Abstract
Background:
African Americans (AA) experience a disproportionally high rate of bladder cancer (BLCA) deaths even though their incidence rates are lower than in other patient groups. We investigated how AA BLCA may differ molecularly from European American (EA) BLCA using a metabolomics approach and examined BLCA patient serum samples with the aim to identify druggable metabolic pathways in AA patients.
Methods:
Targeted metabolomics was applied to measure >300 metabolites in serum samples from two independent cohorts of EA and AA BLCA patients, and healthy EA and AA controls, using LC-MS, followed by the identification of altered metabolic pathways with a focus on AA BLCA. A subset of the differential metabolites was validated using absolute quantification with Biocrates absoluteIDQ p180 kit. The clinical significance of our findings was further examined in the TCGA bladder cancer dataset.
Results:
We identified fifty-three metabolites, mainly related to the amino acid and lipid and nucleotide metabolism, showed significant abundance differences between AA and EA BLCA. For example, levels of taurine, glutamine, glutamate, aspartate, and serine were elevated in serum samples of AA patients when compared with EA patients. By mapping these metabolites to genes, we identified significant relationships to regulators of metabolism, like ME3, P3H2 and KDM2A, that predicted patient survival exclusively in AA BLCA patients.
Conclusions:
Moreover, this metabolic profile of serum samples might be used to assess risk progression in AA BLCA. First-in-field findings describe metabolic alterations in AA BLCA and emphasize a potential biological basis for BLCA health disparities.
Keywords: Bladder Cancer, LC-MS, Racial Disparity, Metabolomics, Serum
Precis:
Possible disparity driven metabolic pathways associated with bladder cancer. Disparity genes may used to predict the African American bladder cancer progression.
Introduction
Bladder cancer (BLCA) is the ninth-most common malignancy worldwide1. Disease incidence and survival rates vary based on race. The incidence of BLCA is two times lower in African Americans (AA), but survival is poorer among AA.2,3 The Surveillance, Epidemiology, and End Results (SEER) data analysis from 1975–2013 found that the five-year relative survival of AA BLCA was close to 66% while EA BLCA had a survival rate of close to 80%4. Socioeconomic and disease biological differences may contribute to the disparity in BLCA. Emerging evidence suggests that both genetic and other biological factors play a role in racial disparities associated with BLCA while the poor survival of AA BLCA patients is also due to a late diagnosis and more aggressive tumor stage in AA5.
To support a sustained biomass, tumor cells exhibit increased demands for amino acids which depends on either de novo synthesis or exogenous supply. Amino acids contribute to the biosynthesis of nucleotides, proteins, lipids, and replenishment of TCA cycle intermediates6. They also play a role in signal transduction, and its derivatives act as cofactors for many enzymes which play major roles in cancer progression. Among them is KDM2A, which is an α-ketoglutaric acid-dependent JmjC-containing histone demethylase, which plays a crucial role in tumor progression by suppression of matrix metalloproteinases (MMP 2,9,14 and 15)7. Other enzymes include mitochondrial malic enzyme (ME3) and P3H2 which is an α-ketoglutaric acid dependent dioxygenase, that plays a critical role in collagen chain assembly, the stability by post-translational hydroxylation of proline residues8. Collagen is a major component of the extracellular matrix and dynamically involved in promoting cancer progression9. In contrast, ME3 oxidatively decarboxylates malate to pyruvate and is essential for NADP/NADPH homeostasis10. Because of the importance of the amino acid metabolism pathway of cancer cell proliferation, drugs targeting amino acid metabolism may reduce bladder cancer growth.
Metabolomic studies have now begun to identify disease-specific metabolic signatures and correlate them with clinical outcomes11–13. As markers, metabolites could be used to improve individual risk prediction in BLCA. Mass spectrometry (MS) in combination with various liquid chromatographic methods have been previously applied to study BLCA induced metabolomic alterations in tissues11,14–17, urine18–21, and serum22. Earlier reports identified tryptophan, hypoxanthine, and glycocholic acid as serum metabolites that can delineate BLCA from healthy normal22. Another study of the serum metabolome identified dimethylamine, malonate, lactate, glutamine, histidine, valine as serum metabolites discriminating between low grade and high-grade BLCA23. However, there are no reports available evaluating racial disparities in BLCA with serum metabolomics. The major challenges in addressing disparity-associated metabolic alterations are 1) difficulties in obtaining BLCA samples from AA for validation since the incidence of BLCA is low, and 2) a general lack of understanding of the disparity-driven mechanisms that accompany BLCA progression in AA population.
In this study, we used liquid chromatography-mass spectrometry (LC-MS) to identify metabolites that are possibly altered in AA BLCA and validated a subset of these metabolites by absolute quantification using independent LC-MS analysis. The altered metabolites were mapped to their associated genes to identify ethnicity-specific altered metabolic pathways, which then allow us to relate our observations to the clinical outcome of the AA BLCA patients in The Cancer Genomic Atlas (TCGA) BLCA cohort. We successfully identified novel disparity genes which predicted poor survival in AA BLCA patients. More in-depth studies are needed to further investigate the role of these genes in AA BLCA progression. Together, these results show that altered metabolism in AA BLCA can be detected in serum samples and linked to metabolic pathways and survival markers. These findings provide a basis to explore further the role of these metabolic pathways, which in turn could lead to new therapeutic targets and prognostic markers for AA BLCA.
Methods
Sample preparation for mass spectrometry and metabolomics analysis
Metabolic extraction was carried out as described previously12,13,24–26. Briefly, serum samples were stored at −80°C until the analysis. 40μl of serum was used for metabolite extraction; serum pool was used as quality controls. To each serum sample 750 μl of ice-cold methanol: water (4:1) containing 20 μl spiked internal standard (ISTD) was added. Ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 4:3:2 methanol: chloroform: water. Both the organic and aqueous layers were transferred into a new tube, dried and resuspended with 50:50 methanol: water. The resuspended sample was then deproteinized using a 3kDa molecular filter, and the filtrate was dried under vacuum. Prior to mass spectrometry, the dried extracts were re-suspended in 1:1 ratio of methanol: water and were subjected to LC-MS.
