Abstract
Medical imaging is critical for assessing the response of patients to new cancer therapies. Quantitative lesion assessment on images is time-consuming, and adopting new promising quantitative imaging biomarkers of response in clinical trials is challenging. The electronic Physician Annotation Device (ePAD) is a freely available web-based zero-footprint software application for viewing, annotation, and quantitative analysis of radiology images designed to meet the challenges of quantitative evaluation of cancer lesions. For imaging researchers, ePAD calculates a variety of quantitative imaging biomarkers that they can analyze and compare in ePAD to identify potential candidates as surrogate endpoints in clinical trials. For clinicians, ePAD provides clinical decision support tools for evaluating cancer response through reports summarizing changes in tumor burden based on different imaging biomarkers. As a workflow management and study oversight tool, ePAD lets clinical trial project administrators create worklists for users and oversee the progress of annotations created by research groups. To support interoperability of image annotations, ePAD writes all image annotations and results of quantitative imaging analyses in standardized file formats, and it supports migration of annotations from various propriety formats. ePAD also provides a plugin architecture supporting MATLAB server-side modules in addition to client-side plugins, permitting the community to extend the ePAD platform in various ways for new cancer use cases. We present an overview of ePAD as a platform for medical image annotation and quantitative analysis. We also discuss use cases and collaborations with different groups in the Quantitative Imaging Network and future directions.
Keywords: medical image annotation, biomarker evaluation, feature extraction, AIM (Annotation and Image Markup), DICOM SR (DICOM Structure Report)
Introduction
Advances in molecular medicine are providing many new treatments that promise to be safer and more effective than traditional cytotoxic treatments by targeting the molecular characteristics of each patient's tumor (1–3). As these new targeted treatments enter clinical trials, there is a growing need to derive quantitative characteristics from images of cancer lesions (“quantitative imaging biomarkers”) that accurately assess the clinical benefit of these treatments (surrogate endpoints in clinical trials). Tumor shrinkage is the hallmark of response to traditional cytotoxic cancer therapies (4), and thus linear measurement of target lesions is the imaging biomarker used in most clinical trials using criteria such as Response Evaluation Criteria in Solid Tumors (RECIST) (5–7), Response Assessment in Neuro-Oncology (RANO) (8, 9), and International Harmonization Criteria (10). However, targeted, noncytotoxic therapies may arrest cancer growth and improve progression-free survival without necessarily shrinking tumors (11–14). Simple linear measurement may underestimate treatment response (15–18), in addition to having other limitations (7, 19). Alternative imaging biomarkers may be more promising than linear measurement for assessing response, especially with targeted therapeutic agents, as they can capture specific imaging features related to biological alterations in tumors during treatment (eg, heterogeneity, hypoxia, or changes in tumor microenvironment) (20–24), unlike tumor shrinkage (15, 25–27). Indeed, quantitative imaging biomarkers that reliably detect the results of anticancer agents (as opposed to detecting only change in tumor size) are desirable for all classes of therapeutic agents (28). Such new imaging biomarkers could become surrogate endpoints in clinical trials, as regulatory approval can be based on surrogate endpoints that document clinical benefit (29).
Development of imaging biomarkers follows a life cycle, starting with discovery and validation (“emerging biomarkers”), then translation and incorporation into clinical trials, and eventually to qualification for clinical use as surrogate endpoints for evaluating treatments (“qualified biomarkers”) (30). A number of research groups are working on the discovery/validation of the spectrum and developing new quantitative imaging biomarkers, including the Quantitative Imaging Network (QIN) (31) and the broader community (32–39). On the translation end of the spectrum, many of the new imaging biomarkers are ready to be translated for use in clinical trials, such as tumor volume (40), changes in contrast enhancement on computed tomography (41), radiotracer uptake on positron emission tomography (PET) (32, 42–46), kinetic parameters in dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) (47–49), and spatial maps of such parameters (50, 51); however, very few of these new imaging biomarkers have yet to be incorporated into clinical trials for assessing treatment response.
Current image viewing and annotation tools are limited in their ability to support incorporating new imaging biomarkers into clinical trials in 4 major ways. First, although there are several commercial and open-source tools available to assess cancer lesions (52–55), they generally support very few measures of cancer lesions, such as linear dimension of target lesions, and they cannot be readily extended to deploy novel imaging biomarkers. Newer algorithms for computing imaging biomarkers are generally written in a variety of languages such as Java, Python, and C/C++, or exist within single toolkits [eg, MATLAB and 3D Slicer (56, 57)], which may not be compatible with current image assessment tools. Second, current lesion assessment tools are designed for only tracking cancer lesions in clinical practice, and they generally do not provide workflow management and study oversight features needed for assessing new image biomarkers in clinical trials. Third, there are no decision tools that use new imaging biomarkers for assessing treatment response in patients and overall drug effectiveness in clinical trial cohorts. Such decision-making requires calculating a variety of response measures in patients and across cohorts—tasks generally done by hand, making it difficult to compare multiple alternative imaging biomarkers. Fourth, it is costly and difficult to accrue aggregate data needed to qualify new imaging biomarkers as surrogate endpoints for clinical trials (58). Qualification of new imaging biomarkers requires collecting context-specific assessments of the performance of the biomarker relative to clinical outcomes (59). It is challenging to acquire sufficient data that link imaging biomarker data with clinical outcomes, such as survival (60). Efforts such as the Quantitative Imaging Biomarker Alliance (QIBA) are creating consensus on processes to qualify new imaging biomarkers (61), but their ultimate success depends on expanding public data sets (62) and leveraging many studies from individual laboratories and cooperative groups, which currently cannot be repurposed for this task because image annotations (or biomarker values extracted from them) are not recorded in standardized formats.
We developed ePAD—one of the research projects of the QIN—to address all of these challenges by developing a modular software platform integrating image viewing with computation of emerging and validated quantitative imaging biomarkers, facilitating translation of novel biomarkers into clinical trials as surrogate endpoints. In this paper, we will present ePAD's core architecture and describe the ways in which it meets the foregoing challenges. We also describe active research projects that are leveraging ePAD.
The ePAD Platform
We describe the design of ePAD and its core architecture, presenting this information from 4 different perspectives that address 4 major challenges mentioned above: (1) as a platform enabling the computing of novel imaging biomarkers of cancer treatment response, (2) as a workflow management and study oversight tool enabling the oversight for assessing new image biomarkers in clinical trials, (3) as a clinical decision support tool for the treatment response assessment using current and new imaging biomarkers, and (4) as infrastructure to permit researchers to aggregate evidence needed to show that new imaging biomarkers predict survival, which can be useful in qualifying them as surrogate endpoints in clinical trials.
Image Annotations in ePAD
A key distinguishing feature of ePAD is its support for standardized formats for image annotations, specifically Annotation and Image Markup (AIM) (63) and DICOM segmentation objects (64). AIM is an information model developed by the National Cancer Imaging Program of NCI for storing and sharing image metadata (65–67), such as lesion identification, location, size measurements, regions of interest (ROIs), radiologist observations, anatomic locations of abnormalities, calculations, inferences, and other qualitative and quantitative image features. The image metadata also include information about the image, such as the name of imaging procedure and how or when the image was acquired. AIM supports controlled terminologies, enabling semantic interoperability. In the use case of lesion annotation in cancer, the value of AIM is recording lesion identifiers (enabling unambiguous tracking of lesions across longitudinal images), anatomic locations of lesions, lesion types (target, nontarget, new lesion, or resolved lesion), and study types (baseline or follow up). This semantic information is critical for automating the generation of tabular summaries of lesions, and it also enables automating comparing the response assessment in patients according to different imaging biomarkers (see Section “Clinical Decision Support Tool for Treatment Response Assessment”). AIM has recently been incorporated into DICOM Structured Report (DICOM SR) (68), with specifications for saving AIM in DICOM-SR (69).
Architecture of ePAD
ePAD Components.
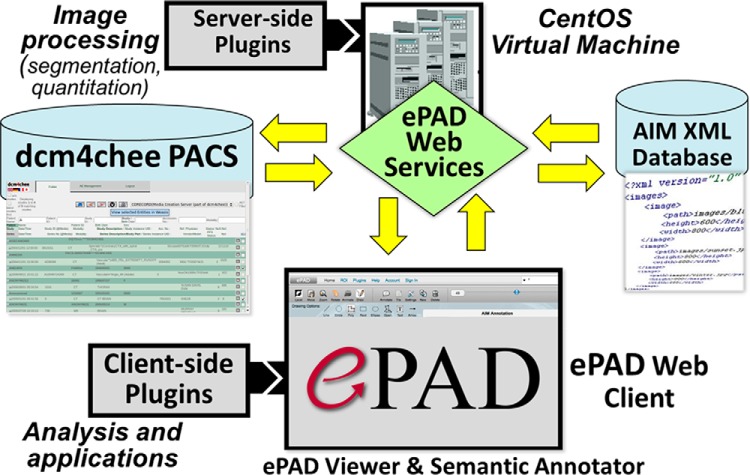
ePAD (70–72) is a freely available quantitative imaging informatics platform (http://epad.stanford.edu) distributed as a virtual machine or as Docker containers. Users can download virtual machine or Docker version of ePAD and host it in their own environment. This enables them to restrict the access to their private networks, typically to the hospital network. These machines generally do not have access to the internet. The core architecture of ePAD is shown in Figure 1. The ePAD platform comprises the following 5 main components: (1) the ePAD viewer, a zero-footprint web image viewer and image annotator, (2) ePAD web services, providing a programming interface to ePAD services, (3) an image database, (4) an image annotation database, and (5) plugin modules (server-side and client-side for extending the ePAD platform). The image database, image annotation database, and ePAD web services comprise the “back end” of ePAD. The ePAD plugin modules extend the functionality of ePAD, and while most of the plugins described below were developed by us, we also describe several developed by the community, and such community engagement will enable ePAD to foster an ecosystem enabling continued evolution of the platform to meet the needs of researchers broadly.
