Abstract
Autism spectrum disorder (ASD) is thought to emerge during early cortical development. However, the exact developmental stages and associated molecular networks that prime disease propensity are elusive. To profile early neurodevelopmental alterations in ASD with macrocephaly, we monitored patient-derived induced pluripotent stem cells (iPSCs) throughout the recapitulation of cortical development. Our analysis revealed ASD-associated changes in the maturational sequence of early neuron development, involving temporal dysregulation of specific gene networks and morphological growth acceleration. The observed changes tracked back to a pathologically primed stage in neural stem cells (NSCs), reflected by altered chromatin accessibility. Concerted overrepresentation of network factors in control NSCs was sufficient to trigger ASD-like features, and circumventing the NSC stage by direct conversion of ASD iPSCs into induced neurons (iPSC-iNs) abolished ASD-associated phenotypes. Our findings identify heterochronic dynamics of a gene network that, while established earlier in development, contributes to subsequent neurodevelopmental aberrations in ASD.
Introduction
Autism spectrum disorder (ASD) is a highly heritable neurodevelopmental condition defined by deficits in social interaction and communication accompanied by restrictive, repetitive behaviors. Considerable genetic and phenotypic heterogeneity has complicated the efforts to find common biological substrates of the syndrome. Meanwhile, recent advances in systems biology approaches have caused a paradigm shift in the field of biomedical sciences from a single gene causation model to pathway perturbation models1,2. Implementing high confidence risk variants discovered in ASD cohorts into a data-driven framework of gene network analysis revealed a convergence on molecular processes that implicate specific perturbations during human fetal cortical development3,4. One complication is that many of the identified genes are involved at multiple time points and in different neural cell types, such as neural stem cells (NSCs), as well as in maturing neurons during human cortical development. Thus, a major challenge in the field is to determine the critical developmental periods, their associated cellular states and the molecular networks that might provide a basis for the emergence of common pathological phenotypes during ASD development.
Here we have performed a time series analysis to follow human idiopathic ASD patient-specific induced pluripotent stem cells (iPSCs) during their early neuronal development to examine when and how the earliest ASD-specific molecular abnormalities and phenotypes arise. Time series transcriptome and cellular phenotype analyses in ASD NSCs and their progeny identified dysregulation of specific transcriptional networks that caused aberrant neuronal maturation of ASD cortical neurons. Temporal reconstruction and network-based screening revealed that coherent neurodevelopmental gene modules were accelerated in ASD and appeared to be enriched with several ASD risk genes. Surprisingly, we found that dysregulation of these modules arose even before the neuronal stage, as module-specific signatures were already present during the NSC stage. Genome-wide comparisons of chromatin accessibility revealed a prevalent pathologically primed state in ASD NSCs that precedes the occurrence of aberrant gene network dynamics at the consecutive neuronal stage. Concerted transgenic expression of subsets of module genes was used to mimic ASD-associated gene network-related changes in healthy controls and was sufficient to recapitulate the occurrence of aberrant neurodevelopmental growth trajectories at the subsequent neuronal stage. We further demonstrated that skipping the NSC stage by direct iPSC-to-neuron conversion (iPSC-iN) also circumvented the critical period for the ASD signature to establish itself, thereby preventing neuronal ASD phenotypes from manifesting. Our study provides a thorough temporal analysis of the neurodevelopmental regulatory gene networks that become impaired in ASD and shows that some of these changes are triggered during early developmental periods.
Results
Time series-based transcriptome analysis
All iPSC lines used in this study were obtained from a cohort of eight idiopathic ASD patients with early developmental brain overgrowth and five unaffected individuals as controls. Children with such signs of early brain enlargement have been shown to be part of a group of patients with high probability of receiving diagnosis5–8. Primary skin fibroblasts were harvested and reprogrammed to iPSCs using retroviral vectors expressing the four Yamanaka factors9 (Extended Data Fig. 1a and Supplementary Table 1). Our previous study showed that neuronal cultures derived from these patients bear an impaired neuronal network connectivity at later, more mature neuronal stages. Other iPSC studies have also reported aberrant features of neuronal connectivity as a phenotypic hallmark for various forms of autism9–11. However, whether dynamic neurodevelopmental aberrations exist that precede the appearance of such advanced phenotypic disease states remains unresolved. Integrative bioinformatics approaches suggest that ASD pathologies may arise from combined deficiencies during distinct phases of human cortical development3,4. To model these early periods, we differentiated the iPSCs into NSCs and then further into functional excitatory MAP2+ neurons (Extended Data Fig. 1b-d) using a reported protocol9,12. All patient-derived cells adopted a cortical fate (Extended Data Fig. 1e-h and 2a and b), and global transcriptional comparison to the BrainSpan dataset13 revealed high correspondence to the dorsal telencephalon (cerebral cortex with dorsolateral, medial and orbitofrontal cortical areas) during early human fetal cortical development (Extended Data Fig. 2c-e). Furthermore, continuous electrophysiological recordings showed a consecutive transition through stages of functional maturation, including the firing of multiple evoked action potentials after 22 days of differentiation (Extended Data Fig. 2f).
One major aim of this study was to model the temporal dimension of shifting biological processes by treating the disorder as an evolving, dynamic system. We reasoned that a tight recapitulation of the consecutive developmental stages of early neuronal development in ASD and control cases would allow us to reconstruct time-related maturational processes. We first used fluorescence-activated cell sorting (FACS) to purify a homogeneous population of NSCs based on the expression of the cell-surface markers CD184+/CD271−/CD44−/CD24−/CD15+ 14. To trace ASD and control neurons over time, we performed a series of retroviral lineage-tracing experiments to isolate the progenies of dividing NSCs using a retroviral vector expressing a membrane-tagged enhanced green fluorescent protein (eGFP) (CAG::LckN-eGFP). Because differentiating neurons express PSA-NCAM on the cell surface (Extended Data Fig. 3a and b), we established a FACS-based protocol for purification of defined subpopulations of retrovirally labeled eGFP+/PSA-NCAM+ double-positive neurons after 2, 4, 7 and 14 days of differentiation (Fig. 1a), which allowed us to capture several consecutive maturational stages during lineage progression. We next performed RNA sequencing (RNA-seq) analysis over the time course of neuronal differentiation and observed dramatic gradual changes in gene expression (Extended Data Fig. 3c). As expected, neuronal genes such as MAP2, MAPT and DCX, voltage-gated channels, synaptic proteins, and neurotransmitter receptors such as GRIN3a and GRIA3 became gradually upregulated, whereas cell cycle-associated genes were consecutively downregulated (Extended Data Fig. 3c, left panel and Supplementary Table 2). Gene Ontology (GO) term analysis of differentially upregulated genes revealed highly significant enrichment for biological processes that control neuron generation and function (Extended Data Fig. 3c, right panel). A highly significant enrichment for factors that control cell cycle and proliferation was present in the list of the most strongly decreased genes (Extended Data Fig. 3c, right panel and Supplementary Table 3), indicating efficient purification of maturing neurons from defined time points during neuronal differentiation.
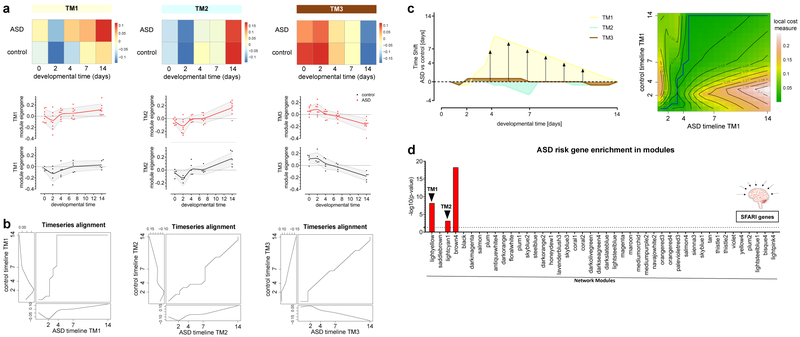
Figure 1: Gene network analysis of time series-based RNA sequencing identifies dynamic developmental gene networks.
a, Schematic of FACS-based purification of NSCs and defined subpopulations of eGFP+/PSA-NCAM+ neurons over the time course of in vitro differentiation. b, WGCNA cluster dendrogram of all 65 time series samples groups genes into 44 distinct modules (top row). Bottom row shows strong differential expression relationships for developmental time (days of in vitro differentiation post lineage tracing). c, Heatmap showing relationships between modules, disease and developmental time. Relationship assignments are based on signed Pearson correlation coefficients indicating positive (red) or negative (blue) correlation of modules with trait and developmental time; n=65 time series samples; ASD: n=8 independent cell lines at 5 time points, control: n=5 independent cell lines at 5 time points. d, Time-based significance measure reveals three modules highly enriched for genes that change with developmental time. This module significance measure was defined as the absolute Pearson correlation between time (days of in vitro differentiation post lineage tracing) and module eigengenes; TM1: n=1,530 genes, TM2: n=294 genes, TM3: n=401 genes. Student p-values for associations with time were adjusted for multiple comparisons by using the false discovery rate (FDR) approach.
Identification of aberrant gene network dynamics in ASD
We next sought to decipher the dynamics of regulatory gene networks to identify deviations in time-critical aspects during ASD development. To cluster genes with similar expression patterns in an unbiased manner, we applied weighted gene co-expression network analysis (WGCNA)15,16 to all 65 time series samples (five control and eight ASD lines at five time points) and identified 44 modules of co-expressed genes (Fig. 1b, Supplementary Table 4 and Methods). Here, modules with high gene numbers mostly corresponded to changes within developmental time (Fig. 1b), whereas cell stage-specific events appeared to be a more subtle transcriptional feature reflected in a number of smaller modules (Fig. 1c and Extended Data Fig. 4a). As developmental progression is based on a sequence of events that changes over time, we were particularly interested in those modules that showed enrichment for genes holding dynamic temporal profiles. Defining a time-based module significance measure (Methods) allowed us to identify three modules that showed a highly significant correlation with the progression of neurodevelopmental timing, referred to as Time Modules (TM) 1, 2 and 3 (Fig. 1d). Alternative network construction strategies (supplementary methods) further demonstrated the robustness in identifying all three TMs, suggesting a high preservation within our gene network (Extended Data Fig. 4d and e).
