ± 69.0 pA/pF (homozygous p.Arg36His-KCNE1 overexpression, n = 10) and to 336.7 ± 79.3 pA/pF (heterozygous p.Arg36His-KCNE1 overexpression, n = 10) (Figure 3B).
Figure 3.
Transient QT prolongation-associated p.Arg36His-KCNE1 variant reduces IKs current density in vitro. A: Whole cell IKs current representative tracings from TSA201 cells expressing KCNQ1+KCNE1-WT or KCNQ1+KCNE1−R36H or KCNQ1+KCNE1−WT+KCNE1−R36H determined from a holding potential of −80 mV and testing potentials from −40 mV to +80 mV in 10-mV increments with 4-second duration. B: Current-voltage (I-V) relationship for KCNQ1+KCNE1−WT (n = 10), KCNQ1+KCNE1−R36H (n = 10), and KCNQ1+KCNE1−WT+KCNE1−R36H (n = 10). All values given as mean ± SEM. *P <.05 vs KCNQ1+KCNE1−WT. WT = wild-type.
Discussion
LQT5-Lite: Phenotype of p.Asp85Asn-KCNE1-positive patients without additional rare variants in LQTS-susceptibility genes presenting to a dedicated Genetic Heart Rhythm Clinic
After the discovery that rare KCNE1 variants cause LQTS type 5 (LQT5), now denoted as KCNE1-LQTS,12 subsequent research has shown that common KCNE1 variants, most notably p.Asp85Asn-KCNE1, may confer susceptibility for a proarrhythmic phenotype, typically in the setting of known exogenous and endogenous QT-aggravating factors.13
Although p.Asp85Asn-KCNE1 is present in 0.8%–1.4% of the general population, it has been associated with longer baseline QTc values in the general population14,15 and an increased risk of acquired LQTS when an individual’s “repolarization reserve” is challenged by QT-prolonging medications, electrolyte abnormalities, or structural heart disease.10,13 Furthermore, p.Asp85Asn-KCNE1 may function as a weak/low penetrant congenital LQT5-type pheno-type in certain genetic backgrounds9 and may modify the disease severity associated with disease-causative variants in the canonical LQTS-susceptibility genes.16
In the present study, p.Asp85Asn-KCNE1 was overrepresented in patients presenting to a dedicated Genetic Heart Rhythm Clinic for evaluation of suspected LQTS (MAF 2.6% vs 1.2%; P = .0001). In the absence of additional variants in LQTS-susceptibility genes, the presence of p.Asp85Asn-KCNE1 was associated with a mild female-predominant form of LQTS characterized by transient QT prolongation with marked QTc variability (mean 50 ms between shortest and longest recorded intervals) we have termed “LQT5-Lite.” Although the observed female predominance suggests an additive effect of p.Asp85Asn-KCNE1 to the estrogen-mediated repolarization reserve deficit present in females,17 by virtue of their referral to a dedicated Genetic Heart Rhythm Clinic, the aggregate phenotype associated with p.Asp85Asn-KCNE1 in this study may reflect phenotypic extremes present in the ~1% of the general Caucasian population that is heterozygous for p.Asp85Asn-KCNE1. Therefore, as a result of this inherent selection bias, the p.Asp85Asn-KCNE1 patients enrolled in this study may be enriched for repolarization reserve-deficient genetic backgrounds and/or additional QT-prolonging risk factors that were not assessed in the current study. As such, a random sample of p.Asp85Asn-KCNE1-positive individuals from the general population may not manifest the same degree of aggregate phenotype observed in this study.
Nevertheless, this study suggests that a subset of p.Asp85Asn-KCNE1-positive individuals as well as those with comparatively less common KCNE1 variants such as p.Arg36His-KCNE1, even in the absence of other disease-contributing variants, may manifest a mild/weak form of LQTS that we denote as “LQT5-Lite.” Although one cannot assume that the majority of patients with so-called LQT5-Lite–causative variants will manifest similar mild/weak yet discernible QT phenotypes, this study does provide further evidence that mild loss-of-function KCNE1 variants, including p.Arg36His-KCNE1 and p.Asp85Asn-KCNE1 (ie, KCNE1 functional risk alleles), are not benign and at a minimum likely confer potential for LQT5-Lite status in settings in which repolarization reserve is endogenously (genetic background, effect of female hormones, etc) or exogenously (QT-prolonging medications, electrolyte abnormalities, etc) impaired. Therefore, until the precise role of genetic background in the genesis of LQT5-Lite can be elucidated, it is necessary to properly identify patients with p.Asp85Asn-KCNE1 and other LQT5-Lite–causative variants who are potentially at risk for LQT5-Lite in order to advise and institute simple but potentially lifesaving QT precautionary measures, such as drugs to avoid whenever possible/feasible (www.crediblemeds.org).