Figure 1.
Architecture of the ePAD platform, which comprises an image database (dcm4chee PACS as a cache for images), a database of image annotations (AIM XML database), the ePAD Viewer (a web application), ePAD Web Services that communicate data between the image and annotation databases and the ePAD Viewer, and back-end (server-side) and front-end (client-side) plugins enabling the community to extend the ePAD platform.
1. ePAD Viewer.
The ePAD viewer is a web application providing the look and feel of a Picture Archiving and Communication System (PACS) to the user, who can browse patient studies and open them to view images. To display images, the ePAD viewer queries an embedded PACS database [dcm4chee (73)] and stores image annotations in ePAD's annotation database. The ePAD viewer was written using HTML5 (74), Java, JavaScript, and the Google Web Toolkit (http://www.gwtproject.org), which supports image rendering with controls for image display (eg, zooming, panning, and window/level) within the Web browser. Drawing and editing image annotations are accomplished with HTML5 Scalable Vector Graphics (SVG).
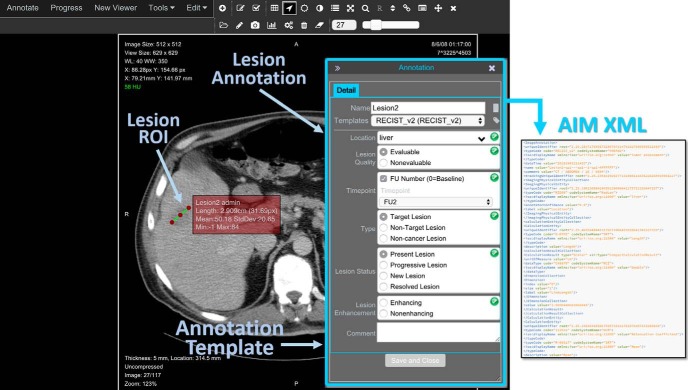
An important component of the ePAD viewer is its image annotation window (Figure 2). The ePAD viewer ensures the minimum information necessary to create a meaningful image annotation is collected from the user: the lesion name, the lesion type (target, nontarget, new lesion, or resolved lesion) and the anatomic location of the lesion, and the study time point (baseline or follow-up). The ePAD viewer automatically labels each lesion with a name (eg, “Lesion1”) to enable unambiguous determination of the same lesion on serial imaging studies (75). To specify the content of annotations, ePAD uses AIM templates (76) that are created by a separate freely available application. AIM templates specify the data elements to be provided by the user when making image annotations. All answer choices in ePAD templates are controlled terminology lists such as RadLex (77). The ePAD viewer prompts the user if certain values in the templates are inconsistent or incomplete (66). The ePAD viewer permits creating 2 types of ROI, coordinate based and pixel map based. The former is saved as coordinates in the AIM file (63), and the latter is saved as a DICOM segmentation object (64).
Figure 2.
ePAD viewer and annotation window. Images are displayed in the ePAD web viewer, and the user records image annotations in using drawing tools (eg, to create an ROI, shown on the left) and an annotation window (to record qualitative image features, shown on right).
2. ePAD Web Services.
The ePAD viewer uses a set of RESTful Web services (78) to communicate with the back end of ePAD to retrieve images and save image annotations, as well as authenticating user credentials and invoking image calculation methods that need to be executed on the server. The ePAD Web Services provides programmatic access to the image database and the annotation database that are components of the ePAD back end (Figure 1). The ePAD Web Services is typically hosted on a server that resides within an institution's firewall so that all traffic between the ePAD viewer and the ePAD Web Services resides within the institution's Intranet. Thus, users can use ePAD to evaluate image data containing protected health information, provided the network on which ePAD is hosted is secure. Another model for hosting ePAD is a centralized, hosted version, which could provide publicly available images (which should be deidentified for public dissemination). The ePAD Web Services are used by plugin developers to extend ePAD's functionality, either as client-side or as server-side plugins (Figure 1). Plugin developers can use the ePAD Web services to access annotations and images in their own applications or to provide extensions to the ePAD platform.
3. Image Database.
Medical images in DICOM format are managed by an open-source PACS called dcm4chee (73). This PACS contains a DICOM image receiver and a programming interface that permits the ePAD Web Services to manage imaging studies within ePAD. The DICOM image database provides a temporary storage depot for images for image display and annotation in ePAD. The AIM annotations and DICOM segmentation objects in ePAD are saved indefinitely, however, as these annotations comprise the user-generated data in ePAD. Because DICOM images are large, the ePAD back end converts them into a lossless compressed PNG image object (“packed PNG”) that takes each 16-bit pixel in a DICOM image and packs it into a PNG color channel before returning it to the ePAD viewer, where it is unpacked. This approach significantly reduces the volume of data provided by the server and speeds performance of the ePAD viewer. To further speed image display performance, ePAD supports the Web-Accessible DICOM Objects [WADO (79)] protocol to retrieve lossy JPG images, while the lossless packed PNGs are initially loading.
4. Annotation Database.
As the user makes annotations on images in ePAD viewer, it creates AIM files. All AIM annotations are stored in an XML database [eXist (80)]. The AIM annotation database is accessible via functions in the ePAD Web Services, and it is the key resource that ePAD queries for lesion tracking and summarizing longitudinal changes in cancer treatment response, as described in Section “Clinical Decision Support Tool for Treatment Response Assessment.”
5. Plugin Modules.
Developers can create server-side and client-side plugins to access the data collected by ePAD to provide a new functionality. The server-side code can be written in a variety of languages, such as MATLAB, python, C/C++, or Java. We and other groups have created plugins to build a variety of features to address the challenges of (1) computing novel imaging biomarkers of cancer treatment response, (2) providing workflow management and study oversight features for assessing new image biomarkers, (3) creating clinical decision support tools for treatment response assessment using current and new imaging biomarkers, and (4) permitting researchers to aggregate evidence needed to show that new imaging biomarkers predict survival, which can be useful in qualifying them as surrogate endpoints in clinical trials.
Plugins currently available in ePAD are listed in the following sections.
JJVector Feature Extraction Plugin.
JJVector is a 2D feature extraction plugin we developed that analyses closed-shape annotations and extracts 2D radiomics features based on the intensity values from the ROI and the surrounding tissue of its associated organ (81). The plugin saves the calculated feature values in an AIM file that can be downloaded in different formats from ePAD, such as an excel summary sheet to be used in other applications such as training machine learning models.
ADLA Biomarker Plugin.
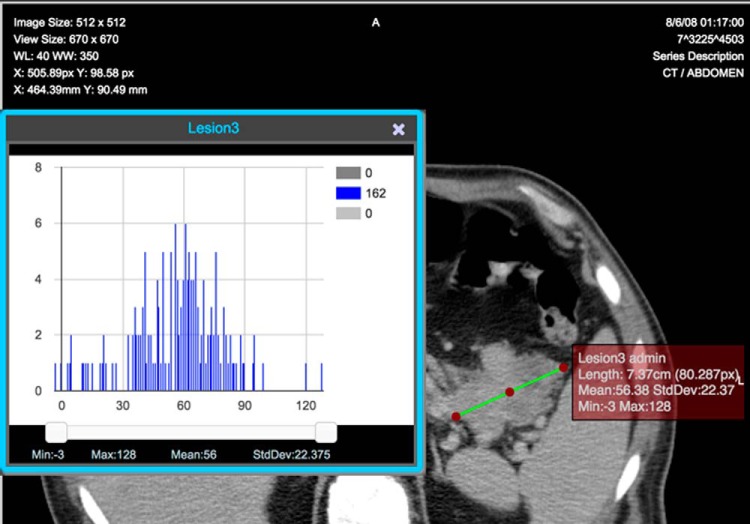
The Attenuation Distribution across the Long Axis (ADLA) plugin implements the ADLA semiquantitative imaging biomarker for assessing treatment response in solid malignancies and a measure of intralesional heterogeneity. We built this plugin in collaboration with prior works that created it (82, 83). ePAD calculates the standard deviation along the long axis to compute ADLA and saves it in the AIM file to be used for analyses such as response assessment as an alternative imaging biomarker. ePAD also generates an ADLA histogram of pixel values within the ROI when the long axis is selected (Figure 3).
Figure 3.
ADLA histogram on a line annotation on a cancer lesion created and visualized in ePAD.
Perfusion Analysis Plugin.