Separation of Metabolites
We used two different analytical methods (A and B) to target more than 300 metabolites. A) ESI positive mode, the HPLC column Waters X-bridge Amide 3.5 μm, 4.6 × 100 mm was used. Mobile phase A and B were 0.1% formic acid in water and acetonitrile respectively. Gradient: 0–3 min-85 % B; 3–12 min-30 % B, 12–15 min-2 % B, 16 min-95%-B, followed by re-equilibration at the end of the gradient 23 min to the initial starting condition 85% B. Flow rate: 0.3 ml/min. B) ESI negative mode the HPLC column used was Waters X-bridge Amide 3.5 μm, 4.6 × 100 mm. Mobile phase A and B were 20 mM ammonium acetate in water with pH 9.0 and 100% acetonitrile respectively. Gradient:0–3 min-85% B, 3–12 min-30% B, 12–15 min-2%-B, 15–16 min-85% B followed by re-equilibration end of the gradient 23 min to the initial starting condition of 85% B. Flow rate: 0.3 ml/min.
Data acquisition through LC/MS Analysis
LC-MS analysis was performed using 6490 triple quadrupole mass spectrometer coupled to an Agilent 1290 series HPLC system using single reaction monitoring (SRM). An Agilent 1290 series HPLC system equipped with a degasser, binary pump, thermostatted auto sampler and column oven. This SRM based measurement of relative metabolite levels used normal phase chromatographic separation. 10 μl of suspended samples were injected and analyzed using source parameters as follows: gas temperature-250 °C; gas flow-14 l/min; nebulizer - 20psi; sheath gas temperature - 350 °C; sheath gas flow-12 l/min; capillary - 3000 V positive and 3000 V negative; nozzle voltage-1500 V positive and 1500 V negative. Approximately 8–11 data points were acquired per each detected metabolite.
Quantification of metabolites using the Biocrates AbsoluteIDQ kit p180
Metabolite concentrations were obtained using the AbsoluteIDQ kit p180 according to manufacturer’s instructions on a QTRAP 6500 LC/MS/MS System. Briefly, 10 μl of the ISTD solution was added to each well of the 96-well plate, 10 μl of the serum samples, quality control samples, blank, and calibration standard were added to the appropriate wells. The plate was then dried, the samples were derivatized with phenyl isothiocyanate for the amino acids and biogenic amines. The samples were dried using GeneVac Vaccum system at 37°C and eluted with 5 mM ammonium acetate in methanol and then were diluted with water for LC-MS.
Statistical Analysis
Agilent MassHunter Workstation Software - Quantitative Analysis was used for manual review of chromatograms and peak area integration was accessed based on the retention time. The normalization of each metabolite peak area was done by the peak area of the spiked internal standard, and then the data were log2 transformed, per method basis and combined the data. For every metabolite in the normalized dataset, two sample t-tests were conducted to compare expression levels between AA and EA BLCA. Differential metabolites were identified by adjusting the p-values for multiple testing at a False Discovery Rate (FDR) threshold of <0.25. PCAs were generated using sklearn package in python, and the heat maps were generated using R program.
Pathway and survival analysis
To identify the altered metabolic pathways involved during AA BLCA development, the differential metabolites were subjected to MetaboAnalyst 3.0, and significant altered pathways were identified. Based on the differentially expressed metabolites integrative metabolic pathways were derived. The survival plots were generated using the R survival package, and gene expression was dichotomized into high or low group quantile of the gene expression value.
Results
We characterized the metabolome of AA BLCA (25%; n=18) and EA BLCA (74%; n=54) and matched age case-control AA and EA sera. The cohort included patients gender (Male and Female), and smoking status (Table 1). We obtained self-reported race/ethnicity from medical records which allowed for a comparison of the metabolic profiles of AA and EA BLCA. Targeted metabolomics was applied for the identification of serum metabolites in BLCA serum samples procured from two different independent cohorts from the National Cancer Institute (NCI), Augusta University Georgia Cancer Center and in serum samples from healthy control from Baylor College of Medicine (Table 1).
Table 1.
Patient characteristics of BLCA and case control serum samples and tissues used for this study (from three cohorts National Cancer Institute (NCI), Augusta university (AU), Georgia Cancer Center and Baylor College of Medicine (BCM).
| BLCA Serum samples | |
| BLCA Patients Characteristics | |
| Ethnicity | AA (n=18; 25%) |
| EA (n=54; 74%) | |
| Unknown (n=1; 1%) | |
| Smoking status | Never smokers (n=26; 36%; AA =4; EA=21; Unknown=1) |
| Former-smokers (n=31; 43%; AA=5; EA=26) | |
| Current smokers (n=10; 15%; AA=8; EA=2) | |
| Unknown (n=6; 6%) | |
| Gender | Male (n= 51; 70%; AA=11; EA=39; Unknown=1) |
| Female (n=22; 30%; AA=7; EA=15) | |
| Cohorts | NCI (n= 65; 89%) |
| AU (n =8; 11%) | |
| Case Control serum samples | |
| Ethnicity | AA (n=14; 70%) |
| EA (n=3; 15%) | |
| Unknown (n=3; 15%) | |
| Smoking status | Never smokers (n=9; 45%; AA=6; EA=1; Unknown =2) |
| Former-smokers (n=7; 35%; AA=4; EA=2; Unknown =1) | |
| Current smokers (n=4; 20%; AA=4; EA= None) | |
| Gender | Male (n= 10; 50%; AA=10; EA=0) |
| Female (n=10; 50%; AA=4; EA=3; Unknown =3) | |
| Cohort | BCM (n=20; 100%) |
| BLCA Tissues Characteristics | |
| Ethnicity | AA (n=6; 50%) |
| EA (n=6; 50%) | |
| Smoking status | Never smokers (n=4 (33 %; AA=2; EA=2) |
| Smokers (n=8; 67%; AA=4; EA=4) | |
| Gender | Male (n= 9; 75%; AA=4; EA=5) |
| Female (n= 3; 25%; AA=2; EA=1) | |
| Cohort | AU (n =12; 100%) |
AA: African American; EA: European American; NCI: National Cancer Institute; BCM: Baylor College of Medicine; AU: Augusta University
The metabolome was separated using normal aqueous phase (ANP) chromatography and examined using quadrupole mass spectrometers employing positive (+) and negative (−) electrospray ionization (ESI) with SRM. We targeted >300 metabolites representing many metabolic pathways and could detected 190 of them. Detected metabolites fell into 16 classes of endogenous biomolecules and xenobiotic compounds (Table 2). An overview of the study is shown in Figure. 1
Table 2.
