Abstract
The use of rCBV as a response metric in clinical trials has been hampered, in part, due to variations in the biomarker consistency and associated interpretation across sites, stemming from differences in image acquisition and post-processing methods. This study leveraged a dynamic susceptibility contrast magnetic resonance imaging digital reference object to characterize rCBV consistency across 12 sites participating in the Quantitative Imaging Network (QIN), specifically focusing on differences in site-specific imaging protocols (IPs; n = 17), and PMs (n = 19) and differences due to site-specific IPs and PMs (n = 25). Thus, high agreement across sites occurs when 1 managing center processes rCBV despite slight variations in the IP. This result is most likely supported by current initiatives to standardize IPs. However, marked intersite disagreement was observed when site-specific software was applied for rCBV measurements. This study's results have important implications for comparing rCBV values across sites and trials, where variability in PMs could confound the comparison of therapeutic effectiveness and/or any attempts to establish thresholds for categorical response to therapy. To overcome these challenges and ensure the successful use of rCBV as a clinical trial biomarker, we recommend the establishment of qualifying and validating site- and trial-specific criteria for scanners and acquisition methods (eg, using a validated phantom) and the software tools used for dynamic susceptibility contrast magnetic resonance imaging analysis (eg, using a digital reference object where the ground truth is known).
Keywords: DSC-MRI, relative cerebral blood volume, standardization, multisite consistency, reproducibility
Introduction
The relative cerebral blood volume (rCBV), derived from dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI), is an established biomarker of glioma status that can aid in diagnosis (1), detecting treatment response (2, 3), guiding biopsies (4, 5), and reliable differentiation of post-treatment radiation effects and tumor progression (6–10). It is also increasingly leveraged as a biomarker of early therapeutic response in clinical trials (11, 12). However, variations in image acquisition and postprocessing methods (PMs) can limit rCBV reproducibility, potentially diminishing its clinical utility. To promote rCBV reproducibility across institutions, many national initiatives are underway to standardize DSC-MRI acquisition and PMs, including National Cancer Institute's Quantitative Imaging Network (QIN), Radiological Society of North America's Quantitative Imaging Biomarkers Alliance (QIBA), and the National Brain Tumor Society's Jumpstarting Brain Tumor Drug Development Coalition. Recent imaging protocol (IP) recommendations by the American Society of Functional Neuroradiology (ASFNR) has served as the first step in standardizing DSC-MRI protocols for clinical applications (13).
To aid in this effort, 12 institutions within the QIN aimed to investigate and determine the current rCBV reproducibility using a recently developed and validated in silico digital reference object (DRO) that is representative of a wide range of possible glioma magnetic resonance signals (14). Leveraging this DRO enables us as a community to determine the multisite consistency in rCBV owing to varying permutations of imaging acquisition parameters and postprocessing steps. In specific, our goals are to characterize rCBV consistency under conditions where there exist: (1) variations in the site-specific imaging acquisition parameters (PMs held constant), (2) variations in only site-specific PMs (IP held constant), and (3) variations owing to site-specific imaging and postprocessing protocols. Results from this community-based challenge will help steer standardization of DSC-MRI rCBV protocols with the hope that it can be successfully translated to the clinical setting.
Materials and Methods
This National Cancer Institute QIN DSC-DRO challenge project was proposed and organized by the investigators at Barrow Neurological Institute (BNI). Eleven centers participated in this project: BNI (the managing center), Brown University (BU), Massachusetts General Hospital (MGH), Mayo Clinic Arizona (Mayo AZ), Mayo Clinic Minnesota (Mayo MN), Medical College of Wisconsin (MCW), University of Michigan (UM1), The University of Texas Health at San Antonio (UTSA), University of Texas at Austin (UT), University of Texas Southwestern Medical Center at Dallas (UTSW), University of Washington (UW), and Washington University (WashU). Unless specifically named, these participating sites have been anonymized, in no particular order, and will be referred to as sites 01–12 as seen in Table 1.
Table 1.
