Abstract
The pre-mRNA branch point sequence (BPS) anneals with a pseudouridine-modified region of the U2 small nuclear (sn)RNA, and offers a 2′ hydroxyl group of a bulged adenosine as the nucleophile for the first catalytic step of pre-mRNA splicing. To increase our structural understanding of branch site selection, we characterized a duplex containing a BPS sequence that is common among multicellular eukaryotes (5´-UACUGAC-3´) and the complementary U2 snRNA site using NMR. A major conformation of the expected branch site adenosine stacked within the duplex and paired with the conserved pseudouridine of the U2 snRNA strand. In contrast, the guanosine preceding the branch site appeared flexible and had weak contacts with the surrounding nucleotides. Pseudouridine-modified and unmodified U2 snRNA–BPS-containing duplexes remained structurally similar. These results highlight the importance of auxiliary factors to achieve the active bulged conformation of the branch site nucleophile for the first step of pre-mRNA splicing.
Keywords: RNA structure, pre-mRNA splicing, 3´ splice site, branch point sequence, U2 snRNA, pseudouridine
Graphical Abstract
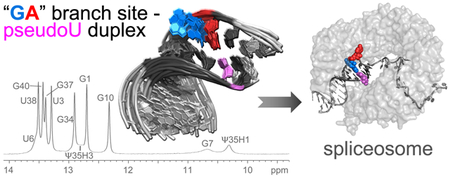
1. Introduction
The highly regulated process of pre-mRNA splicing offers an important source of transcript diversity [1] and is often dysregulated in cancers and hematologic malignancies [2]. The spliceosome assembles through the actions of protein co-factors coupled with dynamic base-pairing among small nuclear (sn)RNAs and specific pre-mRNA sites. At the active site for catalysis, a post-transcriptionally modified region of the U2 snRNA forms a short, base-paired duplex with the branch point sequence (BPS) of pre-mRNA introns [3]. The pre-mRNA splicing reaction then proceeds in two sequential catalytic steps: First, an adenosine within the BPS typically offers a 2′ hydroxyl group for nucleophilic attack on the 5′ splice site [4]. The products of this reaction are a 2´−5´-linkage at the “branch site” adenosine and 3′-hydroxyl group of the upstream 5´-exon. Second, this 3′ hydroxyl of the exon attacks the downstream 3′ splice site, which releases the 2′−5′ branched intron lariat from the spliced exon-exon junction of the mRNA.
Despite emerging views of spliceosome intermediates, selection of the BPS nucleophile remains only partially understood. The BPS of human introns are relatively degenerate (YUNAY where Y is pyrimidine and N any nucleotide) compared to Saccharomyces cerevisiae BPS (5′-UACUAAC-3′) [5, 6]. In principle, either the branch site or preceding nucleotide could base-pair with a highly conserved pseudouridine (Ψ35) in the complementary region of the U2 snRNA, leaving the other nucleotide unpaired. The second adenosine (underlined) typically serves as the branch site nucleophile, yet several findings indicate that the branch site choice has some plasticity. In human nuclear extracts, branch site mutations typically activate a nearby cryptic branch site rather than preventing pre-mRNA splicing [7, 8], and substitution of the canonical branch site with deoxy-adenosine induces efficient branching from the preceding nucleotide [4]. Branch site selection appears to be more stringent in yeast than humans, since pyrimidine mutations of the branch site adenosine in S. cerevisiae reduce splicing efficiency at the mutated nucleotide [9]. Nevertheless, systematic use of an orthogonal U2 snRNA – BPS system [10] demonstrates that a “bulged” conformation of the branch site adenosine is as an important criterion to produce the intron lariat in both S. cerevisiae and humans,
The potential for base-pairing between Ψ35 and either the branch site adenosine or preceding nucleotide raises the question of how the “bulged” conformation of the nucleophile is selected for the first step of splicing. The Ψ35-modification (compared to unmodified U) promotes bulging of the branch site adenosine in NMR characterization of a duplex containing the yeast consensus U2 snRNA and “AA”-containing BPS sequences [11]. On the other hand, crystal structures containing pseudouridine-modified U2 snRNA duplexes with “GA”- or “AA”-containing BPS sequences capture either the expected branch site or the preceding nucleotide in an extrahelical position [12]. Here, we investigate the solution conformation of a “GA”-containing BPS – U2 snRNA duplex, which is a common sequence among splice sites of multicellular eukaryotes. Our results show that a predominant conformation of the expected branch site adenosine (underlined) of a 5´-UACUGAC-3´ BPS pairs with the pseudouridine in the U2 snRNA-containing duplex, whereas the preceding guanosine appears to be unstacked. Comparison of pseudouridylated or unmodified duplexes further shows the conserved Ψ35-modification of the U2 snRNA makes few detectable changes in the BPS conformation of this sequence context.