List of different classes of differential metabolites in AA BLCA disparity
| Class | Number of Metabolites |
|---|---|
| Amino acid and derivatives | 37 |
| Fatty acid | 20 |
| Organic compound | 19 |
| TCA/Glycolysis/PPP | 13 |
| Prostaglandins | 9 |
| Tryptophan associated metabolites | 8 |
| Benzenoids | 7 |
| Nucleic acids/Nucleotide | 7 |
| Carnitine | 6 |
| Steroids and steroid derivatives | 6 |
| Flavonoids | 5 |
| Methylated/Acetylated compounds | 5 |
| Purines/Pyrimidines and derivatives | 4 |
| Neurotransmitter | 3 |
| Other metabolites | 41 |
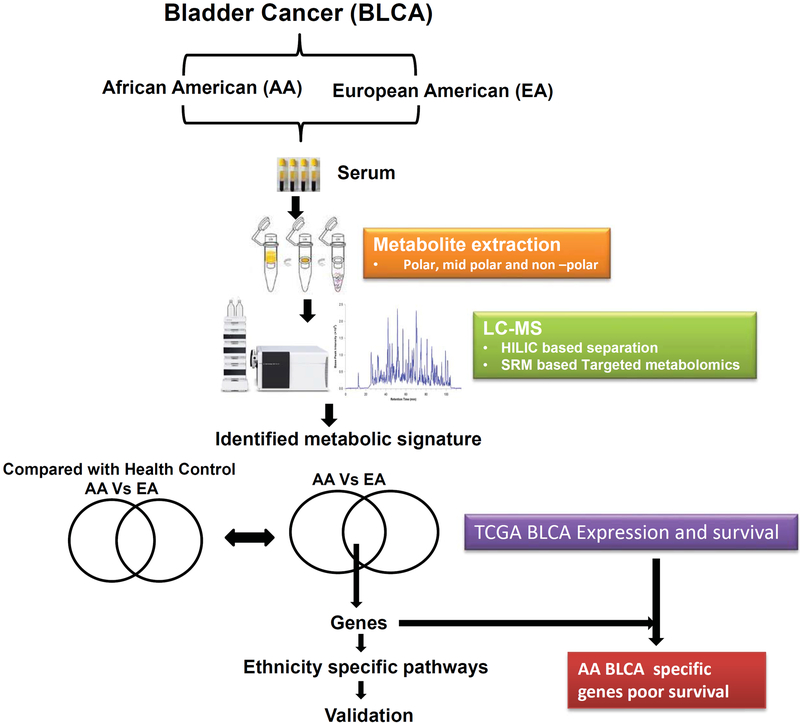
Figure 1.
Overview of strategy used to profile and characterize serum metabolites and associated pathways in AA and EA BLCA patients.
AA BLCA specific metabolic signature and pathways
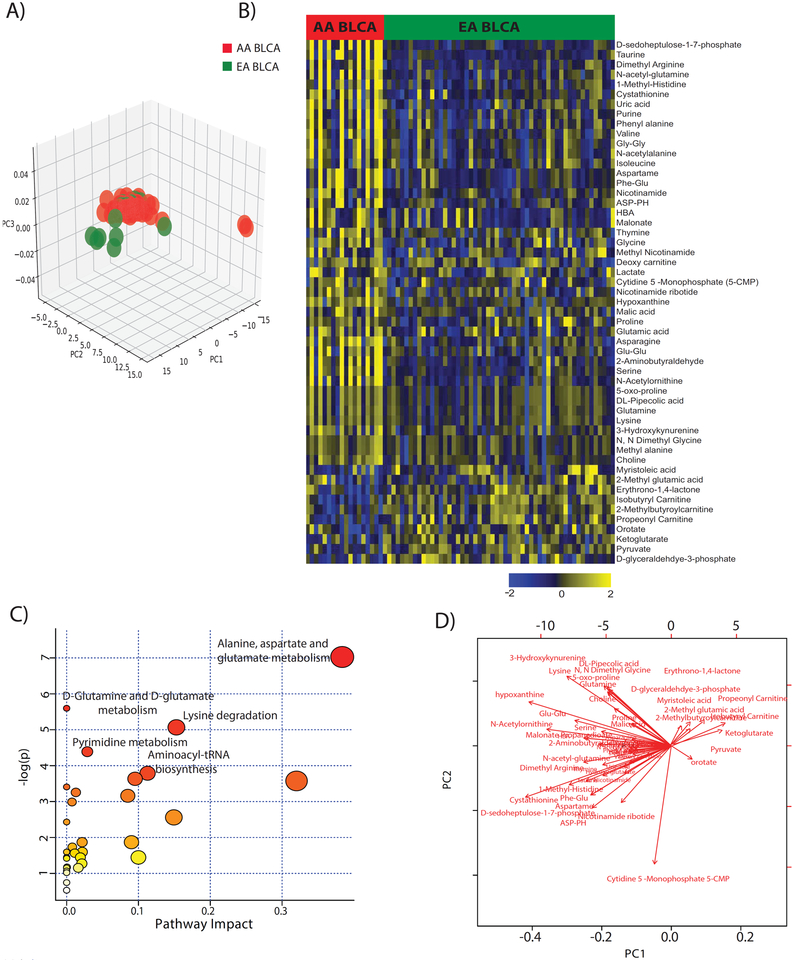
To examine the targeted metabolic profiles in the AA and EA BLCA sera, principal component analysis (PCA) was performed with the metabolome data. The separation between EA and AA BLCA serum indicated the existence of distinct metabolic profiles in AA and EA BLCA (Figure 2A). A total of 53 (out of the 190 identified metabolites) showed a differential abundance comparing AA with EA BLCA sera after multiple comparison adjustment at the false discovery rate (FDR) < 0.25. For example, taurine, glutamine, glutamic acid, phenyl alanine, proline, serine, valine, asparagine, glycine, isoleucine, lysine were all up-regulated while orotate, ketoglutarate and glyceraldehyde 3-phosphate were all down-regulated in AA serum when compared to EA BLCA (Figure 2B). However, we could not find these same differences when we analyzed sera from healthy AA and EA controls (Supplementary Figure 1A and B), indicating a disease-related pattern of the detected metabolic differences between the AA and EA BLCA patients (Figure 2A and Supplementary Figure 2A and B). Serum profiles of previous studies show the increased levels of malonate, histidine, lactate, glutamine, histidine valine decreased levels