To identify temporal deviations within the developmental sequence between ASD and control neurons, we compared temporal gene expression dynamics on the level of the identified gene modules. To overcome the lack of sensitivity to distortion in the time axis when using the Euclidean distance metric, we applied dynamic time warping (DTW) algorithms to compare temporal module dynamics between ASD and control neurons. DTW is a variety of time series alignment algorithms designed to measure similarities between two temporal sequences that may vary in speed17,18. Module Eigengenes (ME) (first principal component of a given module) were used as a representative of the gene expression profiles in the three TMs (Fig. 2a). DTW was then applied to compare the module-based dynamics between ASD and control neurons, revealing distinct deviations between the two time series in TM1 (Fig. 2b left). The two other TMs showed only subtle initial timeline deviations (Fig. 2b middle and right panels, Extended Data Fig. 4b and c), indicating that the temporal sequences in TM2 and TM3 were almost similar between ASD and control neurons. The temporal progression of TM1, however, appeared markedly accelerated in ASD neurons, with earlier time points in ASD neurons corresponding to later time points in control neurons (Fig. 2c), highlighting the emergence of an intrinsic molecular heterochronicity.
Figure 2: Gene network analysis and dynamic time warping identify ASD-specific heterochronicity in a time-related module.
a, Module eigengene (ME) dynamics of the three notable time-related modules averaged across developmental time. Upper panel: Heatmap showing average MEs across time. Blue, low expression: Red, high expression. Lower panel: Plot of TM1, TM2 and TM3 showing trajectories of module eigengenes across time. b, Three-way plot of the alignment for TM1, TM2 and TM3 between control (reference) and ASD (query). This plot allows display of alignments and places the query time series (ASD) in the small lower panel and the reference time series (control) vertically on the left; the large inner panel holds the warping curve. Left: Earlier time points in the query index (ASD time series) of TM1 correspond to later time points in the reference index (control time series), indicating an accelerated progression of TM1. c, Left: DTW alignment curves superimposed for all three TMs, showing the magnitude and onset of time shift. Right: Density plot showing alignment of TM1 between control (reference) and ASD (query). The average cost per step is displayed as a density distribution with contours superimposed. d, Module-specific enrichment for ASD-related risk factors based on the SFARI gene list (see Supplementary Table 5).
To further evaluate the identified developmental gene networks, we computed the relative enrichment for disease-related risk factors in each module based on a set of 859 genes (Supplementary Table 5) evaluated by the knowledgebase SFARI19. Remarkably, TM1 and TM2 showed specific enrichment for ASD risk genes, with TM1 and TM2 featuring highly significant enrichment scores as compared to other modules (Fig. 2d). As further evidence for this confined enrichment, gene expression changes identified in post-mortem ASD cortex studies20,21 and during human cortical development3, as well as genes with de novo variants associated with intellectual disability (ID)22, showed significant enrichment in both TMs (Extended Data Fig. 4f). This confined enrichment in TMs suggests an involvement of autism risk factors in early phases of cortical neuron development, consistent with other recent studies3,4, and relates our findings to relevant ASD-associated changes that are present at later stages.
Characterization of the accelerated ASD TM1
GO term analysis of the genes assigned to TM1 revealed highly significant enrichment for biological processes that control specific aspects of nervous system development (P = 2.8×10−28) such as neuron differentiation (P = 2.7×10−18), the development of neuronal projections (P = 3.4×10−15), cell morphogenesis (P = 3.1×10−15) and synaptic signaling (P = 2.6×10−20) (Fig. 3a, Supplementary Table 6). As TM1 increases expression with time, the large neurodevelopmental signature of this module suggests that these genes reflect gradually increasing neuronal differentiation. Highly connected genes (‘hub genes’) such as BRSK1, CACNB1, RAB3A and GPC2, together with several other interconnected hub genes, are involved in neuron generation and synapse organization (Fig. 3b). To further characterize TM1, we calculated individual gene significance (GS) measures for the top 60 hub genes by correlating their expression with developmental time and condition. As expression of these genes increases with developmental time, GS measures reach highly positive values at later, more mature neuronal stages (Fig. 3c). When comparing the hub gene trajectories of ASD and control neurons, a pronounced shift of GS values markedly emerged at four days after differentiation and continued to be prominent at later time points, although differences were already present at the NSC stage (Fig. 3c), suggesting an ASD-specific priming of gene expression already in pre-neuronal cells. Thus, given the large neurodevelopmental signature of TM1, the presence of such a marked time shift suggests aberrant neurodevelopmental gene network dynamics that could directly disturb the maturational sequence in developing ASD neurons.
Figure 3: Characterization of the heterochronic TM1.
a, GO term analysis of the genes assigned to TM1 reveals specific neurodevelopmental signature. Point sizes correspond to the number of genes assigned to each category (Supplementary Table 6). b, Top gene-gene connections for TM1 are shown, highlighting their involvement in selected biological categories and ASD risk. Different point sizes correspond to the number of time points that show increased expression levels. c, Gene Significance (GS) values of the top 60 hub genes across developmental time and between conditions. GS measures are represented as mean ± s.d.; n=60 genes.
Maturing ASD neurons show aberrant neurodevelopmental growth dynamics
We next explored the extent to which certain morphological aspects of ASD neuron development deviate from the ‘normal’ trajectory, in addition to the marked time shift observed at the transcriptional level. Therefore, a series of structural and morphometric analyses was performed on defined subpopulations of eGFP+/DCX+/PSA-NCAM+ neurons using a retrovirus-based lineage tracing approach (Fig. 4a and b, Extended Data Fig. 5a). Strikingly, and consistent with the shift observed in TM1, systematic reconstruction of individual morphological growth trajectories revealed a significant developmental acceleration in differentiating neurons from ASD patients as compared to control neurons (Fig. 4c and d and Extended Data Fig. 5b). In line with this finding, ASD neurons consistently featured branching patterns that were significantly more complex than those of control neurons (Fig. 4e and Extended Data Fig. 5c). Thus, given the marked time shift in TM1, these data highlight the presence of aberrant maturational dynamics, pertaining to the growth patterns of neuronal projections in developing ASD neurons. As the development of neuronal projections is a time-critical process, we further asked whether this growth acceleration might be accompanied by differences in the states of neuronal maturity. To query intrinsic maturational properties, we designed a retroviral tracing system expressing eGFP under the control of the human Dcx promoter (hDcx::eGFP; Fig. 4f and Extended Data Fig. 5d). Flow cytometry-based reporter analysis revealed a premature activity state of the introduced hDcx promoter in differentiating neurons from ASD patients when compared to the trajectory of control neurons (Fig. 4g and h and Extended Data Fig. 5e), indicating differences in the initiation and the dynamics of the neuronal program. Additional measures of maturational features furthermore revealed significant deviations in the temporal progression of important developmental pathways (Extended Data Fig. 6a-d).
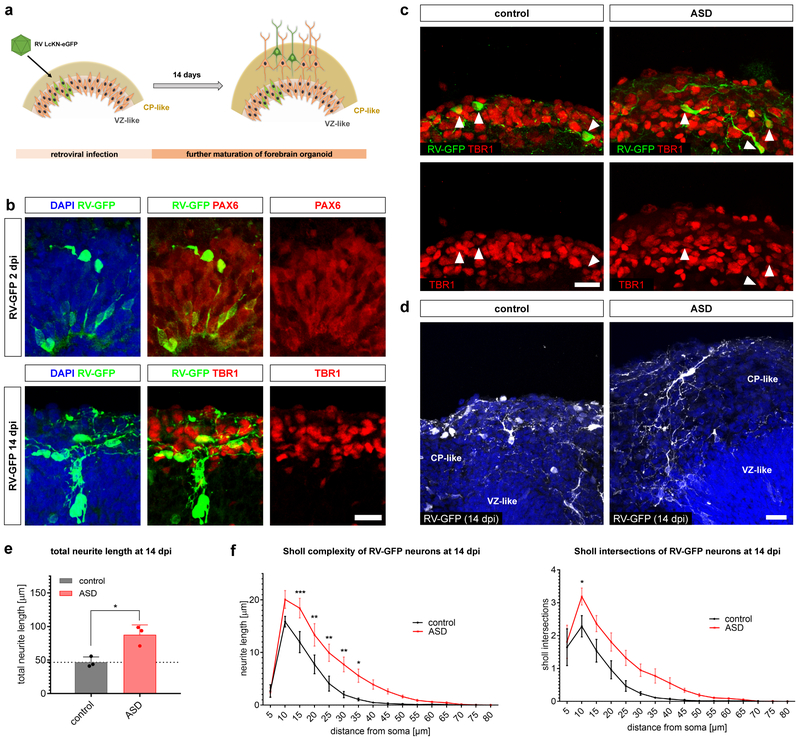
Figure 4: Aberrant neurodevelopmental growth dynamics in maturing ASD neurons.
