Abstract
The NAC family is one of the largest plant-specific transcription factor families, and some of its members are known to play major roles in plant development and response to biotic and abiotic stresses. Here, we inventoried 488 NAC members in bread wheat (Triticum aestivum). Using the recent release of the wheat genome (IWGS RefSeq v1.0), we studied duplication events focusing on genomic regions from 4B-4D-5A chromosomes as an example of the family expansion and neofunctionalization of TaNAC members. Differentially expressed TaNAC genes in organs and in response to abiotic stresses were identified using publicly available RNAseq data. Expression profiling of 23 selected candidate TaNAC genes was studied in leaf and grain from two bread wheat genotypes at two developmental stages in field drought conditions and revealed insights into their specific and/or overlapping expression patterns. This study showed that, of the 23 TaNAC genes, seven have a leaf-specific expression and five have a grain-specific expression. In addition, the grain-specific genes profiles in response to drought depend on the genotype. These genes may be considered as potential candidates for further functional validation and could present an interest for crop improvement programs in response to climate change. Globally, the present study provides new insights into evolution, divergence and functional analysis of NAC gene family in bread wheat.
Introduction
As sessile organisms, plants have developed diverse strategies to modulate their development and growth according to environmental signals such as day/night alternation or daily temperature variations [1]. At the molecular and cellular levels, the early mechanisms of response to these stimuli include signal perception by receptors, followed by signalling cascades involving changes in membrane permeability, protein phosphorylations by protein kinases or regulations of the target protein expression by transcription factors (TFs) [2]. About 6–8% of the plant genome is allocated to the coding of more than 1,500 TFs [3] of which 45% belong to plant-specific families [4]. Among more than 80 TF families, AP2/EREBP, bZIP, MYB/MYC, NAC and WRKY families are known to be strongly involved in the response to biotic or abiotic stresses in plants [5–16]. The NAC (NAM (No Apical Meristem)–ATAF (Arabidopsis Transcription Activation Factor)–CUC (Cup-shaped Cotyledons)) family is one of the largest groups of plant specific TFs, and has been described in several plant species. In the model plants, Arabidopsis and rice, this family includes 117 and 151 members, respectively [17]. NAC family members have also been reported in Brachypodium distachyon (118 members) [18], soybean (101 genes) [19], maize (157 members) [20], durum wheat (168 members) [21], and more recently in Fragaria × ananassa fruits (112 members) [22].
A typical NAC protein contains a highly conserved DNA-binding domain, measuring ~150 amino acids, and located in the N-terminal region. This domain is subdivided into five subdomains named A to E [23]. Subdomains A, C, and D are highly conserved. Subdomains C and D are probably involved in the binding of the NAC protein to its DNA target [24]. An additional poorly conserved transcription regulatory region is present in the C-terminal region of the protein and acts as a transcriptional activator [25] or repressor [26] of target genes. The C-terminal regulatory region often contains simple amino acid repeats and regions rich in serine and threonine, proline and glutamine, or acidic residues [27]. While the majority of the NAC TFs were found as soluble proteins in the nucleus, some NAC have been identified as membrane-bound TFs. They are classified as membrane-related and named NTL (NTM1-like or ‘NAC with transmembrane motif 1’-Like) TFs. For example, nine NTL were identified in durum wheat [21] and at least 85 and 45 in Arabidopsis and rice respectively [28].
Expression of NAC family genes is modulated during plant development and in response to biotic and abiotic stresses [27]. NACs are one of the largest families of genes with very diverse functions. The first NAC gene was described as related to the shoot apical meristem development and the position determination of meristems and primordia [29]. The NAC family regulates many developmental and growth processes including flowering [30], root development [25], leaf senescence [31], secondary cell wall biosynthesis [32], cell division [33], and seed development [34]. The NAC family is also related to grain yield and quality. For example, a reduced expression of OsNAP in rice and TaNAC-S in wheat increases the grain yield by delaying leaf senescence [35–38]. Moreover, NAC proteins have also been shown to be involved in response to biotic stresses such as pathogen infection [39], hypersensitive response-like cell death [40], and abiotic stresses including salt, drought, and temperature [41–43]. Due to their key role in abiotic stress signalling in plants, NAC TFs could be used to improve crop plant tolerance to unfavourable environmental conditions. This is particularly important for bread wheat (Triticum aestivum) because the yield of this major crop, which constitutes the diet basis of millions of people, is constrained by abiotic stresses.
Bread wheat is a hexaploid species. Its genome is composed of three subgenomes AA (originated from T. urartu), BB (from Aegilops speltoides) and DD (from Ae. tauschii) [44]. In a recent publication, Borrill et al. [45] carried out an inventory of the TF coding genes in wheat using published TGAC gene models [46], with an emphasis on the NAC family. They identified 574 (453 high and 121 low-confidence) NAC genes belonging to eight subfamilies. More recently, a high-quality version of the wheat genome (cv Chinese Spring) assembly was released by the International Wheat Genome Sequencing Consortium (IWGSC) [47]. In this study, we used this last version to analyse the genomic organization and the expansion of the NAC family. We identified 488 (460 high and 28 low-confidence) NAC genes, named TaNAC, in the three subgenomes A, B, and D of bread wheat. We established the correspondence (one-to-one) between the two annotations (488 vs 574) yielding a consensus subset containing 361 identical sequences. The expression profiles of these 361 identical TaNAC genes were monitored in different tissues (grains, spikes, leaves and roots) or in response to different abiotic stresses (heat, drought, and heat + drought combination), based on the data available on the www.wheat-expression.com platform [48]. In addition, 23 representative TaNAC genes were selected to investigate their response to drought stress using two contrasting wheat cultivars at two developmental stages (220 and 450 degree days (°Cd) after anthesis). Our results provide valuable information about TaNAC candidate genes to improve drought stress tolerance in wheat.
Materials & methods
Meta-analysis of the NAC family in bread wheat
Survey of TaNAC in the wheat sequence
For TaNAC retrieval, the annotated proteins in the database of the International Wheat Genome Sequencing Consortium (IWGS RefSeq v1.0) [47] were searched using the HMMER2 program implemented in Unipro UGENE 1.25 [49] and by querying the NAM motif (PF02365, generated by alignment of 587 seed sequences) downloaded from Pfam.
The full-length sequences obtained with an E-value cut-off e-1 were scanned for NAC domain superfamily (IPR036093) using InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search, S1 Table) [50]. The combination of HMMER2 and InterProScan searches led to a final set of 488 TaNAC proteins, whose 460 are high-confidence and 28 are low-confidence. The TaNAC genes were mapped on the wheat pseudomolecule from the IWGS Consortium to determine their chromosomal coordinates and visualized using MapGene2Chromosome V2 (http://mg2c.iask.in/mg2c_v2.0/). Homoeologous genes were determined using their chromosomal coordinates.
To create a high quality consensus set of TaNAC genes, a reciprocal homology search was performed between the TGACv1 [46] and the IWGS RefSeq v1.0 [47] cDNA databases (including high and low-confidence sequences), using Usearch program [51]. This query led to the identification of common sequences between the TaNAC gene set of Borrill et al. [45] extracted from TGACv1 cDNA database and the set of 488 TaNAC sequences extracted from IWGS RefSeq V1.0. To minimize the fortuitous detection of homology, the final set of protein-coding sequences in both annotations was used as a query. When reciprocal homology search identified the same pair of TaNAC genes in both directions, they were considered as homologs.
The NAC family structuration in bread wheat
To study the structuration of the TaNAC family and confirm the homoeologous groups, a phylogenetic study was performed. The maximum likelihood method available in MEGA7 software [52] was used to construct an unrooted phylogenetic tree based on the alignment of the amino acid sequences of the common set of sequences established. Alignment was performed using the MUSCLE alignment algorithm [53]. The tree obtained was the consensus of 500 single trees provided by bootstraps, gaps/missing data treatment: partial deletion, model/method LG model, rates among sites: gamma distributed with invariant sites (G).
To explore the contribution of gene duplication to the expansion of the NAC family in bread wheat, we focused on flanking sequences of each TaNAC gene. For each locus containing a TaNAC gene, 25 Kb on either side of the gene were collected. When several TaNAC genes are separated by less than 25 Kb, the window for sequence retrieval was extended to the 25 Kb beyond the leftmost and rightmost TaNAC genes. Homologies between nucleotide sequences were obtained using YASS, a Blast-like tool [54] with an e-value threshold of e-30. The homology search results were then parsed and sequences with at least 80% identity over more than 2 Kb were considered duplicates. Figures showing these duplications were generated using CIRCA (OMGenomics, http://omgenomics.com/circa/).
To monitor the correlation between expression profiles of duplicated genes, Pearson correlation coefficients between TaNAC genes were calculated using the R software (http://www.R-project.org, R development Core Team, 2008).
Expression of TaNAC genes
The expression profiles of TaNAC genes were monitored in different tissues (grains, spikes, leaves, and roots) [55] or in response to different abiotic stresses in leaves (heat shock, polyethylene glycol-induced drought, and a combination of both at a young stage) [56], based on the RNA seq data available on the www.wheat-expression.com platform [48]. Briefly, raw expression data were downloaded and analysed using the R package limma [57]. Raw expression data corresponding to the entire wheat genome were first filtered to remove weakly expressed genes (genes are retained if they are expressed at a counts-per-million (CPM) > 0.5 in at least two samples). The expression of retained genes were then normalized using TMM and Voom methods. Finally, expressions were compared between samples and those genes that displayed an absolute log Fold Change (logFC) >1 with an adjusted p-value < 0.05 were deemed Differentially Expressed and noted DEG. Finally, TaNAC genes were retrieved from this set of DEG genes. Hierarchical Clustering of TaNAC DEG was conducted using Mev software [58] with euclidean distance and average linkage.
