Abstract
Mitochondrial ATP synthase produces the majority of ATP in eukaryotic cells and its dimerization is necessary to create the inner membrane folds, or cristae, characteristic of mitochondria. Proton translocation through the membrane-embedded F0 region turns the rotor that drives ATP synthesis in the soluble F1 region. While crystal structures of the F1 region have illustrated how rotation leads to ATP synthesis, the lack of an experimental atomic model for the FO region has prevented understanding how proton translocation produces rotation. We determined the structure of the dimeric FO complex from Saccharomyces cerevisiae at 3.6 Å resolution by electron cryomicroscopy. The structure reveals the proton path through the complex, how the complex dimerizes, and suggests how it bends the membrane to produce cristae.
One sentence summary
The structure of mitochondrial ATP synthase FO complex explains dimerization, membrane bending, and proton translocation.
During mitochondrial respiration, the protein complexes of the electron transport chain pump protons from the mitochondrial matrix to the intermembrane space (IMS). The resulting proton motive force across the mitochondrial inner membrane powers the adenosine triphosphate (ATP) synthase, a membrane-embedded multi-protein complex. Saccharomyces cerevisiae ATP synthase has a subunit composition similar to the mammalian enzyme (1). The complex comprises a soluble catalytic F1 region consisting of subunits α3β3γδε and a membrane-embedded FO region containing subunits a, b, e, f, g, i/j, k, l, 8, and a ring of ten c-subunits (Fig. 1A). During ATP synthesis, proton translocation from the IMS to the matrix through the FO region, at the interface of subunit a and the c-ring, drives rotation of the central rotor subcomplex (subunits γδεc10). This rotation induces conformational changes in the α3β3 hexamer of the F1 region that lead to synthesis of ATP from adenosine diphosphate (ADP) and phosphate (2). Proton translocation is thought to occur through two offset half-channels, one opening to the IMS and the other to the mitochondrial matrix (3, 4). Protons from the IMS half-channel neutralize conserved Glu residues in the c10-ring (E59 in S. cerevisiae), travel through the lipid bilayer as the ring rotates, and are delivered to the matrix half-channel. The α3β3 complex is prevented from rotating along with the central rotor by the peripheral stalk subcomplex, consisting of subunits OSCP, d, h, and the soluble region of subunit b. Mammals lack subunits i/j, k, and l, but possess the additional subunits DAPIT and 6.8L (5). Mitochondrial ATP synthase complexes assemble into long ribbons of dimers (6, 7) that, due to the presence of a domain containing subunits e and g in the FO region, bend the mitochondrial inner membrane (7–11). These bends are needed to form cristae, highly folded membrane structures that maximize the surface area available for respiration, giving mitochondria their characteristic appearance (6, 7). X-ray crystallography has revealed the atomic structures of most of the soluble subunits of mitochondrial and bacterial ATP synthase (2). However, most of the membrane-embedded region of the complex (subunits a, b, e, f, g, i/j, k, l, and 8) has resisted high-resolution structural characterization. Despite recent advances, the resolution in electron cryomicroscopy (cryo-EM) maps of intact ATP synthases has been insufficient to allow construction of atomic models (9,12–15).
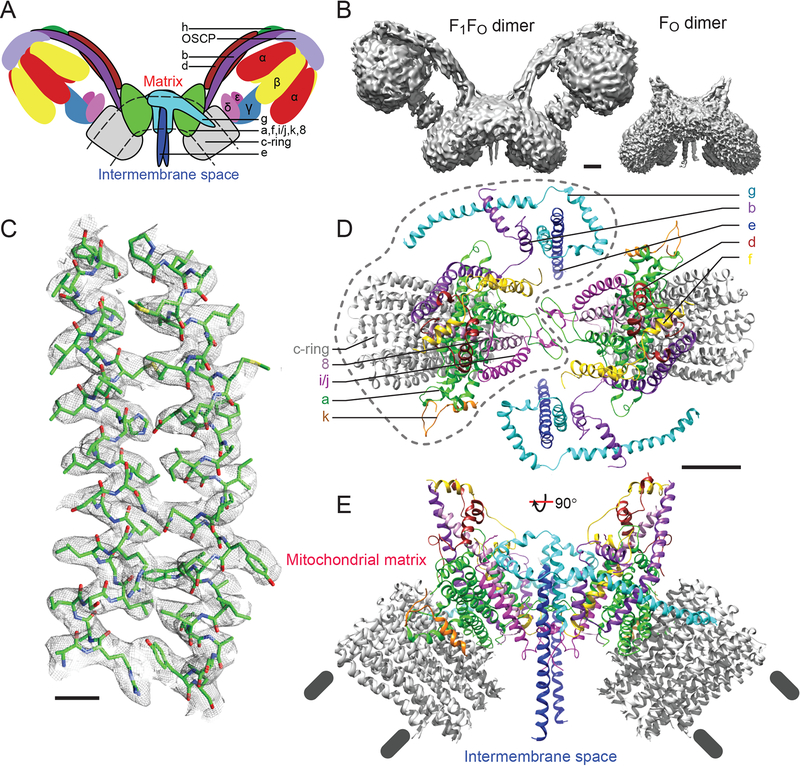
Figure 1. Overall structure of the FO complex.