a, Schematic showing experimental design for lineage tracing-based morphometric assessment. b, Representative confocal images of developing neurons seven days post retroviral infection (dpi) expressing green fluorescent protein (GFP) and DCX and showing immunoreactivity for axonal filaments (Smi312). Scale bars 50 μm. Immunohistochemistry was performed in all 13 patient lines and 2 cell culture replicates per time point with similar results (also see Extended Data Fig. 5a). c, Reconstructions of retrovirus (RV)-labeled neurons derived from ASD and unaffected control individuals over time. Scale bar 50 μm. Experiments were performed in all 13 patient lines and 2 cell culture replicates per time point. Results are shown in d and e. d, ASD neurons showed accelerated growth properties and were significantly longer at 4, 7 and 14 dpi (two-way ANOVA with Sidak correction, 4 dpi: ***P=0.0002, 7 dpi: **P=0.006, 14 dpi: ****P<0.0001). Values show means ± s.d.; ASD (n=8; 320 technical tracing replicates total), control (n=5; 244 technical tracing replicates total); n refers to biologically independent patient lines. e, Sholl analysis of neurite length from ASD and control neurons at 14 dpi. Total sholl neurite length complexity was larger in the ASD group as compared with controls (two-way ANOVA with Sidak correction, *P<0.05, ***P<0.001, ****P<0.0001). Values show means ± s.e.m.; ASD (n=8; 320 technical tracing replicates total), control (n=5; 244 technical tracing replicates total); n refers to biologically independent patient lines. f, Schematic showing retroviral reporter construct designed to assess hDCX promoter activity and the flow cytometry-based gating strategy for the assessment of hDCX:GFP mean fluorescence intensities (MFIs). g, Representative histograms showing MFIs of the hDCX:GFP reporter through the time course of differentiation. Experiments were repeated in all 13 patient lines at 5 different time points; numerical values represent percentages. h, Relative hDCX promoter activities in differentiating neurons. ASD neurons showed premature activation of the inserted hDCX promoter with significantly elevated levels at the NSC stage and at 16 dpi (two-way ANOVA with Sidak correction, NSC stage: **P=0.0076, 16 dpi: **P=0.0059). The data are normalized to the maximum activity over time within each line. Values represent means ± s.e.m.; ASD (n=8), control (n=5); n refers to biologically independent patient lines. Also see Extended Data Figure 5.
To further explore the developmental implications of this intrinsic neurodevelopmental heterochronicity, we used two different protocols to generate a three-dimensional (3D) model of cortical development23,24. As monolayer cultures prove limited to reveal spatial effects on early cortical development, we used a subset of our cohort (Supplementary Table 7) to assess the formation of the cortical plate (CP) in situ. While no major shift in specification of TBR1+ neurons was present in the monolayer system, cerebral organoids as well as TBR1+ cortical regions of forebrain organoids revealed an increased thickness of the CP in the ASD condition (Extended Data Fig. 6f-j), consistent with the hypothesis of increased neuronal numbers and proliferation6,9. These results further emphasize the advantage of 3D models to dissect spatial developmental aberrations.
To exclude the possibility of cell heterogeneity and to study the aberrant cell-intrinsic trajectories, we labeled dividing radial-glia-like cells (RGLs) in the ventricular-like zones of four-week-old forebrain organoids using a GFP-expressing retrovirus (Fig. 5a). This approach allowed us to follow the progenies of labeled RGLs (Fig. 5b, top panel) as they migrated and integrated into the evolving CP (Fig. 5b, bottom panel). In line with our findings from the NSC monolayer system, assessment of developing neurons revealed no difference in overall cell-fate commitment, as both ASD and control RGLs gave rise mostly to TBR1+ deep-layer neurons (Fig. 5c and Extended Data Fig. 6k and l). However, morphological assessment of early-born cortical neurons showed marked differences between the control and ASD groups (Fig. 5d). Quantitative assessment of their early branching patterns showed that ASD neurons harbored aberrantly complex neurite structures at 14 days post infection (dpi) (Fig. 5e-f). Together with our findings from the monolayer system and in line with the observed transcriptional dysregulation, these data highlight the existence of intrinsic ASD-associated perturbations in the sequence of cortical neuron development.
Figure 5: Aberrant neurodevelopmental growth dynamics of early-born neurons in a three dimensional model of cortical development.
a, Schematic showing experimental design for lineage tracing-based morphometric assessment in forebrain organoids. b, Representative confocal images of retrovirally labeled radial glia-like (RGL) cells (upper panel). Over time, retrovirally labeled cells migrate into cortical plate (CP)-like regions and differentiate into neurons that integrate into the evolving cortical layer (bottom panel). Scale bars 20 μm. Experiments were repeated in 6 patient lines and 3 different organoid batches with similar results. c, Representative confocal images of retrovirally labeled neurons at 14 dpi in CP-like regions expressing TBR1. Arrowheads indicate GFP+ neurons co-expressing TBR1 (also see Extended Data Fig. 6k and l). Scale bar 20 μm. Immunohistochemistry was repeated in 6 patient lines and 3 different organoid batches with similar results. d, Representative confocal images of retrovirally labeled neurons in CP-like regions at 14 dpi between ASD and control forebrain organoids. Scale bar 20 μm. Immunohistochemistry was repeated in 6 patient lines and 3 different organoid batches with similar results. e, Developing cortical neurons of ASD patients showed significantly accelerated growth properties at 14 dpi (*P=0.0128, unpaired two-tailed Student’s t-test); Values represent mean ± s.e.m.; ASD: n=3 (9 organoids with 122 tracing replicates total); control: n=3 (9 organoids with 142 tracing replicates total); n refers to biologically independent patient lines. f, Sholl analysis of subtle branching patterns at 14 dpi between ASD and control cortical neurons. Sholl neurite length complexity (left) and intersections (right) were larger in the ASD group as compared with controls (two-way ANOVA with Sidak correction, *P<0.05, **P<0.001, ***P<0.001). Values represent mean ± s.e.m.; ASD: n=3 (12 organoids with 122 tracing replicates total); control: n=3 (12 organoids with 142 tracing replicates total); n refers to biologically independent patient lines.
TM1-specific priming causes aberrant neurodevelopmental growth dynamics
As the timing of neurogenesis is thought to be largely controlled by cell-intrinsic mechanisms encoded within the cortical lineage25,26, we asked whether the disruption of the neotenic neurodevelopmental gene program in ASD neurons could have been primed during an earlier pre-neuronal stage in the timeline. Differential gene expression analysis indicated early aberrant gene expression dynamics in the ASD group with an upregulation of twice as many TM1 genes within the first four days of differentiation (Supplementary Table 8), pointing to the possible involvement of priming during preceding developmental stages. To assess the stage-specific co-expression of genes in TM1, we excluded network genes weakly interconnected with TM1 (kME<0.7) and calculated the significance of gene-wise correlations to the preceding NSC stage using a signed GS measure (Extended Data Fig. 7a). As expected, a high number of TM1-specific genes featured highly significant negative correlations with the NSC stage in the control group (FDR adjusted P < 0.05; Extended Data Fig. 7b), indicating that these genes were downregulated in comparison to all other stages. Remarkably, almost 80% of these downregulated genes (189 of 237 negatively correlated TM1 genes) lacked significant negative GS values in ASD NSCs (Extended Data Fig. 7c and Supplementary Table 9). This difference between groups was also reflected in a marked shift in GS values for these 189 TM1 genes (Extended Data Fig. 7c), suggesting a subtle primed state of the neuronal network program in ASD NSCs. Among the top negatively correlated TM1 genes, the F-box protein 2 (FBXO2) gene is involved in regulating the postmitotic neuronal state and the initiation of synapse formation27,28, whereas other genes encoding for the small GTP-binding protein RASD2 or the transmembrane protein ISLR2 have been implicated in schizophrenia29 or have important functions in neurite extension and regulating forebrain connectivity30,31 (Extended Data Fig. 7d-h). We tested whether an early, dynamically aberrant overrepresentation of TM1-related genes is sufficient to induce ASD-specific morphological and maturational changes. To assess their functional implication, we performed controlled overexpression of selected TM1 genes using FBXO2 alone as well as in combination with additional network genes (Extended Data Fig. 7i and j). As expected, priming the expression of FBXO2 at the NSC stage caused a dynamically aberrant initiation of the subsequent neurodevelopmental program, as measured by neurite outgrowth after four days of differentiation (Extended Data Fig. 7k and l). This effect was even more pronounced when combining FBXO2 with two additional TM1-specific network genes (Extended Data Fig. 7l and m), pointing to synergistic gene network-wide effects. These findings highlight the possibility that the ASD-specific neurodevelopmental dysregulation is controlled by genes identified in TM1 and could arise even before the neuronal stage through an aberrant priming of network-related genes.
Circumventing the pathologically primed NSC stage restores ASD neuron development
To evaluate whether these ASD-associated changes are indeed primed during early periods of neural development, we sought to optimize a technology that could be used to force iPSCs to circumvent early NSC-like stages. We thus generated neurons directly from iPSCs by optimizing a Neurogenin2 (Ngn2)-based induced neuron (iN) protocol32. IPSCs were lentivirally transduced to express rtTA and a 2A-peptide linked transcript coding for Ngn2 and eGFP (N2AG), resulting in transgenic but silent and expandable N2AG iPSCs (Fig. 6a and Extended Data Fig. 8a). Following iPSC-iN conversion, iPSCs underwent marked morphological changes by establishing neuronal projections as early as within four days (Fig. 6b and Extended Data Fig. 8b). As seen with neurons derived from NSCs through directed differentiation, forced expression of Ngn2 also resulted in the generation of functional neurons with high correspondence to the dorsal telencephalon during human fetal cortical development (Fig. 6c, Extended Data Fig. 8c-i and Supplementary Table 10). However, stringent assessment of early trans-differentiation events revealed that iPSC-iN conversion did not require proliferation (Fig. 6d) and bypassed the emergence of NSC-specific signatures (Extended Data Fig. 9a-e), indicating that this iPSC-iN method indeed generated neurons that had never been in an NSC-like state.
