Abstract
The structural features that govern broad-spectrum activity of broadly neutralizing, anti-ebolavirus antibodies (Abs) outside of the internal fusion loop epitope are currently unknown. Here we describe the structure of a broadly neutralizing human monoclonal Ab (mAb), ADI-15946, which was identified in a human survivor of the 2013–2016 outbreak. The crystal structure of ADI-15946 in complex with cleaved Ebola virus glycoprotein (EBOV GPCL) reveals that binding of the mAb structurally mimics the conserved interaction between the EBOV GP core and its glycan cap β17-β18 loop to inhibit infection. Both endosomal proteolysis of EBOV GP and binding of mAb FVM09 displace this loop, thereby increasing exposure of ADI-15946’s conserved epitope and enhancing neutralization. Our work also mapped the paratope of ADI-15946 thereby explaining reduced activity against Sudan virus (SUDV), which enabled rational, structure-guided engineering to enhance binding and neutralization against SUDV while retaining the parental activity against EBOV and Bundibugyo virus (BDBV).
Introduction:
EBOV and related members of the family Filoviridae cause outbreaks of highly lethal disease in humans. Ab therapeutics such as ZMapp1, mAb1142, and a three-mAb cocktail from Regeneron Pharmaceuticals3 have recently been proposed for emergency use against Ebola virus disease4. However, the activity of these therapies is limited to EBOV and does not extend protection to the related virulent ebolaviruses BDBV and SUDV. Both BDBV and SUDV have caused sizeable outbreaks in the past and their potential for re-emergence remains unknown. No therapeutics are currently available for the treatment of BDBV and SUDV which lends urgency to discovery and characterization of broadly active mAbs. The key obstacle for generation of such mAbs stems from limited amino acid sequence conservation among the glycoproteins of ebolaviruses, with only 50% amino acid identity shared between EBOV and SUDV, the two most prevalent ebolaviruses. Detailed characterization of known broadly neutralizing antibodies (bNAbs) and their modes of action will be critical to the design of next-generation broadly protective Ab cocktails and vaccines that elicit broadly protective responses.
All Ab therapeutics currently under development for Ebola virus disease target the ebolavirus surface glycoprotein, GP, which mediates viral entry into host cells by catalyzing viral membrane fusion in host cell endosomes1,2,5. During biogenesis, GP is post-translationally processed to yield GP1 and GP2 subunits (Fig. 1a), held together by a single disulfide bond, which associate into a trimer of GP1,2 heterodimers6,7. GP1 mediates host cell attachment and receptor recognition, whereas GP2 mediates fusion of the viral and host membranes8-12. The GP2 amino acid sequence, which includes the internal fusion loop (IFL), is highly conserved among ebolaviruses6 while the glycan cap and mucin like domains of GP1 show higher sequence diversity. During infection, EBOV GP undergoes host-programmed disassembly mediated by endosomal cysteine proteases (cathepsins B and L), which shed the steric bulk of the extensively glycosylated glycan cap and mucin-like domains and generate the cleaved GP intermediate (GPCL)13,14. This in turn unmasks the receptor-binding site (RBS) within GP1 and reveals other previously inaccessible regions of the GP core12,15,16. Engagement of the intracellular entry receptor Niemann-Pick C1 (NPC1) by the viral RBS is proposed to induce conformational rearrangements in GPCL that culminate in viral membrane fusion12,16-19. Antibodies targeting the functionally critical and conserved viral fusion machinery offer an efficacious mode of viral entry inhibition and have been shown to be strongly protective20-24.
Figure 1.
Structure of ADI-15946 in complex with Ebola virus GPCL. (a) Domain architecture of EBOV GP full length and the construct crystallized here, which is the ectodomain of an enzymatically cleaved GP (GPCL) resembling the form of GP generated in endosomes during viral entry lacking the glycan cap. IFL, internal fusion loop; HR1 and HR2, heptad repeat 1 and heptad repeat 2, respectively. Glycosylation sites are represented above the domains. (b) Crystal structure of the trimeric EBOV GPCL–ADI-15946 complex. The inset table shows the contribution of each CDR to the buried surface area (BSA) on the surface of GPCL. (c) The interaction bridges the fusion loop and other portions of GP2 primarily via light chain and CDR H3 contacts. CDRs H1 and H2 are not involved. (d) The footprint of ADI-15946 (orange) is shifted up and to the left compared to that of the base binding mAb, KZ52 (pink).
The only pan-ebolavirus neutralizing Abs reported thus far target overlapping epitopes in the viral internal fusion loop21,23-26. In contrast, low-resolution negative stain reconstructions of the recently isolated human survivor mAb, ADI-15946, suggest that it may recognize a distinct footprint in the base region of GP, crosslinking the GP1 and GP2 subunits20. ADI-15946 potently neutralizes EBOV and BDBV but lacks neutralizing and protective activity against SUDV20. Accordingly, we sought to define the structural determinants of ADI-15946 activity in the context of its cognate antigen, EBOV GP, in order to characterize features of both its epitope and paratope that confer broad reactivity while investigating its limited activity against SUDV.
Results:
Crystal structure of EBOV GPCL-ADI-15946 complex.
We determined the crystal structure of ADI-15946’s fragment antigen binding (Fab) complexed with EBOV GPCL to 4.1 Å resolution (Fig. 1a-b and Table 1)20. The structure was solved by molecular replacement using the previously published EBOV GPCL structure as a search model (PDB 5HJ3) and was refined to an Rwork/Rfree of 26.2/28.1% (Table 1)16. The ADI-15946 Fab targets the base of a single GP1/GP2 protomer within the GP trimer at an approximately 45° angle relative to the surface of GP, with its constant domains directed downward towards the viral membrane (Fig. 1b).
Table 1.
Data collection and refinement statistics (molecular replacement)
| EBOV GPCL–ADI-15946 Fab (PDB 6MAM) |
||
|---|---|---|
| Data collection | ||
| Space group | P4322 | |
| Cell dimensions | ||
| a, b, c (Å) | 182.27 182.27 262.01 | |
| α, β, γ (°) | 90 90 90 | |
| Resolution (Å) | 48.54 - 4.10 (4.25 - 4.10)a | |
| Rmerge | 0.20 (1.83) | |
| Rpim | 0.05 (0.46) | |
| I/σ(I) | 14.6 (1.8) | |
| CC1/2 | 1.00 (0.70) | |
| Completeness (%) | 99.6 (100.0) | |
| Redundancy | 13.1 (13.4) | |
| Refinement | ||
| Resolution (Å) | 48.54 - 4.10 (4.25 - 4.10) | |
| No. reflections | 35,153 (3,458) | |
| Rwork / Rfree | 0.262 / 0.281 | |
| No. atoms | ||
| Protein | 16525 | |
| Ligands (Glycans) | 172 | |
| B factors | ||
| Protein | 114 | |
| Ligands (Glycans) | 107 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 0.61 | |
Diffraction data were collected from a single crystal.
