Abstract
Double-exclusive liquid repellency (double-ELR) is an extreme wettability phenomenon in which adjacent regions selectively and completely repel immiscible liquids with different surface chemistries on a non-textured substrate (i.e., a substrate in absence of micro/nano-structures). Under double-ELR conditions, each liquid exhibits no physical contact (contact angle of 180°) with its non-preferred surface chemistry, thus enabling complete partitioning of adjacent fluidic volumes (e.g., between water and oil). This enables a new type of cell culture-based assay, where cell loss from common failure modes (e.g., biofouling from inadvertent cell adhesion, detrimental moisture loss/gain, and liquid handling dead volumes) is significantly mitigated. Importantly, the principles of double-ELR were leveraged to achieve underoil sweep patterning, a no-loss, robust and high-throughput distribution of sub-microliter volumes of aqueous media (and cells). In addition to high-efficiency distribution via sweep patterning, double-ELR can be used to construct “modular” (i.e., easily implemented and/or linked together with spatial and temporal control) higher-order architectures for in vitro imitation of physiologically relevant microenvironments that are of particular interest within the cell assay community, including multi-phenotype cultures with excellent spatial and temporal control, three-dimensional layered multi-phenotype cultures, cultures with selective mechanical cues of extracellular matrix (i.e., collagen fiber alignment), and spheroid cultures. Together, these features of double-ELR uniquely facilitate culture and high content analysis of limited cellular samples (e.g., a few hundred to a few thousand cells).
Graphical abstract
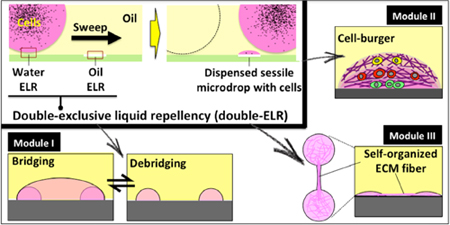
Complex cell-culture microenvironments with both spatial and temporal resolutions are enabled by double-exclusive liquid repellency and underoil sweep patterning.
Introduction
Analytical assays in cell biology have variable sample size requirements, with many traditional assays requiring a large number of cells to ensure robust performance. However, there is an existing need to manipulate and analyze small (i.e., a few hundred to a few thousand) populations of cells, particularly for the analysis of clinical samples. For instance, recent studies of heterogeneity in small clinical samples (e.g., liquid biopsies, fine needle aspirates, bronchoalveolar lavage) have revealed new biological insights.1,2 While such samples hold potentially critical information, particularly in light of the emergence of precision medicine, they place additional demands on assay technologies, as assay artifacts will likely affect small samples more significantly. These demands are further compounded when measurements of sample heterogeneity (i.e., multiple parallel measurements of a single or a few cells) are required.3,4 Specifically, it is critical to minimize or eliminate cellular and/or molecular loss due to adsorption, biofouling, mishandling, or assay dead volumes while simultaneously facilitating high throughput measurement. Furthermore, these assay criteria should be obtained while maintaining relative operational simplicity in order not to impede assay adoption in clinics and other locations where they are likely to generate the highest impact.
To address this important need, we present a cell culture-based assay platform with the following characteristics: (I) The ability to robustly process small volumes (less than a few microliters), thus facilitating efficient manipulation of small samples (e.g., those consisting of less than a few thousand cells);4,5 (II) Passive-yet-accurate metering of samples to ensure efficient sample utilization;6 (III) Safeguards against detrimental evaporation or condensation, which can affect biological relevance (e.g., cellular stress responses triggered by non-isotonic conditions);7,8 (IV) Elimination of adsorption/biofouling phenomena, which has been demonstrated to negatively affect cell signaling;9 (V) Facilitation of the construction of complex architectures incorporating multiple cell phenotypes and/or ECM structures, which will enable more relevant biological assays [e.g., three-dimensional (3D) layered cell culture]; (VI) Phenotypes of interest remain easily retrievable, to facilitate additional downstream analyses;10–12 (VII) Phenotype screening is high throughput,13 without the need for specialized components; (VIII) Low barriers to adoption, including low cost manufacturing, minimal infrastructure requirements, simple operation, and robust (non-fragile) components.14,15
Historically, researchers have employed microfluidics to attempt to address these needs.15–17 However, the ability to merge high relevancy assays (Points I-V from the prior paragraph) and broad-scale accessibility (Points VI-VIII from above) is an area that would benefit from further improvements. In a recent publication, Walsh and co-workers developed a system termed “Freestyle Fluidics” where aqueous media and cell samples are printed on various substrates (e.g., plastic or glass) and then overlaid by a layer of immiscible liquid (e.g., oil).18 This design offers both highly flexible physical accessibility to samples by external tools (e.g., for loading and retrieving cells of interest simply by pipetting through the oil layer) and minimized evaporation. The system was successfully used to study the inflammatory response of human cells and chemotaxis in bacterial biofilms. This study addressed many of the aforementioned requirements, but lacked some features that would further enable high content analysis of rare cells. To achieve our goal of a high-performance assay platform meeting all of our defined criteria, a thorough consideration of the physics in control of liquid repellency and improved cell sample processing and manipulation in a multi-liquid-phase system are needed.
Recently, our group has developed an open multi-liquid-phase technology for rare cell culture and single cell processing.19 As illustrated by Walsh et al., multiphase systems offer unique advantages that address many of the unmet needs in rare sample assay technology. Here, we further streamline assay loading and increase assay robustness and versatility through the application of ELR “double lock” (or double-ELR), a condition where a heterogeneous surface (defined here as a surface with multiple regions of different surface chemistries) alternatively and absolutely repels one of two liquids. For example, on a heterogeneous surface of glass and polydimethylsiloxane (PDMS), a system involving silicone oil (SO) and water will exhibit complete repellency [ELR; with contact angle (CA) of 180°] of water over the PDMS regions under oil and ELR of SO over the glass regions under water. Thus, in double-ELR, each liquid will only contact one of the surfaces. This concept provides a number of practical advantages, including: (I) Elimination of inadvertent adsorption/biofouling due to residual contact [even “superhydrophobic” surfaces with CA > 150° still have some liquid contact with the surface, which often negatively affects repellency of biosamples (e.g., proteins, biosurfactants, cells and/or microorganisms) on the surface, especially from long-term contact];20–22 (II) The long term stability of double-ELR enables “underoil sweep patterning” for advanced biosample [e.g., cells and/or extracellular matrix (ECM)] handling and versatile cell culture-based assays; (III) Double-ELR facilitates the formation of more complex 3D structures that allow for more comprehensive phenotypic interrogations.