2. Materials and methods
2.1. NMR Sample Preparation.
HPLC-purified RNA oligonucleotides (BPS, 5′-GCUACUGACGA; U2, 5′-CGUAGUAGCA, or Ψ-U2, 5′-CGΨAGUAGCA) (GE Healthcare Dharmacon, Inc.) were dissolved in a filter-sterilized solution of 10 mM potassium phosphate, 0.25 mM ethylenediaminetetraacetic acid at pH 6.8 in RNase-free 90% water/10% D2O. The strands were mixed in equal molarity at a concentration of 1.1 mM, annealed by incubating at 60 °C for 15 min followed by gradual cooling to 4 °C, and transferred to a thin-walled Shigemi tube (Allison Park, PA). To acquire spectra in D2O, the oligonucleotides were lyophilized and resuspended thrice in 99.9% D2O (Cambridge Isotope Lab, Inc.). For samples at higher ionic strengths, the oligonucleotides were lyophilized and resuspended in 80 mM NaCl, then MgCl2 was added.
2.2. NMR Spectroscopy.
NMR spectra were acquired using a Varian Inova spectrometer at 600 MHz and a triple-resonance, triple-gradient probe. The one-dimensional imino proton spectra were acquired using either an S-pulse or a watergate-pulse for water signal suppression. Exchangeable imino and amino protons were assigned from water-suppressed two-dimensional NOESY spectra in 95% H2O/5% D2O at 0 °C with mixing times of 50, 100 and 200 ms, at 15 °C with mixing times of 75, 175, and 300 ms, and at 25°C with mixing times of 75, 100, 175, and 400 ms. The spectral width was 15 kHz in the directly-detected dimension, and 8.24 or 7.6 kHz in the indirect dimension (with imino peaks wrapped). Watergate TOCSY and 1H-13C HSQC spectra also were acquired at 0 and 15 °C. Additional two-dimensional spectra acquired at −3°C include an S-pulse NOESY with 125 ms mixing time and a natural abundance 1H-15N HSQC to confirm assignment of the G and U imino protons.
One-dimensional spectra of pseudouridylated U2 snRNA–BPS duplex in 100% D2O were acquired at temperatures ranging from 0 to 60 °C. Two-dimensional NOESY (mixing times of 75, 175, and 400 ms), DQF-COSY, TOCSY (mixing times of 12, 32, and 80 ms), 1H-13C HSQC, and 1H-31P HETCOR spectra were acquired at 25 °C, with additional NOESY, 1H-13C HSQC, and TOCSY spectra at 15 °C. The DQF-COSY, TOCSY, and 1H-13C HSQC spectra were all wrapped in the indirect dimension to maximize resolution. NMRpipe software was used to process the two-dimensional spectra [13]. Protons were assigned using standard procedures [14], using Sparky to assign resonances and analyze NOE cross-peak volumes [15]. All NOE cross-peak volumes for distance information were measured from NOESY spectra with mixing times of 75 msec. Volumes of non-exchangeable protons in 100% D2O samples were converted to distances using 1/r6 scaling by comparison to H1’–H2’ cross-peaks from stem residues which were assumed to represent 2.75 Å. The NOE volumes of non-exchangeable protons in 95% H2O/5% D2O were referenced to the 100% D2O samples by normalization to the averages of (n-1)H2’-nH8/6 and H1’-H8/H6 cross-peaks in the stem (nucleotides 1–6, 9–10, 33–34, 36–41). Proton chemical shifts are reported relative to 4,4-dimethyl-4-silapentane-1-sulfonic acid by referencing to H2O or HDO (Tables S2–S4). We found that simulated annealing models of the Ψ-U2 snRNA/BPS duplex fail to accurately reflect all the spectral information near the branchpoint, which is unsurprising considering the conformational dynamics and few restraints for G7 (see Results). The chemical shifts of the pseudouridine-modified and unmodified RNA duplexes are deposited in the Biological Magnetic Resonance Bank under accession numbers 27770 and 27769.