On the basis of this meta-analysis, 23 TaNAC genes which displayed large fold changes in their expression were selected to assess their response in wheat plants cultivated in field conditions and submitted to drought.
Expression analysis of 23 TaNAC candidate genes in field-grown stressed wheat plants
To gain insights into the function of wheat NAC genes in response to abiotic stresses, we decided to assess the expression patterns of 23 selected TaNAC genes in plants subjected to drought stress in field.
Plant material and field drought treatment
Field drought treatment was performed on the Pheno3C plant phenotyping platform. This four automatic rain shelters facility is able to generate contrasted water regimes by protecting four “stressed” plots from rainfall, while four “control” plots are managed under natural weather and irrigated if necessary to prevent water stress. The eight plots are distributed over a homogeneous three hectares area and minimum distance between two plots is 30 m. Each plot (22 m x 25 m) is subdivided into 96 micro-plots (1.9 m x 1.2 m) where different genotypes were sown. Among those genotypes and based on local expert knowledge and previous trials, two modern bread wheat genotypes (Allez-y (2011, Limagrain) and Altigo (2007, Limagrain) were selected as stress sensitive and stress tolerant cultivars respectively on the basis of yield impact under dry conditions. Soil volumetric water content (SWC) was recorded hourly with CS 655 (Cambell Scientific, Logan, UT, USA) sensors installed at four depth (10, 35, 50 and 75 cm) at three locations per plot. Total soil water content was calculated by integrating sensor values up to 100 cm depth (average soil depth of the plots). Average soil field capacity and available water capacity are respectively 570 and 280 mm. From those values and daily measurements of SWC, a water stress factor (Ks) was calculated from Allen et al. (1998) [59]. A Ks value below 1 is considered as an indicator of water stress that increases in terms of severity when the Ks value decreases.
All plots were sown on 11/04/2016, and managed following local agronomic practices. The stress was applied starting on 03/24/2017 by sheltering the stressed plots. At anthesis (05/20/2017), control plots had received 130 mm more rain than stressed plots, leading to a Ks of 0.85 and 0.36 in the control and stressed plots respectively, defining a very moderate stress in the control and a severe stress in the stress plots. Converted degree-days (°Cd) were calculated as the sum of mean daily temperature (base 0) from anthesis. For the present experiment, three micro-plots per genotype and per hydric conditions (biological triplicates) located in two adjacent blocks were used at both sampling dates. The samplings occurred at 250°Cd after anthesis (end of grain cell division) and 450°Cd after anthesis (middle of the linear phase of grain filling). At 250°Cd, water stresses were very moderate (Ks = 0.91) and very severe (Ks = 0.27) in the control and stressed plots respectively. At 450°Cd, water stresses were moderate (Ks = 0.6) and very severe (Ks = 0.2) in the control and stressed plots respectively. For each sample, three spikes and three leaves were taken randomly and frozen immediately in liquid nitrogen prior to subsequent RT-qPCR analysis. Yield components of each plot were also assessed. Spike number per m2, which is determined before anthesis [60], is a good indicator of the plant stress response before flowering, reflecting the stress impact on the plant tillering. This variable was determined manually by counting all spikes on two 1.9 m rows per micro-plot. Grain yield was measured at maturity with an experimental plot combine harvester.
RNA isolation and relative quantification of candidate genes
To quantify the expression of the 23 candidate genes, a total RNA extraction was performed on the collected samples, using the protocol described by Capron et al. [61] for grain samples, and the TRIzol protocol according to the manufacturer’s instructions (Invitrogen) for leaf samples. Reverse-transcription (RT) was performed on 2.5 μg of total RNA, using the Thermo Scientific Maxima First Strand cDNA Synthesis Kit according to the manufacturer’s instructions. A RNAse H (Thermo Scientific 18021–071) treatment was performed after the reverse transcription.
When selecting the 23 TaNAC genes, only one member per homoeologous group was retained. Then, their specific expression profile was studied using Real Time-quantitative Polymerase Chain Reaction (RT-qPCR) and by designing primers specific to the coding sequence of the considered homoeolog (listed in S2 Table), with PrimerQuest (https://eu.idtdna.com/PrimerQuest). Amplification efficiencies of primer pairs designed were tested to keep only those with an efficiency comprised between 90 and 110% (S2 Table).
Real Time-quantitative Polymerase Chain Reactions (RT-qPCR) were carried out in a 15 μl volume containing 25 ng of cDNA, 7.5 nM gene-specific primers and 7.5 μl of LightCycler 480 SYBR Green I Master (Roche Diagnostics #04887352001), on a LightCycler 480 system (Roche) on the Gentyane platform, INRA de Crouël, France (http://gentyane.clermont.inra.fr/). The thermal cycle used was done as follows: a pre-incubation at 95°C for 10 min, 40 amplifications cycles of 95°C for 10s, optimal temperature of the primer for 15s, and 72°C for 15s.
Relative target gene expression was determined by the 2-ΔΔCt method [62] using Ta2776 (RNase L inhibitor-like protein) for normalization [61]. A principal component analysis was carried out using the expression data of the whole set of 23 genes across all samples and the R package ggfortify [63]. For each gene, a Kruskal-Wallis test was performed to uncover differences between samples.
Results
Inventory and description of the NAC TF family in bread wheat using the IWGS RefSeq v1.0 database
The HMM profile of the Pfam NAC domain (PF02365) was used as a query to identify TaNAC genes in the most recent version of the annotated wheat genome released by the IWGSC RefSeq v1.0, which contains the latest assembly of the wheat cv Chinese Spring pseudomolecule.
The initial HMM search identified 501 putative TaNAC sequences that were manually curated for the presence of the NAC domain (IPR036093) signature using the Interproscan platform. Thirteen sequences were discarded, giving a whole set of 488 full-length sequences (460 high-confidence and 28 low-confidence sequences) containing the NAC domain (S1 Table).
This set was compared with the sequences identified by Borrill et al. [45] using the TGAC database. This analysis provided a robust subset of 361 common sequences, showing a single hit between the two databases (S3 Table). The combination of the two annotations improved the knowledge on the TaNAC genes present in this subset, particularly in terms of chromosomal location. For example, no information on the chromosomal location of the TraesCSU01G137200 gene was available from the IWGS RefSeq v1.0 database, but it was found to be identical to the TRIAE_CS42_2AL_TGACv1_093618_AA0283710 gene in the TGAC database mapped on the chromosome 2A. Similarly, among the 13 non-mapped sequences of the TGAC database, 12 were identical to already mapped IWGS sequences. Finally, 359 TaNAC genes among the 361 consensus sequences have been mapped on the 21 bread wheat chromosomes, the two remaining sequences did not have chromosomal location information. Chromosomes 2 and 7 contain the highest numbers of TaNAC genes with 89 and 81 members respectively, which represent 24.8% and 22.6% of the total. On the contrary, chromosome 1 carries the lowest number of TaNAC sequences, with only 17 genes (4.7%) (S1 Fig). The TaNAC genes are evenly distributed among the subgenomes A, B, and D with a similar proportions (34.5%, 33.5%, and 32.0% respectively) indicating that the expansion of the NAC family predates the bread wheat appearance through interspecific hybridization.
TaNAC genes are mostly distributed next to the telomeric regions vs centromeric one (S1 Fig). Furthermore, most of these genes are tightly linked and lay within clusters. For example, TraesCS4B01G328600, TraesCS4B01G328700, TraesCS4B01G328800, TraesCS4B01G328900, and TraesCS4B01G329100 are close on chromosome 4B within 347 Kb. Similarly, the TaNAC genes denoted TraesCS4A01G419900, TraesCS4A01G420000, TraesCS4A01G420100, TraesCS4A01G420200, TraesCS4A01G420900, TraesCS4A01G421000, TraesCS4A01G421100, TraesCS4A01G421200, and TraesCS4A01G421300 are tightly linked and lay within 796 Kb.
The genomic length of the sequences within the whole set of 488 TaNAC genes, ranged from 273 to 15074 base pairs, corresponding to protein sequences from 91 (TraesCS2B01G556700LC.1) to 1030 amino acids (TraesCS5B01G550800.1) with a mean length of 370 amino acids. One-fifth of the sequences (i.e. 100 sequences out of 488) are intronless. On the other hand, one sequence (TraesCS5B01G550800.1) contains 16 exons. The majority (214 sequences, 44%) contain three exons (S2 Fig). It is interesting to note that eight sequences, representing three homoeologous groups, contain two NAC domains (TraesCS2A01G363700, TraesCS2B01G381700, TraesCS2D01G361500; TraesCS3A01G269900, TraesCS3B01G303800, TraesCS3D01G269600; and TraesCS2A01G328100, TraesCS2D01G334800) (S1 Table).
Evolution by duplication of the TaNAC family
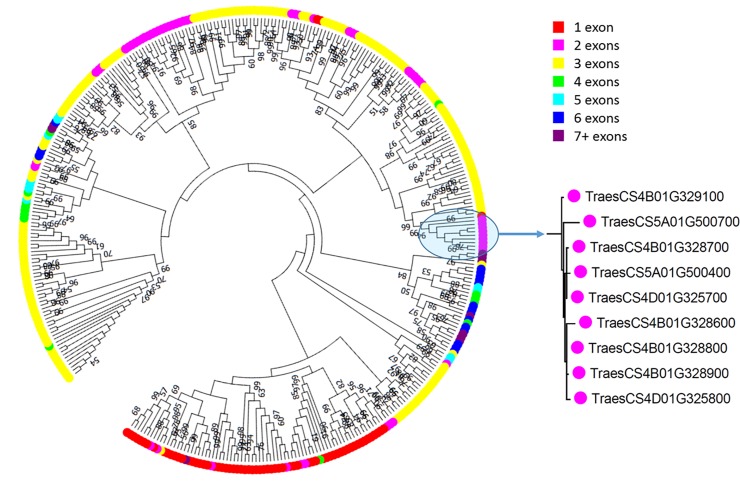
The evolution of the TaNAC family was evaluated by studying the phylogenetic relationships between TaNAC genes. A phylogenetic tree based on multiple sequence alignment of the common set including 361 full-length proteins was constructed (Fig 1), using the Maximum Likelihood method.
