Abstract
Maintenance of biomedical ontologies is difficult. We have previously developed a topological-pattern-based method to deal with the problem of identifying concepts in a reference ontology that could be of interest for insertion into a target ontology. Assuming that both ontologies are parts of the Unified Medical Language System (UMLS), the method suggests approximate locations where the target ontology could be extended with new concepts from the reference ontology. However, the final decision about each concept has to be made by a human expert. In this paper, we describe the universe of cross-ontology topological patterns in quantitative terms. We then present a theoretical analysis of the number of potential placements of reference concepts in a path in a target ontology, allowing for new cross-ontology synonyms. This provides a rough estimate of what expert resources need to be allocated for the task. One insight in previous work on this topic was the large percentage of cases where importing concepts was impossible, due to a configuration called “alternative classification.” In this paper, we confirm this observation. Our target ontology is the National Cancer Institute thesaurus (NCIt). However, the methods can be applied to other pairs of ontologies with hierarchical relationships from the UMLS.
Keywords: Biomedical ontology, Ontology maintenance, Concept Insertion, NCIt
I. Introduction
Biomedical ontologies lay a solid foundation in various healthcare information systems [1, 2], especially for encoding diagnoses, laboratory tests, problem lists in Electronic Health Records [3, 4] and in administrative documents such as billing statements [5]. Moreover, they also play an important role in knowledge management, data integration, and decision support, with their rich semantic relationships between concepts [1]. In Cimino’s “desiderata” of biomedical ontologies in the 21st Century [6], domain completeness is deemed to be the most desirable property of an ontology. With the non-stop growth of medical knowledge, it is essential that curators of a biomedical ontology keep incorporating new concepts to improve its domain coverage [6]. The traditional top-down approach for ontology development involves iterative discussions among ontology developers and domain experts, which is labor intensive and time consuming [7]. Thus, automated and semi-automated ontology learning methods, which may ease the burden of ontology developers and accelerate ontology development, are highly desired [8–12].
The Unified Medical Language System (UMLS) Metathesaurus integrates over 10 million terms from about 207 source vocabularies into 3.6 million concepts, such that terms with the same meaning are assigned the same Concept Unique Identifier (CUI) [13]. In previous research, we have introduced a structural methodology to mine new concepts from a UMLS source for inclusion in another UMLS source where they might be “missing” [14, 15]. This methodology leverages the native term mappings of the UMLS to identify cross-ontology topological patterns that are indicative of a possible import. We found candidate concepts for import into SNOMED CT and domain experts confirmed the validity of this method [14, 15]. We extended this work subsequently with a focus on the National Cancer Institute thesaurus (NCIt), a major reference terminology for cancer [16–18].
Cross-ontology patterns rarely identify a unique location where a reference concept should be inserted. Rather, the patterns provide several choices. A human expert has to determine which, if any, of the choices is correct and desirable. It is of interest to identify the number of possible placement choices, to estimate the difficulty of the task for the human expert. The work by He and Geller [17] was a preliminary analysis of the number of different ways that concepts from a reference ontology, taking part in a cross-ontology topological pattern, can be inserted into a target ontology. The results of He and Geller showed that if the possibility of new cross-ontology synonyms is excluded, the number of possible insertions can be described by a combinatorial formula [17].
However, it was observed that two ontologies might use two different terms to describe the same real-world concept, which makes them synonyms. While the UMLS is an extensive repository of synonyms, cases were observed where two “real world synonymous terms” were not recorded as synonyms, neither in the reference ontology, nor in the target ontology. This may be due to the imperfect semi-automated ontology integration of the UMLS. The previous work [17] dealt only with the situation of inserting a reference concept between target concepts. In this paper, we allow for the possibility of new cross-ontology synonyms and derive a result for the number of choices that a human expert faces in this extended scenario.
When a concept is imported as a synonym, the number of concepts in the target ontology stays the same and more details are added to one target concept, namely the new synonym. One of the surprising results of previous work [14–16] was the large percentage of alternative classifications that were encountered. At this point, we will demonstrate “alternative classifications” with an example. The concept Gastrointestinal Diseases may be shared between two ontologies. In one and only one ontology it has the immediate sub-concept (child) Gallbladder and biliary tree disorders. In the other ontology (and only there), Gastrointestinal Diseases has the sub-concept Gastrointestinal polyps. These two concepts are hierarchically incompatible, because Gallbladder and biliary tree disorders specializes Gastrointestinal Diseases by anatomical location. On the other hand, Gastrointestinal polyps specializes Gastrointestinal Diseases by disease kind. Thus, there is no “clean and easy” way to combine these two children in one ontology. Moreover, if one follows down two separate paths in the two ontologies, a unique, more specialized concept may be encountered that is again shared by both ontologies. The problem of alternative classifications is that they inhibit insertion and cross-ontology synonyms. Thus, there is an interest in better understanding the prevalence and nature of alternative classifications in pairs of ontologies.
Our task has some similarity with ontology integration/alignment [19], which was originated in the database integration area [20]. The goal of database integration is to integrate both the tables and the attributes from different databases. The goal of ontology integration is typically to create one target ontology that contains all the knowledge of two source ontologies [21]. Note that in this work we focus on concept imports, which is different from ontology integration. The goal of import is to “cherry pick” concepts from one ontology that could be deemed missing in another ontology.
