Abstract
BACKGROUND:
Although P1 and Xga are known to be associated with the A4GALT and XG genes respectively, the genetic basis of antigen expression has been elusive. Recent reports link both P1 and Xga expression with nucleotide changes in the promotor regions and with antigen negative phenotypes due to disruption of transcription factor binding.
STUDY DESIGN AND METHODS:
Whole genome sequencing was performed on 113 individuals as part of the MedSeq Project with serologic RBC antigen typing for P1 (n=77) and Xga (n=15). Genomic data were analyzed by two approaches, nucleotide frequency correlation or serologic correlation, to find A4GALT and XG changes associated with P1 and Xga expression.
RESULTS:
For P1, the frequency approach identified 29 possible associated nucleotide changes, and the serologic approach revealed four among them correlating with the P1+/P1− phenotype: chr22:43,115,523_43,115,520AAAG/delAAAG (rs66781836); chr 22:43,114,551C/T (rs8138197); chr22:43,114,020T/G (rs2143918); and chr22:43,113,793G/T (rs5751348). For Xga, the frequency approach identified 82 possible associated nucleotide changes, and among these the serologic approach revealed one correlating with the Xg(a+)/Xg(a–) phenotype: chrX:2,666,384G/C (rs311103).
CONCLUSION:
A bioinformatics analysis pipeline was created to identify genetic changes responsible for RBC antigen expression. This study, in progress prior to the recently published reports, independently confirms the basis for P1 and Xga. Although, this enabled molecular typing of these antigens, the Y chromosome PAR1 region interfered with Xga typing in males. This approach could be used to identify and confirm the genetic basis of antigens, potentially replacing the historical approach using family pedigrees as genomic sequencing becomes common place.
Keywords: red blood cell antigen, blood group, P1, RUNX1, Xga, GATA-1, genomics, whole genome sequencing, WGS, next generation sequencing, NGS, personalized medicine, transfusion medicine
There are over 350 RBC antigens divided into 36 blood group systems. The molecular mechanisms of antigen expression are understood for the vast majority, with >2,000 alleles carrying nucleotide (nt) changes (allelic variations) defined in 45 genes. Despite the advancement in the genotype to phenotype relationship of blood group antigens, the genetic basis of several remain elusive. However, this gap is quickly closing as bioinformatics technologies are applied. For example, recent work has provided evidence for the role of the transcription factors RUNX1 in the expression of P1 antigen1 and of GATA-1 in the expression of Xga.2,3
The P1 antigen was described in 1927 by injecting rabbits with human RBCs and screening for antibodies by testing the sera against human RBCs from different individuals.4 There are ethnic differences in frequency of P1 antigen expression on RBCs of individuals of European Ancestry (EA) and African Ancestry (AA) with RBCs from 79% EA versus 94% AA typing as P1+ (P1 phenotype) and 21% of EA vs 6% of AA typing as P1− (P2 phenotype). In 2001, it was shown that A4GALT encodes a 4-α-galactosyltransferase enzyme which adds α-galactose to paragloboside to create the P1 antigen5. In 2011 and 2014, it was reported that the single nucleotide polymorphisms (SNPs) designated rs81381976, rs21439187, and rs57513487 correlated with P1+/P1− expression, but the mechanism remained unknown. There was uncertainty as to the actual SNP responsible, but recently Westman et al. showed that SNP rs5751348 (nt G>T) is located in a RUNX1 transcription factor binding region which controls the expression of P1 with chromosomal location chr22:43,113,793G associated with the P1+ phenotype and chr22:43,113,793T with P1–.1
The Xga antigen was first described in 1962 in a multiply transfused male of European ancestry whose serum reacted at different frequencies to RBCs from males and females, indicating that the expression of Xga was X-linked, with a gender biased distribution: Xg(a+) 66% males / 89% females, and Xg(a–) 34% males / 11% females.8 In 1994, it was shown that Xga was expressed on the protein product of PBDX (renamed XG) in a manner that suggested that antigen expression was controlled by the presence or absence of Xg protein9, but the mechanism remained unknown. Recently, Moller et al. and Yeh et al. showed that the SNP rs311103 (nt G>C) is located in a GATA-1 transcription factor binding region in intron 1 of XG, and controls the expression of the protein and the Xga antigen with genomic coordinate chrX:26,66,384G associated with Xg(a+) and chrX:26,66,384C associated with a Xg(a–) phenotype.2,3
We recently created and validated an automated blood group antigen typing software (bloodTyper) for translating whole genome sequencing (WGS) data to predict RBC and platelet antigen phenotypes and validated the performance of whole genomes with conventional serologic and SNP typing for the common antigens.10–12 As part of those analyses, we also performed P1 and Xga serologic typing for a subset of samples with the goal of identifying the basis of P1 and Xga expression by correlating the serologic typing with whole genome data. Here we present a bioinformatics analysis of P1 and Xga expression. We also describe updates to our automated typing algorithm to type for the P1 and Xga antigens, including limitations which make it only possible to molecularly type Xga from females only.
