Abstract
Mitosis is an essential process that takes place in all eukaryotes and involves the equal division of genetic material from a parental cell into two identical daughter cells. During mitosis, chromosome movement and segregation are orchestrated by a specialized structure known as the mitotic spindle, composed of a bipolar array of microtubules. The fundamental structure of microtubules comprises of α/β-tubulin heterodimers that associate head-to-tail and laterally to form hollow filaments. In vivo, microtubules are modified by abundant and evolutionarily conserved tubulin post-translational modifications (PTMs), giving these filaments the potential for a wide chemical diversity. In recent years, the concept of a “tubulin code” has emerged as an extra layer of regulation governing microtubule function. A range of tubulin isoforms, each with a diverse set of PTMs, provides a readable code for microtubule motors and other microtubule-associated proteins. This chapter focuses on the complexity of tubulin PTMs with an emphasis on detyrosination and summarizes the methods currently used in our laboratory to experimentally manipulate these modifications and study their impact in mitosis.
Keywords: mitosis, microtubules, mitotic spindle, tubulin post-translational modifications, tubulin code, tyrosination, detyrosination
Introduction
The mitotic spindle is a complex molecular machine composed of microtubules (MTs), motors, and other microtubule-associated proteins (MAPs) whose central function is to accurately segregate chromosomes to two daughter cells during mitosis (Walczak & Heald, 2008). MTs are arranged in a bipolar array with less dynamic minus-ends embedded at the pole and more dynamic plus-ends extending towards the spindle equator and cell cortex. During early mitosis, the nuclear envelope breaks down, MTs invade the nuclear space and attach to chromosomes at the kinetochore, a macromolecular structure comprised of more than a hundred proteins (Hinshaw & Harrison, 2018; Walczak & Heald, 2008). Upon attachment to the kinetochores, MTs promote chromosome congression to the metaphase plate and later segregate the sister chromatids to opposite poles during anaphase (Walczak & Heald, 2008).
MTs are polymers of α/β-tubulin heterodimers that bind head-to-tail to form a linear protofilament. Typically, 13 protofilaments associate laterally to assemble a hollow cylinder with 25 nm of external diameter (Desai & Mitchison, 1997; Evans, Mitchison, & Kirschner, 1985). This organization is responsible for the intrinsic polarity of MTs with β-tubulin subunits facing the plus end and the α-tubulin subunits facing the minus end (Amos & Klug, 1974; Desai & Mitchison, 1997). Another essential property of microtubules is their dynamic instability, a behaviour in which individual MT ends switch from growing to shrinking states, sometimes changing back and forth several times on a time scale of seconds to minutes (T. Mitchison & Kirschner, 1984). Dynamic instability is driven by the hydrolysis of GTP by the β-tubulin subunit during polymerization. Distinct dynamic properties characterize the three populations that exist within the mitotic spindles: astral microtubules, kinetochore microtubules (kMTs) and interpolar-microtubules (ipMTs). Moreover, their dynamic behaviour changes during different stages of mitosis. The diversity of MT populations further results from the incorporation of different tubulin isoforms (also known as isotypes) and from the post-translational modifications (PTMs) of tubulin. How this contributes to confer specificity to MT interactions with MAPs/motors and how it affects the function and dynamics of each MT population during mitotic progression is not completely understood (Barisic & Maiato, 2016).
What is the tubulin code?
The concept of the “tubulin code” was first proposed by Verhey and Gaertig in 2007 as the product of the multiple tubulin PTMs that imprint on MT chemical diversity (Verhey & Gaertig, 2007). Currently, the “tubulin code” is a broader concept that includes not only the tubulin PTMs, but also the differential expression of several α- and β-tubulin genes (Gadadhar, Bodakuntla, Natarajan, & Janke, 2017). In general, tubulin PTMs are the result of the covalent addition or proteolytic cleavage of functional groups. These chemical reactions are carried out by a myriad of enzymes that act on both polymerized and soluble tubulin (Gadadhar et al., 2017; Song & Brady, 2015).
Tubulin isotypes
Numerous α- and β-tubulin isotypes encoded by different genes have been identified in almost all organisms (Luduena, 1998, 2013; Sullivan & Cleveland, 1986). In humans, nine α-tubulin and nine β-tubulin isotypes with tissue-specific expression variability have been identified (for nomenclature see: www.genenames.org/cgi-bin/genefamilies/set/778) (Gadadhar et al., 2017) (Table 1 and Table 2). Currently, little is known about the role of different isotypes in mitosis but there is evidence that some β-tubulin isotypes are more highly expressed in dividing cells than in resting cells (Dumontet et al., 1996; Jouhilahti, Peltonen, & Peltonen, 2008). Significantly, βIII-tubulin (normally expressed at high levels in cells of neuronal origin) was found to be overexpressed in several human cancers and associated with a poor response to microtubule-targeting drugs used in cancer therapy (Person et al., 2017). The amino acid sequence between isotypes of α-tubulin shows a higher level of conservation than in the β-tubulin isotypes. A major site for divergence between tubulin isotypes is the C-terminal tail (CTT), a region that specifies interactions with MAPs (Roll-Mecak, 2015). Interestingly, different isotypes can combine into mosaic MTs with specific chemical properties that may ultimately translate into specialized functions (Joshi & Cleveland, 1989; Lewis, Gu, & Cowan, 1987; Raff, Hoyle, Popodi, & Turner, 2008). Furthermore, tubulin isotype composition may also affect polymer assembly and dynamics. This is supported by recent in vitro studies showing dynamic instability parameters and polymerization properties to be isotype-dependent (Pamula, Ti, & Kapoor, 2016).
Table 1. Human α-tubulin isotypes.
| Gene | Full name | Alternative name | Locus | Transcript variants NCBI Reference Sequence | Number of protein-coding transcribed from this gene https://www.proteinatlas.org/ | Tissue-Expression | Lys 40 | C-terminal | Associated Human Diseases | |
|---|---|---|---|---|---|---|---|---|---|---|
| TUBA1A | Tubulin alpha 1a | B-ALPHA-1 | 12q13.12 | 1- NM_006009.3 2- NM_001270399.1 3- NM_001270400.1 |
7 (50.1, 50.1, 46.3, 12.2, 24.2, 2.7, 24.8 kDa) | Ubiquitous | acetylated | -GEEY* | Lissencephaly type 3 | |
| TUBA1B | Tubulin alpha 1b | K-ALPHA-1 | 12q13.12 | 1-NM_006082.2 | 5 (50.2, 8.3, 5.3, 27.5, 2.7, 8.9 kDa) | Ubiquitous | acetylated | -GEEY* | ND | |
| TUBA1C | Tubulin alpha 1c | TUBA6 | 12q13.12 | 1- NM_001303114.1 2- NM_001303115.1 3- NM_001303116.1 4- NM_001303117.1 5- NM_032704.4 |
4 (49.9, 57.7, 14.4, 36.7 kDa) | Ubiquitous | acetylated | -GEEY* | ND | |
| TUBA3C | Tubulin alpha 3c | TUBA2 | 13q12.11 | 1- NM_006001.2 | 2 (50, 46.1kDa) | Enriched in Testis | acetylated | -GEEY* | Associated with genetic disease Clouston hidrotic ectodermal dysplasia and Kabuki syndrome. | |
| TUBA3D | Tubulin alpha 3d | H2-ALPHA | 2q21.1 | 1- NM_080386.3 | 1 (50kDa) | Enriched in Testis | acetylated | -GEEY* | ||
| TUBA3E | Tubulin alpha 3e | --- | 2q21.1 | 1-NM_207312.2 | 1 (49.9kDa) | Enriched in Testis | acetylated | -GEAY | A missense mutation has been potential linked to microlissencephaly and global developmental delay | |
| TUBA4A | Tubulin alpha 4a | TUBA1 | 2q35 | 1- NM_006000.2 2- NM_001278552.1 |
7 (49.9, 48.3, 16.9, 21.7, 13.1, 19.8, 17.7 kDa) | Ubiquitous | acetylated | -GEE** | ND | |
| TUBA4B | Tubulin alpha 4b | 2q35 | 1- NM_001355221.1 | 1 (27.6) kDa | Ubiquitous low | --- | --- | ND | ||
| TUBA8 | Tubulin alpha 8 | TUBAL2 | 22q11.21 | 1-NM_018943.2 2-NM_001193414.1 |
5 (50.1, 43, 52, 5.3, 31 kDa) | Ubiquitous | Lacks Lys 40; Unusual sequence at positions 35–45 | -GEEF*** | Associated with polymicrogyria and optic nerve hypoplasia |
The C-terminal tyrosine can be removed by a VASH1/2 tubulin carboxypeptidase and added back by TTL.
A tyrosine can be added at the C-terminus to create GEEY.
The phenylalanine cannot be removed by tubulin carboxypeptidase.
ND: Not described.
Table 2. Human β-tubulin isotypes.
| Gene | Full name | Alternative name | Locus | Transcript variants NCBI Reference Sequence | Number of protein-coding transcribed from this gene https://www.proteinatlas.org/ | Tissue-Expression | Associated Human Diseases |
|---|---|---|---|---|---|---|---|
| TUBB | Tubulin beta class I | Tubb5 | 6p21.33 | 1 - NM_001293212.1 2 - NM_178014.3 3 - NM_001293213.1 4 - NM_001293214.1 5 - NM_001293215.1 6 - NM_001293216.1 |
4 (49.7, 41.7, 47.8, 41.7 kDa) | Ubiquitous | Cortical dysplasia, complex, with other brain malformations |
| TUBB1 | Tubulin beta 1 class VI | --- | 20q13.32 | 1 - NM_030773.3 | 1 (50.3 kDa) | Enriched in platelets and megakaryocytes | Autosomal dominant macrothrombocytopenia |
| TUBB2A | Tubulin beta 2A class IIa | TUBB2 | 6p25.2 | 1 - NM_001069.2 2 - NM_001310315.1 |
1 (49.9 kDa) | Ubiquitous | Cortical dysplasia with other brain malformations |
| TUBB2B | Tubulin beta 2B class IIb | MGC8685 | 6p25.2 | 1 - NM_178012.4 | 1 (50.0 kDa) | Ubiquitous | Asymmetric polymicrogyria |
| TUBB3 | Tubulin beta 3 class III | beta-4 | 16q24.3 | 1 - NM_006086.3 2 - NM_001197181.1 |
8 (50.4, 42.4, 5, 20.7, 10.8, 13.6, 18.3, 16.4 kDa) | Ubiquitous | Congenital fibrosis of the extraocular muscles type 3 |
| TUBB4A | Tubulin beta 4A class IVa | TUBB4 | 18p13.3 | 1 - NM_001289123.1 2 - NM_001289127.1 3 - NM_006087.3 4 - NM_001289129.1 5 - NM_001289130.1 6 - NM_001289131.1 |
11 (49.6, 11.8, 17.6, 16.6, 10.6, 11.7, 17.8, 17.4, 12.2, 7.8, 15.8 kDa) | Ubiquitous | Hypomyelinating leukodystrophy-6 and autosomal dominant torsion dystonia-4 |
| TUBB4B | Tubulin beta 4B class IVb | TUBB2C | 9q34.3 | 1 - NM_006088.5 | 1 (49.8 kDa) | Ubiquitous | ND |
| TUBB6 | Tubulin beta 6 class V | MGC4083 | 18p11.21 | 1 - NM_032525.2 2 - NM_001303524.1 3 - NM_001303525.1 4 - NM_001303526.1 5 - NM_001303527.1 6 - NM_001303528.1 7 - NM_001303529.1 8 - NM_001303530.1 |
10 (49.6, 11.8, 17.6, 16.6, 10.6, 11.7, 17.8, 17.4, 12.2, 7.8, 15.8 kDa) | Ubiquitous | ND |
| TUBB8 | Tubulin beta 8 class VIII | bA631M21.2 | 10p15.3 | 1 - NM_177987.2 | 5 (49.8, 7.8, 45.5, 13, 45.7 kDa) | Ubiquitous low | Defects in this gene are a cause of oocyte maturation defect 2 and infertility |
ND: Not described.
Tubulin post-translational modifications
The study of tubulin PTMs started 40 years ago, when it was first shown that the CTT of the α-tubulin is tyrosinated in an RNA-independent manner (Barra, Rodriguez, Arce, & Caputto, 1973). The modifying enzyme was later purified from brain extracts and identified as tubulin tyrosine ligase (TTL) (Ersfeld et al., 1993; Schroder, Wehland, & Weber, 1985). Shortly after, it was found that a cytosolic tubulin carboxypeptidase (TCP) activity, preferentially working on polymerized tubulin, was responsible for the cleavage of the C-terminal tyrosine (Gundersen, Khawaja, & Bulinski, 1987; Hallak, Rodriguez, Barra, & Caputto, 1977). However, the identity of TPC(s) mediating this cleavage remained elusive for more than three decades and only recently were identified by two different groups (Aillaud et al., 2017; Nieuwenhuis et al., 2017). Vasohibin-1 and its homologue Vasohibin-2, previously described has angiogenic factors, were shown to catalyse detyrosination when in complex with a chaperone-like peptide - small vasohibin binding protein (SVBP). In microtubules, detyrosinated-tubulin can undergo further shortening by irreversible removal of the terminal glutamate catalysed by cytoplasmic carboxypeptidases (CCPs). The generated Δ2-tubulin cannot be re-tyrosinated and will no longer contribute to the tyrosination cycle (Paturle-Lafanechere et al., 1994).
Tubulin molecules consist of a predominant globular core and a short unstructured CTT (composed of ~10 and ~20 amino acid residues in α- and β-tubulin respectively) that is negatively charged and decorates the exterior of the MT lattice (Nogales, Whittaker, Milligan, & Downing, 1999). Probably facilitated by their accessibility, CTTs are hotspots for PTMs. Similarly to detyrosination, polyglutamilation and polyglycylation are incorporated at the tubulin CTTs, however they are not exclusive to α-tubulin. Both modifications are catalysed by members of the TTL-like (TTLL) family, which have TTL as the common founding member (Janke et al., 2005). Although TTL and TTLLs share conserved active sites, the surface residues are variable and provide the tubulin binding domains that confer substrate specificities to the multiple enzymes (Roll-Mecak, 2015). Some TTLL glutamylases (TTLL4, 5, and 7) add a single glutamate residue by forming a γ-linked isopeptide bond, while others (TTLL1, 6, 11, and 13) add several glutamates to the γ-linked glutamate through standard peptide linkages to form polyglutamate side chains. Many TTLLs exhibit preference towards α or β tubulins. For instance, TTLL7 preferentially modifies β-tubulin, while TTLL5 and 6 prefer α-tubulin as a substrate (Janke & Bulinski, 2011; Song & Brady, 2015). Three TTLL glycylases work together to generate polyglycylation. While TTLL3 and TTLL8 are initiating glycylases, TTLL10 elongates the polyglycylated chain (Janke, 2014). Glycylation and polyglutamilation share the same substrates and seem to be interdependent (Rogowski et al., 2009; Wloga et al., 2009). However, in contrast to polyglutamylation, polyglycylation is confined to cilia and flagella (Rogowski et al., 2009). A family of six cytoplasmic carboxypeptidases (CCPs) has been shown to catalytically remove glutamate residues from tubulin CTTs of α- and β-tubulin. These enzymes cleave both gene encoded (generating Δ2 and Δ3 tubulin) and post-translationally added glutamates (Aillaud et al., 2016; Gadadhar et al., 2017; Kimura et al., 2010; Pathak, Austin-Tse, Liu, Vasilyev, & Drummond, 2014). So far, no deglycylating enzyme has been identified.
Acetylation of α-tubulin lysine-40 (K40) was first observed more than three decades ago and is one of the best described tubulin PTMs, (L'Hernault & Rosenbaum, 1985). Acetylation occurs at the luminal surface of the MT (Maruta, Greer, & Rosenbaum, 1986; Soppina, Herbstman, Skiniotis, & Verhey, 2012). This unusual localization makes tubulin acetylation one of the few PTMs to occur outside of the CTT. Although several enzymes can acetylate tubulin, α-tubulin acetytransferase (αTAT)/MEC7 is the major K40-modifying enzyme (Akella et al., 2010). Two tubulin deacetylases have been found that remove the acetylation from K40: Histone deacetylase 6 (HDAC6) and sirtuin2 (SIRT2) (Hubbert et al., 2002; North, Marshall, Borra, Denu, & Verdin, 2003). Lysine-252 (K252) on β-tubulin has also been reported as an acetylation site in vivo and in vitro of the acetyltransferase SAN (Chu et al., 2011).