A, Cartoon for the ATP synthase dimer with subunits found in the FO region outlined in black. B, The detergent glyco-diosgenin (GDN) extracts dimeric S. cerevisiae ATP synthase from mitochondrial membranes intact (left) or as the FO complex after sodium bromide treatment of the membranes (right). Scale bar, 25 Å. C, Example of map density that allowed construction of an atomic model (subunit a, residues A168 to F196 and P212 to L242). Scale bars, 5 Å. Top (D) and side (E) views of the FO dimer reveal the arrangement of subunits. One monomer is outlined with a dashed line in part D. Scale bar, 25 Å.
Cryo-EM has shown that rotary ATPases stop in different rotational states when extracted from membranes (9, 13, 15, 16). We hypothesized that this conformational heterogeneity limits cryo-EM resolution and that analysis of dissociated FO regions would reduce heterogeneity, improve resolution, and enable construction of an experimental atomic model. A similar approach was successful with the related Vo complex from the S. cerevisiae V-type ATPase (17). However, while yeast V-ATPases dissociate in response to glucose depletion (18), ATP synthases do not offer a physiological route to separating the F1 region from the FO region. Therefore, we used sodium bromide to dissociate the F1 regions from inside-out submitochondrial particles (19). Similar to the natural product digitonin (9), we found that the recently described synthetic detergent glyco-diosgenin (GDN) (20) extracts the dimeric form of ATP synthase from yeast mitochondrial membranes for both the intact ATP synthase and the FO complex (Fig. 1B). Cryo-EM of the FO complex allowed for determination of the structure to 3.6 Å resolution (Fig. 1C to E, Fig. S1). Side chain detail could be seen for subunits a, f, i/j, 8, the transmembrane α-helix of k, the c-subunits, the transmembrane α-helices of subunit b, and part of subunit d, allowing construction of atomic models for these subunits (Fig. S2, Table S1). Density for subunits e and g allowed construction of poly-alanine models for these proteins. The recently identified subunit l, which has no known function and a sequence similar to subunit k (1), could not be identified in the cryo-EM map. Together with existing atomic models of ATP synthase subcomplexes, the Fo complex structure determined here allows for construction of a nearly complete ‘mosaic model’ (2) of mitochondrial ATP synthase (Fig. S3).
The core subunits of the FO complex, found in both mitochondria and bacteria, are subunits a, b, and c. The fold observed for subunit a (Fig. 2A and B, green) matches the fold deduced from a low-resolution map of the bovine enzyme combined with evolutionary covariance analysis (13). The protein contains five transmembrane α-helices and an amphipathic α-helix (α-helix #2) that lies along the matrix surface of the detergent micelle. α-helices 3 and 4, and 5 and 6 form transmembrane hairpins, with the latter being the long and tilted hairpin that is characteristic of rotary ATPases (12, 13, 15, 16, 21). The simpler bacterial ATP synthase contains two copies of subunit b, each with a single N-terminal transmembrane α-helix and an elongated C-terminal region that constitutes most of the peripheral stalk (15). In contrast, the mitochondrial enzyme has a single subunit b with two N-terminal transmembrane α-helices. The two transmembrane α-helices of the mitochondrial subunit do not pack against each other, as suggested previously (13, 9,14). Instead, subunit b begins with a short, presumably amphipathic, α-helix and a transmembrane α-helix that form a domain with subunits e and g (Fig. 2A, enclosed by a dashed line). A short loop connects this domain to the second transmembrane α-helix of subunit b, which packs against subunit a. The soluble C-terminal portion of subunit b enters the mitochondrial matrix as part of the peripheral stalk (22, 23) and is mostly disordered in the isolated FO complex.