of, isoleucine/leucine, tyrosine, citrate in BLCA compared to healthy control27,28. We further annotated these differential metabolites using online metabolomic data processing, analysis, and interpretation with the annotation tool Metaboanalyst29. The annotation to pathways showed a significant association with the aspartate, alanine, glutamate, glucose, glutamine, and lysine degradation pathways (Figure. 2C). Additional pathway analysis of the differential metabolites also showed relationships to alterations of the purine catabolism, tryptophan, serine, glutamate, and fatty acid metabolism. Significance changes of the differential metabolites illustrated in the biplot (Figure. 2D). A summary of our observations is shown in Figure 3, highlighting the distinct difference in metabolite profiles between AA and EA patients
Figure 2. Racial disparity-associated metabolic pathways in BLCA.
A) PCA plot of the metabolomic profiles in serum samples of 18 AA and 54 EA BLCA patients. B) Heatmap showing hierarchical clustering of the 53 serum metabolites with significant abundance differences between AA and EA patients. Columns and rows represent individual serum samples and the metabolites. Shades of yellow and blue represent higher and lower levels of metabolites, relative to the median metabolite levels (FDR < 0.25). C) Pathway analysis of the 53 differential metabolites between AA and EA BLCA. The node size is proportional to the number of metabolites in the pathway. Red color represents statistical significance (p<0.05). D) Biplot for the 53 differential metabolites.
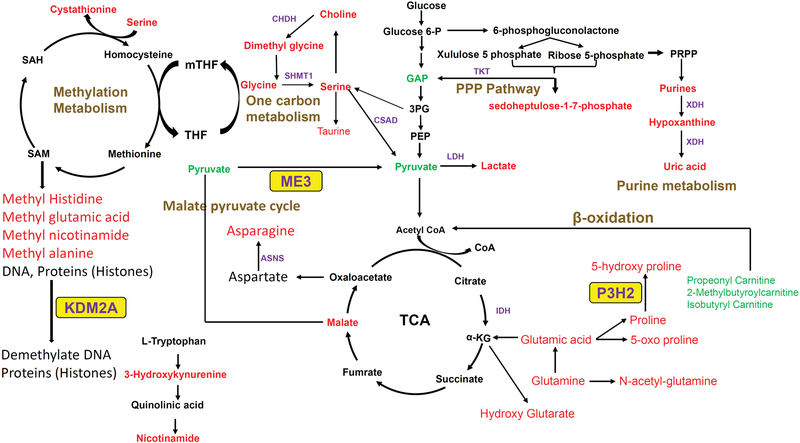
Figure 3. Metabolic pathways and associated key genes in AA BLCA patients.
Metabolites in red and green represent an increase or decrease in levels, respectively, in AA patient serum samples when compared to EA patient serum samples. The disease outcome-related enzymes ME3, KDM2A, and P3H2 are highlighted in yellow.
Validation of serum metabolites using the Biocrates AbsoluteIDQ kit p180 kit.