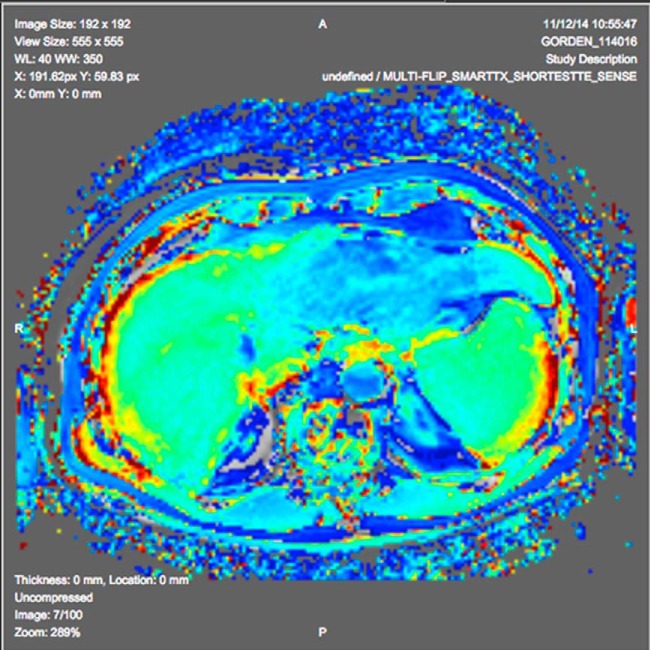
A contributor developed an ePAD plugin deploying an algorithm for computing T1 perfusion maps on dynamic contrast-enhanced studies based on his prior work (84). The plugin analyses the multiframe MRI images having different phases of dynamic contrast enhancement and calculates a T1 map for the imaged volume. The plugin scales the T1 map to 8 bits to save as a standard DICOM object (a probability DICOM Segmentation object) and paints the mask on the image using a color lookup table (Figure 4).
Figure 4.
T1 perfusion map generated by ePAD plugin derived from the algorithm in Jarrett et al.'s study (112), with the map overlaid on magnetic resonance (MR) image using a color lookup table.
Riesz Texture Feature Plugin.
A contributor developed an ePAD plugin that computes image texture features based on Riesz wavelets (85). The latter are a subtype of convolutional approaches that can quantify image derivatives of any order and at multiple scales. The image derivatives are aligned along dominant local orientations, allowing characterization of the local organization of the image direction, with invariance to the local orientation of anatomical structures. These image derivatives have an intuitive interpretation, and the Riesz features have shown to provide valuable imaging measurements in various medical applications.
Quantitative Image Feature Engine (QIFE).
QIFE is an open-source feature-extraction framework we created that computes 3D radiomics features for ROIs that are created as DICOM segmentation objects (86). ePAD stores these image features in an AIM file for further analysis in radiomics studies or as alternative imaging biomarkers of response.
Quantitative Feature Explore (QFExplore) Plugin Suite.
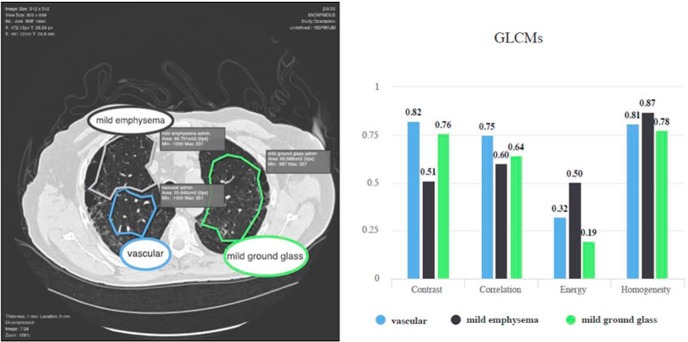
The Quantitative Feature Explore (QFExplore) is a suite of plugins we developed for the ePAD platform, enabling the exploration and validation of imaging biomarkers in a clinical environment (85). Imaging features that can be extracted using QFExplore include histogram bins of Pixel Intensity Distributions (PID), statistical moments of PIDs (ie, mean, standard deviation, skewness, kurtosis), gray-level co-occurrence matrices (GLCMs), and Riesz wavelets (87). Figure 5 illustrates QFExplore plugin suite's feature comparison functions in action. The ROIs are visualized on the left, while color-coded gray-level co-occurrence matrices values are displayed in a chart on the right.
Figure 5.
QF Explore Plugin Suite: gray-level co-occurrence matrix feature extraction and comparison chart. The user can compare the feature values for various regions of interest (ROIs). GLCM contrast and correlation is higher for vascular ROIs (85).
Quantitative Feature Pipeline (QIFP).
We created the QIFP, a cloud-based platform for building processing pipelines of image analysis algorithms (88). It provides a Docker library of image analysis algorithms for preprocessing, segmentation, and feature extraction that can be assembled into pipelines. The QIFP is integrated with ePAD so that any processing pipeline for generating quantitative imaging biomarkers can be executed in ePAD (or ePAD annotations can be consumed and used in QIFP processing pipelines).
ePAD Applications
ePAD includes several applications that are part of the platform and accessible via menu tabs in the ePAD viewer.
Computing and Comparing Imaging Biomarkers
A need that is critical for research is its ability to compute a variety of alternative imaging biomarkers besides linear dimension (used in RECIST and similar criteria). In a given clinical trial, patient response to treatment can be computed using a variety of imaging biomarkers, and a sizeable collection of data can be amassed if this is done across clinical trials that could ultimately be useful in comparing and evaluating alternative imaging biomarkers as secondary endpoints of response. Different imaging biomarker algorithms are written in different languages, and ePAD enables incorporating them into its image analysis workflow through its plugin mechanism described above. These plugins can execute source code modules written in MATLAB, Java, C/C++, or other languages, letting biomarker algorithm developers add their existing code to ePAD easily.
When users make annotations on images, ePAD automatically analyzes each annotation to generate the image biomarkers that the user chooses, and it saves them in AIM format. It also computes the minimum, maximum, standard deviation and mean for all the pixels that are inside the ROI. If the ROI is a line, ePAD calculates the length. If the ROI comprises 2 perpendicular lines, ePAD will calculate the length of the long axis and short axis. Additional features and biomarker candidates can be calculated by various plugins.
Workflow Management and Study Oversight
The ePAD viewer includes an application that provides a summary panel of annotations designed to streamline the task of summarizing for the radiologist all prior measurements and images in prior studies of each patient to convey the list of lesions previously measured, and which need to be measured on the current study. To populate this summary display, the ePAD viewer queries ePAD's annotation database to find all the lesions from the prior exams and list them for the user. This provides the user with a worklist of lesion measurements that need to be made for each imaging study. It also links each measurement to the image from which it was obtained. When the user clicks on a measurement, the corresponding image is retrieved and the measurement is displayed.
ePAD also facilitates oversight and managing image readings for clinical trial researchers and study administrators via user roles, worklists, and study progress monitoring. Project owners and administrators can create users and assign them specific roles to control their access to imaging data and annotations created by other users. Users or study supervisors can create worklists for people and assign to a reader. Using worklists allows the supervisors to divide the readings to multiple readers. A study progress monitoring application module in ePAD monitors the status of image annotations made in clinical trials and summarizes them in a table in the ePAD viewer. Study administrators can follow the image annotations made in multiple studies by group of users assigned to a particular study. The application can also track the progress of the annotation process by identifying which subjects/studies are fully annotated by all the annotators, which annotators have completed the annotation process for each subject and which subjects/studies have not yet been annotated yet (Figure 6). This functionality has been helpful the MGH/HST Martinos Center for Biomedical Imaging used this for MEDICI project (89), which used ePAD.
Figure 6.
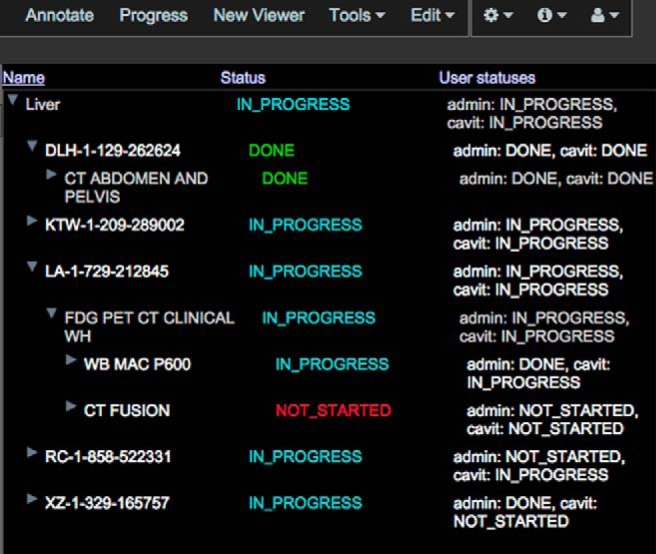
Progress view of ePAD visualizing a particular project (“Liver”) that contains 5 patients. The status column shows the overall status for that series/study or patient, and the user statuses column shows the status of annotations that have been created by each ePAD user associated with that project.
Clinical Decision Support Tool for Treatment Response Assessment
ePAD has applications to assist decision-making based on image biomarker assessments in the following 2 major cancer research tasks: determine treatment response in patients (ePAD longitudinal annotation report) and evaluate treatment effectiveness by determining the cohort-based treatment response (ePAD waterfall plot). We built these applications using ePAD Web services to retrieve AIM annotations and their associated images to track target lesions and compute cancer treatment response according to selected imaging biomarkers.
Longitudinal Annotation Reporting.
