Abstract
Background & Aims
Loss of leucine-rich repeat-containing G-protein–coupled receptor 5–positive crypt base columnar cells provides permissive conditions for different facultative stem cell populations to dedifferentiate and repopulate the stem cell compartment. In this study, we used a defensin α4-Cre recombinase (Defa4Cre) line to define the potential of Paneth cells to dedifferentiate and contribute to intestinal stem cell (ISC) maintenance during normal homeostasis and after intestinal injury.
Methods
Small intestine and enteroids from Defa4Cre;Rosa26 tandem dimer Tomato (tdTomato), a red fluoresent protein, (or Rosa26 Enhanced Yellow Fluorescent Protein (EYFP)) reporter, Notch gain-of-function (Defa4Cre;Rosa26 Notch Intracellular Domain (NICD)-ires-nuclear Green Fluorescent Protein (nGFP) and Defa4Cre;Rosa26reverse tetracycline transactivator–ires Enhanced Green Fluorescent Protein (EGFP);TetONICD), A Disintegrin and Metalloproteinase domain-containing protein 10 (ADAM10) loss-of-function (Defa4Cre;ADAM10flox/flox), and Adenomatous polyposis coli (APC) inactivation (Defa4Cre;APCflox/flox) mice were analyzed. Doxorubicin treatment was used as an acute intestinal injury model. Lineage tracing, proliferation, and differentiation were assessed in vitro and in vivo.
Results
Defa4Cre-expressing cells are fated to become mature Paneth cells and do not contribute to ISC maintenance during normal homeostasis in vivo. However, spontaneous lineage tracing was observed in enteroids, and fluorescent-activated cell sorter–sorted Defa4Cre-marked cells showed clonogenic enteroid growth. Notch activation in Defa4Cre-expressing cells caused dedifferentiation to multipotent ISCs in vivo and was required for adenoma formation. ADAM10 deletion had no significant effect on crypt homeostasis. However, after acute doxorubicin-induced injury, Defa4Cre-expressing cells contributed to regeneration in an ADAM10–Notch–dependent manner.
Conclusions
Our studies have shown that Defa4Cre-expressing Paneth cells possess cellular plasticity, can dedifferentiate into multipotent stem cells upon Notch activation, and can contribute to intestinal regeneration in an acute injury model.
Keywords: Defensin, Paneth Cell, Intestinal Stem Cells, Regeneration, Enteroid, Notch, Chemotherapy
Abbreviations used in this paper: ADAM10, A Disintegrin and Metalloproteinase domain-containing protein 10; APC, adenomatous polyposis coli; CBC, crypt base columnar; CreER, Cre recombinase fused to an estrogen receptor; DAPI, 4′,6-diamidino-2-phenylindole; Defa4, defensin α4; DXR, doxorubicin; EdU, 5-ethynyl-2’-deoxyuridine; EGFP-ires, enhanced green fluorescent protein-internal ribosomal entry site; ENR, epidermal growth factor, noggin and R-Spondin 2; EpCAM, epithelial cell adhesion molecule; EYFP, enhanced yellow fluorescent protein; FACS, fluorescence-activated cell sorter; ISC, intestinal stem cell; Lgr5, leucine-rich repeat-containing G-protein–coupled receptor 5; LRC, label-retaining cell; MUC2, mucin 2; nGFP, nuclear green fluorescent protein; NICD, Notch intracellular domain; OCT, optimum cutting temperature; PAS, periodic acid–Schiff; PBS, phosphate-buffered saline; PFA, paraformaldehyde; RT, reverse-transcription; rtTA, reverse tetracycline transactivator; TA, transit amplifying; tdTomato, tandem dimer Tomato; UEA-1, Ulex Europaeus Agglutinin I; WENR, ENR + Wnt3a
Graphical abstract
See editorial on page 619.
Summary.
Upon leucine-rich repeat-containing G-protein–coupled receptor 5–positive crypt base columnar loss, reserve stem cell populations can repopulate the stem cell niche. This study shows that defensin α4-Cre–expressing cells are fated to become mature Paneth cells during normal homeostasis, but Notch activation or doxorubicin injury causes their dedifferentiation into multipotent stem cells.
The epithelial cell layer lining the small intestine comprises repeating crypt-villus units with cycling leucine-rich repeat-containing G-protein–coupled receptor 5–positive (Lgr5+) crypt base columnar (CBC) stem cells located in the crypt base. These Lgr5+ CBCs undergo constant renewal to replenish differentiated cells lining the intestine, which is required to maintain intestinal homeostasis and tissue integrity. The direct progeny of Lgr5+ CBCs are bipotent transit-amplifying (TA) progenitors, which are short-lived, rapidly cycling progenitors that give rise to all differentiated secretory (eg, Paneth cells, goblet cells, and enteroendocrine cells) and absorptive enterocyte cell types. Paneth cells are positioned between Lgr5+ CBCs at the crypt base and secrete granules containing antimicrobial products including lysozyme and defensins and, as part of the niche, provide important nutrient and stem cell niche signals. Although day-to-day homeostasis largely is maintained by Lgr5+ CBCs, intestinal injury and loss of the Lgr5+ CBC population has been shown to activate other reserve stem cell and progenitor populations that can dedifferentiate and revert to the stem cell state and contribute to crypt regeneration.1, 2
Intestinal regeneration experiments using DNA damaging agents such as irradiation and chemotherapeutics or genetic ablation of Lgr5+ CBCs have shown that many different facultative stem cell populations can dedifferentiate and acquire stem cell behavior.1, 2 Besides slowly cycling cells located about 4 cells up from the crypt base (+4 cells) identified using different Cre recombinase fused to an estrogen receptor (CreER) reporter mice (eg, B lymphoma Mo-MLV insertion region 1 homolog [Bmi1+], Mouse telomerase reverse transcriptase [mTert+], Homeodomain-only protein homeobox [Hopx+]) that can undergo injury-dependent stem cell reversion, there is growing evidence that other progenitor populations as well as more committed cell types from enterocyte, endocrine, and goblet cell lineages show cellular plasticity.3, 4, 5, 6, 7, 8, 9, 10, 11, 12 In addition, distinct label-retaining cells (LRCs) that express Paneth cell and enteroendocrine cell markers also are capable of dedifferentiation in vivo.13 Single-cell profiling of LRCs showed that short-term LRCs are composed of distinct endocrine and Paneth cell progenitor populations with in vitro enteroid growth potential, whereas long-term LRCs are mature Paneth cells.14 Although Lysozyme1 (Lyz1)CreER-marked Paneth cells recently were reported to possess reserve stem cell function in response to radiation-induced damage,15 the exact contribution of Paneth cell progenitors or mature Paneth cells to other forms of intestinal injury have not been defined.
Notch signaling is critical for the maintenance and survival of Lgr5+ CBCs and regulates cell fate specification of TA progenitors into enterocyte and secretory lineages. Active Notch signaling promotes an absorptive cell fate and generation of enterocytes, whereas loss of Notch signaling causes an increase in secretory cell types and loss of proliferative CBCs.16, 17 A Disintegrin and Metalloproteinase domain-containing protein 10 (ADAM10) is the α-secretase responsible for initiating Notch signaling and is expressed in all intestinal epithelial cells. We previously reported that ADAM10-mediated Notch signaling is required for the survival and maintenance of Lgr5+ CBCs.18 ADAM10 deletion in Lgr5+ CBCs places stress on the stem cell compartment, thereby creating permissive conditions for a regenerative response. Intriguingly, Notch activation enhances crypt regeneration under these permissive conditions.18 Indeed, Notch activity is enhanced by DNA damaging agents in several cell types including Paneth cells, and Notch inhibition reduced intestinal regeneration after irradiation, indicating that increased Notch activity was required after injury.15, 19, 20, 21, 22 These results suggest a possible role for active Notch signaling in the mobilization and/or dedifferentiation of specific facultative reserve stem cell populations in response to acute intestinal injury.
In the current study, we show that defensin α 4-Cre (Defa4Cre)-expressing Paneth cells are capable of dedifferentiation to a stem-like state upon Notch activation and can contribute to regeneration after acute doxorubicin (DXR)-induced intestinal injury, in a Notch-dependent manner.
Results
Defa4Cre-Expressing Cells Do Not Contribute to Intestinal Stem Cell Maintenance During Normal Intestinal Homeostasis In Vivo
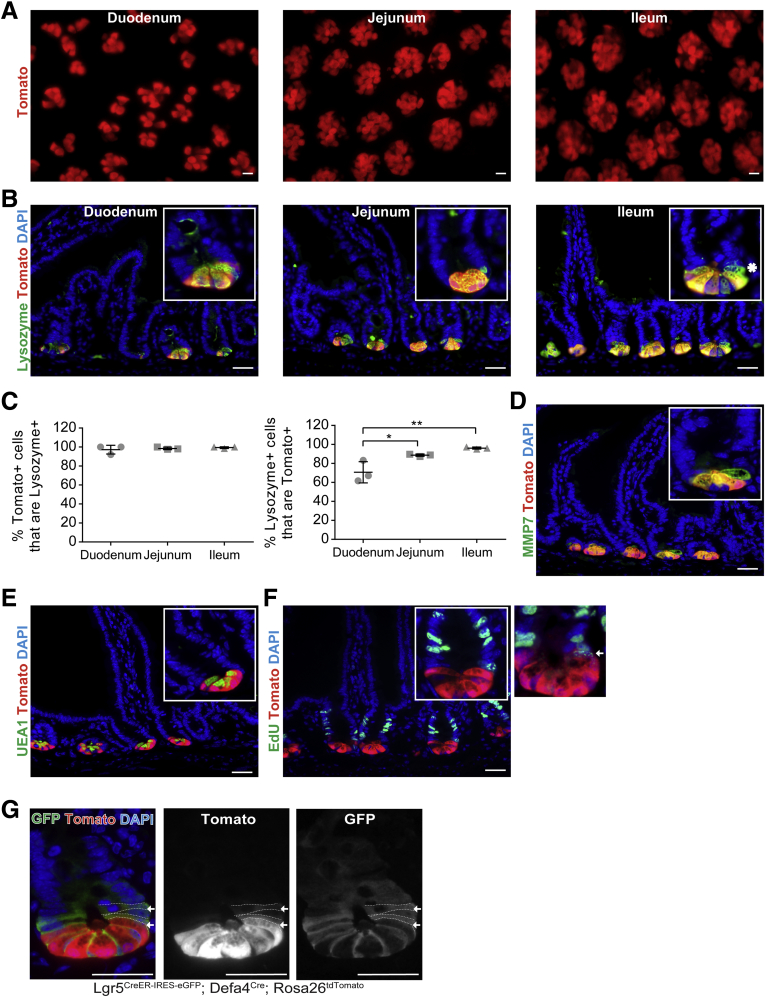
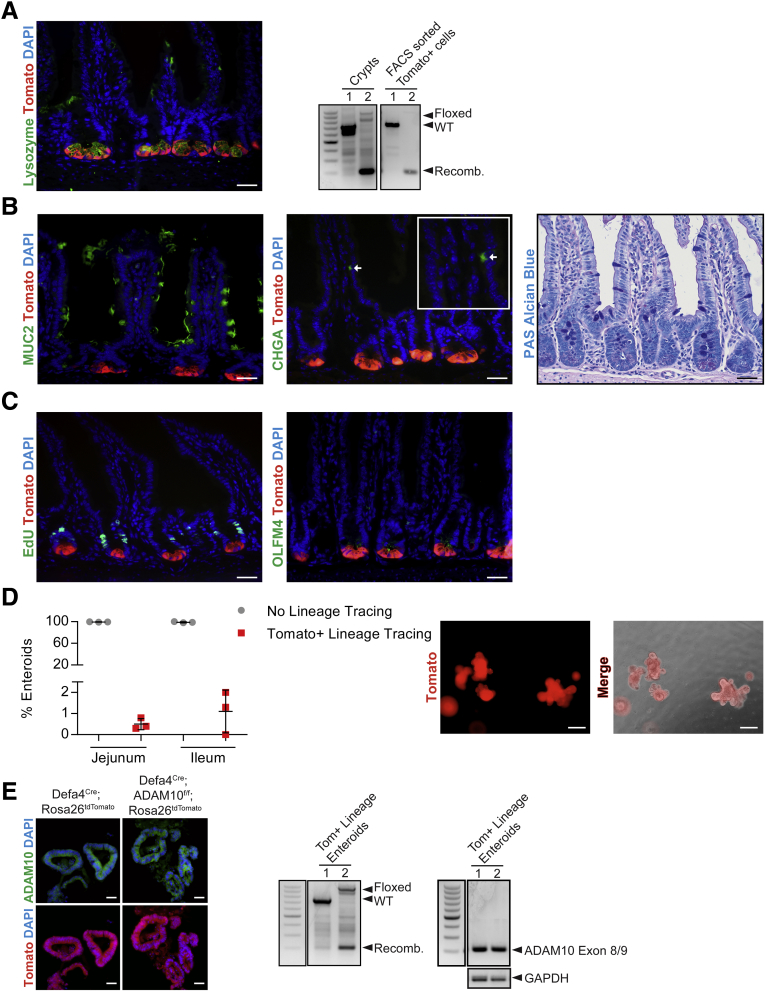
