Abstract
Prostate cancer is the most common noncutaneous cancer in men in the United States. The current paradigm for screening and diagnosis is imperfect, with relatively low specificity, high cost, and high morbidity. This study aims to generate new image contrasts by learning a distribution of unique image signatures associated with prostate cancer. In total, 48 patients were prospectively recruited for this institutional review board–approved study. Patients underwent multiparametric magnetic resonance imaging 2 weeks before surgery. Postsurgical tissues were annotated by a pathologist and aligned to the in vivo imaging. Radiomic profiles were generated by linearly combining 4 image contrasts (T2, apparent diffusion coefficient [ADC] 0-1000, ADC 50-2000, and dynamic contrast-enhanced) segmented using global thresholds. The distribution of radiomic profiles in high-grade cancer, low-grade cancer, and normal tissues was recorded, and the generated probability values were applied to a naive test set. The resulting Gleason probability maps were stable regardless of training cohort, functioned independent of prostate zone, and outperformed conventional clinical imaging (area under the curve [AUC] = 0.79). Extensive overlap was seen in the most common image signatures associated with high- and low-grade cancer, indicating that low- and high-grade tumors present similarly on conventional imaging.
Keywords: prostate Cancer, radiomics, rad-path, radio-pathomics
Introduction
Prostate cancer is the most frequently diagnosed noncutaneous cancer in men in the United States, accounting for ∼1 in 5 new cancer diagnoses (1). Increased screening efforts, early aggressive therapy for high-risk disease, and the relatively indolent nature of the disease in most patients have resulted in an overall 5-year survival rate of 99% for organ-confined prostate cancer (1). The current paradigm for prostate cancer diagnosis centers on obtaining tissue diagnosis before definitive therapy, either through conventional 12-core systematic transrectal ultrasound-guided biopsy systems or newer magnetic resonance imaging (MRI)-fusion targeted biopsies. These data are typically combined with clinical information (ie, prostate-specific antigen [PSA], PSA density, and clinical T stage) and implemented into various nomograms to predict disease “risk” status.
While PSA screening has been shown to reduce mortality (2–4), PSA alone has relatively low specificity for prostate cancer diagnosis and is insufficient in stratifying disease risk status, leading to an abundance of low-risk patients undergoing an invasive biopsy (5). The conventional paradigm for prostate cancer diagnosis and staging has been challenged in recent years, with data showing that nontargeted biopsies can lead to under-sampling, inaccurate risk stratification, or missing the target cancer all together (6, 7). As a result, noninvasive imaging with multiparametric MRI (MP-MRI) of the prostate is increasingly being used as a tool for prostate cancer detection, preoperative staging, active surveillance, targeted biopsy, and guidance for definitive focal therapy. Several recent prospective trials have shown that using MP-MRI in the prebiopsy setting to identify target lesions for targeted biopsy outperforms systematic 12-core biopsy, leading to a higher rate of diagnosis for clinically significant cancers and a fewer clinically insignificant cancers (8, 9). The incorporation of MR-guidance, however, requires a radiologist to identify and label potential targets. With the clinical standard of care shifting toward image-guided biopsies, an increased burden will be placed on radiologists to correctly and efficiently identify prostate tumors for biopsy.
A typical clinical prostate MRI protocol includes T2-weighted imaging to delineate structure and zone anatomy, multi b-value diffusion-weighted imaging to identify areas of diffusion restriction, and dynamic contrast-enhanced (DCE) imaging to identify early or contemporaneous focal enhancement. The exams are interpreted by a radiologist according to the PI-RADS v2 (10) that assesses the probability of clinically significant cancer. The lack of specificity inherent to PSA screening and MP-MRI means that a definitive diagnosis still requires a biopsy procedure which often leads to the overtreatment of low-risk prostate cancer (11). Patients who do not necessarily have cancer undergo invasive biopsy procedures to mitigate the uncertainty in the screening tests. The clinical barrier to overcome is to appropriately stratify patients near the boundary between intervention and active surveillance before biopsy. The line between active surveillance and treatment is histologically Gleason 3 vs Gleason 4, which roughly correlates to PI-RADS 3 vs PI-RADS 4 on MP-MRI.
Computer-aided diagnostic tools informed by postoperative tissue may provide an opportunity to address this clinical barrier (12–14). Predictive models made from aligned whole mount tissue and in vivo imaging provide opportunity to bring additional information into image space, increasing the overall specificity of a nonspecific test. Radiomics and machine-learning based approaches have been a great success over the past decade (15–18) and improved sensitivity yet decreasing the specificity. However, there is a need for further improvement of this technology.