Summary of Participating Teams' IPs and PMs
| Site Number | Imaging Protocol (IP) | CA | Processing Method (PM) | ID Tag for Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scan Protocol | Dose Protocol | |||||||||||
| Field Strength | TR (ms) | TE (ms) | Flip | Preload (mmol/kg) | Injection (mmol/kg) | Time Between (min) | Site IP w/Constant PP | Constant IP w/Site PP | Site IP w/Site PP | |||
| 01 | 01:3.0 T | 1500 | 30 | 60 | 0.05 | 0.10 | 3 | Gadobenate | 01: In-house processing | S01_IP01 | S01_PM01 | S01_IP01_PM01 |
| 02:1.5 T | 1500 | 30 | 60 | 0.05 | 0.10 | 3 | Gadobenate | S01_IP02 | S01_IP02_PM01 | |||
| 02 | 01:3.0 T | 1600 | 30 | 60 | 0 | 0.1 | n/a | Gadobenate | 01: IB Neuro | S02_IP01 | S02_PM01 | S02_IP01_PM01 |
| 03 | 01:3.0 T | 1500 | 31 | 90 | 0.05 | 0.15 | 6.5 | Gadoterate | 01: 3DSlicer | S03_IP01 | S03_PM01 | S03_IP01_PM01 |
| 02:3.0 T | 1500 | 31 | 90 | 0.1 | 0.1 | 6.5 | Gadoterate | 02: nordicICE | S03_IP02 | S03_PM02 | S03_IP01_PM02 | |
| 03: PGUI | S03_PM03 | S03_IP01_PM03 | ||||||||||
| S03_IP02_PM01 | ||||||||||||
| S03_IP02_PM02 | ||||||||||||
| S03_IP02_PM03 | ||||||||||||
| 04 | 01:3.0 T | 1500 | 30 | 80 | 0.10 | 0.10 | 5 | Gadobutrol | 01: IB Neuro | S04_IP01 | S04_PM01 | S04_IP01_PM01 |
| 02:3.0 T | 1500 | 2, 35 | 80 | 0 | 0.10 | n/a | Gadobutrol | S04_IP02 | n/a | |||
| 03:1.5 T | 1500 | 30 | 72 | 0.10 | 0.10 | 5 | Gadobutrol | S04_IP03 | S04_IP03_PM01 | |||
| 04:1.5 T | 1500 | 2, 35 | 72 | 0 | 0.10 | n/a | Gadobutrol | S04_IP04 | n/a | |||
| 05 | 01:3.0 T | 1300 | 30 | 60 | 0.025 | 0.10 | 5 | Gadobutrol | 01: IB Neuro (Integration limits 1) | S05_IP01 | S05_PM01 | S05_IP01_PM01 |
| 02:1.5 T | 1300 | 30 | 60 | 0.025 | 0.10 | 5 | Gadobutrol | 02: IB Neuro (Integration limits 2) | S05_IP02 | S05_PM02 | S05_IP01_PM02 | |
| S05_IP02_PM01 | ||||||||||||
| S05_IP02_PM02 | ||||||||||||
| 06 | 01:3.0 T | 1500 | 30 | 75 | 0.10 | 0.10 | 5 | Gadoteridol | 01: PGUI (rCBV definition 1) | S06_IP01 | S06_PM01 | S06_IP01_PM01 |
| 02: PGUI (rCBV definition 2) | S06_PM02 | S06_IP01_PM01 | ||||||||||
| 07 | 01:3.0 T | 1500 | 30 | 65 | 0.025 | 0.075 | 6 | Gadobenate | 01: In-house processing | S07_IP01 | S07_PM01 | S07_IP01_PM01 |
| 08 | 01:3.0 T | 1500 | 21 | 60 | 0.10 | 0.05 | 6 | Gadobenate | 01: In-house processing | S08_IP01 | S08_PM01 | S08_IP01_PM01 |
| 09 | 01:3.0 T | 1500 | 18 | 60 | 0.05 | 0.05 | 6 | Gadobenate | 01: IB Neuro | S09_IP01 | S09_PM01 | S09_IP01_PM01 |
| 10 | 01:3.0 T | 1900 | 36 | 90 | 0 | 0.10 | n/a | Gadoterate | 01: In-house processing | S10_IP01 | S10_PM01 | S10_IP01_PM01 |
| 02:3.0 T | 1900 | 36 | 90 | 0.10 | 0.10 | 5 | Gadoterate | S10_IP02 | S10_IP02_PM01 | |||
| 11 | 01:3.0 T | 2560 | 71 | 90 | 0.025 | 0.10 | 2 | Gadopentetate | n/a | S11_IP01 | n/a | n/a |
| 12 | 01:3.0 T | 1757 | 30 | 90 | 0.033 | 0.067 | 8 | Gadobutrol | 01: Philips ISP (rCBV definition 1) | S12_IP01 | S12_PM01 | S12_IP01_PM01 |
| 02: Philips ISP (rCBV definition 2) | S12_PM02 | S12_IP01_PM02 | ||||||||||
| 03: Philips ISP (rCBV definition 3) | S12_PM03 | S12_IP01_PM03 | ||||||||||
| Standard Protocol | 01:3.0 T | 1500 | 30 | 60 | 0.10 | 0.10 | 5 | Gadopentetate | n/a | SIP | n/a | n/a |
| Totala: 12 | 19 | 17 | 25 | |||||||||
a Excludes the standard protocol.