To study the cellular plasticity of Paneth cells, we used a Defa4Cre knock-in mouse line in which an ires-Cre cassette was inserted 23 bp downstream of the translational stop sequence within exon 2.23 To validate that the Defa4Cre mouse line constitutively expressed Cre in Paneth cells, Defa4Cre mice were crossed with different Rosa26 reporter mice (ie, Rosa26tandem dimer Tomato (tdTomato) or Rosa26Enhanced Yellow Fluorescent Protein (EYFP)). Whole-mount imaging of the small intestine from Defa4Cre;Rosa26tdTomato mice showed Tomato+ cells located specifically in the crypt base in a pattern consistent with increasing numbers of Paneth cells found in crypts along the duodenal–ileal axis (Figure 1A). To confirm Paneth cell–specific expression of Tomato+ cells, co-staining with the Paneth cell marker lysozyme was performed. Tomato expression co-localized with lysozyme and the percentage of Tomato+ cells that were lysozyme+ was greater than approximately 98% throughout the small intestine (Figure 1B and C). However, when the percentage of lysozyme+ cells that were Tomato+ was assessed, only approximately 71% co-stained in the duodenum, indicating that a significant number of Paneth cells only expressed lysozyme in this region. In the jejunum and ileum these percentages increased to approximately 89% and approximately 96%, respectively; a profile comparable with endogenous Defa4 gene expression along the duodenal–ileal axis.24, 25, 26 Similarly, Tomato expression co-localized with other Paneth cell–specific markers, matrix metalloproteinase 7 and lectin Ulex Europaeus Agglutinin I (UEA-1) (Figure 1D and E). Importantly, no Tomato+ cells were 5-ethynyl-2’-deoxyuridine (EdU)+ within any crypts of the entire mouse small intestine (Figure 1F) with the exception of one crypt in which a single Tomato+, EdU+ cell was located at the edge of the Paneth cell zone. To further assess the relationship between Tomato+ Paneth cells and Lgr5+ CBCs, Defa4Cre;Rosa26tdTomato mice were bred with Lgr5Enhanced Green Fluorescent Protein (EGFP)- Internal Ribosomal Entry Site (IRES)-CreER mice. Immunofluorescence staining showed that TomatoHi+ Paneth cells were a distinct cell population located between Green Fluorescent Protein (GFP)Hi+ Lgr5+ CBCs in the crypt base as reported previously.27 Interestingly, in rare GFP+ crypts, double-positive TomatoLow+/GFPLow+ cells were detected immediately above the TomatoHi+ Paneth cell zone (Figure 1G). Taken together, these results indicate that Defa4Cre-expressing cells are primarily postmitotic Paneth cells. Significantly, no Tomato+ crypt–villus lineage tracing was detected in Defa4Cre;Rosa26tdTomato (or Defa4Cre;Rosa26EYFP) mice at baseline, indicating that all Defa4Cre-expressing cells were fated to become Paneth cells and these Tomato+ Paneth cells do not contribute to intestinal stem cell (ISC) maintenance during normal intestinal homeostasis.
Figure 1.
Defa4Cre-expressing cells are postmitotic Paneth cells. (A–F) Whole-mount and immunohistochemical analysis of small intestine from Defa4Cre;Rosa26tdTomato (N = 8) mice. (A) Representative whole-mount images of Tomato+ cells in the small intestine. (B) Immunofluorescent staining of Tomato+ cells with lysozyme. *Lysozyme+ cell that is Tomato-. (C) Quantification of percentage of Tomato+ that are Lysozyme+ cells and the percentage of lysozyme+ that are Tomato+ cells (N = 3 mice). (D–F) Immunofluorescent staining of Tomato+ cells with matrix metalloproteinase 7 (MMP7), lectin UEA-1, and EdU, respectively. (F) White arrow indicates Tomato+/EdU+ cell. (G) Immunofluorescent staining of Tomato+ cells with GFP in Lgr5EGFP-ires-CreER;Defa4Cre;Rosa26tdTomato crypts (N = 4 mice). White arrows indicate TomatoLow+/GFPLow+ cells. Scale bars: 20 μm. *P ≤ .05 and **P ≤ .01.
Enteroids Generated From Defa4Cre; Rosa26tdTomato Jejunal and Ileal Crypts Can Undergo Sporadic Tomato+ Lineage Tracing
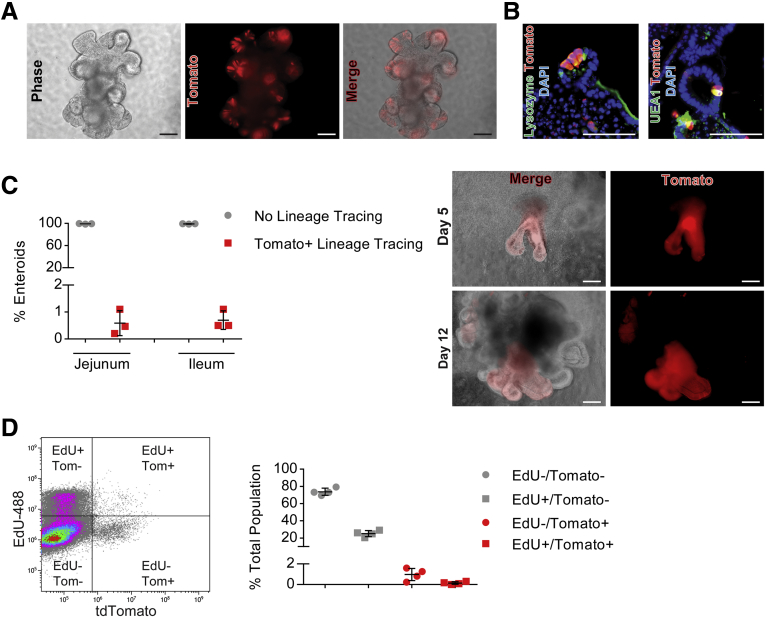
The majority of enteroids generated from Defa4Cre;Rosa26tdTomato jejunal and ileal crypts express Tomato+ cells within bud structures in which individual Tomato+ cells are interspersed between Tomato- cells in a Paneth cell pattern analogous to their crypt distribution in vivo (Figure 2A). In addition, these Tomato+ cells retained their Paneth cell identity as they co-expressed Paneth cells markers lysozyme and lectin UEA-1 (Figure 2B). Interestingly, buds from jejunal enteroids express fewer Tomato+ cells than their ileal counterparts, suggesting that regional Paneth cell programming is conserved in vitro as reported by others (data not shown).28 Although no Tomato+ lineage tracing was detected in vivo (Figure 1), sporadic Tomato+ lineage tracing in enteroids was observed, suggesting that Tomato+ cells can dedifferentiate to multipotent stem cells in vitro. To further understand the origin of these lineage events, freshly isolated jejunal and ileal crypts were cultured in media containing epidermal growth factor (EGF), Noggin and R-spondin 2 (ENR media) and the number of crypts that produced Tomato+ lineage traced enteroids was counted. Overall, approximately 0.5%–0.7% of all jejunal and ileal crypts underwent spontaneous Tomato+ lineage tracing (Figure 2C). As shown in Figure 2C, individual crypts could undergo Tomato+ lineage tracing either at the initiation of enteroid growth or it occurred within individual buds of developing enteroids. Upon passaging, these Tomato+ lineage traced buds would produce enteroids composed entirely of Tomato+ cells (data not shown). Furthermore, when hand-picked enteroids showing the Paneth cell pattern were continuously cultured, they also underwent sporadic Tomato+ lineage tracing at a rate similar to cultured crypts (data not shown). Flow cytometric analysis based on Tomato and EdU expression in jejunal enteroids showed that Tomato- and Tomato+ cell populations represented approximately 8.8% and approximately 1.2% of the total enteroid cell population, respectively. Of the total enteroid population, approximately 25% of cells were EdU+/Tomato-, but a small population of cells were EdU+/Tomato+ (∼0.15%) (Figure 2D). This low but detectable level of Tomato+/EdU+ cells in enteroids indicates that the proliferative status of Tomato+ cells had increased in vitro as compared with in vivo. Because sporadic lineage tracing occurred from cultured crypts and hand-picked Paneth cell enteroids, it suggests that the organoid culture conditions can promote cellular plasticity of Defa4Cre-expressing cells.
Figure 2.
Enteroids generated from small intestinal crypts of Defa4Cre-expressing cells are capable of sporadic Tomato+ lineage tracing. (A) Phase-contrast and immunofluorescent images of jejunal enteroid expressing Tomato+ cells within bud structures. (B) Immunofluorescence staining showing co-localization of Tomato expression with lysozyme and UEA-1 in enteroids. (C) Quantification of crypts generating enteroids showing only Paneth cell expression (ie, no lineage tracing) vs Tomato+ lineage tracing (N = 3 mice). Right: top panels, 5-day-old crypt culture undergoing complete Tomato+ lineage tracing. Bottom panels, Tomato+ lineage tracing within individual buds of a 12-day-old enteroid. (D) Flow cytometric analysis of different Tomato and EdU-expressing cell populations isolated from enteroids. Right: Quantification of individual gated cell populations (N = 4). Scale bars: 100 μm. Tom, Tomato.
Fluorescence-Activated Cell-Sorted Tomato+ Cells Isolated From Defa4Cre;Rosa26tdTomato Crypts Are Capable of Clonogenic Enteroid Growth
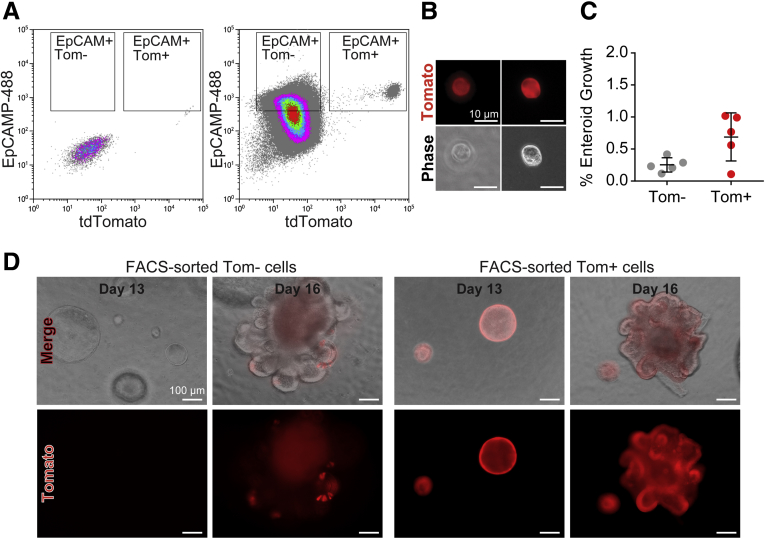
We next set out to test whether fluorescence-activated cell sorter (FACS)-sorted Tomato+ cells obtained from freshly isolated jejunal crypts of Defa4Cre;Rosa26tdTomato mice were capable of clonogenic enteroid growth. Epithelial cell adhesion molecule (EpCAM)+ epithelial cells were sorted based on Tomato expression and the cultured in ENR media or ENR + Wnt3a (WENR) media as described in the Materials and Methods section. Flow cytometric analysis of the EpCAM+/Tomato+ cell population showed a major cell population of EpCAM+/TomatoHi+ cells, and a smaller diverse population of EpCAM+/TomatoLow+ cells (Figure 3A). Both TomatoLow+ and TomatoHi+ cells could be visualized after sorting (Figure 3B). Clonogenic enteroid growth of Tomato- and Tomato+ cells occurred only in WENR media with an efficiency of approximately 0.25% and approximately 0.69%, respectively (Figure 3C). Interestingly, EpCAM+/Tomato- cells generated spherical enteroids that initially were Tomato-, but upon further culturing and bud development, re-expressed Tomato+ cells in a normal Paneth cell distribution within these bud structures (Figure 3D). By contrast, FACS-sorted Tomato+ cells directly produced spherical enteroids that were completely Tomato+ and these continued to express Tomato throughout the entire enteroid upon bud development. These results indicate that Tomato+ cells isolated directly from crypts are capable of clonogenic enteroid growth in vitro. Thus, freshly isolated Tomato+ Paneth cells show cellular plasticity and capacity for dedifferentiation to a stem cell state in an enteroid growth assay.