Figure 6: Transgenic overexpression of proneural transcription factors in iPSCs (iPSC-iNs) circumvents proliferative NSC-like stages.
a, Schematic of lentiviral system for inducible overexpression of Ngn2-2A-eGFP (top). Direct conversion of iPSCs into induced neurons (iNs) by forced expression of an inducible Ngn2 transgene may bypass early NSC-like stages (bottom). b, Representative images of patient-derived iPSC-iNs at different stages in the course of neuronal differentiation. Scale bar 200 μm. Experiments were repeated in all 13 patient lines at 4 different time points. c, Immunocytochemical characterization of iPSC-iNs after two weeks of conversion. Scale bar represents 25 μm. Immunohistochemistry was performed in all 13 patient lines at least once with similar results d, BrdU-labeling experiment with unselected iPSC-iNs during conversion (arrowheads highlight GFP-expressing iPSC-iNs). Scale bar 50 μm. Values represent mean ± s.d.; n=4 independent control cell lines. e, Representative reconstructions of patient-derived iPSC-iNs at the stages indicated in b. Scale bar 100 μm. Experiments were repeated in all 13 patient lines at 4 different time points. Also see Extended Data Figures 8 and 9.
We reasoned that applying this technology in the context of ASD could circumvent the critical NSC-associated priming period for the ASD signature to establish itself. A series of in-depth analyses focusing on maturation and structural development throughout early neuronal growth periods were performed to re-evaluate the differences between control and ASD iPSC-iNs during early neuronal differentiation. Both ASD and control iPSC-iNs underwent marked morphological changes over the time course of differentiation (Fig. 6e). However, quantitative assessment of neuronal differentiation revealed no difference between iPSC-iNs from ASD patients and those generated from unaffected control individuals, as measured by neuronal commitment and neurite branching patterns (Fig. 7a and b and Extended Data Fig. 9f and g). Further, in sharp contrast to the ASD-specific differences in the growth of neuronal projections observed in NSC-derived neurons (Fig. 4), in-depth morphometric analysis of iPSC-iNs derived from the same iPSC clones did not show this aberrant trajectory (Fig. 7c). To compare the developmental gene network dynamics during iPSC-iN maturation with those identified in NSC-derived neurons, we performed time-series RNA-seq analysis at different stages during iPSC-iN conversion (Extended Data Fig. 10a). We applied WGCNA to identify modules of co-expressed genes, similar to the approach that was used for NSC-derived neurons (Fig. 7d, Extended Data Fig. 10b and Supplementary Table 11). Comparing the networks of NSC-derived neurons (Fig. 1) with those of iPSC-iNs revealed a significant conservation of TM1 (Extended Data Fig. 10c-e). In sharp contrast to the NSC-differentiation data, DTW revealed no trajectorial difference in the progression of the TM1-equivalent gene network between control and ASD in iPSC-iNs (Fig. 7e and Extended Data Fig. 10f and g). Furthermore, the prominent ASD-associated transcriptional shift among the top 60 TM1 hub genes, which was prevalent in NSC-derived neurons, appeared to be abolished in IPSC-iNs (Fig. 7f).
Figure 7: Bypassing the NSC state restores early neurodevelopmental aberrations.
a, A significant enrichment of PSA-NCAM+ cells was present in ASD NSC-derived neurons (NSC-N) at 4 dpi as compared to their respective controls (**P=0.0016, Mann-Whitney U-test), whereas no difference was observed in iPSC-iNs (P=0.805, Mann-Whitney U-test, not significant). Box plots show median (center line), mean (‘+’) and interquartile range (IQR), with whiskers representing the minimum and maximum of data points; ASD: n=8 biologically independent patient lines (42 technical replicates total), control n=5 biologically independent patient lines (22 technical replicates total). b, Sholl analysis of subtle branching patterns in patient-derived iPSC-iNs at 14 days post conversion. Values represent mean ± s.e.m.; ASD: n=8 biologically independent patient lines, control: n=5 biologically independent patient lines. c, Left: Total neurite growth assessment of converting iPSC-iNs. Values represent mean ± s.d.; ASD: n=8 biologically independent patient lines (482 tracing replicates total), control: n=5 biologically independent patient lines (264 tracing replicates total). Right: ASD NSC-Ns had significantly longer neurites at 14 dpi as compared with their respective controls (control NSC-N: 421.8 ± 14.14 μm, ASD NSC-N: 552.32 ± 24.86 μm, **P=0.0062, Mann-Whitney U-test), whereas no difference was observed in iPSC-iPSC-iNs at 14 days post conversion (control iPSC-INs: 442.39 ± 23.93 μm, ASD iPSC-iNs: 439.07 ± 14.51 μm, P=0.9433, Mann-Whitney U-test, not significant). Values represent mean ± s.e.m.; ASD: n=8 biologically independent patient lines, control: n=5 biologically independent patient lines. d, WGCNA cluster dendrogram of all 52 iPSC-iN time series samples groups genes into distinct modules (top row). The middle row shows strong differential expression relationships for developmental time (days of iPSC-iN conversion). TM1 genes (bottom row; Fig. 2) show high module preservation in the iPSC-iN gene network (also see Extended Data Fig. 10c-e). e, Density plot showing the temporal ME alignment for the TM1-equivalent iPSC-iN module blue between control (reference) and ASD (query) during iPSC-iN maturation. The average cost per step is displayed as a density distribution with contours superimposed. f, GS values of the top 60 TM1 hub genes for ASD and control individuals in NSC-derived neurons (left) or when bypassing the NSC stage (iPSC-iN technology, right) after 14 days. Violin plots show median (center line), IQR (box), 95% confidence interval and the kernel probability density at different values; n=60 genes. Also see Extended Data Figures 8, 9 and 10.
Thus, circumventing early NSC-like stages by using the iPSC-iN technology rescued ASD-specific phenotypes and restored aberrant transcriptional signatures in ASD neurons, suggesting a pathological priming period during early neurodevelopmental periods.
Aberrant gene network dynamics at early neuronal stages are associated with changes in chromatin accessibility at preceding NSC stages
Given the contribution of early NSC stages in the subsequent ASD-associated phenotype, we sought to further characterize possible mechanisms underlying this pathologically primed state. We reasoned that the assay for transposase-accessible chromatin using sequencing (ATAC-seq) in NSCs would allow us to obtain a depiction of preceding changes in the chromatin landscape. We thus isolated NSCs by FACS using the same surface markers as described above before subjecting them to ATAC-seq. All size-selected ATAC-seq libraries showed the characteristic fragment length distribution (Extended Data Fig. 11a), with the majority of reads falling in the nucleosome-free region (NFR) (Extended Data Fig. 11b). We assessed differential accessibility (DA) between the two conditions and discovered 833 peaks with greater accessibility in ASD NSCs and 760 peaks with decreased accessibility in the ASD group as compared to control NSCs (Fig. 8a and b, FDR adjusted P < 0.05). Hierarchical clustering based on the correlation of accessibility of DA sites separated the samples into two groups based on their condition (Fig. 8c). Similarly, the identified DA peaks could be used to separate the sample groups along the first and second principal component of variation, explaining 75% and 16% of variation respectively (Extended Data Fig. 11d).
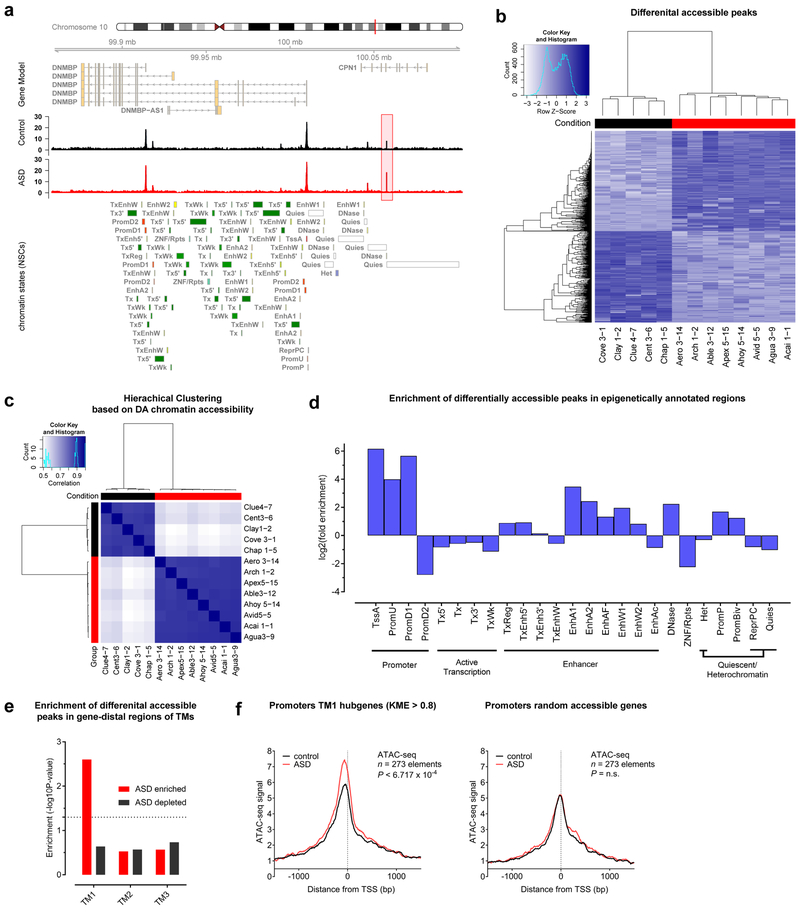
Figure 8: Aberrant gene network dynamics at early neuronal stages are associated with changes in chromatin accessibility at preceding NSC stages.
a, Coverage maps of normalized ATAC-seq signals from ASD and control NSCs showing a differentially accessible (DA) peak (highlighted in red) near the DNMBP gene on chromosome 10. Group-wise sample coverages are displayed and annotated chromatin states are based on the ENCODE 25-state model (see Supplementary Methods). b, Binding affinity heatmap showing normalized accessibilities for DA peaks. c, Correlation heatmap showing hierarchical clustering based on DA peaks (Pearson correlations of peak scores); ASD: n=8 biologically independent patient lines, control: n=5 biologically independent patient lines. d, Log2 fold enrichment of DA peaks in epigenetically annotated regions of the genome (ENCODE 25-state model) shows significant enrichment in promoter and enhancer regions (GAT randomization test53; n=1,593 DA elements; see Supplementary Methods). e, Significance calculations for the enrichment of DA peaks within gene-distal regions (50kbp windows) of TM-associated genes (GAT randomization test53; ASD enriched: n=721 gene-distal DA peaks, ASD depleted: n=755 gene-distal DA peaks; see Supplementary Methods). f, Metagene profiles of normalized ATAC-seq signals at promoters around ±1kb from the transcription start sited (TSS) in NSCs from ASD patients (red) or controls (black). Left: Metagene profiles of promoter regions from TM1 hub genes (kME > 0.8) show higher accessibility in ASD NSCs (P=6.717×10−4, Hotelling’s t test; n=273 elements). Right: Metagene profiles of promoter regions from randomly accessible background genes (P=0.7761, Hotelling’s t test, n.s. – not significant; n=273 elements). Numbers of plotted elements (genes) are similar.