Results
1. Undermedia liquid repellency on non-textured substrates
Double-ELR is successfully achieved when each liquid (e.g., oil, aqueous) is completely repelled (with CA = 180°) by one of two device surfaces (e.g., glass and PDMS). This is in contrast to homogeneous surfaces (substrates with uniform surface chemistry) and more traditional heterogeneous surfaces, where contact with one liquid is favored, but the second is not completely repelled. We term this second, non-ELR regime finite liquid repellency (FLR; with CA < 180°) (Fig. 1A).19,23 The substrates referred to in this work [i.e., glass (O2 plasma-treated), polystyrene (PS), PDMS, glass-PDMS patterned slide, and O2 plasma patterned PS] are all non-textured (see SI Methods), thus enabling: (I) Easy-to-acquire surfaces that are well characterized for cell culture;24 (II) High-quality optical access for maximum resolution imaging;17 (III) Robust mechanical durability. Three oils were used in this study, including SO (5 cSt), the default oil in “underoil” systems unless stated otherwise, mineral oil (MO) and Fluorinert FC-40 (FC).
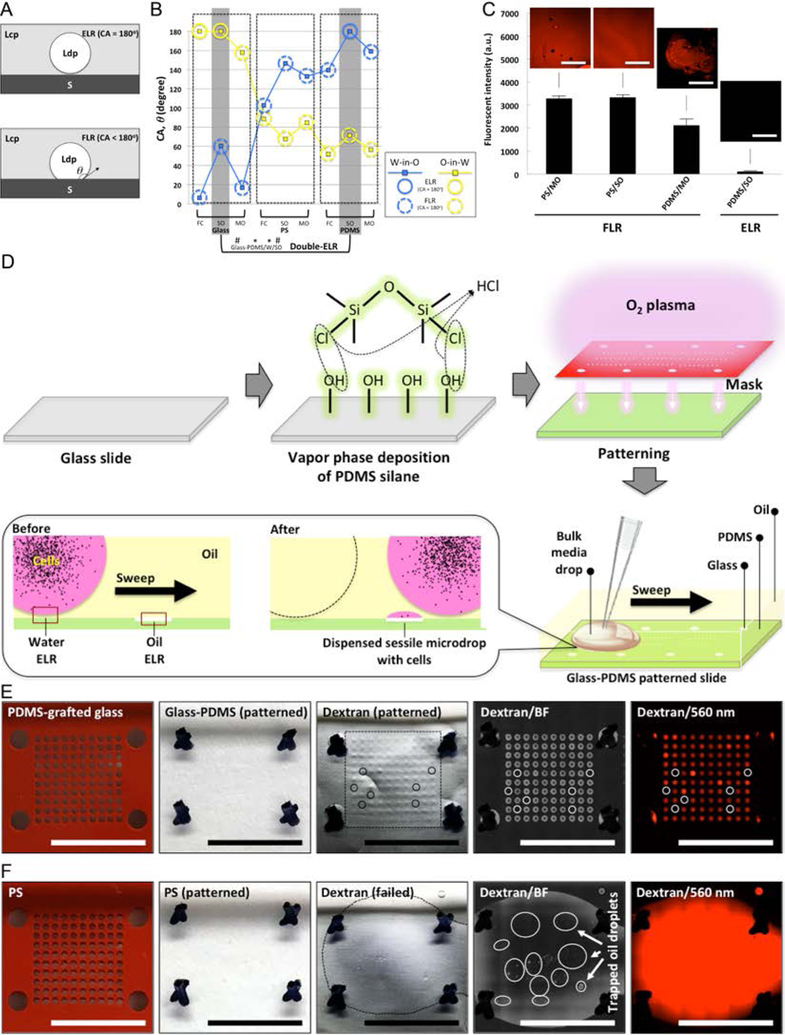
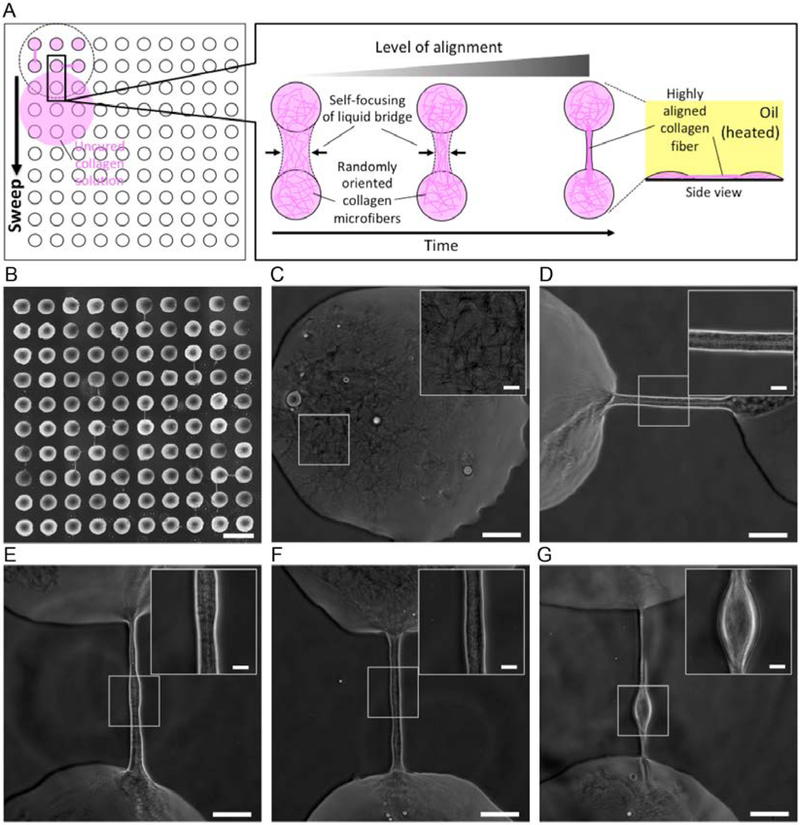
Fig. 1. ELR and FLR in multi-liquid-phase system and double-ELR-enabled underoil sweep patterning.