Figure 3.
FACS-sorted Tomato+ cells are capable of clonogenic enteroid growth. (A) Representative flow cytometric gating of EpCAM+ and Tomato expression used for FACS sorting of single-cell suspensions of isolated jejunal Defa4Cre;Rosa26tdTomato crypts. Left: Control, rat IgG2a–fluorescein isothiocyanate antibody. Right: Rat anti-EpCAM–fluorescein isothiocyanate antibody. (B) Phase-contrast and immunofluorescent images of FACS-sorted TomatoLow+ (left) and TomatoHi+ (right) cells. (C) Quantification of clonogenic growth of FACS-sorted Tomato- and Tomato+ cells grown in Basement Membrane Extract (BME) (N = 5). (D) Phase-contrast and immunofluorescent images show clonogenic organoid growth and Tomato expression in individual FACS-sorted Tomato- and Tomato+ cells. Scale bars: (B) 10 μm; (D) 100 μm. Tom, Tomato.
Notch Activation in Defa4Cre-Expressing Cells Generates Multipotent Stem Cells
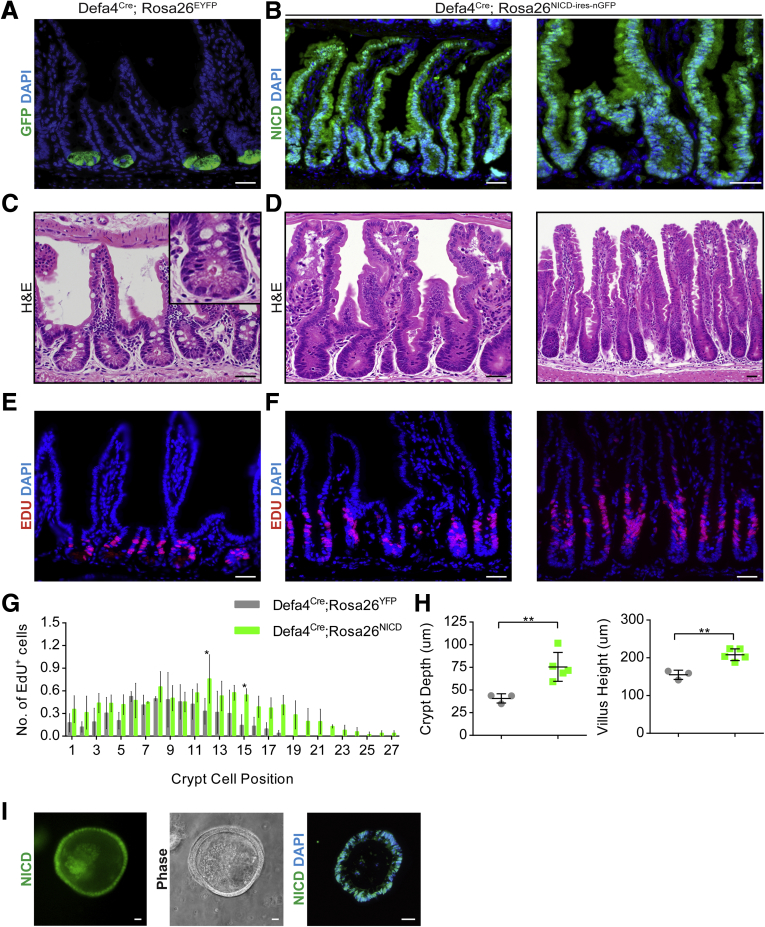
Lgr5+ CBCs and different crypt progenitors have similar chromatin signatures, providing a plausible explanation for the high level of cellular plasticity observed in these stem/progenitor populations. One potential outcome of these findings is that cellular identity/programming of different stem/progenitor cell populations can be altered by changes in transcription factor occupancy.8, 29 We previously showed that ADAM10 deletion in Lgr5+ CBCs leads to ISC failure and crypt loss.18 Intriguingly, Notch activation not only rescues this phenotype, but also enhances the regenerative response, presumably by dedifferentiating Lgr5+ progenitors and increasing the number of multipotent stem cells available to repopulate the stem cell compartment.18 Based on these observations and the cellular plasticity of Tomato+ cells in Defa4Cre;Rosa26tdTomato enteroids, we reasoned that Notch activation may increase the cellular plasticity of Tomato+ Paneth cells directly and allow dedifferentiation to a stem cell state. To test this hypothesis, we generated Defa4Cre;Rosa26Notch intracellular domain (NICD)-ires-nGFP mice, which constitutively express an active NICD.18 Defa4Cre;Rosa26NICD-ires-nGFP mice were healthy and survived beyond ∼18 months of age (data not shown). As predicted, robust NICD+/nGFP+ crypt–villus lineage tracing was detected, particularly within the ileum, indicating that Notch activation had dedifferentiated Defa4Cre-expressing cells into multipotent stem cells (Figure 4A and B). Similar to the profile of lysozyme and Tomato+ co-expression found along the small intestine of Defa4Cre;Rosa26tdTomato mice (Figure 1), we also observed increasing NICD+/nGFP+ lineage tracing along the small intestine. In the duodenum and proximal jejunum, the efficiency of NICD+/nGFP+ lineage tracing events occurred at a low level (∼10%), whereas in the distal ileum the lineage tracing efficiency reached levels greater than 90% (data not shown). Although the explanation for this mosaicism is not known, the long-term viability of these animals likely is owing to sufficient wild-type crypts being present within the duodenum and proximal jejunum to maintain normal intestinal function.
Figure 4.
Notch activation in Defa4Cre-expressing cells induces robust lineage tracing in vivo and in vitro. Immunohistochemical analysis of the ileum from (A, C, and E) Defa4Cre;Rosa26EYFP (N = 3) and (B, D, and F) Defa4Cre;Rosa26NICD-ires-nGFP (n = 5 and n = 2 ≥ 71 wk) mice. (A) Immunofluorescent staining of GFP expression. (B) Immunofluorescent staining of NICD (nGFP+) expression. (C and D) H&E staining. (E and F) Immunofluorescent staining of EdU+ cells. (G) Quantification of EdU+ cells by crypt cell position (N = 3 per genotype). (H) Quantification of crypt depth and villus height (Defa4Cre;Rosa26EYFP, N = 3; Defa4Cre;Rosa26NICD-ires-nGFP, N = 5). (I) Ileal enteroids derived from Defa4Cre;Rosa26NICD-ires-nGFP. Left and middle: Whole-mount fluorescent and phase-contrast images, respectively. Right: Immunofluorescent staining of NICD (nGFP+) in frozen section of enteroid. Scale bars: 20 μm. *P ≤ .05 and **P ≤ .01.
H&E analysis showed that Notch activation had caused crypt enlargement and that the crypt–villus units were lined with relatively undifferentiated cells (Figure 4C and D). Consistent with their morphologic appearance, NICD+/nGFP+ lineage-traced crypts showed increased crypt cell proliferation and EdU+ cells extended further up the crypts (Figure 4E and F). Correspondingly, NICD+/nGFP+ crypt–villus units showed increased crypt depth and villus height compared with controls (Figure 4H). In the distal ileum, some NICD+/nGFP+ crypt–villus units had even more pronounced hyperplastic features (Figure 4D and F). Furthermore, enteroids generated from NICD+/nGFP+ ileal crypts were completely nGFP+, confirming their multipotent stem cell potential (Figure 4I).
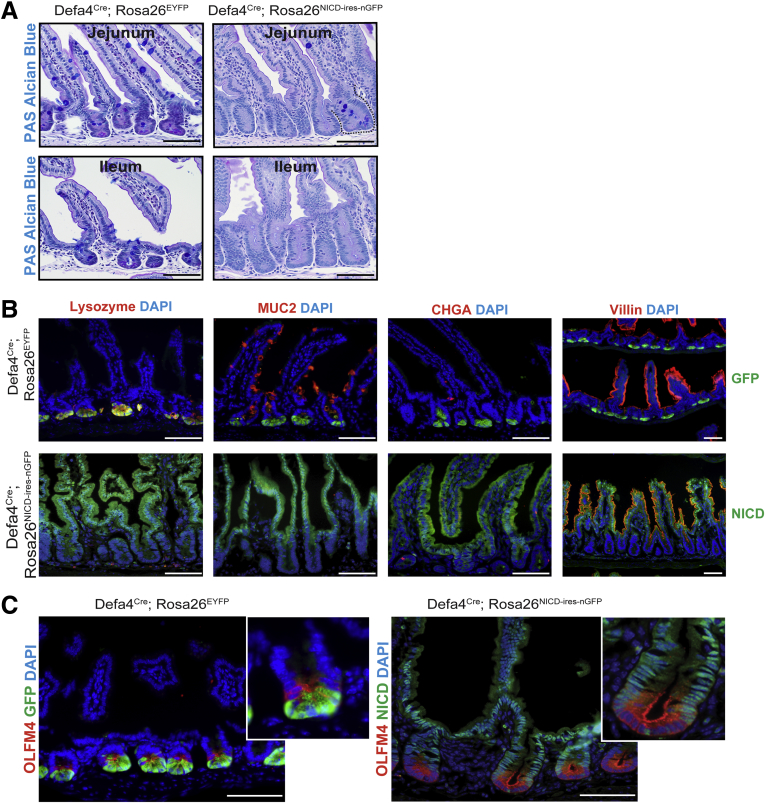
Because Notch activation promotes enterocyte lineage specification at the expense of secretory cell types, there was a complete loss of secretory cell differentiation in NICD+/nGFP+ crypt–villus units as confirmed by periodic acid–Schiff (PAS)/Alcian blue staining (Figure 5A) and absence of lysozyme, mucin 2 (MUC2) (goblet cell), and chromogranin A (enteroendocrine) staining (Figure 5B). By contrast, the enterocyte markers villin and alkaline phosphatase (data not shown) readily were detected at the villus surface of NICD+/nGFP+ crypt–villus units (Figure 5B) consistent with Notch activation being permissive for enterocyte differentiation. To further assess the stem/progenitor cell compartment in NICD+/nGFP+ crypts, we examined the expression of Olfm4, which is a surrogate marker of Lgr5 CBCs30 and a known Notch target.31 Olfm4 was greatly up-regulated in cells of the crypt base of Defa4Cre;Rosa26NICD-ires-nGFP mice, confirming that Defa4Cre-expressing cells had dedifferentiated into multipotent stem cells resembling Lgr5+ CBCs that, because of constitutive Notch activation, were only capable of enterocyte lineage programming (Figure 5C).