Fig 1. Maximum likelihood phylogeny of the 361 common complete protein sequences.
The zoom focused on the clade studied as a case of duplication. Colors indicate the number of exons in each sequence.
The phylogenetic tree showed that, in general, sequences contained in the clades belong to the same homoeologous groups. Based on sequences alignment, 156 TaNAC genes belonging to A, B, and D genomes were grouped into 52 complete homoeologous groups in the phylogenetic tree, representing 43% of the total TaNAC genes. Another 56 incomplete homoeologous groups containing only two members were identified and represent 31% of the total TaNAC genes. Finally, 93 sequences could not be linked to any homoeologous group. It is interesting to note that several sequences with the same exon-intron structuration and clustered on the wheat genome are grouped together in the phylogenetic tree.
In addition, these clades contain genes originating from non-homoeologous chromosomes. These observations suggest that the expansion of the TaNAC family might be the result of both local and interchromosomal duplication events. To test this hypothesis, we retrieved the genomic sequences of 50 Kb loci spanning TaNAC genes and carried out pairwise alignments using the YASS algorithm [54].
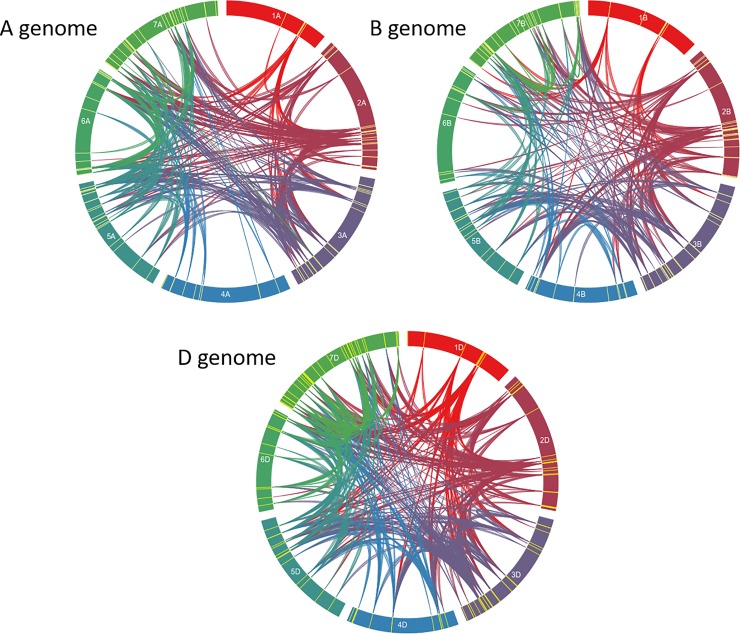
For the purpose of simplicity, we first considered duplications within each subgenome. Fragments displaying at least 90% identity over at least 3 Kb were retained. Using these criteria, 318 duplicated fragments were identified within the subgenome A, with mean-length = 5.75 Kb and median-length = 5.255 Kb. The largest fragment was 13.093 Kb. Concerning the subgenome B, 207 duplicated fragments were detected, with mean-length = 5.546 Kb and median-length = 5.146. The largest duplicated fragment was 10.009 Kb. Concerning the subgenome D, 428 duplicated fragments were detected, with mean-length = 5.639 Kb and median-length = 5.332. The largest duplicated fragment was 9.947 Kb (Fig 2, S4 Table).
Fig 2.
Circos diagram of the seven chromosomes of genomes A, B, and D. Yellow features indicate the presence of a TaNAC. Bands link two chromosomal regions with more than 90% of homology and a minimal length of 3 Kb.
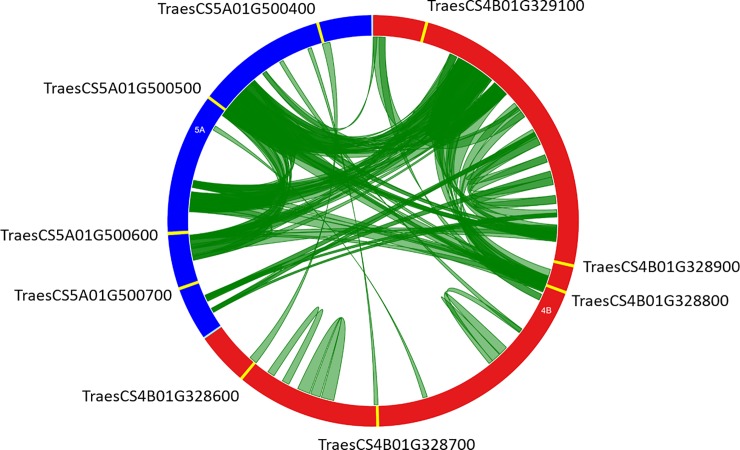
To complete that first analysis and provide also insight into duplications between subgenomes and consequently into the expansion pattern of the TaNAC family, we carefully surveyed one robust clade containing TaNAC genes originating from each wheat subgenome. This clade (pointed out in Fig 1 and boxed in blue in S3 Fig) was chosen because it contained several TaNAC belonging to A (2 sequences from chromosome 5), B (5 sequences from chromosome 4), and D genomes (2 sequences from chromosome 4). Careful examination of regions containing the nine genes of this clade indicated that TaNAC genes from a given chromosome are physically tightly linked (separated by maximum one gene). To avoid homoeology relationships between subgenomes, which may obscure duplications, the subgenome 4D containing two TaNAC genes was not further considered. By using a 25 Kb window around the most distal TaNAC genes within each locus, we retained a first 396.990 Kb long (619563787 to 619960777 bp) locus from the chromosome 4B containing the five TaNAC genes and a second 209.278 Kb long (666499425 to 666708703 bp) locus from the 5A containing four TaNAC genes. Over the four NAC sequences found on this chromosome, only TraesCS5A01G500400 and TraesCS5A01G500700 genes were present in the set of 361 sequences and studied for duplication. A dot plot of these two genomic regions (S4 Fig) highlighted numerous duplicated fragments in sense and antisense orientations. To clarify the nature and the extent of duplications, we identified all the duplicated fragments with a minimal length of 2 Kb and at least 80% identity. A total of 119 non-overlapping duplicated fragments were identified, of which 54 fragments correspond to intrachromosomal duplications (31 on chromosome 4B and 23 on 5A). The remaining 65 duplicated fragments correspond to interchromosomal duplications. The majority of these duplications (66.7%) represents a length less than 4 Kb, but a fragment has the maximum size of 9,006 bp. Duplications between loci of these genomic regions are presented in Fig 3. About 41% of these fragments (49) are duplicated in the sense orientation (S5 Table).
Fig 3. Circos diagram of chromosomes 4B (red, 396.990 Kb) and 5A (blue, 209.278 Kb) genomic regions containing nine TaNAC genes (yellow features).
Green bands link two chromosomal regions with more than 80% of homology and a minimal length of 2 Kb.
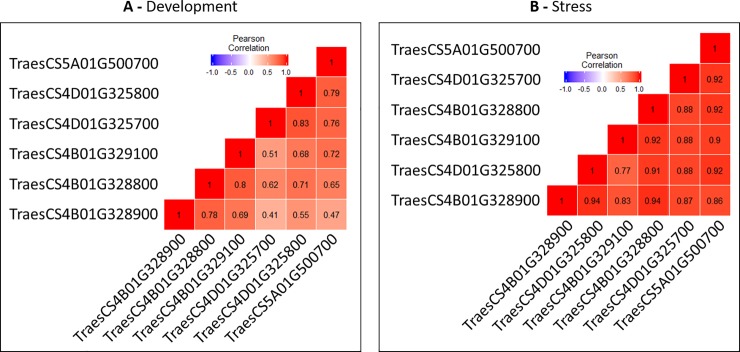
To test whether some duplicated genes could evolve differently, we compared the normalized expression patterns of duplicated genes in five wheat organs during three developmental stages: root (harvested at seedling, three leaves and meiosis), stem (harvested at spike at 1 cm, two nodes and anthesis), leaf (seedling, three tillers avec 50°Cd after anthesis), spike (harvested at two nodes, meiosis and anthesis), and grain (harvested at 50°Cd, 350°Cd and 700°Cd after anthesis [55]. In parallel, the expression profiles of the same genes were compared in response to abiotic stresses (heat, drought, and combined heat-drought) [56]. Only genes that were expressed in both studies were retained for comparison. The correlation (Pearson coefficient) was computed between pairs of genes (Fig 4).
Fig 4.
Pearson correlations between duplicated genes of the studied 4B-4D-7B-5A chromosomal regions during development (A) and stress (B).
During development, the Pearson coefficient ranged from 0.41 (between TraesCS4D01G325700 and TraesCS4B01G328900) to 0.83 (between TraesCS4D01G325800 and TraesCS4D01G325700). In contrast, correlations were higher during responses to stress and ranged from 0.77 to 0.94. These results indicate that the members of this duplication display similar expression profiles in response to the abiotic stresses tested but a substantial variation in expression profiles is observed during development between the same members as shown by the behavior of the two paralogs TraesCS4D01G325700 and TraesCS4B01G328900 (Fig 4).
In a similar way, an examination of a second duplication (boxed in red in the phylogenetic tree provided in S3 Fig) was performed. In this case, the expression profiles of the paralogs were highly correlated during development while they could be weakly correlated during responses to abiotic stresses (i.e. between TraesCS2B01G118500 and TraesCS2A01G101100) (S5 Fig).
