Abstract
While two-component systems (TCSs), composed of a sensor histidine kinase (HK) and a response regulator, are the main signaling pathways in bacteria, global TCS activity remains poorly described. Here, we report the kinetic parameters of the HK autophosphorylation reaction using previously uncharacterized γ-phosphate-modified ATP analogues to further elucidate their utility as activity-based probes for global TCS analysis. Given the increased stability of thiophosphorylated histidine in comparison to that of the native phosphoryl modification, which is attributed to the decreased electrophilicity of this moiety, we anticipated that ATPγS may be turned over much more slowly by the HKs. Surprisingly, we found this not to be the case, with the turnover numbers decreasing <1 order of magnitude. Instead, we found that alkylation of the thiophosphate had a much more dramatic effect on turnover and, in one case, the binding affinity of this substrate analogue (BODIPY-FL-ATPγS).
graphical abstract

Two-component systems (TCSs) are the main signal transduction pathways in bacteria. Composed of a transmembrane histidine kinase (HK) and its cognate cytoplasmic response regulator (RR), TCSs are involved in many cell processes, including growth and development, virulence, and antibiotic resistance.1–3 When stimulated by environmental changes such as the ion concentration, the pH, or the presence of antibiotics, the HK initiates an autophosphorylation event. First, ATP binds to the catalytic CA domain of the HK in the ATP binding pocket, and then the γ-phosphate of ATP is transferred to a conserved histidine in the dimerization-histidine phosphotransfer (DHp) domain.4–6 This phosphoryl group is subsequently transmitted to an aspartic acid on its cognate RR, eliciting a downstream response, typically by acting as a transcription factor for gene expression (Figure 1a).4,7
Figure 1.

(a) HK autophosphorylation event. When activated by an extracellular signal, the conserved histidine on the HK is phosphorylated by ATP. The phosphoryl group is transferred to an aspartic acid on an RR, which triggers a cellular response. (b) Structure of BODIPY-FL-ATPγS, highlighting the portion of the probe that is transferred to the conserved histidine on the HK. (c) Structure of the modified histidine residue.
TCSs are ubiquitous in bacteria, with most organisms expressing 30 TCSs on average, but are not found in higher eukaryotes such as humans.8 As TCSs are structurally similar within and across species, especially in their active sites, there has been great interest in targeting them for novel therapeutics.9–14 While numerous individual TCSs have been well characterized, a gap in our understanding related to global TCS regulation and the conditions under which many TCSs are stimulated remains.15 To improve our understanding, all of an organism’s TCSs must be analyzed simultaneously. Ideally, profiling of global TCS activity could be achieved by utilizing small molecule probes detecting phosphorylation of the HK. While eukaryotic kinases (serine, threonine, and tyrosine) have been previously well characterized, difficulty in profiling bacterial HKs remains because of the instability of the phosphohistidine (P~His) species, particularly at low pH.16,17 Thus, many of the methods used to study eukaryotic kinases cannot be employed as they typically involve acidic steps. However, it has been shown that formation of a thiophosphoramidate rather than a phosphoramidate decreases the rate of hydrolysis because of the increased stability that the less electronegative sulfur confers on the P—N bond.18,19 Taking advantage of the increased stability afforded by formation of a thiophosphoramidate product, our lab previously developed the first nonradioactive activity-based probe for HKs, BODIPY-FL-ATPγS (B-ATPγS), which enables fast and quantitative readout of the autophosphorylation event using fluorescence (Figure 1b,c).17
While B-ATPγS was shown to competitively label several HK proteins, including HK853 from Thermotoga maritima (ID Q9WZV7) and VicK from Streptococcus pneumoniae (ID A0A0U0DLI4), we were unable to perform pull-down assays using biotin-ATPγS with avidin agarose beads for enrichment, leading to the hypothesis that there may be a low rate of turnover of ATPγS-modified analogues by the HKs.20,21 To investigate this possibility, we sought to compare the kinetic parameters of the autophosphorylation reaction of HK853 and VicK using radiolabeled native substrate [γ-33P]ATP versus [γ-35S]ATP and B-ATPγS.