Figure 5.
Notch activation in Defa4Cre-expressing cells leads to loss of secretory cell differentiation and increased expression of the stem cell marker Olfm4. Histologic and immunofluorescent staining of the small intestine from Defa4Cre;Rosa26EYFP and Defa4Cre;Rosa26NICD-ires-nGFP mice. (A) PAS/Alcian blue staining. Dashed line denotes wild-type crypt in jejunum of Defa4Cre;Rosa26NICD-ires-nGFP intestine. (B) Immunofluorescent staining of GFP or NICD (nGFP+) with cell type–specific markers, lysozyme, MUC2, chromogranin A, and villin. (C) Immunofluorescent staining of GFP or NICD (nGFP+) with the stem cell marker Olfm4. Scale bars: 50 μm.
Inducible Notch Activation in Adult DefaCre-Expressing Cells Generates Crypt–Villus Lineage Tracing
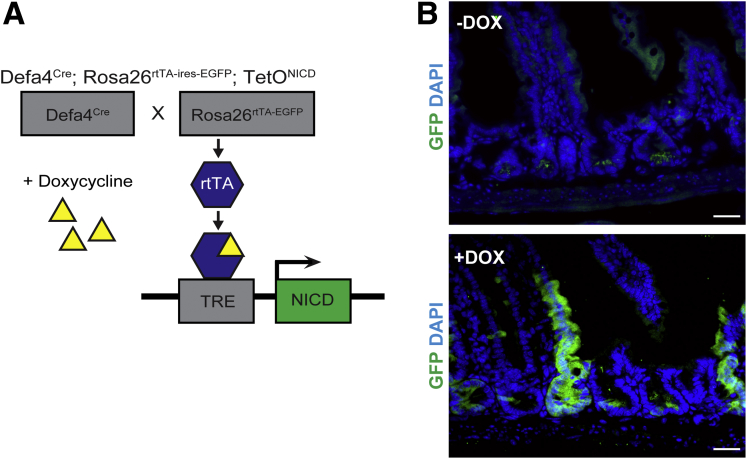
In the earlier-described Notch activation studies, NICD was expressed constitutively using the Defa4Cre knock-in allele, raising the possibility that NICD expression and subsequent dedifferentiation may have occurred before weaning.24, 25, 32 To test if Notch activation in adult Defa4Cre-expressing cells could induce lineage tracing, we generated Defa4Cre;Rosa26reverse tetracycline transactivator (rtTA)-ires-EGFP;TetONICD mice in which NICD expression was doxycycline-inducible33 (Figure 6A). Adult Defa4Cre;Rosa26rtTA-ires-EGFP;TetONICD mice were treated with doxycycline in drinking water for 2 weeks and then analyzed. Immunofluorescent staining showed robust GFP+ crypt–villus units within the small intestine (Figure 6B), clearly showing that adult Defa4Cre-expressing cells can undergo dedifferentiation to multipotent stem cells upon Notch activation.
Figure 6.
Inducible Notch activation in adult Defa4Cre-expressing cells allows for dedifferentiation and lineage tracing. (A) Schematic of inducible Notch activation in adult Defa4Cre-expressing cells. In the presence of doxycycline, reverse tetracycline transactivator will bind the tetracycline response element (TRE) in TetONICD allele and activate NICD expression in Defa4Cre-expressing cells. Defa4Cre;Rosa26rtTA-ires-EGFP;TetONICD mice (N = 5) received 2 mg/mL doxycycline in water for 2 weeks. (B) Immunofluorescent staining with GFP antibody. Top: No doxycycline (N = 4). Bottom: Two weeks on doxycycline (N = 5). Scale bars: 20 μm.
Notch Activation Rapidly Induces Crypt Hyperplasia and Adenoma Formation Upon Biallelic Loss of Adenomatous Polyposis Coli in Defa4Cre Mice
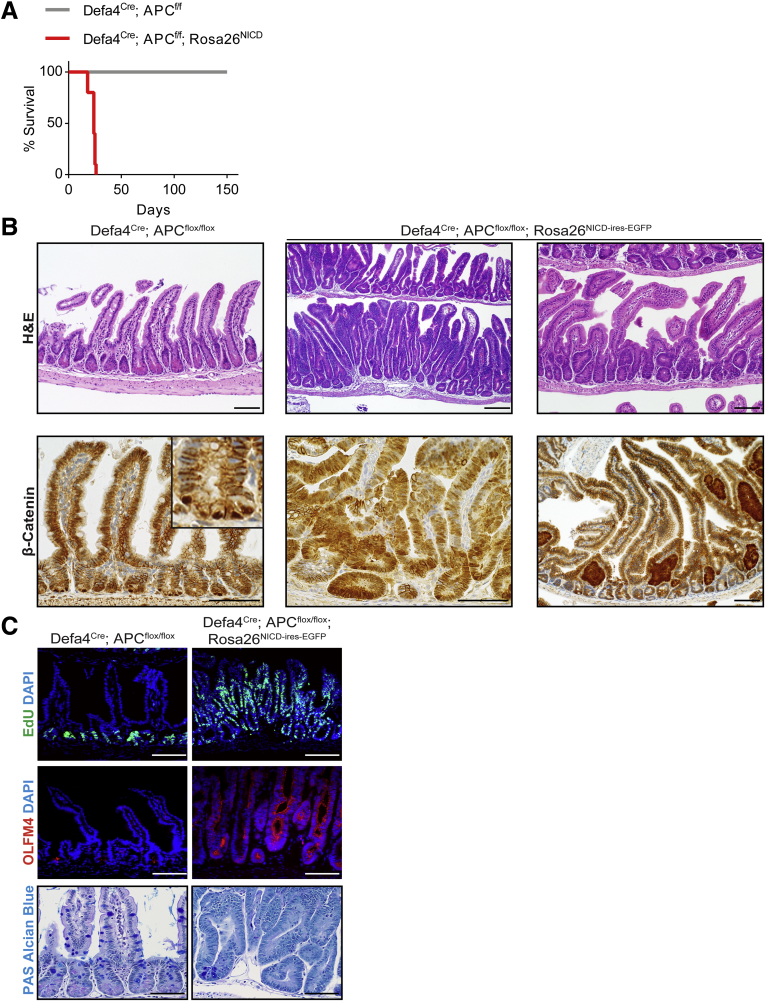
Recent analysis of Wnt-dependent adenoma models has suggested that only cells with stem/progenitor-like properties are susceptible to adenoma formation.34, 35 To further validate the ability of Notch activation to dedifferentiate Defa4Cre-expressing cells into multipotent stem cells, adenoma formation was studied in a model of biallelic APC inactivation using Defa4Cre;APCflox/flox mice and Defa4Cre;APCflox/flox;Rosa26NICD-ires-nGFP mice. Notably, Defa4Cre;APCflox/flox mice were healthy and survived beyond 5 months of age, whereas Defa4Cre;APCflox/flox;Rosa26NICD-ires-nGFP mice rapidly died and no mice survived beyond postnatal day 26 (Figure 7A). The morphologic appearance of the small intestine from Defa4Cre;APCflox/flox mice was normal. By contrast, severely dysplastic crypts and early adenoma formation were observed upon Notch activation and similar to the pattern of NICD+/nGFP+ lineage tracing described earlier, and adenoma formation was more pronounced in the distal ileum (Figure 7B). Despite strong nuclear β-catenin staining in the Paneth cells of Defa4Cre;APCflox/flox mice, suggesting APC inactivation and increased Wnt activity, normal crypt proliferation and secretory differentiation as well as normal Olfm4 expression was observed in crypts from these mice (Figure 7B). These results indicate that APC inactivation and increased Wnt activity alone in Defa4Cre-expressing cells is unable to transform these cells. However, upon Notch activation, we observed dysplastic crypts and adenomas with strong nuclear β-catenin staining associated with a marked increase in cell proliferation, a loss of secretory differentiation, and robust expansion of Olfm4-expressing cells (Figure 7C). These results clearly show that Notch activation had dedifferentiated Defa4Cre-expressing cells into stem/progenitor-like cells that were now susceptible to adenoma formation by APC inactivation.
Figure 7.
Notch activation induces crypt hyperplasia and adenoma formation in Defa4Cre; APCflox/floxmice. (A) Survival curve for Defa4Cre;APCflox/flox (N = 7) and Defa4Cre;APCflox/flox;Rosa26NICD-ires-nGFP (N = 10) mice. (B) H&E analysis and β-catenin staining. Left and middle: Ileum. Right: proximal jejunum. (C) Immunofluorescent staining of cell proliferation (EdU) and Olfm4 within the ileum. Upper right: PAS/Alcian blue staining. Scale bars: 50 μm.
ADAM10 Deletion in Defa4Cre-Expressing Cells Does Not Alter Normal Crypt Homeostasis
ADAM10 is the α-secretase responsible for ligand-dependent Notch activation in the intestine.17, 18 Studies using intestine-specific ADAM10-deficient mice have shown that ADAM10 is required for ISC maintenance and cell fate specification in vivo and in enteroids in vitro and is a surrogate method to study global Notch loss-of-function.18 To study the effects of ADAM10 deletion in Paneth cells, we generated Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice. Analysis of isolated crypts and FACS-sorted Tomato+ cells confirmed efficient ADAM10 recombination in these Tomato+ Paneth cells (Figure 8A). Immunofluorescence staining with lysozyme showed a normal distribution of Tomato+ Paneth cells within crypts throughout the small intestine (Figure 8A). Additional immunofluorescent and histologic staining showed no changes in the expression of other cell type–specific markers, MUC2, chromogranin A, alkaline phosphatase, and villin (enterocyte, data not shown), or PAS/Alcian blue staining (Figure 8B). Importantly, no changes in proliferation or Olfm4 expression were detected in Defa4Cre;ADAM10-deficient mice (Figure 8C). Similar to Defa4Cre;Rosa26tdTomato mice, no Tomato+ lineage tracing was observed in ADAM10-deficient mice at baseline. Taken together, these results suggest that ADAM10 loss in Defa4Cre-expressing cells does not perturb the ISC compartment, crypt proliferation, or cell fate specification during normal intestinal homeostasis.
Figure 8.
ADAM10 deletion in Defa4Cre-expressing does not alter Paneth cell localization or intestinal homeostasis. Analysis of small intestine and enteroids from Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice (N = 3). (A) Immunofluorescent staining of Tomato+ cells with lysozyme. Right: Analysis of ADAM10 recombination in isolated crypts and FACS-sorted Tomato+ cells. Mouse genotypes: (1) wild-type Defa4Cre;Rosa26tdTomato and (2) Defa4Cre;ADAM10flox/flox;Rosa26tdTomato. (B) Immunofluorescent staining of Tomato+ cells with MUC2 and chromogranin A (CHGA). Right: PAS/Alcian blue staining. (C) Immunofluorescent staining of Tomato+ cells with EdU and Olfm4. (D) Quantification of jejunal crypts generating enteroids showing only Paneth cell expression (ie, no lineage tracing) vs Tomato+ lineage tracing (N = 3). Right: Enteroids showing complete Tomato+ lineage tracing. (E) Immunofluorescent staining of Tomato+ lineage traced enteroids with ADAM10. Middle: Analysis of ADAM10 recombination in Tomato+ lineage traced enteroids. Right: RT-PCR analysis of ADAM10 exon 8–9 expression. Mouse genotypes: (1) wild-type Defa4Cre;Rosa26tdTomato and (2) Defa4Cre;ADAM10flox/flox;Rosa26tdTomato. Scale bars: (A–C and E) 20 μm; (D) 100 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WT, wild-type.
