SUMMARY
Drosophila teissieri and D. yakuba diverged approximately 3 million years ago and are thought to share a large, ancestral, African range [1–3]. These species now co-occur in parts of continental Africa and in west Africa on the island of Bioko [1,4]. While D. yakuba is a human commensal, D. teissieri seems to be associated with Parinari fruits, restricting its range to forests [4–6]. Genome data indicate introgression, despite no evidence of contemporary hybridization. Here, we report the discovery of D. yakuba-D. teissieri hybrids at the interface of secondary forests and disturbed, open habitats on Bioko. We demonstrate that hybrids are the F1 progeny of D. yakuba females and D. teissieri males. At high temperatures like those found on Bioko, D. teissieri females are generally less receptive to mating; and in combination with temperature effects on egg lay and egg-to-adult viability, this decreases the potential for gene flow between ♀D. teissieri and ♂D. yakuba relative to the reciprocal cross. Field and laboratory experiments demonstrate that F1 hybrids have a maladaptive combination of D. yakuba behavior and D. teissieri physiology, generating additional barriers to gene flow. Nevertheless, analysis of introgressed and non-introgressed regions of the genome indicate that, while rare, gene flow is relatively recent. Our observations identify precise intrinsic and extrinsic factors that, along with hybrid male sterility, limit gene flow and maintain these species. These data contribute to a growing body of literature that suggests the Gulf of Guinea may be a hotspot for hybridization.
Keywords: Hybridization, Introgression, Reproductive isolation, Speciation, Thermal adaptation
eTOC Blurb
Cooper et al. document hybridization between Drosophila yakuba and D. teissieri on Bioko in west Africa. Temperature-dependent reproductive isolation limits the production of hybrids produced by D. teissieri females. A maladaptive combination of D. yakuba behavior and D. teissieri physiology in hybrids generates additional barriers to gene flow.
RESULTS AND DISCUSSION
Drosophila yakuba and D. teissieri hybridize along forest edges on Bioko
During three separate collection expeditions between 2009 and 2016, we sampled Bioko for D. yakuba-clade individuals. Using morphological variation to classify genotypes in the field (STAR Methods, Figure S1), we discovered that D. teissieri occurs mostly in higher altitude forests; in contrast, D. yakuba occurs primarily in open areas at all altitudes (200 – 2,020 m), but is most common at lower altitudes (Table S1). This suggests D. teissieri and D. yakuba are largely geographically separated on Bioko. However, we identified putative D. yakuba-D. teissieri hybrids near the center of the forest-open habitat ecotone at 1,200–2,020m. Hybridization has been long hypothesized between these two species [7,8], but this discovery is striking given that these species are twice as divergent (Ks = 11%, the synonymous substitutions per synonymous site) as the other naturally hybridizing pairs in the genus [9,10].
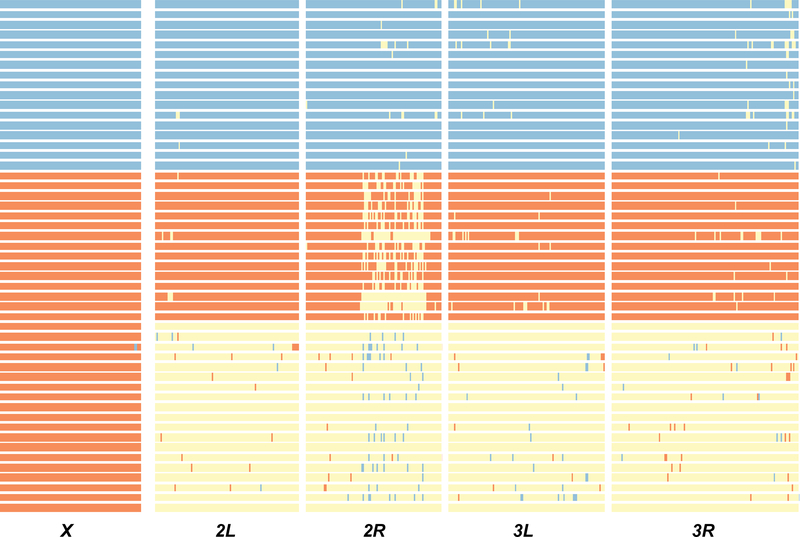
To confirm our discovery of hybrids based on morphology in the field, we sequenced the genomes of D. yakuba, D. teissieri, and putative-hybrid males. All 19-sequenced, putative-hybrid males were heterozygous for D. yakuba and D. teissieri ancestry across their autosomes and hemizygous for the D. yakuba X chromosome (Figure 1). This indicates that all hybrids were the F1 sons of D. yakuba females and D. teissieri males [denoted F1(♀yak × ♂tei)]. Interestingly, the strength of reproductive isolation (RI) between reciprocal D. yakuba-D. teissieri crosses does not differ under standard laboratory conditions [11,12], leaving an open question about our inability to sample hybrids produced by D. teissieri females [denoted F1(♀tei × ♂yak)] on Bioko (see below).
FIGURE 1. Chromosome view of inferred ancestries of wild-caught male D. yakuba (orange), D. teissieri (blue), and predicted hybrids based on morphology (lower third).
Chromosomes are denoted at the bottom of the plot. All individuals classified as hybrids in the field were heterozygous across the autosomes for both parental genotypes (yellow). Hemizygous D. yakuba ancestry on the X chromosome indicates that hybrids were F1 progeny of D. yakuba females and D. teissieri males. Small regions for which genotypes could not be confidently called are colored gray.
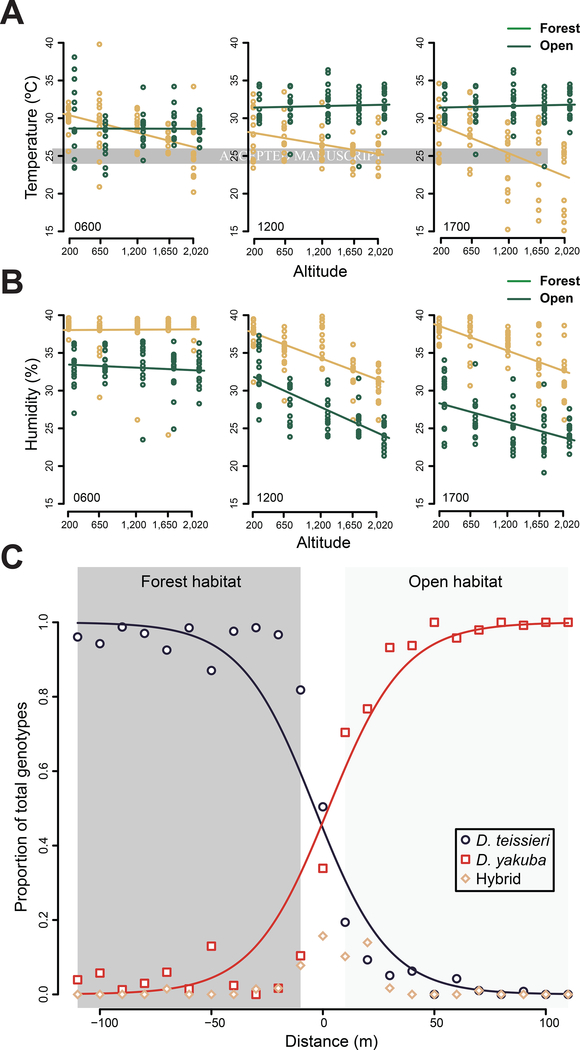
To understand habitat preferences, we performed mark-recapture field experiments on Bioko. We recaptured 8.45% (N = 91) of the D. teissieri, 11.31% (N = 137) of the D. yakuba, and 30% (N = 43) of the hybrid males that we released. In all cases, recaptured hybrid males did not have motile sperm, confirming our genotype assignments [11]. The estimated mean probability of choosing forest habitat differed among D. teissieri (0.97), D. yakuba (0.08), and hybrids (0.32) (LRT: P < 0.0001). Approximately 3% of D. teissieri, 15% of D. yakuba, and 19% of hybrids were recaptured at the center of the forest-open habitat ecotone. Because parental species and hybrids differ in their habitat preferences, we measured the temperature and humidity of each habitat on Bioko across several altitudes—these factors are known to influence the physiology and geographical distributions of Drosophila [13,14]. Both temperature and humidity decreased with altitude (Table S2, Figure 2A and 2B), and forest sites had lower temperatures than open areas at all times of day except at 0600hrs at low altitudes (Figure 2A). Forests were also more humid (Figure 2B). Together, these data indicate that D. yakuba and hybrids prefer warmer and drier conditions than those preferred by D. teissieri.