Other tubulin PTMs include phosphorylation, palmitoylation, S-nitrosylation, polyamination, ubiquitylation, sumoylation, glycosylation and methylation. These modifications are poorly characterized and their function is unclear. Phosphorylation of both α- and β-tubulin has been reported on several serine residues (Eipper, 1972; Peters, Furlong, Asai, Harrison, & Geahlen, 1996). More recently, it was shown that phosphorylation of serine 172 (S172) by Cdk1 in mitosis inhibits polymerization due to the close proximity to the exchangeable nucleotide-binding site. Thus, this phosphorylation seems to be fundamental for MT remodelling during mitosis (Fourest-Lieuvin et al., 2006). Tubulin palmitoylation consists of the covalent binding of a fatty acid group to a cysteine residue and has been reported to occur primarily at cysteine 376 (C376) of α-tubulin in Saccharomyces cerevisiae, where it is involved in nuclear positioning during anaphase (Caron, Vega, Fleming, Bishop, & Solomon, 2001; Ozols & Caron, 1997). However in mammals, the role of tubulin palmitoylation is unclear and the fatty acyltransferase (PAT) remains elusive. Tubulin S-nitrosylation is the non-enzymatic addition of nitric oxide to various cysteine residues of α- and β-tubulin and their in vivo function is unknown (Jaffrey, Erdjument-Bromage, Ferris, Tempst, & Snyder, 2001). Tubulin polyamination consists of the irreversible covalent binding of a polyamine to various glutamine residues on α- and β-tubulin by a transglutaminase (Mehta, Fok, & Mangala, 2006). This is the only PTM described to date that adds positive charges to the tubulin subunits. Studies using rat brain extracts revealed that polyamination is required for MT stability in neurons (Song et al., 2013). Ubiquitination involves the formation of an amide linkage between ε-amine of a lysine target and the C-terminus of ubiquitin (Hershko & Ciechanover, 1998). Tubulin is multiubiquitinated by several ubiquitin ligases (Xu, Paige, & Jaffrey, 2010). More recently, it was shown that loss of the ubiquitin E3 ligase activity of MGRN1 causes spindle misorientation and decreased α-tubulin polymerization, suggesting a role for MGRN1 in regulation of MT stability. The same work proposed a further role in mitotic spindle orientation (Srivastava & Chakrabarti, 2014). Sumoylation is another regulatory system, similar to ubiquitination, in which a SUMO protein is added to lysine residues. α- and β-tubulins have been identified as candidates for sumoylation in global sumoylation screens, however the biological function of this modification to MTs is yet to be discovered (Rosas-Acosta, Russell, Deyrieux, Russell, & Wilson, 2005; Wohlschlegel, Johnson, Reed, & Yates, 2004). Tubulin glycosylation consists of the reversible enzymatic addition of O-linked β-N-acetylglucosamine (O-GlcNAc) to serine/threonine residues in the tubulin sequence (Love & Hanover, 2005). It has been reported that O-GlcNAcylation inhibits dimerization and that O-GlcNAcylated tubulin does not incorporate into MTs (Ji et al., 2011). Methylation was the last tubulin PTM to be identified. α-tubulin is also methylated at K40 by a dual-function histone and microtubule methyltransferase called SET-domain-containing 2 (SETD2). The same study reported that methylation varies between different MT populations. Moreover, acute loss of SETD2 function caused mitotic and/or cytokinesis defects (Park et al., 2016).
How is the tubulin code read?
The myriad of tubulin PTMs display a patterned distribution among the many MT subpopulations (Yu, Garnham, & Roll-Mecak, 2015). In mitosis, detyrosination also distributes stereotypically among the MT subpopulations that compose the mitotic spindle. Several studies have consolidated the hypothesis that these epigenetic marks affect the activity of molecular effectors working on MTs. It has been reported that detyrosination regulates kinesin-1 and kinesin-2 processivity and decreases the depolymerizing activity of kinesin-13 (Dunn et al., 2008; Peris et al., 2009; Sirajuddin, Rice, & Vale, 2014). Furthermore, polyglutamilation enhances kinesin-1 and kinesin-2 motility, whereas kinesin-13 and dynein are insensitive to this modification (Kaul, Soppina, & Verhey, 2014; Konishi & Setou, 2009; Sirajuddin et al., 2014). Dynein is not directly affected by detyrosination but the initiation of its processive movement in complex with dynactin and BicD2 is affected by detyrosination, as well as the recruitment of MT plus-end tracking proteins such as CLIP170 (McKenney, Huynh, Vale, & Sirajuddin, 2016; Peris et al., 2006). The first demonstration of tubulin PTMs impacting on mitosis came from the discovery that CENP-E preferentially moves along detyrosinated MTs to guide chromosomes towards the spindle equator during chromosome congression (M. Barisic et al., 2015). At the entry into mitosis, cyclin-dependent kinase 1 (CDK1) is activated and triggers a cascade of phosphorylation events that ultimately regulate the activity of MAPs and motors (Cassimeris, 1999; Ramkumar, Jong, & Ori-McKenney, 2018). The functional shift of this complex machinery leads to the reconfiguration of the MT landscape in mitosis and thus requires specific methodologies to investigate the implications of tubulin PTMs during this process.
Methods
In this chapter, we provide an overview of the methods currently used in our laboratory to investigate tubulin PTMs and their roles in mitosis, focusing on detyrosination. We address 3 main topics: 1) analysis of the levels and distribution of detyrosination in perturbed and unperturbed cells; 2) study of the effect of detyrosination on MT dynamics during mitosis; and 3) purification of proteins that bind to tyrosinated- or detyrosinated-enriched MTs from mitotic cells.
Section 1 - Modulation of the detyrosination/tyrosination cycle in mammalian cells
Immunofluorescence studies suggest that astral microtubules are mostly (or totally) made of tyrosinated tubulin, while kMTs are comprised of both tyrosinated and detyrosinated forms. In kMTs the detyrosination levels are higher at the minus ends and gradually decrease with increasing distance from the spindle pole (Barisic, Aguiar, Geley, & Maiato, 2014; Bobinnec et al., 1998; Gundersen & Bulinski, 1986) (Fig.1, Fig.2). However, the discrimination between kMTs and ipMTs is difficult and their precise tubulin composition at each stage of mitosis remains unclear. Moreover, immunofluorescence analysis to detect detyrosinated α-tubulin has revealed continuous and discontinuous regions along the MT length, suggesting the existence of alternate stretches of detyrosinated and tyrosinated tubulin, whose functional meaning remains unknown (Geuens et al., 1986; Zink et al., 2012).
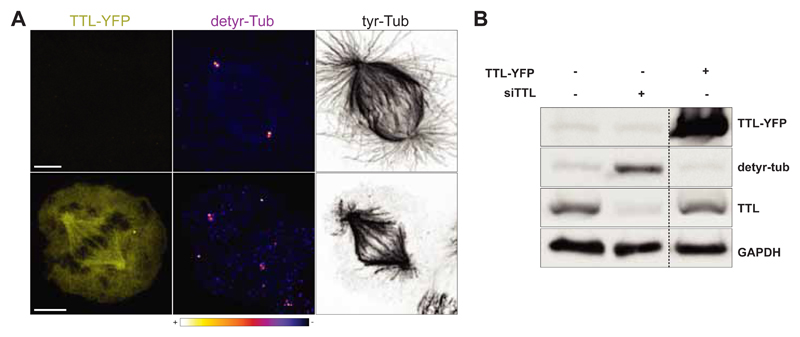
Fig. 1.
A. Confocal image of a representative U2OS cell transiently overexpressing TTL-YFP (bottom panel). A non-transfected U2OS cell (upper panel) is represented as control. Scale bar, 5 μm. B. Western-blot analysis of U2OS cell lysate transiently overexpressing TTL-YFP.
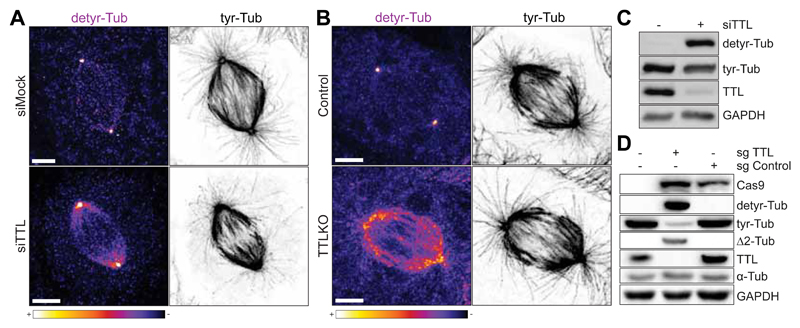
Fig. 2.
A. Confocal image of U2OS cells transfected with non-targeting siRNA (siMock) or with siRNA for TTL (siTTL). B. Confocal image of U2OS control and TTL KO cells. Scale bar, 5 μm. C. Western-blot analysis of U2OS cell lysates before and after siRNA-mediated depletion of TTL. D. Western-blot analysis of U2OS cell lysates in control (sgcontrol) and TTL KO cells (sgTTL).
The heterogeneous distribution of tyrosination along single MTs is probably determined by variations of the expression levels of the different α-tubulin isotypes, by the total amount of free tubulin molecules available and by the regulation of the enzymes that are responsible for this reversible PTM (Aiken et al., 2014; Dumontet et al., 1996). In this section, we describe fixed and live-cell fluorescence microscopy methods to study mitotic phenotypes after perturbation of the detyrosination/tyrosination cycle. We detail protocols for the depletion of TTL, VASH1/2 and endogenous α-tubulins and for the exogenous overexpression of TTL, VASH1/2 and modified forms of α-tubulin in human U2OS cells. We also describe a protocol for TTL inhibition with parthenolide (Fonrose et al., 2007). These protocols can be adapted to study other tubulin PTMs by using other tubulin modifying enzymes and/or small molecule inhibitors (Table 3). When designing and interpreting experiments the promiscuity of the enzymes as well as the inhibitor selectivity should be taken into consideration.
Table 3. Tubulin post-translational modifications modulators.
| Modification | Catalysing enzyme | α/β tubulin site | Modulators | Mechanism |
|---|---|---|---|---|
| Detyrosination -GEE |
VASH1 VASH2 |
Last tyrosine of the CTT of α-tubulin | TTL depletion (siRNA)/knockdown VASH1/2 overexpression |
(Barisic et al., 2015; Erck et al., 2005) (Nieuwenhuis et al., 2017) |
| Tyrosination -GEEY |
TTL | TTL overexpression EpoY, EpoEY, EpoEEY Parthenolide |
(Prota et al., 2013) (Aillaud et al., 2017) (Fonrose et al., 2007; Siedle et al., 2004) |
|
| Δ2 detyrosination -GE |
CCP1, 2, 3, 4, 6 | Penultimate glutamate of the CTT of α-tubulin | CCP1, 2, 3, 4, 6 overexpression | (Aillaud et al., 2016; Rogowski et al., 2010) |
| Δ3 detyrosination -G |
CCP1, 4, 5, 6 | Antepenultimate glutamate of the CTT of α-tubulin | CCP1,4, 5, 6 overexpression | (Aillaud et al., 2016; Rogowski et al., 2010) |
| Acetylation K40 | αTAT1 (MEC7) | K40 at the tubulin body (facing the MT lumen) | αTAT1 overexpression HDAC6 depletion (siRNA)/knockdown Sirt2 depletion (siRNA) Trichostatin A (TSA) AGK2 Tubacin |
(Shida, Cueva, Xu, Goodman, & Nachury, 2010) (Hubbert et al., 2002; Zhang et al., 2008) (North, Marshall, Borra, Denu, & Verdin, 2003) (Hubbert et al., 2002; Yoshida, Kijima, Akita, & Beppu, 1990) (Outeiro et al., 2007; Rumpf et al., 2015) (Haggarty, Koeller, Wong, Grozinger, & Schreiber, 2003) |
| Deacetylation K40 | HDAC6 Sirt2 |
HDAC6 overexpression Sirt2 overexpression αTAT1 depletion (siRNA)/knockdown |
(Hubbert et al., 2002) (North et al., 2003) (Akella et al., 2010; Kalebic et al., 2013; Shida et al., 2010) |
|
| Glutamylation (initiation) | TTLL4 TTLL5 TTLL7 |
preference for β preference for α preference for β |
Overexpression on TTLL4, 5, 7 CCPs depletion/knockdown |
(Janke et al., 2005; Rogowski et al., 2010; van Dijk et al., 2007) |
| Polyglutamylation (Elongation) | TTLL1* TTLL6 TTLL7 TTLL11 TTLL13 |
α/β, preference for α preference for α β preference for α |
TTLL 6, 7,11,13 overexpression CCPs depletion/knockdown |
(Rogowski et al., 2010; van Dijk et al., 2007; Wloga et al., 2010) |
| Deglutamylation (branching point) | CCP5 CCP1** |
α/β | CCPs overexpression TTLL 4, 5, 7 depletion/knockdown Phospinic acid – tested inhibitor for TTLL7 |
(Liu, Garnham, Roll-Mecak, & Tanner, 2013) |
| Deglutamylation (shorten polyglu side chains) | CCP1 CCP4 CCP6 |
α/β | CCPs overexpression TTLL 7, 11, 13 depletion/knockdown Phospinic acid – selective inhibitor for TTLL7 |
(Liu et al., 2013) |
| Monoglycylation | TTLL3 TTLL8 |
α/β preference for β |
TTL3, 8 and 10 overexpression | (Rogowski et al., 2009) |
| Polyglycilation | TTLL10 | |||
| Phosphorylation S172 | CDK1 | β | Overexpression of β-tubulinS172E or β-tubulinS172D mutants | (Fourest-Lieuvin et al., 2006) |
| Polyamination | Transglutaminase | α/β | --- | (Song et al., 2013) |
| Deamynation | unknown | IR072 (irreversible inhibitor of transglutaminase 2) | ||
| Methylation | SETD2 | α | STED2 overexpression | (Park et al., 2016) |
| Deamynation | unknown | SETD2 depletion/knockdown |
Blue, modifications in polymerized tubulin; Red, modifications in soluble tubulin; Black, unclear preference for polymerized or soluble tubulin; Green shade, common features between different modifications.
Overexpression of TTLL1 does not increase the levels of polyglutamylation both in mammalian cells and Tetrahymena thermophila (Janke et al., 2005; van Dijk et al., 2007).
Deglutamylates at branching points glutamates added by TTLL6, but not the ones added by TTLL4 (Rogowski et al., 2010).
A. Cell culture
Human U2OS cells are cultured in DMEM supplemented with 10% FBS (complete growth medium) at 37ºC and 5% CO2. Cell manipulation procedures are performed in a sterile laminar flow hood. U2OS cells are selected due to their relatively high levels of detyrosination, as compared for example with HeLa cells.
B. Transient overexpression of TTL
One day before transfection seed 5x105 cells in 2 mL complete growth medium on a 6 well plate.
-
On the day of transfection prepare solution A and B and incubate for 5 min.
-
A:
250 μl of Opti-MEM + 5 μl of Lipofectamine 2000
-
B:
250 μl of Opti-MEM + 5 μg of TTL-YFP
Add solution A to B and incubate for 30 min.
-
A:
Add the transfection mixture to the well dropwise and incubate for 6 h.
Change medium to complete growth medium and incubate for 12-24 h.
Perform western blot and immunofluorescence analysis (see L2, M4, Fig.1)
C. Depletion of TTL using small interference RNAs (siRNAs)
Purchase siRNA oligonucleotides specific for the mRNA encoding for TTL (Table 7) from a commercial vendor.
One day before transfection seed 2x105 cells per well on a 6 well plate in 1.5 ml of DMEM supplemented with 5 % FBS.
-
On the day of transfection prepare solutions A and B and incubate for 5 min.
-
A:
250 μl of Opti-MEM + 2 μl of Lipofectamine RNAi Max
-
B:
250 μl of Opti-MEM + siRNA oligonucleotides (to a final concentration of 50 nM)
Add solution B to A and incubate for 30 min.
-
A:
Add the transfection mixture to the well dropwise and incubate for 4-6 h.
Change medium to complete growth medium and incubate for 72 h.
Perform western blot and immunofluorescence analysis (see L2, M4, Fig.2).
Table 7. siRNA oligonucleotide sequences.
| siRNA sequence 5’- 3’ | Depletion |
|---|---|
| CAGCCACCAAUCAGUAACU dT | TTL |
| CAAGAAGUCCAAGCUGGAG dT | TubA1A, TubA1B, TubA1C |
| AAGUACAUGGCCUGCUGCA dT | TubA3C, TubA3D, TubA3E, TubA8 |
| AACGAAGCAAUCUAUGACA dT | TubA4A |
D. Knockout of TTL using CRISPR/Cas9
Class 2 Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR), is part of the adaptive immune system in bacteria that has been genetically modified to be used in gene editing (Cong et al., 2013). Due to its simplicity and adaptability, CRISPR has revolutionized genetic engineering. It is based on the co-expression of two components: a short guide RNA (sgRNA), which is a short synthetic nucleotide sequence that is recognized by a CRISPR-associated endonuclease (Cas protein). The sgRNA must target a DNA sequence with ~20 nucleotides that is unique in all genome (to prevent off-targets) and precede an immediately adjacent Protospacer Adjacent Motif (PAM). The Cas recognizes the PAM sequence and cleaves the DNA forming double-strand breaks (DSBs) within the target DNA (~3-4 nucleotides upstream of the PAM sequence). The resulting DSBs is later repaired by non-homologous end-joining. This repair pathway generates small indels, or frameshift mutations that lead to premature stop codons.
Here we describe the production of knockout cell lines for TTL, VASH1 or VASH2 using the CRISPR/Cas9 system and lentiviral transduction expression system. 20 bp sgRNA to target each gene are selected and cloned into lentiviral transfer vectors pLenti-CRISPR-v2 or pLenti-CRISPR-v2 blast. The obtained plasmids contain Cas9, sgRNA and a selection marker (puromycin or blasticidin). The sgRNA in complex with the Cas9 protein targets genomic sequences homologous to the sgRNA (the full CRISPR guide can be accessed at http://www.addgene.org/crispr/guide/).
-
Purchase oligos
Purchase TTL oligo#1 and TTL oligo#2 (Table 10) from a commercial vendor.
Dilute each oligo to 100 μM in nuclease-free sterile water.