Although the kinetic parameters of a handful of HKs have been previously reported using [γ-32P]ATP, analysis of other ATP analogues is lacking. Traditionally, HK kinetic assays are performed by running sodium dodecyl sulfate—polyacrylamide gel electrophoresis (SDS—PAGE) followed by autoradiography.22,23 Ueno et al. have also successfully developed a modified filter paper binding assay classically used for mammalian kinases by quenching with small amounts of phosphoric acid (5.77 mM), resulting in minimal cleavage of the phosphohistidine bond.24,25 The kinetics of HK autophosphorylation are comparable to those of eukaryotic kinases with turnover ranging from 10−2 s−1 for classical HKs such as CheA from Escherichia coli to 10−5 s−1 for dual-functioning HKs with both kinase and phosphatase activity such as Vibrio harveyi HK (ID 555316).24,26–28 On average, HKs have a relatively poor Km value in the range of tens to hundreds of micromolar.22,24,26,29–32 Examining the effects that γ-phosphate-modified ATP analogues have on the autophosphorylation reaction is critical for the design of activity-based probes, which will be essential for the efficient study of the activity of these proteins and, ultimately, for the development of HK-targeting inhibitors with potential as antibacterial agents.
MATERIALS AND METHODS
Commercially available molecules and other chemicals were obtained from Invitrogen, Sigma-Aldrich, PerkinElmer, and Thermo Fisher Scientific at >97% purity. All commercially available chemicals were used without further purification.
Protein Overexpression and Purification.
Total gene synthesis for HK853 was performed as described by Wilke et al.20 The VicK protein construct was received as a gift from the laboratory of M. Winkler (Indiana University) and can be found in ref 22. The gene corresponding to the cytoplasmic portion of HK853 from T. maritima was ligated into the pHisparallel 1 vector with a His tag at the N-terminus and VicK [WalKSpnΔN35 (N)-Sumo] from S. pneumoniae into pSumo with a His tag at the N-terminus. The recombinant plasmids were transformed into E. coli DH5α and then in E. coli BL21(DE3)Rosetta/pLysS overexpression cells and plated on LB agar containing 34 μg/mL chloramphenicol and either 100 μMg/mL ampicillin (HK853) or 30 μg/mL kanamycin (VicK). Plates were incubated at 37 °C overnight. A single colony was transferred to 10 mL of LB medium supplemented with antibiotics as noted above. Cultures were incubated at 37 °C overnight while being shaken at 220 rpm. Media (1 L) containing antibiotics were inoculated with 10 mL of the previous culture and grown to an OD600 of ~0.6. Cultures were induced with 0.22 and 0.1 mM isopropyl β-D-1-thiogalactopyr-anoside for HK853 and VicK, respectively. HK853 cultures were incubated at 20 °C for 16 h while being shaken at 220 rpm. VicK cultures were incubated at 16 °C for 24 h while being shaken at 220 rpm. Cultures were centrifuged at 8000 rpm and 4 °C for 20 min to collect the pellets.
Pellets were resuspended in lysis buffer [25 mM, 1 M NaCl, 10% glycerol (pH 8)] supplemented with 1 μg/mL DNase, 2 mM DTT, 1 mg/mL lysozyme, and one complete mini EDTA-free protease inhibitor tablet (Roche) per 2 g pellet. Approximately 30 mL of lysis buffer was used per 2 g pellet. The pellet was disrupted with a Dounce homogenizer, and the resuspended cells were sonicated on ice for 2 min every 5 min for 30 min (Branson Sonifier 250). Once lysed, the suspension was centrifuged at 10000 rpm and 4 °C for 20 min, and the supernatant was decanted and filtered (0.22 μm).