FIGURE 2. The preferred habitats of D. yakuba and D. teissieri.
A. Temperature conditions (°C) at different altitudes vary between forests and open habitats on Bioko (|t-value|= 2.357; P = 0.019) B. Humidity (%) is generally higher in forests than in open habitats (|t-value|= 2.634; P = 0.009). C. D. teissieri occurs in forests, D. yakuba occurs in open areas, and hybrids occur in a very narrow region centered on the forest-open habitat ecotone. Fitted lines from logistic regressions are plotted for D. yakuba and D. teissieri. For both A and B each panel reports conditions for a specific time of the day labeled in the bottom left corner of each plot. Regression lines for effects of altitude on each environmental condition are plotted. See also Tables S1 and S2.
We next estimated the prevalence of D. yakuba, D. teissieri, and their hybrids across the forest-open habitat ecotone. Including all flies visiting our traps, we captured 951 D. teissieri, 1,172 D. yakuba, and 59 hybrid males during the recapture portion of our mark-recapture experiment. Ninety-eight percent of the captured hybrids traveled less than 30m from the ecotone, and hybrids were never sampled more than 70m from the center of the ecotone (Figure 2C). D. teissieri was mostly absent from open habitat and became more common with distance into forest habitat (z = −22.147, P < 0.0001). In contrast, D. yakuba was mostly absent from forests and became more common with distance into the open habitat (z = 22.267, P < 0.0001; Figure 2C). These data indicate a sharp transition in the distributions of D. teissieri and D. yakuba at the center of the forest-open habitat ecotone where hybrids occur in a very narrow range.
Opposing patterns of dominance between parental alleles in hybrids contribute to the paucity of advanced-generation hybrids on Bioko
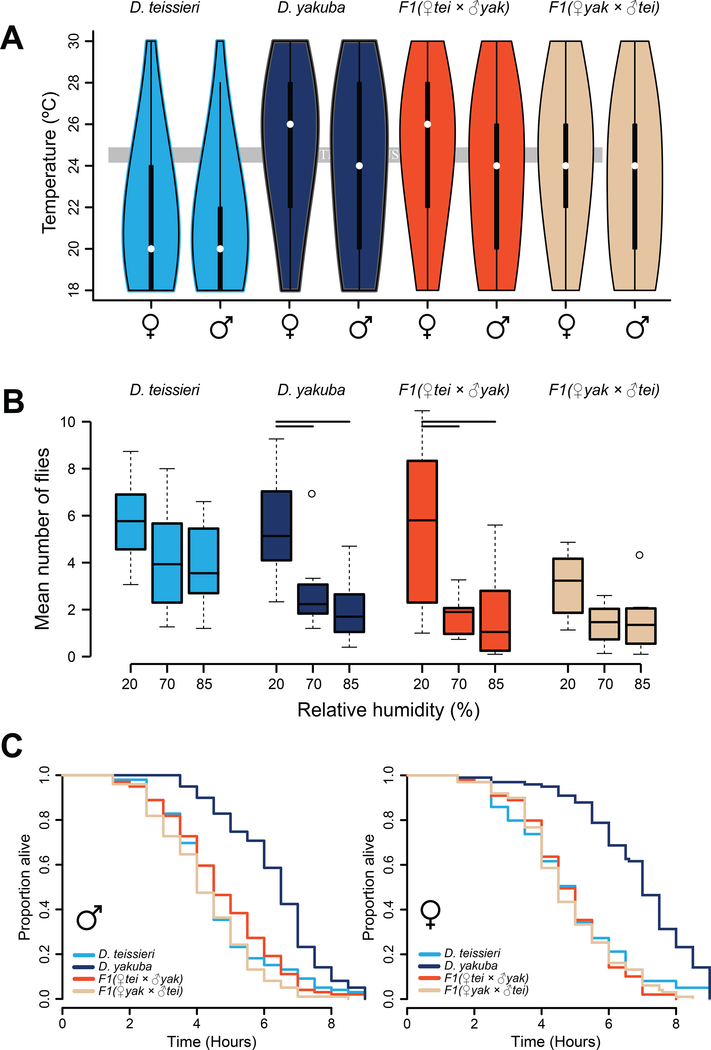
Our field measurements indicate that D. yakuba and hybrid males prefer warmer temperatures than D. teissieri males. To confirm this, we measured the temperature preferences of parental species (as in [11]) and F1 hybrids in the laboratory (Figure 3A). The mean temperature preference of D. teissieri (21.29°C ± 2.91 SD) is 13% lower than D. yakuba (24.51°C ± 3.68 SD), 12% lower than F1(♀tei × ♂yak) (24.12°C ± 3.72 SD), and 10% lower than F1(♀yak × ♂tei) (23.74°C ± 3.82 SD) preferences. Across all genotypes females prefer higher temperatures (23.87°C ± 3.70 SD) than males (22.95°C ± 3.78 SD) (F1,4792 = 57.361, P < 0.0001). F1(♀tei × ♂yak) males (23.59°C ± 3.83 SD), which we did not observe on Bioko, prefer temperatures nearly 12% higher than those preferred by D. teissieri males (21.09°C ± 2.77 SD)—this preference is statistically indistinguishable from D. yakuba (23.91°C ± 3.87 SD) (t = −1.458, df = 2396, P = 0.464) and F1(♀yak × ♂tei) (23.21°C ± 3.87 SD) (t = −1.839, df = 2396, P = 0.2551) preferences. These results indicate that in hybrids D. yakuba’s preference for warm temperatures is dominant or semi-dominant to D. teissieri’s preference.
FIGURE 3. D. yakuba-D. teissieri hybrids have a maladaptive combination of pure species traits.
A. D. yakuba and hybrid genotypes prefer warmer temperatures than D. teissieri, as measured in a laboratory thermocline. Tukey’s HSD post hoc comparisons revealed the temperature preference of F1(♀tei × ♂yak) hybrids differs from D. teissieri (P < 0.0001), but only marginally from D. yakuba (P = 0.05). In contrast, F1(♀yak × ♂tei) hybrids that occur on Bioko differ from both D. teissieri (P < 0.0001) and D. yakuba (P < 0.0001). These patterns stem from a slight, but statistically significant, difference between the two reciprocal hybrids (P = 0.03). B. Like D. yakuba, F1 hybrids tend to prefer lower relative humidity. Horizontal bars represent significant pairwise contrasts (P < 0.01) between humidity conditions fit separately for each genotype (see text for details). C. Both male and female D. yakuba individuals have higher desiccation tolerance than D. teissieri and hybrids. Across sexes F1(♀yak × ♂tei) have the lowest desiccation tolerance indicating that these hybrids are ill-equipped to cope with the dry environments they prefer on Bioko. See also Table S3 (Summary statistics for males and females) and Table S6.
Our field measurements indicate that D. yakuba and hybrids prefer dryer conditions than D. teissieri. To confirm this, we measured the relative humidity (RH) preferences of parental species and F1 hybrids in the laboratory (Figure 3B). Males and females do not differ in RH preference (χ2 = 0.25, P = 0.62); however, D. yakuba and both types of F1 hybrids tend to prefer lower RH (D. yakuba, χ2 = 15.2, P < 0.001; F1(♀yak × ♂tei), χ2 = 7.8, P = 0.02; F1(♀tei × ♂yak), χ2 = 24.4, P < 0.0001), although F1(♀yak × ♂tei) hybrids have weaker preference than do F1(♀tei × ♂yak) hybrids. In contrast, D. teissieri do not prefer a specific RH in the lab (χ2 = 4.1, P = 0.13). These data indicate that in hybrids D. yakuba’s preference for low humidity is dominant or semi-dominant to D. teissieri’s lack of preference.