Values in parentheses are for highest-resolution shell.
The heavy and light chains each contribute roughly half of the overall ADI-15946 binding interface: ~55% from the heavy chain (HC) complementarity-determining region 3 (CDR H3), and the remaining 45% from the combined contributions of the light chain (LC) framework region 3 (FRL3) and CDRs 1, 2, and 3 (L1, L2, and L3 respectively) (Fig. 1b-c). Notably, CDRs 1 and 2 of the heavy chain (H1 and H2, respectively) do not participate in the Fab-GPCL interface (Fig. 1b-c). Instead, HC recognition of GPCL is mediated exclusively by the 22-amino-acid-long CDR H3 which buries approximately 465 Å2 of surface area upon binding (Fig. 1b). The footprint of ADI-15946 partially overlaps those of the EBOV-monospecific binders KZ52, c2G4 and 4G7 (Supplementary Note 1)6,27. However, ADI-15946 likely gains cross-reactivity from an upward shift of its footprint which allows it to target a highly conserved pocket above the upper boundary of the KZ52 epitope as suggested previously (Fig. 1d)20.
Modeling ADI-15946 interactions with GP2 and the glycan cap.
ADI-15946 HC contacts include the strictly conserved lysine at position 510 in the GP2 N terminus—where a substitution to glutamic acid, K510E, was previously shown to give rise to escape from BDBV viral neutralization20 We have confirmed that the same amino acid substitution in EBOV GP also leads to the loss of binding and neutralization phenotypes (data not shown). The electrostatic surface of ADI-15946 is complementary to that of GP, and our sidechain modeling suggests that the K510E substitution introduces a charged/steric clash with residues D100C and/or L100F of CDR H3 thereby explaining the loss of antiviral activity (Fig. 2a-c). We tested ADI-15946 variants containing alanine substitutions at either the D100C or the L100F positions for binding and neutralization and observed loss of both activities against recombinant vesicular stomatitis virus (rVSV) particles bearing BDBV GP K510E (Fig. 2d). Additionally, the ADI-15946 D100CA variant lost activity to wild-type rVSV-BDBV GP likely due to the loss of stabilizing interactions between the D100C sidechain and the backbone of CDR H3 (Fig. 2d). These results suggest that interaction with the conserved K510 residue is a key contributor to ADI-15946’s breadth and that overcoming the K510E escape mutation would be difficult without significant mutagenesis of the antibody paratope.
Figure 2.
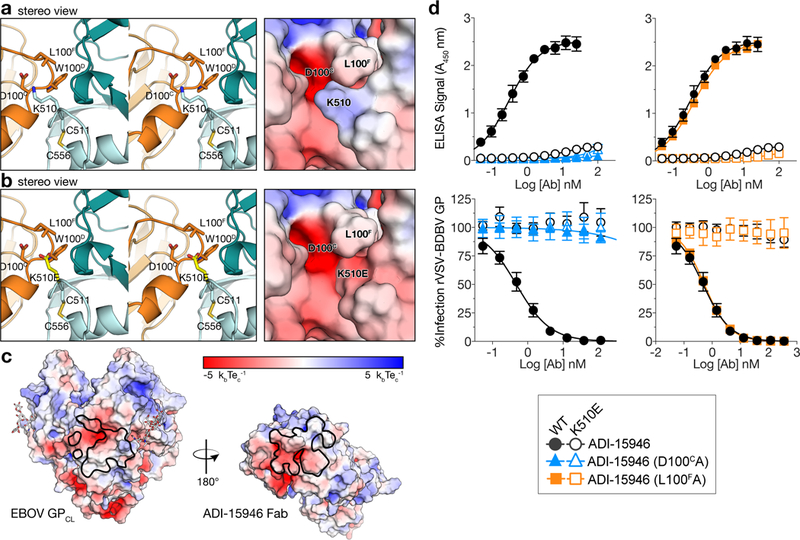
The K510E escape mutation likely clashes with ADI-15946 CDR H3. (a) Stereoview of the EBOV GPCL–ADI-15946 complex (left; ADI-15946 in orange, and GPCL in dark and light teal for GP1 and GP2, respectively) and electrostatic surface potential (right; color scale shown in panel c) showing that residue K510 of GP2 binds into a negatively charged pocket created by ADI-15946 CDR H3. (b) Similar views to (a), with modeling of an escape mutant of ADI-15946, GP K510E, suggesting that K510E clashes with CDR H3 and introduces conflicting negative charge into the CDR H3 pocket. (c) Open-book representation of EBOV GPCL and ADI-15946 showing electrostatic surface potential colored according to included scale. GPCL is shown on the left with the epitope outlined in black. ADI-15946 is shown on the right with the paratope outlined in black. (d) Binding and neutralization assays showing the capacity of ADI-15946 variants containing either the D100CA or the L100FA mutation to bind to rVSV-BDBV GP (WT or K510E) in an ELISA (top, mean± s.d., n=4 biologically independent samples) and neutralize infection by these viruses (bottom, mean± s.d., n=6 biologically independent samples). Electrostatic surface potentials in a and c were generated using the APBS plugin with Pymol.
Wec et al. have shown that ADI-15946 inhibits GP proteolysis in vitro, a key step in unmasking of the viral receptor binding site that is indispensable for infection. The left boundary of ADI-15946’s footprint, formed by LC FRL3, is in immediate proximity to the β13-β14 loop that passes over the IFL and is cleaved by endosomal cathepsins during viral entry (Supplementary Fig. 1)13,14. Overlay of our structure with uncleaved GP (PDB 5JQ3) shows that ADI-15946 binding likely impedes protease access to this loop, thus explaining ADI-15946 inhibition of GP proteolysis (Supplementary Fig. 1)20.
Competition with, and mimicry of, the β17-β18 loop.