Material and Methods
Serologic Typing
With approval from the Partners HealthCare Human Research Committee (IRB) and informed consent from participants, blood samples for RBC isolation were collected in EDTA and RBC serologic antigen typing was performed according to standard tube methods,13 and as previously described.10–12 Commercially available serologic typing reagents were used to type for P1 (Bio Rad, Hercules, Calif) and human source anti-Xga was used to type for Xga.
Whole Genome Sequencing
With approval from the Partners HealthCare Human Research Committee (IRB) and informed consent from participants, blood samples for DNA isolation were collected in PAXgene tubes (PreAnalytiX GmbH, Feldbachstrasse, Switzerland) and genomic DNA was isolated from WBCs by standard methods. For quality control, a genotyping array was performed in parallel to confirm identity and lack of sample inversion during WGS workflow. Another blood sample was also genotyped to serve as an independent verification of identity.
PCR free WGS was performed by the Clinical Laboratory Improvement Amendments (CLIA)-certified, College of America Pathologists (CAP)-accredited Illumina Clinical Services Laboratory (San Diego, CA) using paired-end 100 base pair (bp) reads of DNA fragments with an average length of 300 bp on the Illumina HiSeq platform and sequenced to at least 30× average depth of coverage.14 Sequence read data was aligned to the human reference sequence (GRCh37/hg19) using Burrows-Wheeler Aligner 0.6.1-r104.15 Data from sequencing was analyzed, interpreted, reported and returned to participants as part of the MedSeq Project, a randomized clinical trial of whole genome sequencing in primary care practice.16–19
Whole Genome Based Blood Typing
P1 and Xga antigen nucleotide changes were added to our allele database (http://bloodantigens.com) and the custom typing software (bloodTyper)12 was used to predict P1 and Xga phenotypes from the whole genomes analyzed as part of the MedSeq Project. In brief, variant calls for XG and A4GALT genes and promoter regions were made using Genomic Analysis Tool Kit (GATK) version 2.3–9-gdcdccbb and saved as a variant calling format file (.vcf) showing differences between the WGS data and the reference genome.20 Sequencing coverage was extracted from the alignment file using BEDTools v2.17.0.21 The Integrative Genomics Viewer (IGV)22 was used as needed to verify coverage and sequence identity. Antigen typing using bloodTyper was performed at the relevant genome positions using a 4× sequence read depth of coverage calling cutoff.
Software and Data Availability
The MedSeq Project genomes are available through dbGaP under study accession phs000958. The curated RBC antigen allele database used in this study is available at http://bloodantigens.com. The code used to search the variant calling files for the antigen associated changes using both the frequency and serology approaches is available at http://lanelab.org/data.
Results
Genetic Search Approaches
The genetic basis for P1 and Xga expression were identified using 113 whole genomes from the MedSeq Project with paired serologic typing (Figure 1). Two different, but complementary approaches (frequency and serology) were used to search the the A4GALT and XG genes, including coding exons, introns, and upstream promoter regions for single nucleotide changes and small insertion/deletions which correlated with antigen expression.
Figure 1.
WGS Based approach for determination of molecular basis for P1 and for Xga.