Because hybrids have D. yakuba’s behavioral preference for warm and dry conditions, we assessed whether hybrids also have desiccation tolerances similar to D. yakuba. Body size did not influence desiccation tolerance (F3, 392 = 2.434, P = 0.07), but genotype did (χ2 = 143.55, df = 3, P < 0.0001) (Figure 3C, Table S3). The desiccation tolerance of D. teissieri (4.73 hr ± 1.69 SD) was 40% lower than D. yakuba (6.63 hr ± 1.54 SD), consistent with the distribution of these species in nature. Strikingly, hybrids had approximately 28% lower desiccation tolerance than did D. yakuba [F1(♀yak × ♂tei), 4.50 hr ± 1.47 SD; F1(♀tei × ♂yak), 4.76 hr ± 1.50 SD], despite preferring similarly dry conditions. These results indicate that D. teissieri’s low desiccation tolerance is dominant or semi-dominant to D. yakuba relatively high tolerance. We predict this leaves hybrids physiologically ill-equipped to cope with their preferred conditions in the lab and on Bioko.
Although we did not sample advanced-generation hybrids on Bioko, genome data provide evidence for some introgression [1,8]. We used the distribution of haplotype lengths and possible generation times to estimate the timing of admixture. We estimate admixture between D. teissieri and D. yakuba 611 to 1,124 generations ago assuming a single pulse model and 1,243 to 2,356 generations ago assuming a continuous pulse model [15](Figure S2). Using the temperature dependence of development time to convert generations to absolute ages, we estimate admixture occurred between 23 and 320 years ago (Table S4). Previous simulations using introgressed haplotype size estimated admixture between 1,600 and 8,100 generations ago, assuming very low rates of migration [1]. These prior estimates correspond to admixture between about 60 and 1,100 years ago, which overlaps with our estimates here. Our calculations using the distribution of haplotype sizes assume all haplotypes are neutral (i.e., no hybrid incompatibilities) which will overestimate the age of admixture; further, selection will purge incompatibilities making haplotypes shorter than expected by purely neutral introgression [16].
Intermediate traits in hybrids are a common reason for fitness reduction [17,18], but our results indicate that opposing patterns of dominance between parental alleles decouple behavior and physiology in hybrids (see [19] for a review). A similar situation, but involving only a single trait, has been observed in Heliconius butterflies where hybrids show a combination of wing pattern elements from both parental species and thus do not belong to any of the parental mimicry rings, which leads to higher predation [20]. How often such maladaptive combinations of pure species traits contribute to RI will depend on the genetic bases of the traits and the extent to which the ecological niches of parental species differ. The number of genes involved in deleterious traits specific to hybrids increases with parental divergence time and has been demonstrated for hybrid inviability [21,22] and hybrid sterility [23]. However, a systematic assessment of the number of potential incompatible traits, and their genetic basis, in multiple species is needed to determine the prevalence and rate of evolution of these incompatibilities [24].
Temperature-dependent asymmetries in the strength of RI contributes to the paucity of F1(♀tei × ♂yak) hybrids on Bioko
Under standard laboratory conditions, non-ecological RI is equally strong between ♀D. teissieri × ♂D. yakuba and ♀D. yakuba × ♂D. teissieri crosses, and reciprocal hybrids do not differ in their survival rate [11,12,25]. This presents a conundrum; namely, what barriers might limit the prevalence and/or production of F1(♀tei × ♂yak) hybrids in nature that are not revealed in the laboratory? Both species are infected with maternally transmitted Wolbachia bacteria that cause weak cytoplasmic incompatibility (CI) when uninfected females mate with infected males. However, the strength of CI does not differ between reciprocal crosses, and Wolbachia seem to have no influence on RI in this clade [12], indicating that Wolbachia do not contribute to the paucity of F1(♀tei × ♂yak) hybrids.
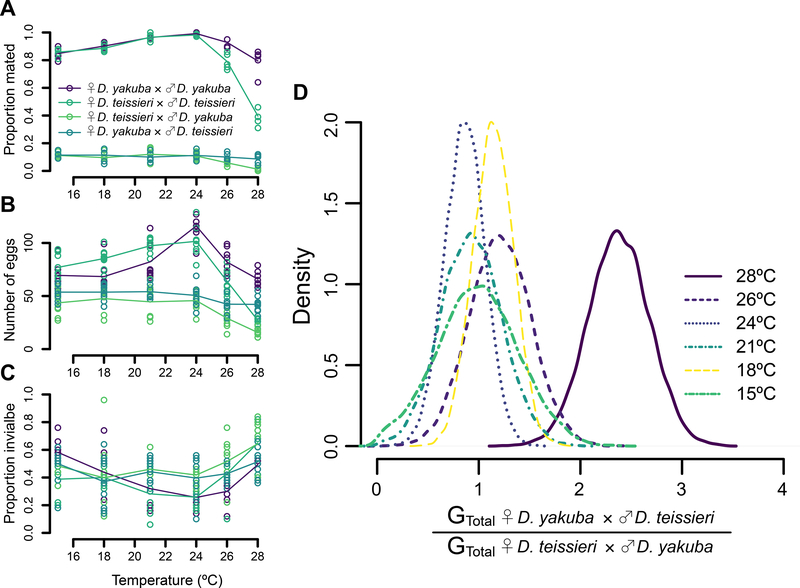
Because D. yakuba and D. teissieri differ in their thermal preferences, we evaluated temperature effects on mating (Figure 4A), egg lay (Figure 4B), and egg-to-adult viability (Figure 4C) for conspecific and heterospecific pairings. This enabled us to estimate the potential for gene flow (GTotal) for each cross at several temperatures. In agreement with past analyses, our estimates of GTotal at 24°C are similar for both crosses (♀D. yakuba × ♂D. teissieri = 0.017 and ♀D. teissieri × ♂D. yakuba = 0.019) with the ratio of GTotal for ♀D. yakuba × ♂D. teissieri to GTotal for ♀D. teissieri × ♂D. yakuba near 1 (Figure 4D). However, at higher temperatures (28°C) like those found on Bioko (Figure 2A), this ratio is more than two times higher at 28°C (2.38) than at the coolest temperatures (15°C = 0.98, 18°C = 1.14, 21°C = 0.92, and 24°C = 0.86) and nearly two times higher than at 26°C (ratio = 1.22) (Figure 4D). This difference is largely caused by adverse effects of high temperature on D. teissieri females in conspecific and heterospecific pairings. At 28°C ♀D. yakuba × ♂D. teissieri matings are seven times more likely to occur than are ♀D. teissieri × ♂D. yakuba matings (8.6% vs 1.2% respectively). In combination with temperature effects on egg lay (Figure 4B) and egg-to-adult viability (Figure 4C), we predict this reduces the potential for gene flow between ♀D. teissieri and ♂D. yakuba crosses on Bioko (Figure 4D).
FIGURE 4. Temperature influences female mating, egg lay following a single mating, and egg-to-adult viability for conspecific and heterospecific crosses.
A. The proportion of mated females paired with conspecific or heterospecific matings for 24 hours depends on cross type (X2 = 8,087.5, P < 0.0001), temperature (X2 = 166.4, P < 0.0001), and their interaction (X2 = 135.3, P < 0.0001) with D. teissieri females being less receptive to conspecific and heterospecific mates at high temperatures. B. The number of eggs laid by females over 10 days following a single mating depends on cross type (F3,192 = 221.96, P < 0.0001), temperature (F5,192 = 61.025, P < 0.0001), and their interaction (F15,192 = 12.81, P < 0.0001) with D. teissieri egg lay reduced at high temperatures found on Bioko. C. The egg-to-adult viability depends on cross type (F3,216 = 6.51, P = 0.0003), temperature (F5,216 = 20.03, P < 0.0001), and their interaction (F15,216 = 3.70, P < 0.0001) with D. teissieri egg-to-adult viability reduced at high temperatures found on Bioko. D. The ratio of GTotal for ♀D. yakuba × ♂D. teissieri to GTotal for ♀D. teissieri × ♂D. yakuba is about two times higher at 28°C than at cooler temperatures. The legend shown in A also applies to B and C.
Selection on ecologically relevant traits such as coloration (e.g. [26,27]) and habitat choice (e.g.[28,29]) can reduce gene flow between locally adapted populations (reviewed in [30]). More recently, studies have begun to test how selection shapes the distribution of hybrid genotypes found in nature using genome-wide data (e.g., [24,31,32]). By measuring ecologically relevant phenotypes of D. yakuba, D. teissieri, and their hybrids in the lab and the field, our findings extend this knowledge to demonstrate temperature-dependent asymmetries in the strength of RI between reciprocal crosses. Notably, these two species also show other barriers to gene flow [11,12]. The role of biotic and abiotic conditions on facilitating hybridization and introgression remain largely understudied (but see [33]).