3. Results
3.1. NMR characterization of “GA”-Containing BPS – U2 snRNA duplexes.
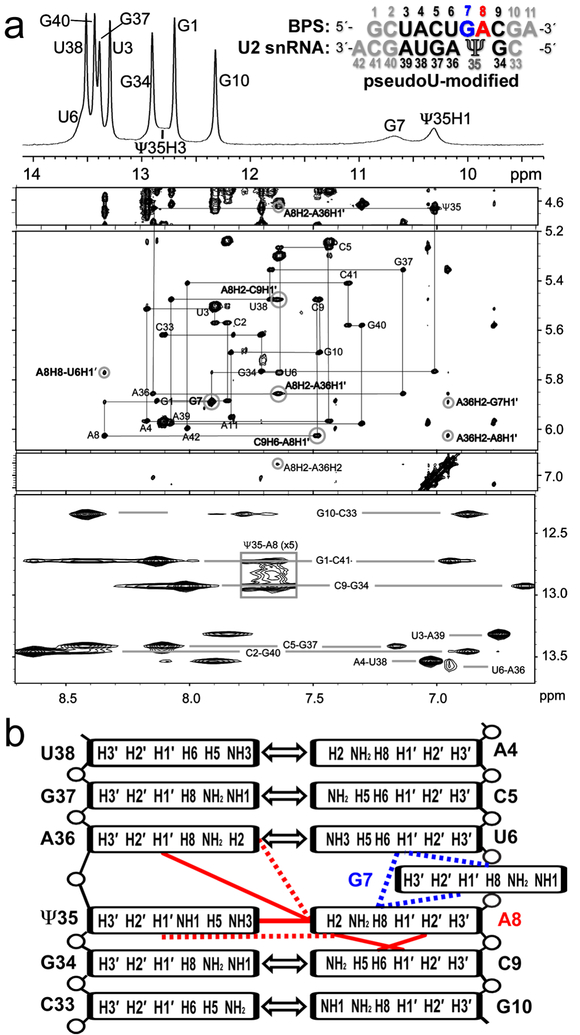
We chose a “GA”-containing sequence with G preceding the expected branch site (A8) (sequence inset in Fig. 1a) for NMR studies of a typical BPS of multicellular organisms [5]. We annealed the BPS with a complementary U2 snRNA region either including a conserved pseudouridine modification or for comparison, the unmodified counterpart (Ψ35 or U35 in 5′-GΨAGUAG-3′ or 5′-GUAGUAG-3′, numbered 34–39). Flanking GC base-pairs and 3′-A overhangs increased duplex stability for NMR measurements [16]. The NOESY spectra of both the pseudouridine-containing and unmodified duplexes indicated that the sequences flanking the branch site (up to and including U6–A36 and C9–G34) form Watson-Crick base-pairs (Fig. 1, Fig. S1, Table S1–S2). Strong G-imino to C-amino and U-imino to A-H2 NOE cross-peaks for these nucleotides were typical of G–C and A–U base-pairs, respectively. The scalar coupling and intra-strand NOE patterns were consistent with A-form sugar puckers, backbone dihedral angles, and base stacking. In the U2 region opposite the branch site, the scalar coupling patterns and NOEs (A36H8 – Ψ35H2′ and Ψ35H6 – G34H2′, strong; A36H8 – Ψ35H3′, medium; G34H1 – Ψ35H1′, medium; A36H1′ – Ψ35H2′, weak; H8/6 – H1′ for nucleotides 34–36, medium), also were typical of A-form backbone conformations and base stacking (Fig. 1a, Fig. S1, Table S1). In the BPS-containing strand, the A8 at the expected branch site, as well as the preceding U6 and G7 nucleotides, appeared to have greater mobility than the stems of the duplex. The JH1′–H2′ values of approximately 4 Hz were consistent with exchange of the U6 and G7 ribose groups between C3′-endo and C2′-endo conformations. Similarly, relatively broad resonances for A8H2, G7H2′, and G7H3′ (Fig. 1, Fig. 2) agreed with dynamic conformations for these nucleotides.
Fig. 1.
NOESY spectra indicate an intrahelical adenosine (A8) at the expected branch site and a partially syn conformer for the preceding guanosine (G7) for the pseudouridine (Ψ)-modified U2 snRNA – BPS duplex. (a) Top panel: One-dimensional spectrum of the imino protons at 0 °C. The RNA sequences are inset (expected branch site A8, red; preceding G7, blue; other consensus sequences, black; terminal nucleotides to facilitate NMR analysis, grey). Four lower panels: Two-dimensional NOESY spectra of the aromatic/amino region of the pseudouridine-containing duplex in 100% D2O at 15 °C with 400 ms mixing time (panels spanning 4.5–7.1 ppm) and in 95/5% H2O/D2O at 0 °C with 50 ms mixing time (bottom panel). Gray lines in the D2O NOESY trace the H8/6-H1′ backbone walk with intra-residue peaks labeled. Cross-peaks between G-imino and C-amino protons and between U-imino and A-H2 protons indicating Watson-Crick base-pairs are connected by light gray lines in the H2O NOESY. Key peaks for defining the branch site conformation are circled and labeled in bold font. Contours in the boxed region of the H2O spectrum are drawn five-fold lower than in the rest of the spectrum to show the Ψ35H3-A8H2 cross-peak. (b) Diagram of major branch site interactions: Double-headed arrows, canonical Watson-Crick base-pairs; blue lines, NOESY interactions used to determine the relative position of G7; red lines, NOESY interactions used to identify the relative position of A8. Dashed lines represent weak NOEs.