In the frequency approach, heterozygous and homozygous nucleotide variations over the A4GALT and XG gene and promoter regions were identified in the genomes. Each change was then individually evaluated by simulating the sample antigen type using the following rules: antigen-negative if homozygous for the change or antigen-positive if heterozygous for this change or homozygous for another change. The resulting antigen-positive and antigen-negative population frequency was then calculated for each change identified. The nucleotide changes were filtered to include only those in the range of the known antigen frequency.
In the serologic approach, serologic typing results were correlated with nt changes over the A4GALT and XG gene and promoter regions, independent of the frequency approach described above. By assuming that antigen-negative individuals resulted from the same recessive homozygous change, a starting list of possible nt changes was created by identifying common homozygous changes. These were then filtered by removing any homozygous changes also present in antigen-positive individuals. To account for the possibility of a serologic typing error or other sample specific issues, the searches were performed multiple times with each sample excluded from the analysis. Therefore, if a serologic typing was incorrect there would be a search in which this erroneous information would not adversely affect the results.
Identification of the basis of P1 from Whole Genomes
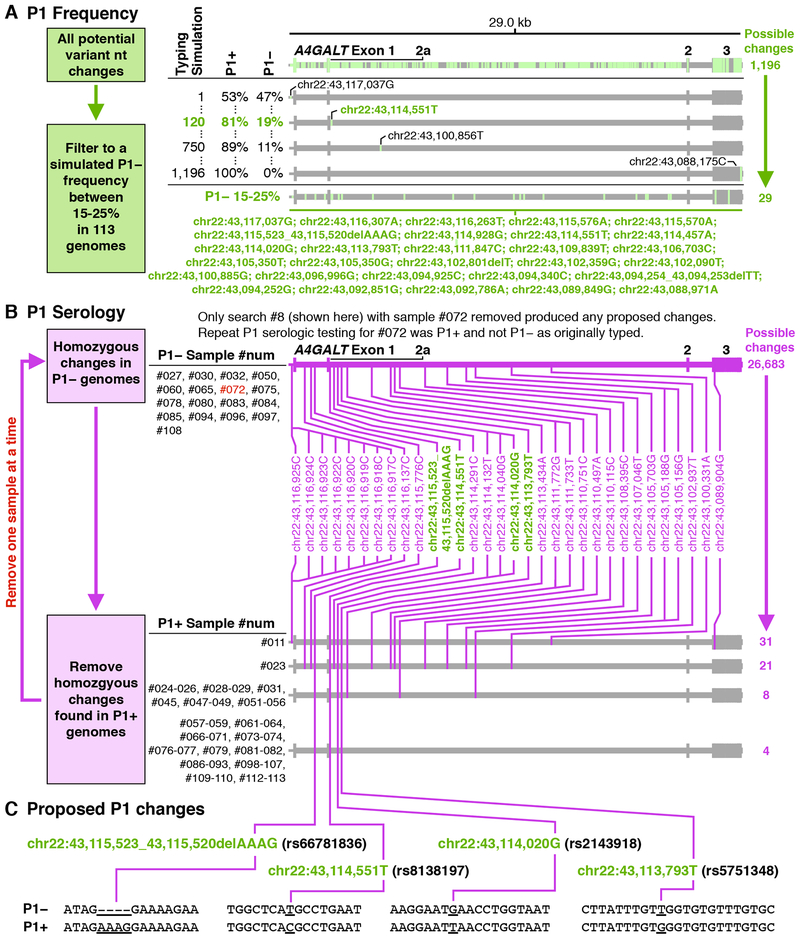
Using a frequency approach, the genetic basis of P1 expression was searched in 113 whole genomes over a 29 kb region (chr22:43,088,126 – chr22:43,117,175) including the A4GALT promoter and gene. The possible P1 associated changes were filtered to include only those with a frequency between 15–25%, close to the known P1− antigen frequency of 21% in individuals of European Ancestry as the recorded ancestry for most MedSeq samples. This reduced the number of possible responsible nucleotide positions from 1,196 to only 29 nucleotide changes (Figure 2A).
Figure 2.
Identification of the P1 Genetic Basis.