CONCLUSIONS
Identifying ecological and intrinsic barriers that limit gene flow is crucial to explain the persistence of species in areas of contact. This is especially true for Drosophila where about half of all closely related species have overlapping geographical ranges [34], and several species pairs show evidence of past introgression [1,35–37]. Our results demonstrate that temperature-dependent asymmetries in the strength of RI and a maladaptive combination of D. yakuba behavior and D. teissieri physiology in hybrids generate barriers to gene flow and contribute to the maintenance of the Bioko hybrid zone. These species are the most divergent pair in Drosophila known to form a hybrid zone; indeed, similarly diverged pairs produce inviable or sterile hybrids in the laboratory (e.g., D. melanogaster-D. simulans hybrids, Ks = 0.10, [22]). At least two cases of stable, insect hybrid zones between species with more extreme levels of divergence have been reported: multiple parallel hybrid zones occur between color pattern races of Heliconius erato and H. melpomene whose genomes differ extensively (KS = 0.15) [38], and two species of termites (Blattodea: Coptotermes) estimated to have diverged 17 million years ago (mya) show little evidence of premating RI and produce hybrid colonies [39]. Hybrid zones between highly diverged vertebrate species seem more common [40,41]. In birds 9 out of 23 hybrid zones are formed between species with divergence times greater than 2 mya ([42] and references therein). The existence of these hybrid zones indicates that the possibility of hybridization, and perhaps introgression, exists after deep divergence between hybridizing species. Finally, our discovery of the Bioko hybrid zone may not be surprising given that hybridization seems to be common in Afronesia (Table S5). Colonial establishment of Bioko occurred in the last 600 years (~1,472), and our inferred interval for the timing of admixture overlaps with the period of time where cocoa plantations and livestock were common in Equatorial Guinea (end of the XIX century-1968; [43,44]). We hypothesize that this hybrid zone may be another example of human disturbance facilitating secondary contact (reviewed in [45]). A systematic exploration of the factors controlling the formation of hybrid zones, and their genomic consequences, is sorely needed.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information and resources should be directed to, and will be fulfilled by, the Lead Contact, Daniel R. Matute (dmatute@email.unc.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Drosophila stocks used for new genomic analyses, and their availability, are described in the Key Resources Table.
METHOD DETAILS
Distinguishing among D. yakuba, D. teissieri, and F1 hybrids using morphology
We sought to heavily sample Bioko for D. yakuba, D. teissieri, and their putative hybrids. However, conducting experiments in the field requires a method for reliably identifying and distinguishing living individuals of each genotype. Based on our experience with D. yakuba-clade flies, we predicted that three male morphological characteristics would enable us to achieve this goal: the number of chitinized spines on anal plates, the number of sex combs on forelegs, and the lengths of tibia. We first measured each trait in a training set of D. yakuba (N = 500) and D. teissieri (N = 500) males, and also in F1(♀tei × ♂ yak) (N = 500) and F1(♀yak × ♂ tei) (N = 500) hybrid males that we created in the laboratory (Figure S1). This provided a distribution of values for each trait within each genotype. We next blindly measured each of the three morphological traits in an additional set of 100 males of each genotype and calculated Mahalanobis distances for each individual [46]. The Mahalanobis distance between a focal individual (i) and the average for a given genotype (i.e., centroids for D. teissieri, D. yakuba, and the two hybrid F1 genotypes estimated using the training set) was calculated as:
| (1) |
where the super-index T denotes matrix transpose, S denotes the covariance matrix of a given dataset, Fi is the vector of phenotypic observations in a focal individual, i, and μLB is the vector of average phenotypic observations of the training set. To estimate the accuracy of this approach, we calculated the proportion of our blind assignments that were correct. We were able to reliably identify pure species (D. yakuba: 100/100, D. teissieri: 100/100) and F1 hybrids (single class, 196/200). We attempted to gain further resolution and assessed our ability to discriminate between F1(♀tei × ♂yak) and F1(♀yak × ♂tei) reciprocal hybrids, but due to their similar morphology our assignments were incorrect 53% of the time. Thus, we sequenced the genomes of a subset of the putative-hybrid males to confirm genotypes.
Sampling and mark-recapture experiments on Bioko
In 2013, we completed a preliminary experiment to first determine the distance flies travel in ~24 hr. D. melanogaster subgroup species males (D. yakuba, D. teissieri, D. simulans, and D. melanogaster) were collected using baited traps, anesthetized with FlyNap, and assigned to one of the four species based on genital morphology [47]. This approach yielded at least 500 male individuals of each species that we dusted with micronized fluorescent powder (Signal green; Day-Glo Color Corporation, Cleveland, OH) and released at once from a single location (1,650 m). Prior to this release, traps that consisted of small buckets with a mixture of banana and yeast were placed every 100 m in six radially arranged transects around the release area (8 traps per transect for a total of 49 traps, including a trap at the intersection of the transects). Traps were sampled between 23 and 25 hr later, and the number of recaptured flies was determined using a UV light. Species were again identified by their genital morphology. These experiments revealed that D. yakuba and D. teissieri flies rarely moved more than 100 m over a period of 24 hr (Table S6).
We next carried out our primary mark-recapture experiment to determine habitat preferences of D. yakuba-clade genotypes. We next lightly dusted D. yakuba, D. teissieri, and hybrid males with one of three types of micronized fluorescent powder (Signal green, Horizon blue, and Fire orange; Day-Glo Color Corporation, Cleveland, OH) and released them from a trap placed at the center of the forest-open habitat ecotone, at an altitude of approximately 1,650m. Buckets baited with a mixture of banana and yeast were evenly spaced by ~10m, with 11 running into the forest and 11 into the open habitat perpendicular to the interface of the two habitats, which our preliminary analysis of fly travel suggested is sufficient (Table S6). After an average of 24hr, we recaptured flies by netting over the buckets. Upon collection, genotypes were identified by dust color under UV light. To further confirm genotype assignments, we dissected and mounted testes from all recaptured males on slides in Ringer’s solution and assessed sperm motility. Hybrid males are sterile and have no motile sperm, while non-hybrids are fertile with motile sperm [11,12].
Distributions of D. yakuba, D. teissieri, and their hybrids across the ecotone
The male individuals that we recaptured during our mark-recapture experiment enabled us to estimate the rate of hybridization and to assess the distributions of each genotype across the ecotone. Flies collected during this experiment included both dusted flies and any other D. yakuba-clade flies visiting our traps at the time of recapture. We combined these data with our original capture data to estimate the prevalence of D. yakuba genotypes across the ecotone. We first calculated the dilution factor, for each genotype, defined as the number of non-dusted flies captured divided by the number of dusted flies recaptured [48]. We then multiplied the number of flies that we originally released by the dilution factor to estimate the census size of each genotype at this single altitudinal sliver of habitat. The estimated census size of hybrids divided by the sum of the three estimated census sizes (D. yakuba, D. teissieri, and hybrids) provides an estimate for hybrid prevalence. To assess variation in the prevalence of D. teissieri and D. yakuba with distance from the center of the ecotone, we fitted logistic models using the ‘MASS’ library [49].
Genomic sequencing of field-sampled hybrids
We sequenced a subset of the male individuals sampled from Bioko in 2013 to confirm our genotype assignments based on Mahalanobis distances in the field. Genomic DNA was extracted from each individual using the Beckman-Coulter DNAdvance magnetic bead protocol for insects (Beckman Coulter, Indianapolis, IN, USA). Libraries were made for each individual using the Tagmentase protocol detailed in Picelli et al. [50], and barcoded for multiplexing (see supplement for barcode sequences). Libraries were then sequenced to low coverage with Illumina 100 bp single end reads (Cornell Genomics Facility). In total, 17 parental D. teissieri, 15 parental D. yakuba, and 19 individuals of putative-hybrid phenotype were sequenced to sufficient coverage (i.e., > 10,000 reads). The mean number of markers per individual was 6.87, and the mean coverage was 2.29X.