The main contribution of this paper is the extended catalog of useful topological patterns for which we show that they can identify more concepts for importing into a target ontology. The other contributions of this paper are as follows. First, we state numeric results about the occurrence of cross-ontology topological patterns found in the UMLS that involve the NCIt as target ontology. Second, we generalize the analysis of He and Geller [17] to include cases where cross-ontology synonyms are allowed. We present a recursive formula that allows us to compute the number of possible insertion/synonym choices into the target ontology, which allows us to estimate the difficulty of the task for a human expert. Third, we present the results of a study by a medical ontology/terminology expert, evaluating two samples of cross-ontology topological patterns that confirm the large percentages of alternative classifications that were observed in previous studies. Last but not least, the uptake of our methods by the curators of major medical ontologies/terminologies would lead to more comprehensive ontologies.
II. Background and Related Work
A. The National Cancer Institute Thesaurus
The NCIt contains over 100,000 concepts that are hierarchically organized in 19 distinct domains related to cancer research, e.g., neoplastic diseases, molecular abnormalities, and genes. It is a central reference ontology of NCI’s Enterprise Vocabulary Services (EVS) [22]. As new concepts are entering healthcare usage, NCIt needs to be extended as needed by its users. NCI EVS exploits internal quality assurance (QA) mechanisms as well as external participation in the QA process of NCIt.
Quality assurance of the NCIt has been conducted by NCI and external researchers [22]. Min et al. constructed a partial-area taxonomy that highlighted potential modeling errors and inconsistencies in NCIt [23]. Cohen et al. conducted an automated comparative audit of the gene hierarchy of NCIt with the Entrez Gene database [24]. Mougin and Bodenreider assessed the consistency of the relationships in NCIt by storing the NCIt concepts in an RDF triple store [25]. Jiang et al. evaluated the data quality of common data elements in the context of cancer research by integrating the NCI Cancer Data Standards Repository, NCIt concepts, and the UMLS Semantic Network with a Semantic Web-based framework for SPARQL-enabled evaluation [26]. An especially notable fact is that the NCI manages its own version of the Metathesaurus, specialized for the needs of cancer research [27].
B. Related Work on UMLS Hierarchical Relationships and Imbalanced Granularity
Previous research on ontology quality assurance (OQA) often emphasized hierarchical/taxonomic relationships (e.g., “IS-A”). As early as in 2003, Bodenreider performed an analysis of redundant hierarchical relationships across families of ontologies in the UMLS and their semantic consistency [28]. Gu et al. [29] detected the transitive structural relationships in the Foundational Model of Anatomy (FMA) and categorized possibly incorrect relationships into five major categories: circular, mutually exclusive, redundant, inconsistent, and missed entries in the FMA. Imbalanced granularity of hierarchical relationships is a barrier for semantic interoperability [30], ontology mapping [7], and data integration [7]; meanwhile, it provides opportunities for ontology enhancement [2, 6, 15]. The existing OQA methods for investigating the imbalanced granularity of hierarchical relationships are limited. Sun and Zhang used string matching to construct rules and further used the rules to identify granularity differences among large biomedical ontologies [31, 32].
Cornet proposed an information-content-based method to improve the conceptual content of an ontology and balance the granularity levels between hierarchies [33]. Our topological-pattern-based method aims to identify useful concepts for importing into an ontology with the use of hierarchical relations and the native term mappings in the UMLS [14–16]. The topological patterns with imbalanced granularity between ontologies can efficiently and effectively highlight potentially useful concepts for ontology enrichment. However, due to the fact that a large percentage of the cases are alternative classifications, the true positive rate for concept import is low.
C. Related Work on Ontology Enrichment and Learning
The development of an ontology is divided into seven tasks, the discovery of 1) terms, 2) synonyms, 3) concepts, 4) hierarchical concepts, 5) relations, 6) hierarchical relations, and 7) axioms [34]. As early as 2004, Smith and Fellbaum used online health information sources to build the Medical WordNet from two corpora, namely Medical FactNet and Medical BeliefNet [35]. Recent work on semi/fully-automated ontology learning focuses on the extraction of terms and predicates (i.e., triples) from a text corpus [12, 36, 37]. For example, our group recently developed an open source text mining tool called simiTerm to identify the terms in a text corpus that are contextually and syntactically similar to existing terms in an ontology [11]. Hoxha et al. [8] developed Ontofier, an unsupervised ontology learning framework that uses agglomerative hierarchical clustering to learn domain taxonomies from biomedical texts. These methods, which focused on building lightweight ontologies, do not intend to enrich existing ontologies with new concepts and cannot identify the locations for them in the existing ontology. Our proposed method, which leverages the taxonomic structure of ontologies and the native term mapping of the UMLS, can fill this gap by identifying new concepts and new synonyms for an existing ontology, as well as the ways in which they should be added.
III. Methods
A. Basic-Cross Ontology Structure
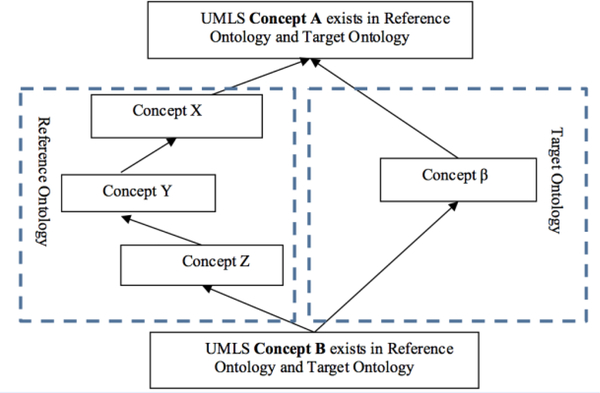
We are operating on topological patterns that span pairs of ontologies within the UMLS. To make these patterns comprehensible to the reader, we provide an abstract example in Figure 1. We are singling out two ontologies within the UMLS, called the reference ontology and the target ontology. The goal of the research is to extend the target ontology with concepts from the reference ontology.