This project comprised 3 phases, summarized in the last 3 columns of Table 1, to evaluate the influence of IPs and/or PMs on multisite consistency:
Phase I (“site IP w/constant PM”) involved each participating site to submit their current clinical DSC IP to the managing center. The managing center then simulated site-specific DROs reflecting the IP parameters provided. Some sites provided >1 IP owing to differences in field strengths (sites 01, 04, and 05), dosing schemes (sites 03 and 10), and acquisition method (site 04). In total, 19 different IPs were submitted. The managing center postprocessed (specific details below in “Site-specific IP and PM”) rCBV maps of each of these submitted site-specific IP DROs to evaluate differences owing to the IP provided.
Phase II (“constant IP w/site PM”) involved analysis of a “standard imaging protocol” (SIP), which represents DSC-MRI data acquired using the IP recommended by ASFNR (13). Each site was asked to process DSC-MRI DRO data derived from the SIP. Some sites choose to use multiple commercially available software packages (site 03) and different rCBV definitions (sites 05, 06, 12), yielding a total of 17 submitted rCBV maps.
Phase III (“site IP w/site PM”) required each site to calculate rCBV maps using their PM of choice and the site-specific DRO data. Combining the possible permutations owing to choice of IP and PM from phases I–II, a total of 25 rCBV maps were submitted.
All sites but 1 completed all 3 phases of the challenge. Site 11 completed only phase I, and these results are included in this study.
DRO Simulations
The DSC-MRI signals for each IP were simulated using a recently developed and validated population-based DRO that was trained to generate realistic signals using in vivo data from >40 000 voxels derived from patient data (14). The resulting DRO, which contains 10 000 unique voxels, reflects the distribution of perfusion, permeability, precontrast T1, T2*, diffusion coefficients, and the vascular and cellular features found in patients with high-grade glioma. Using this DRO, the DSC-MRI signals and resulting rCBV values can be computed for any combination of preload dosing scheme, contrast agent choice (by varying T1 relaxivities specific to the contrast agent), pulse sequence parameters, and postprocessing protocol. For the purposes of this study, the DRO consisted of tumor voxels simulated under two blood-brain-barrier (BBB) conditions to recapitulate DSC-MRI signals from an intact-BBB (Ktrans = 0) and a disrupted-BBB (Ktrans > 0). In addition to the tumor voxels, normal appearing white matter (NAWM) voxels (Ktrans = 0) were simulated to normalize CBV. For the purposes of comparing site-to-site consistency, the SIP that has been postprocessed by the managing center was considered the reference standard where necessary. In our recent study, focused on investigating the influence of IP on CBV fidelity (15), the SIP yielded CBV values, when corrected for contrast agent leakage, that were among the most accurate.
Site-Specific IP and PM Methods
Site-specific IP and PM methods are briefly listed in Table 1. Overall, IPs were similar across sites. Most sites submitted clinical DSC IPs for 3 T with 3 sites that also included a 1.5 T IP. Overall, the following were the imaging parameters [mode (min-max)]: repetition time = 1500 milliseconds (1300–2560 milliseconds), echo time (TE) = 30 milliseconds (18–71 milliseconds), flip angle = 60° (60°–90°), preload dose = 0.05 mmol/kg (0–0.1 mmol/kg), and injection dose = 0.1 mmol/kg (0.05–0.15 mmol/kg). Five different gadolinium contrast agents were used across the 12 sites: gadobenate (n = 5), gadobutrol (n = 3), gadoterate (n = 2), gadoteridol (n = 1), and gadopentetate (n = 1). For PMs, there was a mix of software options used, including in-house-based software scripts (n = 4), IB Neuro (n = 4), 3D Slicer (n = 1), nordicICE (n = 1), PGUI (n = 2), and Philips IntelliSpace Portal (ISP; Philips Healthcare, Best, the Netherlands) (n = 1).