Purification was performed in stages using fast protein liquid chromatography (FPLC; GE, ÄKTA). All buffers, lysates, and proteins were filtered (0.22 μm). Samples were loaded onto a nickel-nitriloacetic acid column (NiNTA; Qiagen) with buffer (25 mM Tris, 1 M NaCl, 10% glycerol, 5 mM imidazole, and 2 mM BME). A reducing agent and a high salt concentration were maintained to prevent protein aggregation. An elution gradient from 5 mM to 1 M imidazole was used, and protein was detected at 215 nm absorbance. The final purification step was size exclusion chromatography using a HiLoad 16/60 Superdex 200 pg column (GE Healthcare). Protein was eluted using storage buffer [10 mM Tris, 0.1 mM EDTA, 0.5 mM NaCl, 12% glycerol, and 2 mM DTT (pH 7.6)]. The concentration of protein after purification was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
[γ-33P]- and [γ-35S]ATP Radioactivity Assays.
Histidine kinase (5.0 μM) was pre-equilibrated in reaction buffer [50 mM Tris, 0.2 M KCl, and 5 mM MgCl2 (pH 7.8)]. The autophosphorylation reaction was initiated by addition of [γ-33P]- or [γ-35S]ATP (specific activity of 2.00 Ci/mmol; PerkinElmer, catalog no. NEG602H or NEG027H, respectively; 0.27 Ci/mmol for assays with 1 mM [γ-33P]ATP). At designated times, 15 μL samples were removed and reactions quenched with 5 μL of 4× SDS-PAGE loading buffer [250 mM Tris, 40% glycerol, 8% (w/v) SDS, 10% 2-mercaptoethanol, and 0.4% (w/v) bromophenol blue (pH 6.8)]. Samples (15 μL) were analyzed without heating by 12% SDS-PAGE for 1 h. After electrophoresis, gels were soaked for 20 min in a fixing solution (40% methanol, 10% acetic acid, and 10% glycerol) and dried for 2 h at 65 °C on a vacuum gel dryer (Bio-Rad). Dried gels were exposed to a storage phosphor screen (GE Healthcare) and analyzed using a Typhoon FLA 9500 scanner (GE Healthcare) and ImageJ software. Data were fit to a Michaelis-Menten plot using GraphPad Prism to determine Km and kcat. After being scanned, gels were rehydrated and stained with Coomassie Brilliant Blue to confirm even protein loading. The amount of (thio)-phosphorylated histidine in each lane was quantified using a standard curve generated by spotting known concentrations of [γ-33P]- or [γ-35S]ATP on filter paper.
B-ATPγS Assays.
Histidine kinase (1.0 μM) was pre-equilibrated in reaction buffer as described above. The autophosphorylation reaction (20 μL) was initiated by addition of B-ATPγS (Invitrogen catalog no. A22184) at varying concentrations. At designated times, reactions were quenched with 4× SDS—PAGE loading buffer and analyzed without heating by 12% SDS—PAGE for 1 h. After electrophoresis, the gel was placed on a Typhoon FLA 9500 scanner (GE Healthcare) with a standard curve generated by spotting 5 μL of known concentrations of B-ATPγS on plastic wrap and scanned by being placed atop the assay gel. The gels were analyzed using ImageJ software, and data were fit to a Michaelis—Menten plot using GraphPad Prism to determine Km and kcat. After being scanned, gels were stained with Coomassie Brilliant Blue to confirm even protein loading. The concentration of the B-ATPγS stock was confirmed using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
Impact of BODIPY on HK853 and VicK Labeling by B-ATPγS.
Histidine kinase (1.0 μM) was pre-equilibrated in reaction buffer as described above. BODIPY (difluoro{2-[(3,5- dimethyl-2H-pyrrol-2-ylidene-N)methyl]-3,5-dimethyl-1H-pyr- rolato-N}boron) was added at different concentrations to reach a final volume of 25 μL and incubated for 30 min. The autophosphorylation reaction was initiated by addition of B- ATPγS (Invitrogen catalog no. A22184; 1 μL, final concentration of 2.0 μM for HK853 and 5.0 μM for VicK), and incubation times were 1 h for HK853 and 2 h for VicK. Reactions were quenched with 4× SDS—PAGE loading buffer (sample not heated) and analyzed by 12% SDS—PAGE for 1 h. After electrophoresis, the gel was scanned on a Typhoon FLA 9500 instrument (GE Healthcare). The gels were analyzed using ImageJ.