Fig. 1.
A Topological Structure in the UMLS, called a Cross-Ontology Diamond. The arrows represent “IS-A” hierarchical relationships.
In the illustrative example in Figure 1, the concepts A, B, β, X, Y, and Z all appear in the UMLS. The concepts X, Y, and Z appear in the reference ontology, but not in the target ontology. The concept β exists in the target ontology, but not in the reference ontology. The concepts A and B exist in both the reference ontology and the target ontology.
The main criteria for selecting a reference ontology and a target ontology are that both must be organized around an IS-A hierarchy backbone and must exhibit a sufficient overlap in content with each other. Furthermore, the target ontology is selected based on scientific or commercial interest in extending it. Due to our own limitations, we consider only English-language ontologies.
Looking at Figure 1, the question arises whether X or Y or Z or all of them should be considered as missing in the target ontology. This is not necessarily the case. It could happen that
X or Y or Z is a synonym of β. In this paper, these are allowable cases that are included in the analysis.
There is an error in the reference ontology or there is an error in the target ontology, meaning that Figure 1 is not a correct reflection of the real world.
It could be that β and X are alternative classifications of the concept A. In other words, in an alternative classification there are two hyponyms (subclass) β and X of the same concept A so that β IS-A A and X IS-A A. However, β is neither a hyponym of X nor is β a hypernym (superclass) of X.
The decision whether X, Y and Z are valid imports into the target ontology or whether one of the other cases applies can only be made by a medical expert with a good knowledge of the underlying domain. We note that this is a two-step decision, as explained below.
Figure 1 is approximately ◊ (diamond) shaped. (In previous work, we referred to this as a “trapezoid.”) In this research, we are mining such diamond structures between pairs of UMLS sources. Every such diamond could indicate the possible import of concepts into the target ontology. We stress that Figure 1 is an example structure, and much more complex diamond structures do exist in the UMLS. Not every diamond is guaranteed to indicate reference concepts that should be imported into the target ontology. Specifically, there are the following considerations.
Some of the diamond structures overlap, which could lead to concepts being suggested twice or multiple times for import. Such duplicate concepts have to be eliminated, because a new concept should be imported only once. This was considered in our algorithms.
As noted, there might be alternative classifications that preclude importing a concept and also exclude concepts from being cross-ontology synonyms.
Semantically valid concepts might be undesirable for import according to the curators of the target ontology, e.g., because they would be without an interesting use case for the owners of the target ontology. Ontology curators do not wish to “clutter up” an ontology with unused concepts, even if those are “correct” concepts within the domain of the ontology.
Because of the above issues, the final decisions have to be made by a domain expert. In this paper, we are attempting to quantify the difficulty of the task of the expert. The more choices an expert has, the more work s/he has to do to decide whether to go ahead with importing a specific concept or not. Thus, we quantify the choices in this paper.
B. Selection Decision and Placement Decision
We analyze cross-ontology diamonds. Figure 1 is an illustrative example. We will use “diamond” to mean “cross-ontology diamond.” Diamonds may be of different sizes.
Definition 1: The two concepts of the cross-ontology diamond that are shared between the reference ontology and the target ontology are called anchor concepts. (A and B).
Definition 2: The concept that is more specific than all other concepts in a diamond is called the bottom anchor (B).
Definition 3: The concept that is more general than all other concepts in a diamond is called the top anchor (A).
Definition 4: All concepts strictly between the bottom anchor and the top anchor that exist in the reference ontology, but not in the target ontology, are called reference concepts (X, Y, Z).
Definition 5: All concepts strictly between the bottom anchor and the top anchor that exist in the target ontology, but not in the reference ontology, are target concepts (β).
Definition 6: The path of all reference concepts including the anchor concepts is called the reference path.
Definition 7: The path of all target concepts including the anchor concepts is called the target path.
Definition 8: A diamond with j reference concepts and l target concepts is called a j/l-diamond. It holds that j > 0 and l > 0 for all diamonds. (Figure 1 shows a 3/1-diamond.)
Definition 9: An alternative classification is a diamond in which the top anchor has one child that is a reference concept (Y) and one child that is a target concept (β) such that for semantic reasons β cannot be made a synonym of Y, nor can it be made a child of Y, nor a parent of Y.
Definition 10: The decision which reference concepts in a diamond should be imported into the target path is called the selection decision.
Definition 11: The decision where, in relation to existing target concepts, the selected reference concepts should be located in the target path is called the placement decision.
Definition 12: When all reference concepts are placed in the target ontology, i.e., when effectively no selection is performed, this is called a full placement. For example, in Figure 1, the selection decision consists of determining whether X alone, Y alone, Z alone, or a subset of {X, Y, Z} should be imported into the target ontology. If all of {X, Y, Z} are placed, this corresponds to full placement.
If the selection decisions are independent from each other then there are n decisions to be made for n reference concepts. In practice, the expert will often not be able to make a decision in isolation. For example, in Figure 1, an expert might decide that Y is too similar to X to warrant its inclusion, but X and Z are needed in the target path.