DA peaks are significantly enriched in previously annotated chromatin states33, including promoters and enhancers, but depleted in regions containing quiescent and actively transcribed regions of the genome (Fig. 8d). Previous studies have shown that regulatory modules as far as 50kbp and more from the transcriptional start site have important contributions to the prediction of a genes’ transcriptional activity34,35. Examining 50kbp windows around genes assigned to any of the three TMs uncovered that DA peaks with increased accessibility in ASD NSCs fall significantly more often in TM1 gene-distal regions than would be expected by chance, whereas gene-distal regions of TM2 and TM3 did not show any enrichment for DA peaks (Fig. 8e and Extended Data Fig. 11 e and f). Remarkably, metagene analysis of gene promoters from TM1 hub genes (kME>0.8) showed a significantly increased amount of nucleosomal depletion in ASD NSCs compared to control NSCs (273 elements, P < 6.717×10−4); this depletion was not present in accessible promoters of randomly selected background genes (Fig. 8f and Extended Data Fig. 11g). These findings highlight the involvement of altered epigenetic chromatin remodeling processes and suggest that, at least in part, increased accessibility in promoter and gene-distal regions may contribute to the aberrant enhancement of transcription that occurs at the subsequent neuronal stage. In line with the finding that the manifestation of the aberrant TM1-specific signature establishes itself during early NSC-like stages, these data provide further evidence for the existence of early pathologically primed states that may trigger later occurring aberrations during ASD development.
Discussion
Our work demonstrates how ASD-associated neurodevelopmental aberrations that disturb the maturational sequence of cortical neuron development are triggered by a pathological priming of gene regulatory networks that evolves during early neural development. These findings contribute to the emerging picture of convergent molecular pathologies in ASD3,4,20 and identify NSC stages as critical developmental periods that lay the groundwork for disease propensity.
Despite etiological heterogeneity, recent large-scale gene expression studies on ASD postmortem brain regions have shown consistent transcriptome changes that evolve during the first decade of brain development20,36,37,21. These ASD-associated changes are thought to represent the consequences of an ongoing process that is triggered largely by genetic and epigenetic factors early in development 1,21,22,38,39. However, little was known about whether this plethora of heritable genetic components might converge on a coherent gene network during specific periods of fetal cortical development. Our current study is based on a carefully selected cohort of macro-cephalic ASD patients representing a highly replicated and significant phenotype within the spectrum8,40,41. Previous ASD postmortem studies have pointed to pathologies in neuronal and micro-columnar organization, spine growth and dendrite morphology6,42–47, suggesting aberrant processes during early brain development. Considering that live neurons from patients are inaccessible and most postmortem tissues are limited to later stages of disease pathogenesis, iPSC-based model systems provide an unprecedented tool to study the progression of early developmental events. Here, we have developed a time series approach that allowed us to follow human idiopathic ASD patient-specific iPSCs during their early cortico-neuronal development. Transcriptome-wide screening revealed an ASD-associated temporal dysregulation within a specific neurodevelopmental gene module that caused marked changes in the maturational sequence of cortical neuron development, including morphological growth acceleration and heterochronic initiation of the neuronal program. It is likely that a disruption of the neotenic gene expression program during cortical neuron development has wide-ranging consequences for a timely, coordinated establishment of interconnected neuronal networks. Experiments in rodents have shown that altering the trajectory of early postnatal cortical development can lead to structural and behavioral features of autism48,49. In line with this finding, human iPSC-based studies have confirmed that neuronal connectivity is affected in various forms of autism9–11. However, while cortical neurons are thought to be particularly susceptible for certain ASD risk genes3,4, it is currently not well understood whether common ASD-associated changes that evolve at the neuronal stage are the consequence of an ongoing process that is primed early in development.
Here we show that the observed neurodevelopmental heterochronicity in ASD neurons originates from functional neurodevelopmental gene networks that become pathologically primed during the pre-neuronal NSC stage. By mimicking this ASD-specific signature in control NSCs using controlled overrepresentation of a subset of network genes, we show that pathological dysregulation in NSCs is sufficient to induce ASD-associated morphological and maturational changes at later neuronal stages. While we only tested a selected subset of network-related genes, it would be interesting to investigate how different combinations of network genes may contribute to the ASD-associated changes in a context- and subject-specific manner.
Trans-differentiation of somatic cells such as fibroblasts into induced neurons (iNs) has been used as an alternative technology to directed differentiation50. More recently this technology has been expanded to enable rapid generation of functional neurons by transgenic overexpression of proneural transcription factors in iPSCs (iPSC-iNs)32. However, it is well established that forced expression of iN pioneer factors during fibroblast-based iN conversion does not require proliferation51. A recent study furthermore demonstrated that the direct programming path diverges into a novel transitional state, bypassing intermediate progenitors, but converging onto the same final state52. In line with our findings, these data indicate that iPSC-iNs indeed generate neurons that have never been in a NSC-like state, thus making them a promising tool to evaluate the implication of early neurodevelopmental states in priming disease progression. Using this technology, we demonstrate that bypassing NSC-like stages by direct conversion of ASD iPSCs into postmitotic neurons prevents the observed neuronal ASD-associated phenotypes from manifesting. Our analysis suggests that some ASD-associated changes are likely to be the consequence of pathological events that are triggered during NSC stages early in development. A genome-wide analysis of the chromatin state further identified a contribution of preceding epigenetic changes to priming disease propensity. Future studies are needed to assess the implication of specific epigenetic aspects on onset and severity of autism-related disease trajectories. Finally, given the growing evidence of specific neurodevelopmental alterations in ASD, these findings and the methodological framework will be valuable for guiding future mechanistic studies aimed at identifying a convergence of causal genetic variations that contribute to ASD risk.
Methods
iPSC reprogramming and maintenance
Primary human dermal fibroblasts were obtained from subjects recruited through the University of California San Diego, Autism Center of Excellence, and reprogramming was performed using the classical retroviral approach54. This study protocol was approved by Salk Institute’s IRB Committee (FWA 00005316) and the Embryonic Stem Cell Research and Oversight (ESCRO) Committee. The Salk Institute is committed to protecting the rights and welfare of human research participants and ensures compliance with all applicable ethical and legal requirements. After a complete description of the study, written informed consent was provided by the parents of the human participants. IPSCs were generated from eight male subjects diagnosed with ASD according to the DSM-IV (average age 13.25 ± 5.67 years) and five male control participants (average age 11.4 ± 5.22 years), which had no history of psychological, genetic or other disorders (Supplementary Table 1). Control subjects were selected randomly from lists of typically developing individuals who had the magnetic resonance imaging (MRI) scan when they were toddlers. ASD subjects were selected from lists of ASD subjects who had been identified and diagnosed with ASD and MRI scanned when they were toddlers. From among these potential ASD subjects, those with larger than normal average total brain volume as compared with typically developing toddlers were selected. The iPSC lines obtained from this cohort have been previously described9 and were reprogrammed in the same facility (Salk Institute for Biological Studies, Laboratory of Genetics) and under the same conditions. Briefly, fibroblasts were transduced with retroviruses containing SOX2, OCT4, KLF4 and MYC to induce overexpression of these genes and were transferred to a co-culture system with murine embryonic fibroblasts. IPSC colonies were identified after around two weeks in this culture system, plated onto Matrigel-coated plates (BD Biosciences) and maintained in mTeSR1 media (Stem Cell Technologies). All cell lines were regularly tested for mycoplasma contamination. Experiments were performed with one or two independent iPSC clones (for details see Supplementary Table7).
Neural differentiation
We differentiated iPSCs into NSCs by following a default differentiation protocol as previously described9,55. Briefly, iPSC colonies were detached by collagenase (Gibco) to form embryoid bodies (EBs) and then cultured in suspension in the presence of 1 μM dorsomorphin (Cayman Chemical). After floating for seven days, EBs were attached on poly-ornithine (10 μg/ml, Life Technologies) and laminin (5 μg/ml, Life Technologies) coated surfaces. At day 16, emerging rosette colonies were manually detached, gently dissociated with accutase (Innovative Cell Technologies) and plated as NSC monolayers onto coated dishes for further neuronal differentiation.
Retroviral lineage tracing and neuronal differentiation
Replication-incompetent retrovirus was produced as described in supplementary methods. For retroviral lineage-tracing experiments, human NSCs were plated at a density of 5×104 cells/cm2 onto coverslips or dishes coated with poly-ornithine (10 μg/ml, Life Technologies) and laminin (5 μg/ml, Life Technologies). The next day, the medium was changed to neuronal differentiation medium (NDM) and sparse labeling of NSCs was achieved by using short time pulse infections with low-titer retroviral particles (for constructs, see supplementary methods). NDM is based on DMEM:F12/Neurobasal (both Gibco) (1:1) and contains the following supplements: N2 supplement (1x, Gibco), B27 supplement (1x, Gibco) and Laminin (1 μg/ml, Life Technologies). After six hrs, the remaining viral particles were washed out and the medium was replaced with NDM. For subsequent analyses, maturing neuronal cultures were followed for 2, 4, 7 and 14 dpi in vitro before being processed for immunocytochemistry, electrophysiology, flow cytometry or FACS-based purification.