RESULTS AND DISCUSSION
Time-dependent assays were performed to determine the linear range of HK activity for both [γ−33P]-and [γ−35S]ATP. Saturation was observed within 10 min for both proteins using [γ−33P]ATP and for [γ−35S]ATP HK853 assays (Figure 2 and Figures S1 and S2). The [γ−35S]ATP VicK reactions were slightly slower, reaching saturation by 30 min (Figure S2). This is consistent with previous work demonstrated by Ueno et al.24 It has been shown that accumulation of ADP inhibits the autophosphorylation reaction, which may contribute to the decrease in P~His observed at later time points.27,33 We selected a quench time of 30 s for subsequent substrate-dependent assays and further confirmed linearity at this time point at a high ATP concentration for both proteins [1 mM (Figures S3 and S4)].
Figure 2.
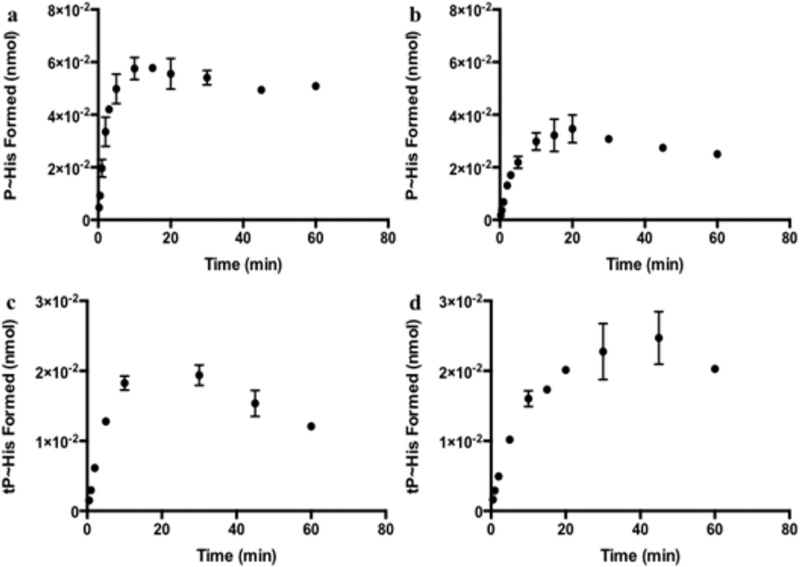
Time-dependent autophosphorylation of HKs used to establish the linear range of HK enzyme activity for substrate-dependent assays. (a) [γ-33P]ATP HK853. (b) [γ-33P]ATP VicK. (c) [γ-35S]ATP HK853. (d) [γ-35S]ATP VicK. All reactions were performed using 6.25 μM ATP at a specific activity of 2.00 Ci/mmol with 5.0 μM protein. Error bars represent the standard error of the mean from at least three trials.
The kinetic parameters of each protein were determined through analysis of phosphorylated and (thio)phosphorylated histidine formed as a function of ATP and ATPγS concentration (Figure 3, Table 1, and Figures S5 and S6). While the autophosphorylation reaction does not strictly follow Michaelis-Menten kinetics as the phosphorylation event decreases the total concentration of the available enzyme, its parameters are useful in comparing the turnover of native ATP versus that of its analogues.29
Figure 3.

[γ-33P]- and [γ-35S]ATP concentration-dependent autophosphorylation of HKs used to calculate Km and kcat as summarized in Table 1. Reactions were quantified as a function of (thio)phosphorylated histidine formed vs ATP. (a) [γ-33P]ATP HK853. (b) [γ-33P]ATP VicK. (c) [γ-35S]ATP HK853. (d) [γ-35S]ATP VicK. All reactions were performed at a specific activity of 2.00 Ci/mmol with 5.0 μM protein. [γ-33P]ATP reactions were quenched at 30 s. [γ-35S]ATP reactions were quenched at 1 min. Error bars represent the standard error of the mean from at least three trials.
Table 1.
