Abstract
Rheumatoid arthritis is characterized by progressive joint inflammation and affects ~1% of the human population. We noted single nucleotide polymorphisms (SNPs) in the apoptotic cell engulfment genes ELMO1, DOCK2, and RAC1 linked to rheumatoid arthritis. As ELMO1 promotes cytoskeletal reorganization during engulfment, we hypothesized that ELMO1 loss would worsen inflammatory arthritis. Surprisingly, Elmo1-deficient mice showed reduced joint inflammation in acute and chronic arthritis models. Genetic and cell biological studies revealed that ELMO1 associates with receptors linked to neutrophil function in arthritis and regulates activation and early neutrophil recruitment to the joints, without general inhibition of inflammatory responses. Further, neutrophils from peripheral blood of human donors that carry the SNP in ELMO1 associated with arthritis display increased migratory capacity, whereas ELMO1 knockdown reduces human neutrophil migration to chemokines linked to arthritis. These data identify ‘non-canonical’ roles for ELMO1 as an important cytoplasmic regulator of specific neutrophil receptors and promoter of arthritis.
Rheumatoid arthritis (RA) affects millions of people worldwide with reduced quality of life and economic costs. RA is characterized by chronic inflammation and progressive joint destruction, with debilitating consequences1. A hallmark of human RA and mouse models of arthritis is the leukocyte influx into the joint synovium, with neutrophils being the most abundant2. Activated neutrophils promote chronic inflammation along with matrix and cartilage degradation2. Although effective therapies have been introduced, a significant fraction of RA patients are refractory to existing therapies3. Genome-wide association studies (GWAS) have identified many genetic loci; however, most of them are single nucleotide polymorphisms (SNPs) in non-coding genetic regions4 with no obvious causality. Therefore, a better understanding of causative and disease contributing factors is needed.
Cell death via apoptosis occurs during homeostasis and tissue inflammation5. While apoptotic cells have been detected in the synovial joints of RA patients6, resistance to apoptosis has also been implied as a contributory factor to chronic disease; therefore, induction of apoptosis has been proposed as a therapeutic avenue7. For these approaches, however, apoptotic cell clearance pathways also need to be considered. Inefficient clearance of apoptotic cells can result in secondary necrosis, and exposure of self-antigens, and cell clearance defects are linked to chronic inflammation and autoimmunity8.
Apoptotic cells expose ‘eat me’ signals on their surface that are recognized by specific receptors on phagocytes9–11. Binding of apoptotic cells to phagocyte recognition receptors results in activation of the engulfment machinery, dynamic changes of the actin cytoskeleton, and corpse uptake9–11. Phagocyte receptors can bind phosphatidylserine exposed on the apoptotic cell surface directly (such as TIM-412 and BAI113) or indirectly, through bridging molecules (such as MerTK14), or recognize cell surface modifications or opsonins bound to apoptotic cells15. The receptor redundancy and the specific signaling pathways downstream of these engulfment receptors are unclear16. One of the better characterized cytoplasmic signaling relays (in both professional and non-professional phagocytes) is the ELMO-DOCK-Rac signaling pathway16. In this mode of signaling, the ELMO-DOCK protein complex acts as a guanine nucleotide exchange factor (GEF) to activate the small GTPase Rac, leading to cytoskeletal rearrangements needed for engulfment17.
In this work, we examined how components of a specific engulfment pathway may link to inflammatory arthritis. Surprisingly, loss of the engulfment signaling protein ELMO1 alleviated disease severity in mouse models of arthritis through ELMO1 regulation of neutrophil recruitment to inflamed joints. Via proteomic and transcriptomic approaches, we uncover an ELMO1-dependent signature in neutrophils and identify a requirement for ELMO1 in signaling downstream of the receptors for arthritis-associated molecules C5a and LTB4. These data suggest a neutrophil-specific ELMO1-dependent signaling nexus that controls different aspects of arthritis.
RESULTS
Engulfment protein ELMO1 is associated with arthritis
To test whether specific engulfment machinery components are associated with human rheumatoid arthritis, we searched publicly available databases for SNPs. We found multiple SNP-Disease associations with human rheumatoid arthritis in ELMO1, as well as in DOCK2 and RAC1 genes (see Methods; Fig. 1a and Supplementary Table 1). In a meta-analysis for common SNPs or gene linkages to both RA and celiac disease (CD), a SNP in human ELMO1 (rs11984075) was discovered18. A previous approach assessing the methylation status of arthritis associated genes also reported that ELMO1 locus was hypomethylated in fibroblast-like synoviocytes (FLS) that line the synovium of the joints19. ELMO1 functions at the interface between the phagocytic receptors and their downstream cytoplasmic signaling activity, leading to corpse internalization13,17,20,21. As apoptotic cell clearance is an anti-inflammatory process, we hypothesized that disruption of ELMO1 might lead to greater joint inflammation.
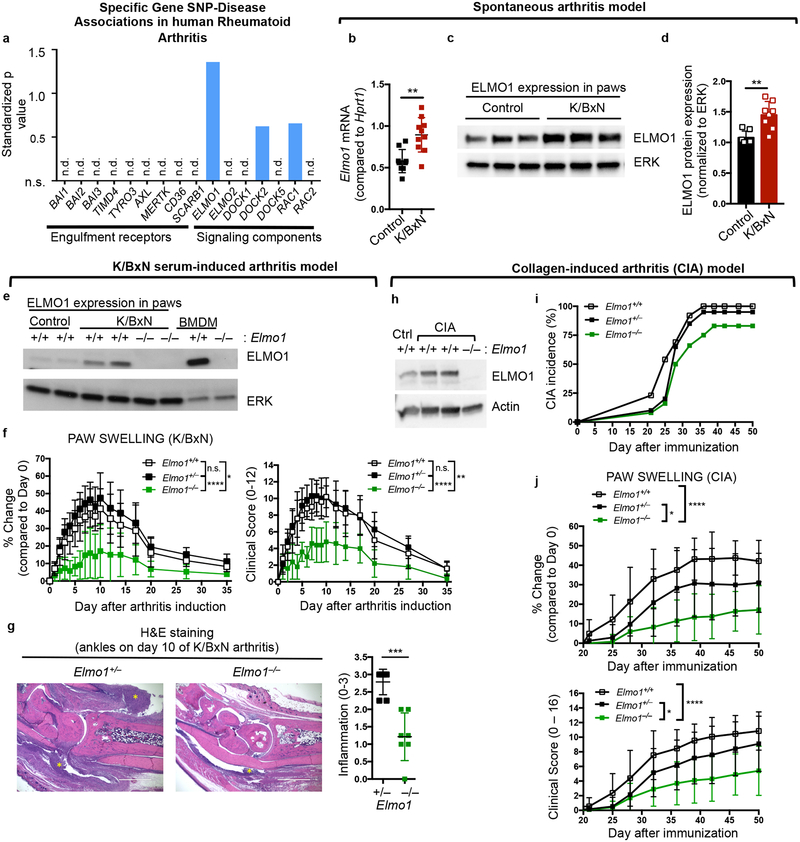
Fig. 1. Engulfment protein ELMO1 contributes to inflammatory arthritis.
a) Disease SNPs in Rheumatoid Arthritis discovered via search of the GWASdb SNP-disease association database. The data are plotted using a standardized p value.
b) Expression of Elmo1 in total paw extracts from K/BxN mice by qRT-PCR. Mean value +/− SD is shown. Each symbol represents an individual animal.
c, d)ELMO1 protein level by immunoblotting (c) and the quantification (d) in total paw extracts from K/BxN mice. The blot was cropped to show relevant bands.
e) ELMO1 protein in the paw extracts of Elmo1+/+ or Elmo1–/– mice either day 0 (Control) or day 10 after K/BxN serum injection. Bone marrow derived macrophages (BMDM) are shown as control.
g) Paw swelling and clinical scores of Elmo1+/+ (n=5, white symbols), Elmo1+/– (n=7, black symbols) and Elmo1–/– (n=5, green symbols) mice injected with 150 μl of K/BxN serum on day 0 and 2.
h) H&E staining of hind paws (see Methods) on day10 after K/BxN serum injection. Yellow asterisks indicate areas of inflammatory cell infiltration. Scale bar = 1 mm. Each symbol represents an individual animal.
i) ELMO1 protein in the paw extracts from healthy mice or on day 50 after CIA induction. The blot was cropped to show relevant bands.
j) Collagen induced arthritis incidence over the indicated time in Elmo1+/+-DBA (n=13, white symbols), Elmo1+/–-DBA (n=20, black symbols) and Elmo1–/–-DBA (n=12, green symbols) mice.
k) Paw swelling and clinical scores of Elmo1+/+-DBA (n=13, white symbols), Elmo1+/–-DBA (n=19, black symbols) and Elmo1–/–-DBA (n=10, green symbols) mice with CIA.
To test whether expression of specific engulfment machinery components might change during joint inflammation in mice, we initially analyzed the K/BxN mouse model of spontaneous arthritis22 (Supplementary Fig. 1a). K/BxN mice develop progressive joint disease resembling rheumatoid arthritis, including autoantibody production, chronic joint inflammation, and bone erosion22. We analyzed mRNA expression of the engulfment receptors Bai1, Bai2, Bai3, Tim4, Mertk and Cd36, as well as the Elmo, Dock and Rac family members in the the paw extracts of 9-week old K/BxN mice. Transcript levels for Elmo1, Rac1 and Rac2 where increased, whereas the expression of engulfment receptors either remained unchanged, or was slightly decreased (Fig. 1b and Supplementary Fig. 1b). ELMO1 protein was also increased in the paw extracts from mice with spontaneous arthritis (Fig. 1c–d). Based on this observation and the linkage of ELMO1 SNPs to human RA, we addressed the role of ELMO1 further.
Elmo1−/− mice show reduced arthritis severity in two different arthritis models
Transferring serum from arthritic K/BxN mice to healthy mice initiates joint disease in many strains of mice23. This K/BxN serum transfer arthritis is thought to model the effector phase of human arthritis (‘inflammatory flare-ups’), due to the transient nature and strict dependence on innate immune system components. Disease induction is rapid, synchronous, and nearly 100% penetrant, with swelling of the paws apparent 1–2 days post K/BxN serum injection23. We tested arthritis development after two injections of the K/BxN serum (day 0 and day 2). Extracts from diseased paws of control mice showed increased ELMO1 protein in the joints (Fig. 1e, lanes 1–4). We next assessed arthritis in Elmo1–/– mice and their control littermates using paw swelling and other disease parameters to assign clinical scores (see Methods). Elmo1–/– mice are generally healthy when housed under specific pathogen free conditions24. Although arthritis was readily observed and comparable in wild type and heterozygous Elmo1+/– mice, homozygous Elmo1–/– mice showed significantly milder disease (Fig. 1f). Histology of ankle sections (day 10) revealed severe inflammatory cell infiltration in controls that was significantly decreased in Elmo1–/– mice (Fig. 1g). The reduction in disease severity in Elmo1–/– mice was also observed with a single injection of K/BxN serum and was not sex-dependent (data not shown). Since two injections of K/BxN serum induced stronger disease (attenuated by ELMO1 deficiency), we used a two-injection regimen in the rest of this work.
We also tested whether ELMO1 plays a role in the collagen-induced arthritis (CIA) model that mimics many features of human rheumatoid arthritis25, and includes involvement of both the innate and adaptive arms of immunity. We back-crossed Elmo1–/– mice to the DBA/1J strain, in which collagen-induced arthritis is most consistently seen25, and immunized them with collagen (see Methods). Again, increased ELMO1 protein levels were seen in the paws of control mice with CIA (Fig. 1h). Although Elmo1+/+DBA mice developed signs of arthritis around day 21, with most mice becoming sick by about 40 days following immunization, about a quarter of Elmo1–/–DBA mice displayed no signs of arthritis throughout the duration of the experiment (Fig. 1i and Supplementary Fig. 1c, 1e). Among those that developed arthritis, Elmo1–/–DBA mice had delayed onset and lower disease severity compared to controls (Fig. 1j and Supplementary Fig. 1d, 1f). This difference persisted through the chronic stages of disease in both sexes (Supplementary Fig. 1c–f). These data collectively suggest that ELMO1 promotes disease severity in arthritis.