Genotypes of the individuals were determined by the Multiplexed Shotgun Genotyping (MSG) pipeline described by Andolfatto et al. [51], which uses a hidden Markov model (HMM) to assign ancestry along a genome with low-coverage read data. Because this approach utilizes linkage disequilibrium on a large physical scale, it is well-suited for assigning genome-wide ancestry in recently formed hybrids (within several generations of backcrossing). The D. yakuba Flybase assembly (version 1.05, [52]) was masked for repeats (RepeatMasker, [53]) and used as the first parental reference. The second parental input for MSG was made by mapping reads from the outbred individual Bioko_cascade_2_2 [1] to the D. yakuba flybase assembly and creating an updated FASTA file with genotype calls from GATK [54,55]. The updated FASTA file uses only single nucleotide polymorphisms (SNPs), and masks all inferred indels plus 5 bases both up and downstream. Some regions of the genome are error-prone in terms of assigning ancestry, due in part to low sequence divergence, poor reference genome assembly, or high polymorphism in the parental species. To reduce the rate of miscalled ancestry, known intermediate frequency SNPs in each parental population (identified by realigning whole-genome Illumina sequencing data to the D. yakuba reference) were masked in the corresponding reference genomes using sequence data from several wild-caught individuals:
Bioko_Cascade_21 (yak), Bioko_Cascade_19_16 (yak), Bioko_NE_4_6 (yak),
Bioko_Balancha_1 (tei), Bioko_cascade_4_3 (tei), Bioko_House_Bioko (tei),
Bioko_cascade_4_2 (tei), Bioko_cascade_4_1 (tei), Bioko_cascade_2_4 (tei),
Bioko_cascade_2_2 (tei), Bioko_cascade_2_1 (tei). The performance of MSG is also influenced by user-specified parameters in the HMM, in particular, those that describe the error-rate of genotypes and the rate at which transitions between ancestries occur (rfac, deltapar1, and deltapar2). To determine the appropriate parameter values for this study, MSG was run iteratively on pure-species individuals while adjusting parameters to achieve the lowest genotyping error rate, assuming that all individuals are homozygous genome-wide for their ancestry. After parameter-tuning, MSG was run on all individuals and ancestry genotypes were called with a posterior probability filter of 0.99.
Approximate timing of introgression
A previous approach had quantified the amount of introgression between D. yakuba and D. teissieri using whole genome data [1]. Using these results, we estimated the age of admixture by converting the physical length of each haplotype to genetic distances, calculating the age of introgression in generations, and inferring the number of generations per year in the field.
Converting physical distances to genetic distances.
We calculated an approximate age of first contact using the mean haplotype size in cM observed in deduced introgression between D. yakuba and D. teissieri. 146 segments have been transferred between the two species: 70 from D. teissieri to D. yakuba, and 76 from D. yakuba to D. teissieri [1]. To convert the haplotype length from physical distances (Mb) to genetic distances (cM) we assumed that the rates of recombination were similar for D. yakuba, D. teissieri, and D. melanogaster at orthologous regions of the genome. We first generated a lift-over table between the three genomes, and we then used this information to identify the orthologous blocks to the introgressed regions in D. melanogaster. Finally, we used the recombination rates experimentally measured for each orthologous interval [56] to convert to genetic distance using [57]. We focused on the Comeron start point rate but other recombination rates yield similar results.
Calculating the age of introgression in generations from genetic distances.
We converted the bootstrapped means to time of admixture. We used two models: a single pulse of introgression and continuous admixture using the expected these two situations derived by [15] where the expected mean haplotype (in cM) is
| (2) |
for a single pulse of admixture, and
| (3) |
for a continuous model of admixture. T is the age of admixture and m is the rate of migration. In both cases, we assumed that m is small (m < 0.05, following simulations from [1] and number of F1 hybrids reported here). The time of admixture then simplifies to
| (4) |
for a single pulse of admixture and
| (5) |
for a continuous admixture model. In both cases T is expressed in number of generations. We used these equations to obtain an admixture time for each inferred introgressed haplotype. This produced a distribution of T values whose mean reflects the approximate time of admixture. We bootstrapped the distribution of obtained T to find confidence intervals of the mean using the library boot (functions boot and boot.ci, [58,59]). Admixture time in generations were converted to years using the average generation time for each of the two species (see Figure S2, Table S4 and below).
Laboratory experiments
Isofemale lines.
For laboratory crosses, we used D. yakuba line cascade21 and D. teissieri line Balacha1. Both lines were collected in the highlands of Bioko in 2013 and are infected with maternally transmitted, endosymbiotic, Wolbachia bacteria. We chose this approach as low titer Wolbachia infections in males, that we might not detect, could in principle generate cytoplasmic incompatibility (CI) when crossed to uninfected females. However, CI is not possible between infected females and infected males, regardless of titer and cross type [11,12].
Temperature preference.
We measured temperature preferences of D. teissieri, D. yakuba, F1(♀tei x ♂ yak) and F1(♀yak x ♂ tei) hybrids in the laboratory. Flies were randomly placed in a thermocline that consisted of a plexiglass chamber (12cm wide × 45cm long × 1cm high) with an aluminum floor. The thermocline was placed in an 18°C room and heat plates (120 VAC Thermo Scientific Cimarec Hot Plate, Thermo Scientific Cimarec, # UX-04600–01, Waltham, MA, USA) were used to generate a thermal gradient ranging from 18°C to 30°C, with a change in temperature of approximately 2°C every 6cm. Flies were allowed to move freely along this gradient over a period of one hour. At the end of each trial we isolated flies into seven chambers, each 10.5 × 6 × 1 cm, by pushing a rod connected to six plexiglass partitions across the width of the chamber. We then recorded the temperature within each partition using a Digi-Sense thermometer equipped with a type-T thermocouple (Cole-Parmer Instrument Co., Chicago, IL; catalog number: 86460–05) and counted the number of flies within each partition [60]. Males and females of each genotype were evaluated separately to avoid sexual attraction that might influence results (N = 3 separate replicates per sex; 400 individuals per replicate).
Humidity preference.
Relative humidity (RH) preferences of D. teissieri, D. yakuba, and F1(♀tei × ♂ yak) and F1(♀yak × ♂ tei) hybrids were evaluated by giving flies the choice of orienting themselves along a humidity gradient. Rows of a 48-well polystyrene tissue culture plate (8 rows by 6 columns; Corning Incorporated, Life Sciences, Tewksbury, MA, USA) were filled with one of three super-saturated salt solutions: LiCl, NaCl, or KH2PO4. Each of these solutions generates a RH of ~20%, ~70%, and ~85%, respectively, in the headspace above the rows. We filled the wells of two adjacent end rows with LiCl, the three following rows with NaCl, and the three remaining rows with KH2PO4. This generates a gradient of RH ranging from ~20% to 85%. The top of each plate was covered with 300 micron nylon netting (MegaView Science Co., Ltd. Taichung, Taiwan) and covered with the culture plate lid on top of the mesh; this left ~1 cm for flies to move freely around the plate. This experimental design is a modified approach from Enjin and colleagues [61]. We lightly anesthetized approximately 50, 4 to 7-day old virgin, males or females of a given genotype and placed them along the long axis of a plate. We ran eight plates simultaneously—one plate for each of the parental and hybrid genotypes, with sexes evaluated separately. Flies were allowed to orient themselves for the first hour after which pictures were taken every 15 min, for an additional two hours. This procedure was repeated on four separate days with the position of each plate and the orientation of the ‘low’ and ‘high’ RH end of the plates relative to the room randomized each day. (This avoids confounding effects of non-uniform lighting and other conditions among days in the laboratory.) In total, we assayed ~200 individuals of each genotype and sex. To score preference, we counted the number of flies over each well of the plates, for each of the eight images generated over the 2 hr assay, and summed counts of flies oriented over wells containing the same super-saturated salt solution. All scoring was double blinded.
Desiccation tolerance in the laboratory.
Desiccation resistance was measured by placing ten, 4-day old virgin, females or males in 30mL empty vials (N = 11 vials per sex), which in turn were placed in a glass desiccator with 200g of Drierite (Sigma Aldrich Catalog number: 7778–18-9; St. Louis, MO) and kept at 21°C (42). The relative humidity was kept under 20% and was measured with a hygrometer. Flies were checked every 30min and the time of death was recorded for each fly.