-
Oligo annealing and cloning into viral transfer vectors
- In a PCR tube prepare the following mixture:
-
-1 μl TTL oligo#1 (100 μM)
-
-1 μl TTL oligo#2 (100 μM)
-
-1 μl 10x T4 Ligation buffer (used to provide ATP to the reaction)
-
-0.5 μl T4 Polynucleotide Kinase
-
-Nuclease-free water up to 10 μlMix the solution and phosphorylate and anneal the oligos in a thermocycler using the following parameters: 37ºC for 5 min, 95ºC for 5 min and then ramp down to 25ºC at 5ºC/min.
-
-
Dilute the phosphorylated double-stranded oligo 200-fold in nuclease-free water.
- Digest lentiviral vector with BsmBI by preparing the following reaction:
-
-10 μg of lentiviral vector
-
-3 μl BsmBI
-
-3 μl Alkaline phosphatase
-
-6 μl 10x Tango buffer
-
-1mM DTT
-
-Nuclease-free water up to 60 μlIncubate the reaction at 37ºC for 1-3 h.
-
-
Run the digestion reaction on 0.7% agarose gel and confirm separation of the 1.9kb filler sequence from the 13kb digested plasmid. Purify the linearized plasmid with the QIAquick gel extraction kit and quantify the DNA concentration.
- Prepare the following ligation reaction:
-
-50 ng digested plasmid
-
-1 μl of diluted oligo duplex from step 2 (or 1 μl of water for the negative control)
-
-1 μl 10x T4 ligation buffer
-
-0.5 μl T4 DNA ligase
-
-Nuclease-free water to 10 μlMix the solution and incubate at RT for 1 h.
-
-
-
Transformation and selection
Add 3 μl of the ligation product from step 2 to 50 μl Stbl3 Chemically Competent E. coli, mix gently and incubate on ice for 30 min. Stbl3 E. coli or other recombination-deficient strains are recommended to be used with lentiviral transfer plasmids containing long terminal repeats (LTRs) to prevent homologous recombination events.
Heat shock the cells for 45 sec at 42ºC followed by incubation on ice for 2 min.
Add 500 μl of pre-warmed LB medium and shake at 37ºC for 1 h at 225 rpm in a shaking incubator.
Spread 100 μl from each transformation on a pre-warmed LB agar plate containing 50 μg/ml ampicillin. Incubate overnight at 37ºC.
Pick colonies into 5 ml LB medium supplemented with 50 μg/ml ampicillin, grow overnight at 37ºC and perform DNA purification using QIAprep Spin Miniprep Kit. Sequence the purified pLenti-CRISPR-v2-TTL with U6 sequencing primer (hU6F, Table 8).
-
Lentivirus production
One day prior to transfection, plate human embryonic kidney (HEK) 293T cells in a 10 cm tissue culture dish to approximately 40% confluence, so that they are 80-90% confluent at the time of transfection. Grow cells in complete growth medium at 37ºC, 5% CO2.
- On the day of transfection prepare the following mixtures:
- A:
-
-2 ml Opti-MEM
-
-17 μg psPAX2 (Gag, Pol, Rev and Tat expressing packaging vector)
-
-6 μg pMD2.G (VSV-G expressing envelope vector)
-
-22 μg lentiviral vector (pLenti-CRISPR-v2-TTL)Mix thoroughly.
- B:
-
-2 ml Opti-MEM
-
-30 μl Lipofectamine 2000Mix thoroughly.
Incubate A and B separately for 5 min at room temperature (RT).
Add A to B and incubate for 30 min at RT to allow for DNA-lipid complexes to form.
Remove medium from HEK293T cells and add the DNA-lipid complex solution dropwise. Gently rock the plate back-and-forth and from side-to-side to achieve even distribution. Incubate the cells at 37ºC, 5% CO2 for 4 h.
Replace cells with 8 ml of complete growth medium and incubate at 37ºC, 5% CO2 for an additional 48-72 h.
Transfer medium to a 15 ml centrifuge tube and centrifuge at 500 g for 10 min. Filter the harvested viral supernatants through a 0.45 μm cellulose acetate filter to remove cellular debris.
Aliquot and store at -80ºC.
-
Transduction of lentivirus to target cells
One day prior to transduction seed cells in a 6-well plate so that they are 80-90% confluent at the time of transduction. Grow in a total volume of 1.5 ml complete growth medium per well at 37ºC, 5% CO2.
Thaw lentiviral aliquots rapidly in a 37°C water bath. Note that each freeze-thaw cycle will decrease virus titre.
To each well add 500 μL of viral particles and 10 μg/mL of polybrene (polybrene is a polycation that reduces charge repulsion between virus and the cellular membrane and is used to improve transduction efficiency). Mix by gentle swirling. Incubate at 37ºC, 5% CO2 for 24h.
Remove the virus-containing transduction medium and add 2 ml per well of fresh complete growth medium.
Incubate the cells for additional 24-48 h.
*Biosafety concerns for lentivirus/retrovirus manipulation: Use BL2+ precautions:- Discard all material and solutions in contact with virus in 100% bleach.
- Disinfect gloves and plates with bleach and discard in the appropriate category waste.
- Clean liquid spills with 100% bleach.
- Clean the flux chamber with 70% alcohol and UV-sterilise for at least 15 min.
-
Selection of knockout cells
24-48 h after transduction start selecting cells by addition of the appropriate antibiotic (2 μg/ml Puromycin or 10 μg/ml Blasticidin S) to the growth medium.
Confirm target protein depletion by western blot and immunofluorescence (L2, M4, Fig.2).
Individual clones might be isolated and sequenced to identify the precise nature of the mutation.
Table 10. CRISPR oligonucleotide sequences.
| Name | Oligonucleotide sequence (5’-3’) | Reference |
|---|---|---|
| TTL oligo1 | CACCGAACAGCAGCGTCTACGCCG | |
| TTL oligo 2 | AAACCGGCGTAGACGCTGCTGTTC | |
| VASH1 oligo1 | CACCGACGGCTTCCAGGCATTTGAT | (Nieuwenhuis et al., 2017) |
| VASH1 oligo2 | AAACATCAAATGCCTGGAAGCCGTC | (Nieuwenhuis et al., 2017) |
sgRNA (highlighted) match a 20 nucleotide target sequence (protospacer sequence) in the genomic DNA and are followed by a protospacer adjacent motif (PAM) sequence of NGG. When annealed, oligos form double stranded DNA with overhangs compatible for cloning into BsmBI site in pLenti-CRISPR-v2 or pLenti-CRISPR-v2 blast.
Table 8. Oligonucleotides sequences.
| Number | Name | Primer Sequence (5' to 3') |
|---|---|---|
| 1 | XbaIF | GCTCTAGAATGGTGAGCAAGGGCGAGG |
| 2 | KpnIR | GGGGTACCTTAGTATTCCTCTCCTTCTTCCTC |
| 3 | Y450*F | CGCGGATCCTTATTATTCCTCTCCTTCTTCCTCA |
| 4 | Y450*R | TGAGGAAGAAGGAGAGGAATAATAAGGATCCGCG |
| 5 | pEGFPC1F | GATCACTCTCGGCATGGACG |
| 6 | TubSEQ | CACTGGCTTCAAGGTTGGCATC |
| 7 | E449*Y450*F | TGAGGAAGAAGGAGAGTAATAATAAGGATCCGCGGCCG |
| 8 | E449*Y450*F | CGGCCGCGGATCCTTATTATTACTCTCCTTCTTCCTCA |
| 9 | CMV-F | CGCAAATGGGCGGTAGGCGTG |
| 10 | Antis_del | GTGGATGGAGATGCACTCACGCATTCTAGAGTCGGTGTCTTC |
| 11 | Sens_del | GAAGACACCGACTCTAGAATGCGTGAGTGCATCTCCATCCAC |
| 12 | EcoRV_R | GCTTTCTCAGCAGAGATGAC |
| 13 | hU6F | GAGGGCCTATTTCCCATGATT |
E. Transient overexpression of VASH1 and VASH2
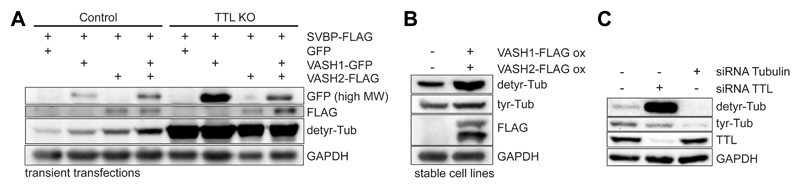
Follow protocol B using 2 μg of pcDNA3.1(-)-VASH1-GFP or pcDNA3.1(-)-VASH2-FLAG together with 2 μg of pcDNA3.1(-)-SVBP-FLAG plasmids and adjust the time of protein overexpression to 24 h (Fig.3 A).
Fig. 3.
Western-blot analysis of U2OS cell lysates. A. Transiently overexpressing VASH1-GFP, VASH2-FLAG and SVBP-FLAG in control and TTL KO cells; B. Stably co-expressing VASH1-GFP and VASH2-FLAG or C. transfected with siRNAs for endogenous tubulins or TTL.
F. Generation of cell lines stably expressing FLAG-VASH1 and FLAG-VASH2
-
Retrovirus production
One day prior to transfection, plate human embryonic kidney (HEK) 293T cells in a 10 cm tissue culture dish to approximately 40% confluency so that they are 80-90% confluent at the time of transfection. Grow cells in complete growth medium at 37ºC, 5% CO2.
- On the day of transfection prepare the following mixtures:
- A:
-
-2 ml Opti-MEM
-
-7 μg pCMV-Gag-Pol (retroviral packaging vector)
-
-6 μg pMD2.G (VSV-G expressing envelope vector)
-
-3.5 μg pAdVantage
-
-22 μg retroviral vectors (pMX-IRES-Blast-VASH1-FLAG and pMX-IRES-Blast-VASH2-FLAG)Mix thoroughly.
- B:
-
-2 ml Opti-MEM
-
-30 μl Lipofectamine 2000Mix thoroughly.
Follow steps D4-8.
-
Transduction of retrovirus to target cells and selection of overexpressing cells
-
1.
Follow D5
-
2.
48h after transduction, select cells by addition of 10 μg/ml Blasticidin S to the growth medium. Grow cells in selective medium for at least 3 weeks.
-
4.
Perform western blot analysis (see L2, Fig.3 B).
-
1.
G. Knockout of VASH1 and VASH2 using CRISPR/Cas9
-
Purchase oligos
Purchase oligos VASH1 oligo#1 and VASH1 oligo#2 (Table 10) from a commercial vendor. Dilute each oligo to 100 μM in nuclease-free sterile water.
-
Oligo annealing and cloning into viral transfer vectors
Clone VASH1 sequence into pLenti-CRISPR-v2-puro and pLenti-CRISPR-v2-blast following steps K2-3.
-
Lentivirus production
Produce lentivirus following steps D4-5 using the lentiviral vector pLenti-CRISPR-v2-blast-VASH2 (Table 5) and pLenti-CRISPR-v2-puro–VASH1 and pLenti-CRISPR-v2-blast-VASH1 produced in G2.
-
Transduction of lentivirus to target cells and selection of knockout cells
Follow step D5.
48 h after transduction start selecting cells by addition of the appropriate antibiotic (2 μg/ml Puromycin or 10 μg/ml Blasticidin S) to the growth medium. Grow cells in selective medium for at least 3 weeks.
Table 5. Material – Reagents and consumables.
| Reagents/consumables | Source |
|---|---|
| General chemical reagents | |
| 10X TGS (Tris-Glycine-SDS) | CAT. GB15.0510, GRiSP Research Solutions |
| 40% Acrylamide/Bis Solution, 29:1 | CAT. 1610146, Bio Rad |
| Acetic Acid glacial | CAT. 1000631000 Millipore/Merk |
| Adenosine 5’-triphosphate magnesium salt | CAT. A9187, Sigma-Aldrich® |
| Ammonium persulfate (APS) | CAT. A3678, Sigma-Aldrich® |
| CaCl2 | CAT. 449709, Sigma-Aldrich® |
| Clarity™ Western ECL Substrate | CAT. 1705061, Bio Rad |
| cOmplete™ Protease Inhibitor Cocktail | CAT. 11697498001, Roche |
| DL-Dithiothreitol | CAT. D9779, Sigma-Aldrich® |
| EDTA | CAT. E5134, Sigma-Aldrich® |
| EGTA | CAT. E4378, Sigma-Aldrich® |
| Ethanol | CAT. 34852- Millipore |
| Formaldehyde solution | CAT. F8775, Sigma-Aldrich® |
| Glycerol ≥99,5% | CAT. G7893, Sigma-Aldrich® |
| Guanosine 5′-triphosphate sodium salt hydrate | CAT. G8877, Sigma-Aldrich® |
| KCl for analysis | CAT. 529552, Millipore/Merk |
| KH2PO4 for analysis | CAT. 529568, Millipore/Merk |
| KOH for analysis | CAT, 1050330500, Millipore/Merk |
| MgCl2 for analysis | CAT. M8266, Sigma-Aldrich® |
| MgSO4 | CAT. 203726, Sigma-Aldrich® |
| N,N,N′,N′-Tetramethylethylenediamine (TEMED) | CAT. T9281, Sigma-Aldrich® |
| Na2HPO4 for analysis | CAT. 567550, Millipore/Merk |
| NaCl for analysis | CAT. 567440, Millipore/Merk |
| Pierce™ Coomassie Plus (Bradford) Assay Kit | 23236, Thermo Fisher Scientific |
| PIPES ≥99% | CAT. P6757, Sigma-Aldrich® |
| Poly-L-Lysine solution, 0.1 % (w/v) in H2O | CAT. P8920, Sigma-Aldrich® |
| Polybrene | CAT. TR-1003, Sigma-Aldrich® |
| Silver nitrate, ACS reagente, 99+% | CAT. 09139-M, Sigma-Aldrich® |
| Sodium acetate anhydrous | CAT. W302406, Sigma-Aldrich® |
| Sodium carbonate | CAT. 791768, Sigma-Aldrich® |
| Sodium Thiosulfate anhydrous | CAT. 72049, Sigma-Aldrich® |
| Sucrose for microbiology | CAT. 1076511000, Millipore/Merk |
| Trichloroacetic acid solution, 6.1N | CAT. T0699, Sigma-Aldrich® |
| Triton™ X-100 | CAT. X100, Sigma-Aldrich® |
| Tween 20 | CAT. P7949, Sigma-Aldrich® |
| Chemical reagents - Fixatives | |
| Methanol for analysis | CAT. 1070182511, Millipore/Merk |
| Paraformaldehyde, 20%, aqueous solution | CAT. 15713, Electron Microscopy Sciences |
| Disposable materials | |
| Square cover glass (22x22mm) | CAT. 2845-22, Corning® |
| Sterile CA filter, ⌀ 25 mm, 0,45 μm pore size | CAT: 1520014, Frilabo |
| Tissue culture dish ⌀ 100 mm | CAT. 83.3902, Starsted |
| Tissue culture plate 6 Well, Standard | CAT. 83.3920.005, Starsted |
| Cell culture reagents | |
| Dulbecco's Modified Eagle Medium (DMEM), high glucose, pyruvate | CAT. 1966052, Gibco™ |
| DMEM Gibco® CO2 Independent Medium | CAT. 21063029, Gibco™ |
| Fetal Bovine Serum (FBS), qualified, heat inactivated, E.U. | CAT. 10500064, Gibco™ |
| Opti-MEM™ I Reduced Serum Media | CAT: 31985054, Gibco™ |
| TrypLE™ Express Enzyme (1X), phenol red | CAT. 12605028, Gibco™ |
| Cell lines | |
| HEK293T | ATCC® CRL-3216™ |
| HeLa | ATCC® CCL-2™ |
| U2OS | ATCC® HTB-96™ |
| U2OS Photoactivatable-TUB/mCherry-TUB | Kindly supplied by Duane Compton |
| Microtubule poisons and cell cycle blocking drugs | |
| MG 132 | CAT. 474790, Calbiochem |
| Nocodazole | CAT. M1404, Sigma-Aldrich® |
| S-Trityl-L-cysteine (STLC) | CAT. 2191, Tocris Bioscience |
| Taxol | CAT. T7191, Sigma-Aldrich® |
| Selection drugs | |
| Ampicillin sodium salt | CAT. A8351, Sigma-Aldrich® |
| Blasticidin S HCl, powder | CAT. R21001, Gibco™ |
| Puromycin, Dihydrochloride | CAT. 540411, Calbiochem |
| Transfection reagents | |
| Lipofectamine™ 2000 Transfection Reagent | CAT. 11668-027, Invitrogen™ |
| Lipofectamine™ RNAiMAX Transfection Reagent | CAT. 13778075, Invitrogen™ |
| Other drugs | |
| Parthenolide | CAT. P0667, Sigma-Aldrich® |
| Molecular Biology | |
| Plasmid vectors | |
| LV-H2B-RFP | #26001, Addgene |
| pIRESneo3-EGFP-TUBA1B | Kindly supplied by Patrick Meraldi |
| pIRESpuro-mRFP-TUBA1B | Kindly supplied by Patrick Meraldi |
| pLenti-CRISPR-v2 | #52961, Addgene |
| pLenti-CRISPR-v2-blast | #98293, Addgene |
| pLenti-CRISPR-v2-blast-VASH2 | Kindly supplied by Thijn Brummelkamp |
| pRRLSIN.cPPT.PGK-GFP.WPRE | # 12252, Addgene |
| psPAX2 | #12260, Addgene |
| pMD2.G | #12259, Addgene |
| pCMV-Gag-Pol | RV-111, Cell Biolabs |
| pAdVantage | E1711, Promega |
| TTL-YFP | Kindly supplied by Carsten Janke |
| pcDNA3.1(-)-VASH1-GFP | Kindly supplied by Thijn Brummelkamp |
| pcDNA3.1(-)-VASH2-FLAG | Kindly supplied by Thijn Brummelkamp |
| pcDNA3.1(-)-SVBP-FLAG | Kindly supplied by Thijn Brummelkamp |
| pMX-IRES-Blast-VASH1-FLAG | Kindly supplied by Thijn Brummelkamp |
| pMX-IRES-Blast-VASH2-FLAG | Kindly supplied by Thijn Brummelkamp |
| Enzymes | |
| Alkaline Phosphatase, Calf Intestinal (CIP) | M0290S, New England Biolabs |
| BsrGI | R0575S, New England Biolabs |
| EcoRV | R0195S, New England Biolabs |
| Esp3I (BsmBI) | ER0451, ThermoFisher Scientific |
| PfuTurbo DNA Polymerase | 600257, Agilent |
| Phusion high-fidelity DNA polymerase | M053S, New England Biolabs |
| T4 DNA ligase | M0202, New England Biolabs |
| T4 Polynucleotide Kinase | M0201S, New England Biolabs |
| Buffers and others chemicals for molecular biology | |
| 100 mM dNTP Set | 10297018, ThermoFisher Scientific |
| 10x CutSmart Buffer | B7204S, New England Biolabs |
| 10X T4 ligation buffer | B0202S, New England Biolabs |
| 10X Tango Buffer | BY5, Thermo Fisher Scientific |
| 5X Phusion HF Buffer | B0518S, New England Biolabs |
| T4 Polynucleotide Kinase Reaction Buffer | B0201S, New England Biolabs |
| Competent cells | |
| DH5alpha Chemically Competent E. coli | 18265017, Thermo Fisher Scientific |
| Stbl3 Chemically Competent E. coli | C7373-03, Thermo Fisher Scientific |
| DNA purification Kits | |
| QIAprep Spin Miniprep Kit | 27104, Qiagen |
| QIAquick Gel Extraction Kit | 28704, Qiagen |
| Imaging equipment | |
| Fixed analysis | |
| AxioImager Z1 | Zeiss |
| CCD camera | ORCA-R2, Hamamatsu |
| Live imaging | |
| Inverted microscope TE2000U | Nikon |
| CSU-X1 spinning-disk confocal head | Yokogawa Corporation of America |
| iXonEM+ EM-CCD camera | Andor Technology |
| General Equipment | |
| ChemiDoc™ XRS+ System | Bio Rad |
| Electrophoresis system: - Mini-PROTEAN® Tetra Cell - PowerPac Basic™ Power Supply |
CAT. 165-8000, Bio Rad CAT. 164-5050, Bio Rad |
| iBlot- IBlot™Dry Blotting System | 25-0911, Invitrogen™ |
| Ultracentrifuge Optima MAX-XP with MLA-130 rotor | Beckman Coulter |
| Ultracentrifuge tubes | CAT. 347287, Beckman Coulter |
H. Depletion of endogenous α-tubulin isotypes using siRNA
Tubulin PTMs can be modulated by the expression of exogenous forms of tubulin as an alternative to the perturbation of the modifying enzymes. However, during the process of polymerization, endogenous and exogenous tubulins will co-assemble, resulting in mosaic MTs. In order to build MTs composed by a single form of α- or β-tubulin, it is essential to deplete the endogenous α- or β-tubulin.