ELMO1 function in neutrophils affects arthritis disease severity
When we assessed uncleared apoptotic cells in the arthritic joints, many cleaved caspase-3 positive cells were seen in the control littermates (Fig. 2a, left panel and inset). Very few such cells were observed in the Elmo1–/– mice (Fig. 2a). Since the fewer apoptotic cells could correlate to reduced inflammation within the joints of Elmo1–/– mice, we focused on the areas of inflammation; even then, there were significantly fewer apoptotic cells (Fig. 2a, right panel). This was not due to compensatory upregulation of the homologs Elmo2 and Elmo3 in the Elmo1–/– mice (Supplementary Fig. 1g). During apoptotic cell clearance, ELMO1 binds to the cytoplasmic tail of the phosphatidylserine receptor BAI1, and via subsequent downstream signaling, promotes corpse engulfment (Supplementary Fig. 1h)13. Bai1–/– mice developed comparable disease to their wild type littermates in K/BxN serum induced arthritis model (Supplementary Fig. 1i). These data suggest that the attenuation of arthritis seen in Elmo1–/– mice is likely not linked to cell clearance per se.
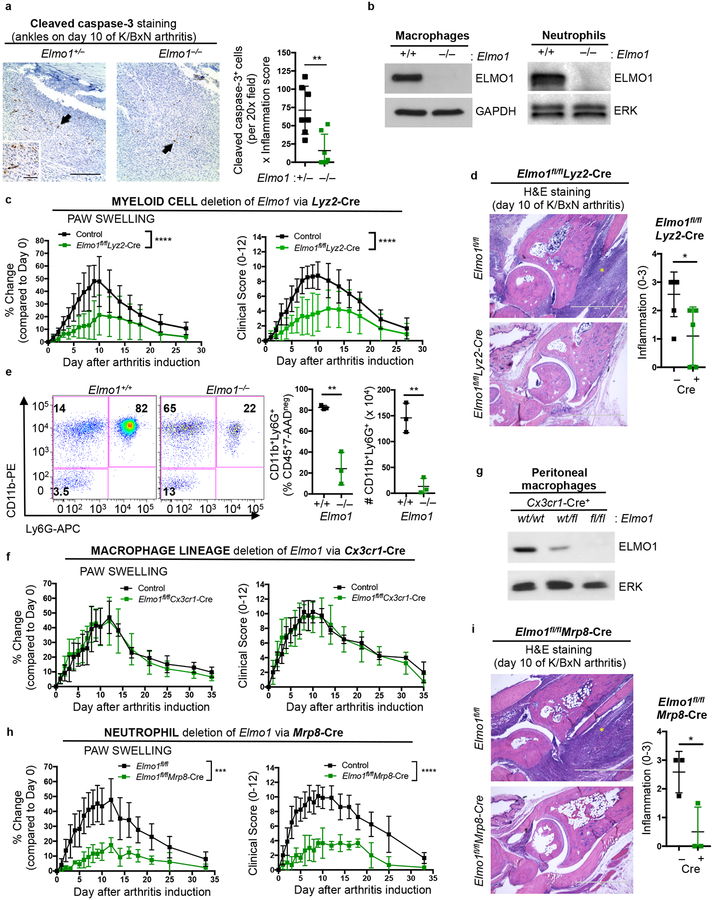
Fig. 2. ELMO1 expression in neutrophils regulates disease severity in arthritis.
a) Hind paw ankle sections were stained with cleaved caspase-3 (CC3) antibody (brown) and hematoxylin (blue) on day 10 after K/BxN serum injection. Apoptotic cells are indicated by arrows. Average CC3 positive cells in two different fields per section was multiplied by the inflammation score. Each symbol represents an individual animal.
b) ELMO1 protein expression in macrophages (left) and neutrophils (right, purified Ly6G+ bone marrow cells). The blots were cropped to show relevant bands.
c) Paw swelling and clinical scores of female Elmo1fl/fl alone or Lyz2-Cre alone (Control, n=9), or Elmo1fl/flLyz2-Cre (n=9) mice injected with K/BxN serum.
d) Representative hind paw ankle sections stained with hematoxylin and eosin on day 10 after K/BxN serum injection. Yellow asterisks indicate areas of inflammatory cell infiltration. Scale bar = 1 mm.
e) Flow cytometric analysis of paws on d10 after K/BxN serum injection. Each symbol represents an individual animal.
f) Elmo1fl/fl alone and Cx3cr1-Cre alone (Control, n=4) or Elmo1fl/flCx3cr1-Cre (n=4) mice were injected with 150 μl of K/BxN serum on day 0 and 2 and scored as in Fig. 2c.
g) ELMO1 immunoblotting of resident peritoneal macrophages. The blot was cropped to show relevant bands.
h) Elmo1fl/fl alone and Mrp8-Cre alone (Control, n=8) or Elmo1fl/flMrp8-Cre (n=3) mice were injected with K/BxN serum on day 0 and 2, and scored as in Fig. 2c.
i) Representative hind paw ankle sections analyzed as in 2d. Scale bar = 1 mm.
We then took a genetic approach to address the cell type(s) in which ELMO1 function was promoting arthritis. Disease development in the K/BxN serum transfer arthritis requires cells of the innate immune system23. As ELMO1 protein expression was detected in both macrophages and neutrophils (Fig. 2b), we generated mice with conditional deletion of Elmo1 broadly in the myeloid lineage by crossing Elmo1fl/fl mice24 with Lyz2-Cre mice26. When injected with K/BxN serum, Elmo1fl/flLyz2-Cre mice showed attenuated disease (Fig. 2c). Histology at disease maximum also revealed reduced inflammation in the ankle joints of Elmo1fl/flLyz2-Cre mice (Fig. 2d), and correlated with ELMO1 protein deletion in myeloid lineage cells (Supplementary Fig. 2a). Of note, Lyz2-Cre expression alone (without floxed Elmo1 alleles) does not alter disease development in the K/BxN serum induced arthritis (Supplementary Fig. 2b). Thus, ELMO1 expression in myeloid cells regulates arthritis severity.
Analyzing the inflammatory infiltrate within the paws, fewer CD11b+Ly6G+ neutrophils were seen at disease maximum in the global Elmo1–/– mice, as well as the Elmo1fl/flLyz2-Cre mice (Fig. 2e, Supplementary Fig. 2c–d). To test the importance of ELMO1 expression in neutrophils versus macrophage/monocytic lineages, we crossed Elmo1fl/fl mice with either Mrp8-Cre mice to delete Elmo1 expression primarily in neutrophils27, or Cx3cr1-Cre mice, which primarily targets monocytes and macrophages28. Elmo1fl/flCx3cr1-Cre mice developed disease comparable to wild type littermates after K/BxN serum injection (Fig. 2f and Supplementary Fig. 2e–f), despite decreased ELMO1 protein in the macrophage lineage (Fig. 2g). In contrast, deletion of Elmo1 in neutrophils in the Elmo1fl/flMrp8-Cre mice led to reduced disease (Fig. 2h and Supplementary Fig. 2g). Histology of ankle joints showed reduced inflammatory cell infiltration in Elmo1fl/flMrp8-Cre mice (Fig. 2i). ELMO1 deletion in neutrophils from Elmo1fl/flMrp8-Cre mice was verified by immunoblotting of purified Ly6G+ cells (Supplementary Fig. 2h). Mrp8-Cre alone does not affect disease severity (Supplementary Fig. 2i and data not shown). Although ELMO1 expression was previously reported in fibroblast-like synoviocytes (FLS)19, even in control mice, ELMO1 protein was barely detectable in the FLS (compared to the strong expression in neutrophils) (Supplementary Fig. 2j). Thus, reduced arthritis induction in Elmo1fl/flMrp8-Cre mice phenocopies the global Elmo1–/– mice (Fig. 2h and 1g).
To address whether ELMO1 functions as a broad inhibitor of inflammation, we tested several models where inflammation is associated with neutrophil infiltration. First, we challenged Elmo1+/+ and Elmo1–/– mice with intranasal administration of lipopolysaccharide (LPS) and monitored the recruitment of neutrophils to the lung (Supplementary Fig. 3a). Eight hours post-LPS administration, recruitment of neutrophils was detected in the bronchoalveolar lavage of control mice and was not significantly altered in Elmo1–/– mice (Supplementary Fig. 3b). Second, to test a bacterial infection in vivo, we subjected Elmo1+/+ and Elmo1–/– mice to fecal-induced peritonitis (FIP), a model of septic shock29, and assessed neutrophil recruitment into the infected peritoneum, as well as clinical parameters of disease development and survival (Supplementary Fig. 3c). Four hours after FIP induction, Elmo1–/– mice had comparable neutrophil infiltration as control mice to the site of infection (Supplementary Fig. 3d). The transient temperature drop, clinical scores, and survival did not differ between Elmo1+/+ and Elmo1–/– mice (Supplementary Fig. 3e–g). Additionally, purified Elmo1–/– neutrophils showed comparable ability to kill a bacterial pathogen Klebsiella pneumoniae, clearance of which potently depends on neutrophil function30 (Supplementary Fig. 3h). Third, as Elmo1–/– mice do not exhibit symptoms associated with autoimmunity under specific pathogen free conditions, we tested them in experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis that involves adaptive immunity and neutrophil function31. Elmo1–/– mice developed EAE symptoms comparable to control animals with similar demyelination (Supplementary Fig. 3i–j and data not shown). These data suggest that ELMO1 deficiency does not generally impair inflammation, and that there is likely a specificity associated with ELMO1 loss in neutrophils and attenuation of arthritis signs.
ELMO1 regulates neutrophil chemotaxis to inflamed joints
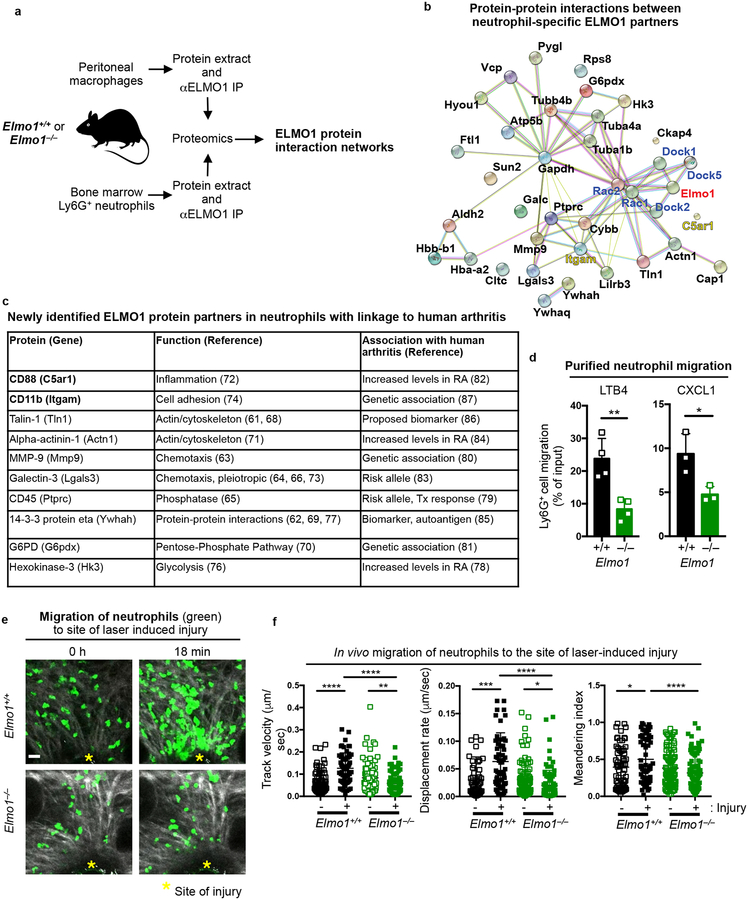
To better understand the differences in arthritis severity due to ELMO1 deletion in neutrophils versus macrophages, and to help decipher how ELMO1 might affect neutrophil function during arthritis, we assessed the ELMO1 protein interactome. We purified Ly6G+ neutrophils from the bone marrow and resident peritoneal macrophages from Elmo1+/+ and Elmo1–/– mice, and performed ELMO1 immunoprecipitation, followed by liquid chromatography mass spectrometry (LC-MS) and proteomics analysis (Fig. 3a). Comparison of the proteomics results between Elmo1+/+ and Elmo1–/– mice helped identify ELMO1-specific protein partners (direct or indirect interaction with ELMO1). Further analysis of ELMO1 protein interactomes in neutrophils versus macrophages (see Methods), helped identify the neutrophil-specific ELMO1 protein interaction network composed of 30 proteins (Fig. 3b and Supplementary Table 2). Neutrophil-specific ELMO1 protein interactome was analyzed by STRING (string-db.org) to reveal protein-protein connections32. The known ELMO1 partners DOCK and RAC were added to provide context (Fig. 3b, highlighted in blue). The newly identified neutrophil-specific ELMO1 partners have a range of distinct functions and several have known associations with human arthritis (Fig. 3c, see Methods for arthritis linkage references).