Radiomics provides a framework for quantifying tumor microenvironment by analyzing images as a mineable database. In addition, by creating a database of aligned pathology or genetics with clinical imaging, it becomes feasible to find radiologic patterns of tumor phenotype which may provide critical predictive information (19–21). Radiomics-based approaches have seen success over the past decade, proving successful across modalities (22, 23) and organ systems, (24–26) by providing a useful means for engineering features amenable to machine learning approaches.
This study uses an aligned rad-path data set to determine whether unique imaging signatures predict the presence of pathologist-identified prostate cancer. We present a method which learns a distribution of unique image characteristics associated with histologic annotations to create voxel-wise predictive maps on a naïve test set.
Methods
Patient Population
Forty-eight patients were recruited prospectively for this institutional review board (IRB)–approved study between June 2014 and February 2017. Written consent was obtained from all patients. Patients' age ranged from 45 to 71 years (mean, 60 years). Inclusion criteria for this study included a scheduled radical prostatectomy and clinical imaging with additional high b value DWI 2 weeks before surgery.
Imaging
MP-MRI was acquired on a 3 T MRI scanner (General Electric, Waukesha, WI) using an endorectal coil. MP-MRI included field of view (FOV)–optimized and –constrained undistorted single shot (FOCUS) DWI, DCE imaging, and T2-weighted imaging. T2 acquisition parameters were as follows: repetition time (TR) = 3370 milliseconds, FOV = 120 mm, voxel dimensions = 0.23 × 0.23 × 3 mm, acquisition matrix = 512, and slices = 26. Diffusion images were collected with 10 b-values (b = 0, 10, 25, 50, 80, 100, 200, 500, 1000, 2000). The DCE images were collected during injection of a gadolinium contrast agent with acquisition matrix = 256, slices = 25, and FOV = 120 mm. All image contrasts used in this study were acquired axially.
MRI Preprocessing
The T2-weighted images were intensity-normalized to the standard deviation within a manually drawn prostate mask (26–29). The B = 0 image was aligned to the T2 using FLIRT (30, 31) and corrected manually if necessary using a freesurfer tool, tkregister2 (surfer.nmr.mgh.harvard.edu). ADC was calculated from 2 combinations of b-value for the purposes of this study, 0 and 1000 and 50 and 2000 (32). The DCE volume with maximal contrast influx was identified using a custom algorithm programmed in Matlab (MathWorks Inc., Natick, MA) and manually aligned to the T2-weighted image using tkregister2. The DCE was intensity-normalized as described above for the T2-weighted images.
Tissue Processing
Following surgery, prostate samples were sectioned using patient-specific tissue slicing molds created from the presurgical T2 images, as previously published (29, 32). Surface models were created from the manually drawn prostate mask using 3D slicer and subtracted from a template-slicing mold matching the T2 slice spacing using Blender. The slicing molds were then 3D-printed using a fifth-generation Makerbot. Tissue sections were paraffin-embedded, whole-mounted, and stained with hematoxylin-eosin. Slides were digitally scanned using a microscope equipped with an automated stage (Nikon Metrology, Brighton MI). The digitized histology was annotated by a fellowship-trained urologic pathologist (KAI) using color codes corresponding to the Gleason grading system. Annotations were drawn on a Microsoft Surface Pro 4 using a predefined color palette. The annotations were saved as a mask overlaid on the high-resolution histology.
Tissue Alignment
Digitized samples were aligned to the T2-weighted images using custom software previously published (29, 32). Control points were manually placed on analogous points on both the histology and the MRI. A nonlinear transform was then calculated to warp the histology into T2 space using the imwarp command in the Matlab image processing toolbox (MathWorks Inc.). The annotations from our pathologist were likewise transformed into T2 space using the same transform and a nearest-neighbor interpolation. The pathologist annotations in MRI space are referred to as “deep annotations” throughout the manuscript.
Stratification by Tumor Volume
A 3-fold cross-validation approach was chosen to test generalizability. A custom sampling algorithm was required to distribute patients into cohorts with balanced tumor burden. Patient-wise tumor burden was calculated as the sum of the number of pathologist-annotated high grade (Gleason 4-5) and low grade (Gleason 3) voxels. Stratification by high grade and low grade was chosen in lieu of individual Gleason grade owing to the relatively limited amount of grade 5 tumor in the data set. The ideal cohort tumor burden was calculated as the total tumor burden divided by the number of cohorts. Random permutations of patients were created, and the difference between the randomly generated tumor burden and the ideal tumor burden was calculated. A separate error metric is calculated for each cohort (n) and then summed to produce a single error score for that permutation as seen in equation 1:
| (1) |
The cost function was empirically set at 3:1, favoring high-grade tumors; this results in a much larger error score if the high-grade tumor volume is unbalanced. There were a larger number of low-grade annotations than high-grade ones in the data set, and balancing high-grade tumor volume was deemed more important than balancing low grade. The permutation producing the lowest error in 10,000 iterations was used as the group assignment for this study. There are approximately 1021 possible combinations, thus sampling 10,000 limits bias but provides relatively balanced cohorts.