Direct conversion of human iPSCs into iNs (iPSC-iN)
IPSCs were transduced with lentiviral particles for pLV-X-UbC:TetOn (UtO) and pLV-X-Ngn2:2A:eGFP (N2AG) and were selected in the presence of puromycin (0.5 μg/ml, Sigma Aldrich) and G418 (100 μg/ml, Life technologies). For iPSC-iN conversion, iPSCs were transferred into PluriPro monolayer conditions (Cell Guidance Systems) and, 24 hrs after plating, were induced with doxycycline (2 μg/ml: Sigma Aldrich) for two days. For further maturation, iNs were switched to Neuron Maturation Medium (NMM) based on DMEM:F12/Neurobasal (both Gibco) (1:1) as used for directed differentiation. NMM contains the following supplements: N2 supplement (1x, Gibco), B27 supplement (1x, Gibco), Laminin (1 μg/ml, Life Technologies) and doxycycline (2 μg/ml: Sigma Aldrich). The medium was changed every other day. For electrophysiological recordings, iNs were dislodged four days after conversion using TrypLE (Gibco), sorted based on eGFP expression and re-plated onto coverslips coated with poly-ornithine (10 μg/ml, Life Technologies) and laminin (5 μg/ml, Life Technologies).
FACS-based purification and RNA isolation
NSCs and maturing neuron cultures as well as iPSCs and iPSC-iNs were detached using accutase (Innovative Cell Technologies) and then washed and stained with fluorophore-coupled antibodies (see Supplementary Methods) for 45 min at 4ºC in phosphate buffered saline (PBS). Cells were washed, suspended in PBS and filtered using a 40-μm cell strainer. The desired cell populations were sorted directly into Trizol-LS (Life Technologies). RNA was isolated according to the manufacturer’s instructions and subsequently digested with TURBO DNase to remove trace quantities of DNA (ambion, Life Technologies). Before RNA-seq library preparation, assessment of RNA integrity numbers (RIN) was performed using the Agilent TapeStation.
RNA-seq and data preprocessing
Libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit according to the manufacturer’s instructions (Illumina). 50 bp single-end (SE) sequencing was performed using the Illumina HiSeq 2500 platform. Adapter trimming and low quality ends removal were conducted using Trimmomatic56 and trimmed reads were mapped to hg38 using STAR57. Expression values, heatmaps and clustering were performed using R 3.3.0 and RStudio 0.99.489 software based on variance stabilizing transformation (VST) and differential gene expression was calculated using the DESeq2 package58.
Weighted gene co-expression analysis (WGCNA)
A signed co-expression network was built using the WGCNA 1.51 package in R. All 65 time series samples were included in the network (five controls and eight ASDs at five time points) and 52 time series samples for the iPSC-iN time line (five controls and eight ASDs at four time points). Network construction was performed by first creating a matrix of pairwise correlations between all pairs of genes across the measured samples. The adjacency matrix was then constructed by raising the co-expression measure (0.5 + 0.5 X correlation matrix) to the power of β = 12 (and β = 14 for the iPSC-iNs time series), which is interpreted as a soft-threshold of the correlation matrix15. Based on the resulting adjacency matrix, we next calculated the topological overlap as a robust and biologically meaningful measure of network interconnectedness59. Average linkage hierarchical clustering was performed to group genes with highly similar co-expression relationships. Using the Dynamic Hybrid Tree Cut algorithm60, we defined modules as branches resulting from the tree cutting of the hierarchical clustering tree. To obtain moderately large and distinct modules, we set the minimal module size to 30 genes and merged modules whose eigengenes were highly correlated (correlation above 0.6). The expression profiles for each module were summarized by representing them as the first principal component (module eigengene, ME), thus explaining the maximum amount of variation of the module expression levels. Module membership measures were defined for each module as the correlation between gene expression values and the module eigengene (module eigengene based connectivity kME). To identify modules that gradually changed over time, a time-based significance measure was defined as the absolute correlation between time (dpi or days of in vitro differentiation post lineage tracing) and the gene expression profiles across all 65 samples. This measure was used to compute the module significances as the average gene significance measure for all genes in a given module (Fig. 1d).
Dynamic Time Warping (DTW)
Expression dynamics of time-related modules were compared using a DTW-based approach 61,62 to calculate the distance between ASD and control time series gene expression data. In brief, we used the expression profiles of the ME calculated by WGCNA and applied smoothing to enhance time resolution. The DTW package62 was then used to create a distance measure by locally compressing or stretching (warping) the query ME profile to best match the reference ME profile. The more warping that is required, the greater the difference between the two timelines. Calculation of the global alignment path used the full data window, symmetric continuity constraint and the Euclidean distance to measure path differences62. The plots were generated by summing the distances of individual aligned elements.
GS measures and network visualization
GS measures were used to incorporate external information into the co-expression network. These measures were defined as correlations between gene expression values and trait-specific cellular stage (time point). The advantage of using correlations to quantify trait relationships is that this measure can take on positive or negative values [−1, 1] and a P value can be easily computed. Thus, GS indicates whether the gene has a positive or negative relationship with the trait-specific time point. In combination with kME, GS was used to assess stage-specific expression or suppression of intramodular genes. Central hub genes of TM1 (genes with highest kME) were depicted using Cystoscape (version 3.3.0)63. In these plots, each point is a gene (or node) and gene-gene connections (or edges) are shown as light yellow or light red lines. Edge width values (gene-gene connection weight) are displayed with increasing thickness. The positioning of the nodes is done by first using an automatic layout and then manually adjusting the positions so that gene names are visible. In the plot showing hub genes of TM1, ASD risk genes are displayed in red and selected GO term memberships as a colored bar graph next to the node. Different point sizes correspond to the number of time points that show increased expression (>0.25 log2FoldChange) for the genes displayed. Significantly suppressed hub genes at the NSC stage are depicted in a second plot. Here, gradually increasing point size of nodes corresponds to GS values at the control NSC stage.
GO analysis
Functional annotation was performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics resource64 and The Gene Ontology Consortium65. For differentially expressed genes and co-expression modules, the background was set to the total list of genes expressed in our data set. In DAVID-based analysis, the reported P values are derived from the Expression Analysis Systematic Explorer (EASE) score probability, a modified Fisher’s exact test that is more stringent than the standard Fisher’s exact test in examining gene lists66. Where indicated, significance of overrepresentation was adjusted to control the false discovery rate (FDR) by means of the Benjamini-Hochberg procedure. The statistical significance threshold for all GO analyses was P<0.01.
Immunocytochemistry
Cells were washed in PBS, fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature and blocked with 5% horse serum in PBS containing 0.1% TritonX-100. Primary antibodies were incubated overnight at 4ºC, washed three times with PBS, incubated with secondary antibodies for two hours at room temperature, stained with DAPI solution and mounted in PVA-DABCO mounting solution (Sigma Aldrich). For confocal imaging, cells were grown on ibidi μ-slides. Antibodies and respective concentrations are listed in the Supplementary Methods.
Imaging
Image data were captured and processed by using a confocal microscope system (Zeiss LSM780) as well as a Zeiss CSU Spinning Disk Confocal Microscope, equipped with a Yokogawa spinning disc scan head with and an EM-CCD camera. Flow cytometry analysis was performed with a BD LSR II (Becton-Dickinson) and the BD InFlux Cytometer (Becton-Dickinson) was used for FACS-based purification of neuronal cell types.
Morphometric analyses
Tiled image stacks were processed using the ZEN Imaging software (Carl Zeiss Microscopy) and exported as TIFF files. Neurolucida version 11 (MBF Bioscience) was used to reconstruct individual retrovirus-labeled neurons over time. In total, 10 to 20 traces per line (eight ASD and five control lines with one or two clones per line, ntotal=564 cells) were analyzed at three different time points (4, 7 and 14 dpi) and plotted as mean for every individual. Identification of individual neurons was based on co-expression of markers PSA-NCAM and DCX. Reconstructions were based on the extent of membrane-bound GFP expression labeled through RV-CAG:LckN-eGFP. IPSC-iNs were traced at the time points indicated post induction (4, 7 and 14 days post doxycycline). In total, 10 to 30 traces per line (eight ASD and five control lines with one or two clones per line, ntotal=746 cells; Supplementary Table 7) were analyzed and plotted as mean for every individual. For Sholl analysis, 5 or 10 μm increments were used to define the gradually increasing radius of concentric circles centered at the centroid of the cell soma. Subsequent analysis of global and subtle branching patterns was performed using the Neurolucida Explorer (MBF Bioscience) and Microsoft Excel.
Generation of cerebral organoids
Cerebral organoids were generated and processed for analysis as described previously23 with minor modifications. Briefly, iPSCs were washed with Dulbecco’s phosphate-buffered saline (D-PBS, Invitrogen), dissociated with EDTA followed by Accutase to generate single cells. A total of 1X104 cells were then seeded into each well of an ultra-low-attachment 96-well plate (Nunc) to form single embryoid bodies (EBs) in medium containing DMEM/F12, 20% KOSR, 3% fetal bovine serum, 2mM GlutaMAX, 1% non-essential amino acids, 50nM b-mercaptoethanol and 4 ng/ml bFGF. ROCK inhibitor Y27632 (40uM) was included for the first 24 hrs. After two days, the medium was changed (same medium but without FGF) and EBs were grown for another three to four days. Medium was then changed to neural induction medium consisting of DMEM/F12, 1X N2 supplement, 1% non-essential amino acids, 2mM GlutaMAX and 1ug/ml heparin (Sigma). On days 10-12, EBs were embedded in 30 ul droplets of Matrigel and allowed to gel at 37°C for 20-30 min. Embedded EBs were subsequently cultured in cerebral differentiation medium (CDM) composed of 50% DMEM/F12, 50% Neurobasal, 0.5X N2 supplement, 1X B27 (without RA) supplement, 2mM GlutaMAX, 2.8 ng/ml human insulin (Sigma), 0.5% non-essential amino acids and 55 µM b-mercaptoethanol. Droplets were cultured in stationary condition in 6-cm suspension culture dishes (Corning) for four days, transferred to an orbital shaker (65-80 rpm) and cultured in CDM +RA media for up to five weeks.