Fig. 3. Neutrophil-specific ELMO1 protein interactome includes regulators of chemotaxis linked to human arthritis.
a) Schematic of neutrophil and macrophage preparation for proteomic analyses.
b) Neutrophil-specific ELMO1 protein interaction network analyzed by STRING (string-db.org). Known protein interactions are indicated by the magenta (experimentally determined) and cyan (curated database indicated) colored lines. Predicted protein interactions are indicated by green (gene neighborhood), olive (text-mining), black (co-expression) and violet (protein homology) colored lines.
c) Linkages of newly identified neutrophil-specific ELMO1 protein partners to human arthritis (see Methods for references). RA – rheumatoid arthritis; Tx – therapy.
d) Ly6G+ bone marrow cells were purified and cell migration to LTB4 (1 nM, n=4) or CXCL1 (25 ng/ml, n=3) evaluated. Each symbol represents an individual animal.
e) Representative images of migration of Elmo1+/+ (n=2) and Elmo1–/– (n=3) neutrophils in vivo after laser-induced injury (see Methods). Damaged area is indicated with yellow asterisks. Second harmonic generation (gray), and neutrophils (green) are shown. Scale bar = 20 μm.
f) Track velocity, displacement rate and meandering index of individual Elmo1+/+ and Elmo1–/– neutrophils before (−) and after (+) laser-induced injury. A total of 66 (−) and 60 (+) neutrophils from Elmo1+/+ (n=2) and 93 (−) and 87 (+) from Elmo1–/– (n=3) mice are shown.
Interestingly, several of the neutrophil-specific ELMO1 protein partners are linked to cell migration (Fig. 3b–c), Initial neutrophil recruitment into the synovium during arthritis is mediated by leukotriene B4 (LTB4) and further sustained by CXCR2 agonists such as CXCL133. As ELMO1 has also been linked to gradient dependent migration20,21, we examined migration of purified neutrophils from wild-type and ELMO1-deficient mice in a Transwell migration assay. Elmo1–/– neutrophils (either purified or in the context of total bone marrow) showed significantly reduced migration toward LTB4 and CXCL1 (Fig. 3d, Supplementary Fig. 4a). To visualize the linkage between ELMO1 expression and neutrophil migration in vivo, we used two-photon microscopy to analyze neutrophil swarming towards the site of laser-induced injury in the mouse ear (which is also LTB4-dependent34). Compared to the increased and directed migration toward the site of injury of control neutrophils, Elmo1–/– neutrophils showed defective migration to the injury site and did not increase the speed of movement (Fig. 3e–f and Supplementary Video 1).
Since neutrophil migration into the joint synovium is a hallmark of acute inflammatory episodes in human rheumatoid arthritis, we asked whether ELMO1 might affect neutrophil migration to the joints. Elmo1+/– and Elmo1–/– mice were injected with K/BxN serum, and neutrophil infiltration into the joints 24h later was imaged via IVIS scanning by the luminescent signal that arises from the myeloperoxidase-mediated conversion of injected luminol35. Compared to control animals, Elmo1–/– mice had much reduced luminescence in the joints (Fig. 4a). This was not due to altered expression of the receptors for the chemokines LTB4 or CXCL1, as Elmo1–/– neutrophils expressed comparable LTB4 receptor (Blt1) transcript to Elmo1+/+ cells (Supplementary Fig. 4b), and the protein levels of the CXCL1 receptor CXCR2 were unchanged on the surface of neutrophils in both blood and bone marrow (Supplementary Fig. 4c).
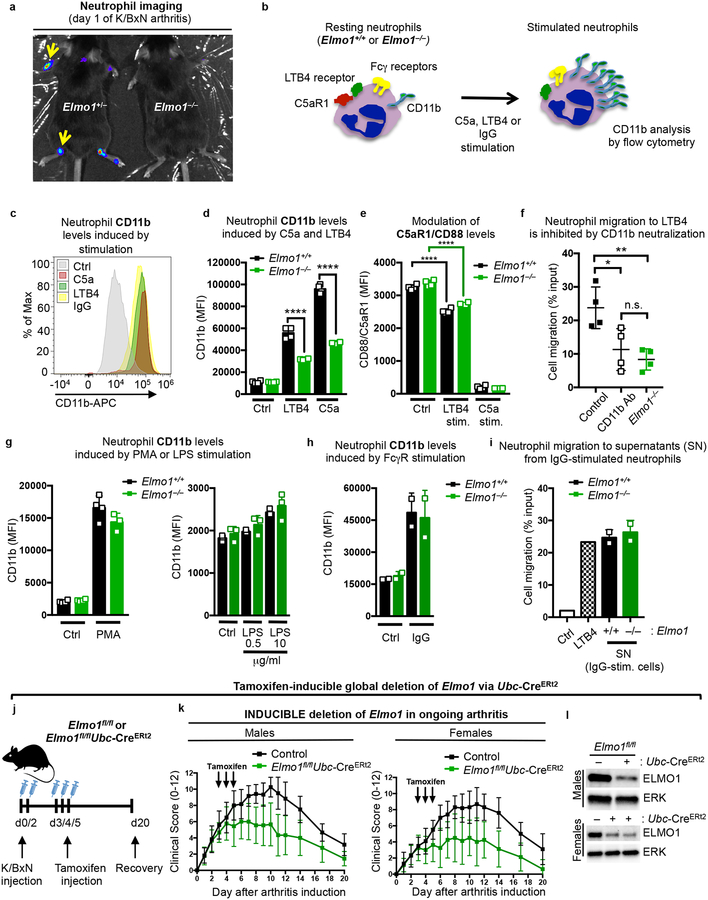
Fig. 4. ELMO1 promotes neutrophil migration to inflamed joints.
a) Mice injected with luminol on d1 of K/BxN serum induced arthritis were imaged via IVIS. Arrows point to areas of neutrophil recruitment in the joints.
b) Schematic for cell surface CD11b up-regulation upon different neutrophil stimuli.
c) Flow cytometric profile of CD11b on neutrophils under unstimulated (Ctrl) or 1h after C5a, LTB4, or IgG-stimulation.
d) CD11b geometric mean fluorescence index (MFI) in resting (Ctrl) and LTB4- or C5a-stimulated neutrophils purified from Elmo1+/+ (black) and Elmo1–/– (green) mice. Each symbol represents an individual animal.
e) C5aR1/CD88 MFI on neutrophils purified from Elmo1+/+ (black) and Elmo1–/– (green) mice, with C5a-stimulation comparably down-modulating surface C5aR1/CD88 levels. Each symbol represents an individual animal.
f) Purified neutrophil migration towards LTB4 (1 nM) with or without CD11b neutralizing antibody (10 μg/ml), compared to Elmo1–/– neutrophils. Each symbol represents an individual animal.
g) CD11b MFI) in resting (Ctrl) and PMA (50 ng/ml) or LPS (right) stimulated neutrophils. Each symbol represents an individual animal.
h) CD11b MFI) in resting (Ctrl) or IgG (plate-bound) stimulated neutrophils.
i) Purified neutrophil migration towards supernatants from IgG-stimulated Elmo1+/+ (black) and Elmo1–/– (green) neutrophils. Migration towards LTB4 (10 nM, checkered) was used as a positive control. Each symbol represents migration to supernatants from an individual animal.
j) Schematic of tamoxifen induced deletion of Elmo1 during ongoing arthritis.
k) Clinical scores of male and female Elmo1fl/flUbc-CreERt2 (n=7–9, green symbols) and littermate control (n=10–12, black symbols) mice with K/BxN serum transfer arthritis.
l) ELMO1 protein level in cells from the peritoneum (males) or bone marrow (females,) from tamoxifen-treated Elmo1fl/flUbc-CreERt2 (+) and littermate control (−) mice on d20 after arthritis induction. The blots were cropped to show relevant bands.
ELMO1 acts in the signaling relay between chemokine receptors and neutrophil activation/migration
The above data demonstrated that ELMO1 regulates neutrophil migration in vitro and in vivo, and in response to chemokines linked to arthritis. As neutrophil migration to other inflammatory stimuli (such as LPS stimulation or bacterial infection) was not affected by loss of ELMO1, one possible mechanistic explanation was that ELMO1 might function downstream of receptor(s) for arthritis-linked chemokines. Our proteomics data (Fig. 3b–c) identified two ELMO1-binding membrane proteins that have been shown to function in inflamed joints - C5aR1 (CD88), receptor for the complement component C5a (also called anaphylatoxin), and the integrin CD11b (Itgam) (Fig. 3b, highlighted in yellow)36,37.
C5a-C5aR1 is referred to as a ‘master switch’ in neutrophil recruitment that drives establishment of clinical signs of disease in arthritis38–40. Of note, C5a-C5aR1 signaling does not appear to regulate disease phenotypes in EAE or LPS challenge models41,42, which we also found are not influenced by ELMO1 deficiency (Supplementary Fig. 3). In arthritis, C5aR1 ligation is uniquely required for initial neutrophil adhesion to the endothelium and firm arrest, and is mediated via upregulation of integrins on the neutrophil surface, such as CD11b (our second ELMO1 binding partner of interest)43. CD11b is also a neutrophil activation marker that is upregulated by different stimuli to regulate migration, including LTB4-dependent neutrophil swarming34. We asked whether CD11b cell surface exposure induced by neutrophil activation with C5a or LTB4 would be altered due to ELMO1 deficiency (Fig. 4b). CD11b levels on wild type neutrophils were strongly increased by C5a or LTB4 stimulation (Fig. 4c–d), and this was significantly stunted in Elmo1–/– neutrophils (Fig. 4d). This suggested that ELMO1 functions in a signaling relay (i.e. inside-out signaling) between neutrophil activation and CD11b integrin-mediated cell adhesion. As control, we did not observe differences in LTB4 or C5a receptor expression, or ligand-dependent C5aR1 internalization in Elmo1–/– neutrophils (Fig. 4e). We then asked whether CD11b is required for neutrophil migration in response to LTB4. Blocking CD11b function with a neutralizing antibody reduced neutrophil migration to the levels observed in Elmo1–/– cells (Fig. 4f), suggesting that ELMO1 regulation of neutrophil migration to LTB4 is in part dependent on CD11b. ELMO1-deficient neutrophils were not inherently deficient in CD11b upregulation, as bypassing receptor-mediated activation by a strong intracellular stimulus such as PMA resulted in comparable CD11b increase between Elmo1+/+ and Elmo1–/– neutrophils (Fig. 4g, left panel).