I. Transient overexpression of tyrosinated, detyrosinated and Δ2 forms of TUBA1B
-
Site-directed mutagenesis of mammalian expression vectors
Use pIRES-puro-mRFP-TUBA1B and pIRES-neo3-EGFP-TUBA1B containing human α-tubulin1B (TUBA1B) cDNA (coding for residue 2 to 452 of NP_006073.2 NCBI reference) in frame with N-terminal mRFP or EGFP tags as templates for the mutagenesis reaction (Table 5).
Purchase oligos Y450*F and Y450*R (Table 8) from a commercial vendor. The set of complementary oligonucleotides contains the mutations that allows for the replacement of Y450 for a stop codon (denoted as *) flanked by unmodified nucleotide sequence.
Dilute each oligo to 100 μM in nuclease-free sterile water.
-
In a PCR tube mix:
-
-10 ng of DNA template (pIRES-puro-mRFP-TUBA1B or pIRES-neo3-EGFP-TUBA1B)
-
-0.5 μl of each primer Y450*F and Y450*R (Table 8)
-
-0.2 mM dNTPs
-
-2.5 μl 10x Pfu DNA polymerase reaction buffer,
-
-0.5 μl PfuTurbo DNA polymerase (2.5 U/μl)
-
-Nuclease-free water up to 25 μl
In parallel perform control PCR reactions in the absence of DNA polymerase. Use the following conditions for PCR: an initial step of denaturation of DNA template at 95°C for 2 min; 18 cycles of 95°C for 1 min, annealing of forward and reverse primers at 55°C for 1 min, and extension of each primer at 68°C for 15 min.
-
-
Place the PCR tubes on ice to cool the reaction.
Digest the amplification products by addition of 1 μl of DpnI (20 U/μl) directly to the tube. Gently and thoroughly mix the reaction by pipetting the solution up and down. Spin down the tubes and immediately incubate at 37ºC for 1 h to digest the parental supercoiled dsDNA.
Transform 2 μl of the treated PCR product into DH5α ultracompetent cells and plate onto LB agar plates containing 50 μg/ml of ampicillin. Pick colonies and grow in LB medium supplemented with 50 μg/ml ampicillin overnight at 37°C.
Isolate plasmid DNA (pIRES-puro-mRFP-TUBA1BY450* and pIRES-neo3-EGFP-TUBA1BY450*) using QIAprep Spin Miniprep Kit and sequence with pEGFPC1F and TubSEQ primers (Table 8).
To generate pIRES-puro-mRFP-TUBA1BE449*Y450* and pIRES-neo3-EGFP-TUBA1BE449*Y450* repeat steps 4-8 using oligos E449*Y450*F and E449*Y450*R (Table 8) and pIRES-puro-mRFP-TUBA1BY450* and pIRES-neo3-EGFP-TUBA1BY450* as templates.
-
Altering the cDNA sequences to confer resistance to siRNA depletion
Obtain from a commercial vendor a synthetic gene coding for residues 2 to 118 of TUBA1B, containing 7 nucleotide silent mutations and 5’ and 3’ regions containing BsrGI and EcoRV restriction sites (pUC57-Kan-TUBA1B2-118 sequence, shown in Table 9).
- Digest pUC57-Kan-TUBA1B2-118 and the mammalian expression plasmids prepared in I1 by mixing:
-
-10 μg plasmid
-
-1.0 μl BsrGI
-
-1.0 μl EcoRV
-
-5.0 μl CutSmart Buffer
-
-Nuclease-free water up to 50 μlIncubate for 1 h at 37ºC.
-
-
Purify the linearized plasmid (6.5kb) and TUBA1B2-118 (679bp) fragment with the QIAquick gel extraction kit and quantify the DNA.
- Prepare the following ligation mixture:
-
-100 ng linearized mammalian expression vector
-
-Digested TUBA1B2-1188 fragment (3:1 to 5:1 molar ratio over vector)
-
-1 μl of 10x T4 DNA ligase buffer
-
-0.5 μl of T4 DNA ligase
-
-Nuclease-free water up to 10 μl
-
-
Gently mix and incubate for 1 h at RT. Also set up a control reaction in which the insert is omitted.
Repeat step I1.7-8 and obtain siRNA resistant versions of pIRES-puro-mRFP-TUBA1BY450*, pIRES-puro-mRFP-TUBA1BE449*Y450, pIRES-neo3-EGFP-TUBA1BY450* and pIRES-neo3-EGFP-TUBA1BY450*.
-
Transient expression of tyrosinated, detyrosinated and Δ2 forms of TUBA1B
Follow protocol B using 3 μg of plamids produced in I and adjusting the time of protein overexpression to 24 h.
Table 9. Synthetic sequence.
| Name | Sequence (5' to 3') |
|---|---|
| TUBA1B2-118 | TCTAGACTGTACAAGTCCGGACTCAGATCTCGAGTGCGTGAGTGCATCTCCATCCACGTTGGCCAGGCTGGTGTCCAGATTGGCAATGCCTGCTGGGAGCTCTACTGCCTGGAACACGGCATCCAGCCCGATGGCCAGATGCCAAGT GACAAGACCATTGGGGGAGGAGATGACTCCTTCAACACCTTCTTCAGTGAGACGGGCGCTGGCAAGCACGTGCCCCGGGCTGTGTTTGTAGACTTGGAACCCACAGTCATTGATGAAGTTCGCACTGGCACCTACCGCCAGCTCTTCC ACCCTGAGCAGCTCATCACAGGCAAGGAAGATGCTGCCAATAACTATGCCCGAGGGCACTACACCATTGGCAAGGAGATCATTGACCTTGTGTTGGACCGAATTCGCAAGCTGGCTGACCAGTGCACCGGTCTTCAGGGCTTCTTGGT TTTCCACAGCTTTGGTGGGGGAACTGGTTCTGGGTTCACCTCCCTGCTCATGGAACGTCTCTCAGTTGATTATGGCAAAAAAAGCAAACTCGAATTCTCCATTTACCCAGCACCCCAGGTTTCCACAGCTGTAGTTGAGCCCTACAACT CCATCCTCACCACCCACACCACCCTGGAGCACTCTGATTGTGCCTTCATGGTAGACAATGAGGCTATTTACGACATCTGTCGTAGAAACCTCGATATCAAGCTT |
J. Generation of a cell line stably expressing H2B-mRFP and tyrosinated, detyrosinated or Δ2 forms of TUBA1B
-
Cloning of TUBA1B cDNA into lentiviral transfer vectors
- PCR amplify human TUBA1B, TUBA1BY450* or TUBA1BE449*Y450* cDNA using flanking primers containing XbaI and KpnI restriction sites (Table 8) from pIRES-neo3-EGFP-TUBA1B expression vectors obtained in I:
-
-10 ng template
-
-1.0 μl dNTPs (10 mM each)
-
-2.5 μl Primer XbaIF (10 μM)
-
-2.5 μl Primer KpnIR (10 μM)
-
-10.0 μl 5x Phusion HF Buffer
-
-0.5 μl Phusion DNA polymerase
-
-Nuclease-free water up to 50 μlIn parallel perform control PCR reactions in the absence of template. Use the following conditions for PCR: an initial step of denaturation of DNA template at 95°C for 1 min; 35 cycles of 95°C for 30 sec, annealing of forward and reverse primers at 65°C for 1 min, and extension of each primer at 72°C for 2 min.
-
-
Subclone the amplified PCR product into the pRRLSIN.cPPT.PGK-GFP.WPRE lentiviral vector (Table 5) following the same procedure described above (I2.4-6).
Isolate plasmid DNA (pRRL-GFP-TUBA1B, pRRL-GFP-TUBA1B- Y450* and pRRL-GFP-TUBA1B- E449*Y450*) using QIAprep Spin Miniprep Kit and sequence with the sequence primers pEGFPC1F and TubSEQ (Table 8).
-
Deletion of EGFP-tag from lentiviral vectors expressing TUBA1B by PCR
The protocol for deletion of EGFP sequence from the lentiviral vectors requires four oligonucleotide primers that are derived partly from the sequence to be deleted and partly from the template, in addition to two outermost flanking primers (CMVF, Antis_del, Sens_del and EcoRVR (Table 8).
- For each template pRRL-EGFP-TUBA1B, pRRL-GFP-TUBA1B- Y450*or pRRL-GFP-TUBA1B- E449*Y450* prepare the following PCR reaction:
-
-10 ng template
-
-1.0 μl dNTPs (10 mM each)
-
-2.5 μl CMVF (10 μM)
-
-2.5 μl Antis_del 10 (10 μM)
-
-10.0 μl 5x Phusion HF Buffer
-
-0.5 μl Phusion DNA polymerase
-
-Nuclease-free water up to 50 μlIn parallel perform control PCR reactions in the absence of template. Use the following conditions for PCR: an initial step of denaturation of DNA template at 95°C for 1 min; 35 cycles of 95°C for 30 sec, annealing of forward and reverse primers at 65°C for 1 min, and extension of each primer at 72°C for 1 min.
-
-
Prepare a second round of PCR by repeating step 1 using the primers (Sens_del and EcoRVR, Table 8).
Electrophoretically resolve the amplified products of the first (150 bp) and second reactions (860 bp) on a 1% agarose gel and purify DNA fragments with the QIAquick gel extraction kit.
- In a PCR tube prepare the following mixture:
-
-17.5 ng PCR product 1 (150 bp)
-
-100 ng PCR product 2 (860 bp)
-
-10 μl 5X Phusion HF Buffer
-
-Nuclease-free water up to 50 μlMix the solution and anneal the two fragments in a thermocycler using the following parameters: 94ºC for 4 min, 50ºC for 2 min and 72ºC for 2 min.
-
-
- Prepare a third round of PCR by adding the following components to the reaction prepared in 5:
-
-1.0 μl dNTPs (10mM each)
-
-1.0 μl CMVF
-
-1.0 μl EcoRVR
-
-0.5 μl Phusion DNA polymerase
-
-Perform PCR using the reaction conditions described in 2.
-
-
Electrophoretically resolve the amplified product (1 kb) on a 1% agarose gel and purify the DNA fragment with the QIAquick gel extraction kit.
Digest the PCR product obtained in 6 and lentiviral vectors prepared in J1 and J2 with BsrGI and EcoRV. Repeat I2.2-2.6 to obtain pRRL-TUBA1B, pRRL-TUBA1B- Y450* and pRRL- TUBA1B- E449*Y450*.
-
Production of lentivirus and transduction to target cells
Follow protocol D4-D5 using LV-H2B-RFP (Table 5) and vectors pRRL-GFP-TUBA1B (coding for tyr-Tub), pRRL-TUBA1B (coding for untagged tyr-Tub), pRRL-GFP-TUBA1B-Y450* (coding for GFP-detyr-Tub), pRRL-TUBA1B-Y450* (coding for untagged detyr-Tub), pRRL-GFP-TUBA1B-E449*Y450* (coding for GFP-Δ2-Tub) and pRRL-TUBA1B-E449*Y450* (coding for untagged Δ2-Tub). In the figures tyr-TUB, detyr-Tub and Δ2-Tub are denoted as –GEEY, -GEE* and -GE* respectively. The untagged versions of tubulin are denoted as NOTAG.
-
Selection of cells expressing H2B-mRFP together with tyrosinated, detyrosinated and Δ2 forms of Tubulin
Select cells expressing H2B-mRFP together with tyrosinated, detyrosinated and Δ2 forms of tubulin by cell sorting.
Perform western blot and immunofluorescence analysis (L2, M4, Fig. 4).
-
Depletion of endogenous α-tubulin isotypes using siRNA from cells stably expressing H2B-mRFP and tyrosinated, detyrosinated and Δ2 forms of TUBA1B
Follow protocol C using siRNA oligonucleotides specific for the mRNAs encoding for the different isotypes of α-tubulins (Table 7) and incubating for 72 h.
-
Generation of cells expressing H2B-mRFP together with tyrosinated, detyrosinated and Δ2 forms of Tubulin, knockout for TTL, VASH1, and VASH2
Follow protocol D using cells produced in J4.
Perform western blot analysis (L2, Fig. 4B)
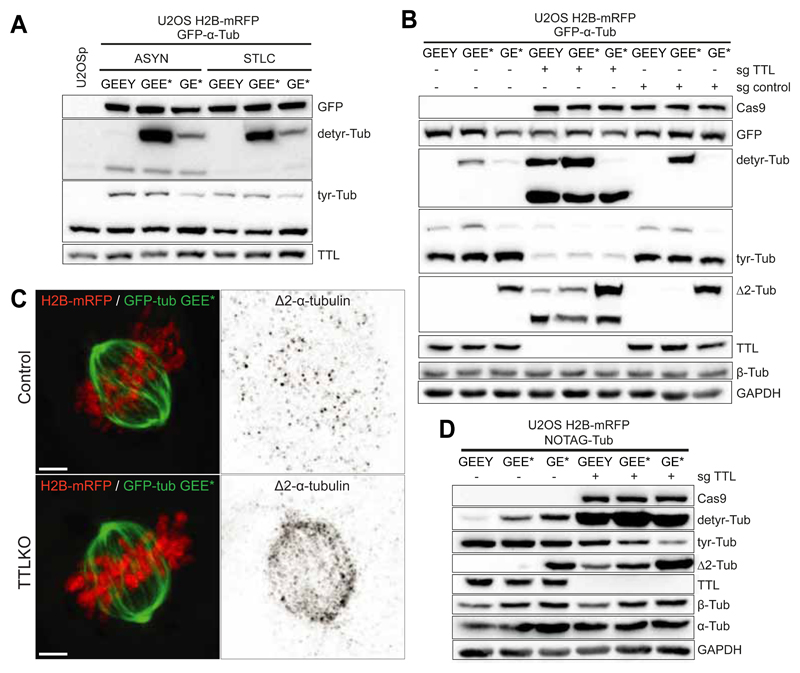
Fig. 4.
A. Western-blot analysis of U2OS cell lysates stably expressing H2B-mRFP and GFP-α-Tub (GEEY, GEE* or GE*) in asynchronous and mitotic populations (cells treated with STLC for 14h). B. Western-blot analysis of U2OS cell lysates stably expressing H2B-mRFP and GFP-α-Tub (GEEY, GEE* or GE*) in control (sgcontrol) and TTL KO cells (sgTTL). C. Deconvolved immunofluorescence showing the cellular distribution of ?2-tubulin in U2OS cells stably expressing H2B-mRFP and GFP-GEE* in the presence or absence of TTL. Scale bar, 5 μm. D. Western-blot analysis of U2OS cells lysates stably expressing H2B-mRFP and NOTAG-α-Tub (GEEY, GEE* or GE*), in control and TTLKO (sgTTL) cells.