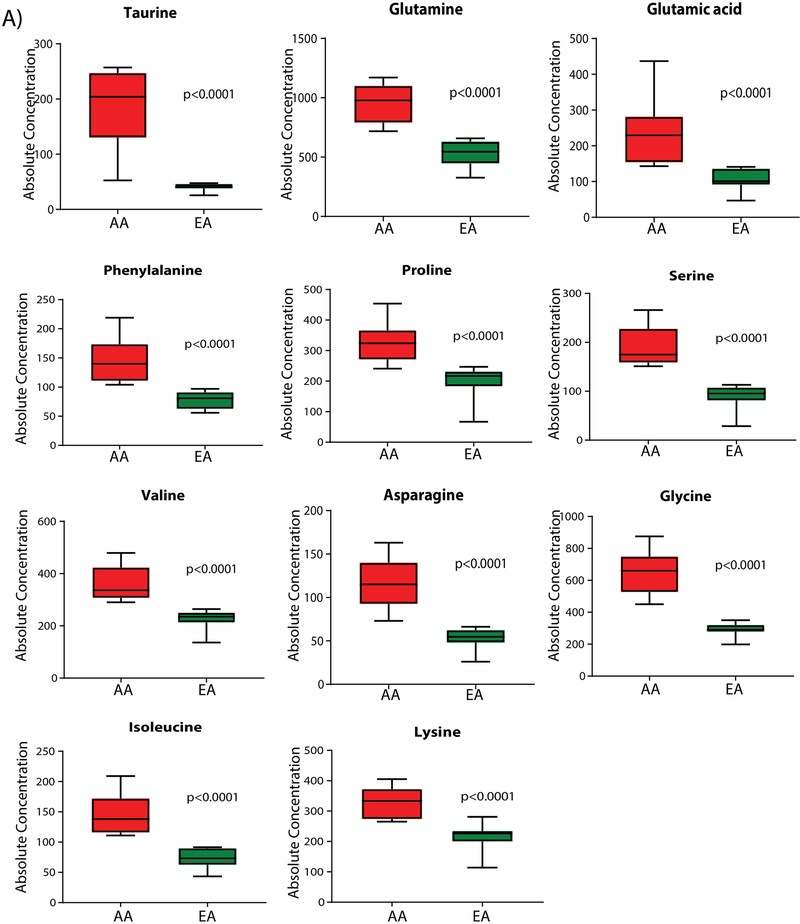
Quantitative validation was performed for a subset of these differential metabolites using a mass-spectrometry-based, reproducible, and quantitative analysis, employing the AbsoluteIDQ p180 kit by Biocrates. This technology can quantify 11 of the 53 differential metabolites using a predetermined metabolite selection. The absolute quantifications of taurine, glutamine, glutamic acid, phenyl alanine, proline, serine, valine, asparagine, glycine, isoleucine, and lysine show significantly higher levels in AA compared to EA BLCA, confirming that these metabolites accumulate to a greater degree in AA with BLCA (Figure. 4).
Figure 4. Absolute quantification of differential serum metabolites in AA and EA BLCA. Shown is the relative abundance of a subset of metabolites with differences between AA and EA patients.
Box plots of absolute concentrations (μM) of 11 elevated metabolites in AA BLCA serum, compared to EA BLCA serum, measured by Biocrates AbsoluteIDQ kit p180.
Identification of serum metabolome pathway genes and their prognosis power in AA BLCA
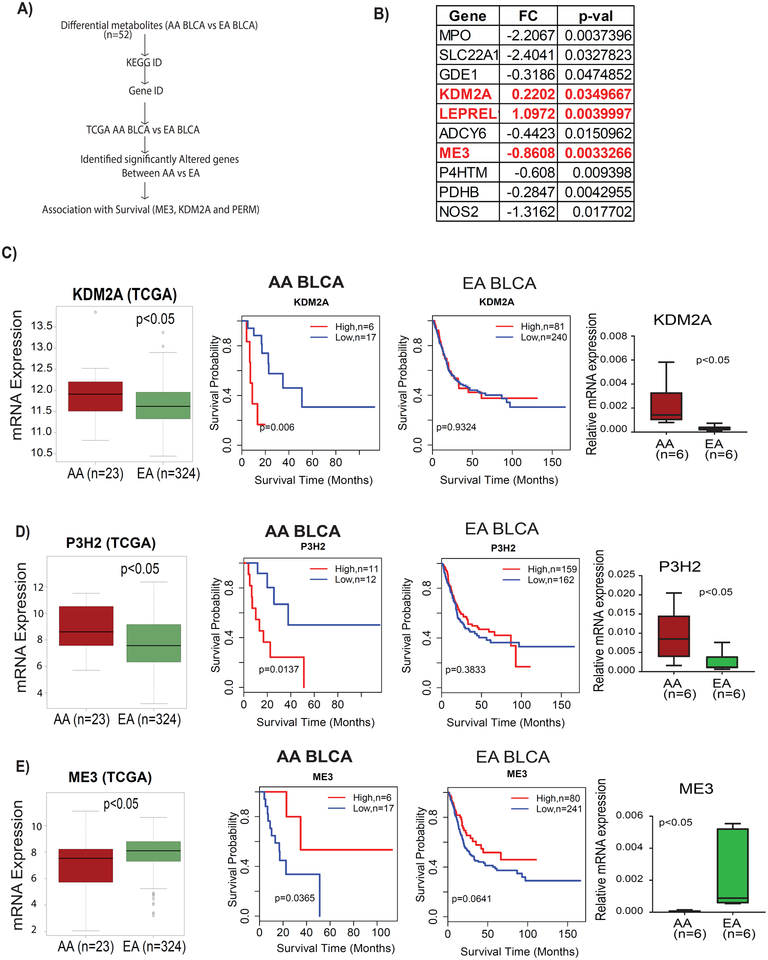
To gain molecular insights, the 53 serum metabolites with different abundance between AA and EA BLCA patients were mapped to their corresponding pathway genes using the Kyoto Encyclopedia of Genes (KEGG) and the Human Metabolic Data Base (HMDB), resulting in the identification of 187 genes which could be associated with the race/ethnic differences in serum metabolite abundance. To test the association of these metabolic genes with survival in AA BLCA, we performed an analysis of TCGA cohort,30 containing gene expressions and clinical outcomes for 23 AA BLCA and 321 EA BLCA. We identified ten genes that were differently expressed between AA and EA BLCA namely: MPO (myeloperoxidase), SLC22A1 (solute carrier family 22 member 1), GDE1 (glycerophospho diester phosphodiesterase 1), KDM2A (lysine demethylase 2A), P3H2 9 (prolyl 3-hydroxylase 2), ADCY6 (adenylate cyclase 6), ME3 (malic enzyme 3), P4HTM (prolyl 4-hydroxylase, transmembrane), PDHB (pyruvate dehydrogenase B), and NOS2 (nitric oxide synthase 2) (Figure 5B). Both KDM2A and P3H2 were up-regulated, whereas ADCY6, ME3, P4HTM, PDHB, NOS2, MPO, SLC22A1, and GDE1 were down-regulated in AAs (Supplementary Figure 3). To obtain additional insights, we further investigated the association of these genes with survival in AA and EA BLCA. We found that 3/10 genes had a significant clinical association in AA BLCA. Kaplan-Meier survival analysis of KDM2A and P3H2 showed that high expression was associated with poor survival in AA but did not show an association with survival among the EA patients (Figure 5C–D). AA patients with low expression of ME3 also showed an increased risk of mortality (Figure 5E). Together, the findings suggest that these genes have roles in the progression of BLCA in AA. We further validated the expression of KDM2A, P3H2, and ME3 in independent EA and AA BLCA tissues and found that elevated mRNA expression of KDM2A and P3H2 and down-regulation of ME3 in AA compared to EA (Figure 5C–E).