Two approaches used to identify A4GALT changes associated with P1 expression. (A) The frequency approach used whole genome sequences of the A4GALT region to correlate nt changes with expected P1 antigen frequency. The percentages represent the antigen phenotype frequencies for each nt change when used to simulate typing for the antigen from whole genomes. (B) The serology approach used whole genome sequences of the A4GALT region to correlate nt changes with serologic typing. Changes also found in the frequency approach shown in A are colored green. (C) Potential nt changes responsible for P1 expression designated by chromosomal coordinates and by rs database numbering. Note: A4GALT is transcribed in reverse to its direction in the reference genome, as such larger genomic positions correspond to smaller gene positions. The nt bases have been reverse complimented from the human reference genome.
Using a serologic approach, P1 expression was correlated between genomic nt changes over the same region as above with the RBC phenotypes of 77 individuals (17 P1− and 60 P1+). Analysis of homozygous nt changes common to P1− individuals identified 26,683 possible nt variants, which were then filtered to remove homozygous changes also found in P1+ individuals (Figure 2B). This initial analysis did not result in any correlated nt changes. However, this could have occurred due to a P1 serologic phenotype error in one or more samples, which could have caused the actual P1 associated nt changes to be incorrectly filtered. As mentioned above, to account for the possibility of serologic testing or transcription error, multiple searches where performed in which each sample was excluded from the analysis. When P1− participant #072 was excluded from the analysis in this manner, four potential P1 associated changes were identified (Figure 2C): chr22:43,115,523_43,115,520AAAG/delAAAG (rs66781836); chr 22:43,114,551C/T (rs8138197); chr22:43,114,020T/G (rs2143918); and chr22:43,113,793G/T (rs5751348). All four of these sites were also identified in the frequency approach above. A follow-up sample from participant #072 was obtained and the RBCs typed as P1+, confirming that the initial P1− serologic data for that individual were incorrect.
Identification of the basis of Xga in Whole Genomes
Because males have only one X chromosome, sequence analysis to identify hemizygous nt changes could theoretically lead to quicker identification of the associated Xga genetic change than analysis of female genomes. Unfortunately, the XG promoter and part of the XG gene are located in the homologous PAR1 region shared by both the X and Y chromosomes.23 This causes male Y chromosome sequences to misalign to the X chromosome, and thus male samples contained regions of misplaced reads which interfered with analysis. In particular, some nt positions incorrectly appeared heterozygous where misplaced Y sequences contain SNPs that differ from the X chromosome. Therefore, only males presenting with homozygous nt positions were considered.
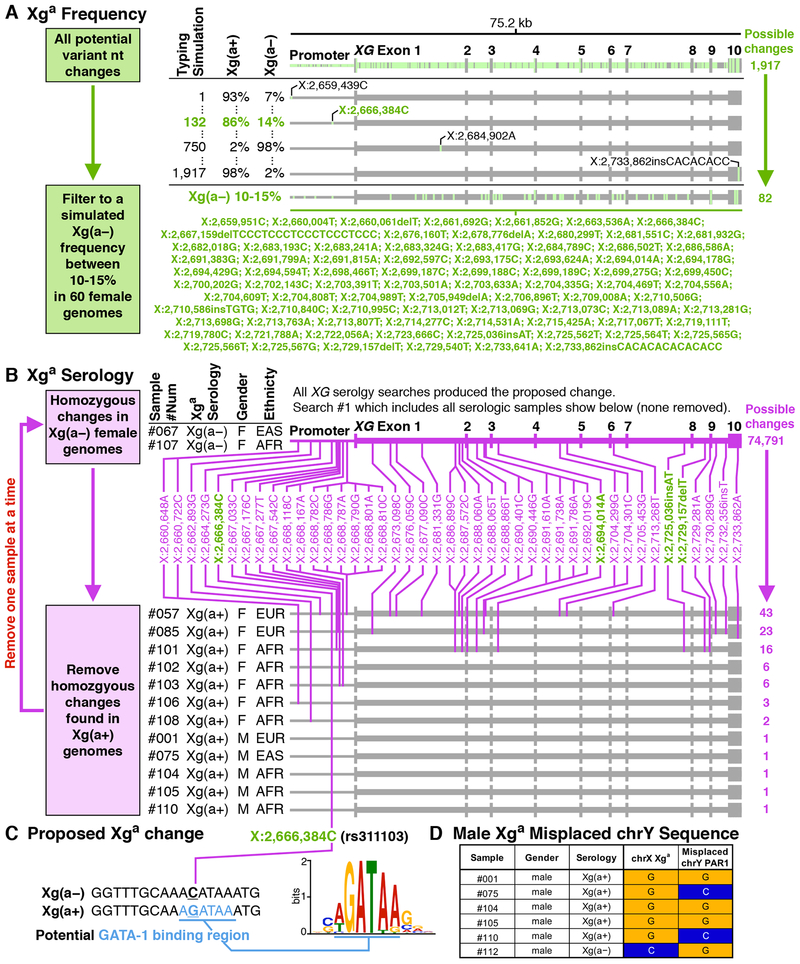
Using the frequency approach, the genetic basis of Xga expression was examined in 60 female genomes over a 75.2 kb region including the XG gene and promotor region (chrX:2,659,351 – chrX:2,734,541). The potential Xga nt associations were filtered to include those that would result in a frequency close to the known 11% female Xg(a–) phenotype frequency, which reduced the number of possible nt variants from 1,917 to only 82 changes (Figure 3A).