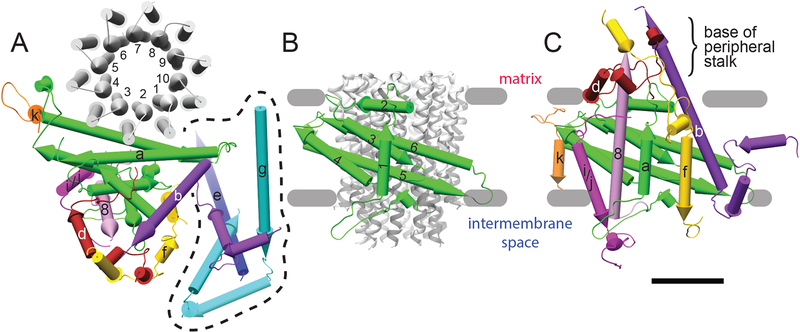
Figure 2. Subunit folds for individual subunits.
A, Matrix view of the FO complex monomer structure, with the domain formed by subunits e and g, and the N-terminal α-helices of subunit b enclosed by a dashed line. B, Fold of subunit a. C, Side view of the FO monomer showing that subunits 8, f, and b, contribute to the base of the peripheral stalk, with subunit d acting as a clip. Scale bar, 25 Å.
Subunits f and 8, which are essential for mitochondrial respiration, previously had no clear function in the ATP synthase. Subunit f (Fig. 2A and C, yellow) consists of a soluble N-terminal sequence of ~50 residues that binds to the base of the peripheral stalk, followed by a single transmembrane α-helix. The transmembrane α-helix of subunit f was mistaken for the first transmembrane α-helix of subunit b in lower-resolution cryo-EM studies of the bovine (13) and yeast (9, 14) enzymes. Subunit 8 (Fig. 2A and C, pink), known as A6L in mammals, is almost entirely α-helical, with the N-terminal part of the helix embedded in the membrane in contact with the first α-helix of subunit a and the C-terminal 14 residues contributing to the base of the peripheral stalk. Subunits f and 8 can therefore be assigned as components of the peripheral stalk. The C-terminal 56 residues of subunit d (Fig. 2A and C, brown), which were absent from earlier crystal structures (22, 23), wrap around subunit 8 and subunit b as they leave the detergent micelle to clamp together the base of the peripheral stalk. The structure shows that the base of the peripheral stalk in the mitochondrial ATP synthase has a significantly more complicated arrangement of subunits than predicted (22, 23) or than the bacterial enzyme (15).
Unlike the dimeric ATP synthase from Polytomella sp., which is held together by subunits specific to algae (12), the yeast dimer is held together by subunits a, i/j, k, and e (Fig 3A to C). The interface formed by subunit i/j, which was mistaken for subunit f in a lower-resolution structure (9), occurs through two short stretches of ~10 residues that pack together in the dimer (Fig. 3A and B, magenta). The lack of homologues for subunit i/j in mammals could explain why yeast ATP synthase dimers are more stable than mammalian dimers (9). Unexpectedly, subunit a also forms a dimer interface, with each monomer contributing two strands of a four-stranded planar structure (Fig. 3A and B, green) with one hydrophobic surface and one hydrophilic surface. This motif superficially resembles an antiparallel β-sheet, with amino acid side chains pointing away from the plane of the motif. The two dimerization motifs are stacked together, with the subunit i/j motif in the IMS and the subunit a motif on the IMS surface of the detergent micelle. In an alignment of subunit a sequences from different species (9), the dimerization motif begins just four residues N-terminal of the first residue in the bovine and human subunit a, which have significantly shorter N-terminal regions than the yeast subunit a. Consequently, the mammalian subunit a dimerization motif likely begins immediately before the subunit’s first transmembrane α-helix. Subunits k and e contribute the final monomer-monomer interaction. Subunit k possesses an N-terminal transmembrane α-helix in the map that is well ordered (Fig. 2, orange). Inspection of an unsharpened version of the map, which allows visualization of lower-resolution features, shows additional density from subunit k (Fig. 3C, orange). This density continues into the IMS to contact subunit e, which extends into the IMS as an α-helix (Fig. 2A and 3C, blue). The lack of this interaction in the bovine ATP synthase monomer may explain why the extended IMS α-helix of subunit e is in different orientations in the monomeric bovine ATP synthase and intact dimeric yeast ATP synthase (13, 9). It has been proposed that dissociation of ATP synthase dimers triggers opening of the mitochondrial permeability transition pore, which can initiate regulated cell death (24). Consequently, the dimer contacts described above warrant further investigation for their role in this phenomenon.