For PM methods, most sites defined rCBV = ∫0t ΔR*2,tumor−BSW/∫0t ΔR*2,NAWM and used the Boxerman–Schmainda–Weiskoff (BSW) method for leakage correction (16). A few sites submitted results that deviated from this postprocessing convention by alternative rCBV definitions (S06, S12) and differences in integration limits as determined by the software (S05). These differences are highlighted in Table 1. Site 06 defined CBV by the area under the curve of the deconvolved residue function. This deconvolved residue function was determined by singular value decompositions (rCBV definition 1) and by oscillating singular value decompositions approach (rCBV definition 2). S12 used 3 different rCBV definitions within the Philips ISP platform: a “model-free” option that integrates the area underneath the signal intensity curve (rCBV definition 1) (17), a “γ-variate” option that integrates the area underneath the signal intensity curve that has been fit to a γ-variate function (rCBV definition 2), and a “leakage correction” option that integrates the area underneath the computed delta R2* curve after a modified BSW leakage correction method is applied (rCBV definition 3). To be clear, the first 2 options of the Philips ISP do not apply any sort of leakage correction algorithm to the data. S05 included CBV maps calculated using the default integration limits set by IB Neuro (integration limits 1) and manually chose all time points in IB Neuro (integration limits 2). A little less than 50% of the submitted rCBV maps were normalized to the NAWM. To compare maps, the managing site normalized tumor CBV to the mean NAWM CBV of all pixels when necessary. Specifics on site-specific postprocessing steps are outlined in Table 2.
Table 2.
Summary of Participating Teams' PMs
| Site Number | Software | CBV Definition | Normalized to NAWM? | Integration Limits | Leakage Correction Method | Comments |
|---|---|---|---|---|---|---|
| 01 | 01: In-house processing | AUC of the ΔR2* time course | No | Time points: 2 to 64 (93 sec) | BSW leakage correction method | Manual inspection of pre- and post- contrast points for rCBV integration |
| 02 | 01: IB Neuro | AUC of the ΔR2* time course | Yes | automatically detected (default option) | BSW leakage correction method | Default IB Neuro settings for rCBV |
| 03 | 01: 3DSlicer | AUC of the ΔR2* time course | No | 118 seconds | BSW leakage correction method | No thresholding |
| 02: nordicICE | AUC of the ΔR2* time course | Yes | Time points: 2 to 121 (178.5 sec) | BSW leakage correction method | ||
| 03: PGUI | AUC of the ΔR2* time course | No | Time points: 2 to 121 (178.5 sec) | BSW leakage correction method | No thresholding, but smoothing applied | |
| 04 | 01: IB Neuro | AUC of the ΔR2* time course | Yes | automatically detected (default option) | BSW leakage correction method | |
| 05 | 01: IB Neuro (Integration limits 1) | AUC of the ΔR2* time course | Yes | automatically detected (default option) | BSW leakage correction method | |
| 02: IB Neuro (Integration limits 2) | AUC of the ΔR2* time course | Yes | 180 seconds (all time points) | BSW leakage correction method | ||
| 06 | 01: PGUI (rCBV definition 1) | Deconvolution of the residue function (SVD) | No | Time points: 5 to 121 (174 sec) | BSW leakage correction method | |
| 02: PGUI (rCBV definition 2) | Deconvolution of the residue function (oSVD) | No | Time points: 5 to 121 (174 sec) | BSW leakage correction method | ||
| 07 | 01: In-house processing | AUC of the ΔR2* time course | No | automatically detected (default option) | BSW leakage correction method | |
| 08 | 01: In-house processing | AUC of the ΔR2* time course | Yes | 90 sec | BSW leakage correction method | |
| 09 | 01: IB Neuro | AUC of the ΔR2* time course | Yes | automatically detected (default option) | BSW leakage correction method | Did not use the entire NAWM ROI - instead used a 6 mm × 6 mm (∼225 pixels) ROI |
| 10 | 01: In-house processing | AUC of the ΔR2* time course | No | 171 sec | BSW leakage correction method | ΔR2* maps were smoothed with a 5 × 5 Gaussian window that had an FWHM value of 3 mm |
| 11 | n/a | |||||
| 12 | 01: Philips ISP (rCBV definition 1) | AUC of the SI time course | No | Based on the characteristics of signal time curves | No leakage correction method | |
| 02: Philips ISP (rCBV definition 2) | AUC of the SI time course fitted to a gamma-variate | No | Based on the characteristics of signal time curves | No leakage correction method | ||
| 03: Philips ISP (rCBV definition 3) | AUC of the ΔR2* time course | No | 180 s | BSW leakage correction method | ||
The managing center postprocessed the site-specific DROs with an in-house script by defining rCBV = ∫0120sec ΔR*2,tumor−BSW/∫0120sec ΔR*2,NAWM, where the conventional ΔR*2 curves in the tumor were corrected for leakage effects using the BSW method. In our recent study, the CBV was found to be the most accurate by using these specific PM steps, and thus was chosen to be used as the reference where applicable (15). No thresholding, smoothing, or quality assessment was done before rCBV calculations when analyzed by the managing center.