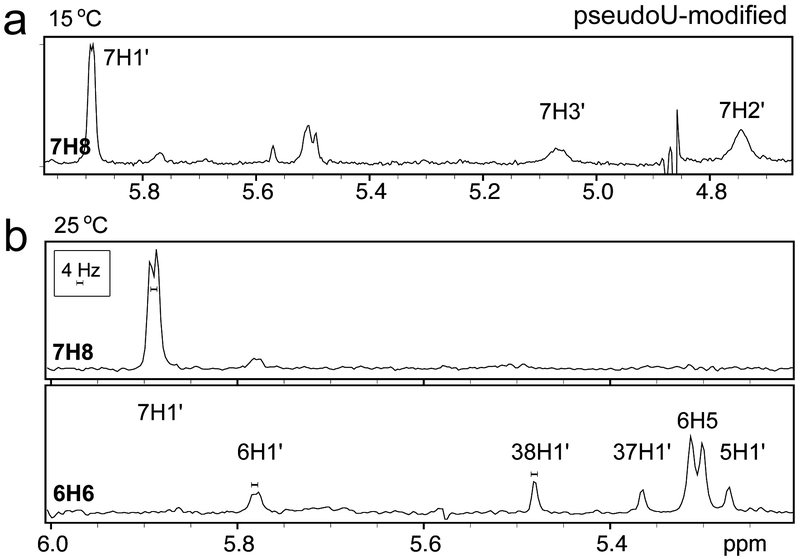
Fig. 2.
One-dimensional slices from two-dimensional NOESY spectra of the pseudouridine-modified U2 snRNA – BPS-containing RNA duplex in 100% D2O with 400 ms mixing times at (a) 15 °C and (b) 25 °C show that residues G7 and U6 are dynamic and exhibit large populations of C2′-endo sugar puckers. Each slice is taken through the aromatic proton indicated in bold type at the left of the slice. In (a), conformational dynamics causes broad resonances for G7H3′ and G7H2′. In (b), the 4 Hz scale bar shows the 4–5 Hz H1′-H2′ scalar coupling for G7 and U6 which can be compared to the negligible H1′-H2′ coupling for other residues.
3.2. The branch site adenosine stacks within the duplex and pairs with pseudouridine.
Several NOESY interactions indicated that the major conformation of the expected branch site A8 stacks with the neighboring C9 in the duplex and base-pairs with pseudouridine Ψ35. A cross-peak between A8H2 and Ψ35H3 is observed at a mixing time of 50 msec and 0 °C. That this peak is observed at such a short mixing time despite the relatively large width of the involved resonances (~60 and ~70 Hz) and the rapid exchange of Ψ35H3 with water, indicates that this is a strong NOE consistent with A8 and Ψ35 forming a canonical base-pair (Fig. 1). The chemical shift of Ψ35H3 (12.8 ppm) was slightly upfield of the ΨH3, as observed for other A–Ψ pairs [17, 18]. The NOE cross-peaks provided further support for an intrahelical conformation of the expected branch site (including A8H2′ – C9H6, strong; A8H2 – A36H1′ and A8H2 – C9H1′, medium; A8H2 – Ψ35H1′ and A8H2 – A36H2, weak) (Fig. 1a, Fig. 3a, Table S1). Accordingly, the distances derived from the A8H2 – A36H1′ and A8H2 – C9H1′ cross-peaks (3.6 Å in both cases) were similar to the equivalent distances in the base-paired stem of this construct (3.6 ± 0.1 Å and 3.4 ± 0.1 Å, respectively). The weak A8H2 – Ψ35H1′ cross-peak also was qualitatively similar to that in the A–U pairs of the stem (Fig. 3a). Consistent with the presence of an intervening A8 stacked between the G7 and C9 nucleotides, no cross-peaks were detected between the surrounding G7 and C9 nucleotides. Altogether, the NMR data supports a conclusion that a major conformation of the canonical branch site A8 is stacked on C9 within the duplex and base-paired with Ψ35 of the U2 snRNA strand.