(A) Schematic of ELR (with CA, ϑ = 180°) and FLR (with ϑ < 180°). S - solid; Ldp - liquid of dispersed phase; Lcp - liquid of continuous phase. (B) CAs measured in water-in-oil (W-in-O, blue line) and oil-in-water (O-in-W, yellow line) conditions on three homogeneous substrates [glass (O2 plasma-treated), PS, and PDMS] with different oils (measurement error, ± 2°). Homogeneous substrate shows only single-ELR (e.g., glass/FC) at best to either water or oil. On heterogeneous substrate (by combining any two of the homogenous substrates, e.g., glass-PDMS/W/SO), double-ELR to both water and oil can be realized (table S2). (C) Fouling test on homogeneous substrates in ELR and FLR with four model systems (PS/MO, PS/SO and PDMS/MO for FLR; PDMS/SO for ELR). 1 mL dextran solution (Texas Red, 10000 MW, 1.0 mM in PBS) was added onto each substrate under oil and then removed shortly (< 10 seconds). Fluorescent intensity was recorded to check the residual of dye on substrate. It is clear only ELR effectively blocked substrate fouling or random sample loss from short contact with the dye solution. (D) Schematic of fabrication of glass-PDMS patterned slides and underoil sweep patterning based on double-ELR. The background (PDMS) shows ELR to water under oil. In contrast, the patterned area (glass) shows ELR to SO under water. A certain volume of culture media with cells (black dots) can be automatically and continuously dispersed onto spots by sweeping a bulk media across the substrate. Sweeping can be executed by dragging a liquid droplet across the surface with a pipet tip. (E) Underoil sweep patterning on a glass-PDMS patterned slide (containing 600 μm spots in a 10 × 10 array). 100 μL dextran solution was added to the spots via sweep then removed by a pipette. Unfilled (or empty) spots were labelled out with solid line circles. Variation of volume distribution measured in mean fluorescent intensity on each spot was shown in fig. S1. (F) Control with O2 plasma patterned PS. Sweep patterning became impractical due to FLR to water on PS under oil. 1 mL dextran solution was added instead to cover all of the spots then removed by a pipette. Discrete oil droplets trapped in the thin layer of aqueous solution were labelled out with solid line circles. In (E) and (F) from left to right: starting substrates (PDMS-grafted glass, PS) with silicone rubber mask (red), O2 plasma patterned substrates with mask removed, dextran-loaded substrates (the frames in black dashed line denote the solution sweeping or loading area on each substrate), media channel [brightfield (BF)] image, and dextran channel (560/607 nm) image. Scale bars: 500 μm in the insets of (C), 1 cm in (E) and (F).
At best, homogeneous surfaces will only exhibit single-ELR to either water or oil, but not both (i.e., double-ELR) within the same device (Fig. 1B). When ELR conditions are absent (i.e., FLR), the liquid will stick wherever it contacts the substrate, which leads to device fouling and/or sample loss from the liquid phase (Fig. 1C). In comparison, double-ELR (i.e., ELR to both water and oil) can be obtained on heterogeneous surfaces with properly tailored surface chemistry contrast (Fig. 1B, table S1, table S2) (generalized “design rules” for ELR and FLR can be found in our earlier publication).19 For example, on a glass-PDMS patterned slide, water and SO will prefer to remain on their “preferred surface” (i.e., the glass “surface” for water, the PDMS “surface” for SO). If either liquid experiences a force away from its preferred surface, it experiences ELR and is completely repelled from adhering on the substrate, thus preventing surface-associated loss.
2. Double-ELR-enabled underoil sweep patterning
Underoil sweep patterning was developed based on double-ELR for no-loss, high throughput distribution of small liquid volumes. Glass-PDMS patterned slides were prepared using a two-step treatment resulting in a glass coated with a patterned molecular layer of PDMS (Fig. 1D). Enabled by double-ELR, water and SO can be robustly confined on their preferred surfaces, as ELR will occur to any liquid outside of its preferred surface (Fig. 1B). Accordingly, a bulk media drop (e.g., 100 μL) can be loaded under oil anywhere and then swept across the substrate. A specific volume of the aqueous media will adhere to its preferred surface (glass in this case) while the oil is repelled via ELR. At the same time the background remains clean in the absence of any randomly attached droplets of aqueous media (Fig. 1E, fig. S2, movie S1), as the aqueous media experiences ELR in this regime (PDMS in this case). In contrast, such underoil sweep patterning completely fails when double-ELR is absent. An O2 plasma-patterned PS dish was prepared by selective O2 plasma treatment to give a heterogeneous surface with ELR-FLR and tested as a control (Fig. 1B, table S2). In this case, the preferred surfaces are the O2 plasma treated spots for water and the untreated PS background for SO. SO shows ELR on its non-preferred surfaces (i.e., the O2 plasma treated spots), which means SO is robustly confined on the untreated PS background and the invasion to the O2 plasma treated spots is inhibited. However, due to the double-FLR of the untreated PS background to both water and SO (table S2), the loaded aqueous media randomly invaded into the oil phase and was pinned on the substrate, leaving randomly scattered, trapped oil droplets across the PS background (Fig. 1F, movie S2).