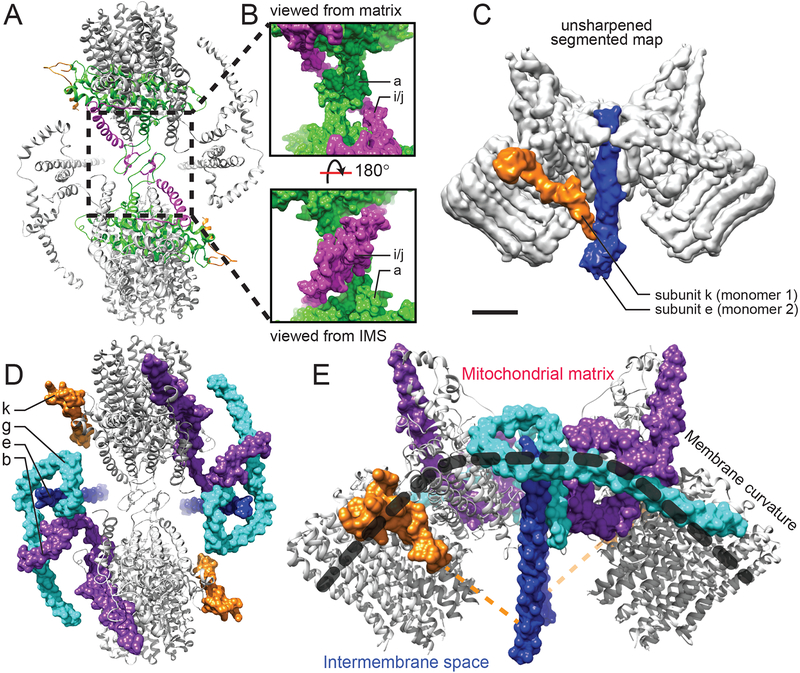
Figure 3. Dimerization and membrane bending.
A, Subunits a and i/j contribute dimerization motifs (enlarged in B). C, The unsharpened map shows that subunit k extends into the IMS to contact subunit e. D and E, Together, subunits b, e, g, and k create the structure that bends the lipid bilayer by almost 90°. The dashed orange line indicates the full length of subunit k. Scale bars, 25 Å.
The unusually-shaped domain containing subunits e, g, and the N-terminal ~50 residues of subunit b (Fig. 3D and E) is responsible for bending the mitochondrial inner membrane (11, 25). Subunit e (Fig. 3D and E, blue) begins with an N-terminal transmembrane α-helix that continues ~40 Å into the IMS. Although this feature is conserved from yeast to mammals, it does not appear to have a role in proton translocation or rotary catalysis. Subunit g has two N-terminal α-helices that lie on the matrix surface of the detergent micelle and a single transmembrane α-helix that interacts with subunit e, probably via the conserved GXXXG motifs of the two proteins (26, 27). The curved structure of the domain formed by subunits e, g, and b offers an explanation for how subunits e and g bend the lipid bilayer and why deletion of the genes for these subunits in yeast leads to defects in cristae formation (8). The combination of membrane-surface and membrane-embedded α-helices from these subunits, with further support from subunit k, creates a curved structure that enforces curvature of the lipid bilayer itself. The membrane surface α-helices of subunits g and b are reminiscent of the structure of BAR domains, which also induce curvature in lipid bilayers through amphipathic α-helices (28).