Summary of HK Autophosphorylation Kinetic Parameters for HK853 and VicK Using [γ-33P]ATP, [γ-35S]ATP, and B-ATPγS
| protein | Km (μM) | kcat (s−1) | Vmax | kcat/Km (M−1 s−1) | |
|---|---|---|---|---|---|
| γ-33S | |||||
| HK853 | 5.178 ± 0.4111 | (4.673 ± 0.105) × 10−3 | (4.678 ± 0.104) × 10−4 nmol/s | 902.5 | |
| VicK | 16.85 ± 1.514 | (4.994 ± 0.146) × 10−3 | (4.998 ± 0.146) × 10−4 nmol/s | 296.4 | |
| γ-35S | |||||
| HK853 | 6.287 ± 0.5376 | (7.360 ± 0.280) × 10−4 | (7.366 ± 0.140) × 10−4 nmol/s | 117.1 | |
| VicK | 9.393 ± 1.171 | (1.170 ± 0.061) × 10−3 | (1.171 ± 0.610) × 10−3 nmol/s | 124.6 | |
| B-ATPγ−S | |||||
| HK853 | 6.273 ± 0.8578 | (1.921 ± 0.073) × 10−5 | (3.842 ± 0.146) × 10−4 nmol/s | 3.062 | |
| VicK | 147.4 ± 20.97 | (5.926 ± 1.022) × 10−6 | (1.186 ± 2.044) × 10−4 nmol/s | 0.040 | |
Overall, calculated parameters for HK autophosphorylation using [γ−33P]ATP were consistent with those previously reported.22,34 While our results showed an expected decrease in catalytic efficiency, surprisingly, the rate of turnover of [γ−33P]- and [γ-35S]ATP by both proteins is within an order of magnitude, between 10−3 and 10−4 s−1, negating the assumption that turnover of B-ATPγS is impeded by the thiophosphate modification. Rather, these data suggested that low levels of labeling were due to either steric hindrance caused by the fluorophore or electronic effects caused by the alkylation of ATPγS. Thus, we sought to quantify the turnover of B-ATPγS.
To determine the kinetic parameters of B-ATPγS, a method similar to the radioactivity assays was used with a few modifications. Following incubation with the probe, the enzyme reaction mixtures were separated using SDS—PAGE and the fluorescently labeled histidine (BtP~His) was quantified with a standard curve generated by spotting known concentrations of B-ATPγS on the gel before analysis (Figure S7c). To the best of our knowledge, this assay is the first use of a fluorescent probe for kinetic analysis of a histidine kinase, making the devised strategy a general method for determination of kinetic parameters without utilizing radioactivity. Previous work had shown that qualitative time-dependent HK853 B-ATPγS assays reach saturation at 60 min using 0.37 μM protein and 2 μM B-ATPγS.20 VicK autophosphorylation with B-ATPγS is much slower, remaining in the linear range throughout a 2.5 h time course (Figure S8). The progress curves used to determine the kinetic parameters of HK autophosphorylation using B-ATPγS are depicted in Figure 4 (Table 1). We further confirmed that these substrate- dependent assays were performed within the linear range of catalysis by quantifying autophosphorylation over time at the highest used probe concentration (Figures S9 and S10). As anticipated, B-ATPγS is turned over much more slowly by both enzymes, with kcat values that are several orders of magnitude smaller than those seen with ATP or ATPγS. To confirm that this was not due to the fluorophore interacting with the protein and affecting its ability to turn over the substrate, we assessed whether BODIPY inhibited either protein. We found that the fluorophore does not inhibit HK853 or VicK (Figures S11 and S12).
Figure 4.

Progress curves of B-ATPγS concentration-dependent autophosphorylation of HKs used to calculate Km and kcat as summarized in Table 1. Reactions were quantified as a function of the fluorescently labeled histidine (BtP~His) formed. (a) B-ATPγS HK853. Reactions performed using 1.0 protein and quenched at 15 min. (b) B-ATPγS VicK. Reactions performed using 1.0 μM protein and quenched at 60 min. Error bars represent the standard error of the mean from at least three trials.