ADI-15946 binds into a hydrophobic pocket formed by residues 71-75 of GP1, which fold into a 310 helix, and is hereafter termed the ‘310 pocket’ (Fig. 3a-b). The 310 helix rearranges in GPCL when bound to NPC1, and stabilization of this region by ADI-15946 CDR H3 may be part of the mAb’s neutralization mechanism (Fig. 3a)12,16,20. In uncleaved GP, the 310 pocket is occupied by the β17-β18 loop (GP1 residues 287-291) that extends down from the glycan cap (Fig. 3a-b)12,28. Interactions of ADI-15946 CDR H3 residues D100C, W100D, and L100E with the 310 pocket mimic the contacts made by β17-β18 loop residues E292, W291, and F290, respectively (Fig. 3b). Correspondingly, we postulated that the viral β17-β18 loop peptide competes for binding with CDR H3 of ADI-15946 in full length GP (GPFL) (Supplementary Fig. 2). To test the effect of β17-β18 loop removal on ADI-15946 binding and neutralization, we measured the apparent binding constant (KDapp) for ADI-15946 interaction with EBOV GPFL and GPCL via Bio-Layer Interferometry (BLI), as well as its neutralization of rVSV-EBOV GPFL and rVSV-EBOV GPCL. Consistent with the previous report, we observed a 10,000-fold improvement in binding and a 100-fold improvement in neutralization potency against EBOV GPCL compared to full-length GP (Fig. 3e and Supplementary Tables 1 and 2)20. Comparison of association (kon) and dissociation (koff) rates of ADI-15946 to EBOV GPFL and GPCL showed that the improved binding to GPCL is primarily driven by over 1,000-fold slower koff and a modest improvement of the association rate (10-fold increased kon) (Fig. 3f, Supplementary Fig. 3, and Supplementary Table 2). Given the sizeable contribution of CDR H3 to the binding interface, the slower dissociation rate against GPCL likely results from its unobstructed access to the 310 pocket.
Figure 3.
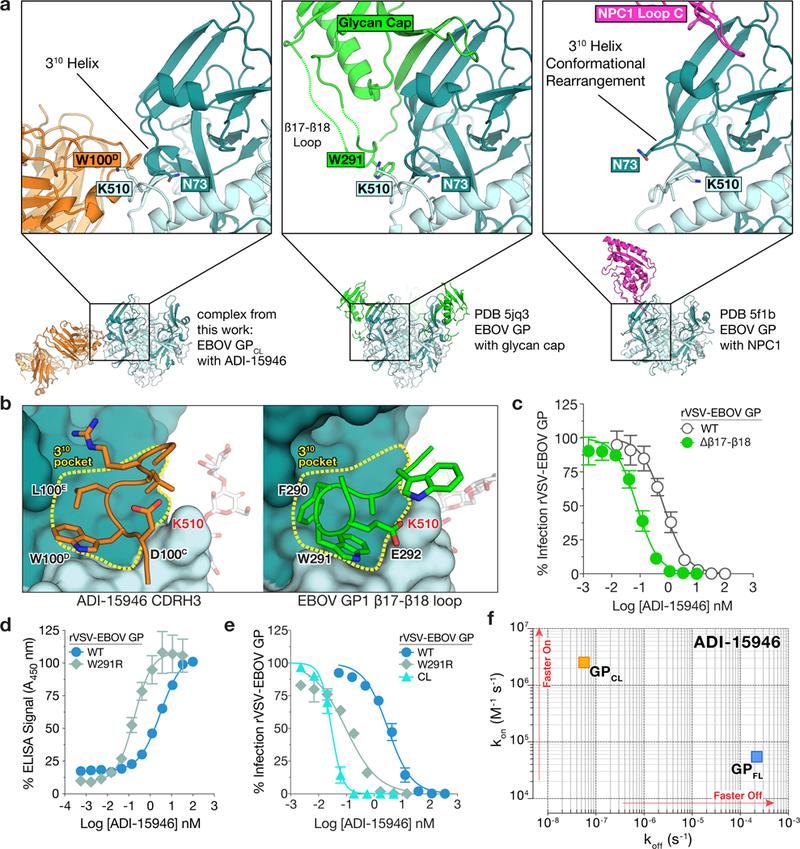
ADI-15946 binds a highly conserved epitope shielded by the mobile β17-β18 loop of the glycan cap. (a) Structures show that ADI-15946 (orange, left) binds the hydrophobic 310 pocket of GP (teal) that is usually occupied by the β17-β18 loop of the glycan cap (green, middle). Right, binding NPC1 loop C (magenta) to GP induces a conformational change in which the GP1 310 helix unwinds and asparagine 73 (N73) becomes solvent exposed while GP2 lysine 510 (K510) inserts into the cavity left behind after the unwinding of the 310 helix. ADI-15946 may prevent these conformational changes from occurring by locking down the 310 helix with residues that mimic those of the β17-β18 loop. (b) An enlarged view of the 310 pocket shows that CDR H3 of ADI-15946 positions similar residues in similar orientations to that of the β17-β18 loop. (c) Neutralization assay showing that ADI-15946 has enhanced neutralization of a GP construct lacking the β17-β18 loop compared to wild type (WT), likely due to increased access to the 310 pocket. Data are mean± s.d., n=6 biologically independent samples. (d) Binding assays showing that a point mutation in the β17-β18 loop (W291R) results in enhanced binding to rVSV-EBOV GP in an ELISA. Data are mean± s.d., n=4 biologically independent samples. (e) Neutralization assays showing that the W291R mutation in the β17-β18 loop or its proteolytic removal (CL) enhance the capacity of ADI-15946 to neutralize rVSV-EBOV GP. Data are mean± s.d., n=6 biologically independent samples. (f) Kinetic binding studies by biolayer interferometry reveal enhanced association rate and slower dissociation rate of ADI-15946 to GPCL compared to uncleaved GP.
To specifically probe the importance of the β17-β18 loop’s hydrophobic packing into the 310 pocket to the shielding of the ADI-15946 epitope, we first tested ADI-15946 binding and neutralization against rVSVs bearing an EBOV GP variant with an arginine substitution at position 291 (rVSV-EBOV GPW291R). Modeling of the arginine sidechain suggested that the W291R substitution would displace the β17-β18 loop from the conserved pocket by introducing charged and steric clashes with the GP2 residue asparagine 512. Consistent with this hypothesis, we observed a 10-fold enhancement in ADI-15946 binding to EBOV GPW291R compared to wild-type EBOV GP (GPWT) presented on rVSV and a >10-fold increase in neutralization efficiency of rVSV bearing EBOV GPW291R relative to EBOV GPWT (Fig. 3d-e and Supplementary Table 1). We then tested ADI-15946 neutralization against rVSVs bearing an EBOV GPΔβ17-β18 variant in which the β17-β18 loop has been deleted (residues 187-198). Removal of the β17-β18 loop had no apparent adverse effects on viral infectivity or replication in vitro (data not shown). We observed a 10-fold increase in neutralization efficiency of rVSV-EBOV GPΔβ17-β18 compared to GPWT ( Fig. 3c). The intermediate neutralization potency of rVSV-EBOV GPW291R compared to GPCL and GPWT, suggests that displacement of the β17-β18 loop from the 310 pocket is the limiting factor for binding of ADI-15946 to EBOV GP (Fig. 3e).
Enhancement of binding and neutralization with mAb FVM09.