Proton translocation in ATP synthase involves subunit a and the c-ring. Remarkably, the c10-ring has a well-defined orientation relative to subunit a (Fig. 4), at least after the rapid cooling that occurs during cryo-EM specimen preparation. This constrained orientation shows that the highly tilted α-helices of subunit a maintain a tight interaction with the ring, although the rotor must still be able to turn during ATP synthesis. Two pores are visible in the FO complex that correspond to the two half-channels required for proton translocation (3, 4). A cavity on the IMS side of the detergent micelle behind the tilted α-helices 5 and 6 forms the IMS half-channel (Fig. 4A, left, circled in red) while an opening on the matrix side of the detergent micelle between subunit a and the c-ring forms the matrix half-channel (Fig. 4A, right, circled in red) (13, 12, 17). The positions of these half-channels are consistent with the proton translocation path proposed from lower-resolution studies of rotary ATPases (13, 9, 12, 16, 17, 21). R176 from subunit a, the only absolutely-required residue of the subunit for enzyme activity (29, 30), extends toward the c-ring (Fig. 4B). X-ray crystallography and simulations of isolated c-rings have shown that under deprotonating conditions the proton-carrying Glu residues adopt an extended conformation with their carboxyl groups facing away from the center of the c-ring (31). Under protonating conditions these residues fold toward the center of the ring in a conformation known as the proton-locked state (32). Although Glu and Asp residues are particularly sensitive to radiation damage and may have low density in cryo-EM maps (33), these residues often remain apparent (34, 35) and the E59 residues closest to the c10-ring are readily visible in the map (Fig. 4B). From the cryo-EM map (Fig. 4B), the conserved E59 residues of the two c-subunits nearest to R176 appear to be in deprotonated (negatively charged) conformations (31) but too far from Ri76 for either Glu to form a direct salt bridge with R176. The remaining c-subunits appear to be in the proton-bound (neutral) conformation (Fig. 4B). Consequently, the resting conformation of the c-ring in the FO structure differs subtly from the V-type enzyme where the Glu residue from one of the c-subunits was close enough to interact with the conserved Arg (17).
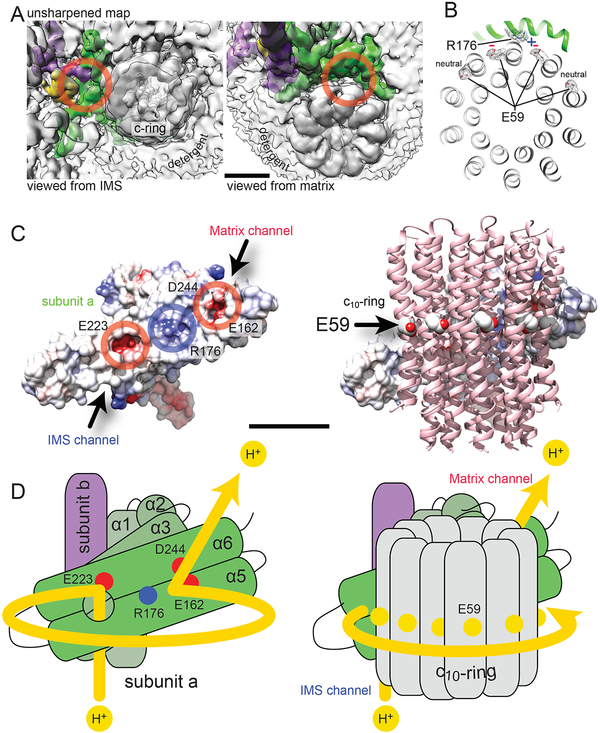
Figure 4. Proton translocation mechanism.
A, The unsharpened map reveals cavities in the complex that correspond to the IMS (left) and matrix (right) half-channels. B, The E59 residues of the two c-subunits nearest to R176 are in extended (deprotonated) conformations. The remaining E59 residues are in proton-bound conformations. C, There are two patches of negative charge on subunit a (left) where it interacts with the c-ring (right) that correspond with expected positions of the IMS and matrix half-channels. D, During proton translocation, protons follow the path indicated by the yellow line (left), entering the IMS half-channel behind transmembrane α-helices 5 and 6 of subunit a and exiting through the matrix half-channel between α-helices 5 and 6 and the c-ring (right). Scale bars, 25 Å.