Generation of forebrain organoids and retroviral labeling
Forebrain organoids were generated as described previously24 with minor modifications. Human iPSC colonies were detached before reaching confluency with collagenase Type IV (Gibco) and transferred to an Ultra-Low attachment 10 cm plate (Corning Costar), containing 10 ml hPSC medium, consisting of DMEM:F12 (Invitrogen), 20% Knockout Serum Replacer (Gibco), 1X Non-essential Amino Acids (Invitrogen), 1X 2-Mercaptoethanol (Gibco), 1X GlutaMAX (Invitrogen), 10 ng/ml FGF-2 (Peprotech) and ROCK inhibitor Y27632 (10μM). Twenty-four hrs later, the medium was replaced with induction medium containing hPSC media without FGF-2, 2 μM dorsomorphin (Cayman Chemical) and 2μM A-083 (Stemgent). At day five the media was replaced with Neural induction medium (NIM) consisted of DMEM:F12 (Invitrogen), 1X N2 Supplement (Invitrogen), 1X Non-essential Amino Acids (Invitrogen), 1X GlutaMAX (Invitrogen), 10 μg/ml Heparin (Sigma), 1X Penicillin/Streptomycin (Gibco), 10 μM CHIR99021 (Cayman) and 1 μM SB-431542 (Stemgent). Seven days after induction, organoids were embedded in 20 μl Matrigel (Trevigen) droplets and continued to grow for an additional week in 6-cm Ultra-Low attachment plates (Corning Costar). From day 14 onwards, organoids were cultured in differentiation medium composed of DMEM:F12 (Invitrogen), 1X N2 and B27 Supplements (Invitrogen), 1X Non-essential Amino Acids (Invitrogen), 1X GlutaMAX (Invitrogen), 1X 2-Mercaptoethanol (Gibco), 1X Penicillin/Streptomycin (Gibco), and 2.5 μg/ml Insulin (Sigma), transferred to an orbital shaker (65-80 rpm). At day 20, residual Matrigel was removed and media changes were performed every 2-3 days using the aforementioned differentiation medium. Labeling with RV-CAG:LckN-eGFP was performed in four-week-old forebrain organoids by using a titer range of approximately 105 - 106 colony forming units/ml. After 12 hrs of incubation, organoids were washed and cultured for up to ten weeks.
Forebrain organoids were stained with TBR1 at six weeks to define CP-like regions and to measure the thickness of the evolving plate. Four independent measurements per ventricular structure of sub-segmented fields (specified in Extended Data Fig. 6g) were averaged and a total of three forebrain organoids with 2–3 ‘VZ-like’ regions per organoid were quantified for every patient line using three ASD and three control individuals (ntotal= 17 organoids; see Supplementary Table 7 for details). Retrovirally labeled newborn neurons were analyzed in forebrain organoids at 14 dpi in regions that showed clearly defined VZ-like and CP-like structures. Three-dimensional reconstructions were based on the extent of membrane-bound GFP (RV-CAG:LckN-eGFP) in newborn neurons located within CP-like regions and analyzed with the software detailed above (morphometric analysis). In total, 30 to 60 traces per line (three ASD and three control lines with three organoid replicates per individual, ntotal=264 cells; Supplementary Table 7) were analyzed and plotted as mean for every individual.
Assay for Transposase-Accessible Chromatin-Sequencing (ATAC-seq).
70,000 FACS-purified NSCs were lysed in 50 μl lysis buffer (10 mM Tris-HCl ph 7.5, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL, CA-630, in water) on ice and nuclei were pelleted by centrifugation at 500 xg for 10 min. Nuclei were then resuspended in 50 µl transposase reaction mix (1x Tagment DNA buffer (Illumina 15027866), 2.5 µl Tagment DNA enzyme I (Illumina 15027865), in water) and incubated at 37°C for 30 min. DNA was then purified with Zymo ChIP DNA concentrator columns (Zymo Research D5205) and eluted with 10 µl of elution buffer. DNA was then amplified with PCR mix (1.25 µM Nextera primer 1, 1.25 µM Nextera index primer 2-bar code, 0.6x SYBR Green I (Life Technologies, S7563), 1x NEBNext High-Fidelity 2x PCR MasterMix, (NEBM0541)) for 8-12 cycles, size selected for fragments (160-500 bp) by gel extraction (10% TBE gels, Life Technologies EC62752BOX) and 125bp paired-end (PE) sequenced on an Illumina HiSeq 2500 platform.
ATAC-seq pre-processing and peak calling.
FASTQ sequencing files from ATAC-seq experiments were mapped to the UCSC genome (hg38) using Bowtie2 with default parameters67. For all data files, optical and PCR duplicates were removed using PicardTools. De-duplicated BAM files from the same sample were merged and PicardTools was again used to remove duplicate reads. Only uniquely mapped and properly paired reads were kept and those reads falling into the mitochondrial genome (chrM), unmapped contigs and blacklisted regions (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/hg38-human/) were removed, resulting in more than 55M high-quality reads on average. All samples showed the characteristic fragment length distribution, with more than 70% of reads falling in the nucleosome-free region (NFR) and a mono-nucleosomal peak representing reads that cut on both sides of a nucleosome (~180-247bp in length). For peak calling of open chromatin, reads were filtered using the deeptools268 command ‘alignmentSieve.py’ with the following parameters: “--ATACshift --minMappingQuality 30 --maxFragmentLength 100”, obtaining only properly paired reads within NFRs (<100bp). MACS2 (version 2.1.1.20160309) was then used to call peaks on filtered bam files using the following command:
ATAC-seq data analysis.
For differential accessibility (DA) analysis, we used DiffBind v2.4.869 to first perform a weighted overlap analysis (dba.count). The number of reads within each peak of the union peakset was counted and normalized for sequencing depth using the total number of non-mitochondrial reads present in the peaks for each sample. The edgeR v3.18.1 package was then used to contrast the control and ASD group using the DiffBind function ‘dba.analyze’ with the settings to use edgeR (method = DBA_EDGER). The DA heatmap (Fig. 8b) displays the normalized peak accessibility scores for each sample. These scores were calculated using the DiffBind default setting ‘DBA_SCORE_TMM_READS_FULL’, which uses the trimmed mean of M-values (TMM) normalization built into the edgeR package using read counts and full library size. Coverage plots (Fig. 8a and Extended Data Fig. 11f) display normalized read counts – the number of reads within a peak divided by the sequencing depth – and were generated with Gviz70 using hg38 NCBI RefSeq gene model annotations downloaded from the UCSC genome browser.
To generate metagene profiles of promoter regions, HOMER was first used to convert aligned and filtered reads into “tag directories” for further analysis71. For the analysis of promoter accessibility, read counts for each sample were normalized to 10 million; then the mean numbers of reads located +/− 1500 bp of gene transcription start sites (TSS) were computed across the regions of the selected genes using HOMER’s “annotatePeaks” command with the “hist” option and a bin size of 10 base pairs. Statistics were calculated in a ±250-bp window surrounding the center of the TSS.
Statistics
All information related to statistical tests is documented in the corresponding figure legends and/or in the main text and in the Supplementary Methods. No statistical methods were used to predetermine sample size but our sample sizes are similar to those reported in previous publications9 that showed significance. No method of randomization was applied, however all biological assays and sequencing samples were randomly assigned to different experimental groups and batches to control for covariates. For data collection and data analysis (counting, tracing, imaging and FACS analysis), investigators were blinded with regard to the group category. All biological assays were performed with iPSCs derived from eight ASD patients and five healthy controls (n=8 ASD patient lines, n=5 control cell lines; all male), with the exception of the organoid validation experiments, for which we randomly selected a subset of three ASD patients and three controls (n=3 ASD patient lines, n=3 control cell lines; Fig. 5 and Extended Data Fig.6f-l). No other collected samples were excluded from the analysis. Statistical comparisons for biological assays were performed in Prism 6.0 (GraphPad Software) using the Mann-Whitney U-test (MWU, one independent variable, single comparison), the one-way ANOVA using Dunnett’s multiple comparisons test (to compare means of three or more sets of unpaired measurements), and the two-way ANOVA using the Sidak multiple comparison test (two independent variables: dpi and neurite length per neuron). Threshold for significance (α) was set at 0.05. Data distribution was assumed to be normal, but this was not formally tested. Means, median, s.d., s.e.m., percentages and confidence intervals of all other quantitative data were calculated using Prism 6.0 and Microsoft Excel.
Differential gene expression was calculated using Wald test statistics as implemented into the DESeq2 package58. Log transformed RPKM values were used for transcriptome correlation analysis (Extended Data Fig. 2 and 8) by using the Pearson correlation coefficient to measure the linear dependence of our samples to the BrainSpan dataset (details see Supplementary Methods). Gene set enrichment analysis was performed using a hypergeometric test and for GO term enrichment, a modified Fisher’s exact test was used66. For GS calculations and other WGCNA related statistics, a two-tailed unpaired Student’s t-test was used and, where indicated, p-values were adjusted by using the FDR approach. For ATAC-seq, Hotelling’s t-square test was used to determine the difference between the transcription start sites (TSS) of the gene sets between ASD and control samples (Fig. 8f), considering n as the number of genes analyzed. Significance calculations for the enrichment of DA peaks within genomic regions was performed using the GAT randomization test53.