Figure 3.
Identification of the Xga Genetic Basis.
Two approaches used to identify XG changes associated with Xga expression. (A) The frequency approach used whole genome sequences of the XG region to correlate nt changes with expected Xga antigen frequency. The percentages represent the antigen phenotype frequencies for each nt change when used to simulate typing for the antigen from whole genomes. (B) The serology approach used whole genome sequences of the XG region to correlate nt changes with serologic typing. Changes also found in the frequency approach shown in A are colored green. (C) Potential nt change responsible for Xga expression. (D) Xga nt changes in males and the corresponding misplaced chromosome Y PAR1 nt changes.
Using the serologic approach, the genetic basis for Xga expression was correlated between nt changes over the same region indicated above with the RBC phenotypes of 15 individuals, two Xg(a–) females and 12 Xg(a+) males and females were included in the analysis. One Xg(a–) male was excluded as uninformative for the reasons described above. Analysis of homozygous nt changes common to Xg(a–) individuals identified 74,791 possibilities (Figure 3B), which were filtered to remove homozygous changes present in Xg(a+) individuals. This identified just one potential variation associated with Xg(a–) phenotype, chrX:2,666,384C (rs311103), located in a GATA-1 binding region (Figure 3C). This nt change was also identified in the frequency approach.
To further investigate and differentiate true X chromosome nt variants and misplaced Y chromosome nt variation, the serologic typing of male samples was correlated with the DNA sequence aligned to position chrX:2,666,384 (Figure 3D). The analysis shows that the Y chromosome PAR1 region sequences indeed misaligned to the X chromosome Xga associated region and can differ in sequence (Figure 3D, #075,110,112). This confounds molecular typing in male samples that appear to be heterozygous. Hence, molecular typing of Xga in males is not readily possible unless they are homozygous at this position.
bloodTyper
We updated our curated antigen allele database (http://bloodantigens.com) with the newly confirmed Xga and P1 alleles, and verified that bloodTyper could correctly type P1 and Xga from the whole genomes analyzed within the MedSeq Project with paired serology. As discussed above, bloodTyper restricts Xga typing to females.
Discussion
We have demonstrated an extensible and reusable framework for antigen allele discovery and verification from whole genomes. As a proof of principle, we used whole genome sequencing data and limited paired serologic typing to identify nt changes correlated with P1 and Xga expression. This analysis was done prior to the recent publications detailing the role of RUNX11 GATA-12,3 and transcription factor binding in expression of these antigens, and is independent confirmation of those reports. Importantly, since the entire genomic sequence was available for analysis, we could rule out the possibility of any other nt changes controlling P1 and Xga expression in the A4GALT and XG genes and upstream promoter regions. As the genetic changes associated with P1 and Xga antigen expression are located outside of the coding exons, a whole genome-based approach was particularly important in determining their identity. By automating the search analysis, we could quickly perform multiple iterative searches with each sample excluded from the analysis to successfully account for any sample discrepancy. We have now updated our curated antigen allele database and companion typing software to provide whole genome-based typing for Xga and P1.