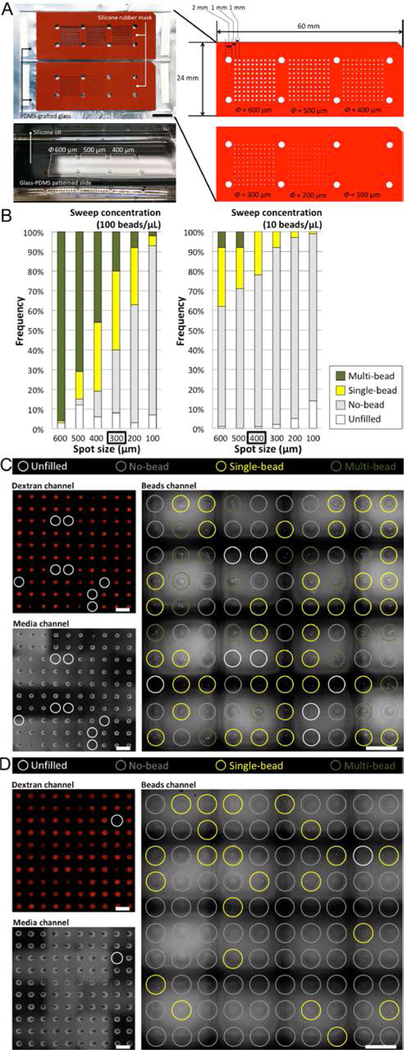
The frequency (or the probability) of a given cell count per spot follows Poisson distribution (Fig. 2A, 2B). The frequency of single-cell spot can be effectively regulated by sweep concentration (i.e., the concentration of cells in the bulk media drop) and the volume dispersed (which is mainly determined by spot size at a given viscosity of cell suspension). As such, stock concentration can be modulated to obtain different outcomes. For instance, maximized single-cell frequency (Fig. 2C) or minimized multi-cell frequency (Fig. 2D) strategies might be employed to achieve maximum single-cell throughput or maximum single-cell usage, respectively. While Poisson distribution is a well-known phenomenon, it can be utilized here to estimate the volume of the sessile droplets from underoil ELR sweep. The volume dispersed can be reversely estimated using the equation of Poisson distribution,25 P = e−λ(λk/k!), where P is the probability (or measured frequency, in this case) of occurrence for k events in interval (i.e., the number of cells per dispersed volume), λ is the average events in interval (i.e., the stock concentration of the solution), and e is the base of natural logarithms. For example, given (I) the theoretical maximum of P (36.8%) is achieved by setting k equal to λ then equal to 1 cell per dispersed volume, and (II) the maximized single-cell frequency (around 40%) in experiment is obtained on the 300 μm spots with a sweep concentration of 100 cells/μL (which is equal to 1 cell/10 nL), the averaged volume per spot dispersed on 300 μm spots can be estimated to be approximately 10 nL. The average volume per spot can be by dividing the total volume distributed by the total number of spots. Alternatively, the individual volume of each spot can be measured by using a laser interferometer to measure liquid height and combining this measurement with the known footprint of the spots (not included in this study).
Fig. 2. Poisson distribution of model particles from underoil sweep patterning.
(A) Silicone rubber masks (red, press-to-seal, 0.5 mm thick) on PDMS-grafted glass and dimension of the arrays of spots. A typical picture showed the distributed sessile droplets of water after underoil sweep patterning. (B) Distribution of model particles [fluorescent beads, 12 μm in diameter, in PBS with 1.0 mM dextran (Texas Red, 10000 MW) and 2 mg/mL collagen I] on glass-PDMS patterned slides containing spots (diameter: 100, 200, 300, 400, 500 and 600 μm in a 10 × 10 array for each size). The bead suspension was stabilized with collagen from settling down over time during sweep. Two sweep concentrations (100 beads/μL vs. 10 beads/μL) were used, aiming to achieve maximized single-cell frequency (left) and minimized multi-cell frequency (right). (C) Microscopic images of maximized single-cell frequency (40% single-bead vs. 20% multi-bead) from 100 beads/μL on 300 μm spots. (D) Microscopic images of minimized multi-cell frequency (22% single-bead vs. 0% multi-bead) from 100 beads/μL on 400 μm spots. In (C) and (D), dextran channel (560/607 nm, upper left), media channel (BF, lower left), and beads channel (485/525 nm, right) were used to check different combinations of the possible outcomes of sample distribution (fig. S2). Scale bars: 1 cm in (A); 1 mm in (C) and (D).
3. Modular applications in cell biology derived from underoil sweep patterning
While droplet arrays containing isolated single-cell phenotypes are valuable for some biological applications,3 the functionality of this assay type is clearly limited. Assays incorporating multiple phenotypes and/or elements of the ECM have recently emerged and have been demonstrated to possess enhanced biological/clinical relevance.26,27 For instance, our lab has extensive experience co-culturing multiple cell types in order to discern the effects of microenvironmental signaling, including assays to predict response to multiple myeloma treatment,28 discover mechanisms of resistance to breast cancer treatment,29 and interrogate new disease progression pathways in prostate cancer.30 Given our experience with these assays, we recognize the value of these assay types and aim to accommodate them in the ELR platform. In particular, our goal is the development of an assay platform that enables physiologically relevant measurement of cellular behavior in rare or limited samples. Fortunately, in addition to the capability of isolating and analyzing single-cell phenotypes, the double-ELR-enabled underoil sweep patterning can be further utilized to construct higher-order and hierarchical architectures considering multiple tiers of components in a physiological microenvironment with both spatial and temporal resolution and thus more assay relevancy without a marked increase in operational complexity. Importantly, each of these architecture “modules” can be implemented via sweep distribution on the standard double-ELR heterogeneous surface. Here, we describe construction of each module and preliminary demonstrations of functionality, in anticipation that readers will leverage the advantages of specific modules (or combinations of modules) to develop new assays that are both relevant and easy-to-use.