C. Placement Decision without Synonyms
In previous work [17], we showed that the total number of placement choices (#PC), assuming that there are no synonyms, is computed by
| (1) |
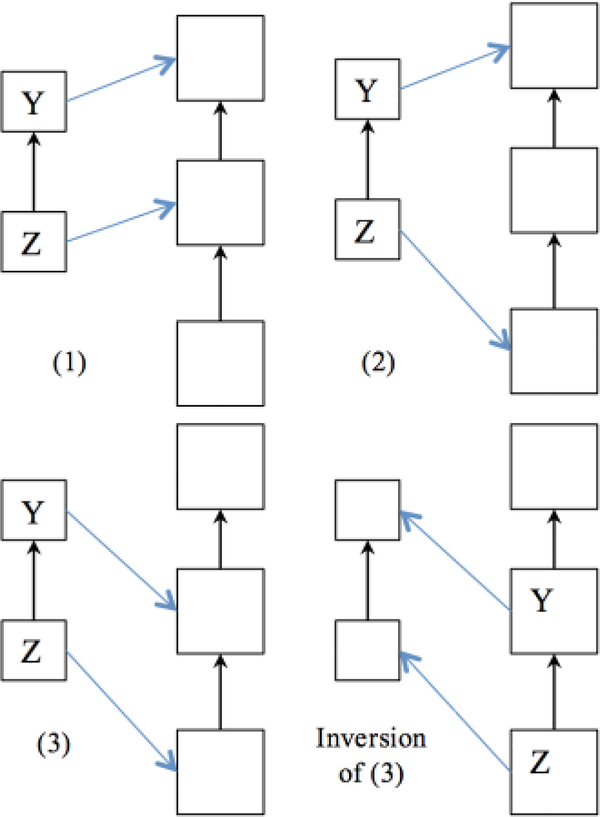
To make this paper self-contained here is a proof sketch. After importing m reference concepts into the target ontology, when there are already k concepts between the anchors on the target path, there will be a total of m + k concepts between the anchor concepts. Let us assume that there are m + k empty positions and we are assigning the m reference concepts first to these empty positions. After this assignment, there will be k empty positions left unfilled. The order of the k target concepts is fixed, because they must be in exactly the same order as before the import, although they might be separated by imported concepts. Thus, there is only one choice how to place the k target concepts after the m reference concepts have been placed. Therefore, the question is reduced to how many ways there are to place the m reference concepts in the m+k spaces. Figure 2 contains four sub-diagrams. The configurations numbered (1), (2), and (3) correspond to the three different possible placements.
Fig. 2.
Three Possible Cases of Placement and Inversion Case (3)
Now we invert the direction of the arrows in Figure 2. (For space reasons, this is only done for configuration 3.). Therefore, the problem is equivalent to the different ways how m elements can be chosen from a set of m + k elements, which is a well-known problem in combinatorics, solved by formula (1).
In our research, we have discovered diamonds with eight reference concepts. Hypothetically, if an expert decides that all should be imported into a specific target path, and there would be two target concepts then there are 45 placement choices, in the worst case.
| (2) |
D. Placement Decisions with Synonyms
We now advance to the case where new cross-ontology synonyms are permitted. Full placement in the example case of Figure 1 allows for the following target paths in Figure 3.
Fig. 3.
The target paths after the full placement in the sample case of Figure 1.
If the target ontology contains two concepts α, β between the anchor concepts, then the number of possible placements increases considerably. Capturing the number of placements requires a recursive definition as follows. The three base cases are:
| (3) |
| (4) |
| (5) |
The recursive case is defined by:
| (6) |
| (7) |
Now in formulas (3)–(7), p(.,.) stands for the number of possible placements. The value n indicates the number of concepts according to the selection decision. Therefore, n is less or equal to the number of reference concepts. The number k counts the target concepts. Several paths with different values of k may exist.
Proof: Base case 1, i.e., Formula (3): k=0 indicates that there are no intermediate concepts between A and B in the target path. Thus there is only one way to perform full placement, independent of the number of concepts between A and B in the reference path, because the order of concepts may not change during import.
Formula (4) is the base case where there are no reference concepts that can be placed in the target ontology. Thus, there is no placement possible and there are 0 choices.
The last base case, Formula (5), assumes that there is only one reference concept. For arguments sake, we will refer to it as X. X can be placed immediately above every one of the k target concepts. Thus, there are k choices. Similarly X can be declared a synonym of every one of the k target concepts. Lastly, X can also be placed right above the bottom anchor, i.e., below the lowest of the k target concepts, in one single way. Combining these three possibilities there are k+k+1 possible cases, as shown in (5).
We are proving the recursive case (6) by structural induction on n. We assume that the number of placements p(n–1, *) is already known, whereby * may be any value from 0 to k. As we have a simple formula for any value of k (Formula (5)) only n needs to be reduced by recursion.
Without loss of generality, we assume that the nth concept (the new concept) is the top concept on the side of the reference ontology. Let this concept be X, as in Figure 1. If X is imported at the top-most possible position, right below A, then all previous placements of n–1 concepts are still possible. This explains the term p(n–1, k) in (7).
Subcase 1: If X is imported as a synonym of the topmost target concept β, then this effectively ties up one target concepts for the n–1 other reference concepts. Thus the previous n–1 concepts can be placed only as if there were k–1 target concepts, explaining the term p(n–1, k–1).