Previous work suggests that a non-neutralizing mAb, FVM09, recognizes and “peels away” the β17-β18 loop from the base, thereby enhancing neutralization by the base-binding mAb c2G429,30. Given the more profound role of the β17-β18 loop in restricting access of ADI-15946 to its base epitope (see above), we evaluated the hypothesis that FVM09 could also work in concert with ADI-15946 to neutralize virus with enhanced potency (Supplementary Fig. 2). We first tested competition between ADI-15946 and FVM09 using BLI-based competitive binding assays and found that both mAbs could bind to EBOV GP simultaneously (Supplementary Fig. 4). We next tested the effect of FVM09 on ADI-15946 neutralization of rVSV-EBOV GP or authentic EBOV in the presence of three fixed concentrations of FVM09. FVM09 potentiated ADI-15946 neutralization in a concentration-dependent manner by >10-fold, but had no effect on neutralization by KZ52 (Fig. 4a). Conversely, titration of FVM09 into a constant, sub-neutralizing concentration of ADI-15946 also showed dose-dependent enhancement of ADI-15946 neutralizing activity (Fig. 4b). In contrast, KZ52 neutralization was somewhat reduced in the same assay, perhaps because KZ52 binds along the surface of the β17-β18 loop whereas ADI-15946 binds beneath the β17-β18 loop (Fig. 4b and Supplementary Fig. 5). We observed similar neutralization enhancement trends with authentic EBOV (Fig. 4c-d), and to a lesser extent, with rVSV-SUDV GP (Fig. 4e-f). The functional significance of the strict conservation of the amino acid sequence in the 310 pocket and the β17-β18 loop among ebolavirus GPs remains unknown, but it may relate to the transduction of conformational changes in GP upon NPC1 binding (Fig. 3a and Supplementary Note 2)12. Our results demonstrate that rare neutralizing antibodies like ADI-15946 can access and exploit this cryptic site of broad ebolavirus vulnerability to achieve neutralization, presumably by interfering with the region’s functional role in viral entry. The enhancement of ADI-15946 binding and neutralization in the presence of β17-β18 loop binders such as FVM09 may reflect a mechanism by which specific combinations of glycan cap- and base-binding mAbs can synergize to interdict viral infection during a natural polyclonal immune response (Fig. 4g).
Figure 4.
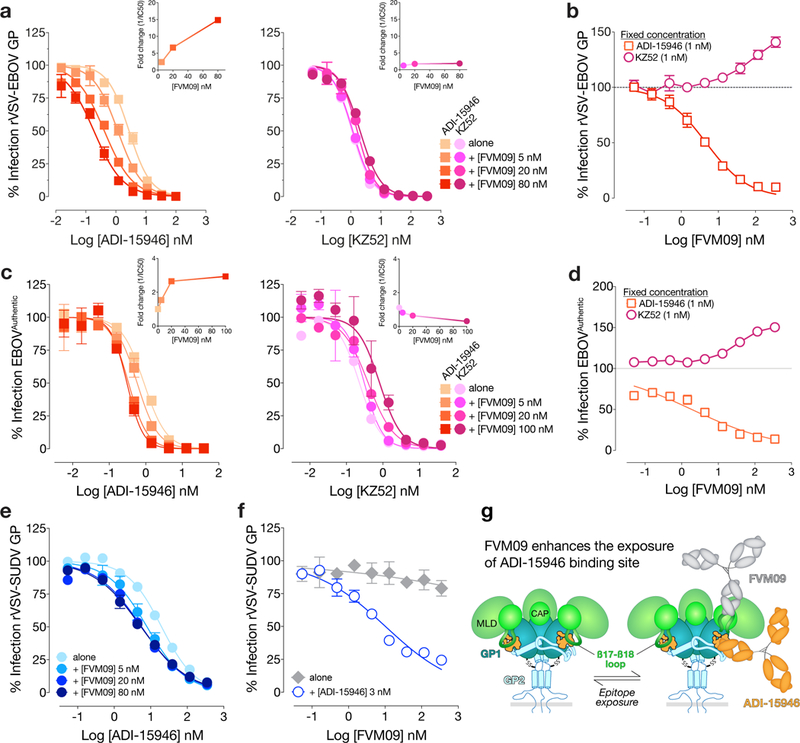
mAb FVM09 potentiates ADI-15946 neutralization of EBOV GP in a dose-dependent manner. (a) Infection assays showing that addition of increasing concentrations of ADI-15946 (left), but not KZ52 (right), to fixed concentrations of FVM09 (5, 20, or 80 nM) promotes rVSV-EBOV GP neutralization. The reciprocal fold change in neutralization IC50 is shown (inset). (b) Infection assays showing that addition of increasing concentrations of FVM09 to a fixed, subneutralizing concentration of ADI-15946 enhances rVSV-EBOV GP neutralization. The same experiment against KZ52 shows inhibition of neutralization as the concentration of FVM09 is increased. (c-f) Infection assays as in a-b, with authentic EBOV (c-d) showing similar trends; this trend is present but less pronounced with rVSV-SUDV GP (e-f). Data in a–f are mean± s.d., n=6 biologically independent samples. (g) A cartoon showing the proposed relationship between FVM09 binding and subsequent exposure of the ADI-15946 binding site.
Molecular determinants for somatic maturation of ADI-15946.
To delineate the molecular basis of ADI-15946’s broad activity, we assigned its light and heavy chain variable domain (VL and VH, respectively) sequences to their most probable inferred germline progenitors (IGL) (Fig. 5a and Supplementary Note 3). We found that reversion of three residues introduced by somatic hypermutation (SHM) in the VH to germline (LCWT:HCIGL; CDR H3 is retained fully mature) had no appreciable impact on binding of full-length or cleaved EBOV, BDBV, or SUDV GP or neutralization of rVSV bearing EBOV, BDBV, or SUDV GP (Fig. 5b-c and Supplementary Fig. 6). These findings agree with our structural observations, since only CDR H3 participates in binding to EBOV GPCL (Fig. 1b).
Figure 5.