Global Thresholding and Segmentation
Each of the 3 cohorts was used as a test set for an algorithm trained on the other 2 cohorts. Three sets of global thresholds were established; for each cohort a global threshold was established using the 2 unused cohorts (32 patients). The contrasts used in this study were ADC (b = 0 and b = 1000), ADC (b = 50 and b = 2000), T2, and DCE. Global thresholds were created using Otsu's method calculated on all voxels in the manually drawn prostate masks for the entire training cohort of 32 patients; thresholds applied to the test set were not generated from these data (33). Global thresholding tests the assumption that all cohorts represent the same probability density function and additionally removes the constraint that each patient expresses each unique image characteristic. Images were segmented using the calculated thresholds into dark, intermediate, and bright intensities represented by values of 1, 2, and 3 respectively.
Generation of Radiomic Profiles
A unique code was created for each voxel by linearly combining the segmented image contrasts. Radiomic profiles were created by multiplying the segmented contrasts by ascending powers of 10 such that each digit represents the segmentation value of that individual contrast. With 4 image contrasts with 3 color values each a total of 81 radiomic profiles are possible; a voxel encoded with 1133 contains dark ADCshort, dark ADClong, bright T2, and bright DCE. A schematic demonstrating the generation of the radiomic profiles is shown in Figure 1 (26).
Figure 1.
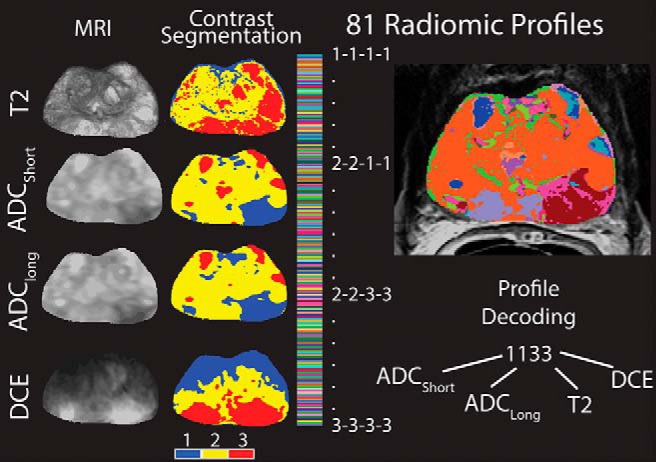
Generation of Radiomic profiles. Left: The 4 contrasts included in this study and the resulting segmentations created using Otsu's method. Right: 81 unique image characteristics created by linearly combining the segmented image contrasts. Each voxel receives a 4-digit code representative of the segmented image contrasts. Code 1133 indicates dark ADCshort, dark ADClong, bright T2, and bright DCE.
Training Set Independence
To determine the training set independence, imaging from an additional 5 patients not included in the previous analysis was processed. Three sets of radiomic profiles were generated using each of the 3 sets of thresholds calculated prior. Each pixel within the prostate mask was analyzed to quantify the overlap among the 3 sets of images. Pixels where the radiomic profile was identical across all images were labelled 3, and pixels where only 2 images matched were labelled as 2. A high overlap score (a large percentage of voxels with a value of 2 or 3) would indicate stable performance regardless of training set.
Gleason Probability Maps Generation
A probability table was generated by analyzing the distribution of each unique image signature within the pathologist-annotated regions. The distribution of each profile in the training set is recorded for low grade, high grade, and benign atrophy (ie, profile 1111 contains benign atrophy 75% of the time). The probability distribution is then propagated to the test set, where each profile is replaced by its respective percentage value, creating 4 maps depicting low grade, high grade, benign, and cancer likelihood. Figure 2 shows the generation of the probability table and Gleason probability maps.
Figure 2.