Finally, we noted that these results are consistent with previous work that demonstrated a decreased level of labeling of VicK by B-ATPγS compared to that seen with HK853.20 As the kinetic parameters between HK853 and VicK are comparable for [γ-35S]ATP, the difference between the parameters for the proteins using B-ATPγS is likely due to a lower level of recognition of the probe by VicK (Km = 6.3 vs 147 μM). To determine whether the difference in binding affinity is due to steric or electronic interference, further studies with ATP and ATPγS analogues modified with alkylated chains of varying lengths could be performed.
CONCLUSIONS
In reporting the kinetic parameters for the autophosphorylation reaction of two HKs using γ-phosphate-modified ATP analogues, this paper further elucidates the utility of small molecule ATPγS-based probes for potential characterization of global TCS activity. Given the stability afforded by the thiophosphate imparted by the less electronegative sulfur atom, it was reasonable to expect that the probes might react more slowly with the nucleophilic histidine residue of the HK.
Surprisingly, this presumption does not appear to be the case as the turnover numbers are similar between ATP and the sulfur-containing analogue. However, we did determine that a bulky moiety attached to the thiophosphate dramatically affected the binding affinity and catalytic efficiency in one instance. This information will be critical in aiding the future design of activity-based probes for the HKs to enable continued exploration of their roles in antibiotic resistance and virulence to ultimately develop new antimicrobial therapies.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank M. Winkler at Indiana University for providing the VicK construct.
Funding
This work was supported by the University of Minnesota, the University of Minnesota National Institutes of Health (NIH; T32GM008347) Biotechnology Training Grant (A.E.), NIH Grant DP20D008592, and an Indiana University Quantitative and Chemical Biology Training Fellowship (K.E.W.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.8b00485.
Additional figures, including [γ-33P]- and [γ-35S]ATP and B-ATPγS time-and substrate-dependent data and evaluation of BODIPY as a potential inhibitor of HK853 and VicK activity (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Hong HJ, Hutchings MI, and Buttner MJ (2008) Biotechnology and Biological Sciences Research Council, U.K., Vancomycin resistance VanS/VanR two-component systems. Adv. Exp. Med. Biol. 631, 200–213. [DOI] [PubMed] [Google Scholar]
- (2).Hong HJ, Hutchings MI, Hill LM, and Buttner MJ (2005) The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280, 13055–13061. [DOI] [PubMed] [Google Scholar]
- (3).Prost LR, and Miller SI (2008) The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell. Microbiol. 10, 576–582. [DOI] [PubMed] [Google Scholar]
- (4).Stock AM, Robinson VL, and Goudreau PN (2000) Two- component signal transduction. Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed] [Google Scholar]
- (5).Stock JB, Stock AM, and Mottonen JM (1990) Signal transduction in bacteria. Nature 344, 395–400. [DOI] [PubMed] [Google Scholar]
- (6).Casino P, Rubio V, and Marina A (2009) Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139, 325–336. [DOI] [PubMed] [Google Scholar]
- (7).Capra EJ, and Laub MT (2012) Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schaller GE, Shiu SH, and Armitage JP (2011) Two- component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21, R320–330. [DOI] [PubMed] [Google Scholar]
- (9).Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, and Utsumi R (2010) Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13, 232–239. [DOI] [PubMed] [Google Scholar]
- (10).Bem AE, Velikova N, Pellicer MT, Baarlen P, Marina A, and Wells JM (2015) Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem. Biol. 10, 213–224. [DOI] [PubMed] [Google Scholar]