The surface of subunit a that contacts the c-ring is mostly hydrophobic with a patch of positive charge from R176 (Fig. 4C, blue), and two patches of negative charge (red) at the expected positions of the half-channels. Near the matrix surface, these negative patches are due to E162 and D244, while near the IMS surface the negative charge is due to E223, all of which are conserved (9) and likely pass protons between the half-channels and the Glu residues of the c-ring. Numerous other conserved and functionally characterized residues of subunit a surround the locations of the two half-channels (Fig. S4). The positions of the half-channels and the orientations of the Arg and Glu residues support an emerging model for proton translocation: donation of a proton from the aqueous IMS half-channel via E223 neutralizes the E59 residue of a c-subunit. Neutralization of the Glu residue allows the c-ring to rotate due to Brownian motion (3, 4), counter clockwise when viewed from F1 towards FO (Fig. 4D), placing the residue in the hydrophobic environment of the lipid bilayer. Rotation of the c-ring brings a neutral E59 residue from a different c-subunit into the matrix half-channel where it is stripped of its proton by R176 of subunit a. This proton is accepted by E162 or D244, which in turn lose the proton to the mitochondrial matrix. Loss of the proton from the E59 residue in the matrix half-channel resets the motor for the next proton. The directionality and force of rotation in this model is governed by the difference in probability of a c-subunit binding a proton from either half-channel, which depends on the ΔG of proton translocation established by the proton motive force.
Supplementary Material
Acknowledgments
JLR conceived the project and experimental approach and supervised the research. SAB made the yeast strain and did some of the preliminary protein purification experiments. HG developed the final protein purification strategy, made cryo-EM specimens, collected cryo-EM data, calculated the cryo-EM maps, and built and refined the atomic models. JLR and HG wrote the manuscript and prepared the figures.
We thank David Mueller for advice on yeast growth and dissociation of the yeast ATP synthase, Richard Henderson for telling us about the detergent GDN, Bridget Carragher and Clint Potter for facilitating access to the Titan Krios, Hui (Alex) Wei for help with Titan Krios data collection, and Jean-Philippe Julien and Voula Kanelis for a critical reading of the manuscript. This work was supported by Canadian Institutes of Health Research operating grant MOP 81294. Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the NIH National Institute of General Medical Sciences (GM103310) and the Simons Foundation (349247). JLR was supported by the Canada Research Chairs program. HG was supported by a University of Toronto Excellence Award. Cryo-EM maps are deposited in the Electron Microscopy Data Bank under accession numbers EMD-7036 (FO dimer with C2 symmetry applied), EMD-7037 (FO dimer with C1 symmetry applied), and EMD-7067 (F1FO dimer with C2 symmetry applied); atomic models are deposited in the Protein Data Bank under accession numbers 6B2Z (FO dimer model) and 6B8H (F1FO dimer model).
Footnotes
Supplementary materials:
Materials and Methods
Supplementary results section (explanation of how densities were identified)
References
- 1.Liu S et al. , The purification and characterization of ATP synthase complexes from the mitochondria of four fungal species. Biochem J. 468, 167–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker JE, The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans 41, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Vik S, Antionio BJ, A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem 269, 30364–9. (1994). [PubMed] [Google Scholar]
- 4.Junge W, Lill H, Engelbrecht S, ATP synthase: an electrochemical transducer with rotary mechanics. Trends Biochem. Sci 22, 420–423 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Runswick MJ, Carroll J, Fearnley IM, Walker JE, Association of two proteolipids of unknown function with ATP synthase from bovine heart mitochondria. FEBS Lett. 581, 3145–3148 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Allen RD, Schroeder CC, Fok AK, An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J. Cell Biol 108, 2233–40 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W, Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 27, 1154–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paumard P et al. , The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn A et al. , Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol. Cell 63, 445–456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiko C et al. , Bovine F1FO ATP synthase monomers bend the lipid bilayer in 2D membrane crystals. Elife. 4, e06119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL, Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proc. Natl. Acad. Sci. U. S. A 109, 11675–11680 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allegretti M et al. , Horizontal membrane-intrinsic alpha-helices in the stator a-subunit of an F-type ATP synthase. Nature. 521, 237–240 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Zhou A et al. , Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. Elife. 4, e10180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinothkumar KR, Montgomery MG, Liu S, Walker JE, Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. Proc. Natl. Acad. Sci 113, 12709–12714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobti M et al. , Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. Elife. 5, e21598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, Benlekbir S, Rubinstein JL, Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 521, 241–245 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Mazhab-Jafari MT et al. , Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature. 539, 118–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kane PM, Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem 270, 17025–17032 (1995). [PubMed] [Google Scholar]