We also tested the specificity of ELMO1 for CD11b upregulation and migration in response to arthritis-associated chemokines LTB4 and C5a. Poor induction of CD11b by LPS was noted in neutrophils from control or Elmo1–/– mice (even at high LPS concentrations) (Fig. 4g, right panel). We also examined CD11b induction with aggregated IgG (to mimic immune complexes), and found that CD11b was comparably upregulated in WT and Elmo1–/– neutrophils (Fig. 4h). Immune complex mediated neutrophil degranulation and release of chemotactic factors was also not impaired by ELMO1 deficiency, as determined by migration of wild type neutrophils to the chemotactic mediators present in the supernatants from IgG-activated Elmo1–/– neutrophils (Fig. 4i). It is worth noting that in Elmo1–/– neutrophils from blood and bone marrow we found no alterations in the surface levels of neutrophil receptors FcγRIII and FcγRIV35,38,44, or the protein tyrosine kinase Syk required for Fcγ receptor signaling45 (Supplementary Fig. 4d–f). Together, these data suggest that ELMO1 is part of the signaling relay that occurs between specific arthritis-associated chemokines (such as C5a and LTB4) and the neutrophil activation marker CD11b linked to neutrophil migration.
Induced deletion of Elmo1 after disease onset reduces arthritis severity
To test whether deletion of Elmo1 after induction of arthritis can alleviate disease parameters, we crossed Elmo1fl/fl mice with Ubc-CreERt2 mice, in which the Cre-recombinase is expressed ubiquitously; however, Cre stays functionally inactive due to its fusion with an ER-domain until the administration of tamoxifen, which can be done in adult mice at the desired time46. After verifying that Elmo1 deletion in Elmo1fl/flUbc-CreERt2 mice does not occur prior to tamoxifen administration (data not shown), we first induced arthritis in Elmo1fl/flUbc-CreERt2 mice and littermate controls by injection of K/BxN serum on days 0 and 2 (Fig. 4j). Starting on day 3, we administered tamoxifen (see Methods) to induce Elmo1 deletion, and monitored disease parameters. Tamoxifen administration in arthritic Elmo1fl/flUbc-CreERt2 mice resulted in the rapid arrest of further disease development and enhanced recovery in both sexes (Fig. 4k and Supplementary Fig. 5a), compared to tamoxifen-treated control mice. Since only a partial loss of ELMO1 protein was observed in peritoneal and bone marrow cells from Elmo1fl/flUbc-CreERt2 mice (Fig. 4l), this suggests that even partial reduction in ELMO1 during acute phases of arthritis could be of benefit. Importantly, Elmo1fl/flUbc-CreERt2 mice did not have reduced disease if tamoxifen was not administered and deletion of Elmo1 was not induced (Supplementary Fig. 5b).
We also deleted Elmo1 during the peak of K/BxN serum induced arthritis, after neutrophil mobilization has already occurred (Supplementary Fig. 5c). When induced by this ‘late’ regimen, loss of ELMO1 did not significantly improve signs of disease (Supplementary Fig. 5d), supporting the critical role for ELMO1 in the early recruitment of neutrophils to the joint.
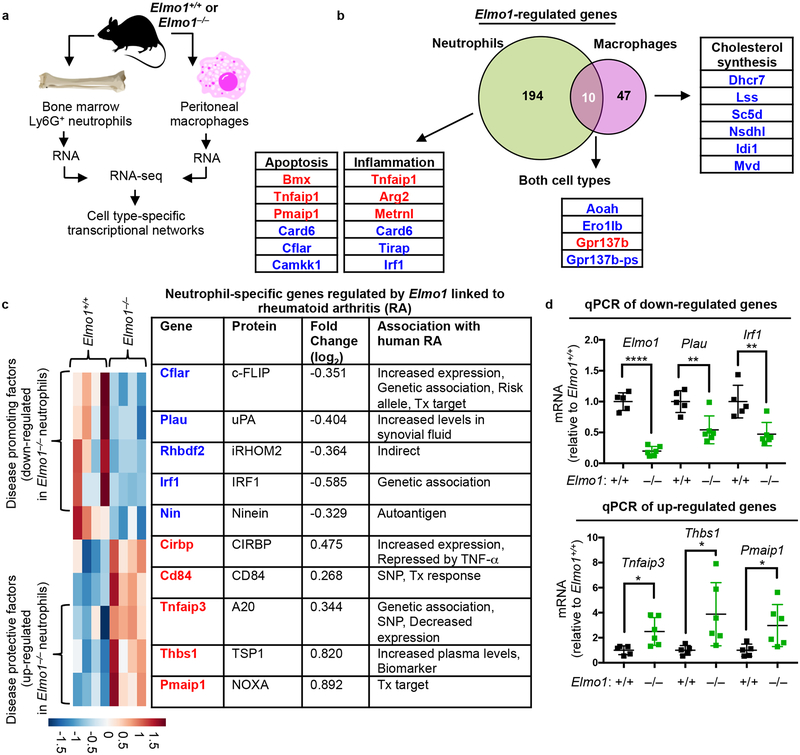
Elmo1 regulates cell type-specific transcriptional programs
Since Rac1, which functions downstream of ELMO1, can regulate gene transcription, we next tested whether ELMO1 modulates transcriptional networks by RNA-seq analysis of cells with or without ELMO1. We also compared the transcriptomes of neutrophils and macrophages to identify neutrophil-specific ELMO1-dependent transcriptional signatures (Fig. 5a). In both cell types, modulators of actin cytoskeleton dynamics and cell adhesion/migration were regulated by Elmo1 (Supplementary Table 3). However, we noted a clear difference in the prominent classes of functionally related transcripts between neutrophils and macrophages. There was an overall paucity of shared genes and transcriptional programs regulated by Elmo1, suggesting a previously unappreciated cellular specificity in Elmo1-dependent gene networks. Elmo1-regulated transcriptomes in neutrophils and macrophages overlapped in only ten genes (including Elmo1 itself) (Fig. 5b and Supplementary Table 3). We found that Elmo1 deficiency in neutrophils associated with decreased expression of arthritis-promoting factors and increased expression of protective modulators (Fig. 5c–d), with many of the Elmo1-dependent neutrophil-specific genes previously linked to arthritis disease progression (Fig. 5b–d, see Methods for arthritis linkage references). Moreover, neutrophils lacking ELMO1 exhibited an anti-inflammatory signature (Fig. 5b and Supplementary Table 3), exemplified by the marked up-regulation of the gene Tnfaip1 (encoding A20), a potent inhibitor of inflammatory signaling and arthritis in mice47. Collectively, these data suggest that Elmo1 could modulate neutrophil function in inflammatory arthritis via mechanisms beyond the regulation of migration.
Fig. 5. Elmo1 function regulates cell type-specific transcriptional programs.
a) Schematic for preparation of neutrophils and macrophages for RNA isolation and RNA-seq experiments.
b) Venn diagram of Elmo1-regulated transcripts in neutrophils and macrophages. Examples of shared and cell-type specific Elmo1-regulated transcripts are shown. Compared to Elmo1+/+ cells, those transcripts down-regulated or upregulated in Elmo1–/– cells are indicated in blue and red text, respectively.
c) Heatmap (each column represents an animal, n=4) and list of Elmo1-regulated neutrophil-specific transcripts with associations to human Rheumatoid Arthritis (RA). SNP – single nucleotide polymorphism; Tx – therapy.
d) Quantitative PCR validation of some neutrophil-specific arthritis-associated genes in c). Each symbol represents a mouse.
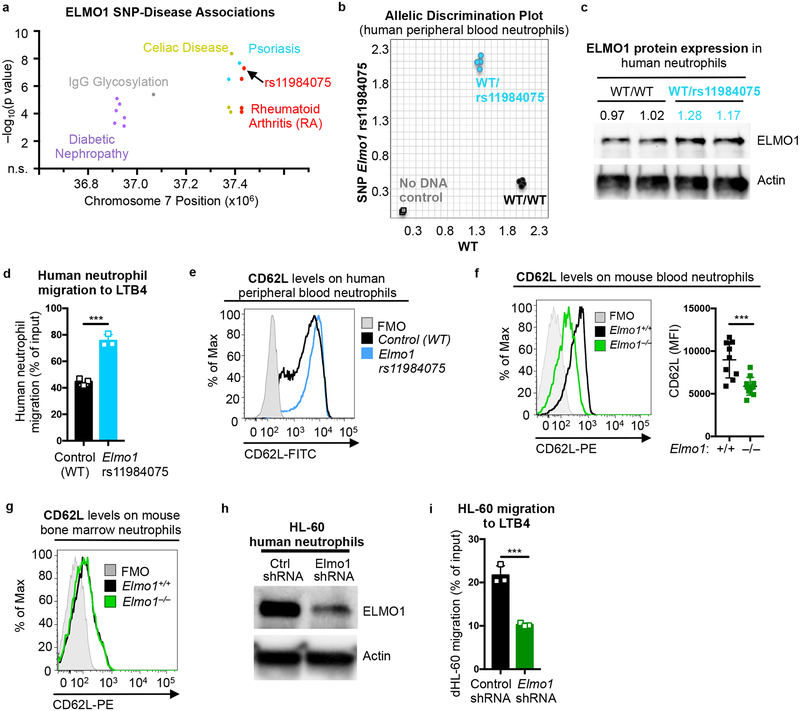
ELMO1 SNP rs11984075 alters neutrophil migration
Within the human ELMO1 locus, we found multiple SNPs correlating with diabetic nephropathy, IgG glycosylation, celiac disease, and psoriasis, in addition to rheumatoid arthritis (Fig. 6a). Although none of the RA-associated SNPs in ELMO1 are known to cause changes in mRNA or protein expression, Elmo1 levels were altered in a mouse model of diabetic nephropathy48. ELMO1 protein expression is readily detected in human peripheral blood neutrophils (Supplementary Fig. 5e). To test whether the presence of SNPs in ELMO1 linked to RA alter ELMO1 protein levels and neutrophil functions, we screened 15 healthy blood donors for ELMO1 rs11984075 and ELMO1 rs10488029 (see Supplementary Table 1). Two of the fifteen donors were heterozygous for ELMO1 rs11984075 (Fig. 6b), whereas none had ELMO1 rs10488029 (not shown). ELMO1 protein expression in neutrophils from the two ELMO1 rs11984075 donors was elevated ~20% compared to controls (Fig. 6c), suggesting that this intronic mutation could have additional effects on gene expression49. Indeed, neutrophils from the two ELMO1 rs11984075 donors displayed increased migration toward LTB4 in a Transwell assay (Fig. 6d) and had higher L-selectin (CD62L), a leukocyte receptor that mediates early stage adhesion to the vascular endothelium during neutrophil extravasation and contributes to arthritis development50 (Fig. 6e). This was particularly interesting, as CD62L on the surface of circulating neutrophils in Elmo1–/– mice was significantly reduced (Fig. 6f), suggesting an additional avenue for ELMO1 regulation of neutrophil recruitment. Since the bone marrow neutrophils from Elmo1–/– mice had comparable levels of CD62L (Fig. 6g), loss of ELMO1 likely regulates this adhesion molecule in fully matured neutrophils.