Defa4Cre;ADAM10-deficient enteroids were viable and showed a similar Paneth cell distribution of Tomato+ cells within bud structures as wild-type Defa4Cre;Rosa26tdTomato enteroids (data not shown). However, similar to wild-type Defa4Cre;Rosa26tdTomato crypts, sporadic Tomato+ lineage tracing also was observed in enteroids generated from jejunal and ileal crypts from Defa4Cre;ADAM10-deficient mice (Figure 8D). Because ADAM10 loss causes enteroid growth failure,18 we examined whether these Tomato+ lineage tracing events had escaped ADAM10 recombination. Indeed, the nonrecombined ADAM10 floxed allele and normal Adam10 messenger RNA levels readily were detected. In addition, ADAM10 immunofluorescence staining showed robust expression throughout these Tomato+ enteroids (Figure 8E). These data indicate sporadic Tomato+ lineage tracing events in Defa4Cre;ADAM10-deficient enteroids had escaped ADAM10 deletion.
Defa4Cre-Expressing Cells Contribute to the Regenerative Response After Doxorubicin-Induced Intestinal Injury
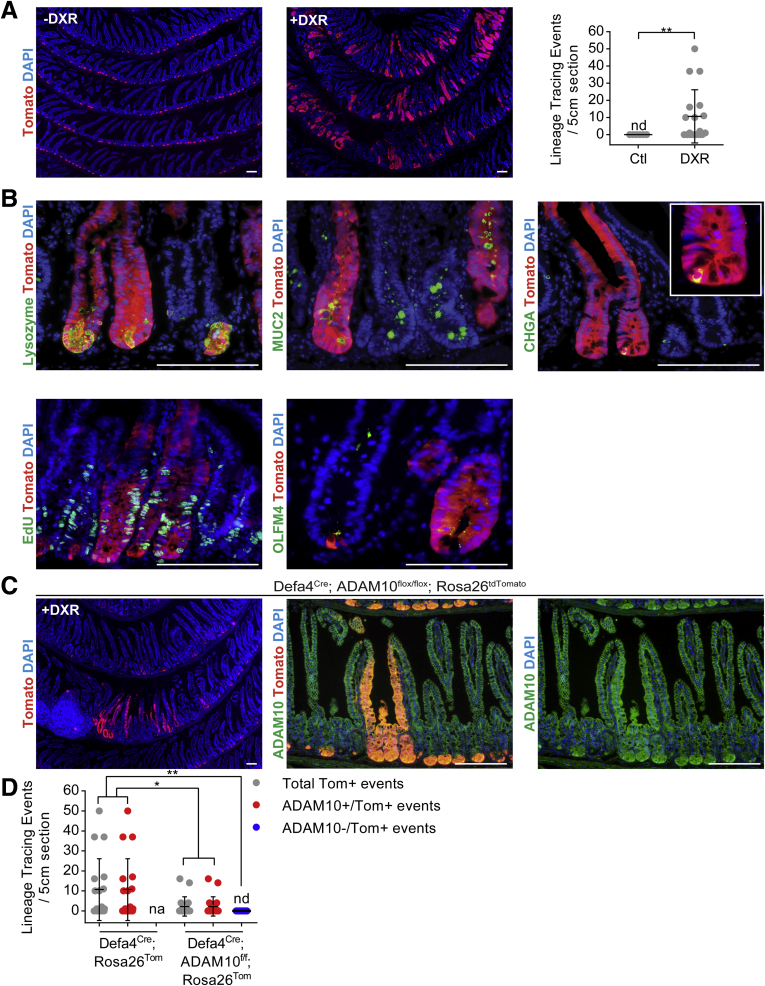
The intestinal epithelium is extremely sensitive to DNA-damaging agents such as chemotherapy and ionizing radiation, but rapidly can regenerate through crypt cell proliferation and crypt fission to repopulate the intestine.1, 2 Lineage tracing studies have shown that several different stem and progenitor cell populations can be mobilized and contribute to this regenerative response.1, 2 To test if Defa4Cre-expressing cells also can contribute to the regenerative response after acute intestinal injury, Defa4Cre;Rosa26tdTomato mice were treated with DXR, a topoisomerase II inhibitor that blocks DNA replication causing cell apoptosis. Robust but variable Tomato+ lineage tracing was observed 7 days after DXR treatment, whereas untreated mice had no such events (Figure 9A). Although Tomato+ lineage tracing was observed throughout the small intestine, the majority of these lineage tracing events were located in the duodenum and proximal jejunum where the most severe intestinal injury was observed (data not shown). Immunofluorescent staining confirmed that Tomato was co-expressed with multiple intestinal cell types including lysozyme (Paneth cell), MUC2 (goblet cell), chromogranin A (enteroendocrine cell), and villin (enterocyte, data not shown) in these Tomato+ crypt–villus units (Figure 9B). These Tomato+ lineage-traced crypts also were hyperproliferative as noted by increased EdU+ cells within the crypts. In addition, Olfm4 still was expressed at the crypt base, indicating that Defa4Cre-marked cells had dedifferentiated in Lgr5+-like stem cells (Figure 9B). Furthermore, increased lineage tracing events were observed when jejunal enteroids from Defa4Cre;Rosa26tdTomato mice were treated with DXR (data not shown). Overall, these results indicated that Defa4Cre-expressing cells can dedifferentiate in multipotential stem cells and contribute to the regenerative response after DXR-induced injury in vivo and in vitro.
Figure 9.
Defa4Cre-expressing cells can dedifferentiate and contribute to regeneration after DXR-induced injury. Analysis of DXR-treated (A and B) Defa4Cre;Rosa26tdTomato and (C) Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice 7–10 days after treatment. (A) Tomato+ lineage tracing in the jejunum of untreated and DXR-treated Defa4Cre;Rosa26tdTomato mice. Right: Quantification of Tomato+ lineage tracing events within a 5-cm region of proximal jejunum (control, N = 8; DXR, N = 18). (B) Immunofluorescent staining of Tomato+ lineage tracing events with cell type–specific markers: lysozyme (Paneth cells), MUC2 (goblet cells), chromogranin A (CHGA) (enteroendocrine cells), and proliferation (EdU) and stem cell markers, Olfm4. (C) Tomato+ lineage tracing in jejunum of DXR-treated Defa4Cre;ADAM10flox/flox;Rosa26tdTomato co-stained with ADAM10. Right: Immunofluorescent staining of ADAM10 shown as a single channel from the middle panel. (D) Quantification of Tomato+ lineage tracing events within a 5-cm region of proximal jejunum from Defa4Cre;Rosa26tdTomato (N = 18) mice and Defa4Cre;ADAM10flox/flox;Rosa26tdTomato (N = 18) mice. Scale bars: 100 μm. *P ≤ .05 and **P ≤ .01. Ctl, control; NA, not applicable; ND, not detected.
ADAM10 Deletion in Defa4Cre-Expressing Cells Inhibits the Regenerative Response After Doxorubicin
To determine if Notch activity was responsible for DXR-induced dedifferentiation of Defa4Cre-expressing cells, we used Defa4Cre;ADAM10flox/flox mice as a model of Notch loss-of-function. As shown in Figure 9C and D, the number of DXR-induced Tomato+ lineage tracing events was reduced upon ADAM10 deletion (Figure 9C and D). More importantly, all Tomato+ crypt–villus units detected had retained ADAM10 expression, indicating that all Tomato+ lineage tracing events had escaped ADAM10 recombination and no ADAM10-deficient Tomato+ lineage tracing was present (Figure 9C and D). These findings indicate that ADAM10-dependent Notch signaling was required for DXR-induced mobilization and dedifferentiation of Defa4Cre-expressing cells into multipotent stem cells.
Discussion
Lgr5+ CBCs are required for normal day-to-day intestinal homeostasis, however, other facultative reserve stem cell populations within the crypt are capable of repopulating the stem cell niche and restoring the Lgr5+ stem cell pool upon injury.1, 2 In this study we have used Defa4Cre-expressing Paneth cells to investigate the cellular plasticity of Paneth cells and their contribution to ISC maintenance during intestinal homeostasis and regeneration after DXR-induced injury. We showed that Defa4Cre-marked cells normally are fated to become postmitotic Paneth cells and do not contribute to ISC maintenance under normal homeostatic conditions. However, upon Notch activation, Defa4Cre-expressing Paneth cells can efficiently dedifferentiate into multipotent stem cells in vivo. Additional evidence for the ability of active Notch signaling to dedifferentiate Defa4Cre-marked Paneth cells was attained through studies of APC-dependent adenoma formation. Biallelic loss of Apc in Defa4Cre-expressing cells had no effect on the intestinal epithelia and no adenoma initiation was observed. In contrast, when Notch activation was combined with Apc loss, a rapid onset of crypt hyperplasia and adenoma formation was observed. This result indicated that Notch activity had dedifferentiated Defa4Cre-expressing cells into a stem/progenitor-like state that was susceptible to WNT pathway hyperactivation and tumor formation. DNA damaging agents such as chemotherapeutics and high-dose γ radiation, which efficiently kill cycling cells, have been used extensively to assess the contribution of different facultative reserve stem cell populations in intestinal regeneration. In the DXR injury model, we detected robust but variable lineage tracing, indicating that DefaCre-expressing Paneth cells had undergone reversion to a stem cell state. ADAM10 is the α-secretase responsible for initiating ligand-dependent Notch activation within the intestine. Genetic deletion of Adam10 in Defa4Cre-marked cells had no effect on the ISC compartment or crypt homeostasis. However, in the DXR-injury model, loss of Adam10 blocked the ability of these cells to revert to multipotent stem cells, implying that Notch signaling is required for their dedifferentiation and contribution to the regenerative response. Taken together, these data argue that Defa4Cre-expressing Paneth cells do not contribute to ISC maintenance during normal intestinal homeostasis but can act as injury-resistant facultative reserve stem cells that require Notch activation to dedifferentiate into multipotent stem cells.
Complex interactions between Wnt and Notch signaling pathways are required for ISC maintenance and to regulate differentiation required for intestinal homeostasis. Notch signaling is active in Lgr5+ CBCs and is required for stem cell proliferation and survival in an Atoh1-independent manner. Separately, Notch controls cell fate decisions of short-lived, bipotent TA progenitors by regulating Atoh1 expression.16 Notch activation induces Hes1, which transcriptionally represses Atoh1 expression in TA progenitors driving differentiation toward the enterocyte lineage, whereas adjacent progenitors that escape Notch activation express Atoh1 and are fated to the secretory lineage. Several transcription factors downstream of ATOH1, including Sterile alpha motif pointed domain containing E26 transformation-specific transcription factor and growth factor independent 1 are important for differentiation of Paneth and goblet cells.16, 36, 37, 38 In addition, β-catenin/Wnt signaling and its downstream transcription factor Sox9 also are required for Paneth cell development and differentiation, highlighting the interdependency of Wnt and Notch signaling in Paneth cell programming and function.39, 40
Cellular plasticity and dedifferentiation within the intestine can be explained, in part, by broadly permissive chromatin found in different stem cell, enterocyte, and secretory progenitor cell populations. In addition, more committed enteroendocrine and goblet cell lineages can undergo chromatin reorganization, allowing dedifferentiation to an ISC-like chromatin state.8, 29 The extracellular signals that regulate these plasticity events at the level of transcription factor activity and chromatin reorganization are poorly defined, but these results imply that cellular identity/programming of different progenitor cell populations can be defined by transcription factor occupancy within this open chromatin.8, 29 ADAM10-mediated Notch signaling is crucial for survival and maintenance of Lgr5+ CBCs. Analogous to other Lgr5+ CBC injury models, we showed previously that ADAM10 deletion in Lgr5+ CBCs leads to an imbalance within the ISC compartment that promotes permissive conditions for regeneration. Notch activation under these permissive conditions enhances the repopulation of the ISC compartment, presumably by driving the dedifferentiation of ADAM10-deficient Lgr5+ progeny back to a multipotent stem cell state.17, 18 In the present study, we showed that Defa4Cre-expressing Paneth cells, which are differentiated postmitotic secretory cells that lack Notch activity, can upon either constitutive or inducible Notch activation dedifferentiate into multipotent stem cells in vivo and in vitro. Sustained Notch activation lead to increased expression of Olfm4, which is a surrogate marker of Lgr5+ CBCs and a Notch gene target, indicating that Defa4Cre-marked cells had dedifferentiated into Lgr5+ CBC-like cells. In addition, because Notch activation blocks secretory lineage specification, no secretory cell types including Paneth cells were detected in NICD+/nGFP+ lineage-traced crypts. However, as expected, villin and alkaline phosphatase still were expressed, indicating that Notch activation had driven fate specification toward enterocyte progenitors and enterocytes. Thus, Notch activation had dedifferentiated Defa4Cre-expressing cells into multipotent stem cells that expressed features of Lgr5+ CBCs and were capable of fate specification into the enterocyte lineage.