LQT5-Lite–causative KCNE1 variants: Monogenic determinants or polygenic drivers?
As shown in this study, functional risk alleles such as p.Asp85Asn-KCNE1, whose MAFs in public exomes substantially exceed disease prevalence in public exomes, seem to be capable, albeit inconsistently, of manifesting QTc values comparable to those observed in individuals with bona fide congenital LQTS. Furthermore, comparatively rarer KCNE1 genetic variants, such as the functional p.Arg36His-KCNE1 (0.03%) variant described in this study and even classic LQT5-causative variants such as p.Asp76Asn-KCNE1 (0.02%),12 are present at MAFs in public exomes that approach the prevalence of all LQTS sub-types combined (1:2500 [0.04%]) and substantially exceed the anticipated prevalence for 1% LQTS subtypes such as LQT5 (1:250,000 [0.0004%]).
Despite their frequency in public exomes, p.Asp85Asn-KCNE1,9 p.Asp76Asn-KCNE1,12 and now, thanks to the current study, p.Arg36His-KCNE1 have been associated in isolation with a phenotype consistent with congenital LQTS. However, the discordance between the genotype prevalence of these KCNE1 variants and the estimated prevalence of the KCNE1-LQTS/LQT5 subtype as well as similar discordances observed for dozens of other functional variants previously deemed to be congenital LQTS-causative remain unexplained. Fortunately, there seem to be several plausible explanations for these genotype-phenotype discordances.
First, KCNE1-LQTS/LQT5 may account for a larger proportion of congenital LQTS and/or the widely accepted congenital LQTS prevalence of 1:2500 may be an underestimate. Given this scenario, KCNE1-mediated LQT5-Lite is viewed as a relatively common monogenic form of LQTS that resides on the mild end of the congenital LQTS disease severity spectrum (with the severe end comprising fully penetrant disorders caused by the canonical LQTS genes, CALM1–3, TRDN, etc). Although this would likely explain why the functional p.Arg36His-KCNE1 and p.Asp76Asn-KCNE1 variants are overrepresented in gnomAD, it fails to adequately explain why the common p.Asp85Asn-KCNE1 functional variant, whose cumulative effect on QTc in a prior population-based genome-wide association meta-analysis was ~7 ms,18 is associated with a discernible “LQT5-Lite” phenotype in the present study.
For p.Asp85Asn-KCNE1, and perhaps to a lesser extent p.Arg36His-KCNE1 and p.Asp76Asn-KCNE1, the possibility of oligogenic or polygenic models of disease provides a second plausible explanation. Several studies have demonstrated that the aggregate effect of multiple QT-influencing common variants, as assessed by weighted-effect genetic risk scores, can predict both interindividual variability in baseline QTc duration and an exaggerated QTc response/torsades de pointes risk after exposure to a medication with known QT-prolonging potential.19,20 As such, it stands to reason that those p.Asp85Asn-KCNE1-positive individuals who ultimately were referred to our tertiary medical center possess p.Asp85Asn-KCNE1 and a repolarization reserve-deficient background driven by QT-prolonging variants such as those in NOS1AP,21,22 the 3′ untranslated region of KCNQ1,23 and those shown through genome-wide association studies to prolong QTc duration in health.24
Unfortunately, the majority of p.Asp85Asn-KCNE1-positive individuals in this study underwent commercial LQTS genetic testing, so the genetic substrate needed to test this polygenic hypothesis was not immediately available. As such, additional genomic studies and potentially a function-guided induced pluripotent stem cell–derived cardiomyocyte approach akin to that recently described by Chai et al25 to describe the modification of LQT2 disease severity by a common variant in REM2 will be needed to fully appreciate the role of p.Asp85Asn-KCNE1 in the generation of the LQT5-Lite phenotype observed in this study.