3.1. Module I—Underoil reconfigurable co-culture with liquid bridge
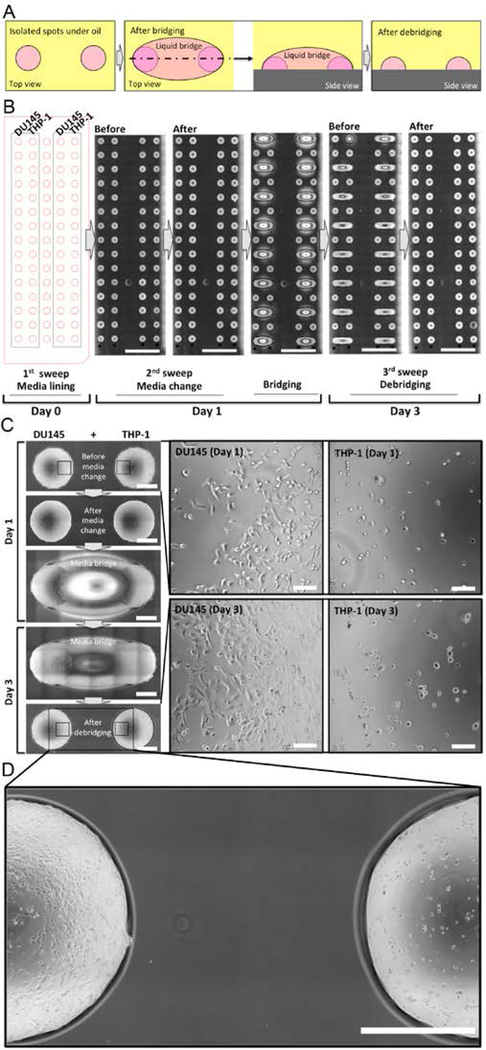
The stability of double-ELR enables us to achieve “reconfigurable co-culture”, where specific cells or cell populations can be repeatedly connected and disconnected with high spatial and temporal resolution (Fig. 3A). With double-ELR, aqueous media and oil can be easily and reliably confined on their preferred surfaces in a multi-liquid-phase system. The “invading” liquid remains completely repelled from touching the neighboring surface on substrate. In addition, it cleanly recedes to its pool after the external perturbation or driving force is removed. For example on a glass-PDMS patterned slide, two adjacent spots can be connected by adding extra volume of media in between to give a liquid “Cloud Gate” (a well-known sculpture at Millennium Park in Chicago, Illinois), i.e., a three-dimensional liquid structure with which crosstalk is enabled between the two connected spots through a liquid bridge, without compromising repellency between the liquid bridge and the substrate surface underneath (movie S3). Such a unique configuration allows reversible connection and disconnection of any two or more neighboring spots by simply adding and removing the liquid bridge (movie S4), thus providing flexibility to manipulate multiple phenotypes and their interactions with both spatial and temporal resolution. Combined with underoil sweep patterning, reconfigurable co-culture of different cell types can be envisioned. A human prostate cancer cell line (DU145) and a human acute monocytic leukemia cell line (THP-1) were chosen to demonstrate this concept and workflow (Fig. 3B). The two cell types were seeded under oil in adjacent rows of the spots by sweeping a small volume (e.g., 20 μL) of one of the cell stocks in each row at a time and given 24 hours to attach onto their spots. THP-1 cells were seeded with phorbol 12-myristate 13-acetate (PMA) to promote adhesion. The tumor spots and monocyte spots were bridged by adding culture media between the spots. After 48 hours, the co-cultured spots were debridged by removing the extra volume in the liquid bridge. Based on microscopic images, the cells were all seen sharply confined in their own spots, showing normal cell morphology during 2-day co-culture (Fig. 3C, 3D). In summary, Module I provides impeccable spatial and temporal resolution, thus enabling study of complex cellular processes (e.g., the invasion of tumor cells into surrounding tissues and distant organs).
Fig. 3. Underoil reconfigurable co-culture with liquid bridge.
(A) Schematic of bridging and debridging on two spots under oil. (B) Layout and workflow of underoil co-culture of tumor cells (DU145) and monocytes (PMA-treated THP-1) for 3 days. (C) High- magnification microscopic images from a pair of DU145/THP-1 spots at different time points. (D) High-magnification close up showing the edges of the spots and the area between the spots after debridging illustrating that cells were successfully confined within their own spots. Scale bars: 1 cm in (B); 1 mm in (C) and (D); 100 μm in the callouts of (C).
3.2. Module II—Underoil layer-by-layer cell stacking and 3D growth of single-cell derived spheroids
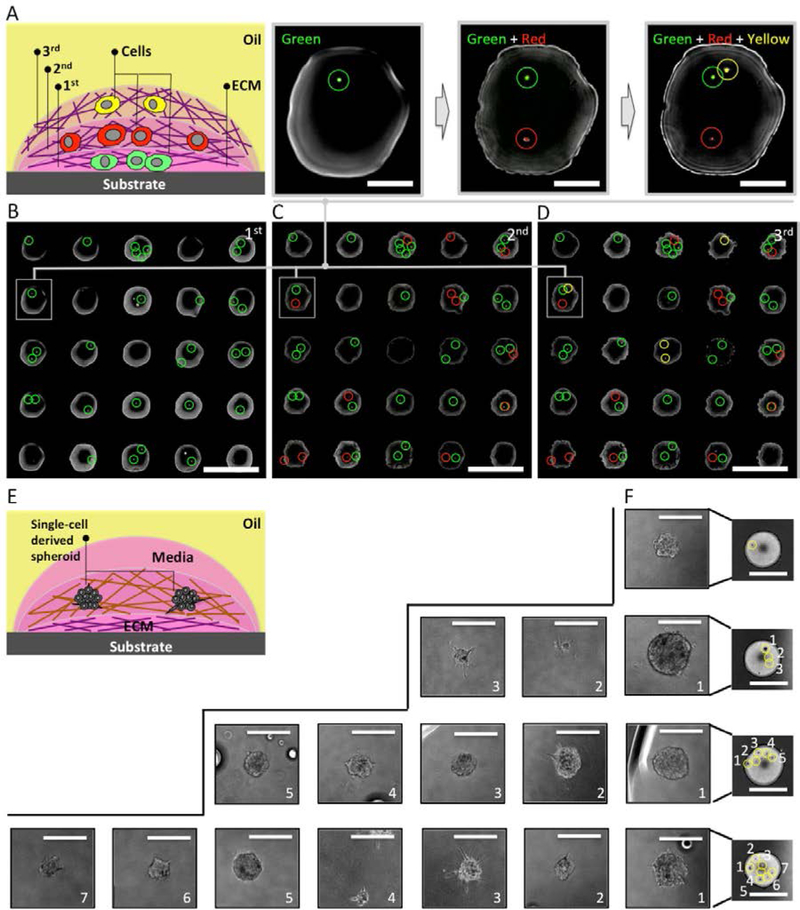
We further expanded underoil sweep patterning to create multi-phenotype microenvironments in cell culture via layer-by-layer cell stacking (Fig. 4A). As extensively demonstrated (by our group as well as others), different phenotypes (along with ECM) can be combined to simulate in vivo spatial distribution of cells and to study cell-to-cell and cell-to-ECM interactions.26,27 To execute layer-by-layer cell stacking, we first need to confirm that sweep distribution of additional cell/ECM layers would not disrupt the cells in the former layers. We determined that cells were securely retained within the intended layer without disruption or loss when the cells were embedded in collagen that is cured in situ after deposition of each layer (fig. S3). Retention of cell sample in ECM enables multiple sweeps for operations such as replenishment of culture media, collection of conditioned media with metabolites, and layer-by-layer cell stacking. As a proof-of-concept, THP-1 stained with cell tracker dyes (green, red and yellow) was used to represent three different cell types (Fig. 4B–4D). Each sweep added one type of cell, resulting in a three-layered “cell-burger”. The distribution of the cells in the former layer remained intact after the next layer was added. In Fig. 4D, distribution was governed by the Poisson statistics, illustrating how layer-by-layer cell stacking can be applied to rapidly construct co-cultures of all different cell combinations. Furthermore, long-term underoil 3D cell culture was introduced to grow single-cell derived tumor spheroids (from a prostate cancer cell line, C4–2) (Fig. 4E). The C4–2 cells confined in collagen were distributed in single-cell resolution at a cell density of 10 cells/μL on 2 mm-diameter spots by underoil sweep patterning. The cells were allowed to grow and proliferate into spheroids from a single mother cell for approximately two weeks (Fig. 4F), thus demonstrating the ability to harbor long-term culture in the double-ELR system. This experiment was later performed with primary epithelial cells obtained from a prostate cancer patient biopsy, thereby demonstrating successful culture of primary tissue (fig. S5). Module II enables complex interrogations of multiple cell phenotypes within an ECM, as encountered in in vivo microenvironments.26
Fig. 4. Underoil layer-by-layer cell stacking and 3D growth of single-cell derived spheroids.