In silico analysis of the expression of TaNAC genes during development or in response to abiotic stresses
In order to monitor the expression of wheat TaNAC genes during development and in response to several abiotic stresses, we took advantage of valuable resources publicly available. In particular, we used a compendium of RNA-seq reads corresponding to various experiments and mapped to the wheat genome by Borrill et al. [45]. Raw counts corresponding to five organs at three developmental stages [55] or to abiotic stress treatments [56] were downloaded, normalized using TMM and voom methods, then Differentially Expressed Genes (DEG) were identified using an absolute logFC = 1 and an adjusted p = 0.05 as thresholds (R package limma) [57].
Responses of TaNAC genes to abiotic stresses
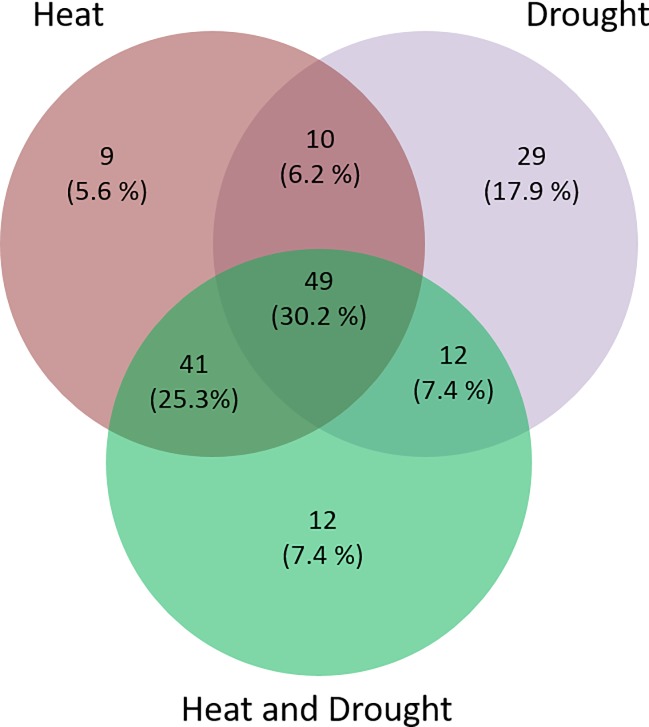
We identified differentially expressed wheat genes in one or several comparisons. The Table 1 shows the details of this analysis where DEG are denoted up- or down-regulated genes in comparison with the non-treated control sample. Among the whole set of DEG genes, 162 were annotated as TaNAC genes and dispatched as follows: 109 unique TaNAC genes were identified in heat treatments, 100 in drought treatments and 114 in heat-and-drought treatments. Some of them are common in two or more conditions (Fig 5). Table 1 and S6 Table show the distribution of these differentially expressed TaNAC genes alongside their logFC and the associated adjusted p-value.
Table 1. Number of differentially expressed genes in response to two abiotic stresses.
| Heat stress (40°C) 1h | Heat stress (40°C) 6h | Drought 1h | Drought 6h | Heat + Drought 1h | Heat + Drought 6h | |
|---|---|---|---|---|---|---|
| Number of down-regulated genes | 6763 | 11420 | 339 | 6426 | 7018 | 11288 |
| Number of down-regulated TaNAC genes | 24 | 39 | 5 | 28 | 28 | 41 |
| Number of up-regulated genes | 9837 | 11195 | 3117 | 6352 | 11796 | 11629 |
| Number of up-regulated TaNAC genes | 43 | 31 | 40 | 54 | 43 | 43 |
Fig 5. Venn diagram of the 162 differentially expressed TaNAC genes in response to heat, drought, and heat + drought stress.
Expression of TaNAC genes during development
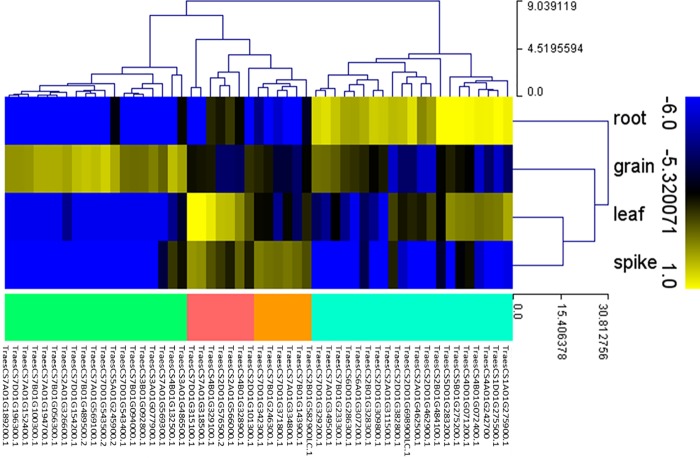
The TaNAC genes have been shown to be involved in plant development with expression profiles that can vary between organs and during organ development. In order to detect preferentially expressed TaNAC genes in certain wheat organs and at certain stages, we used the RNA-seq data of Choulet et al. [55], comprising five organs and three developmental stages per organ. We then also selected the most differentially expressed TaNAC genes compared to other tissues using a logFC threshold of +12. Using these relatively stringent criteria, we identified 52 TaNAC genes with a highly preferred expression pattern distributed as follows: 19 in the grain, 7 in the leaf, 20 in the root, 6 in the spike and none in the stem (Table 2, Fig 6).
Table 2. Number of differentially expressed genes (A) and list of preferentially expressed TaNAC genes (B) in five wheat organs.
| A | Grain | Leaf | Spike | Stem | Root |
|---|---|---|---|---|---|
| Number of down-regulated genes | 6505 | 5815 | 6533 | 6356 | 6362 |
| Number of up-regulated genes | 6862 | 5508 | 4468 | 5117 | 8447 |
| B | |||||
| Transcript | Organ with a preferential expression of the gene | logFC | Adjusted P.Value | ||
| TraesCS2D01G576500.2 | Leaf | 18.178 | 0.0009 | ||
| TraesCS4B01G329100.1 | Leaf | 17.573 | 0.0121 | ||
| TraesCS2D01G101300.1 | Leaf | 17.472 | 0.0231 | ||
| TraesCS4B01G328900.1 | Leaf | 17.299 | 0.0298 | ||
| TraesCS7A01G318500.1 | Leaf | 16.701 | 0.0074 | ||
| TraesCS7D01G315100.1 | Leaf | 16.4 | 0.0356 | ||
| TraesCS2A01G566000.1 | Leaf | 16.21 | 0.0377 | ||
| TraesCS7B01G489500.2 | Grain | 24.77 | 0.0026 | ||
| TraesCS7D01G543500.2 | Grain | 24.394 | 0.007 | ||
| TraesCS7A01G569100.1 | Grain | 24.269 | 0.0038 | ||
| TraesCS2A01G326600.1 | Grain | 24.025 | 0 | ||
| TraesCS7B01G100300.1 | Grain | 23.802 | 0.0025 | ||
| TraesCS7A01G189200.1 | Grain | 23.73 | 0.0002 | ||
| TraesCS7B01G056300.1 | Grain | 23.723 | 0.005 | ||
| TraesCS7D01G154200.1 | Grain | 23.719 | 0.007 | ||
| TraesCS7A01G194700.1 | Grain | 23.663 | 0.0059 | ||
| TraesCS7D01G196300.1 | Grain | 23.432 | 0.0031 | ||
| TraesCS7B01G094000.1 | Grain | 23.334 | 0.0005 | ||
| TraesCS7A01G152400.1 | Grain | 23.258 | 0.0009 | ||
| TraesCS3B01G092800.1 | Grain | 23.077 | 0.0007 | ||
| TraesCS4B01G132500.1 | Grain | 22.886 | 0 | ||
| TraesCS7D01G543400.1 | Grain | 22.879 | 0.0004 | ||
| TraesCS5A01G245900.2 | Grain | 22.67 | 0.0147 | ||
| TraesCS7A01G569300.1 | Grain | 21.729 | 0.0011 | ||
| TraesCS3A01G486500.1 | Grain | 20.566 | 0 | ||
| TraesCS3A01G077900.1 | Grain | 20.52 | 0.0329 | ||
| TraesCS2D01G309800.1 | Root | 23.596 | 0 | ||
| TraesCS2A01G462500.1 | Root | 23.17 | 0 | ||
| TraesCS2A01G311500.1 | Root | 22.266 | 0 | ||
| TraesCS1D01G275500.1 | Root | 21.745 | 0 | ||
| TraesCS2D01G462900.1 | Root | 21.084 | 0.0002 | ||
| TraesCS6A01G307200.1 | Root | 20.627 | 0 | ||
| TraesCS2B01G328300.1 | Root | 20.483 | 0 | ||
| TraesCS2B01G484100.1 | Root | 20.365 | 0.0002 | ||
| TraesCS6D01G286300.1 | Root | 20.325 | 0 | ||
| TraesCS2D01G698900LC.1 | Root | 19.923 | 0 | ||
| TraesCS7D01G329200.1 | Root | 19.261 | 0.0007 | ||
| TraesCS7B01G233300.1 | Root | 19.259 | 0.0001 | ||
| TraesCS7A01G349500.1 | Root | 19.001 | 0.0009 | ||
| TraesCS1A01G275900.1 | Root | 18.528 | 0.0001 | ||
| TraesCS4A01G242700 | Root | 18.32 | 0.0008 | ||
| TraesCS4B01G072400.1 | Root | 18.291 | 0.0003 | ||
| TraesCS5B01G275200.1 | Root | 18.04 | 0.0021 | ||
| TraesCS5D01G283200.1 | Root | 17.985 | 0.0045 | ||
| TraesCS2D01G382800.1 | Root | 17.401 | 0.0022 | ||
| TraesCS4D01G071200.1 | Root | 16.877 | 0.0136 | ||
| TraesCS7D01G371800.1 | Spike | 19.526 | 0.0002 | ||
| TraesCS7B01G143900.1 | Spike | 18.488 | 0.0025 | ||
| TraesCS7A01G334800.1 | Spike | 18.372 | 0.0042 | ||
| TraesCS7B01G246300.1 | Spike | 18.219 | 0.0053 | ||
| TraesCS2B01G582900LC.1 | Spike | 17.862 | 0.005 | ||
| TraesCS7D01G342300.1 | Spike | 17.198 | 0.0151 | ||
A TaNAC gene is preferentially expressed in a tissue if i) its logFC is the highest in this tissue vs the others and ii) the logFC threshold is ≥ 12.