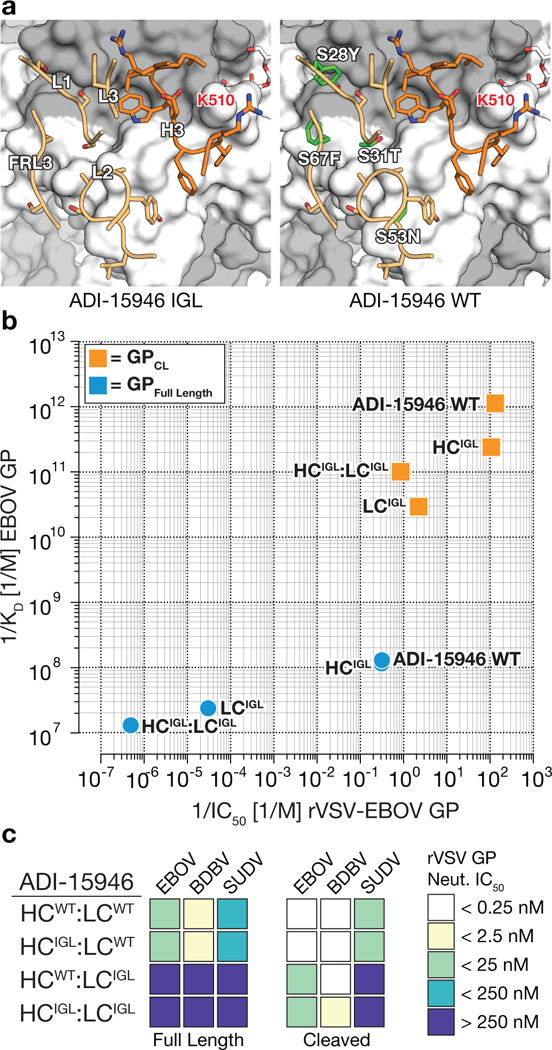
Genesis of ADI-15946. (a) Models showing differences between the ADI-15946 IGL sequence (left) and the mature antibody (WT, right). GP1 is shown in grey and GP2 in white. (b) Comparison of the apparent equilibrium dissociation constant (1/KDapp; higher value is tighter binding) for binding of ADI-15946 variants (WT, IGL, and WT:IGL chimeras) to GPCL to their capacity to neutralize rVSV-EBOV GP infection (1/IC50; higher value is more potent neutralization). (c) Heat maps for neutralization of rVSVs bearing ebolavirus GP and GPCL proteins by the indicated ADI-15946 variants.
Reversion of ten SHM-introduced residues in the VL outside of CDR L3 (LCIGL:HCWT; residues 3, 4, 27, 28, 31, 53, 67, 72, 87, 104) had a profound impact on neutralization of rVSVs bearing EBOV, BDBV, or SUDV GP (Fig. 5b-c and Supplementary Fig. 6). Both the LCIGL:HCWT and the combined LCIGL:HCIGL Ab variants were non-neutralizing against rVSV-EBOV GPFL despite retaining a fully mature CDR H3 and CDR L3, suggesting that LC contacts outside of CDR L3 make key contributions to GP recognition (Fig. 5c). The LCIGL:HCWT and the LCIGL:HCIGL germline-approximating Ab variants retained some neutralizing activity against EBOV GPCL and BDBV GPCL, likely due to improved access of CDR H3 to the 310 pocket (Fig. 5c and Supplementary Table 1). However LCIGL:HCIGL did not neutralize rVSV-SUDV GPCL (Supplementary Fig. 6). Our germline-approximating variants LCIGL:HCWT and the LCIGL:HCIGL neutralized the rVSV-EBOV GPW291R variant more potently than rVSV-EBOV GPCL which suggests that additional LC contacts that are absent in GPCL, likely between the Ab and the glycan cap, contribute to ADI-15946’s activity against GPFL (Supplementary Fig. 1, 6, and Supplementary Table 1).
Enhancement of ADI-15946 activity against SUDV.
Our analysis of the conservation within ADI-15946’s epitope shows that limited SUDV reactivity and lack of activity against Reston virus (RESTV) likely arise from the amino acid sequence divergence on the edges of its footprint (Fig. 6f and Supplementary Note 2). There are only five non-conserved residues across all five ebolavirus GPs involved in the interface with ADI-15946, all located in GP2: N506, N514, H516, L547, and H549. These are predominantly contacted by CDR L2 and FRL3 (Fig. 6f). However, CDR H3 contacts one key residue in EBOV GP (Asn 506) that is not conserved in SUDV GP (Arg 506). Using this information, we separately mutated nearby residues in ADI-15946 CDR H3, R100 and Y100A, to alanine to reduce steric and charge clashes with SUDV R506. The ADI-15946 mutant Y100AA showed decreased binding to both SUDV and BDBV GP compared to wild-type ADI-15946 (Supplementary Fig. 7). However, the mAb variant containing R100A, called 46M1, showed increased binding to SUDV GP ectodomain by ELISA and slightly improved neutralization of authentic virus compared to the parental antibody (Fig. 6b-e and Supplementary Fig. 7).
Figure 6.
Structure-guided affinity maturation of ADI-15946. (a) Molecular models showing the locations of mutations in ADI-15946 variants 46M1, 46M2, and 46M3 in relation to the surface of GP. CDRs are illustrated in dark orange for the heavy chain and light orange for the light chain respectively; engineered side chains that differ from wild-type are colored in green. GP1 and GP2 are shown as a grey and white surface respectively. (b) Binding assays of recombinant EBOV, BDBV, and SUDV GP ectodomains by the indicated ADI-15946 variants determined by ELISA. Data are mean± s.d., n=3 biologically independent samples. (c) Neutralization assays of authentic EBOV, BDBV and SUDV by the indicated ADI-15946 variants. Data are mean± s.d., n=6 biologically independent samples. (d-e) Heat maps for neutralization potency (IC50) of each ADI-15946 variant against rVSVs (d) and authentic filoviruses (e). In panel d, neutralization of rVSVs bearing full-length GPs and cleaved GPs is shown on the top and bottom, respectively. (f) Molecular surface of EBOV GPCL with the ADI-15946 footprint outlined in orange. Differences at five sites are listed on the left. The panel on the right shows the which CDRs are in proximity to these nonconserved sites. (g) Neutralization assays of rVSV-EBOV GP by 46M3 in the presence of increasing concentrations of FVM09 (0–100 nM). Increasing amounts of FVM09 improved 46M3 neutralization. Data are mean± s.d., n=6 biologically independent samples.
Structural alignment of additional Fab-GP complexes enabled comparison of the binding features of the ADI-15946 paratope with those of EBOV monospecific mAbs KZ52, c2G4, and c4G7, which have overlapping epitopes with that of ADI-15946 (Supplementary Fig. 5 and Supplementary Note 1)6,27. We observed that the HC paratope of c4G7 places a tyrosine residue in a similar position and orientation to that of the ADI-15946 LC residue F67 and another tyrosine residue proximal to ADI-15946 LC residue S65. We then attempted to mimic this double tyrosine motif by incorporation of light chain S65Y and F67Y mutations into ADI-15946. The construct bearing heavy chain R100A and light chain S65Y is called variant 46M2, and ADI-15946 bearing the R100A/S65Y pair plus LC substitution F67Y is called variant 46M3 (Fig. 6a). The 46M2 and 46M3 antibodies showed enhanced binding of SUDV, while maintaining parental binding to EBOV and BDBV GPs (Fig. 6b). 46M2 and 46M3 also exhibited a 16- to 33-fold improvement in their capacity to neutralize SUDV GP-bearing rVSV and authentic SUDV respectively, while retaining neutralization of EBOV and BDBV (Fig. 6c-e and Supplementary Fig. 7). Slight variations were observed in the neutralization profiles of the affinity variants versus the parent ADI-15946: we observed an approximately 2-fold increase in neutralization for the affinity variants against rVSV-EBOV GP compared to wild-type ADI-15946 and an approximately 2-fold decrease in neutralization for the affinity variants against authentic EBOV compared to wild-type ADI-15946 (Fig. 6d-e and Supplementary Tables 1 and 3). Further, the variants and the parental antibody similarly recognize the conserved 310 pocket, as all are equally enhanced by β17-β18 loop deletion and, like wild-type ADI-15946, 46M3 neutralization is enhanced in the presence of FVM09 (Fig. 6g and Supplementary Fig. 8). We show that relatively few mutations introduced in the 46M2 and 46M3 variants resulted in improved binding and neutralization of SUDV and confirmed the importance of the LC FR3 region to the binding interface.