- (11).Wilke KE, Francis S, and Carlson EE (2015) Inactivation Of Multiple Bacterial Histidine Kinases By Targeting The ATP-Binding Domain. ACS Chem. Biol. 10, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Goswami M, Wilke KE, and Carlson EE (2017) Rational Design of Selective Adenine-Based Scaffolds for Inactivation of Bacterial Histidine Kinases. J. Med. Chem. 60, 8170–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Francis S, Wilke KE, Brown DE, and Carlson EE (2013) Mechanistic Insight Into Inhibition Of Two-Component System Signaling. MedChemComm 4, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wilke KE, Fihn CA, and Carlson EE (2018) Screening serine/threonine and tyrosine kinase inhibitors for histidine kinase inhibition. Bioorg. Med. Chem, DOI: 10.1016/j.bmc.2018.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, and Ramos JL (2010) Bacterial sensor kinases: diversity in the recognition or environmental signals. Annu. Rev. Microbiol. 64, 539–559. [DOI] [PubMed] [Google Scholar]
- (16).Klumpp S, and Krieglstein J (2002) Phosphorylation and desphosphorylation of histidine residues in proteins. Eur. J. Biochem. 269, 1067–1071. [DOI] [PubMed] [Google Scholar]
- (17).Kee JM, and Muir TW (2012) Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem. Biol. 7, 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pirrung MC, James KD, and Rana VS (2000) Thiophosphorylation of histidine. J. Org. Chem. 65, 8448–8453. [DOI] [PubMed] [Google Scholar]
- (19).Lasker M, Bui CD, Besant PG, Sugawara K, Thai P, Medzihradszky G, and Turck CW (1999) Protein histidine phosphorylation: increased stability of thiophosphohistidine. Protein Sci 8, 2177–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wilke KE, Francis S, and Carlson EE (2012) Activity-based probe for histidine kinase signaling. J. Am. Chem. Soc. 134, 9150–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wilke KE (2015) Chemical probes for histidine kinase protein profiling and inhibitor discovery Ph.D. Dissertation, Indiana University, Bloomington, IN. [Google Scholar]
- (22).Gutu AD, Wayne KJ, Sham LT, and Winkler ME (2010) Kinetic characterization of the WalRKSpn (VicRK) two- component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J. Bacteriol. 192, 2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Foster JE, Sheng Q, McClain JR, Bures M, Nicas TI, Henry K, Winkler ME, and Gilmour R (2004) Kinetic and mechanistic analyses of new classes of inhibitors of two-component signal transduction systems using a coupled assay containing HpkA- DrrA from Thermatoga maritima. Microbiology 150, 885–896. [DOI] [PubMed] [Google Scholar]
- (24).Ueno TB, Johnson RA, and Boon EM (2015) Optimized assay for the quantification of histidine kinase autophosphorylation. Biochem. Biophys. Res. Commun. 465, 331–337. [DOI] [PubMed] [Google Scholar]
- (25).Hastie CJ, McLauchlan HJ, and Cohen P (2006) Assay of protein kinases using radiolabeled ATP: a protocol. Nat. Protoc. 1 , 968–971. [DOI] [PubMed] [Google Scholar]
- (26).Tawa P, and Stewart RC (1994) Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry 33, 7917–7924. [DOI] [PubMed] [Google Scholar]
- (27).Jiang P, Peliska JA, and Ninfa AJ (2000) Asymmetry in the autophosphorylation of the two-component regulatory system transmitter protein nitrogen regulator II of Escherichia coli. Biochemistry 39, 5057–5065. [DOI] [PubMed] [Google Scholar]
- (28).Wang ZX, and Wu JW (2002) Autophosphorylation kinetics of protein kinases. Biochem. J. 368, 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Marina A, Mott C, Auyzenberg A, Hendrickson WA, and Waldburger CD (2001) Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism. J. Biol. Chem. 276, 41182–41190. [DOI] [PubMed] [Google Scholar]
- (30).Kenney LJ (1997) Kinase activity of EnvZ, an osmoregulatory signal transducing protein of Escherichia coli. Arch. Biochem. Biophys. 346, 303–311. [DOI] [PubMed] [Google Scholar]
- (31).Noriega CE, Schmidt R, Gray MJ, Chen LL, and Stewart V (2008) Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J. Bacteriol 190, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, and Whiteley JM (1998) Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry 37, 1365–1375. [DOI] [PubMed] [Google Scholar]
- (33).Casino P, Miguel-Romero L, and Marina A (2014) Visualizing autophosphorylation in histidine kinases. Nat. Commun. 5, 3258. [DOI] [PubMed] [Google Scholar]
- (34).Marina A, Waldburger CD, and Hendrickson WA (2005) Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24, 4247–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