K. Reducing tubulin detyrosination using parthenolide
Small molecule inhibitors frequently offer several advantages over protein depletion by siRNA or knockout of the encoding gene, especially when the efficiency of these strategies is a limiting factor. Small molecule inhibitors act by disrupting protein-protein interactions and are highly penetrant across the cell population. One of the biggest advantages of the small molecule inhibitors is to allow temporal control, which is particularly important in fast processes, such as mitosis. Additionally, these drugs can be combined with other treatments and are usually reversible (Weiss, Taylor, & Shokat, 2007).
The identification of novel detyrosination inhibitors has provided valuable tools for addressing the effects of reducing detyrosinated tubulin (Bocca, Gabriel, Bozzo, & Miglietta, 2004; Fonrose et al., 2007; Miglietta, Bozzo, Gabriel, & Bocca, 2004). The sesquiterpene lactones Parthenolide and Costunolide were originally identified as inhibitors of the NF-κB pathway (Bork, Schmitz, Kuhnt, Escher, & Heinrich, 1997), but their roles in modulating the tyrosination-detyrosination cycle have also recently been described (Barisic & Maiato, 2016; Fonrose et al., 2007; Whipple et al., 2013). In the case of Parthenolide, it has notable anticancer properties and its activity has been linked to several cellular processes including apoptosis (Gopal, Arora, & Van Dyke, 2007), DNA methylation (Liu et al., 2009), p21 signalling (Ghantous et al., 2012) and the regulation of TNF-α (Zhang et al., 2017), amongst others. Since Parthenolide has highly reactive groups that confer high levels of non-specificity, its careful use at the correct dose, as well as proper storage and avoidance of freeze-thaw cycles is highly recommended. As with all inhibitors, using titrated inhibitor concentrations can reduce off-target effects and cellular toxicity without jeopardizing specificity and/or penetrance (Arrowsmith et al., 2015).
Protocol for reducing Tubulin detyrosination using Parthenolide
Grow U2OS cells until 80% confluence in complete growth medium.
Incubate cells with increasing concentrations of parthenolide (0-80 μM) for 1 h.
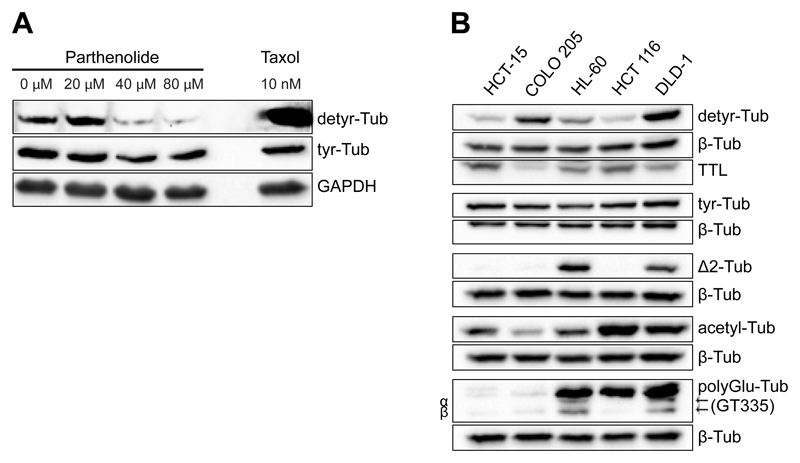
Determine the optimal experimental settings for specifically reducing detyrosinated tubulin by performing western blot and immunofluorescence analysis (see L2, M4, Fig. 5A).
Fig. 5.
A. Western-blot analysis of U2OS cells treated with the indicated concentrations of Parthenolide for 1h. Treatment with 10nM taxol for 1h was used as a control. B. Analysis of the expression profile of tubulin PTMs in the indicated cell lines by Western blot. β-tubulin was used as loading control.
L. Analysis of the expression profile of tubulin PTMs by western blot
-
Antibodies against tubulin PTMs
Numerous antibodies are available for the study of tubulin PTMs. An updated list adapted from (Magiera & Janke, 2013) is provided in Table 4.
-
Western-blotting
Preparation of cell lysates
Grow cells until 90% confluence in complete growth medium.
Harvest the cells by centrifugation at 1200 rpm for 5 min.
Wash pellets once with warm PBS.
Resuspend the pellets in ice-cold lysis buffer (Table 6) freshly supplemented with protease inhibitor cocktail (approximately 500 µl lysis buffer per ~1x107 cells). Incubate on ice for 30 min. To avoid protein degradation, keep tubes on ice from this step onwards. Optional: snap freeze by immersion in liquid nitrogen.
Clarify the lysate by centrifugation at 20,000 g for 8 min at 4ºC.
Sample preparation and electrophoresis
Determine total protein concentration using the Bradford assay.
Denature protein samples in Laemmli sample buffer at 95ºC for 5 min.
Separate 25-50 µg of total proteins by 10% (v/v) SDS-PAGE gel electrophoresis at 80 V through the stacking gel and increase to 120 V for the resolving migration. The amount of protein is determined by the nature of PTM to be detected (e.g. detyr-Tub is usually expressed at very low levels, thus 50 µg of total protein is usually necessary to detect a clear band).
Transfer proteins to a nitrocellulose membrane using a dry blotting system.
To evaluate transfer efficiency, incubate the membrane with Ponceau S solution for 2-5 min.
Immunodetection
Block non-specific binding sites by incubating membranes in blocking solution (TBST supplemented with 5% nonfat dry milk) for 1.5 h at RT with gentle agitation.
Add the primary antibody (diluted in TBST supplemented with 1 % nonfat dry milk) to the membrane and incubate for 1 h at RT or overnight at 4°C.
Wash the membrane 3x in TBST for 10 min.
Add the HRP-conjugated secondary antibody (diluted in TBST supplemented with 1 % nonfat dry milk) to the membrane and incubate for 1 h at RT (gentle agitation).
Wash the membrane 3x in TBST for 10 min.
Incubate the membrane for 1–2 min in enhanced chemiluminescence (ECL) mixture (prepared following manufacturer´s instructions).
Detect and acquire the chemiluminescence signal using an imaging system (Fig. 5B).
Table 4. Primary and secondary antibodies for detection of tubulin PTMs and modifying enzymes.
| Tubulin PTMs | |||||
|---|---|---|---|---|---|
| Modification | Name | Type | Production | References | Commercial availability |
| All α-tubulin isoforms | B-5-1-2 | Mouse Monoclonal |
Raised against Sarkosyl-resistant filaments from sea urchin sperm axonemes | (Piperno, LeDizet, & Chang, 1987) | T5168 Sigma Aldrich® |
| All β-tubulin isoforms | TUB 2.1 | Mouse Monoclonal |
Raised against rat brain tubulin; Recognizes an epitope in the C-terminal part of all five isoforms of β-tubulin (between amino acids 281-446) | (Gozes & Barnstable, 1982) | T4026 Sigma Aldrich® |
| C-terminal -GEEEGEEF and -GEEEGEEY on α-tubulin | Y1/2 | Rat Monoclonal |
Initially raised against purified yeast tubulin (-EEF) | (Kilmartin, Wright, & Milstein, 1982) | MAB1864 Millipore |
| C-terminal –GEEEGEE on α-tubulin | Anti-detyr-tubulin | Rabbit Polyclonal |
Raised against -GEEEGEE | (Gundersen, Kalnoski, & Bulinski, 1984; Paturle-Lafanechere et al., 1991) | AB320 Milipore |
| C-terminal –GEEEGEE on α-tubulin | Anti-detyr-tubulin | Rabbit Polyclonal |
Raised against 10 residue synthetic peptide of the C-terminal domain of human α-tubulin | (Berezniuk et al., 2012) | ab48389 abcam |
| C-terminal -GEEEGE on α-tubulin | Δ2-α-tubulin detyrosination | Rabbit Polyclonal |
Raised against -EGEEEGE | (Gundersen et al., 1984; Paturle-Lafanechere et al., 1994) | AB3203 Milipore |
| C-terminal – GEEEG on α-tubulin | 3EG | Rabbit Polyclonal |
Raised against -GEGEEEG | (Aillaud et al., 2016) | Not available |
| γ-Linked En side chain (n=1,2,3…) on modified E | GT335 | Mouse Monoclonal |
Raised against octapeptide EGEGE*EEG, modified by the addition of two glutamyl units onto the fifth E | (Wolff et al., 1992) | AG-20B-002 AdipoGen |
| C-terminal - En (n≥3) | 1D5 | Mouse Monoclonal |
Raised against the peptide –VDSVEGEGEEEGEE; Recognizes both detyrosinated and polyglutamylated α and β-tubulin with a minimum side chain length of 3 glutamyl residues | (Wehland & Weber, 1987) (Rudiger, Rudiger, Wehland, & Weber, 1999) |
302011 Synaptic Systems |
| C-terminal - En (n≥3) | PolyE | Rabbit Polyclonal |
Recognizes elongated side chains | (Rogowski et al., 2010; Shang, Li, & Gorovsky, 2002) | AG-25B-0030-C05 AdipoGen |
| γ-Linked G side chain (n = 1) on modified E | TAP952 | Mouse Monoclonal |
Raised against Paramecium axonemal tubulin | (Bre et al., 1996; Bre, Redeker, Vinh, Rossier, & Levilliers, 1998; Callen et al., 1994) | MABS277 Millipore |
| γ-Linked Gn side chain (n≥3) on modified E | AXO49 | Mouse Monoclonal |
Raised against Paramecium axonemal tubulin | (Bre et al., 1996; Bre et al., 1998; Callen et al., 1994) | MABS276 Millipore |
| C-terminal polyG chains | polyG | Rabbit Polyclonal |
Recognizes long polyglycylation side chains | (Rogowski et al., 2009; Shang et al., 2002) | Not available |
| Acetylation K40 on α-tubulin | 6-11B-1 | Mouse monoclonal |
Raised against flagellar tubulin of sea urchin | (LeDizet & Piperno, 1991); (Piperno & Fuller, 1985) | MABT868 Millipore |
| Phosphorylation S172 on β-tubulin | Anti-phospho-peptide P172 | Rabbit Polyclonal |
Raised against the peptide Ac-VVPpSPKVSDTVVEC-CONH2 | (Fourest-Lieuvin et al., 2006) | Not available |
| Methylation K40 on α-tubulin | α-TubK40me3 | Rabbit Polyclonal |
Raised against trimethylated K40 peptide (Ac-GQMPSD-Kme3- TIGGGDC-amide) | (Park, Chowdhury, et al., 2016; Park, Powell, et al., 2016) | Not available |
| Modifying enzymes | |||||
| Modification | Name | Type | Production | References | Commercial availability |
| Tubulin tyrosination | 2E5F8 | Mouse monoclonal |
Raised against TTL fusion protein Ag4708 | Unpublished data | 66076-1-Ig Proteintech |
| Tubulin tyrosination | Anti-TTL | Rabbit Polyclonal |
Raised against TTL fusion protein Ag4526 | (Barisic et al., 2015) | 13618-1-AP Proteintech |
| Polyglutamylation | Anti-TTLL1 | Ginea pig Polyclonal |
Raised against purified recombinant TTLL1 | (Ikegami, Sato, Nakamura, Ostrowski, & Setou, 2010) | Several commercial vendors |
| Polyglutamylation | Anti-TTLL4 | Rabbit Polyclonal |
Raised against TTLL4 peptide (aa 516-653) | (Xia et al., 2016) |
PAB22002 Abnova |
| Polyglutamylation | Anti-TTLL5 | Rabbit Polyclonal |
Raised against TTLL5 peptide (aa 1000-1088) | (Sergouniotis et al., 2014) |
PAB22614 Abnova |
| Polyglutamylation | Anti-TTLL6 | Rabbit Monoclonal |
Raised against a human TTLL6 peptide using ARM Technology | (Xia et al., 2016) | H00284076-K Abnova |
| Polyglutamylation | Anti-TTLL7 | Rabbit Polyclonal |
Raised against maltose-binding-fused TTLL7 △370; aa 371-609) | (Ikegami et al., 2006) | Several commercial vendors |
| Deglutamylation | CCP1 (LM-1A7) | Mouse Monoclonal |
Raised against recombinant AGTPBP1 protein of human origin | (Xia et al., 2016) | sc-134251 Santa Cruz Biotechnology |
| Deglutamylation | CCP2 (S-13) | Rabbit Polyclonal |
Raised against a peptide mapping near the C-terminus of CCP2 of human origin. | (Xia et al., 2016) | sc-138193 Santa Cruz Biotechnology |
| Deglutamylation | CCP5 (N-18) | Rabbit Polyclonal |
Raised against human CCP5 | (Wu, Wei, & Morgan, 2017) |
Ab118621 abcam |
| Deglutamylation | CCP6 (N-14) | Rabbit Polyclonal |
Raised against a peptide mapping near the N-terminus of CCP6 of human origin | (Li et al., 2016; Ye et al., 2014) | Discontinued, Santa Cruz Biotechnology |
| Acetylation | Anti-αTAT1 | Rabbit Polyclonal |
Recognizes residues 1-236 of αTAT1 | (Shida, Cueva, Xu, Goodman, & Nachury, 2010) | Other peptides-Several commercial vendors |
| Deacetylation | Anti-HDAC6 | Rabbit Polyclonal |
Recognizes residues 1031-1215 of HDAC6 | (Hubbert et al., 2002) | 07-732 Millipore |
| Deacetylation | Anti-SIRT2 | Rabbit Polyclonal |
Raised against synthetic peptide corresponding to Sirt2 amino acids 341-352 | (Chopra et al., 2012) | S8447 Sigma Aldrich® |
| Phosphorylation | A17 | Mouse monoclonal |
Raised against Xenopus p34cdc2 protein (C-terminal two-thirds) | (Goodger, Gannon, Hunt, & Morgan, 1996) | ab18 abcam |
| Methylation | Anti-SETD2 | Rabbit Polyclonal |
Raised against synthetic peptide from within residues 500 - 600 of human SETD2 | (Park, Powell, et al., 2016) | ab31358 abcam |
Table 6. Buffers and solutions.
| Buffer/Solution | Composition |
|---|---|
| Cytoskeleton buffer with sucrose (CBS) | 137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 4 mM EGTA, 4 mM MgCl2, 10mM PIPES. Adjust pH to 6.1. Autoclave and keep at 4ºC. Add sterile-filter sucrose to a final concentration of 10mM. |
| Mounting medium | 20 mM Tris pH 8, 0,5% N-propyl gallate and 90% glycerol. To dissolve the N-propyl gallate warm up the solution to 37ºC-50ºC while stirring. Aliquot and store at -80ºC. Working aliquots can be store at 20ºC. Discard if the colour changes |
| Phosphate buffer saline (PBS) | 80 g/L NaCl, 2 g/L KCl, 14.4 g/L Na2HPO4, 2.4 g/L KH2PO4 Adjust pH to 7.4 and autoclave |
| LB medium | 10 g/L Tryptone, 5 g/L Yeast Extract, 10 g/L NaCl. Adjust pH to 7.0 and autoclave |
| LB agar | LB medium + 15 agar/L |
| Lysis Buffer | 50mM Tris HCl pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 0.5% NP40, 0.5% Triton™ X-100. |
| Laemmli Sample Buffer (4×) | 250mM Tris HCL pH 6.8, 8% SDS, 40% Glycerol, 20% β-Mercaptoethanol, 0.02% Bromophenol blue. Aliquot and store at -20ºC |
| Ponceau solution | 3% acetic acid (v/v), 0.2% Ponceau S (w/v) |
| Tris-buffered saline (TBS) | 6.05 g/L Tris base, 8.76 g/L NaCl. Adjust pH to 7.6 |
| K-PIPES buffer | 100 mM PIPES pH6.9, 1 mM EGTA, 1 mM MgSO4, 1 mM DTT, 0.1 mM GTP. Adjust pH to 6.9 with KOH. |
| 10% sucrose cushion | 100mM PIPES pH6.9, 1mM EGTA, 1mM MgSO4, 1mM DTT, 0.1mM GTP, 10μM taxol, 10% sucrose. Adjust pH to 6.9 with KOH. Pre-warm to 37ºC before use. |
M. Analysis of the cellular distribution of tubulin (de)tyrosination in mitotic cells using fixed material
Fixation
MTs are labile structures, highly sensitive to thermal and chemical fluctuations. Therefore, the speed of the fixation reaction is a key aspect to the successful preservation of the MT structure. The selection of a given fixative is a compromise between the structural preservation and epitope accessibility. To visualize MTs alone, glutaraldehyde (GTA) allows the best structural preservation and causes less artefacts (Whelan & Bell, 2015). However, it masks the majority of the antigen epitopes. Methanol fixation is usually the best option to co-immunolabel MTs with other proteins. Although it induces some structural artefacts, such as “wavy” MTs, tubulins and tubulin modifications are successfully detected with reduced background. Paraformaldehyde (PFA) fixation is probably the worst option to preserve the morphology of MTs but it is an alternative when methanol or GTA fixations are not appropriate.
Permeabilization
Permeabilization after fixation with aldehydes is required to allow large molecules, such as dyes or antibodies to cross the cellular membranes. Detergents such as Triton X-100 and Tween-20 are generally used. Permeabilization prior to fixation is sometimes used to decrease the cytosolic staining of some proteins while improving the visualization of subcellular structures. This pre-extraction step is beneficial for the staining of KT proteins but disrupts the structure of MTs.