Statistics
To evaluate the consistency of rCBV across sites owing to differences between IP and PM, the intraclass correlation coefficient (ICC) was calculated. Furthermore, to evaluate the agreement of rCBV between sites and a reference (SIP), the 95% limits of agreement (LOA) were extracted from a Bland–Altman analysis. Variability of rCBV was assessed across a distribution of rCBV values by calculating the covariance (CV) across sites. Lastly, Lin's correlation coefficient was calculated for rCBV between the intact-BBB and disrupted-BBB DROs for each permutation of IP and PM to determine the agreement of rCBV after leakage correction was applied. All statistical calculations were done in MATLAB R2018a (The MathWorks Inc., Natick, MA) by the managing center.
Results
In general, the ICC decreases when Ktrans > 0, that is, disrupted-BBB (Table 3) for all the 3 phases of this study. High agreement is observed across sites when a constant PM is applied to site-specific IP (ICC = 0.879). However, when site-specific PMs are applied to either a constant IP or to their site-specific IP, the agreement is quite poor (ICC = 0.439 and 0.380, respectively).
Table 3.
Intraclass Correlation Coefficient Results for Each Phase of the Study for Computed rCBV from the Simulated Intact-BBB and Disrupted-BBB DRO
| Site-Specific IP w/Constant PM | Constant IP w/Site-Specific PM | Site-Specific IP w/Site-Specific PM | |
|---|---|---|---|
| Intact-BBB | 0.970 | 0.690 | 0.641 |
| Disrupted-BBB | 0.879 | 0.439 | 0.380 |
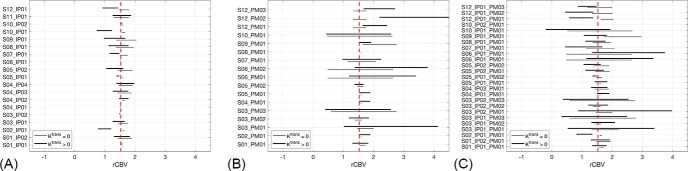
Figure 1 shows consistency in rCBV measurements for all 3 phases of this study when compared with the reference. For each site, the 95% LOA of both Ktrans = 0 (gray lines, intact-BBB) and Ktrans > 0 (black lines, disrupted-BBB) are indicated in comparison to the reference (see Table 1 for site ID descriptions). For phase I (Figure 1A), the 95% LOA are generally fairly narrow and centered around the mean rCBV of the reference for the Ktrans > 0 case. A few exceptions (S02_IP01, S10_IP01, and S12_IP01) show larger 95% LOA and a negative bias compared with the other sites. The first 2 sites (S02 and S10) did not use a contrast agent preload unlike the other sites, while the third site (S12) used 1/3 standard dose for a preload. Sites S09_IP01 and S10_IP01, although centered around the reference's mean rCBV, also express wider ranges of 95% LOA compared with other sites. These 2 sites have markedly lower TE and use less than a full standard dose compared with the other sites. Much larger LOA are seen for phase II in Figure 1B) than for that in Figure 1A. Large 95% LOA are observed for even the Ktrans = 0 case, where no leakage correction is applied during postprocessing. The analysis software that show the smallest 95% LOA with the reference are in-house processing scrips (S01_PM01, S08_PM01), IB Neuro (S02_PM01, S04_PM01, S05_PM01, S05_PM02, S09_PM01), nordicICE (S03_PM02), and the “model-free option” in Philips ISP (S12_PM01). For phase III (Figure 1C), 9 out of the 24 sites show a tight 95% LOA and relatively no bias when compared to the SP reference (S01_IP01_PM01, S01_IP01_PM02, S03_IP02_PM02, S04_IP01_PM01, S04_IP03_PM01, S05_IP01_PM01, S05_IP01_PM02, S05_IP02_PM01, S05_IP02_PM02) for the Ktrans > 0 case. These 4 sites implemented nordicICE, IB Neuro, and an in-house postprocessing script.