Although the combined frequency and serologic approach for P1 expression identified four nt changes, this could simply be due to linkage disequilibrium which, in theory, could be addressed with a larger sample size. Indeed, a recent publication identified these same four nt changes associated with P1, but one (rs66781836) was later excluded in an uncommon sample with haplotype recombination between the possible nt changes.7 To optimize exclusion among strongly linked SNPs, mining of large scale genomic data sources for examples where one or more of the linked nt changes occur independently and performing targeted follow-up serologic typing could be employed. Importantly, the potential P1 associated nts found here include rs5751348 which has recently been reported to contain a RUNX1 transcription factor binding region controlling P1 expression,1 along with three other nt changes located outside of the RUNX1 binding region, but in linkage disequilibrium with it.
The combined frequency and serologic approach was successful in determining the molecular basis of Xga expression as it identified just one potential nt change, rs311103. This is the same change recently reported to contain a GATA-1 transcription factor binding region controlling Xga expression.2,3 We determined that genotyping of Xga from males will prove difficult due to the homologous PAR1 region shared by chromosomes X and Y. Determination of Xga in males by genomic methods will require long read sequencing or long range PCR of a 33.2 kb region spanning the Xga SNP position (chrX:2,666,384) and chromosome X positions located outside of the PAR1 region (around chrX:2,699,625).
Historically, identifying the molecular basis of new blood groups was accomplished using genetic segregation of nt changes with antigen phenotype in related individuals. In this study, we present a new paradigm using whole genomes with paired, and often limited, serologic typing data as illustrated by identification the basis of Xga with less than 14 typed individuals combined with sequence data of 77 unrelated individuals. Although, the frequency approach was only able to filter to 29 A4GALT and 82 XG nt change, even this level of enrichment could be of value. For example, if RBCs had not been available for serologic typing, the nt changes identified by the frequency approach could have been used to design inexpensive targeted SNP assays and tested on samples with paired RBC serologic testing. This type of analysis will accelerate as large-scale genomic sequencing becomes common place, by allowing sequence data analysis from tens of thousands of individuals at present, and millions within the next decade.
As new antigens are described, it is critical to have adequate evidence for allele-antigen associations, but this is challenging since expression systems are not readily available for many blood groups. In addition, bioinformatic analysis can be used to determine if potential molecular changes are located in putative transcription factor binding regions or other regulatory regions along with evaluation of expression using predictive models or large-scale experimental protein expression datasets. This would provide a potential robust hypothesis for association of the SNP with the phenotype, however direct experimental confirmation should be done when possible. As such, large-scale sequencing data sets with paired serologic typing could provide a powerful tool to rapidly identify the effect of novel allelic changes on serologic phenotype.
Since it is already common for large biobanks to archive DNA and serum, they should partner with blood bank and donor centers to freeze RBCs for future antigen-allele association discovery and confirmation. For example, the Xga analysis performed here was only possible because a subset of genomic sequencing participants had RBC samples frozen for future serologic typing. If frozen RBCs for serologic typing were available from a sufficiently large genomic dataset it might be possible to find the molecular basis of a low or high frequency antigen if even just one high frequency antigen negative or low frequency antigen positive individual could be identified.
In summary, we have developed an analysis pipeline to identify the nt changes responsible for RBC antigen expression from whole genomes with paired serologic typing. This approach independently confirms the recently reported nt changes responsible for P1 and Xga antigen expression. Additionally, this approach should allow for rapid and comprehensive antigen discovery as large-scale genomic sequencing data sets are created over the coming years.
Acknowledgements
The MedSeq Project was supported by the National Human Genome Research Institute U01-HG006500. RCG is supported by grant funding from NIH, the Broad Institute and the Department of Defense. RCG receives compensation for advising the following companies: AIA, Helix, Ohana, OptraHealth, Prudential and Veritas; and is co-founder of Genome Medical, Inc, a nationwide telemedicine service providing expert advice in genetics. WJL was additionally supported by the Brigham and Women’s Hospital Pathology Department Stanley L. Robbins M.D. Memorial Research Fund Award. The authors thank the staff and participants of the MedSeq Project, as well as the staff of the Brigham and Women’s Blood Bank.