Immunofluorescence
Immunofluorescence is one of the most informative techniques in cell biology. It allows imaging of the distribution of single molecular species solely based on the properties of fluorescence emission. It relies on the antigen-antibody highly specific binding to identify proteins within the cell. The primary antibody that binds the antigen against which the antibody was raised, binds to a secondary antibody conjugated with a fluorophore. Most of the fluorescence microscopes are equipped with 4 different filters, so that different channels can be used to identify different proteins. Besides immunolabelling, dyes with fluorescence emission after excitation, such as 4',6-diamidine-2'-phenylindole (DAPI) are used to stain different molecules and structures within the cell.
-
Preparation of poly-L-lysine coated coverslips
Prepare 0.01% poly-L-lysine solution in sterile water.
Incubate 22 mm x 22 mm glass coverslips in 0.01% poly-L-lysine for 5 min with gentle agitation.
Wash coverslips with sterile water for 5 min.
Allow to dry and UV-sterilise for 15 min.
-
Fixation with paraformaldehyde
Prepare PFA 4% by diluting commercial PFA 20% in cytoskeleton buffer with sucrose (CBS).
Discard cell medium and add 2 ml of PFA 4% for 10 min (keep cells at 37ºC to avoid temperature fluctuations).
Wash twice with CBS for 5 min.
Wash with PBS for 5 min.
-
Fixation with cold methanol
Prepare the plate for methanol fixation: Add 2 ml of ice-cold methanol per well and keep at -20ºC for at least 30 min.
Transfer coverslips to the cold methanol and incubate for 3 min at -20ºC (perform this step as gently as possible to prevent cell dislodgement from the coverslip).
Prepare the plate for rehydration: Add 3-4 ml of CBS to a new 6-well plate.
Transfer coverslips from the methanol fixation to the rehydration plate.
Wash twice in CBS for 5 min.
Wash in PBS for 5 min.
-
Protocol for immunofluorescence detection of detyrosinated and tyrosinated and Δ2 tubulins
Fix cells with cold methanol (protocol M3).
Wash twice with PBS containing 0.01% Triton (PBST) for 5 min.
Incubate cells in blocking solution (PBST containing 10% FBS and/or 1% bovine serum albumin) in a humidified chamber for 1 h at RT, to block non-specific binding of the antibodies.
Incubate with primary antibodies against tyr-tubulin, detyr-tubulin and/or Δ2-tubulin (Table 4) diluted in blocking solution in a humidified chamber for 1h at RT or overnight at 4°C.
Wash the cells 3x in PBST for 5 min.
Incubate cells with the fluorescent-conjugated secondary antibodies diluted in blocking solution in a humidified chamber for 1 h at RT in the dark (Table 4).
Wash 3x in PBST for 5 min.
Mount coverslips with a drop of mounting medium (Table 6).
Seal coverslips with nail polish to prevent drying and movement under the microscope.
Acquire images using an inverted fluorescence microscope (e.g. Zeiss AxioImager Z1, Table 5).
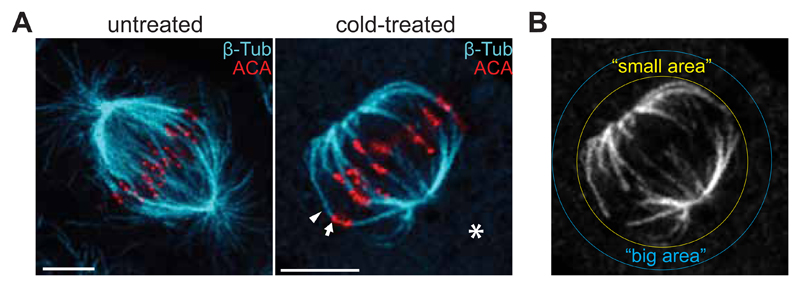
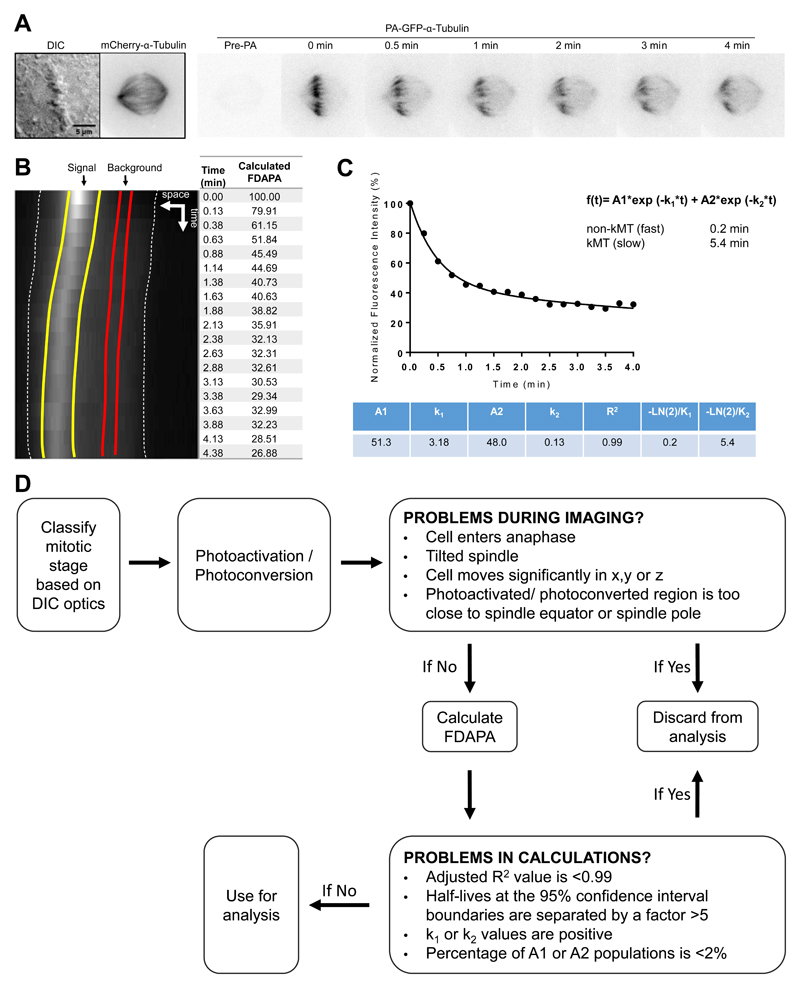
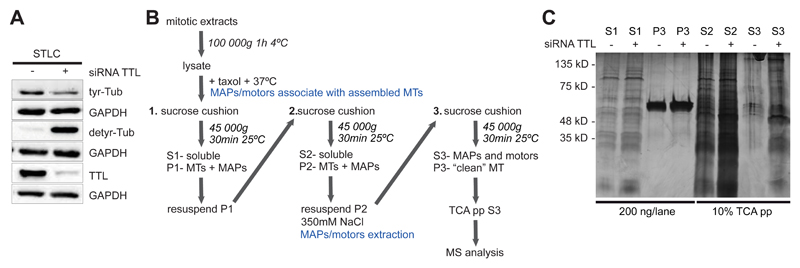
Section 2 - Analysis of microtubule dynamics in mitosis
Stability is a common hallmark for post-translationally modified MTs (Yu et al., 2015). In interphase cells, detyrosinated tubulin is enriched in stable microtubules. However, detyrosination was proposed not to cause stabilization, per se, but rather to be the consequence of MT stabilisation (Infante, Stein, Zhai, Borisy, & Gundersen, 2000; Webster, Wehland, Weber, & Borisy, 1990). Accordingly, cells with very long-lived microtubules additionally accumulate Δ2-tubulin (Paturle-Lafanechere et al., 1994). Conversely, detyrosinated-enriched MTs from TTL knockout MEFs were shown to be resistant to depolymerisation and to present a higher frequency of rescues than wild-type MEFs (Peris et al., 2009). Therefore, the relationship between tyrosination and microtubule stability is still controversial. Imaging single microtubule dynamics in mitosis is particularly difficult due to the high MT density of the mitotic spindle. Also challenging is the unambiguous differentiation between different populations of MTs (kMTs vs ipMTs, tyrosinated-enriched vs detyrosinated-enriched MTs).
In vivo, MT dynamics have been addressed using several imaging techniques including photobleaching, photoactivation, photoconversion and fluorescent speckle miscroscopy (Carminati & Stearns, 1997; T. J. Mitchison, 1989; Waterman-Storer, Desai, Bulinski, & Salmon, 1998). In these techniques fluorescent or non-fluorescent tags are fused to tubulin or tubulin binding proteins to label MTs in a uniform or localized manner. The emergence of fluorescence recovery after photobleaching (FRAP) in the 1970’s allowed quantitative measurements of the tubulin turnover at microtubule plus-ends in mitotic cells (Axelrod, Koppel, Schlessinger, Elson, & Webb, 1976). Later, with the development of photoactivatable (PA) and photoconvertible (PC) proteins (Fernandez-Suarez & Ting, 2008; Sample, Newman, & Zhang, 2009) it became possible to determine tubulin turnover on kMTs and ipMTs, as well as to measure poleward MT flux.
Semi-quantitative methods are still commonly used to assess the dynamic properties of spindle microtubules where stable and dynamic microtubules are identified on the basis of their sensitivity to cold, calcium or to microtubule depolymerizing agents, such as nocodazole (Akera et al., 2017; Brinkley & Cartwright, 1975; Rieder, 1981; Schliwa, Euteneuer, Bulinski, & Izant, 1981; Schulze & Kirschner, 1987). Cold treatment studies performed in the 80’s found no correlation between stability and tyrosination in mitosis and indicated that kMTs and ipMTs are both composed of tyr and detyr-tubulin (Gundersen & Bulinski, 1986). Based on the incorporation of hapten-labeled tubulin into MTs, the same authors later suggested that tyr-MTs turnover faster than detyr-MTs, but these studies were performed in the presence of endogenous tubulins and are therefore difficult to interpret (Webster, Gundersen, Bulinski, & Borisy, 1987).
We describe in this section methods for cold, calcium and nocodazole treatment. All are technically simple to perform but show low reproducibility between experiments. A protocol for turnover determination is also provided. Briefly, using a stable cell line expressing PA-GFP-α-tubulin, a region of the mitotic spindle (usually a transversal strip over one half of the mitotic spindle) is activated with a 405 nm laser and the fluorescence dissipation is measured over time. The resulting curve follows a double exponential, which reflects the presence of two MT populations with different dynamics: a population with fast fluorescence decay (likely corresponding to ipMTs), and a second population with a slower decay (likely kMTs). From the double exponential equation both the relative percentages of stable versus unstable MTs and their respective half-lives (t1/2) is extracted (Zhai, Kronebusch, & Borisy, 1995).
A. Cold-induced MT depolymerisation
This method is based on the differential resistance of the spindle MT subpopulations to low temperatures (Brinkley & Cartwright, 1975). The gentle cooling of cells (to 0-4ºC) induces depolymerisation of cold-sensitive astral and ipMTs while cold-stable kMTs remain unaffected (Rieder, 1981). This simple protocol combined with immunofluorescence analysis using antibodies against different tubulin PTMs can be used to quickly evaluate MT stability in specific conditions.
Protocol for cold-treatment
Seed cells in sterile glass coverslips until they reach 80-90% confluence. Grow cells in complete growth medium at 37ºC, 5% CO2.
In a separate plate, add 3 ml per well of 4ºC cold-complete medium.
Quickly transfer the coverslips to the cold medium, mix gently and incubate on ice for 10 min.
Fix cells with cold methanol or with cold PFA (protocol Section 1, M3, M2). Use the latter only if methanol is incompatible with the subsequent immunofluorescence protocol.
Perform immunofluorescence protocol (Section 1, M4) using antibodies against α- and β-tubulin and/or against other centromeric markers (such as CENP-A or CREST) if visualization of the kMT interface is required (Fig. 6).
Acquire and analyse data as described in D.
Fig. 6.
A. Immunofluorescence of U2OS cells before and after cold-induced MT destabilization. B. Sum projection of the tubulin channel showing the mitotic spindle circumscribed by two ROIs (small area and big area) used to quantify the fluorescence intensity at the spindle region.
B. Calcium-induced MT depolymerisation
In vivo, increase of intracellular concentrations of calcium (nanomolar doses) during mitosis plays a role in the regulation of chromosome condensation, spindle formation and kMT stability (Phengchat, Takata, Uchiyama, & Fukui, 2017). Yet, in vitro, short-term treatment of mammalian cells with micromolar doses of calcium has long been used to inhibit tubulin assembly and to promote microtubule depolymerisation (Schliwa et al., 1981; Weisenberg & Deery, 1981). Calcium treatment is used, as an alternative to cold-treatment, to study MT stability by destabilizing ipMTs prior to fixation.
Protocol for calcium-treatment
Plate cells in glass coverslips until they reach 80-90% confluency. Grow cells in complete growth medium at 37ºC, 5% CO2.
Permeabilize cells for 3 min in 100 mM PIPES (pH 6.8), 1 mM MgCl2, 0.1 mM CaCl2, and 0.1% Triton X-100 (T. Mitchison, Evans, Schulze, & Kirschner, 1986).
Repeat A4-6.
C. Nocodazole-induced MT depolymerisation
A third alternative to study MT stability uses the tubulin-sequestering drug nocodazole. A nocodazole shock, i.e. the treatment of cells with relatively high concentrations of nocodazole for a short period of time, is used to promote rapid MT disassembly (Gayek & Ohi, 2014). In comparison with cold and calcium treatment this method has the advantage of being more specific for MTs and thereby interfering less with other cellular processes.
Protocol for nocodazole shock
Grow cells in glass coverslips in complete growth medium at 37ºC, 5% CO2.
Incubate cells in complete medium with 5 μM of nocodazole for 5 min.
Repeat A 4-6.
D. Data acquisition and analysis
Image fixed cells using a 60x or 100x objective on a fluorescence microscope (e.g. Zeiss AxioImager Z1). Collect series of z-planes covering the entire volume of the mitotic spindle using 200 nm step-size. Quantify the fluorescence intensity of α- or β-tubulin (corresponding to kMTs that remain stable at low temperatures or that are resistant to calcium or nocodazole induced depolymerisation).
Open the tubulin channel on FIJI (Schindelin et al., 2012) and execute the sum projection of the total number of Z-stack images. Fiji > Image > Stack > -Sum projection
Generate a circular region of interest (ROI) that includes the entire mitotic spindle. This ROI corresponds to the “small area” (Fig. 6B). Oval selection + shift (this will allow to draw a perfect circle) > press “T” to save the ROI in the ROI manager.
Draw around the “small area” a second bigger circle around the “small area”. This circle is defined as the “big area”. Save selection in the ROI manager by pressing “T”.
Determine the integrated fluorescence intensity and area of both ROIs. Select both ROIs and press “measure” at the ROI manager window.
Calculate background signal according to equation 1.
Calculate the corrected fluorescence intensity following equation 2.
Equation 1:
Background signal = (integrated fluorescence intensity of “big area” – integrated fluorescence intensity of “small area”)/(“big area” – “small area”).
Equation 2:
Corrected fluorescence intensity = integrated fluorescence intensity of “small area” – (background signal × “small area”).
E. Measuring MT dynamics through photoactivation or photoconversion
-
Preparation of cells
Seed cells stably expressing PA or PC-GFP-α-tubulin ≥24 h before the experiment in coverslips so that they are at 60-80% confluence on the day of the experiment (regular growth media should be used; both uncoated and poly-L-lysine coated coverslips can be used).
Immediately before transferring cells to the microscope, replace with fresh, phenol-red-free cell culture media with 10% FBS. Use CO2-independent media if microscope setup does not include CO2 control. (Optional: 5 μM MG-132 can be added to prevent mitotic exit without affecting MT dynamics (Kabeche & Compton, 2012, 2013; Orr, Talje, Liu, Kwok, & Compton, 2016).
-
Defining optical settings
Set temperature to 37ºC.
If microscope is equipped with a source of CO2, set CO2 levels to 5%.
Adjust 405 nm laser intensity and exposure times (intensity >90% and 250-500 ms exposures are optimal).
Adjust laser intensities for channel used for measuring PA or PC signal intensity (50-75% intensity and 25-50 ms exposure times are optimal).
Seven 1 μm separated z-planes centred at the middle of the mitotic spindle are captured every 10-15 sec for 4.5 min (a pre-PA/PC image should always be acquired as well).
-
Photoactivation / Photoconversion
Identify mitotic cells using DIC optics.
Acquire a DIC image to define the mitotic stage (i.e. prometaphase or metaphase) based on chromosome alignment.
Adjust focus (if using a fluorescent tag, make sure both spindle poles are in the same focal plane and define this plane as the center; avoid performing measurements on tilted spindles).
Define a thin stripe of 1 μm width that spans one half-spindle in an area close to the spindle equator.
Acquire a pre-PA/PC fluorescent image.
Perform PA/PC by pulsing the cell with near UV-irradiation (405 nm laser) at the defined region.
Upon PA/PC, acquire z-stack images every 10-15 sec for 4.5 min.
If treating cells with MG132-free media, confirm that the cell did not enter anaphase during image acquisition.
-
Calculating MT turnover rates
Align spindle poles horizontally and generate whole-spindle, sum-projected kymographs (sum projections generated using ImageJ and kymographs generated as previously described in (Pereira & Maiato, 2010).
Quantify fluorescent intensities for the PA/PC spindle region for each time-point manually (or using a custom-written routine in Matlab) and normalize intensities to the first time-point after PA/PC following background subtraction (background values obtained from quantifying the non-activated other half-spindle).