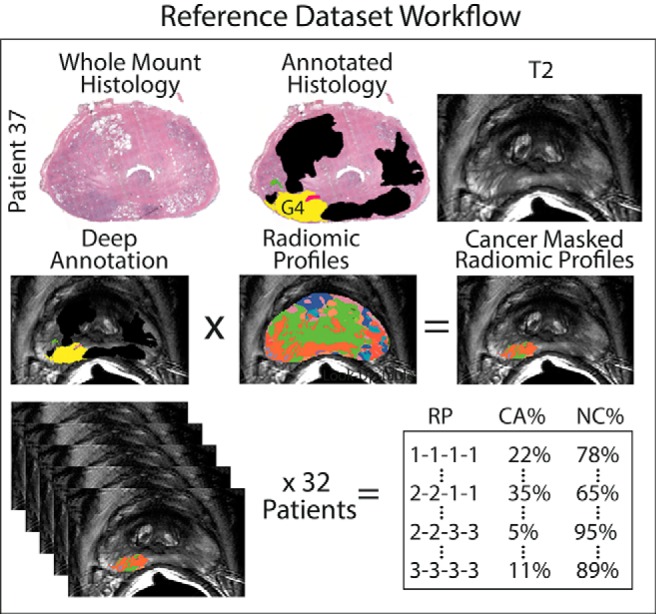
Generation of Gleason probability maps from a training data set. Top: Hematoxylin and eosin stained whole mount histology and the corresponding pathologist annotations and T2 slice. Middle: Radiomic profiles are masked by the pathologist annotations and the distribution of the radiomic profiles. Bottom: The distribution of radiomic profiles within high grade, low grade, and benign regions are analyzed over 32 patients. The resulting probability values are applied to the radiomic profiling images on naïve data to create Gleason probability maps.
Zone Dependence
The imaging signatures of prostate cancer are known to be zone-dependent. Lesions are evaluated via the PI-RADS scale using primarily the T2-weighted images in the transition zone and DWI in the peripheral zone. To test the robustness of the Gleason probability maps to tumor location, additional probability tables were created stratified by zone (ie, profile 3333 contains high-grade tumor in 3% of the voxels located in the peripheral zone and 0% of the voxels in the transition zone) and applied to the test set. These new maps were evaluated using a receiver operator characteristic (ROC) and compared to the nonzone-dependent maps.
Evaluation of Gleason Probability Maps and Comparison to Clinical Imaging
The Gleason probability maps were evaluated lesion-wise using a ROC analysis. High grade was compared to all other tissue, cancer to all other tissue, and high grade to low grade. In addition, the intensity normalized image contrasts were evaluated for their ability to distinguish high-grade cancer from all other tissue and high grade from low grade.
Results
Patient Stratification
Patients were stratified pseudorandomly, attempting to match an empirically determined ideal tumor burden using a custom sampling algorithm. The high-grade tumor and low-grade tumor burdens were, on average, 7.3% and 19.4% off the calculated ideal tumor burden.
Training Set Independence
Radiomic profile maps generated using thresholds from 3 patients cohorts were compared for overlap on data from 5 patients not included in the study. The individual radiomic profile maps and the overlap map can be seen in Figure 3. At least 2 cohorts had identical radiomic profile values on 98.6% of the pixels in the additional subject (red and yellow areas), and all 3 cohorts agreed on 76.3% of voxels (yellow only).
Figure 3.
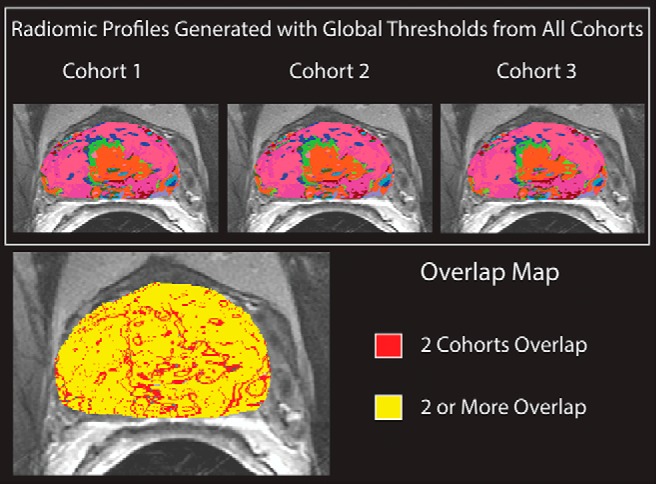
Top: Radiomic profiles generated with global thresholds calculated on 3 different training sets, applied to a patient not otherwise included in the analysis. Bottom: Overlap map: yellow pixels have an identical image signature across all cohorts, red and yellow pixels have identical image signatures in 2 cohorts. Blank pixels have no overlap.
Zone Independence
The zone dependence of the technique was tested using a ROC. The resulting AUCs can be seen in Table 1. The algorithms' performance was nearly identical when zone information was included; however, the addition of zone information requires manually drawn zone masks. Figure 4 shows the high-grade Gleason probability map on the same patient with and without zone information included.
Table 1.
Comparison of ROC AUC in Gleason Probability Maps Made With and Without the Inclusion of Zone Information in the Probability Table
| Zones | No Zones | |
|---|---|---|
| High Grade vs All | 0.76 | 0.77 |
| Cancer vs All | 0.79 | 0.77 |
| Normal vs All | 0.79 | 0.79 |
Figure 4.