Fig. 6. ELMO1 rs11984075 SNP promotes migration of human neutrophils.
a) Unique SNPs in ELMO1 associated with indicated diseases. SNPs are plotted as the negative log of the p value significance by chromosomal position.
b) Allelic discrimination plot of ELMO1 and rs11984075 SNP alleles in the genomic DNA from healthy human donors in duplicates. Two donors were heterozygous for rs11984075 (blue). Two donors with wild type allele (WT) donors are shown. Controls (no DNA) are shown in gray.
c) Immunoblotting of ELMO1 protein in peripheral blood neutrophils from human donors carrying the indicated alleles. Protein abundance was normalized to actin and presented as fold-WT. The blot was cropped to show relevant bands.
d) Transwell migration to LTB4 (15nM) by purified human peripheral blood neutrophils from one control and one Elmo1 rs11984075 donor.
e) Representative flow cytometry plot of cell surface L-selectin/CD62L on purified human peripheral blood neutrophils from donors of the indicated alleles. FMO, fluorescence minus one.
f) Representative flow cytometry plots of CD62L levels on the surface of Ly6G+ peripheral blood neutrophils (left) and quantification of MFI (right) in Elmo1+/+ (n=9) and Elmo1–/– (n=11) mice. Each symbol represents a mouse.
g) Flow cytometry plots of CD62L levels on the surface of Ly6G+ neutrophils from bone marrow.
h) ELMO1 protein expression in HL-60 cells stably transfected with non-targeting control (Ctrl) or ELMO1 targeting shRNA. Actin was used as a loading control. The blot was cropped to show relevant bands.
i) Migration to LTB4 (100nM) was evaluated in HL-60 clones expressing control or ELMO1 targeting shRNA as described in the Methods.
As a corollary to this ‘gain of function’ in migration with rs11984075, we tested whether loss of ELMO1 in human neutrophils might affect their migration to arthritis-associated chemokines. Since ELMO1 depletion in primary human neutrophils is not feasible (due to their short life span), we utilized the human neutrophil cell line HL-60 with knockdown of ELMO1 expression. ELMO1-deficient HL-60 (Fig. 6h) differentiated into neutrophils exhibited reduced migration to LTB4 compared to controls (Fig. 6i). Collectively, these observations demonstrate that ELMO1 modulates human neutrophil chemotaxis to soluble mediators linked to arthritis.
DISCUSSION
Although a few recent biologics targeting specific inflammatory components have revolutionized the control of arthritis symptoms, they have been effective in only a subset of patients3. This work, based on several lines of evidence, identifes the ELMO1-dependent molecular signaling nexus with potential new targets for therapeutic intervention. First, the enhanced expression of ELMO1 protein and enhanced migration of neutrophils from healthy donors carrying the SNP rs11984075 imply a key linkage of SNPs in ELMO1 to human arthritis at a functional and genetic level. Although this clearly needs to be further validated, screening for such SNPs could be a potential biomarker for arthritis susceptibility. Second, based on K/BxN serum transfer and collagen-induced arthritis models, ELMO1 can affect both acute and chronic phases of the disease. Third, many of the genes and proteins regulated in an ELMO1-dependent manner in neutrophils are genetically linked to human arthritis (via SNPs, altered expression, or biomarker status), or are functionally linked to experimental arthritis. Fourth, as deletion of ELMO1 after arthritis initiation could decrease signs of disease in Elmo1fl/flUbc-CreERt2 mice, targeting ELMO1 in ongoing human arthritis may be of benefit. As our data suggest that ELMO1 deficiency does not impair neutrophil response to bacterial challenge, targeting ELMO1 function may not indiscriminately suppress the immune system.
Interestingly, in the transcriptomics of neutrophils and macrophages, only ten core genes were commonly regulated in both cell types due to ELMO1. This goes against the ‘general assumption’ that the same genes/proteins likely perform similar functions in different cell types, especially those within the myeloid lineage. Thus, the potential benefit of targeting common pathways in specific instances needs to be explored.
Much of the literature suggests that the apoptotic cell clearance process is generally anti-inflammatory. The data here advance a concept that evolutionarily conserved apoptotic cell engulfment machinery components can have both canonical and non-canonical roles in inflammation, depending on the cell type and context. Since induction of apoptosis in rheumatoid tissues has been proposed as a potential therapeutic approach7, consideration of non-canonical and less expected roles of engulfment proteins is warranted.
METHODS
Mice
C57BL/6J, DBA/1J, NOD, Mrp8-Cre, Cx3cr1-Cre, Lyz2-Cre, and Ubc-CreERt2 mice were obtained from Jackson Laboratories. Elmo1fl/fl and Elmo1–/– mice have been described previously24. To generate mice with deletion of Elmo1 in myeloid cells, neutrophils or macrophages, Elmo1fl/fl mice were crossed to Lyz2-Cre, Mrp8-Cre or Cx3cr1-Cre mice, respectively. To generate mice with inducible deletion of Elmo1, Elmo1fl/fl mice were crossed to Ubc-CreERt2. In these mice, Elmo1 deletion was induced by three daily intraperitoneal injections of 40 mg/kg tamoxifen dissolved in corn oil. To generate Elmo1–/–DBA mice, Elmo1–/– mice were backcrossed onto the DBA/1J background for at least 5 generations. KRN TCR transgenic mice22 were a gift from Dr. Diane Mathis at the Harvard Medical School, and were bred to NOD mice to obtain the K/BxN mice23, which develop progressive spontaneous arthritis. K/BxN serum was collected from 9-week old K/BxN mice by cardiac puncture. Age- and sex-matched littermate control animals were used for all experiments, and both males and females were assessed. In Elmo1fl/flLyz2-Cre mice, reduced disease severity was observed in female, but not in male mice (data not shown). All animal procedures were approved by and performed according to guidelines of the Institutional Animal Care and Use Committee (IACUC) at the University of Virginia.
K/BxN serum transfer induced arthritis
Mice were injected with 150 μl of serum from K/BxN mice on day 0 (one dose K/BxN) or on days 0 and 2 (two dose K/BxN). In experiments with inducible deletion of Elmo1, tamoxifen was administered on days 3, 4 and 5. Paw swelling was measured at indicated time points using a caliper (Fisher). Clinical scores were assigned for each paw as follows: 0 – no paw swelling or redness observed, 1 – redness of the paw or a single digit swollen, normal V shape of the hind foot (the foot at the base of the toes is wider than the heel and ankle) 2 – two or more digits swollen or visible swelling of the paw, U shape of the hind foot (the ankle and the midfoot are equal in thickness), 3 – reversal of the V shape of the hind foot into an hourglass shape (the foot is wider at the heel than at the base of the toes). A combined clinical score of all paws is presented. Scores were assigned by an investigator blinded to the mouse genotypes.
Collagen induced arthritis (CIA)
Elmo1–/– mice and wild type controls in the DBA background were immunized at the base of the tail by intradermal injection of a 1:1 solution of 10 mM acetic acid containing 100 μg of bovine collagen-II and Complete Freund’s Adjuvant (Sigma) containing 100 μg of heat killed Mycobacterium tuberculosis H37Ra (BD). Mice received a booster immunization on day 21. Paw swelling was measured at indicated time points using a caliper. Clinical scores were assigned for each paw as follows: 0 – no paw swelling or redness observed, 1 – redness of the paw or a single digit swollen, 2 – two or more digits swollen or visible swelling of the paw, 3 – visible swelling of the ankle, 4 – ankylosis of the joint. A combined clinical score of all paws is presented. Scores were assigned by an investigator blinded to the mouse genotypes.
Experimental autoimmune encephalomyelitis (EAE)
EAE was induced in female mice (8 to 12 weeks) by subcutaneous injection of MOG35–55 peptide (100 μg, CSBio) emulsified in complete Freund’s adjuvant. Pertussis toxin (250 ng, List Biologicals) was administered i.p. on the day of and 1d after MOG immunization. For clinical evaluation, mice were scored daily according to the following criteria: 0 - no clinical disease, 1 - limp tail, 2 - hind limb weakness, 3 - hind limb paralysis, 4 - hind limb paralysis and partial front limb paralysis, 5 - moribund. Scores were assigned by an investigator blinded to the mouse genotypes.
Histology
For K/BxN serum transfer induced arthritis, mice were euthanized at the indicated time points and the paws were fixed in 10% formalin (Fisher). Decalcification, sectioning, paraffin embedding and hematoxylin and eosin (H&E) staining was performed by HistoTox Labs (Boulder, CO). Histology scoring was performed by an investigator blinded to the mouse genotypes. For inflammation scoring, the following criteria were used - 0, none; 1, mild; 2, moderate; 3, severe. For bone erosion scoring, the following criteria were used: 0, no bone erosions observed; 1, mild cortical bone erosion; 2, severe cortical bone erosion without the loss of bone integrity; 3, severe cortical bone erosion with the loss of cortical bone integrity and trabecular bone erosion. Cleaved caspase-3 staining was performed at the University of Virginia Biorepository and Tissue Research Facility. For RNA and protein isolation, the paws were snap frozen in liquid nitrogen at indicated time points and subjected to mechanical disruption using a tissue pulverizer (Spectrum Laboratories, Inc.). For EAE experiments, spinal cords were isolated on day 26 after immunization, paraffin embedded and 4 μm sections were cut, stained with Luxol Fast Blue and counterstained with hematoxylin following standard protocols. Extent of demyelination (loss of blue staining) in the white matter of the spinal cord was measured at 4 different levels of the spinal cord.
Flow cytometry of arthritic joints
Mice were injected with the K/BxN serum on day 0 and day 2, as described above. On day 10, mice were euthanized and the skin removed from the hind paws. Ankle tendons were cut and the foot was detached from the tibia and placed in DMEM containing 20 units/ml of collagenase IV (Roche). The soft tissue was cut with a scalpel and the joints opened by gentle pulling of the foot bones. The paws were incubated for 2 hours at 37°C with periodic trituration. Liberated cells were collected, strained though a 70 μm filter (Fisher), washed with DMEM, counted and subjected to flow cytometry on Canto I flow cytometer using 7-aminoactinomycin D (7-AAD) to identify live cells, and anti-CD45 (eBioscience clone 30-F11), anti-CD11b (eBioscience clone M1/70) and anti-Ly6G (clone 1A8, BD Pharmingen) antibodies.
Neutrophil isolation, stimulation and in vitro migration
Femurs were removed from 6–8 week old mice, flushed with sterile serum-free α-MEM and single cell suspensions were prepared using the 70 μm cell strainer. Cells were incubated with anti-Ly6G antibody (clone 1A8, BD Pharmingen) and purified using the anti-PE MACS kit (Myltenyi), following manufacturer’s instructions. Purified neutrophils (Ly6G+) were seeded in 96-well tissue culture U-bottom plates (106 cells per well) and stimulated with 100 ng/ml mouse C5a (R and D Systems), 1–10 nM LTB4, 50 ng/ml PMA (Calbiochem) or 0.5–10 μg/ml LPS (Escherichia coli 0111:B4, Sigma). Alternatively, high-binding 96-well plates (Greiner Bio-One) were coated with 50 μg/ml of mouse IgG (Southern Biotech) in PBS for 16 hours at 4°C, washed with PBS and purified neutrophils were seeded as above to induce FcγR-mediated neutrophil stimulation. Cells were stimulated for 1 hour at 37°C, and the cells were then washed and stained with antibodies specific for CD11b or CD88/C5aR1 (eBioscience clone S5/1) and subjected to flow cytometry analysis on the Canto I (BD Bioscience) or the Attune Nxt (Thermo Fisher) flow cytometer. In some experiments, supernatants from cells stimulated with IgG were collected by centrifugation and removal of cell pellets and used for migration, as described below. For migration experiments, total bone marrow (2 × 106 cells per well) or purified Ly6G+ bone marrow cells (0.2–0.5 × 106 cells per well) were seeded in top chambers of 3 μm polycarbonate Transwell plates (Corning) in 100 μl of HBSS (with calcium and magnesium, no phenol red, Gibco) containing 1% bovine serum albumin (Roche). Bottom chambers contained 600 μl of media without (control) or with the indicated concentrations of leukotriene B4 (LTB4, Tocris Bioscience) or CXCL1 (BioLegend), or undiluted supernatants from IgG-stimulated neutrophils. After 3 hours at 37°C, the cells in the bottom wells were collected and subjected to flow cytometry analysis with counting beads (Spherotech, Inc.). The data is presented as percent of input cells. Alternatively, bone marrow neutrophils were isolated using the EasySep Mouse Neutrophil Enrichment Kit according to manufacturer’s instructions (StemCell Technologies, Vancouver, BC). For some experiments, peripheral blood cells and the bone marrow cells were stained for flow cytometry with anti-Ly6G (clone 1A8, BD Pharmingen), anti-CD11b (eBioscience clone M1/70), anti-CD62L (eBioscience clone MEL-14), anti-CXCR2 (BioLegend clone SA045E1), anti-FcgRIII (R&D Systems clone # 275003), or antiFcgRIV (Sino Biological, clone #012), or for immunoblot with anti-FcgRI (Santa Cruz sc-7642) or anti-Syk (Santa Cruz sc-1077).