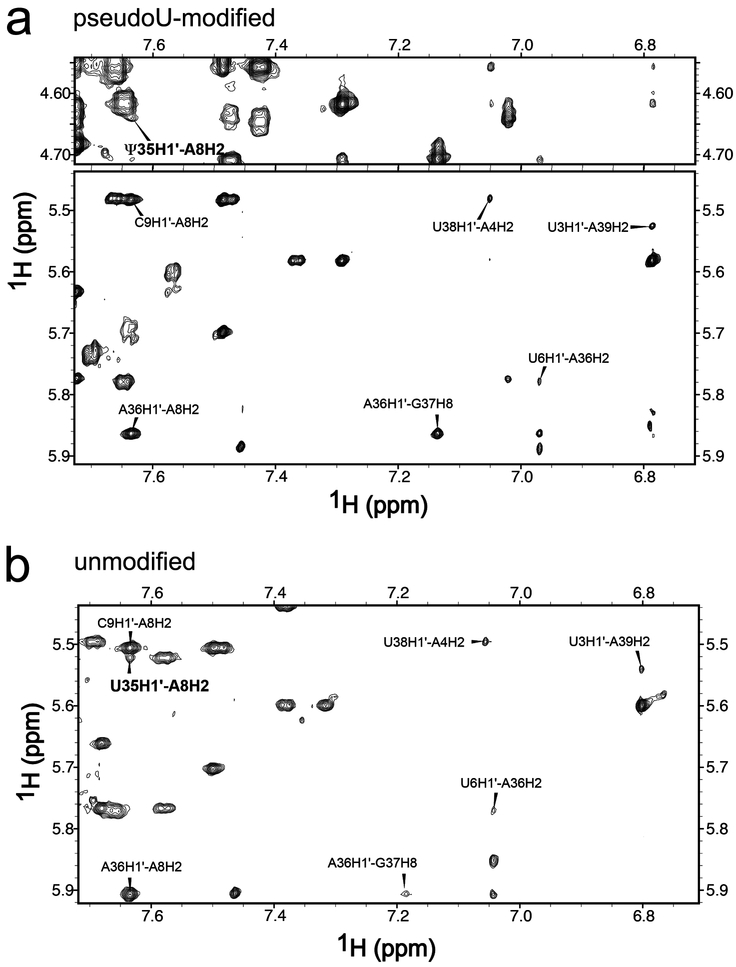
Fig. 3.
Two-dimensional NOESY spectra of (a) pseudouridine (Ψ)-containing and (b) unmodified duplexes in 100% D2O at 25°C with 400 msec mixing time show that the Ψ35H1′ – A8H2 cross-peaks (bold font) have similar intensities as equivalent NOEs in Watson-Crick A–U pairs. The A8H2 cross-peaks to H1′ of C9 and A36, and the cross-peak G37H8–A36H1′ are indicated.
3.3. An extrahelical syn-conformer of the guanosine precedes the consensus adenosine.
Several pieces of spectroscopic evidence indicated that the major conformation of G7 preceding the expected branch site is unpaired and positioned at least partially outside the duplex. First, numerous NOE cross-peaks between A8 and the U6–A36 base-pair (including A8H8 – U6H2′, A8H8 – U6H1′, A8H1′ – A36H2, A8H2 – A36H1′, and A8H2 – A36H2) (Fig. 1a, Table S1) supported the conclusion that the majority of G7 conformations are extrahelical (although the observed A8 → U6 cross-peaks were weaker than expected if A8 and U6 were always stacked and G7 completely extruded from the helix). Second, the G7 nucleotide lacked detectable NOESY interactions with nucleotides in the opposing U2 snRNA strand and exhibited properties of large imino proton line-width (> 100 Hz versus approximately 15 Hz for other imino protons) and water exchange typical of unpaired G nucleotides (Fig. 1a, Fig. S1). Third, the H8 proton of the guanine base, which is the base proton most proximal to the G7 ribose, exhibited NOE cross-peaks to neighboring nucleotides (U6H2′, medium and corresponding to ~3.4 Å contact; U6H6 and U6H1′, weak; A8H8, very weak). The low amplitudes of the G7H8 – U6H2’/H1’ and A8H8 – G7H2’/H1’ cross-peaks compared to C9H6 – A8H2’/H1’ and equivalent NOEs in the stem are consistent with a G7 conformation that is not as well stacked as A8, although an unusual C2´-endo population for the U6 and G7 sugars (>50%) could produce a similar effect. Finally, a syn conformation of G7 is supported by the intra-nucleotide NOESY interactions and integrated volume of the G7H1′ – G7H8 peak, which indicated a shorter distance than the customary anti orientation (Fig. 4a). Based on the respective H8 to H1′ distances of ~3.7 Å and ~2.6 Å in the anti and syn orientations, the G7H1′ – G7H8 NOE volume suggested that approximately 1/3rd of the G7 ensemble populates a syn-conformation about the glycosidic bond. This conclusion was further supported by the relatively downfield 13C chemical shift of G7’s C8 atom (140.3 ppm), which is at least 3 ppm from the other guanosine C8 shifts of 135.7 to 136.7 ppm (Fig. 4c) and characteristic of syn-guanosines [19–21].
Fig. 4.