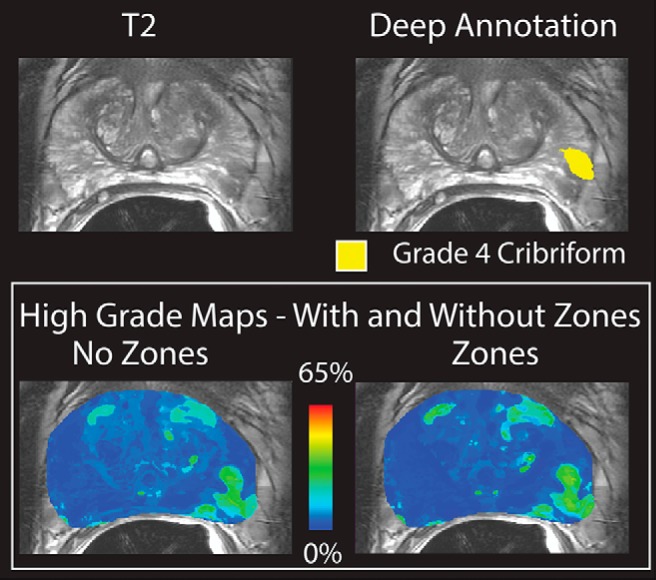
Top: T2-weighted image and deep annotation overlaid on the same slice. A grade 4 cribriform tumor is shown in yellow. Bottom: Gleason probability maps created with and without the inclusion of zone information in the training data set. The images are nearly identical and the tumor is clearly visible.
Receiver Operator Characteristic
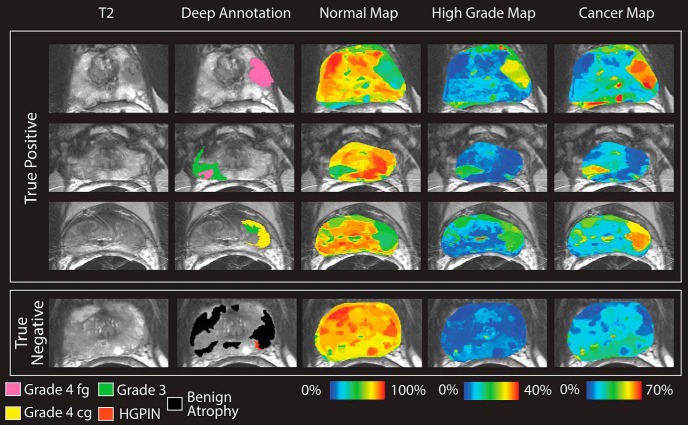
The resulting ROCs can be seen in Table 2 along with the difference between the 2 conditions. The AUC of the clinical contrasts alone ranged from 0.55 to 0.78. The Gleason probability maps achieved an AUC of 0.79, distinguishing cancer from benign atrophy. Figure 5 shows ROCs comparing the Gleason probability maps to the raw image contrasts used in the analysis. Figure 6 shows the resulting Gleason probability maps on 3 true-positive and 1 true-negative case.
Table 2.
Comparison of ROC AUC in Gleason Probability Maps and Clinical Image Contrasts
| Cancer vs Benign | High Grade vs Low Grade | |
|---|---|---|
| T2 | 0.58 | 0.53 |
| ADC 0-1000 | 0.77 | 0.58 |
| ADC 50-2000 | 0.78 | 0.60 |
| DCE | 0.65 | 0.51 |
| Gleason Probability Map | 0.79 | 0.56 |
Both cancer versus benign and high grade vs low grade were tested.
Figure 5.
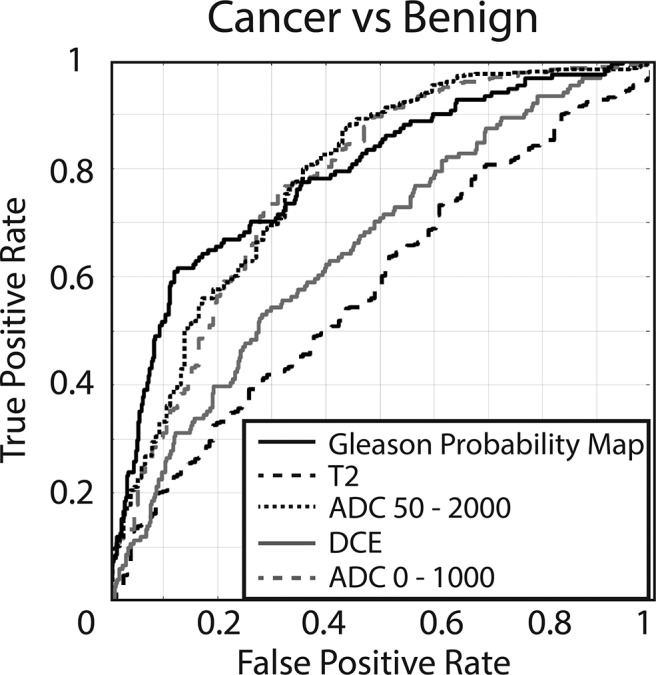
Receiver operator characteristic (ROC) evaluating the performance of the 4 raw image contrasts compared to Gleason probability maps (cancer probability). ADC 50-2000 = 0.78, ADC 0-1000 = 0.77, DCE = 0.65, T2 = 0.57, Gleason probability map = 0.79.