Fig 6. Heat map of 52 TaNAC genes expression profile in four organs (grain, spike, leaf, and root), using the RNAseq data of Choulet et al.
[55]. These 52 TaNAC genes have a highly preferred expression pattern distributed as follows: 19 in the grain (green branches), 7 in the leaf (red branches), 20 in the root (blue branches), and 6 in the spike (orange branches). This figure was generated using MeV software. Hierarchical clustering was performed for the transcript abundance from all conditions.
TaNAC expression analysis in field under drought stress
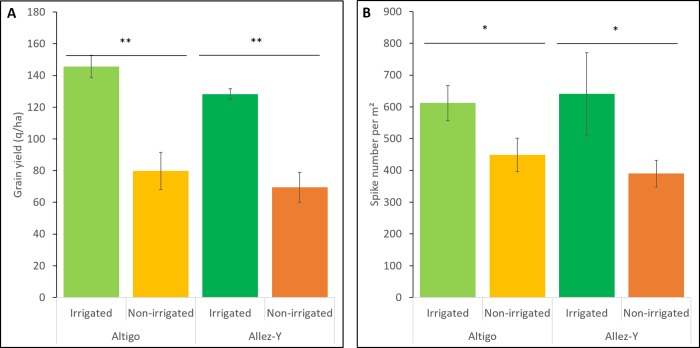
Grain yield and spike number per m2 were used to characterize the effect of the water stress generated in the drought trial. Under non-irrigated treatment, yield losses of 45.2% and 45.8% were observed for Altigo and Allez-y genotypes respectively. In the same way, a decrease of the spike number per m2 was observed (-26.6% for Altigo and -39.1% for Allez-y) (Fig 7). which translates to a high level of water stress that affected plant growth [60].
Fig 7.
Drought effect on the grain yield (q/ha) (A) and spike number per m2 (B) of two genotypes (Altigo and Allez-y). Mean values and standard deviation were obtained from three replicates (Student’s T test, * indicates a P < 0.05 and ** indicates a P < 0.01).
TaNAC gene expression profile under irrigated conditions
The in silico analysis resulted in a set of TaNAC genes tagged as DEG. A subset of 23 TaNAC displaying high fold changes in their expression were selected and analyzed in plants grown in the field under agronomic conditions. Their expression was monitored in two organs (leaf and grain) at two stages representative of grain development: cell division (220°Cd) and grain filling (450°Cd) in Altigo and Allez-y.
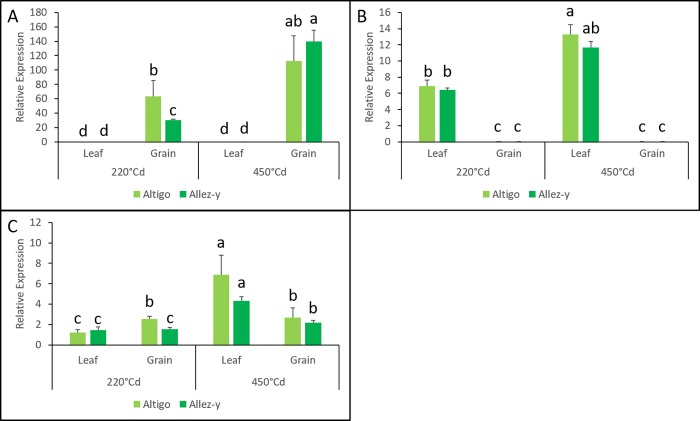
The 23 TaNAC genes tested in irrigated fields exhibited statistically differential transcript levels between organs (leaves and grains), regardless of the genotype or the stage. Seven genes appeared to be preferentially expressed in the grain and 16 in the leaf. The gene TraesCS5A01G245900 showed the highest expression in the grain compared to the leaf (10 018 times more) whereas TraesCS7D01G315100 was the most expressed gene in the leaf (332 times more than in the grain) (Fig 8A and 8B).
Fig 8.
RT-qPCR analysis of three TaNAC genes (TraesCS5A01G245900.2 (A), TraesCS7D01G315100.1 (B), and TraesCS2A01G101100.1 (C)) in leaves and grains at 220°Cd and 450°Cd in irrigated conditions. Mean values and standard deviation were obtained from three replicates. According to a Kruskal-Wallis test, the same letter indicates no significant difference.
In addition, some TaNAC gene expressions varied depending on the developmental stage (220°Cd vs 450°Cd). One gene (TraesCS2A01G566300.1) appeared to be preferentially expressed at 220°Cd while 15 genes were preferentially expressed at 450°Cd. For example, the expression level of TraesCS2A01G101100 gene was found to be 4.19 times more in leaves at 450°Cd compared to 220°Cd (Fig 8C).
Expression of TaNAC genes under drought stress
The expression data of the 23 selected genes in drought field conditions were presented in a principal component analysis (S6 Fig). The first two Principal Components (PC1 and PC2) explained respectively 83.5% and 10.8% of the total variation. Cumulatively, these two PCs contributed to 94.3% of the total variation of gene expression in response to drought, organ, genotype, and stage conditions. The first axis is explained by gene expression in the leaves under drought conditions at the later stage, 450°Cd (S6 Fig). The second axis is mainly explained by an organ x stage effect. Indeed, grain samples at 450°Cd were plotted in the lowest part of the S6 Fig. In the grain subset, the samples corresponding to early and late grain developmental stages (full form– 220°Cd and empty form– 450°Cd) constitute distinct groups along the second axis (PC2). In addition, considering only the data from grain at 450°Cd, it can be noticed that the two genotypes are plotted into two different groups.
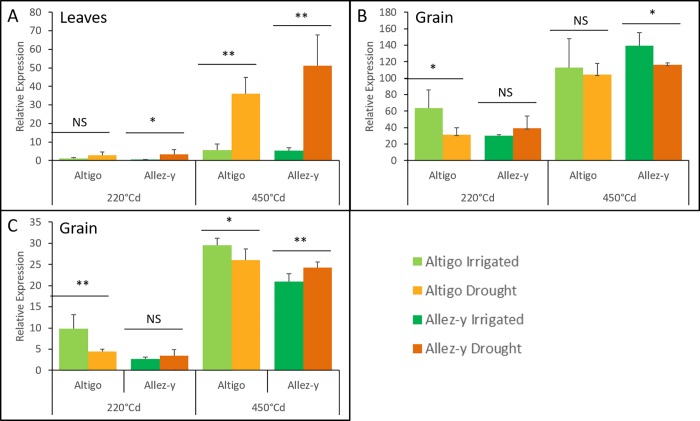
In leaves, the highest difference under drought stress was observed at 450°Cd compared to 220°Cd, regardless of the genotype (Fig 9A). A genotype effect was observed in response to drought only in grains compared to leaves. More precisely, grain-specific genes were differentially expressed at a different stage depending on the genotype. Indeed, a down-regulation under drought treatment was observed at 220°Cd in Altigo for five grain-specific genes. In Allez-y, four of these transcripts (TraesCS3A01G077900.1, TraesCS5A01G245900.2, TraesCS7B01G056300.1 and TraesCS7B01G100300.1) were down-regulated and the last one was up-regulated (TraesCS7A01G569100 .1) only at 450°Cd (Fig 9B, Fig 9C).
Fig 9.
RT-qPCR expression analysis of TraesCS5A01G143200.1 (A), TraesCS5A01G245900.2 (B), and TraesCS7A01G569100.1 (C). The 2-ΔΔCt method was used to calculate the ratio of their relative expression levels in drought conditions with the one in irrigated conditions. Mean values and standard deviation were obtained from three replicates (Student’s T test, * indicates a P < 0.05 and ** indicates a P < 0.01).
Discussion
The sequence availability of the whole bread wheat genome and expression data among different wheat genotypes, organs, and conditions are rich resources for systematic analysis of gene transcriptional regulation. Particularly, these data provide useful information for the identification of candidate genes involved in wheat development and its stress response.
Among transcription factors, the NAC family is one of the largest families and they are studied in a plethora of cereals [18,20,21] particularly in response to abiotic stresses [42,64,65], but only scarce information is available on bread wheat.
However, valuable resources and studies have recently been released. Particularly, Borrill et al. [45] identified 453 high confidence TaNAC retrieved from TGAC database [48]. In addition, large RNA-seq expression data exist on several wheat genotypes, in different organs [55] and abiotic stresses (heat shock or drought) [56] and are available to the scientific community. Based on these data, we studied the organisation of the NAC family in bread wheat and its expansion that could have occurred during evolution through potential duplication events. These resources were also used to identify differentially expressed TaNAC genes in wheat grown under drought conditions in field and to validate the potential specialization of some TaNAC members.
TaNAC genes: A large family
First, we identified 488 members of the NAC family in bread wheat using the IWGS RefSeq v1.0 database [47], whereas only 117 members were identified in Arabidopsis, 151 in rice [17] or 168 in durum wheat [21]. This high number is due to the presence of three subgenomes in bread wheat. After improving our dataset with TGAC sequences retrieved by Borrill et al. [45], we ended up with 361 sequences representing a high quality consensus subset. Considering the hexaploidy of wheat, one copy of each gene is expected to be carried by each of the three genomes (A, B, and D) creating homoeologous groups among the consensus subset. In this family, 43% of the TaNAC members can be gathered in complete homoeologous groups, 31% form incomplete homoeologous groups and 26% are not linked to a group. A divergence of sequences due to several mutations or a lack of sequences present in the subset could explain the incomplete homoeologous groups. This result has already been observed in other TF families, such as the bZIP family [66].