(A) Schematic of a three-layered “cell-burger”. Cells were locked in ECM (matrigel, 5 mg/mL) in each layer. THP-1 cells in three different colors (green, red and yellow, 100 cells/μL) were distributed on a 5 × 5 array of spots (500 μm in diameter) after (B) the first (green), (C) the second (red), and (D) the third (yellow) sweep. (E) Schematic of the layout for a spot with single-cell derived tumor spheroids. Matrigel (5 mg/mL) was applied as the first layer of ECM under oil via sweep, followed by cells (10 cells/μL) embedded in the second layer of ECM (collagen I, bovine, 2 mg/mL). Culture media (5 μL) was added onto the spot at last. (F) Microscopic images (BF) of spots with different number of single-cell derived spheroids, showing heterogeneous growth dynamics and morphological phenotypes. Scale bars: 1 mm in (B)-(D); 200 μm in the callouts of (B)-(D); 2 mm in spot images of (F); 200 μm in the callouts of (F).
3.3. Module III—Underoil formation of self-organized highly aligned collagen fibers
Collagen fiber orientation plays an important role in cell signaling, but modulation of collagen fiber alignment is difficult to achieve in vitro.31 ECM fiber alignment provides directional ‘tracks’ for cancer cells to invade into surrounding tissue.32 For example, increased fiber alignment has been associated with aggressive breast tumor invasion and poor prognosis for breast cancer patients.33,34 Therefore, in vitro models that can reliably recapitulate ECM alignment would have utility for advancing our understanding of cancer progression and improving patient outcomes.35–37 In a double-ELR environment, highly aligned collagen fibers (width, 10–30 μm) can be selectively formed between spots (spacing, 400 μm) when sweeping uncured collagen solution under hot oil (Fig. 5A, fig. S4). Self-focusing of the liquid bridge enabled by a synergistic effect of double-ELR and Rayleigh instability during dewetting of the solution in underoil sweep was proposed as the mechanism.38 A similar mechanism can be found in forming the highly ordered nanofibers by dewetting and air bridging a polymer or polymer composite solution on micropillar arrays.39,40 In our ELR system, fiber formation can be selectively and repeatably induced by modulating the collagen curing conditions to promote curing during sweep distribution. Specifically, we sweep uncured collagen under oil that has been pre-heated (using a dry block heated to 50 °C) to polymerize the collagen in situ.41 In experiment, it turned out the orientation of the fibers between spots largely followed the direction of sweep thus the local dewetting of the collagen solution (Fig. 5B). In the direction of sweep, 19 fibers were formed out of 90 spacings. A couple of fibers were found formed in the direction perpendicular to the sweep but in a much lower rate (i.e., only 4 out of 90 spacings). Overall the yield is low, but that parametric optimization of the fiber formation process [e.g., temperature of the oil, concentration of the collagen solution, geometry of the surface pattern (i.e., size, spacing), and sweep velocity] should improve yield for those applications requiring more predictable fiber placement. It can be noticed that collagen microfibers were formed everywhere when media existed. On spots, the microfibers were randomly oriented, showing no clear alignment (Fig. 5C). In comparison, the microfibers converged into a bundle at the edge of a spot and assembled into a highly aligned fiber, forming a bridge to an adjacent spot (Fig. 5C–5G). Most of the fibers showed a varying width along the length direction and some of the fibers ended up with a spindle-shaped structure, which are likely a result of the competition between Rayleigh instability and the dramatically increased viscosity of the liquid bridge during the self-focusing process. To extend the functionality, Module III can be further combined with Module I (i.e., underoil reconfigurable co-culture with liquid bridge) and Module II (i.e., underoil layer-by-layer cell stacking) to create a complete microenvironment with both molecular and mechanical cues. For example, single cellular phenotypes can be built into cell- burgers, which can be further interconnected into higher-order sub-colonies via either the reconfigurable liquid bridge or the highly aligned collagen fibers.
Fig. 5. Underoil formation of self-organized highly aligned collagen fibers.
(A) Schematic of self-focusing of liquid bridge and alignment of collagen microfibers during underoil sweep. (B) A glass-PDMS patterned slide [containing a 10 × 10 array of spots (600 μm in diameter, 400 μm in edge-to-edge spacing)] after underoil sweep of collagen I solution (rat tail, 2 mg/mL in RPMI 1640). (C) Randomly oriented microfibers (width, < 1 μm) on spot. (D)-(G) Highly aligned collagen fibers (width, 10 to 30 μm) between spots in different orientations (horizontal/vertical). (insets) High-magnification close ups showing a typical contrast of the highly aligned microfibers in the bundle. The spindle-shaded structure found in (G) was likely a typical result of Rayleigh instability during self-focusing of the liquid bridge. Scale bars: 1 mm in (B); 100 μm in (C)-(G); 20 μm in the insets of (C)-(G).