Conspecific and heterospecific mate preference.
To establish whether temperature influences the magnitude of female choice, we conducted no-choice mating experiments en masse [62–64] at six different temperatures (15°C, 18°C, 21°C, 24°C, and 26°C and 28°C). We collected virgin males and females within eight hours of eclosion from D. teissieri and D. yakuba, and maintained each sex in isolation for four days. In the morning of the fifth day, we paired groups of approximately 100 virgin females with approximately 100 males in vials with cornmeal food, so that each vial contained 200 flies—this was completed for all four possible pairings (♀D. yakuba × ♂ D. teissieri, ♀ D. teissieri × ♂ D. yakuba, ♀ D. teissieri × ♂D. teissieri, ♀ D. yakuba × ♂ D. yakuba). These males and females were kept together for 24 hours. After 24 hours, all individuals were anesthetized with CO2. We extracted the reproductive tracts from females and then mounted them in chilled Ringer’s solution. If we observed sperm on either the seminal receptacle or any of the spermathecae, females were considered mated. We repeated this assay five times per temperature for a total of 30 experimental blocks and ~3,000 females. We calculated the proportion of mated heterospecific to conspecific females as:
| (6) |
Conspecific and heterospecific fecundity.
We evaluated fecundity after conspecific and heterospecific crosses at the six focal temperatures. For all four possible crosses (♀D. yakuba × ♂ D. teissieri, ♀ D. teissieri × ♂ D. yakuba, ♀ D. teissieri × ♂D. teissieri, ♀ D. yakuba × ♂ D. yakuba), one virgin female and one virgin male were housed in a 20mL vial with corn meal. We watched each pair and assess whether mating occurred. We obtained 60 females per cross by watching copulating pairs for a period of eight hours. Once copulation finished, we removed the males from the vial (each vial contained only one female). Females were then randomly assigned to one of six batches which were then placed at the six different temperatures. Every 24 hours, we counted the number of eggs laid. Females were then transferred to a fresh vial. The procedure was repeated for ten days. We calculated the proportion of eggs laid for heterospecific to conspecific crosses as:
| (7) |
Conspecific and heterospecific egg-to-adult viability.
We scored the proportion of embryos from conspecific and heterospecific crosses that molt successfully into adults. To score embryo viability, we housed virgin individuals from the two sexes in the four possible combinations (♀D. yakuba × ♂ D. teissieri, ♀ D. teissieri × ♂ D. yakuba, ♀ D. teissieri × ♂D. teissieri, ♀ D. yakuba × ♂ D. yakuba) in oviposition cages (Flystuff, San Diego, CA, USA) with apple juice agar at 24°C. From these cages, we collected freshly deposited eggs (less than six hours after oviposition). Eggs were then collected in batches of 50, placed into black filter paper and housed in 30mL plastic vials containing cornmeal food. For each cross, we obtained 3,000 embryos which were then divided into five batches. Each batch was placed into the six different temperatures listed above. Vials were tended daily. Once marks of larvae were seen on the side of the food, we added a Kimwipe (Kimberly Clark, Delicate Task) dampened with 0.5% propionic acid solution as a pupation substrate. Adult flies were counted as they hatch every two days. There were ten replicates per cross per temperature (500 individuals). We calculated hybrid inviability as:
| (8) |
Hybrid sterility.
F1 hybrids between D. yakuba and D. teissieri are sterile at 24°C [11,25]. We scored whether hybrid males were also sterile at different temperatures. Hybrid males were produced as described above at six different temperatures (Section ‘Conspecific and heterospecific egg-to-adult viability’). Four to eight day old males from conspecific and heterospecific crosses were then lightly anesthetized with CO2. Their testes were then extracted with forceps and mounted in Ringer’s solution; we assessed sperm motility for 100 males per cross (2 conspecifics and 2 heterospecifics). We assumed all hybrid females were fertile. The index of sterility (‘hybrid male sterility’ in equation 9) was the number of sterile hybrid males.
Temperature dependence of development times.
Since the temperature in Bioko varies substantially, we measured generation times at four different temperature regimes. We collected 40 batches of similarly aged embryos (within 3 hours of deposition) for each of the two species. Each batch was placed in a corn meal vial and randomly assigned to one of four temperature regimes: 11°C/16 °C, 16°C/21 °C, 21°C/26 °C, and 26°C/31 °C. The first temperature corresponds to the night (dark cycle) temperature and the second one to the day (light cycle) temperature. Temperatures were controlled by housing all experiments in incubators (Percival DR-36VL with a H3X pan-type humidifier). Vials were tended daily by adding hydration (propionic acid 0.05%), and added a pupation substrate (Kimwipe). We scored the time of emergence of each fly and calculated the mean time of emergence for each vial. The mean generation time for D. teissieri was significantly longer than D. yakuba (F1,736 = 305.710, P < 0.0001). Moreover, the life history of the two species also differs; D. teissieri is mostly associated to Parinari fruits, which means the species is most likely to breed during the dry period, when Parinari fruits are commonly found in the ground (6 months out of the year, [65,66]). On the other hand, D. yakuba is found in a multitude of substrates and is likely to be collected year round. The number of generations per year is thus likely to differ for the two species. We used both numbers as the generation time to convert the bootstrapped distribution of T. We thus obtained sixteen different mean values—with their respective confidence intervals—for the time of admixture (2 admixture models × 4 temperature regimes × 2 life histories).
QUANTIFICATION AND STATISTICAL ANALYSIS
Distributions of D. yakuba, D. teissieri, and their hybrids across the ecotone
We modeled the probability of male D. teissieri, D. yakuba, and their hybrids choosing forest habitat over open habitat. We fitted a generalized linear mixed model with binomially distributed error using the ‘lme4’ library [67]. The full model included genotype (either D. teissieri, D. yakuba, or their hybrids) and the random effect of block (N = 3 blocks). We calculated mean values using parameter estimates from the most likely model, and took the inverse logit of these values to calculate the estimated mean probabilities of choosing forest habitats. To estimate P-values, we fitted the same model again using the mixed function within the ‘afex’ library [68]. This approach first fits the full model, and then individual fixed effects are removed to compare the reduced model to the full model. P-values were calculated using the likelihood ratio test (LRT) method within mixed. These analyses and all others were conducted using the R Statistical Package (version 3.3.1; [69]).
Variation in temperature and humidity in the field
To assess variation in temperature and humidity, we fitted two linear models, one for temperature and one for humidity. The two models followed the same form and assessed the effect of altitude, type of habitat, and time of the day on each of the two environmental factors. We also included all possible interactions. We compared temperature and humidity at open versus forest habitats using the Tukey’s Honest Significant Difference (HSD) post hoc pairwise comparisons in the ‘multcomp’ library [70].
Temperature preference, humidity preference, and desiccation tolerance in the laboratory
Because temperature preference was not normally distributed, and remained non-Gaussian after both log (Shapiro Wilk test, W = 0.914, P < 0.0001) and square root (Shapiro Wilk test, W = 0.919, P < 0.0001) transformations, we used the ‘ARTool’ library to complete analyses of variance on aligned rank transformed data [71]. We specifically assessed the effects of genotype, sex, and their interactions on temperature preference. We then used lsmeans to conduct post hoc pairwise comparisons [72].
Because there was no effect of sex on the number of flies choosing a given RH (LRT: χ2 = 0.25; P = 0.62), sexes were analyzed together. We first tested whether humidity preference varied across genotypes by modeling the mean number of individuals choosing a given RH as a function of genotype, RH, and the interaction between genotype and RH. We assumed a Poisson distribution of error. To explicitly test whether different genotypes displayed preferences for different RH, we also modeled the mean number of individuals choosing a given humidity, for each genotype separately, as a function of RH (fixed effect) and plate identity (random effect). We tested for variation in humidity preference using LRTs comparing these genotype-specific models to those lacking the fixed effect of RH.
Differences in desiccation resistance between genotypes were analyzed using a survival analysis and a Cox regression (‘rms’ library, [73]) using the cph function. Plots were generated with the ‘survplot’ function [73]. To assess any effects of body size on desiccation tolerance, we measured the thorax length of 4-day old males and females (N = 50 of each sex from each genotype) reared at 24°C. We then fitted a linear model to evaluate the fixed effects of sex and genotype, and the interaction between these factors, using the function lm function in the ‘stats’ package [69].