Subcase 2: If X is imported immediately under the top-most target concept, there are again p(n–1, k–1) placement choices, because the concept X does not interfere with any other placements below X. Theoretically all n–1 reference concepts could be included between X and β. This is exactly the same situation as in Subcase 1.
Summing Subcase 1 and Subcase 2 explains the term 2 * p(n–1, k–1). The same pattern of reasoning can now be repeated by assigning X as synonym of the second-to-top-most target concept, or right below it. Repeating all these steps down to the bottom anchor constitutes the remaining terms of (7).
We observe that p(n, k) grows very quickly (Table I).
TABLE I.
Growth of p(n, k).
| n | K | p(n, k) |
|---|---|---|
| 2 | 2 | 13 |
| 2 | 3 | 25 |
| 3 | 2 | 25 |
| 3 | 3 | 63 |
| 2 | 4 | 41 |
| 4 | 2 | 41 |
| 4 | 3 | 129 |
| 3 | 4 | 129 |
E. Counting Cross-Ontology Diamonds
In this work, we used the 2015AA version of the UMLS Metathesaurus. The NCIt is used as the target ontology. The NCIt version in the UMLS is 2014 03E. The main criteria for selecting a reference ontology include: 1) the reference ontology must be in English; 2) the reference ontology must be organized with an IS-A hierarchy backbone; 3) the reference ontology must exhibit sufficient overlap in content with NCIt.
We first identified seven English source terminologies with “PAR” (parent-child) relationships and “INVERSE_IS_A” relationship attributes, including MEDCIN, Gene Ontology (GO), Anatomical Therapeutic Chemical Classification System (ATC), Medical Entities Dictionary (CPM), Current Procedural Ontology (CPT), SNOMED CT, Veterinary Extension of SNOMED CT (SNOMED VET), and Foundational Model of Anatomy Ontology (FMA). The University of Washington Digital Anatomist (UWDA) would fit the three criteria given above, however, it is part of FMA and was therefore excluded.
We first identified all the diamonds in the UMLS that involve the above reference ontologies with NCIt as target ontology. The UMLS may contain cycles of IS-A relationships [38]. We eliminated any cycles in the diamonds by detecting repeating Concept Unique Identifiers (CUIs) in the IS-A paths. The numbers of diamonds will be reported in the Result Section. A diamond with j reference concepts and l target concepts is called a j/l-diamond. With increasing values of j and l, the runtime of the mining program may grow to the point where no results are returned within our experimental time frame. Thus, only the following diamonds were mined: 1/2, 1/3, 1/4, 1/5, 1/6, 2/1, 2/2, 2/3, 2/4, 2/5, 2/6, 2/7, 2/8, 2/9, 3/1, 3/2, 3/3, 3/4, 3/5. In general, the number of diamonds goes down with growing j and l. Thus, the loss of limiting these two parameters is minimal.
After all existing j/l -diamonds were identified; we chose random samples of 50 3/1 and 50 1/3-diamonds based on the limited availability of the human expert. The medical ontology expert YC investigated the content of both the reference ontology and the target ontology, and assessed the relationships between the reference concepts and the target concepts. The ontology expert chose one of the following options: 1) One of the reference concepts can be imported into NCIt; 2) One of the reference concepts is a synonym of a target concept in NCIt; 3) the reference concepts and the target concepts define alternative classifications of the top concept; and 4) There is an error in the reference ontology or there is an error in the target ontology. For the options 1), 2), and 4), the ontology expert was also asked to give rationales for making such a choice.
When embarking on this research program [14], alternative classifications were assumed to be outliers. However, previous studies [15–18] resulted in surprisingly large percentages of alternative classifications. The results of this paper confirm these previous findings with 68% and 60% cases being alternative classifications in the sample of 50 1/3 and 50 3/1 samples respectively.
IV. Results
A. Mining the Diamonds
Due to our special interest in the NCIt, we are limiting ourselves to this one target ontology. We have found the following numbers of diamond structures with the NCIt as target ontology as given in Table II. The column OTHERS sums up the number of diamonds for all other reference ontologies except MEDCIN, GO and SNOMED CT. We discovered 170 diamonds with one reference concept and three target concepts (1/3-diamonds) between the whole SNOMED CT and the NCIt (Table II). In total, we discovered 251 1/3-diamonds. In a 1/3-diamond there is only one selection choice. If the decision is made to include the reference concept, then there are seven placement choices (Formula 5).
TABLE II.
THE NUMBERS OF DIAMOND STRUCTURES WITH THE NCIT AS THE TARGET ONTOLOGY IN THE UMLS.
| Diamond | Reference Ontology | Total | |||
|---|---|---|---|---|---|
| MEDCIN | GO | SNOMED CT | OTHERS | ||
| 1/2 | 119 | 3 | 464 | 36 | 622 |
| 1/3 | 71 | 1 | 170 | 9 | 251 |
| 1/4 | 52 | 0 | 65 | 0 | 117 |
| 1/5 | 3 | 0 | 30 | 0 | 33 |
| 1/6 | 0 | 0 | 3 | 0 | 3 |
| 2/1 | 90 | 25 | 1682 | 16 | 1813 |
| 2/2 | 106 | 22 | 825 | 21 | 974 |
| 2/3 | 64 | 11 | 364 | 9 | 448 |
| 2/4 | 63 | 4 | 189 | 0 | 256 |
| 2/5 | 0 | 0 | 98 | 0 | 98 |
| 2/6 | 0 | 0 | 28 | 0 | 28 |
| 2/7 | 0 | 0 | 5 | 0 | 5 |
| 2/8 | 0 | 0 | 2 | 0 | 2 |
| 2/9 | 0 | 0 | 1 | 0 | 1 |
| 3/1 | 19 | 10 | 769 | 19 | 817 |
| 3/2 | 16 | 30 | 1582 | 25 | 1653 |
| 3/3 | 16 | 13 | 856 | 6 | 891 |
| 3/4 | 19 | 3 | 483 | 2 | 507 |
| 3/5 | 0 | 0 | 359 | 0 | 359 |
| Total | 638 | 122 | 7975 | 143 | 8878 |
Combining the results from Table I with the findings in Table II for SNOMED CT, the aggregate numbers of placement decisions are given in Table III.