Implications for clinical genetic test reporting
The 2015 ACMG/AMP guidelines specifically avoided guidance on how common variants, which may serve as modifiers for heritable diseases or cause concealed/subclinical but clinically relevant forme fruste heritable diseases, should be classified and reported. As a result, those clinical laboratories that adopted ACMG/AMP-based classification strategies were given latitude to interpret (or misinterpret) how relatively common variants, including p.Asp85Asn-KCNE1, typically classified as “benign variants” or “likely benign variants” on the basis of MAF, are reported (or not reported) on diagnostic LQTS genetic tests.8 Unfortunately, the prevailing interpretation has resulted in the exclusion or sequestration of these potentially clinically relevant common genetic variants (ie, the functional risk alleles), such as p.Asp85Asn-KCNE1, to supplemental reports that must be requested by the ordering health care providers. Therefore, critical genetic information may not be disclosed to the patient and the health care provider(s).
As illustrated by the case of the p.Asp85Asn-KCNE1-positive transient QT prolongation pedigree described in this study, the failure to report p.Asp85Asn-KCNE1 almost certainly resulted in unnecessary diagnostic ambiguity. Had the presence of p.Asp85Asn-KCNE1 been disclosed on the initial commercial genetic testing report, the mild transient QT prolongation phenotype observed in all 3 individuals could have been attributed easily to this well-known “functional risk allele” and the costly and time-consuming exercise of sequencing and analyzing this family’s exomes avoided. Instead, the probable primary genetic driver for the family’s phenotype was discovered nearly 2 years after the initial decision to pursue diagnostic LQTS genetic testing and only after costly research laboratory-based WES was instituted with the goal of elucidating a novel genetic substrate. Unfortunately, this example highlights the pressing need to implement mechanisms that ensure that so-called functional risk alleles such as p.Asp85Asn-KCNE1 are reported to patients and their health care provider(s) by means that minimize the risk of potential diagnostic miscues.
To this end, a common variant classification mechanism, based on existing criteria within the 2015 ACMG/AMP guidelines, was proposed recently to differentiate so-called functional risk alleles, such as p.Asp85Asn-KCNE1, from ambiguous VUS, likely benign variants, and benign variants.8 Regardless of how clinical genetic testing laboratories ultimately elect to communicate the presence/absence of functional risk alleles such as p.Asp85Asn-KCNE1, if such variants are encountered on diagnostic LQTS or broader pan-cardiac gene panel testing for suspected sudden cardiac death–predisposing syndromes, they should be disclosed and the patient and the family afforded every opportunity to receive appropriate counseling regarding relevant QT precautionary measures. Furthermore, if p.Asp85Asn-KCNE1 is identified through clinical whole exome/genome sequencing of diagnostic odyssey cases, appropriate precautionary measures should be advised and a surveillance ECG obtained to determine whether the patient is exhibiting an LQT5-lite phenotype. Presently, rare variants in KCNE1 in general and p.Asp85Asn-KCNE1 in particular do not reside on ACMG’s list of “reportable” incidental findings in clinical exome/genome sequencing but probably should for the reasons given herein.26 Finally, it remains to be seen whether it is time to proactively identify the ~1% of otherwise healthy individuals who are p.Asp85Asn-KCNE1-positive, thereby facilitating pre-prescription genotyping before exposure to medications with known QT-prolonging/torsadogenic potential.
Study limitations
The patients included in this study were assessed in the context of a referral institution with expertise in heritable cardiac channelopathies. Because of this referral bias, included patients may represent the more severe end of this mild p.Asp85Asn-KCNE1 phenotype. Additional work is needed to determine whether the mild shared transient QT prolongation phenotype associated with isolated p.Asp85Asn-KCNE1 in this referral cohort is generalizable or whether the LQT5-Lite patients who presented clinically in this study did so because of the presence of p.Asp85Asn-KCNE1 in an otherwise repolarization-deficient genetic background.
Conclusion
The common variant p.Asp85Asn-KCNE1 is associated with a female-predominant phenotype of transient QT prolongation, with milder symptomatology but remaining susceptibility to adverse events stemming from QT prolongation. Clinical laboratories that routinely conduct diagnostic LQTS, pan-cardiac, and clinical whole exome/genome sequencing are urged to implement mechanisms to ensure the appropriate classification and reporting of p.Asp85Asn-KCNE1 and other functional risk alleles. In doing so, patients and ordering health care providers can be assured they have received as accurate and complete a depiction of genetic risk as possible and can be afforded every opportunity to have simple precautionary measures instituted in order to mitigate the small but increased risk of arrhythmia associated with the presence of functional risk alleles such as p.Asp85Asn-KCNE1.