Figure 6.
Gleason probability maps. Top: True-positive cases. High-grade tumors are shown on the deep annotation in pink (cribriform) and yellow (not cribriform). Low-grade tumors are shown in green. Images are scaled to reflect the maximum probability in the training data set. Bottom: True negative. The displayed slide has only benign atrophy, and thus, no hot spots occur in the Gleason probability maps.
Image Signatures Unique to Low- and High-Grade Tumors
The 10 most common profiles by volume seen in both high- and low-grade cancer can be seen in Table 3. Seven of the top 10 most frequently seen profiles in high-grade cancer also frequently appear in low-grade cancer. Approximately 13,000 voxels containing high-grade tumor are explained by profile 1122, making it the most frequently seen high-grade image signature. That profile is the fifth most common low-grade profile, but the volume is nearly identical at 12,000 voxels, resulting in a similar probability that a voxel containing 1122 in the test set is high grade or low grade.
Table 3.
Top 10 Most Common Radiomic Profiles in High- and Low-Grade Lesions Ordered by Volume
| Low Grade | High Grade | ||
|---|---|---|---|
| Volume | Profilea | Volume | Profile |
| 22 179 | 2222 | 13 572 | 1122 |
| 17 900 | 1123 | 12 146 | 1112 |
| 16 302 | 1112 | 9871 | 1132 |
| 13 609 | 2212 | 8470 | 1123 |
| 13 060 | 1122 | 8356 | 1111 |
| 12 651 | 2223 | 8323 | 1121 |
| 9159 | 1111 | 7800 | 2211 |
| 8972 | 1121 | 5873 | 2221 |
| 8780 | 2221 | 5452 | 1133 |
| 8712 | 2211 | 5157 | 2222 |
a Profiles that are common between the two are shown in italics on the low-grade profile list.
The similarity in image characteristics between high- and low-grade tumors occurs independent of lesion size. Normal and benign regions, on average, exhibit image signature 2222. Lesions less than 200 contiguous voxels (<12.5 mm2 in-plane) exhibit an identical image signature—these lesions are indistinguishable from normal tissue. Large lesions exhibit profile 1122 regardless of final Gleason grade.
Discussion
This study translated a technique developed as a risk stratification model in glioblastoma (26) to identify unique image signatures associated with prostate cancer. The technique outperforms the diagnostic capacity of each of the clinical images individually (Figure 5) and brings histologic data in the form of a learned probability distribution of unique image signatures into image space on naive data. Patients were successfully stratified pseudorandomly into cohorts with roughly equivalent volumes of high- and low-grade prostate cancer. Gleason probability mapping produces nearly identical results independent of training cohort and functions without requiring zone information.
Other prostate radiomics approaches have seen success in detecting prostate cancer using MP-MRI (34, 35). These tools rely either on confirmed pathologically radiologist ROIs or aligned, annotated whole mount slides and frequently combine first-order histogram features with texture and volume features to create a single risk score rather than applying global thresholds. Many volume- and histogram-based features require an a priori ROI in the test set, whereas pixel-wise approaches can provide both disease severity and detection. These tools have trended toward aligned whole mount histology which allows voxel-wise predictions and eliminates the need for a radiologist to manually draw ROIs. Reported AUC varies by technique and ranges from 0.76 to 0.99; however, all published techniques distinguish cancer (G3+) from benign or healthy tissue. Notably, to the best of our knowledge, no technique has been published to date that is capable of distinguishing G3 and G4 patterned lesions, which is where the clinical decision is often made, as it pertains to choosing active surveillance versus definitive therapy. The method introduced in this study, Gleason probability mapping, likewise performs poorly at distinguishing high-grade tumor from low-grade tumor, likely because of the limitations of the imaging techniques themselves. Future studies focused specifically on differentiating G3 from G4+ need to occur.
Transition zone lesions are identified primarily by the T2-weighted images in PI-RADS v2 because benign prostatic hyperplasia nodules often exhibit diffusion restriction and early/contemporaneous enhancement (mimicking cancer) but appear morphologically different from significant cancers, which may not be captured fully by a segmentation-based method. Currently, no prostate CAD tool reads a prostate exam like a radiologist—techniques are either contrast-based and well suited to identify peripheral zone lesions or texture-based and well suited to identify transition zone lesions. It is plausible that the most effective method of identifying prostate tumors distinguished by zone and uses vastly different techniques depending on the lesions location.