Intravital imaging of neutrophil motility
To visualize migration of neutrophils, intravital imaging was performed using FV1000-AOM multiphoton system (Olympus) equipped with a 25x NA1.05 water immersion objective. For two-photon excitation, a Mai-Tai HP Ti:Sa Deep See laser system (Spectra-Physics) was tuned to 900nm for imaging. Neutrophils labeled with CellTracker™ Green CMFDA (Thermo Fisher Scientific Inc.) were intradermally injected into ear skin 2h before in vivo imaging. Mice were anesthetized by intraperitoneal injection of pentobarbital (Nembutal Sodium solution, Oak Pharmaceuticals, Inc., IL) and anesthetic condition was maintained using isoflurane. Hair of the mouse ear was removed using a commercial hair remover (Nair, Princeton, NJ). The anesthetized mice were laid in a ventral recumbent position on a custom-designed stage to expose the dorsal side of the ear pinna for imaging. Micropore™ (3M health care, MN) tapes were placed to immobilize the ear skin. Body temperature of the mice was controlled with heat pad and the ear immersed in a drop of glycerol/saline (40:60 v/v) and covered with a coverslip. Laser injury was induced by focusing the multiphoton laser tuned to 800 nm at a region within the ear dermis for 5 seconds. To track and analyze the movements of the neutrophils, Volocity software (Improvision/Perkin-Elmer, Waltham, MA) and ImageJ (National Institutes of Health, Bethesda, MD) were used.
LPS challenge and bacterial killing
Elmo1+/+, Elmo1–/–, Elmo1fl/fl and Elmo1fl/flMrp8-Cre mice were anesthetized with isoflurane and 20 μg of LPS was administered intranasally in a 50 μl volume. Eight hours later, mice were euthanized by anesthetic overdose. Post-euthanasia, an incision was made in the trachea and the bronchoalveolar spaces were lavaged with 750 – 1,000 μl of PBS. The bronchoalveolar lavage (BAL) fluid was subjected to flow cytometry using 7-AAD (BD Bioscience), anti-Ly6G-APC (eBioscience clone 1A8), anti-Ly6C-FITC (BD Biosciences clone AL-21), and anti-CD11b-PE-Cy7 (eBioscience clone M1/70). Data was analyzed on the Canto I (BD) or the Attune Nxt (Thermo Fisher) flow cytometer. Counting beads (Spherotech ACBP-50–10) were used to calculate the absolute number of neutrophils in the BAL fluid. For neutrophil-mediated bacterial killing assays, neutrophils from Elmo1+/+ and Elmo1–/– mice were purified from the bone marrow using anti-Ly6G antibodies, as described above. Klebsiella pneumoniae strain 43816 (ATCC) was opsonized with heat inactivated, sterile FBS for 30 min at 37°C. Bacteria were washed twice and 200,000 bacteria were added to 100,000 purified neutrophils in a 96-well tissue culture plate for 1 hour at 37°C. In some wells, extracellular bacteria were killed by 1 μg/ml of gentamicin for 10 min at 37°C. Number of surviving bacteria was determined by hypotonic lysis of neutrophils in cold water and serial dilution and culture on blood agar plates.
Fecal-induced peritonitis (FIP)
Fecal material was isolated from the caecum of donor wild type mice, resuspended in saline and passed through a 70μM strainer to remove large particles. Eight-week old male Elmo1+/+ and Elmo1–/– mice were injected intraperitoneally with an LD50 dosage of 1.5mg fecal content/g mouse weight29,51. Core body temperature was measured using a MicroTherma 2T handheld thermometer (Braintree Scientific, TW2–107) and mice were scored for clinical signs according to a published murine sepsis severity scale51. Some mice were euthanized 4 hours after FIP induction and peritoneal cells were flushed out with PBS containing 5% FBS. Cells were stained with Fixable Yellow (to identify viable cells, Thermo Fisher) and anti-Ly6G (eBioscience clone 1A8) and subjected to flow cytometry on the Attune Nxt flow cytometer (Thermo Fisher).
Human peripheral blood neutrophils
Human neutrophils were kindly provided by Dr. Alison Criss and Dr. Asya Smirnov (University of Virginia). Materials used in this study were considered discarded materials from other studies and were exempt from human subjects oversight. Briefly, neutrophils were collected from venous blood of de-identified healthy human donors using dextran sedimentation followed by Ficoll gradient sedimentation and hypotonic lysis as described52,53. Buffy coats were also collected and consist of the non-granulocytic cells collected from the Ficoll gradient. Genomic DNA from purified neutrophils was prepared using the MagMax DNA kit (Applied Biosystems) and the status of the ELMO1 rs11984075 SNP was determined using the TaqMan SNP genotyping assay (Applied Biosystems), following manufacturer’s instructions. For Transwell migration assays, 5 × 105 cells were seeded in top chambers of 3 μm polycarbonate Transwell plates in 100 μl of HBSS containing 1% bovine serum albumin, as described for mouse neutrophils. Bottom chambers contained 600 μl of 15 nM leukotriene B4. After 2 hours at 37°C, the cells in the bottom wells were collected and subjected to flow cytometry analysis with counting beads (Spherotech, Inc.), TO-PRO-3 iodide (to identify viable cells, Life Technologies), anti-CD16 (eBioscience clone 3G8), anti-CD11b (BD Pharmingen) and anti-CD62L (BioLegend clone DREG-56) on Canto I flow cytometer (BD Bioscience). The data is presented as percent of input cells.
Analysis of human HL-60 cells
HL-60 cell line (ATCC) was grown in IMDM containing 20% FBS (Gemini Bio-Products) and 1% penicillin/streptomycin/glutamine (Gemini Bio-Products) and routinely tested negative for Mycoplasma contamination. For differentiation into neutrophil-like cells, HL-60 cells were treated with 1.25% DMSO (Sigma) for 4 days54 and then cultured without DMSO for 16h. For generating Elmo1 knockdown cells, HL-60 cells were transfected with 10 μg of human Elmo1 targeting or control shRNA pGFP-V-RS expression vectors (OriGene Technologies, Inc.) using electroporation using a 250 V, 25 ms pulse. After 24h, stably transfected cells were selected with 0.2 μg/ml puromycin (Gibco) and single cell cloning was performed by serial dilution. A total of three control and four Elmo1 shRNA clones were analyzed. Differentiated cells (1 × 105 cells per well) were seeded in top chambers of 3 μm polycarbonate Transwell plates in 100 μl of HBSS containing 1% bovine serum albumin, as described for neutrophils. Bottom chambers contained 600 μl of 100 nM leukotriene B4. After 1 hour at 37°C, the cells in the bottom wells were collected and subjected to flow cytometry analysis, as described above. The data is presented as percent of input cells.
Macrophage cultures
For resident peritoneal macrophage preparation, peritoneal cells were flushed out with PBS containing 5% FBS. Collected cells were spun down, re-suspended in XVIVO-10 (Lonza) containing 1% penicillin/streptomycin/glutamine and cultured on tissue culture dishes at 37°C and 5% carbon dioxide for 16h. For bone marrow derived macrophage cultures, femurs were removed from 6–8 week old mice, flushed with sterile serum-free α-MEM (Corning) and single cell suspensions were prepared using the 70μm cell strainer (Fisher). Cells were cultured on petri dishes in α-MEM containing 10% FBS and 1% penicillin/streptomycin/glutamine with added 10% L929 cell (ATCC) conditioned media as a source of macrophage-colony stimulating factor at 37°C and 5% carbon dioxide for 7 days.
Fibroblast-like synoviocytes
Fibroblast-like synoviocytes (FLS) were prepared following established protocols55. Briefly, skin was removed from the hind paws of euthanized 6–8 week old mice, the tendons of the ankles were cut, the paws dislodged from the tibias and placed into DMEM containing 1 mg/ml of collagenase IV, 10% FBS and 1% penicillin/streptomycin/glutamine. The joints of the foot were opened using gentle pulling of the foot bones. After a 1-hour incubation at 37°C, the liberated cells were collected, strained though a 70 μm filter (Fisher), and washed with DMEM containing 10% FBS and 1% penicillin/streptomycin/glutamine. The cells were seeded into 10 cm tissue culture coated dishes and cultured at 37°C and 5 % CO2. Media was exchanged every three days and the cells were passaged using 0.25 % trypsin with 2.21 mM EDTA at near confluency. Cells were used for experiments after three passages. Culture purity was confirmed by flow cytometry analysis using anti-CD90.2 (eBioscience clone 53–2.1), anti-VCAM1 (eBioscience clone 429) and anti-CD11b (eBioscience clone M1/70) antibodies. FLS had the spindle-like shape of fibroblasts and were CD90.2+VCAM1+CD11bneg.
Microscopy
All images were taken on an EVOS FL Auto (Fisher) and analyzed using the accompanying software.
Quantitative RT-PCR
Total RNA was isolated from cultured cells or pulverized paws using the RNA Easy kit (Qiagen) and cDNA prepared using the QuantiTect kit (Qiagen) according to manufacturer’s instructions. Quantitative gene expression for target and housekeeping genes was done using Taqman probes (Applied Biosystems) run on a StepOnePlus Real Time PCR System (Applied Biosystems).
Immunoblotting
Protein extracts were prepared from cultured cells or pulverized paws using RIPA lysis buffer with added protease inhibitors cocktail (Calbiochem), 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma) and 1 mM sodium orthovanadate (Sigma). Equal numbers of purified human neutrophils and buffy coat cells were directly lysed in SDS-PAGE sample buffer containing protease inhibitors cocktail, PMSF, sodium orthovanadate and 100 mM dithiothreitol. Equal amounts of protein extract were loaded on TGX Precast gels (Bio-Rad), subjected to SDS-PAGE and transferred onto PVDF membranes using the Trans-Blot Turbo transfer system (Bio-Rad). Immunoblotting was performed using anti-ELMO1 rabbit polyclonal antibody17, or anti-ERK2 (Santa Cruz, goat polyclonal), anti-GAPDH-HRP (Sigma clone GAPDH-71.1) or anti-beta-ACTIN-HRP (Sigma clone AC-15) as a loading control. Blots were exposed using the Western Lightning Plus ECL kit (Perkin Elmer) on the ChemiDoc Touch imaging system (Bio-Rad).