Supplementary Material
Footnotes
Appendix
Supplementary data
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.hrthm.2018.03.038.
References
- 1.Giudicessi JR, Ackerman MJ. Genotype- and phenotype-guided management of congenital long QT syndrome. Curr Probl Cardiol 2013;38:417–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giudicessi JR, Kullo IJ, Ackerman MJ. Precision cardiovascular medicine: state of genetic testing. Mayo Clin Proc 2017;92:642–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm 2011;8:1308–1339. [DOI] [PubMed] [Google Scholar]
- 4.Ackerman MJ, Tester DJ, Jones GS, Will ML, Burrow CR, Curran ME. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc 2003;78:1479–1487. [DOI] [PubMed] [Google Scholar]
- 5.Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm 2004;1:600–607. [DOI] [PubMed] [Google Scholar]
- 6.Ackerman MJ. Genetic purgatory and the cardiac channelopathies: exposing the variants of uncertain/unknown significance issue. Heart Rhythm 2015; 12:2325–2331. [DOI] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giudicessi JR, Roden DM, Wilde AAM, Ackerman MJ. Classification and reporting of potentially proarrhythmic common genetic variation in long QT syndrome genetic testing. Circulation 2018;137:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishio Y, Makiyama T, Itoh H, et al. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J Am Coll Cardiol 2009; 54:812–819. [DOI] [PubMed] [Google Scholar]
- 10.Kaab S, Crawford DC, Sinner MF, et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ Cardiovasc Genet 2012;5:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boczek NJ, Best JM, Tester DJ, Giudicessi JR, Middha S, Evans JM, Kamp TJ, Ackerman MJ. Exome sequencing and systems biology converge to identify novel mutations in the L-type calcium channel, CACNA1C, linked to autosomal dominant long QT syndrome. Circ Cardiovasc Genet 2013;6:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet 1997;17:338–340. [DOI] [PubMed] [Google Scholar]
- 13.Weeke P, Mosley JD, Hanna D, et al. Exome sequencing implicates an increased burden of rare potassium channel variants in the risk of drug-induced long QT interval syndrome. J Am Coll Cardiol 2014;63:1430–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouas L, Nicaud V, Chaouch S, Berthet M, Forhan A, Tichet J, Tiret L, Balkau B, Guicheney P. Confirmation of associations between ion channel gene SNPs and QTc interval duration in healthy subjects. Eur J Hum Genet 2007;15:974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marjamaa A, Newton-Cheh C, Porthan K, et al. Common candidate gene variants are associated with QT interval duration in the general population. J Intern Med 2009;265:448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahtinen AM, Marjamaa A, Swan H, Kontula K. KCNE1 D85N polymorphism—a sex-specific modifier in type 1 long QT syndrome? BMC Med Genet 2011; 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varró A, Baczkó I. Cardiac ventricular repolarization reserve: a principle for understanding drug-related proarrhythmic risk. Br J Pharmacol 2011;164:14–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arking DE, Pulit SL, Crotti L, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 2014;46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noseworthy PA, Havulinna AS, Porthan K, Lahtinen AM, Jula A, Karhunen PJ, Perola M, Oikarinen L, Kontula KK, Salomaa V, Newton-Cheh C. Common genetic variants, QT interval, and sudden cardiac death in a Finnish population-based study. Circ Cardiovasc Genet 2011;4:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss DG, Vicente J, Johannesen L, Blinova K, Mason JW, Weeke P, Behr ER, Roden DM, Woosley R, Kosova G, Rosenberg MA, Newton-Cheh C. Common genetic variant risk score is associated with drug-induced QT prolongation and torsade de pointes risk: a pilot study. Circulation 2017;135:1300–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ, George AL Jr. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation 2009;120:1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomas M, Napolitano C, De Giuli L, Bloise R, Subirana I, Malovini A, Bellazzi R, Arking DE, Marban E, Chakravarti A, Spooner PM, Priori SG. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J Am Coll Cardiol 2010;55:2745–2752. [DOI] [PubMed] [Google Scholar]
- 23.Amin AS, Giudicessi JR, Tijsen AJ, et al. Variants in the 3’ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J 2012;33:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res 2013; 161:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai S, Wan X, Ramirez-Navarro A, Tesar PJ, Kaufman ES, Ficker E, George AL Jr, Deschenes I. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest 2018;128:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.