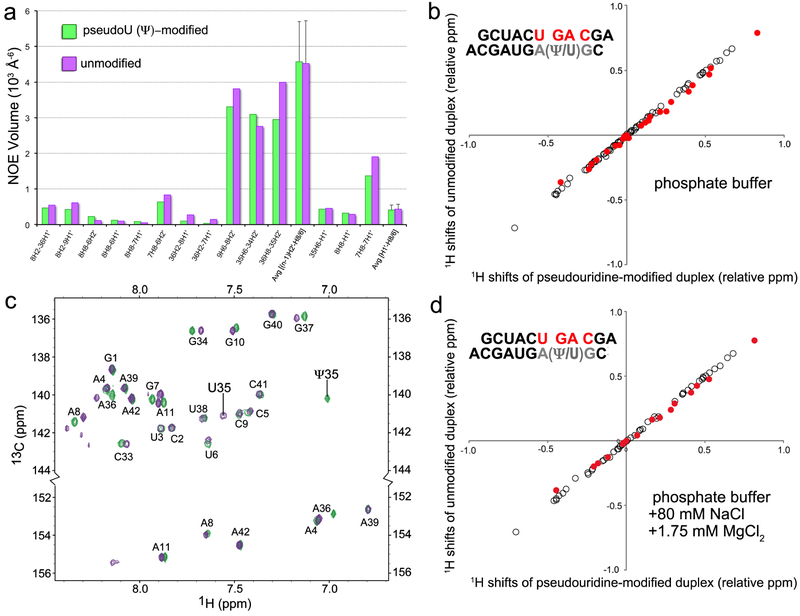
Comparison of pseudouridine-modified and unmodified U2 snRNA – BPS-containing duplexes. (a) NOESY cross-peak volumes from the branch site region of the pseudouridine-modified (green) and unmodified (purple) RNA duplexes. The average H1′–H6/H8 and H2′– (n+1)H8/6 NOE volumes of the base-paired stem regions (nucleotides 1–6, 9–10, 33–34, 36–41) are given for comparison with the branch site nucleotides. (b) Correlation of pseudouridine-modified versus unmodified proton shifts (branch site-region nucleotides U6-C9, filled red; other nucleotides 1–5, 10–11, 33, 37–42, unfilled). To expand the region of interest, the shifts of H6/H8/H2, H5/H1´, and H2´/H3´/H4´ protons are expressed relative to 7.5, 5.5, and 4.5 ppm, respectively. The assigned chemical shifts are listed in Table S2. (c) Overlay of natural abundance [13C,1H]-HSQC spectra of pseudouridine-modified (green) and unmodified (purple) duplexes including the C6/C8 (top) and adenine C2 regions (bottom). (d) Proton shift correlations in the presence of 80 mM NaCl and 1.75 mM MgCl2, plotted as in (b). Samples for (a) and (b) were in 100% D2O. Samples for (c) and (d) were in 95% H2O/5% D2O.
3.4. Subtle effects of the pseudouridine modification on branch site conformation.
To test the effect of the pseudouridine-modification on the branch site structure, we compared chemical shifts and NOESY interactions of the pseudouridine-modified RNA duplex with those of an unmodified counterpart (Fig. 3, Fig. 4, Tables S1–S2). The NOESY peak volumes of the modified and unmodified duplexes were similar, as shown in Fig. 4a (see also Table S1), which implies similar interatomic distances between the atoms involved. The evidence that the expected branch site A8 is primarily intrahelical and paired with Ψ35 (now U35), whereas the preceding G7 is primarily extrahelical, remains similar to the discussion above for the pseudouridine-containing duplex. One exception is the lack of a detectable A8H2 – U35H3 cross-peak; nonetheless, the U35H3 – water cross-peak was evident at the expected downfield position (Fig. S2). The volumes of the A8H2 – A36H1′, A8H2 – C9H1′, and A8H8 – U6H1′/H2′ cross-peaks were comparable between the modified and unmodified duplexes (Fig. 4a), which indicates that the pseudouridine (as compared to uridine) does not detectably influence the overall orientation of G7 and A8. The weak A8H2 – U35H1′ cross-peak is consistent with pairing of A8 and U35 (Fig. 3b). A relatively large G7H8 – H1′ cross-peak and downfield shift of G7C8 provided evidence that the population of the syn-G7 conformation remains similar in the unmodified duplex (Fig. 4a,c). Likewise, the chemical shift and width of the G7 imino resonance (Table S2, Fig. S3) again were consistent with solvent-exposure as opposed to base-pair formation. The NMR spectra also indicated that the dynamic properties of nucleotides U6, G7, and A8, including JH1’-H2’ ~ 4 Hz for residues U6 and G7, and broad resonances for G7H3´, G7H2´, and A8H2, remained nearly identical in the pseudouridine-modified and unmodified duplexes.