TABLE III.
Aggregate numbers of placement decisions
| Kind of Diamond | SNOMED CT Diamonds of this Kind | Placement Decisions per Diamond | Total Placement Decisions |
|---|---|---|---|
| 2/2 | 825 | 13 | 10725 |
| 2/3 | 364 | 25 | 9100 |
| 3/2 | 1582 | 25 | 39550 |
| 3/3 | 856 | 63 | 53928 |
| 2/4 | 189 | 41 | 7749 |
| 3/4 | 483 | 129 | 62307 |
| Total | 183359 |
Let us reaffirm the meaning of the total number in Table III. A human expert who is given suggestions by an algorithm how to insert SNOMED CT concepts and who is also given target paths in NCIt for those insertions nevertheless has to make a staggering number of decisions how to actually perform such an import. Naturally, there is a difference between the theoretical worst case placement decisions and the actual decisions of a human expert. In many cases, intuition and understanding of the medical context will allow an experienced user to quickly eliminate whole sets of possible insertions, even if they are semantically valid. It should also be noted that in many cases no placement decisions need to be made at all, as many diamonds constitute alternative classifications. Thus the staggering number of 183,359 would never be reached in practice.
B. Expert Analysis of Two Samples
We randomly selected a sample of 50 1/3-diamonds, meaning that there was one reference concept from a reference ontology (e.g., SNOMED CT) and there were three target concepts in NCIt. A human expert (YC) determined that this sample contained 34 cases (68%) of alternative classifications, 10 (20%) cases where an intermediate concept could be inserted into NCIt, and six (12%) cases of synonyms. The synonyms, however, were not unique. The same synonym was recognized in several diamonds. Thus, the number of unique synonyms was three. For example, considering SNOMED CT as the reference ontology, the domain expert suggested that the concept Malignant epithelial tumor of ovary in SNOMED CT could be inserted between the concepts Malignant neoplasm of ovary AND Malignant Ovarian Surface Epithelial-Stromal Tumor in NCIt. As for synonyms the concept Congenital atresia of intestinal tract present in SNOMED CT was identified to be a synonym of Intestinal Atresia in NCIt.
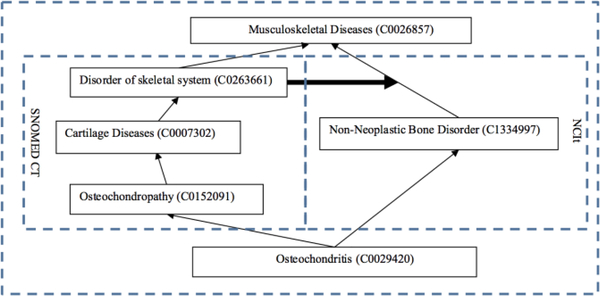
We also randomly selected a sample of 50 3/1-diamonds. YC determined that this sample contained 30 cases (60%) of alternative classifications, 11 (22%) cases where an intermediate concept could be inserted into NCIt and 9 (18%) cases of synonyms. The unique numbers of insertion and synonym cases are 7 and 9, respectively. In the 3/1 diamond shown in Figure 4, the ontology expert YC suggested that the reference concept Disorder of skeletal system (C0263661) can be inserted as the parent of the target concept Non-Neoplastic Bone Disorder (C1134997).
Fig. 4.
A concrete example of a 3/1-diamond in which a reference concept in SNOMED CT can be inserted as a parent of the target concept in NCIt. The bold horizontal arrow indicates the point of insertion.
V. Discussion
Alternative classifications were initially assumed to be outliers in this research program. We found that they were not outliers, but in some cases constituted the majority of a sample. With 483 3/4-diamonds between SNOMED CT and NCIt and 129 possible placements when allowing insertion of reference concepts and new cross-ontology synonyms a staggering number of 62,307 choices becomes possible.
One limitation of this work was given by computing resources. It is possible that with more powerful computational resources and/or smarter algorithms a small number of diamonds could be found that are larger than the 2/9 and the 3/5-diamonds that defined the limits in this study. Another limitation of this work is that ontology experts are required to make decisions on the relationships between reference concepts and target concepts. In future work, we can use some extrinsic sources such as WordNet to help identify the relationships between concepts (e.g., synonyms).
In a diamond, if one reference concept is a synonym of the target concept, the descendants of the reference concept may also be imported, namely as descendants of the target concept. This creates its own set of complications and is left for future work.
All previous work [14–18] and the current paper rely on vertical densities to define topological structures. An initial study of horizontal density differences has been conducted (in press) [39]. An analysis of 64 (30+34) alternative classifications has indicated that IS-A links might carry additional information that is never made explicit. Several cases each of IS-A “by disease kind,” “by procedure kind” and “by anatomical location” were identified. A quantitative analysis of these different kinds of novel annotations for IS-A links will remain a topic of future research. Lastly, it would be interesting to compare the predictions made by the formula of placement decisions with the real time taken by an expert to perform the actual insertions.