There are known sources of error associated with rad-path studies that may reduce accuracy. Our sample includes patients with cancer: other confounding diseases are unlabeled and may thus contribute to error. While we have previously validated our control point–warping technique, there is still error involved in the process. This technique used global thresholds generated from 1 set of acquisitions on similar magnets. Future studies should validate these thresholds on acquisitions from magnets by other manufacturers. This study is limited to endorectal coil images, but future studies may quantify the generalizability to a population imaged without an endorectal coil.
Conclusions
Gleason probability mapping stratifies cancer tissue from normal prostate tissue independent of zone and training set. The technique performs better than traditional image contrasts alone and provides a voxelwise map which may be potentially useful for biopsy guidance and reading clinical scans. Additional research is necessary to further classify regions of tumor among the different Gleason patterns.
Acknowledgments
We would like to thank the patients who graciously participated in our study and Eugenia Westfall for helpful conversations.
This study was funded by The State of Wisconsin Tax Check-off Program for Prostate Cancer Research, RO1CA218144, R01CA113580, National Center for Advancing Translational Sciences, NIH UL1TR001436 and TL1TR001437, 1R21CA231892-01.
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- MRI
- Magnetic resonance imaging
- PSA
- prostate-specific antigen
- MP-MRI
- multiparametric MRI
- DCE
- dynamic contrast-enhanced
- FOV
- field of view
- ROC
- receiver operator characteristic
- AUC
- area under the curve
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Hugosson J, Carlsson S, Aus G, Bergdahl S, Khatami A, Lodding P, Pihl CG, Stranne J, Holmberg E, Lilja H. Mortality results from the Goteborg randomised population-based prostate-cancer screening trial. Lancet Oncol. 2010;11:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schroder FH, Roobol MJ. Defining the optimal prostate-specific antigen threshold for the diagnosis of prostate cancer. Curr Opin Urol. 2009;19:227–231. [DOI] [PubMed] [Google Scholar]
- 4. Schroder FH, van den Bergh RC, Wolters T, van Leeuwen PJ, Bangma CH, van der Kwast TH, Roobol MJ. Eleven-year outcome of patients with prostate cancers diagnosed during screening after initial negative sextant biopsies. Eur Urol. 2010;57:256–266. [DOI] [PubMed] [Google Scholar]
- 5. Loeb S, Bruinsma SM, Nicholson J, Briganti A, Pickles T, Kakehi Y, Carlsson SV, Roobol MJ. Active surveillance for prostate cancer: a systematic review of clinicopathologic variables and biomarkers for risk stratification. Eur Urol. 2015;67:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mufarrij P, Sankin A, Godoy G, Lepor H. Pathologic outcomes of candidates for active surveillance undergoing radical prostatectomy. Urology. 2010;76:689–692. [DOI] [PubMed] [Google Scholar]
- 7. Serefoglu EC, Altinova S, Ugras NS, Akincioglu E, Asil E, Balbay MD. How reliable is 12-core prostate biopsy procedure in the detection of prostate cancer? Can Urol Assoc J. 2013;7:E293–E298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG, Freeman A, Kirkham AP, Oldroyd R, Parker C, Emberton M; PROMIS study group. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–822. [DOI] [PubMed] [Google Scholar]
- 9. Kasivisvanathan V, Emberton M, Moore CM. MRI-targeted biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;379:589–590. [DOI] [PubMed] [Google Scholar]
- 10. Vargas HA, Hotker AM, Goldman DA, Moskowitz CS, Gondo T, Matsumoto K, Ehdaie B, Woo S, Fine SW, Reuter VE, Sala E, Hricak H. Updated prostate imaging reporting and data system (PIRADS v2) recommendations for the detection of clinically significant prostate cancer using multiparametric MRI: critical evaluation using whole-mount pathology as standard of reference. Eur Radiol. 2016;26:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenkrantz AB, Taneja SS. Prostate MRI can reduce overdiagnosis and overtreatment of prostate cancer. Acad Radiol. 2015;22:1000–1006. [DOI] [PubMed] [Google Scholar]