Figure 5. Expression of key metabolic genes and their association with survival in AA and EA BLCA patients.
A) Outline of the analysis to identify the 3 key genes B) List of all metabolism-related genes which are significantly differently expressed (p<0.05) between AA and EA BLCA patients. C) Expression and survival of KDM2A in AA and EA BLCA patients in TCGA and UA cohorts D) Expression and survival of P3H2 in AA and EA BLCA patients in TCGA and UA cohorts . E) Expression and survival of ME3 in AA and EA BLCA patients in TCGA and UA cohorts.
Discussion
Dysregulation of metabolic pathways is a hallmark of cancer development and progression,31 and altered metabolites in body fluids are direct indicators of deregulated metabolic pathways. Cancer cells require high quantities of energy and anabolic resources to support their rapid cell division; therefore, their key metabolic pathways are altered. Although cancer is the leading cause of mortality worldwide, prominent racial disparities are observed in the incidence, prevalence, and mortality rates in breast, prostate, lung, colorectal, and bladder cancer32,33.
Overall, our platform was able to identify altered metabolites and associated pathways in AA compared to EA BLCA. The differential metabolites were found to belong to an amino acid, nucleotide, carbohydrate, lipid, and carnitine metabolism. The elevated levels of methyl arginine, methyl histidine, methyl alanine, methyl nicotinamide, serine, and glycine indicate dysregulation of the one-carbon metabolism in AA with BLCA. This metabolic pathway supports several physiological processes that provide one-carbon units for methylation reactions, nucleotide synthesis, amino acid homeostasis, and reductive metabolism, supporting the high proliferation of cancer cells34,35. Notably, we observed that elevated levels of the purine catabolic pathway metabolites hypoxanthine and uric acid were associated with AA with BLCA. Recent studies have demonstrated that elevated levels of uric acid in serum are associated with greater cancer risk, recurrence, and mortality in colorectal, breast, prostate cancers36–38 and its pro-inflammatory properties may play a role in the BLCA pathogenesis in AA. Purine metabolism is associated with numerous biochemical functions, including cell cycle regulation, immune function, and signal transduction39,40. Hnce, its deregulation may be associated with disparity driven cancer progression in BLCA.
Metabolic pathway associations derived from differential metabolites suggested that the metabolisms of glutamate, glutamine, alanine, lysine degradation, and the nucleotide are particularly altered in AA BLCA. Glutamine, the most abundant amino acid, is a nutrient used by cancer cells to support cancer progression under metabolic stress conditions. Glutamine metabolism fuels the TCA cycle and fatty acid synthesis by activating mTOR signaling to promote protein synthesis41,42. Studies have shown glutamine metabolism reprogramming in a tissue-dependent and tumor-heterogeneous manner43. its role in disparity remains to be investigated. Lysine catabolism is important in the early stages of liver metastasis in colorectal cancer, but it is not essential for growth in liver metastases44.
We also examined the expression of metabolic genes and their survival in AA BLCA from TCGA and found significant expression differences and clinical correlations. There was no clinical association of pathway genes in EA BLCA that associated with survival in AA BLCA. These results suggest that the identified metabolic genes may activate cancer progression in AA BLCA. This is the first study to report disparities related to metabolism-related genes associated with poor survival among AA BLCA. High expression of KDM2A and P3H2is associated with poor survival among AA BLCA patients. Low expression of ME3 is associated with poor survival among AA BLCA. Interestingly, the same genes didn’t show clinical significance in EA BLCA suggesting the importance of these genes in AA BLCA. Others have shown that KDM2A is a histone demethylase that targets mono- and di-methylated H3K36 of histone H345 and depends on amino acid metabolism for its enzymatic activity. In breast cancer, KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes46. The up-regulated epigenetic regulator KDM2A might be involved in the suppression of disparity-associated genes in AA BLCA. KDM2A activity depends on aminoacid metabolism On the other hand, P3H2 is an α-ketoglutarate and iron-dependent dioxygenase enzyme, and it catalyzes the post-translational hydroxylation of proline residues in collagens47. Collagens play a critical function for tumor cell migration and metastasis48,49 as being part of the extracellular matrix. The higher hydroxylation provides unique structural alterations in collagen which may drive tumor towards the muscle-invasive BLCA in AA. Reduced expression of ME3 is associated with poor survival in AAs with BLCA. The mitochondrial malic enzyme ME3 regulates the pyruvate-malate cycle in which oxaloacetate is reduced to malate by mitochondrial malate dehydrogenase and then is converted to pyruvate by oxidative decarboxylation catalyzed by ME350. This pathway is crucial for NADPH regeneration and reactive oxygen species homeostatis51. Recent studies in pancreatic cancer revealed that ME3 depletion selectively kills ME2-null cancer cells, conferring its role in cancer10. The metabolic role of ME3 in the progression of AA BLCA needs further investigation.