Proteomics and Mass Spectrometry
Cells extracts were prepared in Triton X-100 lysis buffer (20 mM Tris, pH 8.0, 137 mM NaCl, 10% glycerol, 0.2% Triton X-100, protease inhibitors cocktail (Calbiochem), 1 mM PMSF and 5 mM sodium orthovanadate) for 1h on ice. Lysates were cleared by centrifugation for 10 min at 13,200 rpm. ELMO1 was immunoprecipitated using the mouse anti-ELMO1 monoclonal antibody17, and immune complexes were washed and subjected to SDS-PAGE. The gels were stained with SimplyBlue Safe stain (Invitrogen) and destained with water. The gel bands were excised and frozen at −80 °C. After drying in a vacuum centrifuge, the gel bands were digested with 100 μL of trypsin (Promega) (20 μg/mL) at 37 °C for 16 h. The digested peptides were extracted with 100 μL 70% acetonitrile, 5% formic acid in a sonication bath. Each peptide sample was dried in a vacuum centrifuge and resuspended in 16 μL 0.1% TFA. Each gel band digest was analyzed by nanoflow liquid chromatography (LC) (Easy-nLC1000, ThermoFisher Scientific Inc.) coupled online with a an Orbitrap Fusion Lumos Tribrid MS (Thermo Fisher Scientific), samples were loaded on a C18 nano trap column, (Acclaim PepMap100 C18, 2 cm, nanoViper, Thermo Scientific) and resolved on a C18 Easy-Spray column (Acclaim PepMap RSLC C18, 2 μm, 100 Å, 75 μm × 500 mm, nanoViper, Thermo Scientific) with a linear gradient of 2% mobile phase B (95 % acetonitrile with 0.1% formic acid) to 32% mobile phase B within 60 min at a constant flow rate of 250 nL/min. The C18 Easy-Spray column was heated at 50 °C during the analysis. The most intense molecular ions in each MS scan within 3s were sequentially selected for high-energy collisional dissociation (HCD) using a normalized collision energy of 35%. The mass spectra were acquired at the mass range of m/z 400–1600. The Easy-Spray ion source (Thermo Scientific) capillary voltage and temperature were set at 1.9 kV and 275 °C, respectively. Dynamic exclusion (30 s) was enabled during the MS/MS data acquisition to minimize redundant peptide fragmentation events. The RF lens was set to 30% during the MS analysis and both MS1 and MS2 spectra were collected in profile mode. Data was searched against a Swiss-Prot human protein database (http://www.uniprot.org/uniprot/) using Proteome Discoverer (v.2.1.1.21, Thermo Fisher Scientific) via Mascot (v. 2.5.1, Matrix Science Inc.) with the automatic decoy search option set followed by false-discovery rate (FDR) processing by Percolator. Data was searched with a precursor mass tolerance of 10 ppm and a fragment ion tolerance of 0.05 Da, a maximum of two tryptic miscleavages and dynamic modifications for oxidation (15.9949 Da) on methionine residues. Resulting peptide spectral matches (PSMs) were filtered using an FDR of ≤ 1% (Percolator q-value ≤ 0.01). For determination of cell type-specific ELMO1 protein partners, only proteins that were present in all runs were analyzed. We calculated the normalized ratio of the Elmo1–/– spectral counts (Elmo1–/–/Elmo1+/++ Elmo1–/–). This calculation gives a value ranging from 0 to 1 with defined properties, where anything below 0.5 indicates a protein present in Elmo1+/+ cells but lost in Elmo1–/– cells. We considered values at least one standard deviation and 2-fold decreased (e.g. values less than 0.36) to be meaningful ELMO1 binding partners. Cross-comparison of normalized ratios afforded us the ability to then cross-compare ELMO1 binding partners between cell types. Neutrophil-specific ELMO1 partners identified in this study have a range of distinct functions56–72, and several have known associations with human arthritis36, 73–81.
IVIS Imaging
At the indicated time points during K/BxN serum transfer induced arthritis, mice were anesthetized using isoflurane, injected intraperitoneally with 200 μl of 50 mg/ml luminol (Sigma) in sterile saline solution, and scanned/analyzed using the IVIS Spectrum (Perkin Elmer), following manufacturer’s instructions.
RNA-Seq analysis
Neutrophils were purified from the bone marrow using anti-Ly6G antibody, as described above. Peritoneal macrophages were prepared as described above. Total RNA was extracted and an mRNA library was prepared using Illumina TruSeq platform and followed by transcriptome sequencing using an Illumina NextSeq 500 cartridge. Cultures from four mice per group were sequenced. The statistical software package R (version 3.3.2) was used for all analyses. The Bioconductor package DESeq2 was used for differential gene expression analysis of RNA-seq data. Heatmaps of neutrophil-specific Elmo1-dependent genes with associations to human arthritis82–94 were created using the R package gplots via the heatmap.2 package.
GWAS and experimental linkage analysis
We searched publicly available genome-wide association studies of human disease for SNP by gene interactions. The list of curated databases of published studies that were used is summarized in Supplementary Table 1. Significance of a given SNP was determined in the original study. SNP by phenotype interactions were determined using the GWASdb SNP-phenotype association database. The data are plotted using a standardized p value as reported95. For determination of experimental linkage, a search of curated databases such as the DISEASES experimental gene-disease association database was performed (Supplementary Table 1). Significance of the experimental linkage association was determined via a previously established aggregated score96–100.
Statistical analysis.
Statistical significance was determined using GraphPad Prism 5 or 6 using unpaired Student’s two-tailed t-test or two-way ANOVA. Variance was similar between groups. No inclusion/exclusion criteria were pre-established. A p-value of <0.05 (indicated by one asterisk), <0.01 (indicated by two asterisks), <0.001 (indicated by three asterisks), and <0.0001 (indicated by four asterisks) were considered significant. Details are available in the Statistics and Reproducibility file.
Data availability
The data that support the findings from this study, including the R code used for bioinformatics analysis and heatmap generation, are available from the corresponding authors upon reasonable request. Sequencing data have been deposited in the GEO database under the accession number GSE122412. Reagents used in this study are also listed in the Life Sciences Reporting Summary.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Ravichandran laboratory for discussions and critical reading of the manuscript, A. Criss and A. Smirnov for human neutrophils, S. T. Fleury for assistance with neutrophil purification, and K. Koster for assistance with bone marrow preparations. This work is supported by grants to K.S.R. from NIGMS R35GM122542, NIMH (MH096484), NHLBI (P01HL120840), NICHD (HD07498), and the Center for Cell Clearance/University of Virginia School of Medicine, and the Odysseus Award from the FWO, Belgium, and to M.R.E. from NIAID (AI114554). Additional support was provided by the Philip S. Magaram, Esq. Research Award from the Arthritis Foundation to S.A., and K.K.P. is supported by an NHLBI F30 award (F30 HL126385) and previously by the NIH T32 Immunology Training Grant (T32 AI007496).
Footnotes
ACCESSION CODES
Sequencing data supporting the findings of this study have been deposited in the GEO database under the accession number GSE122412.
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.McInnes IB & Schett G The pathogenesis of rheumatoid arthritis. The New England journal of medicine 365, 2205–2219, doi: 10.1056/NEJMra1004965 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Wright HL, Moots RJ & Edwards SW The multifactorial role of neutrophils in rheumatoid arthritis. Nature reviews Rheumatology 10, 593–601, doi: 10.1038/nrrheum.2014.80 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Moots RJ & Naisbett-Groet B The efficacy of biologic agents in patients with rheumatoid arthritis and an inadequate response to tumour necrosis factor inhibitors: a systematic review. Rheumatology (Oxford) 51, 2252–2261, doi: 10.1093/rheumatology/kes217 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Ding J, Eyre S & Worthington J Genetics of RA susceptibility, what comes next? RMD Open 1, e000028, doi: 10.1136/rmdopen-2014-000028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arandjelovic S & Ravichandran KS Phagocytosis of apoptotic cells in homeostasis. Nature immunology 16, 907–917, doi: 10.1038/ni.3253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Highton J, Hessian PA, Kean A & Chin M Cell death by apoptosis is a feature of the rheumatoid nodule. Annals of the rheumatic diseases 62, 77–80 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pope RM Apoptosis as a therapeutic tool in rheumatoid arthritis. Nature reviews. Immunology 2, 527–535, doi: 10.1038/nri846 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Nagata S, Hanayama R & Kawane K Autoimmunity and the clearance of dead cells. Cell 140, 619–630, doi: 10.1016/j.cell.2010.02.014 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Elliott MR & Ravichandran KS The Dynamics of Apoptotic Cell Clearance. Developmental cell 38, 147–160, doi: 10.1016/j.devcel.2016.06.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erwig LP & Henson PM Clearance of apoptotic cells by phagocytes. Cell death and differentiation 15, 243–250, doi: 10.1038/sj.cdd.4402184 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Gregory CD & Pound JD Microenvironmental influences of apoptosis in vivo and in vitro. Apoptosis: an international journal on programmed cell death 15, 1029–1049, doi: 10.1007/s10495-010-0485-9 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Miyanishi M et al. Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439, doi: 10.1038/nature06307 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Park D et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434, doi: 10.1038/nature06329 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lemke G & Rothlin CV Immunobiology of the TAM receptors. Nature reviews. Immunology 8, 327–336, doi: 10.1038/nri2303 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savill J Recognition and phagocytosis of cells undergoing apoptosis. British medical bulletin 53, 491–508 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Penberthy KK & Ravichandran KS Apoptotic cell recognition receptors and scavenger receptors. Immunological reviews 269, 44–59, doi: 10.1111/imr.12376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brugnera E et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature cell biology 4, 574–582, doi: 10.1038/ncb824 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Zhernakova A et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet 7, e1002004, doi: 10.1371/journal.pgen.1002004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitaker JW et al. Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets. PloS one 10, e0124254, doi: 10.1371/journal.pone.0124254 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimsley CM et al. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. The Journal of biological chemistry 279, 6087–6097, doi: 10.1074/jbc.M307087200 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gumienny TL et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Kouskoff V et al. Organ-specific disease provoked by systemic autoimmunity. Cell 87, 811–822 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Korganow AS et al. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity 10, 451–461 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Elliott MR et al. Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 467, 333–337, doi: 10.1038/nature09356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brand DD, Latham KA & Rosloniec EF Collagen-induced arthritis. Nature protocols 2, 1269–1275, doi: 10.1038/nprot.2007.173 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Clausen BE, Burkhardt C, Reith W, Renkawitz R & Forster I Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research 8, 265–277 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Passegue E, Wagner EF & Weissman IL JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119, 431–443, doi: 10.1016/j.cell.2004.10.010 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Yona S et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91, doi: 10.1016/j.immuni.2012.12.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis AJ, Seymour CW & Rosengart MR Current Murine Models of Sepsis. Surg Infect (Larchmt) 17, 385–393, doi: 10.1089/sur.2016.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong H et al. Distinct Contributions of Neutrophils and CCR2+ Monocytes to Pulmonary Clearance of Different Klebsiella pneumoniae Strains. Infect Immun 83, 3418–3427, doi: 10.1128/IAI.00678-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierson ER, Wagner CA & Goverman JM The contribution of neutrophils to CNS autoimmunity. Clin Immunol, doi: 10.1016/j.clim.2016.06.017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research 45, D362–D368, doi: 10.1093/nar/gkw937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou RC et al. Lipid-cytokine-chemokine cascade drives neutrophil recruitment in a murine model of inflammatory arthritis. Immunity 33, 266–278, doi: 10.1016/j.immuni.2010.07.018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lammermann T et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375, doi: 10.1038/nature12175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancardi DA et al. Cutting Edge: The murine high-affinity IgG receptor FcgammaRIV is sufficient for autoantibody-induced arthritis. J Immunol 186, 1899–1903, doi: 10.4049/jimmunol.1003642 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Hornum L et al. C5a and C5aR are elevated in joints of rheumatoid and psoriatic arthritis patients, and C5aR blockade attenuates leukocyte migration to synovial fluid. PloS one 12, e0189017, doi: 10.1371/journal.pone.0189017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park-Min KH et al. Negative regulation of osteoclast precursor differentiation by CD11b and beta2 integrin-B-cell lymphoma 6 signaling. J Bone Miner Res 28, 135–149, doi: 10.1002/jbmr.1739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H et al. Arthritis critically dependent on innate immune system players. Immunity 16, 157–168 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Monach PA et al. Neutrophils in a mouse model of autoantibody-mediated arthritis: critical producers of Fc receptor gamma, the receptor for C5a, and lymphocyte function-associated antigen 1. Arthritis and rheumatism 62, 753–764, doi: 10.1002/art.27238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wipke BT & Allen PM Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol 167, 1601–1608 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Morgan BP, Griffiths M, Khanom H, Taylor SM & Neal JW Blockade of the C5a receptor fails to protect against experimental autoimmune encephalomyelitis in rats. Clin Exp Immunol 138, 430–438, doi: 10.1111/j.1365-2249.2004.02646.x (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rittirsch D et al. Functional roles for C5a receptors in sepsis. Nature medicine 14, 551–557, doi: 10.1038/nm1753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyabe Y et al. Complement C5a Receptor is the Key Initiator of Neutrophil Adhesion Igniting Immune Complex-induced Arthritis. Sci Immunol 2, doi: 10.1126/sciimmunol.aaj2195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corr M & Crain B The role of FcgammaR signaling in the K/B × N serum transfer model of arthritis. J Immunol 169, 6604–6609 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Kiefer F et al. The Syk protein tyrosine kinase is essential for Fcgamma receptor signaling in macrophages and neutrophils. Molecular and cellular biology 18, 4209–4220 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruzankina Y et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126, doi: 10.1016/j.stem.2007.03.002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matmati M et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nature genetics 43, 908–912, doi: 10.1038/ng.874 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Hathaway CK et al. High Elmo1 expression aggravates and low Elmo1 expression prevents diabetic nephropathy. Proceedings of the National Academy of Sciences of the United States of America 113, 2218–2222, doi: 10.1073/pnas.1600511113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cooper DN Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics 4, 284–288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szanto S, Gal I, Gonda A, Glant TT & Mikecz K Expression of L-selectin, but not CD44, is required for early neutrophil extravasation in antigen-induced arthritis. J Immunol 172, 6723–6734 (2004). [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 51.Shrum B et al. A robust scoring system to evaluate sepsis severity in an animal model. BMC Res Notes 7, 233, doi: 10.1186/1756-0500-7-233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyum A Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl 97, 77–89 (1968). [PubMed] [Google Scholar]
- 53.Stohl EA, Criss AK & Seifert HS The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol Microbiol 58, 520–532, doi: 10.1111/j.1365-2958.2005.04839.x (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins SJ, Ruscetti FW, Gallagher RE & Gallo RC Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proceedings of the National Academy of Sciences of the United States of America 75, 2458–2462 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Armaka M, Gkretsi V, Kontoyiannis D & Kollias G A standardized protocol for the isolation and culture of normal and arthritogenic murine synovial fibroblasts. (2009).