Correct values for photobleaching by normalizing to the values obtained from the quantification of fluorescence loss of whole-cell (including cytoplasm), sum projected images (i.e. each cell has its own bleaching constant). Alternatively, normalize to averages obtained from the quantification of fluorescence loss of defined spindle regions of cells treated with 1-5 μM Taxol to fully stabilize MTs (Orr et al., 2016). For each of these methods, do not use background-subtracted values, since these expose non-linear photobleaching kinetics that may be observed during the first 1-2 min of imaging.
To calculate MT turnover, fit the normalized intensity values at each time point (corrected for photobleaching) to a double exponential curve A1*exp(-k1*t) + A2*exp(-k2*t) using Matlab (Mathworks), in which t is time, A1 represents the less stable (ipMTs) population and A2 the more stable (kMT) population with decay rates of k1 and k2, respectively (avoid using cells that display an R squared value <0.99).
From these curves, obtain the rate constants and the percentage of MTs for the fast (typically interpreted as the fraction corresponding ipMTs) and the slow (typically interpreted as the fraction corresponding K-MTs) processes.
The half-life is calculated as ln2/k for each population of microtubules.
Use the Student’s two-tailed t-test to perform statistical analysis on the results and make sure to discriminate between prometaphase and metaphase cells (that have different MT dynamics) (Fig. 7).
Fig. 7.
Calculating MT turnover through photoactivation. A. DIC and time-lapse fluorescent images of a representative metaphase U2OS cell expressing PA-GFP-α-tubulin and mCherry-α-tubulin. The mitotic spindle is visualized by mCherry fluorescence. Fluorescent images are inverted for better visualization of the photoactivated GFP molecules. B. Sum-projected, whole-spindle kymograph generated to quantify the Fluorescence Dissipation After Photo-Activation (FDAPA). Dashed white lines indicate the spindle poles; yellow lines indicate the boundaries used to quantify the signal generated from PA; red lines indicate the boundaries used for determining background levels. C. Normalized fluorescence intensity. Once fitted as a double exponential curve, the values obtained allow for the calculation of the dynamics of fast and slow MT populations. D. Photoactivation troubleshooting flowchart used to determine the exclusion criteria for calculating MT turnover using PA.
Section 3 - Identification of microtubule-associated proteins (MAPs) and motors binding to (de)tyrosinated microtubules
The tubulin code proposes that different tubulin genes and/or PTMs confer microtubule diversity (Sirajuddin et al., 2014), regulate interactions of tubulin with specific MAPs and motors and mediate different MT functions in the cell. The functionally specialization of MTs is supported by studies showing that the presence or absence of the α-tubulin C-terminal Tyr is implicated in the regulation of several motor proteins and MAPs (Badin-Larcon et al., 2004; M. Barisic et al., 2015; Dunn et al., 2008; Konishi & Setou, 2009; Liao & Gundersen, 1998; Peris et al., 2006; Peris et al., 2009). In mitosis, (de)tyrosination is important for spindle position (Peris et al., 2006) and CENP-E dependent chromosome congression (Marin Barisic et al., 2015). In this section, we describe a protocol to identify MAPs and motors that bind to tyrosinated or detyrosinated taxol-stabilized microtubules isolated from mitotic cells (Fig. 8).
Fig. 8.
A. Western blot analysis of mitotic HeLa cells lysates after treatment with siRNA for TTL. B. Schematic diagram of the protocol used for the purification of MAPs and motors from mitotic extracts. Protocol adapted from (Sloboda, 2015). C. Silver staining analysis of fractions obtained following protocol B.
A. Cell culture
HeLa cells are cultured in DMEM supplemented with 10% FBS (complete growth medium) at 37ºC and 5% CO2.
B. Isolation of MAPs and motors from mitotic cells
-
Preparation of mitotic extracts
Grow HeLa cells in T75 flasks until they reach ~70% confluence.
Incubate with 5 μM of S-trityl-l-cysteine (STLC) for 14h. (STLC is a reversible tight binding inhibitor of the plus-end directed motor Eg5/kinesin-5 that is used to increase the proportion of mitotic cells (DeBonis et al., 2004). Treatment with STLC causes monopolar spindles, in which the chromosomes are arranged radially around the centrosome with the centromeres oriented towards the centre. By preventing mitotic spindle bipolarity cells are arrested in mitosis for several hours.)
Shake-off mitotic cells by gentle shaking or tapping the flask. (During mitosis cells round up and are less firmly attached to the flask, so that gentle shaking will detach mitotic cells.)
Pellet cells at 500 g for 5 min.
Wash once with PBS.
Resuspend the cell pellet (1 g) in 1.5 ml K-PIPES buffer (Table 6) supplemented with 1 mM Mg-ATP and protease inhibitors at 4ºC. Incubate on ice for 15 min.
Gentle sonicate and centrifuge at 100.000 g for 1 h at 4ºC.
In parallel, prepare mitotic extracts from 1 g of HeLa cells with enriched levels of detyrosinated tubulin by treatment with 50 nM siRNA against TTL for 72 h (Section 1C) previous to STLC incubation (STLC + siRNA TTL).
Perform western blot analysis to confirm depletion (see Section1 L2, Fig. 8A).
-
Isolation of microtubules with bound MAPs and motors
Transfer supernatant to a new tube; add 20 μM taxol and incubate for 20 min at 37ºC. (Taxol lowers the critical concentration for tubulin polymerization and promotes assembly of microtubules; Mg-ATP promotes the interaction between MTs and motors).
Pellet assembled MTs over 10% sucrose cushion at 45.000 g for 30 min at 25ºC.
Remove supernatant and wash the tube gently with 400 μl with K-PIPES buffer pre-warmed to RT. Save a small aliquot from the supernatant (S1) for later analysis.
Remove cushion and rinse tube walls with 400 μl K-PIPES buffer, being careful not to disturb the pellet.
Resuspend the pellet in 200 μl K-PIPES buffer supplemented with 10 μM taxol pre-warmed to RT.
Pellet MTs over a 10% sucrose cushion at 45.000 g for 30 min at 25ºC. Save a small aliquot from the supernatant (S2) for subsequent analysis.
-
Extraction of MAPs and motors from microtubules
Resuspend the pellet in 100 μl of K-PIPES buffer supplemented with 10 μM taxol and 350 mM NaCl.
Pellet MTs over a 10% sucrose cushion at 45.000 g for 30 min at 25ºC. Save a small aliquot from the supernatant (S3) and from the pellet (P3) for subsequent analysis.
-
Analysis of protein fractions
Determine protein concentration of S1, S2 and S3 using the Bradford assay.
Resuspend the MT pellet P3 in 100 μl of ice-cold K-PIPES buffer supplemented with 10 mM CaCl2 and incubate on ice for 30 min. (Addition of high concentration of Ca2+ induces MT depolymerisation). Determine tubulin concentration by OD280 nm (extinction coefficient of tubulin = 115.000M-1cm-1).
Denature S1, S2, S3 and P3 protein samples in Laemmli sample buffer at 95ºC for 5-10 min.
Separate 50-200 ng of each sample by 10 % (v/v) SDS-PAGE gel electrophoresis.
Fix gel twice in 100 ml 30% ethanol, 10% acetic acid.
Sensitize gel in 0.001% (w/v) Na2S2O3, 30% ethanol, 0.1 M sodium acetate pH 6.8
Wash 3x in water for 10 min.
Stain gel with 0.01% (w/v) AgNO3, 30% ethanol, 0.25% (v/v) CH2O. Incubate until the desired intensity is achieved.
Rinse in water to remove loose silver ions.
Develop gel in 2.5% (w/v) Na2CO3, 0.5% (v/v) CH2O.
Stop the staining reaction by adding 100 ml of 1% (v/v) acetic acid. (Fig. 8C).
-
Precipitation of proteins from supernatants containing MAPs and motors
Add 1 volume of 100% (w/v) trichloroacetic acid to 4 volumes of supernatant S2 and S3.
Incubate 15 min at 4ºC.
Centrifuge at 20.000 g for 30 min at 4ºC.
Remove the supernatant and wash the protein pellet twice with 200 μl cold-acetone.
Centrifuge at 20.000 g for 10 min at 4ºC.
Dry the pellets and identify MAPs and motors by mass spectrometry.
Conclusions and outlook
Over the last decade, the study of knockout animal models for tubulin modifying enzymes, the correlation of abnormally modified MTs with cancer and the myriad of neurodegenerative diseases associated with the disruption of tubulin genes, leave no doubt for the importance of the tubulin code in animal physiology. Most of the functional dissection of the tubulin PTMs and tubulin isotypes have been provided by in vitro systems. However, the relation between specific tubulin modifications and cellular functions can only be achieved using in vivo models. Yet, the lack of spatiotemporal control of the plethora of modifications occurring simultaneously in the cell has been a great limitation. Less is known about the impact of the tubulin code in mitosis. So far, it is known that tubulin PTMs generate spatial cues that guide distinct mitotic motors along spindle MTs. However, how tubulin PTMs modulate the intrinsic properties of spindle MTs and how they contribute to the assembly and dynamics of the mitotic spindle remains unknown. Here, we shed light into the methodologies used in our laboratory to study tubulin PTMs and their role in mitosis. We describe overexpression and CRISPR/Cas9 mediated loss-of-function approaches to modify the normal distribution and composition of PTMs within the mitotic spindle. We also discuss ways of studying the impact of PTMs on the dynamics of different MT populations. Lastly, we present a protocol for the identification of MT-interacting proteins that are capable of “reading” tubulin PTMs, detyrosinatio/tyrosination in particular. Alone or in combination, these methodologies have the potential to help us to characterize the mechanisms behind spindle MT organization and function during mitosis. In the future, it will be important to establish cell biology approaches sensitive enough to identify subtle alterations in MT behaviour. The recent identification of the tubulin carboxypeptidases (TPCs) and the development of more potent and selective inhibitors will be critical to explore the tyrosination/detyrosination cycle and to better understand the impact of these modifications in mitosis.
Acknowledgements
The authors would like to thank Joppe Nieuwenhuis and Thijn Brummelkamp for sharing results and reagents prior to publication. Work in the laboratory of H.M. is supported by CODECHECK grant from the European Research Council and FLAD Life Science 2020.
References
- Aiken J, Sept D, Costanzo M, Boone C, Cooper JA, Moore JK. Genome-wide analysis reveals novel and discrete functions for tubulin carboxy-terminal tails. Curr Biol. 2014;24(12):1295–1303. doi: 10.1016/j.cub.2014.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aillaud C, Bosc C, Peris L, Bosson A, Heemeryck P, Van Dijk J, et al. Moutin MJ. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science. 2017;358(6369):1448–1453. doi: 10.1126/science.aao4165. [DOI] [PubMed] [Google Scholar]
- Aillaud C, Bosc C, Saoudi Y, Denarier E, Peris L, Sago L, et al. Moutin MJ. Evidence for new C-terminally truncated variants of alpha- and beta-tubulins. Mol Biol Cell. 2016;27(4):640–653. doi: 10.1091/mbc.E15-03-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, et al. Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467(7312):218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, et al. Lampson MA. Spindle asymmetry drives non-Mendelian chromosome segregation. Science. 2017;358(6363):668–672. doi: 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos L, Klug A. Arrangement of subunits in flagellar microtubules. J Cell Sci. 1974;14(3):523–549. doi: 10.1242/jcs.14.3.523. [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, et al. Zuercher WJ. The promise and peril of chemical probes. Nat Chem Biol. 2015;11(8):536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16(9):1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badin-Larcon AC, Boscheron C, Soleilhac JM, Piel M, Mann C, Denarier E, et al. Job D. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc Natl Acad Sci U S A. 2004;101(15):5577–5582. doi: 10.1073/pnas.0307917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M, Aguiar P, Geley S, Maiato H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat Cell Biol. 2014;16(12):1249–1256. doi: 10.1038/ncb3060. [DOI] [PubMed] [Google Scholar]
- Barisic M, Maiato H. The Tubulin Code: A Navigation System for Chromosomes during Mitosis. Trends Cell Biol. 2016;26(10):766–775. doi: 10.1016/j.tcb.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M, Silva e Sousa R, Tripathy SK, Magiera MM, Zaytsev AV, Pereira AL, et al. Maiato H. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science. 2015;348(6236):799–803. doi: 10.1126/science.aaa5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic Marin, Silva e Sousa Ricardo, Tripathy Suvranta K, Magiera Maria M, Zaytsev Anatoly V, Pereira Ana L, et al. Maiato Helder. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science. 2015;348(6236):799–803. doi: 10.1126/science.aaa5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra HS, Rodriguez JA, Arce CA, Caputto R. A soluble preparation from rat brain that incorporates into its own proteins (14 C)arginine by a ribonuclease-sensitive system and (14 C)tyrosine by a ribonuclease-insensitive system. J Neurochem. 1973;20(1):97–108. doi: 10.1111/j.1471-4159.1973.tb12108.x. [DOI] [PubMed] [Google Scholar]
- Berezniuk I, Vu HT, Lyons PJ, Sironi JJ, Xiao H, Burd B, et al. Fricker LD. Cytosolic carboxypeptidase 1 is involved in processing alpha- and beta-tubulin. J Biol Chem. 2012;287(9):6503–6517. doi: 10.1074/jbc.M111.309138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39(3):223–232. doi: 10.1002/(sici)1097-0169(1998)39:3<223::aid-cm5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Bocca C, Gabriel L, Bozzo F, Miglietta A. A sesquiterpene lactone, costunolide, interacts with microtubule protein and inhibits the growth of MCF-7 cells. Chem Biol Interact. 2004;147(1):79–86. doi: 10.1016/j.cbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Bork PM, Schmitz ML, Kuhnt M, Escher C, Heinrich M. Sesquiterpene lactone containing Mexican Indian medicinal plants and pure sesquiterpene lactones as potent inhibitors of transcription factor NF-kappaB. FEBS Lett. 1997;402(1):85–90. doi: 10.1016/s0014-5793(96)01502-5. [DOI] [PubMed] [Google Scholar]
- Bre MH, Redeker V, Quibell M, Darmanaden-Delorme J, Bressac C, Cosson J, et al. Levilliers N. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci. 1996;109(Pt 4):727–738. doi: 10.1242/jcs.109.4.727. [DOI] [PubMed] [Google Scholar]
- Bre MH, Redeker V, Vinh J, Rossier J, Levilliers N. Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in paramecium. Mol Biol Cell. 1998;9(9):2655–2665. doi: 10.1091/mbc.9.9.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR, Cartwright J., Jr Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann N Y Acad Sci. 1975;253:428–439. doi: 10.1111/j.1749-6632.1975.tb19218.x. [DOI] [PubMed] [Google Scholar]
- Callen AM, Adoutte A, Andrew JM, Baroin-Tourancheau A, Bre MH, Ruiz PC, et al. Isolation and characterization of libraries of monoclonal antibodies directed against various forms of tubulin in Paramecium. Biol Cell. 1994;81(2):95–119. doi: 10.1016/s0248-4900(94)80002-2. [DOI] [PubMed] [Google Scholar]
- Carminati Janet L, Stearns Tim. Microtubules Orient the Mitotic Spindle in Yeast through Dynein-dependent Interactions with the Cell Cortex. J Cell Biol. 1997;138(3):629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron JM, Vega LR, Fleming J, Bishop R, Solomon F. Single site alpha-tubulin mutation affects astral microtubules and nuclear positioning during anaphase in Saccharomyces cerevisiae: possible role for palmitoylation of alpha-tubulin. Mol Biol Cell. 2001;12(9):2672–2687. doi: 10.1091/mbc.12.9.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L. Accessory protein regulation of microtubule dynamics throughout the cell cycle. Curr Opin Cell Biol. 1999;11(1):134–141. doi: 10.1016/s0955-0674(99)80017-9. [DOI] [PubMed] [Google Scholar]
- Chopra V, Quinti L, Kim J, Vollor L, Narayanan KL, Edgerly C, et al. Ferrante RJ. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington’s disease mouse models. Cell reports. 2012;2(6):1492–1497. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CW, Hou F, Zhang J, Phu L, Loktev AV, Kirkpatrick DS, et al. Zou H. A novel acetylation of beta-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol Biol Cell. 2011;22(4):448–456. doi: 10.1091/mbc.E10-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, et al. Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther. 2004;3(9):1079–1090. [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dumontet C, Duran GE, Steger KA, Murphy GL, Sussman HH, Sikic BI. Differential expression of tubulin isotypes during the cell cycle. Cell Motil Cytoskeleton. 1996;35(1):49–58. doi: 10.1002/(sici)1097-0169(1996)35:1<49::aid-cm4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, Peckham M. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121(Pt 7):1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- Eipper BA. Rat brain microtubule protein: purification and determination of covalently bound phosphate and carbohydrate. Proc Natl Acad Sci U S A. 1972;69(8):2283–2287. doi: 10.1073/pnas.69.8.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, et al. Wehland J. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc Natl Acad Sci U S A. 2005;102(22):7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K, Wehland J, Plessmann U, Dodemont H, Gerke V, Weber K. Characterization of the tubulin-tyrosine ligase. J Cell Biol. 1993;120(3):725–732. doi: 10.1083/jcb.120.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L, Mitchison T, Kirschner M. Influence of the centrosome on the structure of nucleated microtubules. J Cell Biol. 1985;100(4):1185–1191. doi: 10.1083/jcb.100.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9(12):929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- Fonrose X, Ausseil F, Soleilhac E, Masson V, David B, Pouny I, et al. Lafanechere L. Parthenolide inhibits tubulin carboxypeptidase activity. Cancer Res. 2007;67(7):3371–3378. doi: 10.1158/0008-5472.CAN-06-3732. [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell. 2006;17(3):1041–1050. doi: 10.1091/mbc.E05-07-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar S, Bodakuntla S, Natarajan K, Janke C. The tubulin code at a glance. J Cell Sci. 2017;130(8):1347–1353. doi: 10.1242/jcs.199471. [DOI] [PubMed] [Google Scholar]
- Gayek AS, Ohi R. Kinetochore-microtubule stability governs the metaphase requirement for Eg5. Mol Biol Cell. 2014;25(13):2051–2060. doi: 10.1091/mbc.E14-03-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens G, Gundersen GG, Nuydens R, Cornelissen F, Bulinski JC, DeBrabander M. Ultrastructural colocalization of tyrosinated and detyrosinated alpha-tubulin in interphase and mitotic cells. J Cell Biol. 1986;103(5):1883–1893. doi: 10.1083/jcb.103.5.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghantous A, Saikali M, Rau T, Gali-Muhtasib H, Schneider-Stock R, Darwiche N. Inhibition of tumor promotion by parthenolide: epigenetic modulation of p21. Cancer Prev Res (Phila) 2012;5(11):1298–1309. doi: 10.1158/1940-6207.CAPR-12-0230. [DOI] [PubMed] [Google Scholar]