- 19.Tzagoloff A, Assembly of the mitochondrial membrane system III. Function and synthesis of the oligomycin sensitivity conferring protein of yeast mitochondria. J. Biol. Chem 245, 1545–51 (1970). [PubMed] [Google Scholar]
- 20.Chae PS et al. , A new class of amphiphiles bearing rigid hydrophobic groups for solubilization and stabilization of membrane proteins. Chemistry. 18, 9485–9490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schep DG, Zhao J, Rubinstein JL, Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Proc. Natl. Acad. Sci 113, 3245–3250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE, On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 25, 2911–2918 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees DM, Leslie AG, Walker JE, The structure of the membrane extrinsic region of bovine ATP synthase. Proc. Natl. Acad. Sci. U. S. A 106, 21597–21601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonora M et al. , Mitochondrial permeability transition involves dissociation of F1FO ATP synthase dimers and C-ring conformation. EMBO Rep. 18, 1077–1089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arselin G et al. , The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J. Biol. Chem 279, 40392–40399 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Bustos DM, Velours J, Segalen V, Sae S, The Modification of the conserved GXXXG motif of the membrane-spanning segment of subunit g destabilizes the supramolecular species of yeast ATP Synthase. J. Biol. Chem 280, 29004–29010 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Saddar S, Stuart RA, The yeast F1F0-ATP synthase: Analysis of the molecular organization of subunit g and the importance of a conserved GXXXG motif. J. Biol. Chem 280, 24435–24442 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Peter BJ et al. , BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 303, 495–500 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Lightowlers RN, Howitt SM, Hatch L, Gibson F, Cox GB, The proton pore in Escherichia coil FOF1-ATPase: a requirement for arginine at position 210 of the a-subunit. Biochem. Biophys. Acta 894, 399–406 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Cain BD, Simoni RD, Proton Translocation by the F1FOATPase of Escherichia coli -mutagenic analysis of the a subunit. J. Biol. Chem 264, 3292–3300 (1989). [PubMed] [Google Scholar]
- 31.Symersky J et al. , Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol 19, 485–491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogoryelov D, Yildiz O, Faraldo-Gomez JD, Meier T, High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol 16, 1068–1073 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Baker LA, Rubinstein JL, Radiation damage in electron cryomicroscopy. MethodEnzym. 481, 373–390 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Merk A et al. , Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell. 165, 1698–1707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S, Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy Alberto. Proc. Natl. Acad. Sci 111, 11709–11714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benlekbir S, Bueler SA, Rubinstein JL, Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol 19, 1356–1362 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Rubinstein JL, Dickson VK, Runswick MJ, Walker JE, ATP synthase from Saccharomyces cerevisiae: location of subunit h in the peripheral stalk region. J. Mol. Biol 345, 513–520 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Marr CR, Benlekbir S, Rubinstein JL, Fabrication of carbon films with approximately 500 nm holes for cryo-EM with a direct detector device. J. Struct. Biol 185, 42–47 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Meyerson JR et al. , Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci. Rep 4, 7084 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tivol WF, Briegel A, Jensen GJ, An improved cryogen for plunge freezing. Microsc. Microanal 14, 375–379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carragher B et al. , Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol 132, 33–45 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Rubinstein JL, Brubaker MA, Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol 192, 188–95 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Scheres SHW, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol, 5–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Brubaker MA, Benlekbir S, Rubinstein JL, Description and comparison of algorithms for correcting anisotropic magnification in cryo-EM images. J. Struct. Biol 192, 209–15 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–6 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K, Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Adams PD et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barad BA et al. , EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF et al. , UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Brown A, Long F, Robert A, Toots J, Emsley P, research papers Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions research papers. Acta Crystallogr D Biol Crystallogr, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arselin G et al. , The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur. J. Biochem 270, 1875–1884 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Belogrudov GI, Tomich JM, Hatefi Y, Membrane Topography and Near-neighbor Relationships of the Mitochondrial ATP Synthase Subunits e, f, and g*. J Biol Chem. 271, 20340–20345 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.