Discussion
Revisiting the desired platform characteristics defined at the beginning of this manuscript, we have demonstrated that double-ELR is uniquely enabling for studying the behavior of limited biological samples. In particular, double-ELR offers precision in compartmentalization of low cell numbers (i.e., less than a few thousand), such that cells can be cultured, manipulated, and interrogated with minimal loss and excellent control, thus ensuring that the maximum amount of information is extracted from rare/precious samples. Here, we highlight some of the important differences between double-ELR and other existing technologies. (I) The concept of employing patterned surfaces with differential regions of wettability (or surface tension) for dispensing small volumes of liquid has been widely studied.42 Typically, these approaches utilize patterned superhydrophobic or superomniphobic (CA > 150° to water or to both water and oil) surfaces with superhydrophilic/superomniphilic (CA = 0° to water or to both water and oil) domains. While the background of such surfaces do show high repellency to bulk liquid, its intrinsic FLR nature (CA < 180°) can result in either device failure20,21 or sample loss due to biofouling when brought in contact with living organisms (e.g., cells, bacteria), especially for applications where the surface would be subjected to prolonged contact with the biospecimen (which could potentially affect the properties of the surface).22 (II) ELR does not require micro/nano-textured surfaces to achieve functionality. For instance, slippery liquid-infused porous surfaces (SLIPS) were invented by Leslie et al. to successfully inhibit biofouling.43 While effective, these strategies imposed additional fabrication requirements while potentially inhibiting optical clarity, thus preventing acquisition of high-quality imaging endpoints. (III) Several groups have reported patterning of selective aqueous regions in air (i.e., not under oil).42 While effective, these droplets are at increased risk of detrimental evaporation/condensation driven effects, even within a humidified incubator setting. Small perturbations in media component concentration can lead to dramatic stress responses,7,8 further compounding platform-derived artifacts. In contrast, detrimental media loss via evaporation can be greatly mitigated thus long-term cell culture in small volume of media becomes feasible by maintaining media drops (with cells) under oil (Fig. 4E). In the meantime, we have demonstrated permeation of vital gases (e.g., oxygen) in cell culture is allowed through the oil layer thus normoxic/hypoxic conditions can be selectively reached under oil (fig. S6).
While surface-mediated sample loss mechanisms are mitigated by double-ELR, there remains some concern that extraction of hydrophobic (or lipophilic) molecules from aqueous phase to oil phase could lead to molecular artifacts. Based on prior experiments, this may not be a significant issue, as we have demonstrated that ELR-based cell culture can be achieved with identical viability compared to traditional culture.19 However, for molecular endpoints, the absorption of lipophilic moieties into the oil phase might be a concern. In response, we first tested in SO and showed mRNA (admittedly a relatively hydrophilic molecule) extraction and quantification with quantitative reverse transcription polymerase chain reaction (RT-qPCR) can be achieved from varying cell densities (fig. S7). To minimize the possible loss of lipophilic molecules via extraction, we have developed an ELR system compatible with fluorinated oil. While fluorinated oil comes with very low partition coefficient for both hydrophilic and lipophilic molecules (if nonfluorinated), ELR cannot be directly achieved in a combination of a PDMS surface and pure fluorinated oil (Fig. 1B). However, ELR can be obtained under fluorinated oil by adding a small amount of fluorosurfactants (e.g., 0.2 wt% of fluoro-PEG-ylated surfactant). Detailed discussion can be found in our earlier publication.19 Potential inconvenience from using fluorinated oil might be caused by its high density (e.g., 1.86 g/mL for FC). Especially in ELR, water drops will float away to air-oil interface rather than settling down onto substrate. In order to perform sweep patterning under fluorinated oil with fluorosurfactant, we used a thin layer of oil (the thickness of oil layer is less than the height of the hanging drop) to avoid aqueous droplet floating. More oil can be added after without disrupting the distributed aqueous droplets if a thicker layer of oil is required.
While double-ELR is extremely robust, here we discuss specific situations where stability may be affected by kinetic effects and strategies that we have developed to achieve equilibrium/ELR more rapidly. First, one of the two liquids must be added first and this single liquid system will not represent double-ELR. For instance, on glass-PDMS patterned slides where a large volume of oil is added first, water will replace oil on its preferred surface (i.e., the glass spots) to favor a lower system free energy. Due to ELR, negligible oil can stay on the spots when exposed to and replaced by aqueous media, which allows the cells to directly face and attach on the glass surface for two-dimensional (or 2D) culture after sweep. While this is thermodynamically favorable, it still takes time for water to penetrate the oil layer and reach the glass surface. To accelerate dispersal, an anti-static gun (Milty Zerostat 3) was used (fig. S8). Opposite charges can be added alternately on the oil phase, which generates a local perturbation that facilitates penetration of water through the oil layer.44 Second, as described previously, extra aqueous volume can be added between two spots to form a liquid bridge under oil (movie S3). The stability of the liquid bridge to external perturbation (e.g., shaking, handling) can be described by the ratio of the bridge inertia to its surface tension, which is described by the Weber number, written as We = ρν2l/γW/O, where ρ is the density of the liquid, ν is the velocity of a perturbation (e.g., inadvertent shaking), l is the characteristic length (i.e., the length of the liquid bridge), and γW/O is the interfacial tension between water and oil. Therefore, short bridges are more stable, but provide more opportunity for motile cells to unwantedly “cross the bridge”. While understanding this tradeoff is important, a gel bridge can be used to impede cell migration while having little effect on the Weber number.
Conclusions
In summary, double-ELR directly addresses the parallel needs to provide more relevant in vitro assays while simultaneously reducing barriers-to-adoption for microfluidic technologies. In this manuscript, we describe the design conditions required to achieve double-ELR and demonstrate the utility of double-ELR via the demonstration of specific phenomena that are directly enabled by this specific but powerful combination surface chemistry criteria. First, we demonstrate that double-ELR facilitates sweep distribution, where arrays of precise sub-microliter droplets can be distributed with minimal infrastructure and effort. Next, we build upon underoil sweep patterning through the development of several modules, which (with minimal effort) create a number of structures of interest to the contemporary cell-based assay community, including co-cultures with spatial and temporal resolution, complex 3D co-cultures, and the combination of cell culture and directed ECM architecture. Ultimately, our aim with this manuscript is to describe a new open multi-liquid-phase technology and describe the structures and assay types enabled by this technology. Our hope is that this information will provide the cellular assay community with the knowledge base to build innovative assays that leverage the unique benefits of double-ELR.