In conclusion, our results support the notion that serum metabolic detection might be used as a non-invasive approach for assessing the AA BLCA risk progression. We found that high levels of circulating hypoxanthine, uric acid, taurine, and the amino acids glutamine, serine, asparagine, valine, glycine, and lysine have the potential to discriminate between AA and EA BLCA patients. Pathway analysis revealed that one-carbon, tryptophan and nucleotide metabolisms are significantly altered in AA BLCA patients. In addition, the analysis of metabolite-associated genes in the TCGA cohort revealed potential disparity-driving genes for BLCA outcomes in AA patients. Our studies are in line with recent studies in which the importance of metabolic changes in BLCA progression and chemo resistance has been highlighted. Targeting these changes could potentially be used for therapeutic applications52. Future prospective validation using larger cohorts is necessary to confirm these findings, and a further functional investigation is needed to establish the roles of ME3, KDM2A, and P3H2 in the BLCA disparity. Finally, this study highlights many unique metabolites and metabolic pathways that can lead to the identification of new biochemical nodes that contribute to racial disparities in BLCA development. A limitation of this study is the inability to procure a large number of samples from AA patient cohort.
Supplementary Material
Supplementary Figure 1. A) PCA plot of serum metabolomic profiles in 3 EA and 14 AA healthy individuals. B) Heatmap showing hierarchical clustering of undeferential metabolites across EA and AA healthy case controls (FDR < 0.25).
Supplementary Figure 2. Typical total ion chromatograms (TIC) obtained in LC–high-resolution MS for serum samples and abundance differences between AA and EA patients. A) Positive ESI total ion chromatogram with an example of phenylalanine. B) Negative ESI total ion chromatogram with an example of uric acid.
Supplementary Figure 3. Box plots showing differential expression of metabolite related genes in AA and EA BLCA patients in the TCGA cohort (p<0.05).
Acknowledgments
This research was fully supported by NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.) and American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N.P.), partially supported by the following grants: NIH/NCI U01 CA167234 (A.S.K), as well as funds from Alkek Center for Molecular Discovery (A.S.K.). This project was also supported by the Agilent Technologies Center of Excellence (COE) in Mass Spectrometry at Baylor College of Medicine, Metabolomics Core, Population Sciences Biorepository core at Baylor College of Medicine with funding from the NIH (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility (N.P.), (RP170005), and Dan L. Duncan Cancer Center. We want to thank Maharajni Perla for her input on the analysis.
Funding: This research was fully supported by NIH/NCI R01CA220297 (N.P), and NIH/NCI R01CA216426 (N.P.) and American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N.P.), partially supported by the following grants: NIH/NCI U01 CA167234 (A.S.K), Metabolomics Core, Population Sciences Biorepository core at Baylor College of Medicine with funding from the NIH (P30 CA125123) (N.P, A.S.K.), CPRIT Proteomics and Metabolomics Core Facility (N.P.), (RP170005).
References
- 1.Ferlay J et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136, E359–386, doi: 10.1002/ijc.29210 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Bach PB et al. Survival of blacks and whites after a cancer diagnosis. JAMA 287, 2106–2113 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Prout GR Jr. et al. Survival experience of black patients and white patients with bladder carcinoma. Cancer 100, 621–630, doi: 10.1002/cncr.11942 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA Cancer J Clin 66, 7–30, doi: 10.3322/caac.21332 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Lee CT, Dunn RL, Williams C & Underwood W 3rd. Racial disparity in bladder cancer: trends in tumor presentation at diagnosis. J Urol 176, 927–933; discussion 933–924, doi: 10.1016/j.juro.2006.04.074 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Lukey MJ, Katt WP & Cerione RA Targeting amino acid metabolism for cancer therapy. Drug Discov Today 22, 796–804, doi: 10.1016/j.drudis.2016.12.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizwani W, Schaal C, Kunigal S, Coppola D & Chellappan S Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS One 9, e100888, doi: 10.1371/journal.pone.0100888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiainen P, Pasanen A, Sormunen R & Myllyharju J Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem 283, 19432–19439, doi: 10.1074/jbc.M802973200 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Fang M, Yuan J, Peng C & Li Y Collagen as a double-edged sword in tumor progression. Tumour Biol 35, 2871–2882, doi: 10.1007/s13277-013-1511-7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey P et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542, 119–123, doi: 10.1038/nature21052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Rundstedt FC et al. Integrative Pathway Analysis of Metabolic Signature in Bladder Cancer: A Linkage to The Cancer Genome Atlas Project and Prediction of Survival. J Urol 195, 1911–1919, doi: 10.1016/j.juro.2016.01.039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra P et al. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J Clin Invest 128, 323–340, doi: 10.1172/JCI93815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terunuma A et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 124, 398–412, doi: 10.1172/JCI71180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahu D, Lotan Y, Wittmann B, Neri B & Hansel DE Metabolomics analysis reveals distinct profiles of nonmuscle-invasive and muscle-invasive bladder cancer. Cancer Med 6, 2106–2120, doi: 10.1002/cam4.1109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao CH et al. Metabolite marker discovery for the detection of bladder cancer by comparative metabolomics. Oncotarget 8, 38802–38810, doi: 10.18632/oncotarget.16393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan EC et al. Metabonomic profiling of bladder cancer. J Proteome Res 14, 587–602, doi: 10.1021/pr500966h (2015). [DOI] [PubMed] [Google Scholar]