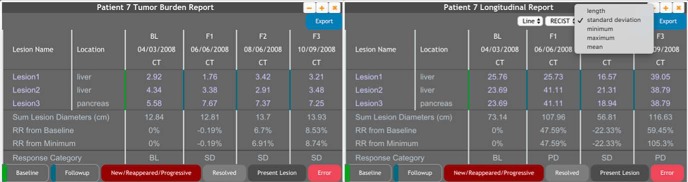
ePAD supports longitudinal annotation tracking, which provides a summary of quantitative image features across time. This is the basis for RECIST and other reports of response assessment. However, ePAD can generate such reports bases on any quantitative imaging biomarker it can collect from image annotations. It analyzes all the annotations of a subject and populates 3 dropdown menus to facilitate selecting them by shape, template, and measurement type (Figure 7). Users can select the basis for the longitudinal annotation report based on the selected measurement types. If a measurement is not present for a particular time point of a lesion, the table display it as a missing value. The summary section of the report will be filled automatically for the measurement type.
Figure 7.
A tumor burden report (using linear measurement as the imaging biomarker and RECIST response criteria) and a longitudinal annotation report of a patient having 4 time points and 3 lesions. This report is automatically generated from ePAD's image annotations and enables clinicians to determine image-based treatment response in the patient.
ePAD can generate a RECIST report by querying the annotations that are of linear type (Figure 7) and calculating sum of lesion dimensions on the images of each time point. RECIST report generation supports line and perpendicular lines annotations, as well as an image-based response rate (the percentage change in the sum of lesion dimensions compared with baseline). ePAD applies the RECIST rules to classify the response rate to determine the response category (ie, stable disease, partial response, complete response, and progressive disease). This information is displayed with the lesion measurements in the ePAD viewer (Figure 7). ePAD also checks the consistency of the annotations to determine if the anatomic location of the lesion is specified consistently on different time points of the patient; otherwise, the measurement will be marked as error to notify the user. The report also marks missing annotations for a lesion as error. The user can open the annotation in the ePAD viewer by clicking on the annotation measurement on a specific time point. The user can also open all annotations of the lesions on all time points by clicking the lesion name.
Besides using longitudinal measurement of lesions, ePAD can generate reports of lesion response based on other imaging biomarkers, such as the ADLA biomarker (82). The report evaluates the progress of the disease using the sum of ADLA scores for each timeline, similar to RECIST.
Waterfall Plots.
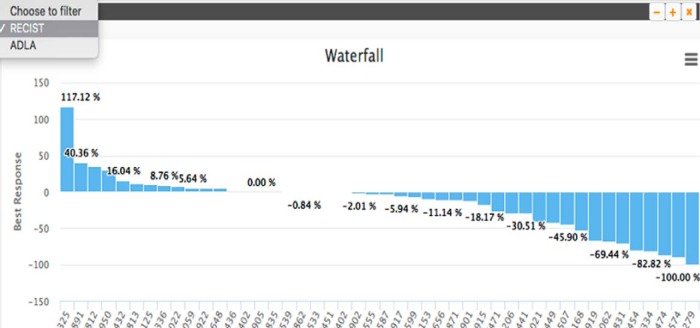
Waterfall plots are bar graphs showing the response of a cohort of patients to the same cancer treatment. The height of the bars represents the best overall response the patient had during the course of treatment, and each bar (patient) is ordered from best to worst response, which resembles a waterfall. These plots are highly useful for seeing how well a patient cohort responded to treatment, with the percentage of patients with positive response indicating effective treatment. ePAD generates waterfall plots of user-specified patient cohorts by computing the longitudinal annotation-based response in each patient in the cohort and ordering the response from best to worst response (Figure 8). The plot can be based on longitudinal measurement of lesions as the basis of evaluating response (ie, RECIST), but importantly, it can also be based on using newer imaging biomarkers of response such as ADLA or other imaging biomarkers that have been recently introduced by researchers. If the user selects to use RECIST, the waterfall plot module analyzes every subject in the cohort, generates the RECIST tables, gets the best response for each subject, and plots it in a decreasing order forming a waterfall plot. If the user selects to use ADLA, a waterfall plot is generated based on an ADLA table that ePAD computes for each subject, using the standard deviation of the line annotations on lesions as the measurement type (82). Then, the best response from the ADLA table for each patient is used to create the waterfall plot. Users can drill down to more granular data within the waterfall plot; the user can access the table that is used to make the best response rate computation by clicking the specific bar in the waterfall plot.
Figure 8.
Waterfall report plot based on linear measurement as the imaging biomarker of response and RECIST as the response criteria, showing the best response score for each patient in the study cohort. This plot enables researchers to assess the effectiveness of cancer treatment in the cohort, and a variety of these plots can be generated using different imaging biomarkers of response (upper left corner).
Application for Aggregating Evidence for Evaluating New Imaging Biomarkers
As noted earlier, ePAD has plugins to compute a variety of image biomarkers. Some of these plugins assume that cancer lesions are circumscribed, and if the images input into these biomarker plugins were annotated using only line annotations (eg, as part of RECIST measurements), ePAD can generate ROIs that circumscribe lesions automatically by executing image segmentation plugins that use the line or point annotations as seeds. In addition to segmentation, there are quantitative image analysis plugins that may operate on the entire image, and ePAD supports those as well. Current automated segmentation plugins and other analysis plugins available in ePAD are listed in the following sections.
Automated Segmentation in PET Images.
The plugin invokes automated segmentation of cancer lesions seen on PET images (90). It is triggered with a seed point ROI within the lesion. It analyzes the image volume to create a 3D ROI enclosing the lesion and creates a DICOM segmentation object marking the volume of the lesion. The DICOM segmentation object is added to ePAD with its associated AIM annotation file and displayed on the image series as a mask.
Automated 2D Lesion Segmentation.
ePAD has a 2D lesion segmentation plugin, LesionSeg. The plugin is triggered with drawing a polygon or a long axis line within a lesion. It analyzes the image and creates a polygon ROI stored as an AIM file containing the contours of the lesion (91).
Automated Image Segmentation of QF Explore Plugin Suite.
The QFExplore plugin suite has a plugin for automatically segmenting lungs in a DICOM image volume (85). The plugin analyses the volume, segments the lung volume, and creates a DICOM segmentation object.
ePAD Usage
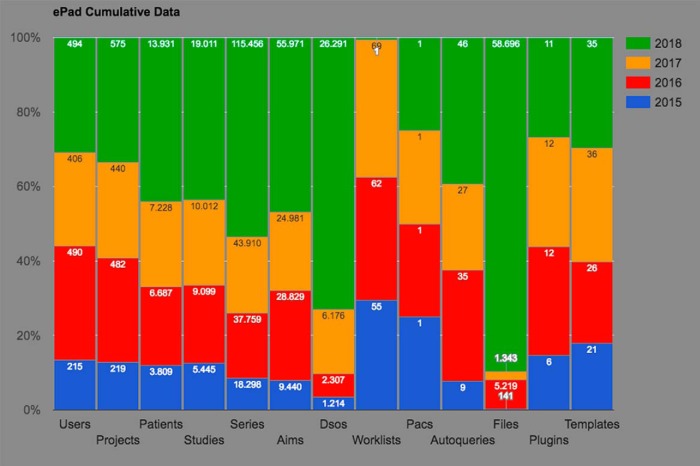
To track usage statistics, ePAD collects anonymous data from all ePAD machines that are connected to internet (if the statistics are not disabled by the user). The statistics consist the number of users, projects, patients, studies, series, AIM annotations, DICOM segmentation objects, plugins, and templates that exist on the ePAD instances. Figure 9 shows the ePAD usage statistics collected from 2015 to 2018. For plugins and templates, the maximum number of entities is reported, as many are the same versions of the plugins and templates across ePAD instances. For all the other entities, the values reported by each ePAD instance are computed by getting the latest reported values for each year and summing them to obtain the total number. For example, in 2018, over 19,000 imaging studies were hosted in various ePAD instances worldwide, and over 55,000 annotations were created in ePAD on those imaging studies. As ePAD collects only the number of entities for privacy purposes, the numbers are cumulative; that is, this does not mean 55,000 annotations were created during 2018, but it means that at the end of 2018, 55,000 annotations existed on ePAD instances. In addition, currently there are a maximum of 11 plugins and 35 templates that are being used across all ePAD instances.
Figure 9.
Cumulative ePAD statistics collected from ePAD instances between 2015 and 2018.
Interoperability
One of the key aims of ePAD is to facilitate collaborations among research sites and repurposing of their existing data, which we achieve by supporting standards and interoperability for images and annotations.
ePAD saves all image annotations that it collects using existing standards, in particular AIM (63) and DICOM segmentation objects (64), for volumetric ROIs. ePAD also supports the DICOM-SR standard via the dcmqi library (92) for volumetric ROI annotations. Recently AIM was harmonized with the DICOM standard, which provided DICOM-SR support of AIM annotation types under Supplement 200 with specifications for saving AIM in DICOM-SR (68, 69). ePAD also supports DICOM radiation therapy (DICOM-RTs) and tiff image files. ePAD analyses the DICOM-RT objects and extracts its ROI contours using the DICOM file interface library developed by MAASTRO (93). It then creates a DICOM segmentation object for each contour and saves it and an AIM file. ePAD also supports uploading tiff files and creates a DICOM image series from them using the patient identification number, patient name, study description, and series description supplied by the user. The file list is analyzed, and a DICOM file is created for each tiff file. The instance numbers of the DICOM files are ordered in the alphabetical order of tiff filenames.
ePAD also has migration tools that were developed in collaboration with various laboratories that enable ePAD to leverage the existing annotations created by other software tools, including ROIs exported from Osirix (94) and Mint Lesion (53). Specifically, ePAD analyzes the exported proprietary file from Osirix via ExportROIs plugin and creates an AIM file for each ROI in the file. ePAD also creates AIM files from JavaScript Object Notation (JSON) objects that are created from the Mint Lesion commercial system.