VI. Conclusion
We have presented progress in the study of cross-ontology topological patterns (“diamonds”) for the purpose of extending the domain coverage of important ontologies such as the NCI thesaurus. We have confirmed the existence of 7975 such patterns between SNOMED CT and NCIt. Altogether, there are 8878 diamonds, with most of the balance supplied by MEDCIN and GO. We derived a recursive formula that expresses the many choices of placing new concepts into a target ontology, either between existing concepts or as new synonyms of existing concepts. The large number of placement choices indicates that the task of ontology curators is difficult. Sufficient human resources should be allocated.
In two samples of 50 diamonds, 34 and 30 alternative classifications, respectively, were identified by a human expert, confirming strongly that alternative classifications are not outliers. SNOMED CT and NCIt use surprisingly different structures within the same general ontology framework to describe information that is also similar in content. In future work, we will investigate the kinds of alternative classifications in pairs of ontologies to identify possible useful cases for ontology enhancement.
Acknowledgments
Sherri do Coronado of NCI has commented on our previous research on extending the NCIt.
Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190779. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Zhe He, School of Information, Florida State University Tallahassee, Florida USA zhe.he@cci.fsu.edu.
Vipina Kuttichi Keloth, Department of Computer Science, New Jersey Institute of Technology, Newark, NJ USA, vk396@nijt.edu.
Yan Chen, Department of Computer Inforamtion Systems, BMCC, CUNY, New York, NY USA, yan.chen222@gmail.com.
James Geller, Department of Computer Science, New Jersey Institute of Technology, Newark, NJ USA, james.geller@njit.edu.
References
- [1].Bodenreider O: ‘Biomedical ontologies in action: role in knowledge management, data integration and decision support’, Yearbook of medical informatics, 2008, pp. 67–79 [PMC free article] [PubMed] [Google Scholar]
- [2].Cimino JJ: ‘High-quality, standard, controlled healthcare terminologies come of age’, Methods Inf Med, 2011, 50, (2), pp. 101–104 [PMC free article] [PubMed] [Google Scholar]
- [3].Rector A, Qamar R, and Marley T: ‘Binding ontologies and coding systems to electronic health records and messages’, Applied Ontology, 2009, 4, (1), pp. 51–69 [Google Scholar]
- [4].Gonzalez C, Blobel Bg Fau - Lopez DM, and Lopez DM: ‘Ontology-based framework for electronic health records interoperability’, Studies in health technology and informatics, 2011, 169, pp. 694–698 [PubMed] [Google Scholar]
- [5].Finnegan R: ‘ICD-9-CM coding for physician billing’, Journal, 1989, 60, (2), pp. 22–23 [PubMed] [Google Scholar]
- [6].Cimino JJ: ‘Desiderata for controlled medical vocabularies in the twenty-first century’, Methods Inf Med, 1998, 37, (4–5), pp. 394–403 [PMC free article] [PubMed] [Google Scholar]
- [7].Weng C, Gennari JH, and Fridsma DB: ‘User-centered semantic harmonization: a case study’, J Biomed Inform, 2007, 40, (3), pp. 353–364 [DOI] [PubMed] [Google Scholar]
- [8].Hoxha J, Jiang G, and Weng C: ‘Automated learning of domain taxonomies from text using background knowledge’, J Biomed Inform, 2016, 63, pp. 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chandar P, Yaman A, Hoxha J, He Z, and Weng C: ‘Similarity-Based Recommendation of New Concepts to a Terminology’, AMIA Annu Symp Proc, 2015, 2015, pp. 386–395 [PMC free article] [PubMed] [Google Scholar]
- [10].Zeng QT, Tse T, Divita G, Keselman A, Crowell J, Browne AC, Goryachev S, and Ngo L: ‘Term identification methods for consumer health vocabulary development’, J Med Internet Res, 2007, 9, (1), pp. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].He Z, Chen Z, Oh S, Hou J, and Bian J: ‘Enriching consumer health vocabulary through mining a social Q&A site: a similarity-based approach’, J Biomed Inform, 2017, 69: pp. 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lossio-Ventura JA, Hogan W, Modave F, Hicks A, Hanna J, Guo Y, He Z, and Bian J: ‘Towards an obesity-cancer knowledge base: Biomedical entity identification and relation detection’, in Editor (Ed.)^(Eds.): ‘Book Towards an obesity-cancer knowledge base: Biomedical entity identification and relation detection’ (2016, edn.), pp. 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bodenreider O: ‘The Unified Medical Language System (UMLS): integrating biomedical terminology’, Nucleic Acids Res, 2004, 32, (Database issue), pp. D267–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].He Z, Geller J, and Elhanan G: ‘Categorizing the Relationships between Structurally Congruent Concepts from Pairs of Terminologies for Semantic Harmonization’, AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science, 2014, 2014, pp. 48–53 [PMC free article] [PubMed] [Google Scholar]
- [15].He Z, Geller J, and Chen Y: ‘A comparative analysis of the density of the SNOMED CT conceptual content for semantic harmonization’, Artif Intell Med, 2015, 64, (1), pp. 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].He Z, Chen Y, de Coronado S, Piskorski K, and Geller J: ‘Topological-Pattern-based Recommendation of UMLS Concepts for National Cancer Institute Thesaurus’, AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium, 2016, 2016, pp. 618–627 [PMC free article] [PubMed] [Google Scholar]