- Goodger NM, Gannon J, Hunt T, Morgan PR. The localization of p34cdc2 in the cells of normal, hyperplastic, and malignant epithelial and lymphoid tissues of the oral cavity. J Pathol. 1996;178(4):422–428. doi: 10.1002/(SICI)1096-9896(199604)178:4<422::AID-PATH497>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Gopal YN, Arora TS, Van Dyke MW. Parthenolide specifically depletes histone deacetylase 1 protein and induces cell death through ataxia telangiectasia mutated. Chem Biol. 2007;14(7):813–823. doi: 10.1016/j.chembiol.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Gozes I, Barnstable CJ. Monoclonal antibodies that recognize discrete forms of tubulin. Proc Natl Acad Sci U S A. 1982;79(8):2579–2583. doi: 10.1073/pnas.79.8.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Bulinski JC. Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J Cell Biol. 1986;102(3):1118–1126. doi: 10.1083/jcb.102.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38(3):779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol. 1987;105(1):251–264. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak ME, Rodriguez JA, Barra HS, Caputto R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977;73(2):147–150. doi: 10.1016/0014-5793(77)80968-x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hinshaw SM, Harrison SC. Kinetochore Function from the Bottom Up. Trends Cell Biol. 2018;28(1):22–33. doi: 10.1016/j.tcb.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Mukai M, Tsuchida J, Heier RL, Macgregor GR, Setou M. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem. 2006;281(41):30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Sato S, Nakamura K, Ostrowski LE, Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc Natl Acad Sci U S A. 2010;107(23):10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante AS, Stein MS, Zhai Y, Borisy GG, Gundersen GG. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113(Pt 22):3907–3919. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206(4):461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12(12):773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, et al. Edde B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308(5729):1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Ji S, Kang JG, Park SY, Lee J, Oh YJ, Cho JW. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids. 2011;40(3):809–818. doi: 10.1007/s00726-010-0698-9. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Cleveland DW. Differential utilization of beta-tubulin isotypes in differentiating neurites. J Cell Biol. 1989;109(2):663–673. doi: 10.1083/jcb.109.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhilahti EM, Peltonen S, Peltonen J. Class III beta-tubulin is a component of the mitotic spindle in multiple cell types. J Histochem Cytochem. 2008;56(12):1113–1119. doi: 10.1369/jhc.2008.952002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol. 2012;22(7):638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502(7469):110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Sorrentino S, Perlas E, Bolasco G, Martinez C, Heppenstall PA. alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat Commun. 2013;4 doi: 10.1038/ncomms2962. 1962. [DOI] [PubMed] [Google Scholar]
- Kaul N, Soppina V, Verhey KJ. Effects of alpha-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophys J. 2014;106(12):2636–2643. doi: 10.1016/j.bpj.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93(3):576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, et al. Setou M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs) J Biol Chem. 2010;285(30):22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12(5):559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. Detection of acetylated alpha-tubulin by specific antibodies. Methods Enzymol. 1991;196:264–274. doi: 10.1016/0076-6879(91)96025-m. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Gu W, Cowan NJ. Free intermingling of mammalian beta-tubulin isotypes among functionally distinct microtubules. Cell. 1987;49(4):539–548. doi: 10.1016/0092-8674(87)90456-9. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24(2):473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- Li C, Wang J, Hao J, Dong B, Li Y, Zhu X, et al. Wang GQ. Reduced cytosolic carboxypeptidase 6 (CCP6) level leads to accumulation of serum polyglutamylated DNAJC7 protein: A potential biomarker for renal cell carcinoma early detection. Oncotarget. 2016;7(16):22385–22396. doi: 10.18632/oncotarget.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Gundersen GG. Kinesin is a candidate for cross-bridging microtubules and intermediate filaments. Selective binding of kinesin to detyrosinated tubulin and vimentin. J Biol Chem. 1998;273(16):9797–9803. doi: 10.1074/jbc.273.16.9797. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu S, Xie Z, Pavlovicz RE, Wu J, Chen P, et al. Chan KK. Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J Pharmacol Exp Ther. 2009;329(2):505–514. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Garnham CP, Roll-Mecak A, Tanner ME. Phosphinic acid-based inhibitors of tubulin polyglutamylases. Bioorg Med Chem Lett. 2013;23(15):4408–4412. doi: 10.1016/j.bmcl.2013.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the "O-GlcNAc code". Sci STKE. 2005;2005(312):re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Luduena RF. A hypothesis on the origin and evolution of tubulin. Int Rev Cell Mol Biol. 2013;302:41–185. doi: 10.1016/B978-0-12-407699-0.00002-9. [DOI] [PubMed] [Google Scholar]
- Luduena RF. Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol. 1998;178:207–275. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- Magiera MM, Janke C. Investigating tubulin posttranslational modifications with specific antibodies. Methods Cell Biol. 2013;115:247–267. doi: 10.1016/b978-0-12-407757-7.00016-5. [DOI] [PubMed] [Google Scholar]
- Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J Cell Biol. 1986;103(2):571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Vale RD, Sirajuddin M. Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 2016;35(11):1175–1185. doi: 10.15252/embj.201593071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K, Fok JY, Mangala LS. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci. 2006;11:173–185. doi: 10.2741/1789. [DOI] [PubMed] [Google Scholar]
- Miglietta A, Bozzo F, Gabriel L, Bocca C. Microtubule-interfering activity of parthenolide. Chem Biol Interact. 2004;149(2–3):165–173. doi: 10.1016/j.cbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45(4):515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol. 1989;109(2):637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis J, Adamopoulos A, Bleijerveld OB, Mazouzi A, Stickel E, Celie P, et al. Brummelkamp TR. Vasohibins encode tubulin detyrosinating activity. Science. 2017;358(6369):1453–1456. doi: 10.1126/science.aao5676. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96(1):79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Orr B, Talje L, Liu Z, Kwok BH, Compton DA. Adaptive Resistance to an Inhibitor of Chromosomal Instability in Human Cancer Cells. Cell Rep. 2016;17(7):1755–1763. doi: 10.1016/j.celrep.2016.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, et al. Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317(5837):516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- Ozols J, Caron JM. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol Biol Cell. 1997;8(4):637–645. doi: 10.1091/mbc.8.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamula MC, Ti SC, Kapoor TM. The structured core of human beta tubulin confers isotype-specific polymerization properties. J Cell Biol. 2016;213(4):425–433. doi: 10.1083/jcb.201603050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IY, Chowdhury P, Tripathi DN, Powell RT, Dere R, Terzo EA, et al. Walker CL. Methylated alpha-tubulin antibodies recognize a new microtubule modification on mitotic microtubules. MAbs. 2016;8(8):1590–1597. doi: 10.1080/19420862.2016.1228505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, et al. Walker CL. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell. 2016;166(4):950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N, Austin-Tse CA, Liu Y, Vasilyev A, Drummond IA. Cytoplasmic carboxypeptidase 5 regulates tubulin glutamylation and zebrafish cilia formation and function. Mol Biol Cell. 2014;25(12):1836–1844. doi: 10.1091/mbc.E13-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paturle-Lafanechere L, Edde B, Denoulet P, Van Dorsselaer A, Mazarguil H, Le Caer JP, et al. Job D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry. 1991;30(43):10523–10528. doi: 10.1021/bi00107a022. [DOI] [PubMed] [Google Scholar]
- Paturle-Lafanechere L, Manier M, Trigault N, Pirollet F, Mazarguil H, Job D. Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J Cell Sci. 1994;107(Pt 6):1529–1543. doi: 10.1242/jcs.107.6.1529. [DOI] [PubMed] [Google Scholar]
- Pereira AJ, Maiato H. Improved kymography tools and its applications to mitosis. Methods. 2010;51(2):214–219. doi: 10.1016/j.ymeth.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, et al. Job D. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174(6):839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechere L, Brocard J, Moore AT, Kozielski F, et al. Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185(7):1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person F, Wilczak W, Hube-Magg C, Burdelski C, Moller-Koop C, Simon R, et al. Jacobsen F. Prevalence of betaIII-tubulin (TUBB3) expression in human normal tissues and cancers. Tumour Biol. 2017;39(10) doi: 10.1177/1010428317712166. 1010428317712166. [DOI] [PubMed] [Google Scholar]
- Peters JD, Furlong MT, Asai DJ, Harrison ML, Geahlen RL. Syk, activated by cross-linking the B-cell antigen receptor, localizes to the cytosol where it interacts with and phosphorylates alpha-tubulin on tyrosine. J Biol Chem. 1996;271(9):4755–4762. doi: 10.1074/jbc.271.9.4755. [DOI] [PubMed] [Google Scholar]
- Phengchat R, Takata H, Uchiyama S, Fukui K. Calcium depletion destabilises kinetochore fibres by the removal of CENP-F from the kinetochore. Sci Rep. 2017;7(1):7335. doi: 10.1038/s41598-017-07777-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101(6):2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104(2):289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, et al. Steinmetz MO. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol. 2013;200(3):259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff EC, Hoyle HD, Popodi EM, Turner FR. Axoneme beta-tubulin sequence determines attachment of outer dynein arms. Curr Biol. 2008;18(12):911–914. doi: 10.1016/j.cub.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkumar A, Jong BY, Ori-McKenney KM. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev Dyn. 2018;247(1):138–155. doi: 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84(1):145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Rogowski K, Juge F, van Dijk J, Wloga D, Strub JM, Levilliers N, et al. Janke C. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137(6):1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, et al. Janke C. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143(4):564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A. Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin Cell Dev Biol. 2015;37:11–19. doi: 10.1016/j.semcdb.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4(1):56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger AH, Rudiger M, Wehland J, Weber K. Monoclonal antibody ID5: epitope characterization and minimal requirements for the recognition of polyglutamylated alpha- and beta-tubulin. Eur J Cell Biol. 1999;78(1):15–20. doi: 10.1016/s0171-9335(99)80003-x. [DOI] [PubMed] [Google Scholar]
- Rumpf T, Schiedel M, Karaman B, Roessler C, North BJ, Lehotzky A, et al. Jung M. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat Commun. 2015;6 doi: 10.1038/ncomms7263. 6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample V, Newman RH, Zhang J. The structure and function of fluorescent proteins. Chem Soc Rev. 2009;38(10):2852–2864. doi: 10.1039/b913033k. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, Euteneuer U, Bulinski JC, Izant JG. Calcium lability of cytoplasmic microtubules and its modulation by microtubule-associated proteins. Proc Natl Acad Sci U S A. 1981;78(2):1037–1041. doi: 10.1073/pnas.78.2.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder HC, Wehland J, Weber K. Purification of brain tubulin-tyrosine ligase by biochemical and immunological methods. J Cell Biol. 1985;100(1):276–281. doi: 10.1083/jcb.100.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, Kirschner M. Dynamic and stable populations of microtubules in cells. J Cell Biol. 1987;104(2):277–288. doi: 10.1083/jcb.104.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergouniotis PI, Chakarova C, Murphy C, Becker M, Lenassi E, Arno G, et al. Plagnol V. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014;94(5):760–769. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158(7):1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A. 2010;107(50):21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedle B, Garcia-Pineres AJ, Murillo R, Schulte-Monting J, Castro V, Rungeler P, et al. Merfort I. Quantitative structure-activity relationship of sesquiterpene lactones as inhibitors of the transcription factor NF-kappaB. J Med Chem. 2004;47(24):6042–6054. doi: 10.1021/jm049937r. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16(4):335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloboda RD. Isolation of microtubules and microtubule-associated proteins using Paclitaxel. Cold Spring Harb Protoc. 2015;2015(1) doi: 10.1101/pdb.prot081190. pdb prot081190. [DOI] [PubMed] [Google Scholar]
- Song Y, Brady ST. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 2015;25(3):125–136. doi: 10.1016/j.tcb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Kirkpatrick LL, Schilling AB, Helseth DL, Chabot N, Keillor JW, et al. Brady ST. Transglutaminase and polyamination of tubulin: posttranslational modification for stabilizing axonal microtubules. Neuron. 2013;78(1):109–123. doi: 10.1016/j.neuron.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V, Herbstman JF, Skiniotis G, Verhey KJ. Luminal localization of alpha-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS One. 2012;7(10):e48204. doi: 10.1371/journal.pone.0048204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Chakrabarti O. Mahogunin-mediated alpha-tubulin ubiquitination via noncanonical K6 linkage regulates microtubule stability and mitotic spindle orientation. Cell Death Dis. 2014;5:e1064. doi: 10.1038/cddis.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Cleveland DW. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc Natl Acad Sci U S A. 1986;83(12):4327–4331. doi: 10.1073/pnas.83.12.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26(3):437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Walczak Claire E, Heald Rebecca. Mechanisms of Mitotic Spindle Assembly and Function. A Survey of Cell Biology. 2008:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8(22):1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, Borisy GG. Differential turnover of tyrosinated and detyrosinated microtubules. Proc Natl Acad Sci U S A. 1987;84(24):9040–9044. doi: 10.1073/pnas.84.24.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DR, Wehland J, Weber K, Borisy GG. Detyrosination of alpha tubulin does not stabilize microtubules in vivo. J Cell Biol. 1990;111(1):113–122. doi: 10.1083/jcb.111.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehland J, Weber K. Turnover of the carboxy-terminal tyrosine of alpha-tubulin and means of reaching elevated levels of detyrosination in living cells. J Cell Sci. 1987;88(Pt 2):185–203. doi: 10.1242/jcs.88.2.185. [DOI] [PubMed] [Google Scholar]
- Weisenberg RC, Deery WJ. The mechanism of calcium-induced microtubule disassembly. Biochem Biophys Res Commun. 1981;102(3):924–931. doi: 10.1016/0006-291x(81)91626-0. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3(12):739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan DR, Bell TD. Image artifacts in single molecule localization microscopy: why optimization of sample preparation protocols matters. Sci Rep. 2015;5 doi: 10.1038/srep07924. 7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple RA, Vitolo MI, Boggs AE, Charpentier MS, Thompson K, Martin SS. Parthenolide and costunolide reduce microtentacles and tumor cell attachment by selectively targeting detyrosinated tubulin independent from NF-kappaB inhibition. Breast Cancer Res. 2013;15(5):R83. doi: 10.1186/bcr3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Dave D, Meagley J, Rogowski K, Jerka-Dziadosz M, Gaertig J. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot Cell. 2010;9(1):184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Webster DM, Rogowski K, Bre MH, Levilliers N, Jerka-Dziadosz M, et al. Gaertig J. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16(6):867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279(44):45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Wolff A, de Nechaud B, Chillet D, Mazarguil H, Desbruyeres E, Audebert S, et al. Denoulet P. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol. 1992;59(2):425–432. [PubMed] [Google Scholar]
- Wu HY, Wei P, Morgan JI. Role of Cytosolic Carboxypeptidase 5 in Neuronal Survival and Spermatogenesis. Sci Rep. 2017;7 doi: 10.1038/srep41428. 41428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, et al. Fan Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol. 2016;17(4):369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28(8):868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Li C, Yang Z, Wang Y, Hao J, Wang L, et al. Fan Z. Cytosolic carboxypeptidase CCP6 is required for megakaryopoiesis by modulating Mad2 polyglutamylation. J Exp Med. 2014;211(12):2439–2454. doi: 10.1084/jem.20141123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265(28):17174–17179. [PubMed] [Google Scholar]
- Yu I, Garnham CP, Roll-Mecak A. Writing and Reading the Tubulin Code. J Biol Chem. 2015;290(28):17163–17172. doi: 10.1074/jbc.R115.637447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131(3):721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu RT, Zhang P, Zhang N, Yang CL, Yue LT, et al. Duan RS. Parthenolide inhibits the initiation of experimental autoimmune neuritis. J Neuroimmunol. 2017;305:154–161. doi: 10.1016/j.jneuroim.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, et al. Matthias P. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28(5):1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink S, Grosse L, Freikamp A, Banfer S, Muksch F, Jacob R. Tubulin detyrosination promotes monolayer formation and apical trafficking in epithelial cells. J Cell Sci. 2012;125(Pt 24):5998–6008. doi: 10.1242/jcs.109470. [DOI] [PubMed] [Google Scholar]