Use Cases for ePAD in QIN and Other Research
Many research studies in that require viewing and annotating radiology images for making measurements of lesions or extracting radiomics features from them could benefit from using ePAD. Clinical trials of cancer treatments can be particularly helped given ePAD's workflow support and multireader support features, its support of interoperability standards, as well as its ability to compute many imaging biomarkers seamlessly within routine image annotation workflow. ePAD has been used by many researchers worldwide to support clinical research and clinical trials, and it has supported many published studies (75, 81, 85, 95–110), and it has been shown to improve the workflow of measuring target lesions (111). We briefly highlight support it has provided several projects in NCI's QIN.
Vanderbilt QIN.
In collaboration with Vanderbilt QIN, “Quantitative MRI for Predicting Response of Breast Cancer to Neoadjuvant Therapy” in which this group developed algorithms for computing quantitative perfusion maps of MRI images to deduce biomarkers of treatment response (112), we deployed their biomarker algorithms as a plugin to ePAD. As these researchers incorporate these perfusion analyses into clinical trials, ePAD will be able to deploy them as part of the image interpretation workflow.
Dana Farber Cancer Institute QIN.
The QIN project at Dana Farber, “Genotype and Imaging Phenotype Biomarkers in Lung Cancer (113),” developed pyRadiomics, a flexible platform that extracts a large panel of predefined features from medical images and is useful in characterizing cancer lesions. We incorporated pyRadiomics into ePAD as a Docker module that runs on the QIFP platform (88) (see above) so that users can invoke generation of these image features as part of image analysis workflows in clinical trials.
ECOG-ACRIN QIN.
The QIN project within the ECOG-ACRIN cooperative group, “ECOG-ACRIN-Based QIN Resource for Advancing Quantitative Cancer Imaging in Clinical Trials,” is leveraging ePAD as a testbed for evaluating the deployment of imaging biomarkers into clinical trials. Currently this project is comparing ability of ePAD to evaluate a variety of quantitative imaging biomarkers as part of the routine workflow of image viewing and annotation in clinical trials.
American College of Radiology (ACR) Core Laboratory.
The ACR has a data archive and research toolkit called DART Portal (114) that operates as a gateway to browse and query data for research, quality improvement, and clinical study operational purposes. They are adding ePAD as an interface to DART to enable collecting image annotations as part of clinical trials in AIM format and storing that in DART.
Discussion
Response assessment in patients with cancer in clinical trials is based on analysis of CT and magnetic resonance images (115). Objective criteria, such as RECIST, are critical to evaluation of response assessment in clinical trials, but lesion measurements vary with user experience, and they are often inconsistent or incomplete (105). There is a pressing need to recognize signals in radiology images that optimally assess and predict response to treatment. Tumor shrinkage is the hallmark of response to cytotoxic cancer therapies (4), and thus, linear measurement of target cancer lesions is the imaging biomarker used in current response criteria such as RECIST and International Harmonization Criteria (10). However, new targeted, noncytotoxic therapies arrest cancer growth and improve progression-free survival without necessarily shrinking tumors (11–14); thus, simple linear measurement may underestimate treatment response (15–18), and may not be the best proxy for tumor activity. To address these limitations, researchers are developing quantitative imaging biomarkers that may better assess the benefit of new treatments, but they have been challenging to introduce into clinical trial workflows.
In the paper, we have presented ePAD from 4 different viewpoints to highlight how it addresses key challenges for incorporating quantitative imaging biomarkers into clinical trials. First, it provides a platform for computing a variety of imaging biomarkers. Second, it provides a workflow management and study oversight tool enabling oversight for assessing new image biomarkers in clinical trials. Third, it provides clinical decision support tools to help clinical researchers assess treatment response using current and new imaging biomarkers. Fourth, ePAD provides infrastructure to permit researchers to aggregate evidence about how well imaging biomarkers predict response, which may help in qualifying them as surrogate endpoints in clinical trials.
There are many existing commercial and freely available tools available for medical image viewing and annotation, and although ePAD provides similar capability in terms of image viewing and drawing shapes on images, it provides many unique features that address many unmet needs in evaluating images clinical research. Osirix (94) and ClearCanvas (55) are 2 medical image annotation applications that provide similar image viewing capabilities, although they depend on a thick client, limiting collaboration as they are platform-dependent, while ePAD is a web-based viewer and requires no installation for users other than hosting a single instance of the ePAD machine for all users. In addition, Osirix saves its annotations in propriety format and supports exporting ROIs using ExportROIs plugin. On the other hand, ePAD and ClearCanvas supports AIM format. ePAD also supports the new DICOM-SR AIM object. Beyond the open-source tools, we recognize that several commercial tools are available for image viewing and analysis to enable response assessment. These tools were developed to enable evaluating established criteria such as RECIST in clinical trials; however, such tools are not optimal for research studies that wish to include novel imaging biomarkers of treatment response (eg, those being developed by NCI's QIN). This gap was a primary motivator for developing the ePAD system.
3D Slicer (56, 116), ImageJ (117), and MIPAV (118) are additional freely available desktop applications. 3D Slicer is a cross-platform open-source software for visualization and image computing. It has a plugin architecture to enable researchers to develop their algorithms via C++ plugins and Python scripted modules. It supports DICOM standard for the volumetric annotations and DICOM Structured Report (DICOM-SR) for the measurements collected from the ROIs. ImageJ and MIPAV are both Java applications that can run on any Java-enabled operating system, and researchers can develop their own plugins using Java language. Imagej2, an extended version of ImageJ, supports writing plugin scripts in various programming languages. ImageJ saves the labels and annotations as modified versions of the images or propriety ROI file formats, and MIPAV uses an Extensible Markup Language (XML) format they introduced in an effort to make their format readable by researchers. Although all 3 applications are cross-platform, they are desktop applications for a single user, which makes multiuser collaboration more difficult.
The Open Health Imaging Foundation (OHIF; http://ohif.org/) is a full-stack Javascript platform, which enables creating a zero-footprint web page and various applications using it. The OHIF Viewer provides web-based image viewing similar to ePAD. The OHIF LesionTracker enables users to annotate and track long-axis and short-axis lesions for oncology workflow; however, it does not save the image annotations in a standard format like AIM.
ePAD was developed to facilitate collecting annotations and measurements on target lesions in compliance with standards in the cancer imaging community. ePAD makes sharing code, data and annotations easy being a web application and saving the collected annotation data in well-documented and standardized formats [DICOM segmentation objects (64) and AIM (63) in particular].
In addition to providing standards-based storage of annotations, ePAD enables user-defined templates for flexible capture of information in the form of data collection templates as part of the annotations. The ePAD platform is also extensible via plugins that lets researchers implement analysis codes as server-side modules in MATLAB or other languages. Many plugins for segmentation and quantitative image biomarker computation are included with ePAD, and users can add additional biomarker modules.
Other functionalities of ePAD that differentiates it from similar existing image viewing applications is that ePAD supports important features unique to image analysis in clinical trial workflow. Specifically, ePAD provides tools enabling oversight of annotations as part of clinical trials, and it lets the users create a collaborative environment by creating projects and assigning users appropriate rights to limit their access facilitating large studies with multiple annotators. ePAD also provides decision support tools—longitudinal annotation summary and waterfall plots—that help researchers evaluate individual patient and cohort population treatment response, respectively. Finally, by computing a variety of image biomarkers on cohorts of patients, ePAD can accumulate a substantial amount of data that can permit studies comparing effectiveness of different imaging biomarkers as indicators of treatment response.
An ultimate metric of the success of ePAD will be increased use of the newer imaging biomarkers in clinical trials. This will require clinical trial groups to include computation of the biomarkers into their study protocols. As the community becomes aware of the potential of these methods and of the facility of tools such as ePAD to include them in clinical trials, we expect these methods will be more commonly used. Certainly, the amount of research studies undertaken to date using ePAD suggests promising future directions.
Acknowledgments
This work was supported in part by grants from the National Cancer Institute, National Institutes of Health, U01CA142555, 1U01CA190214, and 1U01CA187947.
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- ePAD
- Electronic Physician Annotation Device
- QIN
- Quantitative Imaging Network
- PET
- positron emission tomography
- MRI
- magnetic resonance imaging
- ROIs
- regions of interest
- PACS
- Picture Archiving and Communication System
- ADLA
- Attenuation Distribution across the Long Axis
- PID
- Pixel Intensity Distributions
- GLCMs
- gray-level co-occurrence matrices
References
- 1. Garrett MD, Workman P. Discovering novel chemotherapeutic drugs for the third millennium. Eur J Cancer. 1999;35:2010–2030. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 3. NIH Clinical Trials Working Group. Final CTWG report to the National Cancer Advisory Board, “Restructuring the National Cancer Clinical Trials Enterprise”. http://integratedtrials.nci.nih.gov/ict/CTWG_report_June2005.pdf; Accessed: December 21, 2008.