Similar to what has been described in other plant species such as maize [67] and rice [17], the TaNAC gene distribution among the chromosomes was uneven. On the contrary, the three subgenomes carry the same proportion of TaNAC genes, which is consistent with the distribution observed in durum wheat, a diploid species [21]. This result suggests that TaNAC family expansion was likely prior to a polyploidization event in bread wheat.
Expansion of the wheat TaNAC genes family due to small-scale duplications
Several recent studies have proposed different mechanisms to explain the duplication events and gene expansion in plants. These proposals included, for instance, retrotransposition, duplicated DNA transposition, unequal crossing-over, and polyploidization [68–70]. In addition to these mechanisms, Wicker et al. [71] then Glover et al. [72] proposed another mechanism leading to gene movement and expansion. In particular, Glover et al. [72] showed that massive small-scale interchromosomal gene duplications occurred in the bread wheat genome after double-strand DNA break repair. Consequently, many non-syntenic genes have high sequence similarity. Regardless of the mechanisms involved in gene duplication, this may eventually lead to a pseudogenization, a conservation of the gene function, a subfunctionalization or a neofunctionalization of duplicate genes [73].
In this study, we noticed that several clades in the phylogenetic tree contained sequences that were either physically tightly linked on the same chromosome or mapped to a non-homoeologous group, suggesting that both intra and inter-chromosomal duplication events occurred. To test the possibility of small-scale duplications, we focused on three genomic regions on the chromosomes 4B, 4D, and 5A that encompass 11 TaNAC genes, which display highly similar sequences and similar genomic structures (2 exons). By aligning the sequences at the vicinity of these genes, we uncovered a high homology between the genomic regions (Fig 3). This finding support the hypothesis evoked by Glover et al. [72] about the expansion of the multi-genes family that is partially due to small-scale duplication driven by repetitive sequences.
Expansion of the wheat TaNAC genes family due to retroposition
Of the 361 high-quality sequences, 68 TaNAC genes (19%) are intronless, which is a characteristic of retroposons [74]. The subfamily NAC-h includes 65 of these 68 genes that are grouped in the same clade of the phylogenetic tree. However, in addition to an absence of intron, the presence of a poly(A) tract at the 3’end and a TSD (Target Site Duplication) in a gene sequence constitutes a hallmark of a retroposition event [74]. However, we failed to identify any of these signatures in these sequences. These signatures were possibly obscured by multiple mutations during evolution to such extent that they are no longer recognisable. Furthermore, several intronless TaNAC genes of this clade are grouped into homoeologous groups suggesting that retroposition/duplication events probably occurred prior to wheat polyploidization.
Interestingly, the distribution of TaNAC intronless genes among chromosomes was found not to be equivalent to the one observed for the whole TaNAC family. For example, although the gene proportions of the whole TaNAC family on the chromosome 3 and 5 are close (13 and 16% respectively), a higher proportion of intronless TaNAC genes was noticed on chromosome 3 (29% of the total intronless genes) whereas no intronless genes were observed on chromosome 5.
Taken together, these results indicate that both small-scale duplication and retroposition phenomena led to the expansion of this family of genes. Furthermore, these mechanisms could act in combination, i.e. a gene retroposition could be followed by small-scale duplications driven by repetitive sequences at the very vicinity of the gene.
For example, TraesCS2A01G328100, TraesCS2A01G328200, TraesCS2A01G328300 and TraesCS2A01G328500 are intronless and tightly linked genes, and could be an example of the retroposition/duplication events that could have occurred during evolution.
Expression of TaNAC genes under drought field conditions
As a major widely spread environmental stress, drought is known to significantly affect wheat yields worldwide. Interactions between plant development and stress response are complex, especially in the field, where crops are submitted to a combination of environmental variables (such as water availability and temperature). Although a few NAC TFs have been studied in wheat compared with the model plants Arabidopsis and rice, it has been shown that they may also be involved in response to environmental conditions [5]. Some TaNAC genes have already been studied in bread wheat. For example, Arabidopsis overexpressing TaNAC2, TaNAC29, or TaNAC67 display improved tolerance to multiple abiotic stresses [16,42,75]. In the same way, TaNAC2a, TaNAC2D, TaNAC4a, TaNAC6, TaNAC7, TaNAC13, and TaNTL5 were induced by water stress, high salinity, and/or low temperature, at different intensities [76,77]. Recently, TaRNAC1 was found to enhance drought tolerance leading to higher biomass, grain yield, and root length [78]. In addition, transgenic wheat overexpressing TaNAC47 showed a drought-tolerant phenotype that may be induced in response to ABA [79].
Here, we have studied 23 TaNAC genes that we identified as the most up-regulated (p-value < 0.05) in specific organs (leaf or grain) or in response to treatments (drought and heat) (S7 Table). Among the 23 selected genes, 20 belong to the subfamilies NAC-a to d, which have been described as Stress Related NAC containing subfamilies [80] (S3 Table).
Some TaNAC genes exhibit an extreme level of transcript abundance specifically depending on the organ
NAC genes are recognized to be largely involved in many developmental and growth processes, such as the differentiation of major organs [81]. Our study highlighted genes that are preferentially expressed in the leaf (16) or in the grain (7) regardless of the genotype, the grain developmental stage, or the field conditions.
Interestingly, of those genes, seven leaf-specific and five grain-specific genes were found, with an expression level at least 10 times higher than in the other organ (S7 Table), suggesting their specific implication in the leaf or grain development. For example, TraesCS7A01G305200.1 gene up-regulated in leaves either in the earliest grain developmental stage (220°Cd) and the grain filling stage (450°Cd), was found to be similar to TaNAC1/TaNACS already described by Zhao et al. [37]. In this work, the authors highlighted an increase in nitrogen concentration in wheat grains from lines over-expressing the TaNAC-S protein (TaNAC1) and a stay-green phenotype. These authors concluded that NAC TFs, expressed mainly in the leaves, may participate to delay post-anthesis foliar senescence (stay-green phenotype) through a better remobilization of the nutrients during senescence. Another gene responding mainly in leaf in our experiment (TraesCS4D01G176000.1) is homologous to a NAC gene in the rose, RhNAC100, that has been characterized as a repressor of cellular expansion of rose petals [82], suggesting its involvement in the determination of cell size.
Among the seven up-regulated TaNAC grain-specific genes, one gene (TraesCS7A01G569100.1) was found to be homologous to ONAC20 and ONAC026 described in rice [24]. In this latter study, the authors showed that three NAC genes (ONAC020, ONAC026 and ONAC023) are expressed specifically during seed development at a very high level, suggesting a potential role of these genes, via their heteromerization capability, in the regulation of the grain size/weight. Another gene, TraesCS3A01G077900.1, was found to be homologous to a barley HvNAC019 gene which is preferentially expressed in grain, indicating that this gene could regulate seed development in association with two other genes (HvNAC017 and HvNAC018) [83].
Leaf-specific TaNAC genes are up-regulated at the grain filling stage in response to drought
Sixteen genes were found differentially expressed in both leaves and grains between the early grain developmental stage (cell division) and the grain filling stage, and mainly up-regulated at 450°Cd (15 genes).
Drought stress during grain filling stimulates leaf senescence and enhances remobilization of water-soluble carbohydrates to the grains [84]. Interestingly, we found a set of eight TaNAC genes that were preferentially expressed in leaves during grain filling in response to drought. This finding may indicate that these genes could participate in the resource remobilization from the leaves to the grains. For example, the three members of the same homoeologous group, located on chromosome 5 (TraesCS5A01G143200.1 (Fig 9A), TraesCS5B01G142100.1 and TraesCS5D01G148800.1) were found similar to TaNAC69. This gene has been well-characterized as a drought stress regulator [85], and was shown to improve grain yield. However, its expression was found mainly in roots compared to leaves [65,85,86]. It is worthy to note that, in our study, the genome A copy-gene was expressed at a higher level than the two other homoeologous copies in leaves. This finding may suggest that the B and D copies could be related to the root-specific expression that has not been measured in this study. The homologous gene in rice (OsNAC10) has also been demonstrated to be involved in drought adaptation mainly by enhancing the level of stress-protection genes, such as chitinase, ZIM family gene, and glyoxalase I family gene; and increases grain yield significantly under field drought conditions [87]. Also, two members of the same homoeologous group (TraesCS4D01G176000, TraesCS4B01G174000) are homologs to the rice gene ONAC39 (OSCUC1/ONAC039) that has also been described by Jeong et al. [87] as a drought responsive gene (fold change: 9.44). The genes of this homoeologous group in wheat may have similar functions and indicates that these genes may be associated with drought stress.
Another wheat gene (TraesCS2A01G101100, Fig 8C) was up-regulated in leaves under drought at 450°Cd in our field conditions. In contrast, its homolog, TaGRAB2, which was involved in response to biotic stress (a plant DNA virus Geminiviridae), showed an expression pattern barely detectable in leaves [39]. These findings suggest that the same gene could be expressed or not in leaves, depending on the stress.
Finally, no characterized homolog has been found in the literature for TraesCS3B01G303800.1 and TraesCS6D01G390200.1. These genes may be interesting candidates for field drought tolerance and require further investigations.
Grain-specific TaNAC genes respond differentially to drought according to the genotype
Five TaNAC genes (TraesCS3A01G077900.1, TraesCS5A01G245900.2, TraesCS7A01G569100.1, TraesCS7B01G056300.1, and TraesCS7B01G100300.1) are grain-specific. Interestingly, their behavior in response to drought varies according to the genotype. They are down-regulated under drought treatment during the early grain developmental stage (cell division) in the genotype which exhibited the highest number of spikes/m2 (Fig 7B) (Altigo); whereas they are down-regulated (4 genes) or up-regulated later during the grain filling stage (TraesCS7A01G569100.1), in the other genotype (Allez-y) (Fig 9B, Fig 9C). These results suggest that, depending on the genetic background, those genes may harbor different alleles that could affect their drought tolerance.