Discussion:
The crystal structure of ADI-15946 Fab in complex with EBOV GPCL and accompanying biochemistry presented here illuminated antigenic features of GP that dictate access to a broadly conserved but previously unappreciated epitope shared by ebolaviruses. This site of vulnerability is shielded by the descending β17-β18 loop of the glycan cap, removal of which potentiates ADI-15946 activity. Our findings support a mechanism of action whereby ADI-15946 gains enhanced neutralizing activity against the endosomal, cleaved viral GP species generated by host proteases during entry. This feature allows ADI-15946 to neutralize both the extracellular and intracellular viral species. In contrast, other potent neutralizers that target the GP base, such as KZ52 and the ZMapp components c2G4 and c4G7, have been shown to lose their activity against GPCL and therefore have activity only against the extracellular virus6,20,27. The unique mechanism of neutralization by ADI-15946 may involve binding of ADI-15946 in the 310 pocket, which in turn prevents rearrangement of GP1 residues 71-75 upon binding of GPCL to NPC1 by mimicking the hydrophobic interactions of the β17-β18 loop that packs into the pocket in GPFL (Fig. 3a-b)12. In addition, ADI-15946 may also inhibit conformational changes required for membrane fusion by anchoring to the GP1-GP2 interface across the IFL thereby preventing its unraveling from the GP1 core during fusion triggering.
We also uncovered potentially synergistic neutralization between ADI-15946 and FVM09, a non-neutralizing glycan cap-binding antibody. FVM09 was previously shown to enhance binding activity of another glycan cap mAb, m8C4, yet reduce binding of the base-targeting mAb KZ52—the latter being consistent with our own observations30. As with ADI-15946, mutations of the β17-β18 loop also enhanced m8C4’s binding and neutralizing activity in vitro30. The exact mechanism of FVM09 and m8C4 cooperation is difficult to discern without structural details of the binding interface between the two mAbs and GP. In the case of cooperativity between FVM09 and ADI-15946, however, we propose that FVM09 primarily plays a supporting role by exposing the 310 pocket thereby enhancing ADI-15946 binding and neutralization. It is possible that co-administration of FVM09 and ADI-15946 would lead to enhancement in protective efficacy in vivo. However, given that FVM09 is a non-neutralizing and non-protective antibody, viral escape from ADI-15946 neutralization would render the two antibody combination non-protective. Follow up studies will be required to fully evaluate the therapeutic usefulness of antibody combinations of this sort.
Our analysis of the contributions of somatic hypermutation to the binding and neutralization properties of ADI-15946 indicated that only a limited number of amino acid substitutions in the germline antibody sequence were required for binding of this conserved GP site and that many of these mutations occurred in the framework region. Klein et al. show that somatic mutations in the immunoglobulin framework region of antibodies against HIV-1 enhance affinity by decreasing the dissociation rate and are generally necessary to achieve broad neutralization31. We found that, similar to bNAbs against HIV-1, ADI-15946 also required somatic maturation of the framework region, which bestowed the mAb with an improved dissociation rate and broad neutralizing activity of ebolaviruses31. Specifically, as a result of SHM, a serine at position 67 of ADI-15946 FRL3 was replaced with an aromatic phenylalanine sidechain that interacts with GP2 residue H516 (100% conserved between EBOV, BDBV, and SUDV). Interestingly, the non-pathogenic RESTV encodes a tyrosine at position 516 in GP2, which may play a role in the inability of ADI-15946 to neutralize RESTV due to steric hindrance with the antibody phenylalanine 67 (Supplementary Fig. 7e and Supplementary Note 2).
This structure provided the molecular details of the paratope required for rational engineering of the parent mAb to expand its activity against SUDV. We showed that structure-guided substitutions, two in the FRL3 and one in CDR H3, enhanced both binding and neutralization activity against SUDV without loss of activity against EBOV or BDBV. However, follow up work will be required to evaluate additional mAb variants and their biophysical properties in order to ensure favorable in vivo pharmacokinetics and protective efficacy against SUDV, since unlike natural antibodies, mAbs generated through in vitro engineering have not undergone immune tolerance selection. Our work also detailed structural features of GP recognition by a broadly active anti-ebolavirus antibody and suggests specific strategies such as the use of GPCL (lacking the glycan cap) or GP variants with β17-β18 loop deletion to enable targeting of the highly conserved site in the design of vaccines and immunotherapeutics. Like ADI-15946, the pan-ebolavirus mAb ADI-15878 also binds into a cryptic pocket at a distinct site in the GP base, that is typically occupied by the N-terminal tail of GP2 when not bound by the mAb21. Future ebolavirus vaccine designs may strive to enhance the exposure of both of these vulnerable pockets on a single antigen by developing an immunogen that combines β17-β18 loop deletion with a truncated GP2 N-terminus. Such immunogens may help elicit bNAbs like ADI-15946 at a higher frequency while minimizing potential for viral escape by eliciting bNAbs towards multiple broadly conserved sites, both located proximal to the functionally critical region of the GP fusion loop. A broadly reactive vaccine would thus permit increased preparedness against ebolavirus disease regardless of the specific virus responsible for an outbreak.
Online Methods:
Cloning.
Mutants were introduced into parental ADI-15946 by site-directed mutagenesis using QuickChange (Agilent), with all mutations confirmed by sequencing (Eton).
Protein expression & purification.
Expression and purification of EBOV GPCL was performed as described previously16. Briefly, Ebola virus GP (lacking the mucin domain residues 312 to 462) was produced by stable expression in Drosophila melanogaster S2 cells. Effectene (Qiagen) was used to transfect S2 cells with a modified pMT-puro vector plasmid containing the GP gene of interest, followed by stable selection of transfected cells with 6 μg/ml puromycin. Cells were cultured at 27 °C in complete Schneider’s medium for selection and then adapted to Insect Xpress medium (Lonza) for large-scale expression in 2-liter Erlenmeyer flasks. Secreted GP ectodomain expression was induced with 0.5 mM CuSO4, and supernatant harvested after 4 days. Ebola virus GP was engineered with a double Strep-tag at the C terminus to facilitate purification using Strep-Tactin resin (2-1201-010) (Qiagen) and then further purified by Superdex 200 (GE) size exclusion chromatography (SEC) in 10 mM Tris-buffered saline (Tris-HCl, pH 7.5, 150 mM NaCl [TBS]). EBOV GPCL was produced by incubation of 1 mg GP with 0.02 mg thermolysin overnight at room temperature in TBS containing 1 mM CaCl2 and purified using Superdex 200 SEC.