Figure 1.
Bland–Altman limits of agreement (LOA) against the standard imaging protocol (SIP) plotted for site-specific IP w/constant postprocessing method (PM) (A), constant IP w/site-specific PM (B), and site-specific IP w/site-specific PM (C). The vertical dashed line is the mean rCBV across 10 000 voxels for the SIP.
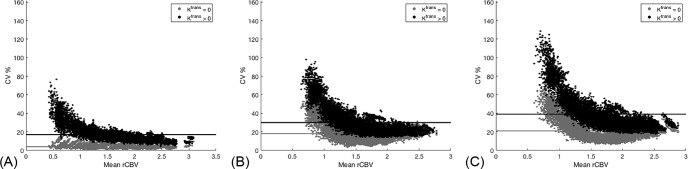
Figure 2 illustrates the CV as a function of rCBV across DROs for all voxels. The covariance across DROs (nPhase I = 19, nPhase II = 17 nPhase III = 25) was calculated in the 10 000 tumor voxels and plotted against the mean rCBV of each voxel across DROs. The DRO simulated with Ktrans = 0 (gray circles) and Ktrans > 0 (black circles) is plotted along with the mean CV (horizontal line plots) across all voxels. This figure does not assume a reference for calculations. In general, the CV increases for each phase when more freedom is allowed in the rCBV calculations for both IP and PM. For phase I (Figure 2A), the average CV is 4% and it remains fairly flat over the rCBV distribution for Ktrans = 0. However, when Ktrans > 0, the average CV rose to 17% and exponentially decreased from roughly 60% to 10% as rCBV increased. For phases II and III (Figure 2, B and C, respectively), the CV is observed to exponentially decrease for both Ktrans cases. For phase II, the average CV is 18% and 30% for Ktrans = 0 and > 0, respectively. As rCBV increases, the CV exponentially decreases from roughly 80% to 20% for both Ktrans cases. For phase III, the average CV is 21% and 39% for Ktrans = 0 and > 0, respectively. As rCBV increases, the CV exponentially decreases from roughly 120% to 35% for both Ktrans cases.
Figure 2.
The covariance (CV%) across all relative cerebral blood volume (rCBV) maps for each of the 10 000 voxels plotted across the mean rCBV of the voxels for site-specific IP w/constant PM (A), constant IP w/site-specific PM (B), and site-specific IP w/site-specific PM (C). Results from the Ktrans = 0 (light gray) and Ktrans > 0 (black) are included with their mean CV% across all 10 000 voxels indicated for the horizontal lines. For all 3 phases of this study, the largest variation in rCBV occurs at the low rCBV range for Ktrans > 0, and CV% increases when more freedom was introduced in the choice of IPs and PMs.
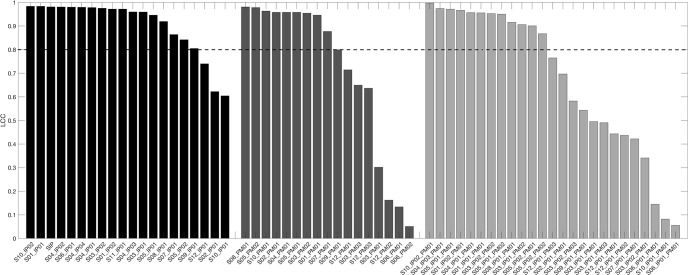
Figure 3 examines the agreement between the intact-BBB (Ktrans = 0) and disrupted-BBB (Ktrans > 0) DRO for each processed rCBV map. The LCC for each analysis combination was sorted from the highest (perfect agreement = 1) to the lowest (no agreement = 0) for each of the 3 phases. A high agreement indicates that the processed CBV from the simulated disrupted-BBB DRO had high accuracy when compared to the simulated intact DRO where no leakage occurs. Site-specific IP with constant PM is shown by the black bars in the bar graph. Note that the third black bar is the SIP and has a high LCC value, which is consistent with previous results (15) and therefore used as reference in Figure 1. Here we observed that most of the sites' IPs are able to accurately compute CBV—most likely because these sites already use IPs similar to the SIP. Three site protocols had an LCCC < 0.8, indicating low rCBV accuracy when leakage effects are introduced: S02_IP01, S10_IP01, and S12_IP01. These protocols also resulted in large LOA and a negative bias as seen in Figure 1. These results indicate that the IP is highly sensitive to contrast agent leakage effects even when a leakage correction PM algorithm is applied. Constant IP with site-specific PM results are indicated in the dark gray bars in the bar graph. Here we observe 10 software programs that clearly show high agreement: in-house scripts (n = 4), IB Neuro (n = 4), nordicICE (n = 1), and “model-free” option in the Philips ISP. Lastly, site-specific IP with site-specific PM resulted in 50% of the rCBV maps with LCC < 0.8, most likely owing to a combination of variations in IPs and postprocessing as deduced from the earlier 2 phases.