Four of these five genes (TraesCS7A01G569100.1, TraesCS5A01G245900.2, TraesCS7B01G056300.1, and TraesCS7B01G100300.1) have been found previously differentially expressed in grain in response to a moderate warming of +8°C applied during the grain cell division stage [88]. These results suggest that these four TaNAC genes may be involved in response to close abiotic stresses (drought and high temperature), such as other NAC genes like MlNAC9, SlNAC3, ONAC022, GmNAC3 and GmNAC4, that were described involved in several environmental stresses [19,89–91].
Conclusion
In conclusion, we performed a comprehensive study of NAC TFs in the bread wheat genome (T. aestivum), cultivar Chinese Spring. We identified a high quality set of 361 TaNAC genes in common with those described in Borrill et al. [45]. We provided evidences as to how small scale duplications and retroposition contributed to the expansion of this family of genes. Finally, the expression patterns of 23 TaNAC genes in two organs at two stages under drought in the field, combined with our study of duplication events, revealed a functional diversification of this gene family’s members. Further functional characterizations of five grain specific TaNAC genes are in process to identify their role in the wheat grain development and in abiotic stress responses.
Supporting information
Using their chromosomal location information, 451 sequences have been mapped on the seven chromosomes of the three wheat subgenomes A (red), B (green), and D (blue). The first seven letters of the sequence names have been removed (e.g. 4B01G328600 refers to TraesCS4B01G328600, located on the chromosome 4B according to the IWGSC annotation). The position of each TaNAC can be estimated using the scale on the left. The two circles show the chromosomal regions studied as an example of duplication.
(TIF)
The percentage of sequences is indicated at the top of each histogram.
(TIF)
The blue and red boxes focused on the clades studied as case of duplication. The number of exons is represented by a red (1 exon), pink (2 exons), yellow (3 exons), green (4 exons), clear blue (5 exons), dark blue (6 exons), or violet (7 and more exons) circle.
(PDF)
Each gene location is indicated on the x- and y-axis by a blue arrow. Green and red dots indicate sense and antisense homologies respectively.
(TIF)
Pearson correlations between duplicated genes of the studied 2A-2B-2D chromosomal regions during development (A) and stress (B).
(TIF)
Expression of the 23 TaNAC genes in leaves and grains of Altigo (square) and Allez-y (circle) genotypes at 220°Cd (full form) and 450°Cd (empty form) in irrigated (blue) and drought (red) conditions.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The preferential expression have been verified by a Student Test, with an e-value threshold at 0.05.
(XLSX)
Acknowledgments
We would like to thank C. Dupuits and the Gentyane Plateform for their technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77. 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- 2.Khan S-A, Li M-Z, Wang S-M, Yin H-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int J Mol Sci. 31 Mai 2018;19(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 11 Févr 2014;111(6):2367–72. 10.1073/pnas.1316278111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 15 Déc 2000;290(5499):2105–10. [DOI] [PubMed] [Google Scholar]
- 5.Gahlaut V, Jaiswal V, Kumar A, Gupta PK. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor Appl Genet. November 2016;129(11):2019–42. 10.1007/s00122-016-2794-z [DOI] [PubMed] [Google Scholar]
- 6.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. Juin 1998;379(6):633–46. [DOI] [PubMed] [Google Scholar]
- 7.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 10 October 2000;97(21):11632–7. 10.1073/pnas.190309197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. Mars 2002;7(3):106–11. [DOI] [PubMed] [Google Scholar]
- 9.Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. The Plant Cell. 1 Janv 2003;15(1):63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. sept 2004;39(6):863–76. [DOI] [PubMed] [Google Scholar]
- 11.Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. sept 2004;16(9):2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran L-SP, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. Janv 2007;49(1):46–63. 10.1111/j.1365-313X.2006.02932.x [DOI] [PubMed] [Google Scholar]
- 13.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol Biol. 1 Janv 2006;60(1):107–24. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- 14.Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. October 2011;62(14):4731–48. 10.1093/jxb/err210 [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Liu S, Meng C, Qin L, Kong L, Xia G. WRKY Transcription Factors in Wheat and Their Induction by Biotic and Abiotic Stress. Plant Molecular Biology Reporter. October 2013;31(5):1053–67. [Google Scholar]
- 16.Huang Q, Wang Y, Li B, Chang J, Chen M, Li K, et al. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 4 November 2015;15:268 10.1186/s12870-015-0644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 1 October 2010;465(1–2):30–44. 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Chen G, Zhu J, Zhu Y, Lu X, Li X, et al. Molecular Characterization and Expression Profiling of NAC Transcription Factors in Brachypodium distachyon L. PLOS ONE. 7 October 2015;10(10):e0139794 10.1371/journal.pone.0139794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinheiro G, Marques C, Costa M, A.B. Reis P, Alves M, Carvalho C, et al. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 1 Juill 2009;444:10–23. 10.1016/j.gene.2009.05.012 [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Sun Q-P, Zhang D-F, Wang T-Y, Pan J-B. Identification of 7 stress-related NAC transcription factor members in maize (Zea mays L.) and characterization of the expression pattern of these genes. Biochem Biophys Res Commun. 26 Juin 2015;462(2):144–50. 10.1016/j.bbrc.2015.04.113 [DOI] [PubMed] [Google Scholar]
- 21.Saidi MN, Mergby D, Brini F. Identification and expression analysis of the NAC transcription factor family in durum wheat (Triticum turgidum L. ssp. durum). Plant Physiol Biochem. Mars 2017;112:117–28. 10.1016/j.plaphy.2016.12.028 [DOI] [PubMed] [Google Scholar]
- 22.Moyano E, Martínez-Rivas FJ, Blanco-Portales R, Molina-Hidalgo FJ, Ric-Varas P, Matas-Arroyo AJ, et al. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLOS ONE. 3 Mai 2018;13(5):e0196953 10.1371/journal.pone.0196953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 31 Déc 2003;10(6):239–47. [DOI] [PubMed] [Google Scholar]
- 24.Mathew IE, Das S, Mahto A, Agarwal P. Three Rice NAC Transcription Factors Heteromerize and Are Associated with Seed Size. Front Plant Sci. 2016;7:1638 10.3389/fpls.2016.01638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 1 Déc 2000;14(23):3024–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Y-J, Wei W, Song Q-X, Chen H-W, Zhang Y-Q, Wang F, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. October 2011;68(2):302–13. 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- 27.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. Févr 2005;10(2):79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 28.Kim S-G, Lee S, Seo PJ, Kim S-K, Kim J-K, Park C-M. Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics. Janv 2010;95(1):56–65. 10.1016/j.ygeno.2009.09.003 [DOI] [PubMed] [Google Scholar]
- 29.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 19 Avr 1996;85(2):159–70. [DOI] [PubMed] [Google Scholar]
- 30.Sablowski RW, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 9 Janv 1998;92(1):93–103. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. Mai 2006;46(4):601–12. 10.1111/j.1365-313X.2006.02723.x [DOI] [PubMed] [Google Scholar]
- 32.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. November 2005;17(11):2993–3006. 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y-S, Kim S-G, Park J-E, Park H-Y, Lim M-H, Chua N-H, et al. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. November 2006;18(11):3132–44. 10.1105/tpc.106.043018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S-G, Lee A-K, Yoon H-K, Park C-M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. Juill 2008;55(1):77–88. 10.1111/j.1365-313X.2008.03493.x [DOI] [PubMed] [Google Scholar]
- 35.Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 24 November 2006;314(5803):1298–301. 10.1126/science.1133649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA. 8 Juill 2014;111(27):10013–8. 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao D, Derkx AP, Liu D-C, Buchner P, Hawkesford MJ. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol (Stuttg). Juill 2015;17(4):904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X, Qu B, Li W, Zhao X, Teng W, Ma W, et al. The Nitrate-Inducible NAC Transcription Factor TaNAC2-5A Controls Nitrate Response and Increases Wheat Yield. Plant Physiol. November 2015;169(3):1991–2005. 10.1104/pp.15.00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Q, Sanz-Burgos AP, Guo H, García JA, Gutiérrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol. Mars 1999;39(4):647–56. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Lin R, Feng J, Chen W, Qiu D, Xu S. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci. 2015;6:108 10.3389/fpls.2015.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA. 29 Août 2006;103(35):12987–92. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao X, Chen S, Li A, Zhai C, Jing R. Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE. 2014;9(1):e84359 10.1371/journal.pone.0084359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang Y, Liao K, Du H, Xu Y, Song H, Li X, et al. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot. November 2015;66(21):6803–17. 10.1093/jxb/erv386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol. Juill 2007;24(7):1506–17. 10.1093/molbev/msm077 [DOI] [PubMed] [Google Scholar]