Paneth cells show strong nuclear β-catenin staining indicative of high Wnt activity and are highly sensitive to changes in Wnt activity within the crypt compartment. Acute loss of Wnt signaling results in depletion of Paneth cells,41, 42, 43 whereas broad activation of canonical Wnt signaling results in the expansion of Paneth cell numbers.41, 42, 43, 44, 45 Surprisingly, activation of Wnt signaling through Apc inactivation in Defa4Cre-expressing Paneth cells had no effect on intestinal homeostasis, and the normal crypt expression of Olfm4 indicated that the stem/progenitor cell compartment also was not perturbed. Because Paneth cells already possess high Wnt activity, it suggests that DefaCre-expressing Paneth cells are unable to respond to additional cell-autonomous Wnt signaling. Beyond regulating canonical Wnt signaling, APC has other distinct functions including RNA binding and regulation and microtubule nucleation.46, 47, 48 It therefore will be important to determine if Apc inactivation in Paneth cells alters other aspects of Paneth cell differentiation and function.
Two primary models for the cellular origin of human colorectal neoplasia have been described: the “bottom up“ model proposes that tumor initiation originates within the crypt stem cell niche whereas the “top down” model proposes that early adenoma formation occurs at the luminal surface; an event physically independent of the crypt stem cell niche.49 In support of the bottom-up model, several mouse studies have shown that Cre-mediated Wnt activation in Lgr5+ CBCs and other reserve stem cell populations (eg, Prom1+, Bmi1+, or Lrig1+) produce rapidly growing adenomas.7, 9, 34, 35 Interestingly, studies combining diphtheria toxin receptor–mediated ablation of Lgr5+ cells with Apc deletion showed that adenoma initiation readily can occur in the Lgr5− cell population.50 However, further evidence supporting the top-down model for tumor initiation has required constitutive Wnt activation to be combined with additional mutations, tissue injury, or changes in the microenvironment such as inflammation.51, 52, 53, 54 Analogous to these studies, we found that Apc inactivation in combination with constitutive Notch activation produced robust crypt hyperplasia and adenoma formation in Defa4Cre-expressing cells in which Olfm4 expression was greatly expanded. Most likely, Notch activation caused Defa4Cre-expressing cells to dedifferentiate into a stem/progenitor-like state that then was responsive to Wnt hyperactivation resulting in adenoma initiation. Importantly, this result further supports the view that Notch activation can induce cellular plasticity and dedifferentiation of Defa4Cre-marked cells.
For injury-resistant reserve stem cell populations, most studies have focused on crypt regeneration in response to DNA damaging agents. DXR, a chemotherapeutic agent, causes rapid apoptosis and cell-cycle arrest of rapidly proliferating cells and loss of Lgr5+ CBCs within the crypt compartment, whereas Paneth cells are resistant to this injury. During the regenerative phase, a Lgr5(-) stem cell population expands and increased numbers of “intermediate“ cells with Paneth and goblet cell markers and mature Paneth cells are observed.55, 56 We showed that robust, albeit variable, lineage tracing of Defa4Cre-marked cells can occur upon DXR treatment in vivo. In these lineage-traced crypts, Olfm4 expression was maintained in the crypt compartment and all differentiated cell types were detected, indicating that Defa4Cre-expressing cells had dedifferentiated into multipotent Lgr5+ CBC-like stem cells. Previous studies have shown that Notch activity is enhanced by DXR in other systems20 and Notch inhibition reduced regeneration after irradiation, signifying that higher Notch activity is necessary after injury.19, 22 Significantly, we showed that Adam10 loss blocked Defa4Cre lineage tracing, indicating that cell-autonomous ADAM10-dependent Notch signaling within these Paneth cells likely is involved in the regenerative response. Because all secretory progenitors including Paneth cell progenitors as well as mature postmitotic Paneth cells are defined by Atoh1+ expression owing to low Notch activity, it suggests that Notch signaling has been up-regulated to confer the dedifferentiation response. Although the mechanism for increased Notch activity is undefined, ADAM10 proteolytic activity can be up-regulated by changes in reactive oxygen species and inflammatory signals that are known to be generated by different injury models and cancer.17, 57
Several groups have shown that LRCs have the capacity to act as reserve stem cells.13, 14, 58 By using a histone H2B-split-Cre reporter, Buczacki et al13 showed the potential of LRCs to undergo lineage tracing after treatment with DNA-damaging agents including DXR. Molecular profiling showed that these LRCs had overlapping expression of both endocrine and Paneth cell markers, suggestive of secretory precursors. More detailed single-cell profiling has shown that LRCs are a heterogenous population that can be categorized into short-term LRCs and long-term LRCs. Short term-LRCs are 2 distinct populations of endocrine and Paneth cell progenitors that have in vitro organoid growth capacity, whereas long term-LRCs are mature Paneth cells.14 Based on constitutive expression of the Defa4Cre knock-in allele and our preliminary identification of rare TomatoLow+/GFPLow+ cells above the main Paneth cell zone in Lgr5EGFP-ires-CreER;Defa4Cre;Rosa26tdTomato crypts, it is possible that Defa4Cre-expressing Paneth cell progenitors have the potential to undergo dedifferentiation. It will be important to perform single-cell gene expression profiling to further define the heterogeneity of the Defa4Cre-expressing population. Nonetheless, this study clearly shows that Defa4Cre-expressing Paneth cells can dedifferentiate and contribute to intestinal regeneration after injury in a Notch-dependent manner.
Materials and Methods
Animal Studies
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Colorado Anschutz Medical Campus. The following mouse strains were used in this study: Defa4Cre,23 Lgr5-EGFP-ires-CreERT2 (JAX stock no. 008875; The Jackson Laboratory, Sacramento, CA),27 Rosa26EYFP (JAX stock no. 006148; The Jackson Laboratory),59 Rosa26tdTomato (JAX stock no. 007909; The Jackson Laboratory),60 Rosa26NICD-ires-nEGFP (JAX stock no. 008159; The Jackson Laboratory),61 TetONICD (provided by Dr Ben Stanger, University of Pennsylvania, Philadelphia, PA),33 Rosa26rtTA-ires-EGFP (JAX stock no. 005670; The Jackson Laboratory),62 Adam10 floxed allele,18 Apc floxed allele (APC 580S; provided by Dr Eric Fearon, University of Michigan, Ann Arbor, MI),63 and mice carrying a deletion for tumor necrosis factor (TNF) AU-rich elements (ARE) (TNFΔARE/+) TNFΔARE/+ (provided by Dr Eoin McNamee, University of Colorado, Aurora, CO) (All JAX mice from The Jackson Laboratory, Sacramento, CA).64 All experimental animals were adult mice, ≥8 weeks of age, represented both sexes, and, when possible, littermate controls were used.
EdU Administration and Tissue Collection
Mice were injected intraperitoneally with EdU (100 μg per mouse, 1 mg/mL in phosphate-buffered saline [PBS], C10337; Invitrogen, ThermoFisher Scientific, Carlsbad, CA) 1 hour before tissue collection. For histologic analysis, intestinal tissue was removed rapidly, flushed with ice-cold PBS, opened longitudinally, and then fixed in cold 4% Paraformaldehyde (PFA) in PBS overnight at 4°C. Tissue then was Swiss-rolled before embedding in optimum cutting temperature (OCT) compound, or paraffin embedded. For RNA analysis, full-thickness intestine or scraped mucosa was collected.
Whole-Mount Imaging of Intestinal Tissue
Freshly collected intestine was prepared as described earlier and then briefly fixed in 4% PFA before being mounted and imaged using an Olympus IX-71 (Olympus, Melville, NY) inverted fluorescent microscope and DP72 digital camera.
Immunofluorescent and Immunohistochemical Staining
Frozen and paraffin tissue sections (4–8 μm) were used for histologic analysis and immunohistochemistry, as previously described.18 Frozen enteroid sections (4–8 μm) were prepared from enteroids fixed in 4% PFA and embedded in OCT. Primary and secondary antibodies, dilutions, and catalog numbers used are listed in Table 1. Alkaline phosphatase staining (SK-5100; Vector Laboratories, Burlingame, CA) and PAS and Alcian blue staining were performed according to the manufacturer’s instructions or by the Morphology and Phenotyping Core, Gates Center of Regenerative Medicine. β-catenin staining was performed on deparaffinized sections according to the manufacturer’s protocol (MP-2400; Vector). EdU-labeled cells were detected using the Click-iT EdU imaging kit (C10337; Invitrogen, ThermoFisher Scientific, Carlsbad, CA). Briefly, OCT slides were permeabilized with 0.1% Triton X-100 (Fisher Scientific, Fair Lawn, NJ) in PBS before detection with the Click-iT reaction and Alexa Fluor 488 or Alexa Fluor 594 according to the manufacturer’s protocol. ProLong Gold Antifade with 4′,6-diamidino-2-phenylindole (DAPI) was used for mounting slides and detection of nuclei (P36931; Invitrogen). Microscopy was performed using an Olympus IX71 inverted fluorescent microscope equipped with a DP72 digital camera and cellSens software.
Table 1.
Antibody Information
| Antibody | Company | Catalog number | Dilution |
|---|---|---|---|
| ADAM10 | R&D Systems, Mineapolis, MN | FAB946P | 1:100 |
| Alkaline phosphatase | Rockland, Limerick, PA | 200-4135S | 1:2000 |
| β-catenin | Cell Signaling, Danvers, MA | 9587S | 1:100 |
| β-catenin | BD Biosciences, San Jose, CA | 610154 | 1:100 |
| Chromogranin A | Abcam, Cambridge, MA | Ab15160-1 | 1:200 |
| E-cadherin | Cell Signaling | 3195S | 1:200 |
| F4/80 | BioLegend, San Diego, CA | 122601 | 1:100 |
| GFP | Invitrogen, ThermoFisher Scientific | A10262 | 1:200–1:400 |
| Ki67 | Abcam | Ab16667 | 1:100 |
| Lysozyme | Dako, Santa Clara, CA | A0099 | 1:600–1:1000 |
| MMP7 | Vanderbilt Antibody and Protein Resource, Nashville, TN | 1:200 | |
| Muc2 | Santa Cruz Biotech, Dallas, TX | Sc-15334 | 1:200 |
| Olfm4 | Cell Signaling | 39141S | 1:200 |
| Sox9 | Millipore, Temecula, CA | AB5535 | 1:200 |
| UEA1 lectin | Sigma, St. Louis, MO | L9006 | 1:100 |
| Villin | Abcam | Ab130751 | 1:1000 |
| Anti-chicken IgY Alexa-488 | Jackson ImmunoResearch, West Grove, PA | 703-545-155 | 1:200 |
| Anti-chicken IgY Cy3 | Jackson ImmunoResearch | 703-165-155 | 1:200 |
| Anti-rabbit IgG Alexa-488 | Jackson ImmunoResearch | 711-545-152 | 1:200 |
| Anti-rabbit IgG Cy3 | Jackson ImmunoResearch | 711-165-152 | 1:200–1:800 |
| Anti-rabbit IgG FITC | Jackson ImmunoResearch | 711-095-152 | 1:200–1:600 |
| Anti-rat IgG Alexa-488 | Jackson ImmunoResearch | 712-545-153 | 1:100 |
| Anti-rat IgG Cy3 | Jackson ImmunoResearch | 712-165-153 | 1:100–1:200 |
| Anti-rat IgG FITC | Jackson ImmunoResearch | 712-095-153 | 1:100 |
| Anti-mouse CD326 (EpCAM) FITC | eBioscience | 11-5791-82 | 1:66 |
| Rat IgG2a K isotype control FITC | eBioscience | 11-4321-82 | 1:66 |
FITC, fluorescein isothiocyanate; MMP7, matrix metalloproteinase 7.