Supplementary Material
Acknowledgements
This work is funded by National Science Foundation grant (EFRI-MKIS), Prostate Cancer Foundation Challenge Award, University of Wisconsin Carbone Cancer Center Cancer Center Support Grant P30 CA014520, NIH R01 EB010039 BRG, NIH R01 CA185251, NIH R01 CA186134, NIH R01 CA181648, and EPA H-MAP 83573701. We thank Dr. Max Gong for the discussion and insightful suggestions on the writing of Module III.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Goodspeed A, Heiser LM, Gray JW and Costello JC, Mol. Cancer Res, 2016, 14, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGranahan N and Swanton C, Cell, 2017, 168, 613–628. [DOI] [PubMed] [Google Scholar]
- 3.Hu P, Zhang W, Xin H and Deng G, Front Cell Dev Biol, 2016, 4, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakadan SM, Shalek AK and Weitz DA, Nat. Rev. Genet, 2017, 18, 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R and Koltay P, Int. J. Mol. Sci, 2015, 16, 16897–16919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singhvi R, Kumar A, Lopez G, Stephanopoulos G, Wang D, Whitesides G and Ingber D, Science, 1994, 264, 696–698. [DOI] [PubMed] [Google Scholar]
- 7.Ho SN, J. Cell. Physiol, 2005, 206, 9–15. [DOI] [PubMed] [Google Scholar]
- 8.Ip WKE and Medzhitov R, Nat. Commun, 2015, 6, 6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET and Beebe DJ, Lab Chip, 2009, 9, 2132–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaigala GV, Lovchik RD and Delamarche E, Angew. Chem. Int. Ed, 2012, 51, 11224–11240. [DOI] [PubMed] [Google Scholar]
- 11.Casavant BP, Berthier E, Theberge AB, Berthier J, Montanez-Sauri SI, Bischel LL, Brakke K, Hedman CJ, Bushman W, Keller NP and Beebe DJ, Proc. Natl. Acad. Sci. U. S. A, 2013, 110, 10111–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Moore JS and Beebe DJ, Science, 2001, 291, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 13.Du G, Fang Q and den Toonder JMJ, Anal. Chim. Acta, 2016, 903, 36–50. [DOI] [PubMed] [Google Scholar]
- 14.Paguirigan AL and Beebe DJ, Bioessays, 2008, 30, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh DI 3rd, Kong DS, Murthy SK and Carr PA, Trends Biotechnol, 2017, 35, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sackmann EK, Fulton AL and Beebe DJ, Nature, 2014, 507, 181–189. [DOI] [PubMed] [Google Scholar]
- 17.Aleklett K, Toby Kiers E, Ohlsson P, Shimizu TS, Caldas VEA and Hammer EC, ISME J, 2017, 12, 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh EJ, Feuerborn A, Wheeler JHR, Tan AN, Durham WM, Foster KR and Cook PR, Nat. Commun, 2017, 8, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Yu J, Schehr J, Berry SM, Leal TA, Lang JM and Beebe DJ, ACS Appl. Mater. Interfaces, DOI: 10.1021/acsami.8b03627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo H, Hwang KW, Kim D, Kiyofumi M, Park HS, Kim MH and Ahn HS, Sci. Rep, 2015, 5, 9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H, Liu T, Ma J, Wang P, Wang Y, Leprince-Wang Y and Jing G, Physical Review Fluids, DOI: 10.1103/physrevfluids.1.053901. [DOI] [Google Scholar]
- 22.Epstein AK, Wong T-S, Belisle RA, Boggs EM and Aizenberg J, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 13182–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas S, Pomeau Y and Chaudhury MK, Langmuir, 2016, 32, 6860–6870. [DOI] [PubMed] [Google Scholar]
- 24.Berthier E, Young EWK and Beebe D, Lab Chip, 2012, 12, 1224–1237. [DOI] [PubMed] [Google Scholar]
- 25.Franken P, Biom. Z, 1970, 12, 66–67. [Google Scholar]
- 26 26.Balkwill FR, Capasso M and Hagemann T, J. Cell Sci, 2012, 125, 5591–5596. [DOI] [PubMed] [Google Scholar]
- 27.Quail DF and Joyce JA, Nat. Med, 2013, 19, 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pak C, Callander NS, Young EWK, Titz B, Kim K, Saha S, Chng K, Asimakopoulos F, Beebe DJ and Miyamoto S, Integr. Biol, 2015, 7, 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang JD, Berry SM, Powers GL, Beebe DJ and Alarid ET, Integr. Biol, 2013, 5, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domenech M, Yu H, Warrick J, Badders NM, Meyvantsson I, Alexander CM and Beebe DJ, Integr. Biol, 2009, 1, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee P, Lin R, Moon J and Lee LP, Biomed. Microdevices, 2006, 8, 35–41. [DOI] [PubMed] [Google Scholar]
- 32.Paul CD, Mistriotis P and Konstantopoulos K, Nat. Rev. Cancer, 2017, 17, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG and Keely PJ, BMC Med, 2008, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A and Keely PJ, Am. J. Pathol, 2011, 178, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provenzano PP, Eliceiri KW, Inman DR and Keely PJ, Curr. Protoc. Cell Biol, 2010, 47, 10.17.1–10.17.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polacheck WJ, Zervantonakis IK and Kamm RD, Cell. Mol. Life Sci, 2012, 70, 1335–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mak M, Spill F, Kamm RD and Zaman MH, J. Biomech. Eng, 2016, 138, 021004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacCuan J, Retardation of Plateau-Rayleigh Instability: A Distinguishing Characteristic Among Perfectly Wetting Fluids, 1997. [Google Scholar]
- 39.Pabba S, Sidorov AN, Berry SM, Yazdanpanah MM, Keynton RS, Sumanasekera GU and Cohn RW, ACS Nano, 2007, 1, 57–62. [DOI] [PubMed] [Google Scholar]
- 40.Guan J, Yu B and Lee LJ, Adv. Mater, 2007, 19, 1212–1217. [Google Scholar]
- 41.Vader D, Kabla A, Weitz D and Mahadevan L, PLoS One, 2009, 4, e5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ras R and Marmur A, Non-wettable Surfaces: Theory, Preparation and Applications, Royal Society of Chemistry, 2016. [Google Scholar]
- 43.Leslie DC, Waterhouse A, Berthet JB, Valentin TM, Watters AL, Jain A, Kim P, Hatton BD, Nedder A, Donovan K, Super EH, Howell C, Johnson CP, Vu TL, Bolgen DE, Rifai S, Hansen AR, Aizenberg M, Super M, Aizenberg J and Ingber DE, Nat. Biotechnol, 2014, 32, 1134–1140. [DOI] [PubMed] [Google Scholar]
- 44.Karbaschi M, Shahi P and Abate AR, Biomicrofluidics, 2017, 11, 044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.