- 4. El-Maraghi RH, Eisenhauer EA. Review of phase II trial designs used in studies of molecular targeted agents: outcomes and predictors of success in phase III. J Clin Oncol. 2008;26:1346–1354. [DOI] [PubMed] [Google Scholar]
- 5. Taylor PT, Haverstick D. Re: New guidelines to evaluate the response to treatment in solid tumors (ovarian cancer). J Natl Cancer Inst. 2005;97:151. [DOI] [PubMed] [Google Scholar]
- 6. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. [DOI] [PubMed] [Google Scholar]
- 7. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–245. [DOI] [PubMed] [Google Scholar]
- 8. Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, Degroot J, Wick W, Gilbert MR, Lassman AB, Tsien C, Mikkelsen T, Wong ET, Chamberlain MC, Stupp R, Lamborn KR, Vogelbaum MA, van den Bent MJ, Chang SM. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. [DOI] [PubMed] [Google Scholar]
- 9. Gallego Perez-Larraya J, Lahutte M, Petrirena G, Reyes-Botero G, Gonzalez-Aguilar A, Houillier C, Guillevin R, Sanson M, Hoang-Xuan K, Delattre JY. Response assessment in recurrent glioblastoma treated with irinotecan-bevacizumab: comparative analysis of the Macdonald, RECIST, RANO, and RECIST + F criteria. Neuro Oncol. 2012;14:667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheson BD. The International Harmonization Project for response criteria in lymphoma clinical trials. Hematol Oncol Clin North Am. 2007;21:841–854. [DOI] [PubMed] [Google Scholar]
- 11. Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, Negrier S, Chevreau C, Solska E, Desai AA, Rolland F, Demkow T, Hutson TE, Gore M, Freeman S, Schwartz B, Shan M, Simantov R, Bukowski RM. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. [DOI] [PubMed] [Google Scholar]
- 12. Ratain MJ, Eisen T, Stadler WM, Flaherty KT, Kaye SB, Rosner GL, Gore M, Desai AA, Patnaik A, Xiong HQ, Rowinsky E, Abbruzzese JL, Xia C, Simantov R, Schwartz B, O'Dwyer PJ. Phase II placebo-controlled randomized discontinuation trial of sorafenib in patients with metastatic renal cell carcinoma. J Clin Oncol. 2006;24:2505–2512. [DOI] [PubMed] [Google Scholar]
- 13. Ratain MJ. Phase II oncology trials: let's be positive. Clin Cancer Res. 2005;11:5661–5662. [DOI] [PubMed] [Google Scholar]
- 14. Ratain MJ, Eckhardt SG. Phase II studies of modern drugs directed against new targets: if you are fazed, too, then resist RECIST. J Clin Oncol. 2004;22:4442–4445. [DOI] [PubMed] [Google Scholar]
- 15. Benjamin RS, Choi H, Macapinlac HA, Burgess MA, Patel SR, Chen LL, Podoloff DA, Charnsangavej C. We should desist using RECIST, at least in GIST. J Clin Oncol. 2007;25:1760–1764. [DOI] [PubMed] [Google Scholar]
- 16. Choi H, Charnsangavej C, de Castro Faria S, Tamm EP, Benjamin RS, Johnson MM, Macapinlac HA, Podoloff DA. CT evaluation of the response of gastrointestinal stromal tumors after imatinib mesylate treatment: a quantitative analysis correlated with FDG PET findings. AJR Am J Roentgenol. 2004;183:1619–1628. [DOI] [PubMed] [Google Scholar]
- 17. Adams VR, Leggas M. Sunitinib malate for the treatment of metastatic renal cell carcinoma and gastrointestinal stromal tumors. Clin Ther. 2007;29:1338–1353. [DOI] [PubMed] [Google Scholar]
- 18. Choi H. Response evaluation of gastrointestinal stromal tumors. Oncologist. 2008;13(Suppl 2):4–7. [DOI] [PubMed] [Google Scholar]
- 19. Therasse P, Eisenhauer EA, Verweij J. RECIST revisited: a review of validation studies on tumour assessment. Eur J Cancer. 2006;42:1031–1039. [DOI] [PubMed] [Google Scholar]
- 20. Cyran CC, Paprottka PM, Eisenblatter M, Clevert DA, Rist C, Nikolaou K, Lauber K, Wenz F, Hausmann D, Reiser MF, Belka C, Niyazi M. Visualization, imaging and new preclinical diagnostics in radiation oncology. Radiat Oncol. 2014;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lemasson B, Galban CJ, Boes JL, Li Y, Zhu Y, Heist KA, Johnson TD, Chenevert TL, Galban S, Rehemtulla A, Ross BD. Diffusion-weighted MRI as a biomarker of tumor radiation treatment response heterogeneity: a comparative study of whole-volume histogram analysis versus voxel-based functional diffusion map analysis. Transl Oncol. 2013;6:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh D, Lee JE, Huh SJ, Park W, Nam H, Choi JY, Kim BT. Prognostic significance of tumor response as assessed by sequential 18F-fluorodeoxyglucose-positron emission tomography/computed tomography during concurrent chemoradiation therapy for cervical cancer. Int J Radiat Oncol Biol Phys. 2013;87:549–554. [DOI] [PubMed] [Google Scholar]
- 24. Teng FF, Meng X, Sun XD, Yu JM. New strategy for monitoring targeted therapy: molecular imaging. Int J Nanomedicine. 2013;8:3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith AD, Lieber ML, Shah SN. Assessing tumor response and detecting recurrence in metastatic renal cell carcinoma on targeted therapy: importance of size and attenuation on contrast-enhanced CT. AJR Am J Roentgenol. 2010;194:157–165. [DOI] [PubMed] [Google Scholar]
- 26. Weber WA. Assessing tumor response to therapy. J Nucl Med. 2009;(50 Suppl 1):1S–10S. [DOI] [PubMed] [Google Scholar]
- 27. Worhunsky DJ, Krampitz GW, Poullos PD, Visser BC, Kunz PL, Fisher GA, Norton JA, Poultsides GA. Pancreatic neuroendocrine tumours: hypoenhancement on arterial phase computed tomography predicts biological aggressiveness. HPB (Oxford). 2013;16:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Workman P, Aboagye EO, Chung YL, Griffiths JR, Hart R, Leach MO, Maxwell RJ, McSheehy PM, Price PM, Zweit J. Minimally invasive pharmacokinetic and pharmacodynamic technologies in hypothesis-testing clinical trials of innovative therapies. J Natl Cancer Inst. 2006;98:580–598. [DOI] [PubMed] [Google Scholar]
- 29. U.S. Department of Health and Human Services Food and Drug Administration. Guidance for Industry: Clinical Trail Endpoints the Approval of Cancer Drugs and Biologics. [cited 2014 Jan 19]. Available from: http://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&cad=rja&ved=0CCkQFjAA&url=http%3A%2F%2Fwww.fda.gov%2Fdownloads%2FDrugs%2FGuidanceComplianceRegulatoryInformation%2FGuidances%2FUCM268555.pdf&ei=n2XcUuPoGdDdoASnooGoAw&usg=AFQjCNGiW-duJ8M5E1pqEEVWyyPafAJCqA&sig2=wsdc99GnjhhUNO4qjyCkMg&bvm=bv.59568121,d.cGU
- 30. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. [DOI] [PubMed] [Google Scholar]
- 31. Levy MA, Freymann JB, Kirby JS, Fedorov A, Fennessy FM, Eschrich SA, Berglund AE, Fenstermacher DA, Tan Y, Guo X, Casavant TL, Brown BJ, Braun TA, Dekker A, Roelofs E, Mountz JM, Boada F, Laymon C, Oborski M, Rubin DL. Informatics methods to enable sharing of quantitative imaging research data. Magn Reson Imaging. 2012;30:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kelloff GJ, Hoffman JM, Johnson B, Scher HI, Siegel BA, Cheng EY, Cheson BD, O'Shaughnessy J, Guyton KZ, Mankoff DA, Shankar L, Larson SM, Sigman CC, Schilsky RL, Sullivan DC. Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin Cancer Res. 2005;11:2785–2808. [DOI] [PubMed] [Google Scholar]
- 33. Munden RF, Swisher SS, Stevens CW, Stewart DJ. Imaging of the patient with non-small cell lung cancer. Radiology. 2005;237:803–818. [DOI] [PubMed] [Google Scholar]
- 34. Johnson JR, Williams G, Pazdur R. End points and United States Food and Drug Administration approval of oncology drugs. J Clin Oncol. 2003;21:1404–1411. [DOI] [PubMed] [Google Scholar]
- 35. Pien HH, Fischman AJ, Thrall JH, Sorensen AG. Using imaging biomarkers to accelerate drug development and clinical trials. Drug Discov Today. 2005;10:259–266. [DOI] [PubMed] [Google Scholar]
- 36. Schnall M, Rosen M. Primer on imaging technologies for cancer. J Clin Oncol. 2006;24:3225–3233. [DOI] [PubMed] [Google Scholar]