Nearly identical proton and carbon chemical shifts further supported the structural similarity of the pseudouridine-modified and unmodified duplexes (Fig. 4b–c, Table S2). Differences for peaks corresponding to position 35 of the U2 snRNA-containing strand (Ψ35 or U35) and its immediate neighbors were expected due to direct effects of the chemical modification. Otherwise, the proton shifts and overlaid CHSQC spectra of the pseudouridine-modified and unmodified duplexes were similar. The average non-exchangeable proton shift differences in the branch point and flanking residues (0.019 ± 0.018 ppm) were only slightly greater than the differences between the stems (0.006 ± 0.006 ppm), and have a maximum difference of 0.06 ppm, which is small compared to the ~1 ppm range of RNA protons (Table S2, Fig. 4b). Subtle differences in the 31P chemical shifts for both duplex strands may reflect altered backbone torsion angles or phosphate oxygen hydrogen bonding. Perhaps related to this observation, the G37H8 and A36H8 peaks of the unmodified duplex are broadened compared to pseudouridine-modified duplex, which can account for the apparent differences in the height of the G37H8-A36H1´ NOE (Fig. 3). Nevertheless, comparison of NOESY volumes and chemical shifts altogether indicate that the pseudouridine-modification has little effect on the overall U2 snRNA – BPS conformation.
3.5. Influence of ionic strength and magnesium on U2 snRNA/BPS conformation.
To test whether the branch point conformation is influenced by ionic conditions, NMR spectra were acquired following addition of NaCl and MgCl2 (Fig. 4d, Fig. S3–S5, Table S3–S4). The internuclear distances did not significantly change according to the NOE volumes (Fig. S5a), including the Ψ35H3 – A8H2 cross-peak of the A–Ψ pair (Fig. S4). The chemical shifts of a number of protons were influenced by ionic strength and magnesium, but these changes were observed in the stem as well as the U2 snRNA – BPS loop (Fig. S5). The shifts induced by MgCl2 were generally smaller and always in the same direction as the NaCl-induced shifts (Tables S3 and S4), suggesting that magnesium does not bind site-specifically. The chemical shifts of both pseudouridine-modified and unmodified duplexes were affected by buffer conditions in similar manners, resulting in high shift correlation between the two duplexes in both low salt and higher ionic strength buffers (Fig. 4b,d, Fig. S5b–c). Finally, the H1’-H2’ scalar coupling was unaffected for all nucleotides, including the ~4 Hz coupling observed for U6 and G7. Altogether, the U2 snRNA – BPS-containing duplex does not appear to interact with magnesium, and the ionic conditions tested here only subtly affected its structure and dynamic properties.
4. Discussion
Our NMR characterization of an isolated, “GA”-containing BPS – U2 snRNA duplex shows that a major conformation of expected branch site adenosine stacks within the duplex and pairs with Ψ35 of the U2 snRNA strand, whereas the preceding guanosine is primarily extrahelical. This base orientation and pairing appears independent of the Ψ35-modification in the U2 snRNA strand. The reason for the contrast between this “GA” branch point and an “AA” branch point sequence, for which pseudouridine modification is thought to promote an extrahelical consensus branch site, may simply arise from greater stability of an A–Ψ pair over a G–Ψ pair, much as an A–U pair is more stable than a G–U pair. Moreover, the syn-G7 conformation, which is more favorable for guanosine-5′-phosphate than for other nucleotides [22], may stabilize its extrahelical placement in the context of the “GA”-containing BPS sequence. Our results clarify that Ψ35 is not always the driving force for an extrahelical nucleotide, nor does it determine the exact position of the nucleotide “bulge” in this sequence context. Accordingly, the functional effects of Ψ35 for U2 snRNP assembly and splicing are subtle [23, 24]. Instead, other mechanisms such as protein chaperones are likely to select the branch site nucleophile in the context of the assembling spliceosome. Additionally, the post-transcriptional modifications of the U2 snRNAs in multicellular eukaryotes are extensive, including base methylations, 2′-O-methylations, and further pseudouridylations [25] that could act synergistically with Ψ35 to influence BPS choice. By analogy, a series of post-transcriptional modifications prepare the tRNALys3 anticodon structure for mRNA binding [26]. Future studies are needed to fully understand the interplay of pre-mRNA splicing factor contacts and snRNA post-transcriptional modifications for pre-mRNA branch site selection.
Supplementary Material
Highlights.
A syn-guanosine precedes the expected branch site of a splice site RNA duplex.
A conserved U2 snRNA pseudouridine lacks detectable effects on the RNA conformation.
The expected branch site adenosine pairs with pseudouridine within this RNA duplex.
Other factors/contexts may extrude the branch point adenosine for pre-mRNA splicing.
Acknowledgments
This work was supported by the National Institutes of Health grants R01GM117005 and R01GM070503. We thank Dr. Samuel E. Butcher for guidance with NMR sample preparation.