Figure 3.
A bar plot of Lin's correlation coefficient (LCC) for each rCBV map for site-specific IP w/constant PM (black), constant IP w/site-specific PM (medium gray), and site-specific IP w/site-specific PM (light gray). Each phase is sorted by the resulting LCC from the highest to the lowest value. A horizontal bar at LCC = 0.8 is placed to evaluate agreement good agreement (LCC > 0.8).
Discussion and Conclusion
Reproducibility in DSC-MRI rCBV is crucial for the success of multisite clinical trials. In this study, we have evaluated rCBV consistency owing to differences in both IPs and PMs across 12 QIN sites using a DRO. The results outlined in this manuscript show that standardization of both is warranted.
Our prior DRO investigation highlighted the significant influence of IPs (including preload dosing and pulse sequence parameters) on CBV accuracy (15). The findings of this study strongly indicate that differences in the PM can also confound multisite CBV consistency and accuracy. High agreement when site-specific IP were processed by the managing center most likely reflects the similarity of the IP parameters across all the sites owing to previous initiatives from the ASFNR that aimed to standardize IPs (13). However, it was observed that when no preload was used in the IP (S02_IP01 and S10_IP01), a systematic negative bias relative to the SIP occurs. Furthermore, a slight negative bias is observed for the sites that administered less than a full standard dose as the main injection (S07_IP01, S08_IP01, S09_IP01, S12_IP01). These 2 findings underscore potential challenges to comparing CBV changes in a clinical trial from sites that use dissimilar preload and bolus dosing protocols. Three sites (S01, S04, S05) provided clinical IPs for 1.5 T. Sites 01 and 05 used the same IP at both field strengths, and it was observed that the LOA did widen when compared to the 3.0 T protocol. Site 04 used a smaller flip angle at 1.5 T than at 3 T; however, a widening of LOA was still observed. Differences here may warrant further investigation into a standardized 1.5 T IP; however, for the scope of this paper, high agreement was observed when both field strengths were compared together.
When each site was asked to postprocess the SIP, agreement decreased substantially as indicated by the ICC and the 95% LOAs. Interestingly, the disagreement across sites is not isolated to differences in the leakage correction method, as poor agreement is also observed with the Ktrans = 0 case. For the Ktrans = 0 case, 1 potential source of disagreement in rCBV arises from whether smoothing is implemented in the software and the CBV definition. Methods 01 and 02 from S12 deviated from the traditional CBV definition, as these methods calculated CBV from the signal intensity curves, potentially losing the biophysics and kinetic properties. For the Ktrans > 0 case, potential sources of disagreement in rCBV may be attributed to smoothing and the algorithms and/or implementation of algorithms used for leakage correction.
It is challenging to compare the results from this current study directly to prior ones since we performed a voxel-wise analysis across the DRO, whereas most other studies, like the recent DSC-MRI challenge (18), report comparisons between mean region of interest tumor values across data that likely exhibits patient-specific rCBV distributions. As seen in Figure 2, there is greater variation across platforms at low rCBV values. These differences most likely average out when hotspot types of analyses are performed. Although most likely sufficient for diagnosis of tumor grade, this might not be ideal for longitudinal assessment of treatment response where voxel-wise analysis and/or CBV difference quantification has shown to be more beneficial (11, 12). Despite this, our results indicating inconsistent CBV values as more freedom is allowed to the IP and processing methods is not surprising. Kelm et al. compared rCBV measurements using 3 software platforms (IB Neuro, FuncTool, and nordicICE) and also found significant variation in rCBV (19).