- [17].He Z, and Geller J: ‘Preliminary Analysis of Difficulty of Important Pattern-Based Concepts into the National Cancer Institute Thesaurus’, Studies in health technology and informatics, 2016, 288, pp. 389–393 [PMC free article] [PubMed] [Google Scholar]
- [18].He Z, Chen Y, and Geller J: ‘Perceiving the Usefulness of National Cancer Institute Metathesaurus for Enriching NCIt with Topological Patterns’, Studies in health technology and informatics, 2017 [PMC free article] [PubMed] [Google Scholar]
- [19].Euzenat J, Meilicke C, Shvaiko P, Stuckenschmidt H, and Trojahn C: ‘Ontology Alignment Evaluation Initiative: six years of experience’, Journal on Data Semantics, 2011, XV (6720), pp. 158–192 [Google Scholar]
- [20].Larson JA, Navathe SB, and Elmasri R: ‘A theory of attributed equivalence in databases with application to schema integration’, IEEE Transactions on Software Engineering, 1989, 15, (4), pp. 449–463 [Google Scholar]
- [21].Huang KC, Geller J, Halper M, Perl Y, and Xu J: ‘Using WordNet synonym substitution to enhance UMLS source integration’, Artif Intell Med, 2009, 46, (2), pp. 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].de Coronado S, Wright LW, Fragoso G, Haber MW, Hahn-Dantona EA, Hartel FW, Quan SL, Safran T, Thomas N, and Whiteman L: ‘The NCI Thesaurus quality assurance life cycle’, J Biomed Inform, 2009, 42, (3), pp. 530–539 [DOI] [PubMed] [Google Scholar]
- [23].Min H, Perl Y, Chen Y, Halper M, Geller J, and Wang Y: ‘Auditing as part of the terminology design life cycle’, Journal of the American Medical Informatics Association : JAMIA, 2006, 13, (6), pp. 676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cohen B, Oren M, Min H, Perl Y, and Halper M: ‘Automated comparative auditing of NCIT genomic roles using NCBI’, J Biomed Inform, 2008, 41, (6), pp. 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mougin F, and Bodenreider O: ‘Auditing the NCI thesaurus with semantic web technologies’, AMIA Annu Symp Proc, 2008, pp. 500–504 [PMC free article] [PubMed] [Google Scholar]
- [26].Jiang G, Solbrig HR, and Chute CG: ‘Quality evaluation of value sets from cancer study common data elements using the UMLS semantic groups’, Journal of the American Medical Informatics Association : JAMIA, 2012, 19, (e1), pp. e129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].https://ncimeta.nci.nih.gov/ncimbrowser/,
- [28].Bodenreider O: ‘Strength in numbers: exploring redundancy in hierarchical relations across biomedical terminologies’, AMIA Annu Symp Proc, 2003, pp. 101–105 [PMC free article] [PubMed] [Google Scholar]
- [29].Gu HH, Wei D, Mejino JL Jr., and Elhanan G: ‘Relationship auditing of the FMA ontology’, J Biomed Inform, 2009, 42, (3), pp. 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Richesson RL, Fung KW, and Krischer JP: ‘Heterogeneous but “standard” coding systems for adverse events: Issues in achieving interoperability between apples and oranges’, Contemporary clinical trials, 2008, 29, (5), pp. 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sun P, and Zhang S: ‘Identifying Granularity Differences between Large Biomedical Ontologies through Rules’. Proc. AMIA Annu Symp Proc 2010 [PMC free article] [PubMed] [Google Scholar]
- [32].Sun P, and Zhang S: ‘Using rules to investigate the differences in partonomy between biomedical ontologies’, IEEE International Conference on Bioinformatics and Biomedicine, 2011, pp. 623–626 [Google Scholar]
- [33].Cornet R: ‘Information-content-based measures for the structure of terminological systems and for data recorded using these systems’, Studies in health technology and informatics, 2010, 160, (Pt 2), pp. 1075–1079 [PubMed] [Google Scholar]
- [34].Cimiano P: ‘Ontology Learning and Population from Text: Algorithms, Evaluation and Applications’ (Springer-Verlag New York, Inc., 2006. 2006) [Google Scholar]
- [35].Smith B, and Fellbaum C: ‘Medical WordNet: a new methodology for the construction and validation of information resources for consumer health’. Proc. Proceedings of the 20th international conference on Computational Linguistics, Geneva, Switzerland2004 [Google Scholar]
- [36].Liu K, Hogan WR, and Crowley RS: ‘Natural Language Processing methods and systems for biomedical ontology learning’, J Biomed Inform, 2011, 44, (1), pp. 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Amith M, Song HY, Zhang Y, Xu H, and Tao C: ‘Lightweight predicate extraction for patient-level cancer information and ontology development’, BMC Med Inform Decis Mak, 2017, 17, (Suppl 2), pp. 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bodenreider O: ‘Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and prevention’, Proceedings. AMIA Symposium, 2001, pp. 57–61 [PMC free article] [PubMed] [Google Scholar]
- [39].Keloth VK, He Z, Chen Y, and Geller J: ‘Leveraging horizontal density differences between ontologies to identify missing child concepts: A proof of concept’. Proc. AMIA 2018 Annual Symposium, 2018, pp. 644–653 [PMC free article] [PubMed] [Google Scholar]