- 12. Chatterjee A, Bourne RM, Wang S, Devaraj A, Gallan AJ, Antic T, Karczmar GS, Oto A. Diagnosis of prostate cancer with noninvasive estimation of prostate tissue composition by using hybrid multidimensional MR imaging: a feasibility study. Radiology. 2018;287:864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Litjens GJ, Elliott R, Shih NN, Feldman MD, Kobus T, Hulsbergen-van de Kaa C, Barentsz JO, Huisman HJ, Madabhushi A. Computer-extracted features can distinguish noncancerous confounding disease from prostatic adenocarcinoma at multiparametric MR imaging. Radiology. 2016;278:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ozer S, Langer DL, Liu X, Haider MA, van der Kwast TH, Evans AJ, Yang Y, Wernick MN, Yetik IS. Supervised and unsupervised methods for prostate cancer segmentation with multispectral MRI. Med Phys. 2010;37:1873–1883. [DOI] [PubMed] [Google Scholar]
- 15. Niaf E, Lartizien C, Bratan F, Roche L, Rabilloud M, Mege-Lechevallier F, Rouviere O. Prostate focal peripheral zone lesions: characterization at multiparametric MR imaging–influence of a computer-aided diagnosis system. Radiology. 2014;271:761–769. [DOI] [PubMed] [Google Scholar]
- 16. Viswanath S, Bloch BN, Rosen M, Chappelow J, Toth R, Rofsky N, Lenkinski R, Genega E, Kalyanpur A, Madabhushi A. Integrating structural and functional imaging for computer assisted detection of prostate cancer on multi-protocol in vivo 3 Tesla MRI. Proc SPIE Int Soc Opt Eng. 2009;7260:72603I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vos PC, Hambrock T, Hulsbergen-van de Kaa CA, Futterer JJ, Barentsz JO, Huisman HJ. Computerized analysis of prostate lesions in the peripheral zone using dynamic contrast enhanced MRI. Med Phys. 2008;35:888–899. [DOI] [PubMed] [Google Scholar]
- 18. Viswanath S, Bloch BN, Chappelow J, Patel P, Rofsky N, Lenkinski R, Genega E, Madabhushi A. Enhanced multi-protocol analysis via intelligent supervised embedding (EMPrAvISE): detecting prostate cancer on multi-parametric MRI. Proc SPIE Int Soc Opt Eng. 2011;7963:79630U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60:5471–5496. [DOI] [PubMed] [Google Scholar]
- 24. Cameron A, Khalvati F, Haider MA, Wong A. MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng. 2016;63:1145–1156. [DOI] [PubMed] [Google Scholar]
- 25. Kickingereder P, Burth S, Wick A, Gotz M, Eidel O, Schlemmer HP, Maier-Hein KH, Wick W, Bendszus M, Radbruch A, Bonekamp D. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280:880–889. [DOI] [PubMed] [Google Scholar]
- 26. McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, Schmainda KM, LaViolette PS. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ellingson BM, Kim HJ, Woodworth DC, Pope WB, Cloughesy JN, Harris RJ, Lai A, Nghiemphu PL, Cloughesy TF. Recurrent glioblastoma treated with bevacizumab: contrast-enhanced T1-weighted subtraction maps improve tumor delineation and aid prediction of survival in a multicenter clinical trial. Radiology. 2014;271:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellingson BM, Zaw T, Cloughesy TF, Naeini KM, Lalezari S, Mong S, Lai A, Nghiemphu PL, Pope WB. Comparison between intensity normalization techniques for dynamic susceptibility contrast (DSC)-MRI estimates of cerebral blood volume (CBV) in human gliomas. J Magn Reson Imaging. 2012;35:1472–1477. [DOI] [PubMed] [Google Scholar]
- 29. McGarry SD, Hurrell SL, Iczkowski KA, Hall W, Kaczmarowski AL, Banerjee A, Keuter T, Jacobsohn K, Bukowy JD, Nevalainen MT, Hohenwalter MD, See WA, LaViolette PS. Radio-pathomic maps of epithelium and lumen density predict the location of high-grade prostate cancer. Int J Radiat Oncol Biol Phys. 2018;101:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. [DOI] [PubMed] [Google Scholar]
- 31. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. [DOI] [PubMed] [Google Scholar]
- 32. Hurrell SL, McGarry SD, Kaczmarowski A, Iczkowski KA, Jacobsohn K, Hohenwalter MD, Hall WA, See WA, Banerjee A, Charles DK, Nevalainen MT, Mackinnon AC, LaViolette PS. Optimized b-value selection for the discrimination of prostate cancer grades, including the cribriform pattern, using diffusion weighted imaging. J Med Imaging (Bellingham). 2018;5:011004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 34. Smith CP, Czarniecki M, Mehralivand S, Stoyanova R, Choyke PL, Harmon S, Turkbey B. Radiomics and radiogenomics of prostate cancer. Abdom Radiol. 2018. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 35. Stoyanova R, Takhar M, Tschudi Y, Ford JC, Solorzano G, Erho N, Balagurunathan Y, Punnen S, Davicioni E, Gillies RJ, Pollack A. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res. 2016;5:432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]