Quantification of % Tomato+ that are lysozyme+ cells and the percentage of lysozyme+ that are Tomato+ cells was determined by counting Tomato+ and lysozyme+ cells within 25 well-oriented crypts in each region (duodenum, jejunum, and ileum) of the small intestine for each mouse (N = 3 mice). The mean values from each region of each mouse were used to calculate the means and SD for each region.
Cell positional distribution of EdU+ cells in crypts was determined as described previously.18 Briefly, frozen sections (4–8 μm) of small intestine were cut, and EdU-labeled cells were detected as described earlier. Well-orientated half-crypts (14–21 crypts per intestine, N = 3 mice/genotype) were scored for the presence of EdU+ nuclei at all the cell positions along the crypt axis, with position 1 being the middle of the crypt base. The frequency of each event at each position was plotted.
Villus Height and Crypt Depth Measurements
For each genotype (N = 3–5 mice), villus height and crypt depth were measured from well-orientated ileal crypts (35–69 crypts per mouse) using Olympus cellSens software. The mean value from each mouse was used to calculate means and SD for each genotype.
Lineage Tracing of Crypt–Villus Units
For analysis of lineage tracing, a crypt–villus unit was defined on histologic cross-section when the crypt lumen was contiguous with the crypt base and both sides of the crypt exited onto the villus as previously described.18 For each genotype, Defa4Cre;Rosa26tdTomato and Defa4Cre;ADAM10flox/flox;Rosa26tdTomato, a total of control untreated (N = 8) and DXR-treated (N = 18) mice were analyzed for lineage tracing. For DXR-treated mice, quantification of Tomato+ lineage tracing events within tissue sections from a 5-cm region of proximal jejunum was performed 7–10 days after treatment. The mean values from each mouse were used to calculate the means and SD for each treatment group.
In Vitro Culture of Intestinal Crypts
Intestinal crypts were isolated from the jejunum (5-cm region located 2 cm from the gastroduodenal junction) and ileum (5-cm region, distal ileum) as previously described.65 Crypts were plated in ice-cold Reduced Growth Factor Basement Membrane Extract (BME) (Cultrex, R&D Systems, Minneapolis, MN) (15 μL) into wells of 48-well plates and grown in 300 μL basal ENR media. The basal ENR culture medium (advanced Dulbecco’s modified Eagle medium/F12 [no. 12634020] supplemented with 1× penicillin/streptomycin [no. 15070063], 10 mmol/L HEPES [no. 15630080], 1× Glutamax [no. 35050061], 1× B27 [no. 17504044]; all from Life Technologies, Carlsbad, CA), and 1 mmol/L N-acetylcysteine (A7250; Sigma-Aldrich, St. Louis, MO) was supplemented with 50 ng/mL murine recombinant epidermal growth factor (315-09; PeproTech, Rocky Hill, NJ), 25 ng/mL murine recombinant Noggin (250-38; PeproTech), and R-spondin 2 (conditioned medium, 5%–10% final volume). When needed, single cells or enteroids were grown in WENR media, in which the earlier-described basal media had recombinant Noggin and R-spondin 2–conditioned media replaced with 50% Wnt3a, Noggin, R-Spondin 3 (WNR)-conditioned media. R-spondin 2 and WNR-conditioned media was produced using HEK293T cells stably transfected with mouse Rspondin2-chimeric fusion tag (Fc)66 and L cells stably transfected with mouse Wnt-3A, R-spondin 3, and Noggin (provided by Dr Stappenbeck, Washington University, St. Louis, MO).67 Media was replaced every 2–3 days.
Epithelial Cell Isolation, Flow Cytometry, and FACS Sorting
Single-cell suspensions for flow cytometric analysis and sorting were generated from isolated jejunal crypts (a 2- to 12-cm segment from the gastroduodenal junction) of Defa4Cre;Rosa26tdTomato and Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice as previously described.68 Briefly, isolated crypts were incubated with dispase (0.3 U/mL, 17105041; LifeTech) and DNaseI (0.1 mg/mL, 17105041; LifeTech), cell suspensions were filtered through a 40-μm cell strainer and then stained with rat anti-EpCAM–fluorescein isothiocyanate IgG2a antibody (epithelial cell marker, 11-5791-82; eBioscience, San Diego, CA) and DAPI (1 μg/mL, live/dead cell marker). Rat IgG2a–fluorescein isothiocyanate (11-4321-82; eBioscience) was used as a control. The MoFlo XDP100 cytometer (Beckman Coulter, Indianapolis, IN) with a 100-μm nozzle was used for single-cell FACS sorting EpCAM+ tdTomato- and EpCAM+ tdTomato+ cell populations. For the clonogenic enteroid assay, FACS-sorted Tomato- and Tomato+ cells were plated in ice-cold Basement Membrane Extract (BME) and grown in either 300 μL of ENR or WENR media containing 10 μmol/L Y27632. Media without Y27632 was replaced every 2–3 days. Enteroid growth was monitored daily by phase-contrast microscopy combined with Tomato+ fluorescent imaging using an Olympus IX71 inverted fluorescent microscope, DP72 digital camera, and cellSens imaging software. In general, budding from clonigenic enteroids occurred after day 7 in culture, however, Figure 3D shows budding that occurred much later, between day 13 and day 16. The percentage of clonogenic enteroid growth = total number of enteroids generated/total number of single cells plated ×100.
For flow cytometric analysis of EdU-labeled enteroids, enteroids grown in Basement Membrane Extract (BME) were labeled in vitro with 10 μmol/L 5-ethyl deoxyuridine solution in ENR media for 1 hour at 37°C. Enteroids were collected in ice-cold 5 mmol/L EDTA–PBS (Invitrogen, ThermoFisher Scientific, Grand Island, NY), washed in PBS, and then incubated in TrypLE (Gibco, ThermoFisher Scientific, Grand Island, NY) containing 0.1 mg/mL DNaseI (Calbiochem, EMD Biosciences, San Diego, CA) to generate a single-cell suspension. Cells then were fixed in 4% PFA, and then permeabilized with 0.1% Triton X-100 in PBS before detection with the Click-iT reaction and Alexa Fluor 488 using the manufacturer’s protocol. Cell suspension was filtered through a 40-μm cell strainer before staining with DAPI (1 μg/mL), and then analyzed using a Yeti flow cytometer (Propel Labs, Fort Collins, CO).
Inducible Notch Activation in Mice
To induce Notch activation, adult Defa4Cre;Rosa26rtTA-ires-EGFP;TetONICD mice were given doxycycline (2 mg/mL, D9891; Sigma) supplemented with 5% sucrose in water for 2 weeks. Tissue was collected as previously described.
DXR Studies
For in vivo DXR experiments, Defa4Cre;Rosa26tdTomato and Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice were injected intraperitoneally with DXR (15 mg/kg body weight, 1 mg/mL in PBS, D1515; Sigma) and tissue was collected at various time points after injection (6 h, 24 h, 4 days, 7 days, and 10 days) and compared with vehicle-injected controls (N = 3–18). The number of Tomato+ crypt–villus units was counted from frozen sections of small intestine and expressed as the total number of Tomato+ crypt–villus units per 5-cm intestinal section (N = 8–18 per genotype).
For in vitro DXR experiments, jejunal enteroids from Defa4Cre;Rosa26tdTomato and Defa4Cre;ADAM10flox/flox;Rosa26tdTomato mice were passaged in Matrigel and grown in ENR containing 10 μmol/L Rho-associated, coiled-coil containing protein kinase (ROCK) inhibitor Y27682 (Reprocell, Beltsville, MD) for 24 hours. The total number of enteroids and spontaneous Tomato+ lineage-traced enteroids (these included complete Tomato+ enteroids and enteroids with Tomato+ lineage-traced buds) were counted. Enteroid cultures then were treated with 7.5 μg/mL DXR in 300 μL ENR media for 3 hours, wells were washed twice, and fresh ENR media added. Enteroid cultures then were re-fed every 2–3 days and, if necessary, were replated for imaging. The total number of surviving enteroids and the number of Tomato+ lineage-traced enteroids was counted.
ADAM10 Recombination and Gene Expression Analysis
In the ADAM10 floxed allele, exon 9, which encodes the catalytic domain, is flanked by loxP sites inserted in introns 8 and 9. Intron 8 forward primer (5’-CAGTGTAAATGTGAACTCACCC-3’) and intron 9 reverse primer (5’-CGTATCTCAAAACTACCCTCCC-3’) will amplify a 955-bp sequence of nonrecombined ADAM10 floxed allele or a 217-bp sequence of the recombined allele from genomic DNA.69
ADAM10 gene expression was determined by reverse-transcription (RT)-PCR with N = 3 mice per group. RNA was isolated from intestinal tissue or enteroids using TRIzol (15596026; Invitrogen) or Buffer RLT (Qiagen, Hilden, Germany), respectively, followed by purification with the RNeasy kit (74104; Qiagen). RT reactions used 0.4 μg RNA and the High Capacity complementary DNA Reverse Transcriptase Kit (438814; Applied Biosystems, ThermoFisher Scientific, Foster City, CA), following the manufacturer's instructions. RT-PCR was performed with ADAM10 exon 8 forward primer (5’-CAGACCGGGATTTTGATGAT-3’) and exon 9 reverse primer (5’-TCCAACTTCATGAGCAAACG-3’) to generate a 217-bp product. Gene expression levels were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase, which remained the same in the various groups.
Statistics
Quantitative data are presented as the mean of independent animals or samples ± SD and analyzed by the 2-tailed Student t test. For cell positional analysis, differences between groups were determined by 2-way analysis of variance with the Bonferroni multiple comparisons test. P < .05 was considered significant. Statistical analysis was performed using Prism 6 software (GraphPad, La Jolla, CA).
Footnotes
Author contributions Jennifer C. Jones and Peter J. Dempsey conceived the study, designed the experiments, supervised the study, and prepared the manuscript; Jennifer C. Jones, Constance D. Brindley, Nicholas H. Elder, and Peter J. Dempsey acquired, analyzed, and interpreted data; Martin G. Myers Jr, Michael W. Rajala, Christopher M. Dekaney, Eoin N. McNamee, Noah F. Shroyer, and Mark R. Frey provided material support and advice on experimental design; and Peter J. Dempsey provided funding. All authors reviewed and approved the submitted manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by National Institutes of Health grants R01-DK093697 (P.J.D.), RNA Bioscience Institute Pilot and Feasibility grant, and the Gastrointestinal and Liver Innate Immunity program Pilot and Feasibility grant from the University of Colorado Medical School (P.J.D.). This work also was supported by the Flow Cytometry Shared Resource, which receives direct funding support from the National Cancer Institute through a Cancer Center support grant (P30CA046934).