- 56.Alon R & Shulman Z Chemokine triggered integrin activation and actin remodeling events guiding lymphocyte migration across vascular barriers. Experimental cell research 317, 632–641, doi: 10.1016/j.yexcr.2010.12.007 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Bridges D & Moorhead GB 14–3–3 proteins: a number of functions for a numbered protein. Sci STKE 2005, re10, doi: 10.1126/stke.2962005re10 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Delclaux C et al. Role of gelatinase B and elastase in human polymorphonuclear neutrophil migration across basement membrane. American journal of respiratory cell and molecular biology 14, 288–295, doi: 10.1165/ajrcmb.14.3.8845180 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Diaz-Alvarez L & Ortega E The Many Roles of Galectin-3, a Multifaceted Molecule, in Innate Immune Responses against Pathogens. Mediators of inflammation 2017, 9247574, doi: 10.1155/2017/9247574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Germena G, Volmering S, Sohlbach C & Zarbock A Mutation in the CD45 inhibitory wedge modulates integrin activation and leukocyte recruitment during inflammation. J Immunol 194, 728–738, doi: 10.4049/jimmunol.1401646 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Gittens BR, Bodkin JV, Nourshargh S, Perretti M & Cooper D Galectin-3: A Positive Regulator of Leukocyte Recruitment in the Inflamed Microcirculation. J Immunol 198, 4458–4469, doi: 10.4049/jimmunol.1600709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glennon-Alty L, Hackett AP, Chapman EA & Wright HL Neutrophils and redox stress in the pathogenesis of autoimmune disease. Free Radic Biol Med 125, 25–35, doi: 10.1016/j.freeradbiomed.2018.03.049 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Gough RE & Goult BT The tale of two talins - two isoforms to fine-tune integrin signalling. FEBS letters 592, 2108–2125, doi: 10.1002/1873-3468.13081 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin J et al. Proteomic, functional, and domain-based analysis of in vivo 14–3–3 binding proteins involved in cytoskeletal regulation and cellular organization. Current biology: CB 14, 1436–1450, doi: 10.1016/j.cub.2004.07.051 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Kletzien RF, Harris PK & Foellmi LA Glucose-6-phosphate dehydrogenase: a “housekeeping” enzyme subject to tissue-specific regulation by hormones, nutrients, and oxidant stress. FASEB journal: official publication of the Federation of American Societies for Experimental Biology 8, 174–181 (1994). [DOI] [PubMed] [Google Scholar]
- 66.Oikonomou KG, Zachou K & Dalekos GN Alpha-actinin: a multidisciplinary protein with important role in B-cell driven autoimmunity. Autoimmunity reviews 10, 389–396, doi: 10.1016/j.autrev.2010.12.009 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Sadik CD, Miyabe Y, Sezin T & Luster AD The critical role of C5a as an initiator of neutrophil-mediated autoimmune inflammation of the joint and skin. Semin Immunol 37, 21–29, doi: 10.1016/j.smim.2018.03.002 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Sano H et al. Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J Immunol 165, 2156–2164 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Springer TA Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76, 301–314 (1994). [DOI] [PubMed] [Google Scholar]
- 70.Thomas DC The phagocyte respiratory burst: Historical perspectives and recent advances. Immunology letters 192, 88–96, doi: 10.1016/j.imlet.2017.08.016 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Wyatt E et al. Regulation and cytoprotective role of hexokinase III. PloS one 5, e13823, doi: 10.1371/journal.pone.0013823 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X et al. Structural basis for protein-protein interactions in the 14–3–3 protein family. Proceedings of the National Academy of Sciences of the United States of America 103, 17237–17242, doi: 10.1073/pnas.0605779103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balakrishnan L et al. Differential proteomic analysis of synovial fluid from rheumatoid arthritis and osteoarthritis patients. Clin Proteomics 11, 1, doi: 10.1186/1559-0275-11-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui J et al. Rheumatoid arthritis risk allele PTPRC is also associated with response to anti-tumor necrosis factor alpha therapy. Arthritis and rheumatism 62, 1849–1861, doi: 10.1002/art.27457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Rooy DP et al. A genetic variant in the region of MMP-9 is associated with serum levels and progression of joint damage in rheumatoid arthritis. Annals of the rheumatic diseases 73, 1163–1169, doi: 10.1136/annrheumdis-2013-203375 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Gheita TA, Kenawy SA, El Sisi RW, Gheita HA & Khalil H Subclinical reduced G6PD activity in rheumatoid arthritis and Sjogren’s Syndrome patients: relation to clinical characteristics, disease activity and metabolic syndrome. Mod Rheumatol 24, 612–617, doi: 10.3109/14397595.2013.851639 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Hu CY, Chang SK, Wu CS, Tsai WI & Hsu PN Galectin-3 gene (LGALS3) +292C allele is a genetic predisposition factor for rheumatoid arthritis in Taiwan. Clin Rheumatol 30, 1227–1233, doi: 10.1007/s10067-011-1741-2 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Huang RY, Huang QC & Burgering BM Novel insight into the role of alpha-actinin-1 in rheumatoid arthritis. Discov Med 17, 75–80 (2014). [PubMed] [Google Scholar]
- 79.Maksymowych WP & Marotta A 14–3–3eta: a novel biomarker platform for rheumatoid arthritis. Clin Exp Rheumatol 32, S-35–39 (2014). [PubMed] [Google Scholar]
- 80.Tsuzaka K, I. Y, S. N and Morishita T. Plasma talin is a new diagnostic and monitoring marker for rheumatoid arthritis. Arthritis and rheumatism S134 (2011). [Google Scholar]
- 81.Warchol T, Lianeri M, Lacki JK, Olesinska M & Jagodzinski PP ITGAM Arg77His is associated with disease susceptibility, arthritis, and renal symptoms in systemic lupus erythematosus patients from a sample of the Polish population. DNA Cell Biol 30, 33–38, doi: 10.1089/dna.2010.1041 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Cottier KE, Fogle EM, Fox DA & Ahmed S Noxa in rheumatic diseases: present understanding and future impact. Rheumatology (Oxford) 53, 1539–1546, doi: 10.1093/rheumatology/ket408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cui J et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet 9, e1003394, doi: 10.1371/journal.pgen.1003394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elsby LM et al. Functional evaluation of TNFAIP3 (A20) in rheumatoid arthritis. Clin Exp Rheumatol 28, 708–714 (2010). [PMC free article] [PubMed] [Google Scholar]
- 85.Howng SL et al. Autoimmunity against hNinein, a human centrosomal protein, in patients with rheumatoid arthritis and systemic lupus erythematosus. Mol Med Rep 4, 825–830, doi: 10.3892/mmr.2011.505 (2011). [DOI] [PubMed] [Google Scholar]
- 86.Issuree PD et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. The Journal of clinical investigation 123, 928–932, doi: 10.1172/JCI66168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin T, Tarkowski A, Carmeliet P & Bokarewa M Urokinase, a constitutive component of the inflamed synovial fluid, induces arthritis. Arthritis research & therapy 5, R9–R17 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lopez M et al. Tumor necrosis factor and transforming growth factor beta regulate clock genes by controlling the expression of the cold inducible RNA-binding protein (CIRBP). The Journal of biological chemistry 289, 2736–2744, doi: 10.1074/jbc.M113.508200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marotte H et al. Blocking of interferon regulatory factor 1 reduces tumor necrosis factor alpha-induced interleukin-18 bioactivity in rheumatoid arthritis synovial fibroblasts by induction of interleukin-18 binding protein a: role of the nuclear interferon regulatory factor 1-NF-kappaB-c-jun complex. Arthritis and rheumatism 63, 3253–3262, doi: 10.1002/art.30583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohyama K et al. Serum immune complex containing thrombospondin-1: a novel biomarker for early rheumatoid arthritis. Annals of the rheumatic diseases 71, 1916–1917, doi: 10.1136/annrheumdis-2012-201305 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Okada Y et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381, doi: 10.1038/nature12873 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perlman H et al. Rheumatoid arthritis synovial macrophages express the Fas-associated death domain-like interleukin-1beta-converting enzyme-inhibitory protein and are refractory to Fas-mediated apoptosis. Arthritis and rheumatism 44, 21–30, doi: (2001). [DOI] [PubMed] [Google Scholar]
- 93.Rico MC et al. Amelioration of inflammation, angiogenesis and CTGF expression in an arthritis model by a TSP1-derived peptide treatment. Journal of cellular physiology 211, 504–512, doi: 10.1002/jcp.20958 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Yoo IS et al. Serum and synovial fluid concentrations of cold-inducible RNA-binding protein in patients with rheumatoid arthritis. Int J Rheum Dis, doi: 10.1111/1756-185X.12892 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Li MJ et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic acids research 40, D1047–1054, doi: 10.1093/nar/gkr1182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX & Jensen LJ DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89, doi: 10.1016/j.ymeth.2014.11.020 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Davis AP et al. The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic acids research 43, D914–920, doi: 10.1093/nar/gku935 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis AP et al. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic acids research 37, D786–792, doi: 10.1093/nar/gkn580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eppig JT, Blake JA, Bult CJ, Kadin JA & Richardson JE The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic acids research 43, D726–736, doi: 10.1093/nar/gku967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blake JA, Richardson JE, Bult CJ, Kadin JA & Eppig JT MGD: the Mouse Genome Database. Nucleic acids research 31, 193–195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings from this study, including the R code used for bioinformatics analysis and heatmap generation, are available from the corresponding authors upon reasonable request. Sequencing data have been deposited in the GEO database under the accession number GSE122412. Reagents used in this study are also listed in the Life Sciences Reporting Summary.