Abbreviations:
- BPS
branch point sequence
- DQF-COSY
double quantum filtered-correlated spectroscopy
- HETCOR
heteronuclear correlation
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- NOESY
nuclear overhauser effect spectroscopy
- TOCSY
Total Correlation Spectroscopy
- U2 snRNA
U2 small nuclear RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data related to this article can be found at (http-ref).
References
- [1].Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB, Alternative isoform regulation in human tissue transcriptomes, Nature, 456 (2008) 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jenkins JL, Kielkopf CL, Splicing factor mutations in myelodysplasias: Insights from spliceosome structures, Trends Genet, 33 (2017) 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ruskin B, Green MR, Role of the 3’ splice site consensus sequence in mammalian pre-mRNA splicing, Nature, 317 (1985) 732–734. [DOI] [PubMed] [Google Scholar]
- [4].Query CC, Moore MJ, Sharp PA, Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model, Genes Dev, 8 (1994) 587–597. [DOI] [PubMed] [Google Scholar]
- [5].Harris NL, Senapathy P, Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis, Nucleic Acids Res, 18 (1990) 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mercer TR, Clark MB, Andersen SB, Brunck ME, Haerty W, Crawford J, Taft RJ, Nielsen LK, Dinger ME, Mattick JS, Genome-wide discovery of human splicing branchpoints, Genome Res, 25 (2015) 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Padgett RA, Konarska MM, Aebi M, Hornig H, Weissmann C, Sharp PA, Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes, Proc Natl Acad Sci U S A, 82 (1985) 8349–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ruskin B, Greene JM, Green MR, Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants, Cell, 41 (1985) 833–844. [DOI] [PubMed] [Google Scholar]
- [9].Jacquier A, Rosbash M, RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point, Proc Natl Acad Sci U S A, 83 (1986) 5835–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Smith DJ, Konarska MM, Query CC, Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast, Mol Cell, 34 (2009) 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Newby MI, Greenbaum NL, Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine, Nat Struct Biol, 9 (2002) 958–965. [DOI] [PubMed] [Google Scholar]
- [12].Lin Y, Kielkopf CL, X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines, Biochemistry, 47 (2008) 5503–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A, NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J Biomol NMR, 6 (1995) 277–293. [DOI] [PubMed] [Google Scholar]
- [14].Bloomfield VA, Crothers DM, Tinoco I, Nucleic Acids: Structures, Properties and Functions, University Science Books, Sausalito, CA, 1999. [Google Scholar]
- [15].Goddard KDG T. D., SPARKY 3, University of California, San Francisco, (2004). [Google Scholar]
- [16].Xia T, SantaLucia J Jr., Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH, Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs, Biochemistry, 37 (1998) 14719–14735. [DOI] [PubMed] [Google Scholar]
- [17].Desaulniers JP, Chang YC, Aduri R, Abeysirigunawardena SC, SantaLucia J Jr., Chow CS, Pseudouridines in rRNA helix 69 play a role in loop stacking interactions, Org Biomol Chem, 6 (2008) 3892–3895. [DOI] [PubMed] [Google Scholar]
- [18].Schroeder KT, Skalicky JJ, Greenbaum NL, NMR spectroscopy of RNA duplexes containing pseudouridine in supercooled water, RNA, 11 (2005) 1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ghose R, Marino JP, Wiberg KB, Prestegard JH, Dependence of 13C chemical shifts on glycosidic torsional angles in ribonucleic acids, J Am Chem Soc, 116 (1994) 8827–8828. [Google Scholar]
- [20].Fares C, Amata I, Carlomagno T, 13C-detection in RNA bases: revealing structure-chemical shift relationships, J Am Chem Soc, 129 (2007) 15814–15823. [DOI] [PubMed] [Google Scholar]
- [21].Fonville JM, Swart M, Vokacova Z, Sychrovsky V, Sponer JE, Sponer J, Hilbers CW, Bickelhaupt FM, Wijmenga SS, Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment, Chemistry, 18 (2012) 12372–12387. [DOI] [PubMed] [Google Scholar]
- [22].Son TD, Guschlbauer W, Gueron M, Flexibility and conformations of guanosine monophosphates by the Overhauser effect, J Am Chem Soc, 94 (1972) 7903–7911. [DOI] [PubMed] [Google Scholar]
- [23].Yang C, McPheeters DS, Yu YT, Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae, J Biol Chem, 280 (2005) 6655–6662. [DOI] [PubMed] [Google Scholar]
- [24].Valadkhan S, Manley JL, Characterization of the catalytic activity of U2 and U6 snRNAs, RNA, 9 (2003) 892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gu J, Patton JR, Shimba S, Reddy R, Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions, RNA, 2 (1996) 909–918. [PMC free article] [PubMed] [Google Scholar]
- [26].Vendeix FA, Murphy FVT, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF, Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism, J Mol Biol, 416 (2012) 467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.