Footnotes
Members of the MedSeq Project (including authors listed above):
Members of the MedSeq Project are as follows: David W. Bates, MD, Carrie Blout, MS, CGC, Kurt D. Christensen, PhD, Allison L. Cirino, MS, Robert C. Green, MD, MPH, Carolyn Y. Ho, MD, Joel B. Krier, MD, William J. Lane, MD, PhD, Lisa S. Lehmann, MD, PhD, MSc, Calum A. MacRae, MD, PhD, Cynthia C. Morton, PhD, Denise L. Perry, MS, Christine E. Seidman, MD, Shamil R. Sunyaev, PhD, Jason L. Vassy, MD, MPH, SM, Erica Schonman, MPH, Tiffany Nguyen, Eleanor Steffens, Wendi Nicole Betting, Brigham and Women’s Hospital and Harvard Medical School; Samuel J. Aronson, ALM, MA, Ozge Ceyhan-Birsoy, PhD, Matthew S. Lebo, PhD, Kalotina Machini, PhD, MS, Heather M. McLaughlin, PhD, Danielle R. Azzariti, MS, Heidi L. Rehm, PhD, Ellen A. Tsai, PhD, Partners Healthcare Personalized Medicine; Jennifer Blumenthal-Barby, PhD, Lindsay Z. Feuerman, MPH, Amy L. McGuire, JD, PhD, Kaitlyn Lee, Jill O. Robinson, MA, Melody J. Slashinski, MPH, PhD, Baylor College of Medicine, Center for Medical Ethics and Health Policy; Pamela M. Diamond, PhD, University of Texas Houston School of Public Health; Kelly Davis, Peter A. Ubel, MD, Duke University; Peter Kraft, PhD, Harvard School of Public Health; J. Scott Roberts, PhD, University of Michigan; Judy E. Garber, MD, MPH, Dana-Farber Cancer Institute; Tina Hambuch, PhD, Illumina, Inc.; Michael F. Murray, MD, Geisinger Health System; Isaac Kohane, MD, PhD, Sek Won Kong, MD, Boston Children’s Hospital.
Declaration of Interests
All authors declare that they have no conflicts of interest relevant to the manuscript.
References
- 1.Westman JS, Stenfelt L, Vidovic K, Moller M, Hellberg A, Kjellstrom S, Olsson ML. Allele-selective RUNX1 binding regulates P1 blood group status by transcriptional control of A4GALT. Blood 2018;131: 1611–6. [DOI] [PubMed] [Google Scholar]
- 2.Moller M, Lee YQ, Vidovic K, Kjellstrom S, Bjorkman L, Storry JR, Olsson ML. Disruption of a GATA1-binding motif upstream of XG/PBDX abolishes Xg(a) expression and resolves the Xg blood group system. Blood 2018;132: 334–8. [DOI] [PubMed] [Google Scholar]
- 3.Yeh CC, Chang CJ, Twu YC, Chu CC, Liu BS, Huang JT, Hung ST, Chan YS, Tsai YJ, Lin SW, Lin M, Yu LC. The molecular genetic background leading to the formation of the human erythroid-specific Xg(a)/CD99 blood groups. Blood Adv 2018;2: 1854–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landsteiner K, Levine P. Further Observations on Individual Differences of Human Blood. Experimental Biology and Medicine 1927;24: 941–2. [Google Scholar]
- 5.Steffensen R, Carlier K, Wiels J, Levery SB, Stroud M, Cedergren B, Nilsson Sojka B, Bennett EP, Jersild C, Clausen H. Cloning and expression of the histo-blood group Pk UDP-galactose: Ga1beta-4G1cbeta1-cer alpha1, 4-galactosyltransferase. Molecular genetic basis of the p phenotype. J Biol Chem 2000;275: 16723–9. [DOI] [PubMed] [Google Scholar]
- 6.Thuresson B, Westman JS, Olsson ML. Identification of a novel A4GALT exon reveals the genetic basis of the P1/P2 histo-blood groups. Blood 2011;117: 678–87. [DOI] [PubMed] [Google Scholar]
- 7.Lai YJ, Wu WY, Yang CM, Yang LR, Chu CC, Chan YS, Lin M, Yu LC. A systematic study of single-nucleotide polymorphisms in the A4GALT gene suggests a molecular genetic basis for the P1/P2 blood groups. Transfusion 2014;54: 3222–31. [DOI] [PubMed] [Google Scholar]
- 8.Mann JD, Cahan A, Gelb AG, Fisher N, Hamper J, Tippett P, Sanger R, Race RR. A sex-linked blood group. Lancet 1962;1: 8–10. [DOI] [PubMed] [Google Scholar]