Conspecific and heterospecific mating, egg laying, and egg-to-adult viability
We modeled the probability of mating by fitting a generalized linear mixed model with binomially distributed error using the lme4 library in the R Statistical Package [67].
Because the number of eggs laid was not normally distributed (Shapiro Wilk test, W = 0.979, P = 0.003), we used the ‘ARTool’ library to complete analyses of variance on aligned rank transformed data [71]. We specifically assessed the effects of cross type (♀D. yakuba × ♂ D. teissieri, ♀ D. teissieri × ♂ D. yakuba, ♀ D. teissieri × ♂D. teissieri, ♀ D. yakuba × ♂ D. yakuba) and temperature on the eggs laid over 10 days.
Because the number of eggs laid was not normally distributed (Shapiro Wilk test, W = 0.985, P = 0.015), we used the ‘ARTool’ library to complete analyses of variance on aligned rank transformed data [71]. We specifically assessed the effects of cross type (♀D. yakuba × ♂ D. teissieri, ♀ D. teissieri × ♂ D. yakuba, ♀ D. teissieri × ♂D. teissieri, ♀ D. yakuba × ♂ D. yakuba) and temperature on egg-to-adult viability.
Potential for gene flow for ♀D. yakuba × ♂ D. teissieri and ♀ D. teissieri × ♂ D. yakuba crosses
We calculated the magnitude of possible gene flow for the two reciprocal crosses using a modification of the serial isolation equation of Coyne and Orr [74] Sobel et al. [75] using mean values for each trait where:
| (9) |
We calculated 12 mean GTotal values, one for each reciprocal cross at the six studied temperatures. We also calculated the minimum and maximum GTotal values for each temperature by finding the lowest and highest value of GTotal from all possible combinations of individual trait values for a total of 450 values per temperature.
DATA AND SOFTWARE AVAILABILITY
All new data reported in this paper are available as described in the Key Resources Table.
Supplementary Material
Highlights.
Drosophila yakuba prefers open areas and D. teissieri prefers forests on Bioko
These species hybridize at the center of the forest-open habitat ecotone
Temperature-dependent reproductive isolation limits gene flow
A maladaptive combination of parental traits in hybrids also limits gene flow
ACKNOWLEDGEMENTS
We would like to thank R.C. Bell, W.R. Conner, P. McLaughlin, B. Miles, G. Ousmane, and the Bioko Biodiversity Protection Program, and the Ministry of Environment for assistance in the field. C.J. Jones, R. Marquez, E.L. Larson, M.W. Hahn, members of the Matute lab, and the University of Montana CHALK group for helpful scientific discussions and comments. Three anonymous reviewers provided thoughtful comments that greatly improved the paper. This work was supported in part by NIH awards R35GM124701 (B.S.C.), F32GM117763 (A.S.) and R01GM121750 (D.R.M.). The authors declare no conflicts of interest.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Turissini DA, and Matute DR (2017). Fine scale mapping of genomic introgressions within the Drosophila yakuba clade. PLoS Genet. 13, e1006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lachaise D, Lemeunier F, and Veuille M (1981). Clinal variations in male genitalia in Drosophila teissieri Tsacas. Am. Nat, 117, 600–608. [Google Scholar]
- 3.Cobb M, Huet M, Lachaise D, and Veuille M (2000). Fragmented forests, evolving flies: molecular variation in African populations of Drosophila teissieri. Mol. Ecol 9, 1591–1597. [DOI] [PubMed] [Google Scholar]
- 4.Comeault AA, Serrato Capuchina A, Turissini DA, McLaughlin PJ, David JR, and Matute DR (2017). A nonrandom subset of olfactory genes is associated with host preference in the fruit fly Drosophila orena. Evol. Lett 63, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lachaise D, Cariou M-L, David JR, Lemeunier F, Tsacas L, and Ashburner M (1988). Historical biogeography of the Drosophila melanogaster species subgroup In Evolutionary biology (pp. 159–225). Springer, Boston, MA. [Google Scholar]
- 6.David JR, Lemeunier F, Tsacas L, and Yassin A (2007). The historical discovery of the nine species in the Drosophila melanogaster species subgroup. Genetics 177, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monnerot M, Solignac M, and Wolstenholme DR (1990). Discrepancy in divergence of the mitochondrial and nuclear genomes of Drosophila teissieri and Drosophila yakuba. J. Mol. Evol 30, 500–508. [DOI] [PubMed] [Google Scholar]
- 8.Bachtrog D, Thornton K, Clark A, and Andolfatto P (2006). Extensive introgression of mitochondrial dna relative to nuclear genes in the Drosophila yakuba species group. Evolution 60, 292–302. [PubMed] [Google Scholar]
- 9.Turissini DA, Comeault AA, Liu G, Lee YCG, and Matute DR (2017). The ability of Drosophila hybrids to locate food declines with parental divergence. Evolution 71, 960–973. [DOI] [PubMed] [Google Scholar]
- 10.Matute DR, and Ayroles JF (2014). Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol 27, 1057–1068. [DOI] [PubMed] [Google Scholar]
- 11.Turissini DA, Liu G, David JR, and Matute DR (2015). The evolution of reproductive isolation in the Drosophila yakuba complex of species. J. Evol. Biol 28, 557–575. [DOI] [PubMed] [Google Scholar]
- 12.Cooper BS, Ginsberg PS, Turelli M, and Matute DR (2017). Wolbachia in the Drosophila yakuba complex: pervasive frequency variation and weak cytoplasmic incompatibility, but no apparent effect on reproductive isolation. Genetics 205, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellermann V, Van Heerwaarden B, Sgrò CM, and Hoffmann AA (2009). Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science 325, 1244–1246. [DOI] [PubMed] [Google Scholar]
- 14.Kellermann V, Overgaard J, Hoffmann AA, Flojgaard C, Svenning J-C, and Loeschcke V (2012). Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc. Natl. Acad. Sci 109, 16228–16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin W, Li R, Zhou Y, and Xu S (2014). Distribution of ancestral chromosomal segments in admixed genomes and its implications for inferring population history and admixture mapping. Eur. J. Hum. Genet 22, 930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muirhead CA, and Presgraves DC (2016). Hybrid Incompatibilities, local adaptation, and the genomic distribution of natural introgression between species. Am. Nat 187, 249–261. [DOI] [PubMed] [Google Scholar]
- 17.Nosil P (2007). Divergent host plant adaptation and reproductive isolation between ecotypes of Timema cristinae walking sticks. Am. Nat 169, 151–162. [DOI] [PubMed] [Google Scholar]
- 18.Delmore KE, and Irwin DE (2014). Hybrid songbirds employ intermediate routes in a migratory divide. Ecol. Lett 17, 1211–1218. [DOI] [PubMed] [Google Scholar]
- 19.Nosil P (2012). Ecological Speciation. Oxford University Press; Oxford, UK. [Google Scholar]
- 20.Merrill RM, Wallbank RWR, Bull V, Salazar PCA, Mallet J, Stevens M, and Jiggins CD (2012). Disruptive ecological selection on a mating cue. Proc. R. Soc. B Biol. Sci 279, 4907–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyle LC, and Nakazato T (2010). Hybrid Incompatibility snowballs between Solanum species. Science 329, 1521–1523. [DOI] [PubMed] [Google Scholar]
- 22.Matute DR, Butler IA, Turissini DA, and Coyne JA (2010). A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329, 1518–1521. [DOI] [PubMed] [Google Scholar]
- 23.Wang RJ, White MA, and Payseur BA (2015). The pace of hybrid incompatibility evolution in house mice. Genetics 201, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumer M, Cui R, Powell DL, Dresner R, Rosenthal GG, and Andolfatto P (2014). High-resolution mapping reveals hundreds of genetic incompatibilities in hybridizing fish species. Elife 3, e02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turissini DA, McGirr JA, Patel SS, David JR, and Matute DR (2018). The rate of evolution of postmating-prezygotic reproductive isolation in Drosophila. Mol Biol Evol 2018 35:312–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallet J, and Barton N (1989). Inference from clines stabilized by frequency-dependent selection. Genetics 122, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nosil P, and Crespi BJ (2006). Experimental evidence that predation promotes divergence in adaptive radiation. Proc. Natl. Acad. Sci 103, 9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Via S (1991). The genetic structure of host plant adaptation in a spatial patchwork: demographic variability among reciprocally transplanted pea aphid clones. Evolution 45, 827–852. [DOI] [PubMed] [Google Scholar]