- 37. Shankar LK, Sullivan DC. Functional imaging in lung cancer. J Clin Oncol. 2005;23:3203–3211. [DOI] [PubMed] [Google Scholar]
- 38. Smith JJ, Sorensen AG, Thrall JH. Biomarkers in imaging: realizing radiology's future. Radiology. 2003;227:633–638. [DOI] [PubMed] [Google Scholar]
- 39. O'Connor JP, Jackson A, Asselin MC, Buckley DL, Parker GJ, Jayson GC. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 2008;9:766–776. [DOI] [PubMed] [Google Scholar]
- 40. Galanis E, Buckner JC, Maurer MJ, Sykora R, Castillo R, Ballman KV, Erickson BJ. Validation of neuroradiologic response assessment in gliomas: measurement by RECIST, two-dimensional, computer-assisted tumor area, and computer-assisted tumor volume methods. Neuro Oncol. 2006;8:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi H, Charnsangavej C, Faria SC, Macapinlac HA, Burgess MA, Patel SR, Chen LL, Podoloff DA, Benjamin RS. Correlation of computed tomography and positron emission tomography in patients with metastatic gastrointestinal stromal tumor treated at a single institution with imatinib mesylate: proposal of new computed tomography response criteria. J Clin Oncol. 2007;25:1753–1759. [DOI] [PubMed] [Google Scholar]
- 42. Fendler WP, Philippe Tiega DB, Ilhan H, Paprottka PM, Heinemann V, Jakobs TF, Bartenstein P, Hacker M, Haug AR. Validation of several SUV-based parameters derived from 18F-FDG PET for prediction of survival after SIRT of hepatic metastases from colorectal cancer. J Nucl Med. 2013;54:1202–1208. [DOI] [PubMed] [Google Scholar]
- 43. Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med. 2009;50(Suppl 1):122S–150S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Downey RJ, Akhurst T, Gonen M, Vincent A, Bains MS, Larson S, Rusch V. Preoperative F-18 fluorodeoxyglucose-positron emission tomography maximal standardized uptake value predicts survival after lung cancer resection. J Clin Oncol. 2004;22:3255–3260. . [DOI] [PubMed] [Google Scholar]
- 45. Rizk N, Downey RJ, Akhurst T, Gonen M, Bains MS, Larson S, Rusch V. Preoperative 18[F]-fluorodeoxyglucose positron emission tomography standardized uptake values predict survival after esophageal adenocarcinoma resection. Ann Thorac Surg. 2006;81:1076–1081. [DOI] [PubMed] [Google Scholar]
- 46. Guermazi A, Juweid ME. Commentary: PET poised to alter the current paradigm for response assessment of non-Hodgkin's lymphoma. Br J Radiol. 2006;79:365–367. [DOI] [PubMed] [Google Scholar]
- 47. O'Connor JP, Jackson A, Parker GJ, Jayson GC. DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. Br J Cancer. 2007;96:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang EY, Li X, Jerosch-Herold M, Priest RA, Enestvedt CK, Xu J, Springer CS Jr., Jobe BA. The evaluation of esophageal adenocarcinoma using dynamic contrast-enhanced magnetic resonance imaging. J Gastrointest Surg. 2008;12:166–175. [DOI] [PubMed] [Google Scholar]
- 49. d'Arcy JA, Collins DJ, Padhani AR, Walker-Samuel S, Suckling J, Leach MO. Informatics in Radiology (infoRAD): Magnetic Resonance Imaging Workbench: analysis and visualization of dynamic contrast-enhanced MR imaging data. Radiographics. 2006;26:621–632. [DOI] [PubMed] [Google Scholar]
- 50. Galban CJ, Chenevert TL, Meyer CR, Tsien C, Lawrence TS, Hamstra DA, Junck L, Sundgren PC, Johnson TD, Ross DJ, Rehemtulla A, Ross BD. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat Med. 2009;15:572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X, Dawant BM, Welch EB, Chakravarthy AB, Freehardt D, Mayer I, Kelley M, Meszoely I, Gore JC, Yankeelov TE. A nonrigid registration algorithm for longitudinal breast MR images and the analysis of breast tumor response. Magn Reson Imaging. 2009;27:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. MIM Software. MIMviewer® and PET Edge™. [cited 2014 Jan 28]. Available from: http://www.mimsoftware.com/products/radnuc
- 53. Mint Medical. Mint Lesion™. [cited 2014 Jan 28]. Available from: http://www.mint-medical.de/productssolutions/mintlesion/mintlesion/
- 54. Siemens Inc. syngo.via for Oncology. [cited 2014 Jan 28]. Available from: http://www.healthcare.siemens.com/medical-imaging-it/syngoviaspecialtopics/syngo-via-for-oncology/syngo-via-for-oncology-follow-up
- 55. ClearCanvas Workstation. In, 2014, Available from: http://clearcanvas.ca
- 56. Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kikinis R, Pieper S. 3D Slicer as a tool for interactive brain tumor segmentation. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:6982–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC, Aerts HJ, Bendriem B, Bendtsen C, Boellaard R, Boone JM, Cole PE, Conklin JJ, Dorfman GS, Douglas PS, Eidsaunet W, Elsinger C, Frank RA, Gatsonis C, Giger ML, Gupta SN, Gustafson D, Hoekstra OS, Jackson EF, Karam L, Kelloff GJ, Kinahan PE, McLennan G, Miller CG, Mozley PD, Muller KE, Patt R, Raunig D, Rosen M, Rupani H, Schwartz LH, Siegel BA, Sorensen AG, Wahl RL, Waterton JC, Wolf W, Zahlmann G, Zimmerman B. Quantitative imaging test approval and biomarker qualification: interrelated but distinct activities. Radiology. 2011;259:875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Institute of Medicine (U.S.). Forum on Drug Discovery Development and Translation. Olson S, Robinson S, Giffin RB. Accelerating the Development of Biomarkers for Drug Safety: Workshop Summary. Washington, DC: National Academies Press; 2009. [Google Scholar]
- 60. Waterton JC, Pylkkanen L. Qualification of imaging biomarkers for oncology drug development. Eur J Cancer. 2012;48:409–415. [DOI] [PubMed] [Google Scholar]
- 61. Buckler AJ, Mozley PD, Schwartz L, Petrick N, McNitt-Gray M, Fenimore C, O'Donnell K, Hayes W, Kim HJ, Clarke L, Sullivan D. Volumetric CT in lung cancer: an example for the qualification of imaging as a biomarker. Acad Radiol. 2010;17:107–115. [DOI] [PubMed] [Google Scholar]
- 62. Buckler AJ, Schwartz LH, Petrick N, McNitt-Gray M, Zhao B, Fenimore C, Reeves AP, Mozley PD, Avila RS. Data sets for the qualification of volumetric CT as a quantitative imaging biomarker in lung cancer. Opt Express. 2010;18:15267–15282. [DOI] [PubMed] [Google Scholar]
- 63. Channin DS, Mongkolwat P, Kleper V, Sepukar K, Rubin DL. The caBIG annotation and image Markup project. J Digit Imaging. 2010;23:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DICOM Standards Committee WG 17 (3D). Supplement 111: segmentation storage SOP class. Digit Imaging Commun Med. 2006;17. [Google Scholar]
- 65. caBIG In-vivo Imaging Workspace. Annotation and Image Markup (AIM). https://cabig.nci.nih.gov/tools/AIM; Accessed: December 26, 2008.
- 66. Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS. Medical imaging on the semantic web: annotation and image markup. In: 2008 AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration Stanford, CA: Stanford University; 2008. [Google Scholar]
- 67. Rubin DL, Mongkolwat P, Channin DS. A semantic image annotation model to enable integrative translational research. Summit Transl Bioinform 2009;2009:106–110. [PMC free article] [PubMed] [Google Scholar]
- 68. DICOM Standards Committee - Working Group 8 - Structured Reporting. Digital Imaging and Communications in Medicine (DICOM); Sup 200 - Transformation of NCI Annotation and Image Markup (AIM) and DICOM SR Measurement Templates. Available from: ftp://medical.nema.org/medical/dicom/Supps/LB/sup200_lb_AIM_DICOMSRTID1500.pdf.
- 69. DICOM Standards Committee. DICOM PS3.21 2017e - Transformations between DICOM and other Representations; A.6 AIM v4 to DICOM TID 1500 Mapping. Available from: http://dicom.nema.org/medical/Dicom/2017e/output/chtml/part21/sect_A.6.html.