Data Availability
RNA-seq datasets are available at EMBL-EBI ArrayExpress with the accession code: E-MTAB-6018. This raw data is associated with the following figures: 1, 2, 3, 7d-f, S3c, S4, S7a-e, S8f-g and S10. Additional data generated or analyzed during this study is included in this published article and its supplementary information files. Supplementary tables are available for the following figures: 1, 2, 3, 7d-f, S3c, S4a, S7a-d, S8f-g and S10 as well as for additional differential expression analysis (Supplementary table 8 and 10). Supplementary table 1 (Supplementary Data and Notes) is providing additional information about all patient-derived iPSC lines used in this study. ATAC-seq datasets are available from the corresponding author upon request.
Code Availability
The analysis routines and R packages used for data analysis are publically available (http://bioconductor.org/) and the parameters and settings used have been described in the Methods and Supplementary Methods. The following R packages were used for data analysis: DESeq2 v1.20.0, WGCNA v1.51, dtw v1.20-1, DiffBind v2.4.8, edgeR v3.18.1, Gviz v1.20.0. Other packages used in this study included: STAR v2.5.2a, Cudadapt v1.6 with Python 3.4.1, FastQC v0.10.1, Bowtie2 v2.2.8, PicardTools MarkDuplicates v2.18.3-SNAPSHOT, samtools v1.6, deeptools2 v2.5.4, macs2 v2.1.1.20160309, Homer, GAT v1.3.5, Cytoscape v3.3.0 and Prism v6.0 (GraphPad Software).
Supplementary Material
Acknowledgments
We thank the patients who participated in this study. We also thank M.L. Gage and M. Pena for editorial comments; E. Courchesne for his contribution to fibroblast collection; L. Moore, A. Mendes, B. Miller, K. Rehder, A. Mar and K. Niedrig for technical assistance; K. Diffenderfer and the Salk STEM core facility for technical support; C. Fitzpatrick and C. O’Connor for help on flow cytometry; and T. Toda for helpful discussions. This work was supported by the Flow Cytometry Core Facility and the NGS core facility of the Salk Institute with funding from NIH-NCI CCSG: P30 014195, the Chapman Foundation and the Helmsley Charitable Trust. We also acknowledge support from The James S. McDonnell Foundation, G. Harold & Leila Y. Mathers Charitable Foundation, JPB Foundation, NIH (Grants MH095741 and MH090258), The Engman Foundation, Annette C. Merle-Smith, The Paul G. Allen Family Foundation and The Leona M. and Harry B. Helmsley Charitable Trust (Grant #2012-PG-MED00). S.T.S. was funded by a fellowship from the German Research Foundation (DFG) and was recently awarded the NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation.
Footnotes
Accession Codes
RNA-seq datasets have been deposited at EMBL-EBI ArrayExpress: E-MTAB-6018.
Competing Interests
The authors declare no competing interests.
References
- 1.de la Torre-Ubieta L, Won H, Stein JL & Geschwind DH Advancing the understanding of autism disease mechanisms through genetics. Nat. Med 22, 345–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parikshak NN, Gandal MJ & Geschwind DH Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet 16, 441–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parikshak NN et al. Integrative Functional Genomic Analyses Implicate Specific Molecular Pathways and Circuits in Autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willsey AJ et al. Coexpression Networks Implicate Human Midfetal Deep Cortical Projection Neurons in the Pathogenesis of Autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courchesne E, Carper R & Akshoomoff N Evidence of brain overgrowth in the first year of life in autism. JAMA 290, 337–344 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Courchesne E et al. Mapping early brain development in autism. Neuron 56, 399–413 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Emerson RW et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci. Transl. Med 9, eaag2882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazlett HC et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 542, 348–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchetto MC et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry (2016). doi: 10.1038/mp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariani J et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shcheglovitov A et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 503, 267–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein JL et al. A Quantitative Framework to Evaluate Modeling of Cortical Development by Neural Stem Cells. Neuron 83, 69–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Home :: BrainSpan: Atlas of the Developing Human Brain. Available at: http://www.brainspan.org/. (Accessed: 11th November 2016)
- 14.Yuan SH et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PloS One 6, e17540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B & Horvath S A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol 4, (2005). [DOI] [PubMed] [Google Scholar]
- 16.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakoe H Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans. Acoust. Speech Signal Process. 26, 43–49 (1978). [Google Scholar]
- 18.Velichko VM & Zagoruyko NG Automatic recognition of 200 words. Int. J. Man-Mach. Stud 2, 223–234 (1970). [Google Scholar]
- 19.Basu SN, Kollu R & Banerjee-Basu S AutDB: a gene reference resource for autism research. Nucleic Acids Res 37, D832–836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voineagu I et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parikshak NN et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature advance online publication, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lancaster MA et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian X et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molyneaux BJ, Arlotta P, Menezes JRL & Macklis JD Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci 8, 427–437 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Toma K, Wang T-C & Hanashima C Encoding and decoding time in neural development. Dev. Growth Differ 58, 59–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erhardt JA et al. A Novel F Box Protein, NFB42, Is Highly Enriched in Neurons and Induces Growth Arrest. J. Biol. Chem 273, 35222–35227 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Atkin G et al. Loss of F-box Only Protein 2 (Fbxo2) Disrupts Levels and Localization of Select NMDA Receptor Subunits, and Promotes Aberrant Synaptic Connectivity. J. Neurosci 35, 6165–6178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vitucci D et al. Rasd2 Modulates Prefronto-Striatal Phenotypes in Humans and ‘Schizophrenia-Like Behaviors’ in Mice. Neuropsychopharmacology 41, 916–927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abudureyimu S et al. Essential Role of Linx/Islr2 in the Development of the Forebrain Anterior Commissure. Sci. Rep 8, 7292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandai K, Reimert DV & Ginty DD Linx Mediates Interaxonal Interactions and Formation of the Internal Capsule. Neuron 83, 93–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y et al. Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron 78, 785–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium RE et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilczynski B, Liu Y-H, Yeo ZX & Furlong EEM Predicting Spatial and Temporal Gene Expression Using an Integrative Model of Transcription Factor Occupancy and Chromatin State. PLoS Comput. Biol 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin F et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow ML et al. Age-Dependent Brain Gene Expression and Copy Number Anomalies in Autism Suggest Distinct Pathological Processes at Young Versus Mature Ages. PLOS Genet 8, e1002592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun 5, 5748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W et al. Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell 167, 1385–1397.e11 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lim ET et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci 20, 1217–1224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hazlett HC et al. Early Brain Overgrowth in Autism Associated With an Increase in Cortical Surface Area Before Age 2 Years. Arch. Gen. Psychiatry 68, 467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumann CM et al. Longitudinal MRI Study of Cortical Development through Early Childhood in Autism. J. Neurosci. Off. J. Soc. Neurosci 30, 4419–4427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauman ML & Kemper TL Neuroanatomic observations of the brain in autism: a review and future directions. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci 23, 183–187 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Belmonte MK et al. Autism and Abnormal Development of Brain Connectivity. J. Neurosci 24, 9228–9231 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carper RA & Courchesne E Localized enlargement of the frontal cortex in early autism. Biol. Psychiatry 57, 126–133 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Casanova MF, Buxhoeveden DP, Switala AE & Roy E Minicolumnar pathology in autism. Neurology 58, 428–432 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Casanova MF et al. Abnormalities of cortical minicolumnar organization in the prefrontal lobes of autistic patients. Clin. Neurosci. Res 6, 127–133 (2006). [Google Scholar]
- 47.Weir RK, Bauman MD, Jacobs B & Schumann CM Protracted dendritic growth in the typically developing human amygdala and increased spine density in young ASD brains. J. Comp. Neurol 526, 262–274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chomiak T, Karnik V, Block E & Hu B Altering the trajectory of early postnatal cortical development can lead to structural and behavioural features of autism. BMC Neurosci 11, 102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chomiak T et al. Auditory-cued sensorimotor task reveals disengagement deficits in rats exposed to the autism-associated teratogen valproic acid. Neuroscience 268, 212–220 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Vierbuchen T et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fishman VS et al. Cell divisions are not essential for the direct conversion of fibroblasts into neuronal cells. Cell Cycle 14, 1188–1196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briggs JA et al. Mouse embryonic stem cells can differentiate via multiple paths to the same state. eLife 6, e26945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heger A, Webber C, Goodson M, Ponting CP & Lunter G GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29, 2046–2048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi K et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Marchetto MCN et al. A Model for Neural Development and Treatment of Rett Syndrome Using Human Induced Pluripotent Stem Cells. Cell 143, 527–539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics bts635 (2012). doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yip AM & Horvath S Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8, 22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langfelder P, Zhang B & Horvath S Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Aach J & Church GM Aligning gene expression time series with time warping algorithms. Bioinformatics 17, 495–508 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Giorgino T Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package. J. Stat. Softw 31, 1–24 (2009). [Google Scholar]
- 63.Shannon P et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang DW, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Ashburner M et al. Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosack DA, Dennis G, Sherman BT, Lane HC & Lempicki RA Identifying biological themes within lists of genes with EASE. Genome Biol 4, R70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramírez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ross-Innes CS et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hahne F & Ivanek R Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol. Biol. Clifton NJ 1418, 335–351 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets are available at EMBL-EBI ArrayExpress with the accession code: E-MTAB-6018. This raw data is associated with the following figures: 1, 2, 3, 7d-f, S3c, S4, S7a-e, S8f-g and S10. Additional data generated or analyzed during this study is included in this published article and its supplementary information files. Supplementary tables are available for the following figures: 1, 2, 3, 7d-f, S3c, S4a, S7a-d, S8f-g and S10 as well as for additional differential expression analysis (Supplementary table 8 and 10). Supplementary table 1 (Supplementary Data and Notes) is providing additional information about all patient-derived iPSC lines used in this study. ATAC-seq datasets are available from the corresponding author upon request.