- 29.Hawthorne DJ, and Via S (2001). Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904–907. [DOI] [PubMed] [Google Scholar]
- 30.Schluter D (2001). Ecology and the origin of species. Trends Ecol. Evol 16, 372–380. [DOI] [PubMed] [Google Scholar]
- 31.Schumer M, Xu C, Powell D, Durvasula A, Skov L, Holland C, Sankararaman S, Andolfatto P, Rosenthal G, and Przeworski M (2017). Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science 360, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenov GA, Scordato ESC, Khaydarov DR, Smith CCCR, Kane NC, and Safran RJ (2017). Effects of assortative mate choice on the genomic and morphological structure of a hybrid zone between two bird subspecies. Mol. Ecol 26, 6430–6444. [DOI] [PubMed] [Google Scholar]
- 33.Matute DR (2014). The magnitude of behavioral isolation is affected by characteristics of the mating community. Ecol. Evol 4, 2945–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turelli M, Lipkowitz JR, and Brandvain Y (2014). On the Coyne and Orr-igin of species: Effects of intrinsic postzygotic isolation, ecological differentiation, x chromosome size, and sympatry on Drosophila speciation. Evolution 68, 1176–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, and Presgraves DC (2012). Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22, 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulathinal RJ, Stevison LS, and Noor MAF (2009). The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5, e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohse K, Clarke M, Ritchie MG, and Etges WJ (2015). Genome-wide tests for introgression between cactophilic Drosophila implicate a role of inversions during speciation. Evolution (N. Y). 69, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadeau NJ, Ruiz M, Salazar P, Counterman B, Medina JA, Ortiz-Zuazaga H, Morrison A, McMillan WO, Jiggins CD, and Papa R (2014). Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 24, 1316–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TRC, Cameron SL, Evans TA, Ho SYW, and Lo N (2015). The origins and radiation of Australian Coptotermes termites: From rainforest to desert dwellers. Mol. Phylogenet. Evol 82, 234–244. [DOI] [PubMed] [Google Scholar]
- 40.Colliard C, Sicilia A, Turrisi GF, Arculeo M, Perrin N, and Stöck M (2010). Strong reproductive barriers in a narrow hybrid zone of west-Mediterranean green toads (Bufo viridis subgroup) with Plio-Pleistocene divergence. BMC Evol. Biol 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dufresnes C, Brelsford A, Crnobrnja-Isailović J, Tzankov N, Lymberakis P, and Perrin N (2015). Timeframe of speciation inferred from secondary contact zones in the European tree frog radiation (Hyla arborea group). BMC Evol. Biol 15, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price TD (2007). Speciation in Birds. W. H. Freeman and company, New York (NY) USA [Google Scholar]
- 43.Howes FN (1946). The early introduction of cocoa to West Africa Afr. Aff (Lond: ). 45, 152–153. [Google Scholar]
- 44.Frynas JG (2004). The oil boom in Equatorial Guinea. Afr. Aff. (Lond) 103, 527–546. [Google Scholar]
- 45.Vallejo-Marín M, and Hiscock SJ (2016). Hybridization and hybrid speciation under global change. New Phytol. 211, 1170–1187. [DOI] [PubMed] [Google Scholar]
- 46.Mahalanobis PC (1936). Mahalanobis distance. Proceedings National Institute of Science of India. 49, 234–256. [Google Scholar]
- 47.Markow TA, and O’Grady P (2006). Drosophila: A Guide to species identification and use. Academic Press, London (UK) [Google Scholar]
- 48.White GC, Anderson DR, Burnham KP, and Otis DL (1982). Capture-recapture and removal methods for sampling closed populations. Los Alamos National Laboratory. [Google Scholar]
- 49.Venables WN and Ripley BD (2002). Modern applied statistics with S. Fourth Edition. Springer, New York (NY), USA. [Google Scholar]
- 50.Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andolfatto P, Davison D, Erezyilmaz D, Hu TT, Mast J, Sunayama-Morita T, and Stern DL (2011). Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21, 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218. [DOI] [PubMed] [Google Scholar]
- 53.Smit A, Hubley R, and Green P (2013). RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org. Available at: http://repeatmasker.org.
- 54.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comeron JM, Ratnappan R, and Bailin S (2012). The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8, e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiston-Lavier AS, Singh ND, Lipatov M, and Petrov DA (2010). Drosophila melanogaster recombination rate calculator. Gene 463, 18–20. [DOI] [PubMed] [Google Scholar]
- 58.Canty A, and Ripley B (2012). boot: Bootstrap R (S-Plus) functions. R Packag. version, 3–7. [Google Scholar]
- 59.Davison AC, and Hinkley DV (1997). Bootstrap methods and their applications. Cambridge University Press, Cambridge (UK). [Google Scholar]
- 60.Matute DR, Novak CJ, and Coyne JA (2009). Temperature-based extrinsic reproductive isolation in two species of Drosophila. Evolution 63, 595–612. [DOI] [PubMed] [Google Scholar]
- 61.Enjin A, Zaharieva EE, Frank DD, Mansourian S, Suh GSB, Gallio M, and Stensmyr MC (2016). Humidity sensing in Drosophila. Curr. Biol 26, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coyne JA, Kim SY, Chang AS, Lachaise D, and Elwyn S (2002). Sexual isolation between two sibling species with overlapping ranges: Drosophila santomea and Drosophila yakuba. Evolution 56, 2424–2434. [DOI] [PubMed] [Google Scholar]
- 63.Matute DR, and Coyne JA (2010). Intrinsic reproductive isolation between two sister species of Drosophila. Evolution 64, 903–920. [DOI] [PubMed] [Google Scholar]
- 64.Matute DR (2010). Reinforcement of gametic isolation in Drosophila. PLoS Biol. 8, e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muhanguzi HDR, Obua J, Oryem-Origa H, and Vetaas OR (2003). Tree fruiting phenology in Kalinzu Forest, Uganda. Afr. J. Ecol 41, 171–178. [Google Scholar]
- 66.Williamson J (1956). Useful plants of Nyasaland Useful plants of Nyasaland. Government Printer, Zomba, Malawi [Google Scholar]
- 67.Bates D, Maechler M, Bolker B, and Walker S (2013). lme4: Linear mixed-effects models using Eigen and S4.
- 68.Mason RL, Gunst RF, and Hess JL (2003). Fractional Factorial Experiments In Statistical Design and Analysis of Experiments, Edited by: Mason RL, Gunst RF, and Hess JL pp. 228–270. John Wiley and Sons, New York, NY (USA) [Google Scholar]
- 69.R Core Team (2016). R Development Core Team. R A Lang. Environ. Stat. Comput 55, 275–286. [Google Scholar]
- 70.Hothorn T, Bretz F, and Westfall P (2008). Simultaneous Inference in Genneral Parametric Models. Biometrical J. 50, 346–363. [DOI] [PubMed] [Google Scholar]
- 71.Kay M, & Wobbrock J (2016). ARTool: aligned rank transform for nonparametric factorial ANOVAs. R package version 0.10, 2.
- 72.Lenth R, and Hervé M (2015). lsmeans: Least-Squares Means. R Packag. version 2.16.
- 73.Harrell F (2017). Regression Modeling Strategies. Version 5.1–0. In http://biostat.mc.vanderbilt.edu/rm.
- 74.Coyne JA, and Orr HA (2004). Speciation. Sunderland, MA. [Google Scholar]
- 75.Sobel JM, Chen GF, Watt LR, and Schemske DW (2010). The biology of speciation. Evolution (N. Y). 64, 295–315. [DOI] [PubMed] [Google Scholar]
- 76.Hothorn T, Hornik K, Van De Wiel MA, and Zeileis A (2006). A lego system for conditional inference. Am. Stat 60, 257–263. [Google Scholar]
- 77.Hothorn T, Hornik K, Van De Wiel MA, and Zeileis A (2008). Implementing a class of permutation pests: the coin package.
- 78.Paradis E, Claude J, and Strimmer K (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 20, 289–290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new data reported in this paper are available as described in the Key Resources Table.