- 70. Hwang KH, Lee H, Koh G, Willrett D, Rubin DL. Building and querying RDF/OWL database of semantically annotated nuclear medicine images. J Digit Imaging. 2017;30:4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moreira DA, Hage C, Luque EF, Willrett D, Rubin DL. 3D markup of radiological images in ePAD, a web-based image annotation tool. In: 2015 IEEE 28th International Symposium on Computer-Based Medical Systems (CBMS), 2015;97–102, Available from: http://ieeexplore.ieee.org/ielx7/7164867/7167433/07167466.pdf?tp=&arnumber=7167466&isnumber=7167433. [Google Scholar]
- 72. Rubin DL, Willrett D, O'Connor MJ, Hage C, Kurtz C, Moreira DA. Automated tracking of quantitative assessments of tumor burden in clinical trials. Transl Oncol. 2014;7:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Warnock MJ, Toland C, Evans D, Wallace B, Nagy P. Benefits of using the DCM4CHE DICOM archive. J Digit Imaging. 2007;20(Suppl 1):125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hoy MB. HTML5: a new standard for the Web. Med Ref Serv Q. 2011;30:50–55. [DOI] [PubMed] [Google Scholar]
- 75. Levy MA, Rubin DL. Computational approaches to assist in the evaluation of cancer treatment response. Imaging Med. 2011;3:233–246. [Google Scholar]
- 76. Mongkolwat P, Channin DS, Rubin VK. Informatics in radiology: an open-source and open-access cancer biomedical informatics grid annotation and image markup template builder. Radiographics. 2012;32:1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26:1595–1597. [DOI] [PubMed] [Google Scholar]
- 78. Bruno EJ. SOA, Web services, and RESTful systems—a framework for building RESTful systems. Dr Dobbs J. 2007;32:32. [Google Scholar]
- 79. Lipton P, Nagy P, Sevinc G. Leveraging Internet technologies with DICOM WADO. J Digit Imaging. 2012;25:646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Meier W. eXist: an open source native XML database. Web Web Serv Database Syst. 2003;2593:169–183. [Google Scholar]
- 81. Napel SA, Beaulieu CF, Rodriguez C, Cui J, Xu J, Gupta A, Korenblum D, Greenspan H, Ma Y, Rubin DL. Automated retrieval of CT images of liver lesions on the basis of image similarity: method and preliminary results. Radiology. 2010;256:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Abramson RG, Lakomkin N, Hainline A, Kang H, Hutson MS, Arteaga CL. The attenuation distribution across the long axis of breast cancer liver metastases at CT: a quantitative biomarker for predicting overall survival. AJR Am J Roentgenol. 2018; 210:W1–W7. [DOI] [PubMed] [Google Scholar]
- 83. Lakomkin N, Kang H, Landman B, Hutson MS, Abramson RG. The Attenuation Distribution Across the Long Axis (ADLA): preliminary findings for assessing response to cancer treatment. Acad Radiol. 2016;23:718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yankeelov TE, Gore JC. Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples. Curr Med Imaging Rev. 2009;3:91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schaer R, Cid YD, Alkim E, John S, Rubin DL, Depeursinge A. Web-based tools for exploring the potential of quantitative imaging biomarkers in radiology intensity and texture analysis on the ePAD platform. In Biomedical Texture Analysis: Fundamentals, Tools and Challenges; 2017:379–410. [Google Scholar]
- 86. Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an open-source, modular engine for 3D quantitative feature extraction from volumetric medical images. J Digit Imaging. 2018;31:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Depeursinge A, Foncubierta-Rodriguez A, Van de Ville D, Muller H. Rotation-covariant texture learning using steerable riesz wavelets. IEEE Trans Image Process. 2014;23:898–908. [DOI] [PubMed] [Google Scholar]
- 88. John S, Rubin DL, Gude D, Echegaray S, Bakr S, Mattonen S, Napel S. QIFP : A web application to support quantitative imaging pipelines on medical images. In: 3rd Annual Scientific Conference on Machine Intelligence in Medical Imaging (C-MIMI) of the Society for Imaging Informatics in Medicine (SIIM) San Francisco, CA; 2018. [Google Scholar]
- 89. Fornaciari G, Vitiello A, Giusiani S, Giuffra V, Fornaciari A, Villari N. The Medici Project first anthropological and paleopathological results of the exploration of the Medici tombs in Florence. Med Secoli. 2007;19:521–543. [PubMed] [Google Scholar]
- 90. Graves EE, Quon A, Loo BW. RT_Image: an open-source tool for investigating PET in radiation oncology. Technol Cancer Res Treat. 2007;6:111–121. [DOI] [PubMed] [Google Scholar]
- 91. Hoogi A, Beaulieu CF, Cunha GM, Heba E, Sirlin CB, Napel S, Rubin DL. Adaptive local window for level set segmentation of CT and MRI liver lesions. Med Image Anal. 2017;37:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Herz C, Fillion-Robin JC, Onken M, Riesmeier J, Lasso A, Pinter C, Fichtinger G, Pieper S, Clunie D, Kikinis R, Fedorov A. dcmqi: an open source library for standardized communication of quantitative image analysis results using DICOM. Cancer Res. 2017;77:E87–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Maastro. Dicom File interface [Internet]. [Cited 2018 Sep 29]. Available from: https://bitbucket.org/maastro/dicomfileinterface
- 94. Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gevaert O, Mitchell LA, Xu J, Yu C, Rubin D, Zaharchuk G, Napel S, Plevritis S. Radiogenomic analysis indicates MR images are potentially predictive of EGFR mutation status in glioblastoma multiforme. In: AACR 103nd Annual Meeting Chicago, IL; 2012. [Google Scholar]
- 96. Gevaert O, Xu J, Hoang C, Leung A, Quon A, Rubin DL, Napel S, Plevritis S. Integrating medical images and transcriptomic data in non-small cell lung cancer. In: AACR 102nd Annual Meeting Orlando, FL; 2011. [Google Scholar]
- 97. Gevaert O, Xu JJ, Hoang CD, Leung AN, Xu Y, Quon A, Rubin DL, Napel S, Plevritis SK. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data-methods and preliminary results. Radiology. 2012;264:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gimenez F, Xu J, Liu Y, Liu TT, Beaulieu C, Rubin DL, Napel S. On the feasibility of predicting radiological observations from computational imaging features of liver lesions in CT scans. In: First IEEE Conference on Healthcare Informatics, Imaging, and Systems Biology (HISB), IEEE Computer Society: San Jose, CA; 2011. [Google Scholar]
- 99. Gimenez F, Xu J, Liu TT, Beaulieu C, Rubin DL, Napel S, Liu Y. Prediction of radiologist observations using computational image features: method and preliminary results. In: Ninety-Seventy Annual Scientific Meeting of the RSNA Chicago, IL; 2011. [Google Scholar]
- 100. Hoang C, Napel S, Gevaert O, Xu J, Rubin DL, Leung A, Merritt R, Whyte R, Shrager J, Plevritis S. NSCLC gene profiles correlate with specific CT characteristics: image-omics. In: American Association for Thoracic Surgery (AATS). Philadelphia, PA; 2011. [Google Scholar]
- 101. Napel S, Hoang C, Xu J, Gevaert O, Rubin DL, Plevritis S, Xu Y, Leung A, Quon A. Computational and semantic annotation of CT and PET images and integration with genomic assays of tumors in non-small cell lung cancer (NSCLC) for decision support and discovery: method and preliminary results. In: Ninety-Seventy Annual Scientific Meeting of the RSNA Chicago, IL; 2011. [Google Scholar]
- 102. Plevritis S, Gevaert O, Xu J, Hoang C, Leung A, Xu Y, Quon A, Rubin DL, Napel S. Rapid Identification of Prognostic Imaging Biomarkers for Non-small Cell Lung Carcinoma (NSCLC) by integrating image features and gene expression and leveraging public gene expression databases. In: Ninety-Seventy Annual Scientific Meeting of the RSNA Chicago, IL; 2011. [Google Scholar]
- 103. Korenblum D, Rubin D, Napel S, Rodriguez C, Beaulieu C. Managing biomedical image metadata for search and retrieval of similar images. J Digit Imaging. 2011;24:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Levy MA, O'Connor MJ, Rubin DL. Semantic reasoning with image annotations for tumor assessment. AMIA Annu Symp Proc. 2009;2009:359–363. [PMC free article] [PubMed] [Google Scholar]
- 105. Levy MA, Rubin DL. Tool support to enable evaluation of the clinical response to treatment. AMIA Annu Symp Proc. 2008:399–403. [PMC free article] [PubMed] [Google Scholar]
- 106. Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N, Napel S. Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging. 2015;2:041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Echegaray S, Nair V, Kadoch M, Leung A, Rubin D, Gevaert O, Napel S. A Rapid segmentation-insensitive “digital biopsy” method for radiomic feature extraction: method and pilot study using CT images of non-small cell lung cancer. Tomography. 2016;2:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, Berry GJ, Plevritis SK, Rubin DL, Napel S, Gevaert O. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology. 2018;286:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bakr S, Echegaray S, Shah R, Kamaya A, Louie J, Napel S, Kothary N, Gevaert O. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham). 2017;4:041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Itakura H, Achrol AS, Mitchell LA, Loya JJ, Liu T, Westbroek EM, Feroze AH, Rodriguez S, Echegaray S, Azad TD, Yeom KW, Napel S, Rubin DL, Chang SD, Harsh GRT, Gevaert O. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015;7:303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rubin DL, Willrett D, O'Connor MJ, Hage C, Kurtz C, Moreira DA. Automated tracking of quantitative assessments of tumor burden in clinical trials. Transl Oncol. 2014;7:23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jarrett AM, Hormuth DA, Barnes SL, Feng X, Huang W, Yankeelov TE. Incorporating drug delivery into an imaging-driven, mechanics-coupled reaction diffusion model for predicting the response of breast cancer to neoadjuvant chemotherapy: theory and preliminary clinical results. Phys Med Biol. 2018;63:105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Yip SS, Kim J, Coroller TP, Parmar C, Velazquez ER, Huynh E, Mak RH, Aerts HJ. Associations between somatic mutations and metabolic imaging phenotypes in non-small cell lung cancer. J Nucl Med. 2017;58:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. American College of Radiology. DART Portal. https://dart.acr.org/https://dart.acr.org/.
- 115. Strosberg JR, Halfdanarson TR, Bellizzi AM, Chan JA, Dillon JS, Heaney AP, Kunz PL, O'Dorisio TM, Salem R, Segelov E, Howe JR, Pommier RF, Brendtro K, Bashir MA, Singh S, Soulen MC, Tang L, Zacks JS, Yao JC, Bergsland EK. The North American Neuroendocrine Tumor Society Consensus guidelines for surveillance and medical management of midgut neuroendocrine tumors. Pancreas. 2017;46:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pieper S, Halle M, Kikinis R. 3D SLICER. IEEE International Symposium on Biomedical Imaging ISBI 2004, 2004:632–635. [Google Scholar]
- 117. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. McAuliffe M. Using MIPAV to label and measure brain components in Talairach space. Tech report. National Institutes of Health. [cited 2018 Sep 15]. Available from: https://mipav.cit.nih.gov/index.php.