ADI-15946 Fab used for crystallization experiments was cloned into a modified pMT-puro vector with a heavy chain C-terminal Strep-tag, and then expressed and purified according to the protocol for GPCL with the exception that SEC was performed with a Superdex 75 column (GE)16.
ADI-15946 IgG used for ELISA and neutralization assays were produced in ExpiCHO cells (ThermoFisher Scientific) and purified via Protein A chromatography according to the standard ThermoFisher ExpiCHO protocol for a 25 mL culture volume. Cells were pelleted 8 days post transfection by centrifugation for 30 minutes at 3000 xg and supernatant was collected for purification of soluble ADI-15946 IgG. Supernatant was flowed over 2 mL of Pierce Protein A Plus Agarose resin (ThermoFisher Scientific). Column was washed with 5 column volumes of DPBS (Gibco) and then IgG was eluted with DPBS supplemented with 25 mM glycine pH 2.2.
Crystallography & structure determination.
Trimeric EBOV GPCL was complexed with ADI-15946 Fab fragments, and the resulting complex was then purified via SEC. The purified EBOV GPCL-ADI-15946 Fab complex was concentrated to 4.2 mg/ml in TBS. The crystal drops consisted of a 1:1 ratio of protein/well solution. Crystals grew over the course of one month in 0.2 M sodium citrate tribasic dihydrate pH 8.2 and 20% polyethylene glycol 3350. Crystals were cryoprotected with 20% glycerol and flash frozen in liquid nitrogen for storage and shipping. Diffraction data was collected remotely on SSRL beamline 12-2 on a pilatus 6M detector32-35. Data was processed using XDS36,37, and the structure was determined using molecular replacement with PHASER38, within the CCP4 suite39, using the structure of EBOV GPCL (PDB 5HJ3) as an initial search model16. Iterative rounds of model building were performed using Coot40, and each round was refined with Phenix41. Five percent of the data was set aside prior to refinement for the Rfree calculations for each data set42. The statistics and stereochemistry of the crystal structure were checked using the MolProbity server42,43. Structural figures were rendered using Open Source PyMOL (PyMOL Molecular Graphics System, version 1.7.0.0; Schrödinger, LLC).
Structural alignment and visualization of ebolavirus glycoproteins.
Alignment was performed using clustalomega on uniprot with the following protein sequences: Zaire ebolavirus: Q05320, Bundibugyo ebolavirus: B8XCN0, Sudan ebolavirus: Q66814, Taï Forest ebolavirus: Q66810, Reston ebolavirus: Q66799. Sequence conservation was numbered according to EBOV GP and visualized using the Espript server (http://espript.ibcp.fr) and colored according to the percent equivalent scoring function with a cutoff of 70%44.
Determination of inferred germline progenitors.
ADI-15946 heavy and light chain sequences were aligned to their nearest predicted germline progenitors using IMGT. Amino acid substitutions in the variable region of the heavy and light chain outside of CDR H3 and CDR L3 of the mature antibody were reverted to germline-encoded amino acids. Germline-reverted sequences of VL and VH were then ordered as gBlocks with overhangs to allow for homologous recombination into Saccharomyces cervisiae.
Anti-GP mAb ELISAs.
High-binding 96-well ELISA plates (Corning) were coated with 50 μL GP antigens in phosphate-buffered saline (PBS) at 4 μg/mL, and allowed to bind for 1 h at room temperature. After washing, the wells were blocked with PBS containing 3% bovine serum albumin (PBSA) for 1 h at room temperature, followed by washing then incubation with ADI-15946 or one of its mutant derivatives in serial dilutions of PBS. A horseradish-peroxidase conjugated anti-human secondary antibody (Santa Cruz Biotechnology) was added and allowed to bind for 1 h at room temperature and then subsequently detected by ultra-TMB (3,3′,5,5′-tetramethylbenzidine) substrate (ThermoFisher Scientific). Optical density was measured at 450 nm, and absorbance readings were subjected to a nonlinear regression analysis (GraphPad Prism software) to generate binding curves and calculate an EC50 value. ELISA assays were performed in triplicate, across seven 5-fold dilutions, beginning at 1000 ng/ml.
Biolayer interferometry assays.
The Octet Red™ system (FortéBio, Pall) was used to determine the binding properties of different IgGs to various forms of EBOV GP. Anti-human Fc (AHC) capture sensors (ForteBio) were used for initial mAb loading at 25 mg/mL in 1X kinetics buffer (PBS supplemented with 0.002% Tween-20 and 1 mg/mL of BSA). Binding to GP was performed across two-fold serial dilutions of EBOV GPΔTM or GPCL. The baseline and dissociation steps were carried out in the 1X kinetics buffer as per the instrument manufacturer’s recommendations. Kinetic binding data in all cases are adequately and accurately described by a 1:1 binding model, but given the bivalent nature of the IgG (immobilized) and the trimeric state of GP (analyte), the association stoichiometry is likely to be more complex. Thus, koff/kon likely reflects an ensemble of binding stoichiometries, and accordingly, we refer to this ratio as apparent KD (KDapp) throughout.
rVSV neutralization assays.
Neutralization of rVSV: Recombinant vesicular stomatitis virus (VSV) expressing both enhanced green fluorescent protein (EGFP) and recombinant surface GP (rVSV-EBOV GP) in place of VSV G are previously described45-47. Vero cells were seeded at 6.0 × 104 cells/well and cultured overnight in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) and 100 I.U./ml penicillin and 100 mg/ml streptomycin at 37°C and 5% CO2. The next day, virus was incubated with serial 3-fold antibody dilutions beginning at 350 nM (~50 μg/ml) in serum free DMEM for one hour at room temperature before infecting Vero cell monolayers in 96-well plates. The virus was incubated with the cells in 50% v/v DMEM supplemented with 2% FBS, 100 I.U./ml penicillin and 100 μg/ml streptomycin at 37 °C and 5% CO2 for 14-16 hours before the cells were fixed and the nuclei stained with Hoescht. rVSV infectivity was measured by counting EGFP-positive cells in comparison to the total number of cells indicated by nuclear staining using a Cellinsight CX5 automated microscope and accompanying software (ThermoFisher Scientific).
Authentic virus neutralization assays.