- 9.Ellis NA, Tippett P, Petty A, Reid M, Weller PA, Ye TZ, German J, Goodfellow PN, Thomas S, Banting G. PBDX is the XG blood group gene. Nat Genet 1994;8: 285–90. [DOI] [PubMed] [Google Scholar]
- 10.Lane WJ, Westhoff CM, Uy JM, Aguad M, Smeland-Wagman R, Kaufman RM, Rehm HL, Green RC, Silberstein LE, MedSeq P. Comprehensive red blood cell and platelet antigen prediction from whole genome sequencing: proof of principle. Transfusion 2016;56: 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baronas J, Westhoff CM, Vege S, Mah H, Aguad M, Smeland-Wagman R, Kaufman RM, Rehm HL, Silberstein LE, Green RC, Lane WJ. RHD Zygosity Determination from Whole Genome Sequencing Data. Blood Disorders & Transfusion 2016;7. [Google Scholar]
- 12.Lane WJ, Westhoff CM, Gleadall NS, Aguad M, Smeland-Wagman R, Vege S, Simmons DP, Mah HH, Lebo MS, Walter K, Soranzo N, Di Angelantonio E, Danesh J, Roberts DJ, Watkins NA, Ouwehand WH, Butterworth AS, Kaufman RM, Rehm HL, Silberstein LE, Green RC, MedSeq P. Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study. Lancet Haematol 2018;5: e241–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung MK, Eder AF, Spitalnik SL, Westhoff CM, editors. Technical Manual, 19th edition. 19th ed.: American Association of Blood Banks (AABB), 2017. [Google Scholar]
- 14.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008;456: 53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26: 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassy JL, Lautenbach DM, McLaughlin HM, Kong SW, Christensen KD, Krier J, Kohane IS, Feuerman LZ, Blumenthal-Barby J, Roberts JS, Lehmann LS, Ho CY, Ubel PA, MacRae CA, Seidman CE, Murray MF, McGuire AL, Rehm HL, Green RC, MedSeq P. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials 2014;15: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaughlin HM, Ceyhan-Birsoy O, Christensen KD, Kohane IS, Krier J, Lane WJ, Lautenbach D, Lebo MS, Machini K, MacRae CA, Azzariti DR, Murray MF, Seidman CE, Vassy JL, Green RC, Rehm HL, MedSeq P. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet 2014;15: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassy JL, Christensen KD, Schonman EF, Blout CL, Robinson JO, Krier JB, Diamond PM, Lebo M, Machini K, Azzariti DR, Dukhovny D, Bates DW, MacRae CA, Murray MF, Rehm HL, McGuire AL, Green RC, MedSeq P. The Impact of Whole-Genome Sequencing on the Primary Care and Outcomes of Healthy Adult Patients: A Pilot Randomized Trial. Ann Intern Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen KD, Vassy JL, Phillips KA, Blout CL, Azzariti DR, Lu CY, Robinson JO, Lee K, Douglas MP, Yeh JM, Machini K, Stout NK, Rehm HL, McGuire AL, Green RC, Dukhovny D. Short-term costs of integrating whole-genome sequencing into primary care and cardiology settings: a pilot randomized trial. Genet Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20: 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26: 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14: 178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Mogharbel N, Graves JAM. X and Y Chromosomes: Homologous Regions 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MedSeq Project genomes are available through dbGaP under study accession phs000958. The curated RBC antigen allele database used in this study is available at http://bloodantigens.com. The code used to search the variant calling files for the antigen associated changes using both the frequency and serology approaches is available at http://lanelab.org/data.