- 45.Borrill P, Harrington SA, Uauy C. Genome-Wide Sequence and Expression Analysis of the NAC Transcription Factor Family in Polyploid Wheat. G3 (Bethesda). 07 2017;7(9):3019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017;27(5):885–96. 10.1101/gr.217117.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium (IWGSC) TIWGS, Investigators IR principal, Appels R, Eversole K, Feuillet C, Keller B, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 17 Août 2018;361(6403):eaar7191 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- 48.Borrill P, Ramirez-Gonzalez R, Uauy C. expVIP: a Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016;170(4):2172–86. 10.1104/pp.15.01667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okonechnikov K, Golosova O, Fursov M, UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 15 Avr 2012;28(8):1166–7. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 50.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 1 Mai 2014;30(9):1236–40. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 1 October 2010;26(19):2460–1. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noé L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 1 Juill 2005;33(Web Server issue):W540–543. 10.1093/nar/gki478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, et al. Structural and functional partitioning of bread wheat chromosome 3 B. Science. 18 Juill 2014;345(6194):1249721. [DOI] [PubMed] [Google Scholar]
- 56.Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, et al. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 20 Juin 2015;15:152 10.1186/s12870-015-0511-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 20 Avr 2015;43(7):e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 15 November 2011;27(22):3209–10. 10.1093/bioinformatics/btr490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen RG, Pereira LS, Raes D, Smith M. Crop evapotranspiration—Guidelines for computing crop water requirements—FAO Irrigation and drainage paper 56:15. [Google Scholar]
- 60.Slafer GA, Savin R, Sadras VO. Coarse and fine regulation of wheat yield components in response to genotype and environment. Field Crops Research. 15 Févr 2014;157:71–83. [Google Scholar]
- 61.Capron D, Mouzeyar S, Boulaflous A, Girousse C, Rustenholz C, Laugier C, et al. Transcriptional profile analysis of E3 ligase and hormone-related genes expressed during wheat grain development. BMC Plant Biol. 14 Mars 2012;12:35 10.1186/1471-2229-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. Déc 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 63.Tang Y, Horikoshi M, Li W. ggfortify: Unified Interface to Visualize Statistical Results of Popular R Packages. 2016;8:12. [Google Scholar]
- 64.Baloglu MC, Oz MT, Oktem HA, Yucel M. Expression Analysis of TaNAC69-1 and TtNAMB-2, Wheat NAC Family Transcription Factor Genes Under Abiotic Stress Conditions in Durum Wheat (Triticum turgidum). Plant Mol Biol Rep. 1 October 2012;30(5):1246–52. [Google Scholar]
- 65.Baloglu MC, Inal B, Kavas M, Unver T. Diverse expression pattern of wheat transcription factors against abiotic stresses in wheat species. Gene. 15 October 2014;550(1):117–22. 10.1016/j.gene.2014.08.025 [DOI] [PubMed] [Google Scholar]
- 66.Li X, Feng B, Zhang F, Tang Y, Zhang L, Ma L, et al. Bioinformatic Analyses of Subgroup-A Members of the Wheat bZIP Transcription Factor Family and Functional Identification of TabZIP174 Involved in Drought Stress Response. Front Plant Sci. 2016. November 16;7:1643 10.3389/fpls.2016.01643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng X, Zhao Y, Li X, Wu M, Chai W, Sheng L, et al. Genomewide identification, classification and analysis of NAC type gene family in maize. J Genet. sept 2015;94(3):377–90. [DOI] [PubMed] [Google Scholar]
- 68.Huo N, Zhang S, Zhu T, Dong L, Wang Y, Mohr T, et al. Gene Duplication and Evolution Dynamics in the Homeologous Regions Harboring Multiple Prolamin and Resistance Gene Families in Hexaploid Wheat. Front Plant Sci. 2018. May 23;9:673 10.3389/fpls.2018.00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ning P, Liu C, Kang J, Lv J. Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ. 4 Mai 2017;5:e3232 10.7717/peerj.3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, et al. Identification and Characterization of Shared Duplications between Rice and Wheat Provide New Insight into Grass Genome Evolution. The Plant Cell. 1 Janv 2008;20(1):11–24. 10.1105/tpc.107.056309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wicker T, Buchmann JP, Keller B. Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. sept 2010;20(9):1229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glover NM, Daron J, Pingault L, Vandepoele K, Paux E, Feuillet C, et al. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 9 September 2015;16:188 10.1186/s13059-015-0754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R. Gene duplication as a major force in evolution. Journal of Genetics. Avr 2013;92(1):155–61. [DOI] [PubMed] [Google Scholar]
- 74.Brosius J. Retroposons—seeds of evolution. Science. 15 Févr 1991;251(4995):753 [DOI] [PubMed] [Google Scholar]
- 75.Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot. Mai 2012;63(8):2933–46. 10.1093/jxb/err462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang Y, Liu M, Gao S, Zhang Z, Zhao X, Zhao C, et al. Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol Plant. Mars 2012;144(3):210–24. 10.1111/j.1399-3054.2011.01539.x [DOI] [PubMed] [Google Scholar]
- 77.Huang Q, Wang Y. Overexpression of TaNAC2D displays opposite responses to abiotic stresses between seedling and mature stage of transgenic Arabidopsis. Front. Plant Sci. 2016. November 23;7:1754 10.3389/fpls.2016.01754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen D, Chai S, McIntyre CL, Xue G-P. Overexpression of a predominantly root-expressed NAC transcription factor in wheat roots enhances root length, biomass and drought tolerance. Plant Cell Rep. Févr 2018;37(2):225–37. 10.1007/s00299-017-2224-y [DOI] [PubMed] [Google Scholar]
- 79.Zhang L, Zhang L, Xia C, Zhao G, Jia J, Kong X. The Novel Wheat Transcription Factor TaNAC47 Enhances Multiple Abiotic Stress Tolerances in Transgenic Plants. Front Plant Sci. 2016. January 18;6:1174 10.3389/fpls.2015.01174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen H, Yin Y, Chen F, Xu Y, Dixon RA. A Bioinformatic Analysis of NAC Genes for Plant Cell Wall Development in Relation to Lignocellulosic Bioenergy Production. BioEnergy Research. Déc 2009;2(4):217–32. [Google Scholar]
- 81.Mathew IE, Agarwal P. May the Fittest Protein Evolve: Favoring the Plant-Specific Origin and Expansion of NAC Transcription Factors. Bioessays. 25 Juin 2018;e1800018 10.1002/bies.201800018 [DOI] [PubMed] [Google Scholar]
- 82.Pei H, Ma N, Tian J, Luo J, Chen J, Li J, et al. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. oct 2013;163(2):775–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christiansen MW, Holm PB, Gregersen PL. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res Notes. 19 Août 2011;4:302 10.1186/1756-0500-4-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang B, Li W, Chang X, Li R, Jing R. Effects of Favorable Alleles for Water-Soluble Carbohydrates at Grain Filling on Grain Weight under Drought and Heat Stresses in Wheat. Ali J, éditeur. PLoS ONE. 18 Juill 2014;9(7):e102917 10.1371/journal.pone.0102917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xue G-P, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant. Juill 2011;4(4):697–712. 10.1093/mp/ssr013 [DOI] [PubMed] [Google Scholar]
- 86.Chen D, Richardson T, Chai S, Lynne McIntyre C, Rae AL, Xue G-P. Drought-Up-Regulated TaNAC69-1 is a Transcriptional Repressor of TaSHY2 and TaIAA7, and Enhances Root Length and Biomass in Wheat. Plant Cell Physiol. October 2016;57(10):2076–90. 10.1093/pcp/pcw126 [DOI] [PubMed] [Google Scholar]
- 87.Jeong JS, Kim YS, Baek KH, Jung H, Ha S-H, Do Choi Y, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. Mai 2010;153(1):185–97. 10.1104/pp.110.154773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Girousse C, Roche J, Guerin C, Le Gouis J, Balzegue S, Mouzeyar S, et al. Coexpression network and phenotypic analysis identify metabolic pathways associated with the effect of warming on grain yield components in wheat. PLoS ONE. 2018;13(6):e0199434 10.1371/journal.pone.0199434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao X, Yang X, Pei S, He G, Wang X, Tang Q, et al. The Miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene. 15 Juill 2016;586(1):158–69. 10.1016/j.gene.2016.04.028 [DOI] [PubMed] [Google Scholar]
- 90.Han Q, Zhang J, Li H, Luo Z, Ziaf K, Ouyang B, et al. Identification and expression pattern of one stress-responsive NAC gene from Solanum lycopersicum. Mol Biol Rep. Févr 2012;39(2):1713–20. 10.1007/s11033-011-0911-2 [DOI] [PubMed] [Google Scholar]
- 91.Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front Plant Sci. 2016;7:4 10.3389/fpls.2016.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia N, Zhang G, Sun Y-F, Zhu L, Xu L-S, Chen X-M, et al. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiological and Molecular Plant Pathology. 1 September 2010;74(5):394–402. [Google Scholar]
- 94.Macovei A, Tuteja N. microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 8 October 2012;12:183 10.1186/1471-2229-12-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, et al. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics. September 2010;284(3):173–83. 10.1007/s00438-010-0557-0 [DOI] [PubMed] [Google Scholar]
- 96.Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. Mai 2014;65(8):2119–35. 10.1093/jxb/eru072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using their chromosomal location information, 451 sequences have been mapped on the seven chromosomes of the three wheat subgenomes A (red), B (green), and D (blue). The first seven letters of the sequence names have been removed (e.g. 4B01G328600 refers to TraesCS4B01G328600, located on the chromosome 4B according to the IWGSC annotation). The position of each TaNAC can be estimated using the scale on the left. The two circles show the chromosomal regions studied as an example of duplication.
(TIF)
The percentage of sequences is indicated at the top of each histogram.
(TIF)
The blue and red boxes focused on the clades studied as case of duplication. The number of exons is represented by a red (1 exon), pink (2 exons), yellow (3 exons), green (4 exons), clear blue (5 exons), dark blue (6 exons), or violet (7 and more exons) circle.
(PDF)
Each gene location is indicated on the x- and y-axis by a blue arrow. Green and red dots indicate sense and antisense homologies respectively.
(TIF)
Pearson correlations between duplicated genes of the studied 2A-2B-2D chromosomal regions during development (A) and stress (B).
(TIF)
Expression of the 23 TaNAC genes in leaves and grains of Altigo (square) and Allez-y (circle) genotypes at 220°Cd (full form) and 450°Cd (empty form) in irrigated (blue) and drought (red) conditions.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The preferential expression have been verified by a Student Test, with an e-value threshold at 0.05.
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.