- 17.Putluri N et al. Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res 71, 7376–7386, doi: 10.1158/0008-5472.CAN-11-1154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kind T et al. Interstitial Cystitis-Associated Urinary Metabolites Identified by Mass-Spectrometry Based Metabolomics Analysis. Sci Rep 6, 39227, doi: 10.1038/srep39227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen C et al. Developing urinary metabolomic signatures as early bladder cancer diagnostic markers. OMICS 19, 1–11, doi: 10.1089/omi.2014.0116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittmann BM et al. Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS One 9, e115870, doi: 10.1371/journal.pone.0115870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberice JV et al. Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. J Chromatogr A 1318, 163–170, doi: 10.1016/j.chroma.2013.10.002 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lin L et al. LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics 12, 2238–2246, doi: 10.1002/pmic.201200016 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Bansal N, Gupta A, Sankhwar SN & Mahdi AA Low- and high-grade bladder cancer appraisal via serum-based proteomics approach. Clin Chim Acta 436, 97–103, doi: 10.1016/j.cca.2014.05.012 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Dasgupta S et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature 556, 249–254, doi: 10.1038/s41586-018-0018-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purwaha P et al. Unbiased Lipidomic Profiling of Triple-Negative Breast Cancer Tissues Reveals the Association of Sphingomyelin Levels with Patient Disease-Free Survival. Metabolites 8, doi: 10.3390/metabo8030041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putluri N et al. Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia 16, 390–402, doi: 10.1016/j.neo.2014.05.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao M, Zhao L, Chen H, Xue W & Lin D NMR-based metabolomic analysis of human bladder cancer. Anal Sci 28, 451–456 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Bansal N et al. Low- and high-grade bladder cancer determination via human serum-based metabolomics approach. J Proteome Res 12, 5839–5850, doi: 10.1021/pr400859w (2013). [DOI] [PubMed] [Google Scholar]
- 29.Xia J, Sinelnikov IV, Han B & Wishart DS MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res 43, W251–257, doi: 10.1093/nar/gkv380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322, doi: 10.1038/nature12965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamin DI, Cravatt BF & Nomura DK Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab 16, 565–577, doi: 10.1016/j.cmet.2012.09.013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esnaola NF & Ford ME Racial differences and disparities in cancer care and outcomes: where’s the rub? Surg Oncol Clin N Am 21, 417–437, viii, doi: 10.1016/j.soc.2012.03.012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs BL et al. Disparities in bladder cancer. Urol Oncol 30, 81–88, doi: 10.1016/j.urolonc.2011.08.011 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Newman AC & Maddocks ODK One-carbon metabolism in cancer. Br J Cancer 116, 1499–1504, doi: 10.1038/bjc.2017.118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducker GS & Rabinowitz JD One-Carbon Metabolism in Health and Disease. Cell Metab 25, 27–42, doi: 10.1016/j.cmet.2016.08.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorge T et al. Metabolic syndrome and breast cancer in the me-can (metabolic syndrome and cancer) project. Cancer Epidemiol Biomarkers Prev 19, 1737–1745, doi: 10.1158/1055-9965.EPI-10-0230 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Giovannucci E Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am J Clin Nutr 86, s836–842, doi: 10.1093/ajcn/86.3.836S (2007). [DOI] [PubMed] [Google Scholar]
- 38.Rose DP, Haffner SM & Baillargeon J Adiposity, the metabolic syndrome, and breast cancer in African-American and white American women. Endocr Rev 28, 763–777, doi: 10.1210/er.2006-0019 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Pedley AM & Benkovic SJ A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci 42, 141–154, doi: 10.1016/j.tibs.2016.09.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Virgilio F & Adinolfi E Extracellular purines, purinergic receptors and tumor growth. Oncogene 36, 293–303, doi: 10.1038/onc.2016.206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altman BJ, Stine ZE & Dang CV From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 16, 749, doi: 10.1038/nrc.2016.114 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Hensley CT, Wasti AT & DeBerardinis RJ Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 123, 3678–3684, doi: 10.1172/JCI69600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cluntun AA, Lukey MJ, Cerione RA & Locasale JW Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 3, 169–180, doi: 10.1016/j.trecan.2017.01.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Z et al. TPO-Induced Metabolic Reprogramming Drives Liver Metastasis of Colorectal Cancer CD110+ Tumor-Initiating Cells. Cell Stem Cell 17, 47–59, doi: 10.1016/j.stem.2015.05.016 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Tsukada Y et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816, doi: 10.1038/nature04433 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Chen JY, Luo CW, Lai YS, Wu CC & Hung WC Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis 6, e369, doi: 10.1038/oncsis.2017.71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes RJ, Farnand AW, Traeger GR, Weis MA & Eyre DR A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens. J Biol Chem 286, 30662–30669, doi: 10.1074/jbc.M111.267906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilera KY et al. Collagen signaling enhances tumor progression after anti-VEGF therapy in a murine model of pancreatic ductal adenocarcinoma. Cancer Res 74, 1032–1044, doi: 10.1158/0008-5472.CAN-13-2800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Provenzano PP et al. Collagen density promotes mammary tumor initiation and progression. BMC Med 6, 11, doi: 10.1186/1741-7015-6-11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pongratz RL, Kibbey RG, Shulman GI & Cline GW Cytosolic and mitochondrial malic enzyme isoforms differentially control insulin secretion. J Biol Chem 282, 200–207, doi: 10.1074/jbc.M602954200 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Jiang P, Du W, Mancuso A, Wellen KE & Yang X Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693, doi: 10.1038/nature11776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woolbright BL, Ayres M & Taylor JA 3rd. Metabolic changes in bladder cancer. Urol Oncol 36, 327–337, doi: 10.1016/j.urolonc.2018.04.010 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A) PCA plot of serum metabolomic profiles in 3 EA and 14 AA healthy individuals. B) Heatmap showing hierarchical clustering of undeferential metabolites across EA and AA healthy case controls (FDR < 0.25).
Supplementary Figure 2. Typical total ion chromatograms (TIC) obtained in LC–high-resolution MS for serum samples and abundance differences between AA and EA patients. A) Positive ESI total ion chromatogram with an example of phenylalanine. B) Negative ESI total ion chromatogram with an example of uric acid.
Supplementary Figure 3. Box plots showing differential expression of metabolite related genes in AA and EA BLCA patients in the TCGA cohort (p<0.05).