References
- 1.Beumer J., Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 2.Yousefi M., Li L., Lengner C.J. Hierarchy and plasticity in the intestinal stem cell compartment. Trends Cell Biol. 2017;27:753–764. doi: 10.1016/j.tcb.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., van Oudenaarden A., Clevers H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 5.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., Martens A.C.M., Barker N., van Oudenaarden A., Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S., Breault D.T. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jadhav U., Saxena M., O'Neill N.K., Saadatpour A., Yuan G.C., Herbert Z., Murata K., Shivdasani R.A. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell. 2017;21:65–77 e5. doi: 10.1016/j.stem.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., Guo Y., Shyr Y., Aronow B.J., Haigis K.M., Franklin J.L., Coffey R.J. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Landeghem L., Santoro M.A., Krebs A.E., Mah A.T., Dehmer J.J., Gracz A.D., Scull B.P., McNaughton K., Magness S.T., Lund P.K. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–G1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan K.S., Gevaert O., Zheng G.X.Y., Anchang B., Probert C.S., Larkin K.A., Davies P.S., Cheng Z.F., Kaddis J.S., Han A., Roelf K., Calderon R.I., Cynn E., Hu X., Mandleywala K., Wilhelmy J., Grimes S.M., Corney D.C., Boutet S.C., Terry J.M., Belgrader P., Ziraldo S.B., Mikkelsen T.S., Wang F., von Furstenberg R.J., Smith N.R., Chandrakesan P., May R., Chrissy M.A.S., Jain R., Cartwright C.A., Niland J.C., Hong Y.K., Carrington J., Breault D.T., Epstein J., Houchen C.W., Lynch J.P., Martin M.G., Plevritis S.K., Curtis C., Ji H.P., Li L., Henning S.J., Wong M.H., Kuo C.J. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21:78–90 e6. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 14.Li N., Nakauka-Ddamba A., Tobias J., Jensen S.T., Lengner C.J. Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastroenterology. 2016;151:298–310 e7. doi: 10.1053/j.gastro.2016.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu S., Tong K., Zhao Y., Balasubramanian I., Yap G.S., Ferraris R.P., Bonder E.M., Verzi M.P., Gao N. Paneth cell multipotency induced by notch activation following injury. Cell Stem Cell. 2018;23:46–59 e5. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demitrack E.S., Samuelson L.C. Notch regulation of gastrointestinal stem cells. J Physiol. 2016;594:4791–4803. doi: 10.1113/JP271667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones J.C., Rustagi S., Dempsey P.J. ADAM proteases and gastrointestinal function. Annu Rev Physiol. 2016;78:243–276. doi: 10.1146/annurev-physiol-021014-071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai Y.H., VanDussen K.L., Sawey E.T., Wade A.W., Kasper C., Rakshit S., Bhatt R.G., Stoeck A., Maillard I., Crawford H.C., Samuelson L.C., Dempsey P.J. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834 e13. doi: 10.1053/j.gastro.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mei H., Yu L., Ji P., Yang J., Fang S., Guo W., Liu Y., Chen X. Doxorubicin activates the Notch signaling pathway in osteosarcoma. Oncol Lett. 2015;9:2905–2909. doi: 10.3892/ol.2015.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potten C.S., Booth C., Tudor G.L., Booth D., Brady G., Hurley P., Ashton G., Clarke R., Sakakibara S., Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 22.Qu D., May R., Sureban S.M., Weygant N., Chandrakesan P., Ali N., Li L., Barrett T., Houchen C.W. Inhibition of Notch signaling reduces the number of surviving Dclk1+ reserve crypt epithelial stem cells following radiation injury. Am J Physiol Gastrointest Liver Physiol. 2014;306:G404–G411. doi: 10.1152/ajpgi.00088.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burger E., Araujo A., Lopez-Yglesias A., Rajala M.W., Geng L., Levine B., Hooper L.V., Burstein E., Yarovinsky F. Loss of Paneth cell autophagy causes acute susceptibility to Toxoplasma gondii-mediated inflammation. Cell Host Microbe. 2018;23:177–190 e4. doi: 10.1016/j.chom.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darmoul D., Brown D., Selsted M.E., Ouellette A.J. Cryptdin gene expression in developing mouse small intestine. Am J Physiol. 1997;272:G197–G206. doi: 10.1152/ajpgi.1997.272.1.G197. [DOI] [PubMed] [Google Scholar]
- 25.Darmoul D., Ouellette A.J. Positional specificity of defensin gene expression reveals Paneth cell heterogeneity in mouse small intestine. Am J Physiol. 1996;271:G68–G74. doi: 10.1152/ajpgi.1996.271.1.G68. [DOI] [PubMed] [Google Scholar]
- 26.Ouellette A.J., Darmoul D., Tran D., Huttner K.M., Yuan J., Selsted M.E. Peptide localization and gene structure of cryptdin 4, a differentially expressed mouse Paneth cell alpha-defensin. Infect Immun. 1999;67:6643–6651. doi: 10.1128/iai.67.12.6643-6651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 28.Kraiczy J., Nayak K.M., Howell K.J., Ross A., Forbester J., Salvestrini C., Mustata R., Perkins S., Andersson-Rolf A., Leenen E., Liebert A., Vallier L., Rosenstiel P.C., Stegle O., Dougan G., Heuschkel R., Koo B.K., Zilbauer M. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68:49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T.H., Li F., Ferreiro-Neira I., Ho L.L., Luyten A., Nalapareddy K., Long H., Verzi M., Shivdasani R.A. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Flier L.G., Haegebarth A., Stange D.E., van de Wetering M., Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 31.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud A., Grosse A.S., Gumucio D.L., Ernst S.A., Tsai Y.H., Dempsey P.J., Samuelson L.C. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouellette A.J., Hsieh M.M., Nosek M.T., Cano-Gauci D.F., Huttner K.M., Buick R.N., Selsted M.E. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62:5040–5047. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanger B.Z., Datar R., Murtaugh L.C., Melton D.A. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Gibson P., Currle D.S., Tong Y., Richardson R.J., Bayazitov I.T., Poppleton H., Zakharenko S., Ellison D.W., Gilbertson R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregorieff A., Stange D.E., Kujala P., Begthel H., van den Born M., Korving J., Peters P.J., Clevers H. The ets-domain transcription factor Spdef promotes maturation of goblet and Paneth cells in the intestinal epithelium. Gastroenterology. 2009;137:1333–1345 e1-3. doi: 10.1053/j.gastro.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 37.Noah T.K., Kazanjian A., Whitsett J., Shroyer N.F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp Cell Res. 2010;316:452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shroyer N.F., Wallis D., Venken K.J., Bellen H.J., Zoghbi H.Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastide P., Darido C., Pannequin J., Kist R., Robine S., Marty-Double C., Bibeau F., Scherer G., Joubert D., Hollande F., Blache P., Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori-Akiyama Y., van den Born M., van Es J.H., Hamilton S.R., Adams H.P., Zhang J., Clevers H., de Crombrugghe B. SOX9 is required for the differentiation of Paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Andreu P., Colnot S., Godard C., Gad S., Chafey P., Niwa-Kawakita M., Laurent-Puig P., Kahn A., Robine S., Perret C., Romagnolo B. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 42.Andreu P., Peignon G., Slomianny C., Taketo M.M., Colnot S., Robine S., Lamarque D., Laurent-Puig P., Perret C., Romagnolo B. A genetic study of the role of the Wnt/beta-catenin signalling in Paneth cell differentiation. Dev Biol. 2008;324:288–296. doi: 10.1016/j.ydbio.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 43.Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayase E., Hashimoto D., Nakamura K., Noizat C., Ogasawara R., Takahashi S., Ohigashi H., Yokoi Y., Sugimoto R., Matsuoka S., Ara T., Yokoyama E., Yamakawa T., Ebata K., Kondo T., Hiramine R., Aizawa T., Ogura Y., Hayashi T., Mori H., Kurokawa K., Tomizuka K., Ayabe T., Teshima T. R-spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J Exp Med. 2017;214:3507–3518. doi: 10.1084/jem.20170418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sansom O.J., Reed K.R., Hayes A.J., Ireland H., Brinkmann H., Newton I.P., Batlle E., Simon-Assmann P., Clevers H., Nathke I.S., Clarke A.R., Winton D.J. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mili S., Moissoglu K., Macara I.G. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preitner N., Quan J., Nowakowski D.W., Hancock M.L., Shi J., Tcherkezian J., Young-Pearse T.L., Flanagan J.G. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–382. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith K.J., Levy D.B., Maupin P., Pollard T.D., Vogelstein B., Kinzler K.W. Wild-type but not mutant APC associates with the microtubule cytoskeleton. Cancer Res. 1994;54:3672–3675. [PubMed] [Google Scholar]
- 49.Huels D.J., Sansom O.J. Stem vs non-stem cell origin of colorectal cancer. Br J Cancer. 2015;113:1–5. doi: 10.1038/bjc.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metcalfe C., Kljavin N.M., Ybarra R., de Sauvage F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Asfaha S., Hayakawa Y., Muley A., Stokes S., Graham T.A., Ericksen R.E., Westphalen C.B., von Burstin J., Mastracci T.L., Worthley D.L., Guha C., Quante M., Rustgi A.K., Wang T.C. Krt19(+)/Lgr5(-) cells are radioresistant cancer-initiating stem cells in the colon and intestine. Cell Stem Cell. 2015;16:627–638. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis H., Irshad S., Bansal M., Rafferty H., Boitsova T., Bardella C., Jaeger E., Lewis A., Freeman-Mills L., Giner F.C., Rodenas-Cuadrado P., Mallappa S., Clark S., Thomas H., Jeffery R., Poulsom R., Rodriguez-Justo M., Novelli M., Chetty R., Silver A., Sansom O.J., Greten F.R., Wang L.M., East J.E., Tomlinson I., Leedham S.J. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat Med. 2015;21:62–70. doi: 10.1038/nm.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwitalla S., Fingerle A.A., Cammareri P., Nebelsiek T., Goktuna S.I., Ziegler P.K., Canli O., Heijmans J., Huels D.J., Moreaux G., Rupec R.A., Gerhard M., Schmid R., Barker N., Clevers H., Lang R., Neumann J., Kirchner T., Taketo M.M., van den Brink G.R., Sansom O.J., Arkan M.C., Greten F.R. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Westphalen C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D.J., Nuber A.H., Brandtner A., Setlik W., Remotti H., Muley A., Chen X., May R., Houchen C.W., Fox J.G., Gershon M.D., Quante M., Wang T.C. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124:1283–1295. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dekaney C.M., Gulati A.S., Garrison A.P., Helmrath M.A., Henning S.J. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am J Physiol Gastrointest Liver Physiol. 2009;297:G461–G470. doi: 10.1152/ajpgi.90446.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King S.L., Mohiuddin J.J., Dekaney C.M. Paneth cells expand from newly created and preexisting cells during repair after doxorubicin-induced damage. Am J Physiol Gastrointest Liver Physiol. 2013;305:G151–G162. doi: 10.1152/ajpgi.00441.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanderson M.P., Abbott C.A., Tada H., Seno M., Dempsey P.J., Dunbar A.J. Hydrogen peroxide and endothelin-1 are novel activators of betacellulin ectodomain shedding. J Cell Biochem. 2006;99:609–623. doi: 10.1002/jcb.20968. [DOI] [PubMed] [Google Scholar]
- 58.Roth S., Franken P., Sacchetti A., Kremer A., Anderson K., Sansom O., Fodde R. Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 2012;7:e38965. doi: 10.1371/journal.pone.0038965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., Lein E.S., Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S.E., Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shibata H., Toyama K., Shioya H., Ito M., Hirota M., Hasegawa S., Matsumoto H., Takano H., Akiyama T., Toyoshima K., Kanamaru R., Kanegae Y., Saito I., Nakamura Y., Shiba K., Noda T. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 64.Kontoyiannis D., Pasparakis M., Pizarro T.T., Cominelli F., Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 65.Mahe M.M., Aihara E., Schumacher M.A., Zavros Y., Montrose M.H., Helmrath M.A., Sato T., Shroyer N.F. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol. 2013;3:217–240. doi: 10.1002/9780470942390.mo130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bell S.M., Schreiner C.M., Wert S.E., Mucenski M.L., Scott W.J., Whitsett J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–1058. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- 67.Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magness S.T., Puthoff B.J., Crissey M.A., Dunn J., Henning S.J., Houchen C., Kaddis J.S., Kuo C.J., Li L., Lynch J., Martin M.G., May R., Niland J.C., Olack B., Qian D., Stelzner M., Swain J.R., Wang F., Wang J., Wang X., Yan K., Yu J., Wong M.H. A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305:G542–G551. doi: 10.1152/ajpgi.00481.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibb D.R., El Shikh M., Kang D.J., Rowe W.J., El Sayed R., Cichy J., Yagita H., Tew J.G., Dempsey P.J., Crawford H.C., Conrad D.H. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207:623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]