Neutralization at BSL-4 was tested against replication-competent infectious EGFP-expressing EBOV and chimeric EBOV/BDBV-GP and EBOV/SUDV-GP constructs (referred as EBOV, BDBV and SUDV, respectively) in HTS format, as previously described48. The neutralization assays were performed using Vero-E6 cells obtained from ATCC and maintained in Minimal Essential Medium (MEM) (ThermoFisher Scientific) supplemented by 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin at 5% CO2, 37°C. Neutralization assays were performed in triplicate, across twelve 4-fold dilutions, beginning from 200 μg/ml.
Authentic virus cooperativity assays.
The authentic Ebola virus/H.sapiens- tc/COD/1995/Kikwit-9510621 (EBOV/Kik-9510621; ‘EBOV-Zaire 1995’)49, was used in this study. Antibodies were diluted to indicated concentrations in culture media and incubated with EBOV for 1 h. Vero cells were exposed to antibody/virus inoculum at an MOI of 0.2 plaque-forming unit (PFU)/cell for 1 h. Antibody/virus inoculum was then removed and fresh culture media was added. At 48 h post-infection, cells were fixed with formalin, and blocked with 1% bovine serum albumin. EBOV-infected cells and uninfected controls were incubated with EBOV GP-specific mAb KZ5250. Cells were washed with PBS prior to incubation with goat anti-human IgG conjugated to Alexa 488. Cells were counterstained with Hoechst stain (Invitrogen), washed with PBS and stored at 4°C. Infected cells were quantitated by fluorescence microscopy and automated image analysis. Images were acquired at 20 fields/well with a 20× objective lens on an Operetta high content device (Perkin Elmer, Waltham, MA). Operetta images were analyzed with a customized scheme built from image analysis functions available in Harmony software.
Supplementary Material
Acknowledgements:
X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory. SSRL is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). Opinions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense. We acknowledge National Institutes of Health grants U19 AI109762 (EOS, KC, JMD), R01 AI132256 (KC), U19 AI109711 (AB), R01 AI132204 (EOS, MJA), R01 AI126587 (MJA), Defense Threat Reduction Agency HDTRA1-13-1-0034 (AB), and the Viral Hemorrhagic Fever Immunotherapeutic Consortium for support. This is manuscript number 29630 from The Scripps Research Institute.
Footnotes
Competing interests:
M.J.A. has stocks in Integrated Biotherapeutics, a company developing antibody therapeutics for ebolavirus disease. A.Z.W., E.G. and L.M.W. are employees and equity holders of Adimab, LLC. The remaining authors declare no competing interests.
Data availability:
Coordinates and structure factors have been deposited in the Protein Data Bank under accession number PDB 6MAM. Other data are available from corresponding author upon reasonable request.
References:
- 1.Qiu X et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corti D et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351, 1339–1342 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Pascal KE et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in non-human primates. J. Infect. Dis. 218, S612–S626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox E et al. Notes for the record: consultation on monitored emergency use of unregistered and investigational interventions for Ebola virus disease. World Health Organization; (2018). Available at: http://www.who.int/emergencies/ebola/MEURI-Ebola.pdf?ua=1 (Accessed: 17th August 2018) [Google Scholar]
- 5.Sivapalasingam S et al. Safety, pharmacokinetics, and immunogenicity of a co-formulated cocktail of three human monoclonal antibodies targeting Ebola virus glycoprotein in healthy adults: a randomised, first-in-human phase 1 study. Lancet Infect. Dis. 18, 884–893 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Lee JE et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volchkov VE, Feldmann H, Volchkova VA & Klenk HD Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. 95, 5762–5767 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory SM et al. Structure and function of the complete internal fusion loop from Ebola virus glycoprotein 2. Proc. Natl. Acad. Sci. 108, 11211–11216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J et al. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. 114, E7987–E7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissenhorn W, Carfí A, Lee KH, Skehel JJ & Wiley DC Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Malashkevich VN et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9 Å resolution. Proc. Natl. Acad. Sci. 96, 2662–2667 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H et al. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell 164, 258–268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schornberg K et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80, 4174–4178 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandran K, Sullivan NJ, Felbor U, Whelan SP & Cunningham JM Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hood CL et al. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 84, 2972–2982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bornholdt ZA et al. Host-primed Ebola virus GP exposes a hydrophobic NPC1 receptor-binding pocket, revealing a target for broadly neutralizing antibodies. MBio 7, e02154–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Côté M et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carette JE et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller EH et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31, 1947–1960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wec AZ et al. Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell 169, 878–890.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West BR et al. Structural basis of pan-ebolavirus neutralization by a human antibody against a conserved, yet cryptic epitope. MBio 9, e01674–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saphire EO et al. Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174, 938–952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuyama W et al. Discovery of an antibody for pan-ebolavirus therapy. Sci. Rep. 6, 20514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milligan JC et al. Structural characterization of pan-ebolavirus antibody 6D6 targeting the fusion peptide of the surface glycoprotein. J. Infect. Dis. 219, 415–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bornholdt ZA et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X et al. Immunization-elicited broadly protective antibody reveals ebolavirus fusion loop as a site of vulnerability. Cell 169, 891–904.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pallesen J et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat Microbiol 1, 16128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y et al. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 535, 169–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keck Z et al. Macaque monoclonal antibodies targeting novel conserved epitopes within filovirus glycoprotein. J. Virol. 90, 279–291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howell KA et al. Cooperativity enables non-neutralizing antibodies to neutralize Ebola virus. Cell Rep. 19, 413–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein F et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references:
- 32.Russi S, Song J, McPhillips SE & Cohen AE The Stanford automated mounter: pushing the limits of sample exchange at the SSRL macromolecular crystallography beamlines. J. Appl. Crystallogr. 49, 622–626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen AE, Ellis PJ, Miller MD, Deacon AM & Phizackerley RP An automated system to mount cryo-cooled protein crystals on a synchrotron beam line, using compact sample cassettes and a small-scale robot. J. Appl. Crystallogr. 35, 720–726 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soltis SM et al. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr. D Biol. Crystallogr. 64, 1210–1221 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McPhillips TM et al. Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brünger AT Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robert X & Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng M et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann-Pick C1. Virology 468-470, 637–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong AC, Sandesara RG, Mulherkar N, Whelan SP & Chandran K A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84, 163–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wec AZ et al. A ‘Trojan horse’ bispecific-antibody strategy for broad protection against ebolaviruses. Science 354, 350–354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilinykh PA et al. Chimeric filoviruses for identification and characterization of monoclonal antibodies. J. Virol. 90, 3890–3901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jahrling PB et al. Evaluation of immune globulin and recombinant interferon-alpha2b for treatment of experimental Ebola virus infections. J. Infect. Dis. 179 Suppl 1, S224–34 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Maruyama T et al. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 73, 6024–6030 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