A limitation to our study is that the ROIs for brain tumor and NAWM have been clearly outlined and predetermined for analysis. In the context of patient data, allowing users to define ROIs would likely contribute to greater rCBV inconsistency. Schmainda et al. showed high mean CBV agreement when ROIs were predetermined (18). In addition, sites were not required to determine an AIF for the CBV calculations within this manuscript.
Results from this study and our prior DRO analysis, which focused on IP optimization (15), highlight the IPs and PMs that maximize rCBV accuracy and multisite consistency. First, IPs that yield the highest rCBV accuracy and multisite concordance utilize a full-dose contrast agent preload and a full-dose bolus injection, low (30°) or moderate (65°) flip angle, ∼30 millisecond TE, and a ∼1.5 millisecond TR. In both studies, the use of lower bolus dose injections (eg, 1/2 dose) were found to substantially reduce both consistency and accuracy, likely owing to the lower CNR. Second, the 2 studies further show that, even with optimized IPs, leakage correction should be applied to DSC-MRI data in brain tumors. Further, the correction algorithms should be based on the underlying biophysics and kinetics, such as the BSW correction, as they maximize both accuracy and precision. Generic leakage correction algorithms (like gamma variate fitting) that arbitrarily modify the shape of DSC-MRI data to remove T1 and/or T2* leakage effects are not recommended. It should be noted that in the IP optimization study (15), a low flip angle approach (30° with a 30 millisecond TE) with a full-dose bolus injection, no preload, and application of BSW leakage correction provided accuracy slightly less than that using the ideal protocol. Studies are currently underway to validate the clinical potential of this protocol as it could be a compelling single-dose option for routine surveillance scans and in clinical trials.
Although great efforts have been made to standardize DSC-MRI imaging acquisition protocols, this study highlights that poor CBV agreement can arise when there are variations in processing platforms. Highest agreement is observed when site-specific CBV maps are processed by 1 managing center, as might be expected in a clinical trial setting where acquisition and PMs are predetermined, and/or raw data are sent to a single site for analysis. However, differences in CBV, especially at low values, as would be expected with effective therapy, arise when different platforms are used. This finding has important implications for comparing CBV values across trials, where variability in trial-specific PMs could confound the comparison of therapeutic effectiveness and/or any attempts to establish thresholds for categorical response (eg, predetermined percent changes in rCBV values that could be used to refine RANO criteria). To overcome these challenges and to ensure the successful use of rCBV as a clinical trial biomarker, it is critical that the DSC-MRI community establish qualifying and validating criteria, similar to that in the RSNA DCE-MRI Profile (20), for scanners and acquisition methods to be used in clinical trials (eg, using a validated phantom) and the software used for DSC-MRI analysis (eg, using a DRO where the ground truth is known).
Acknowledgments
NIH/NCI R01CA213158 (LCB, NS, CCQ); NIH/NCI U01CA207091 (AJM, MCP); NIH/NCI U01CA166104 and P01CA085878 (DM, TLC); NIH/NCI U01CA142565 (CW, AGS, TEY, NR); NIH/NCI U01 CA176110 (KMS, MAP).
Disclosure: No disclosures to report.
Conflicts of Interest: The authors have no conflicts of interest to declare.
Footnotes
- rCBV
- Relative cerebral blood volume
- PMs
- postprocessing methods
- IPs
- imaging protocols
- DSC-MRI
- dynamic susceptibility contrast magnetic resonance imaging
- QIN
- Quantitative Imaging Network
- ASFNR
- American Society of Functional Neuroradiology
- DRO
- digital reference object
- SIP
- standard imaging protocol
- ICC
- intraclass correlation coefficient
- LOA
- limits of agreement
- CV
- covariance
- TE
- echo time
- NAWM
- normal appearing white matter
References
- 1. Fink JR, Muzi M, Peck M, Krohn KA. Multimodality brain tumor imaging: MR imaging, PET, and PET/MR imaging. J Nucl Med. 2015;56:1554–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boxerman JL, Ellingson BM, Jeyapalan S, Elinzano H, Harris RJ, Rogg JM, Pope WB, Safran H. Longitudinal DSC-MRI for distinguishing tumor recurrence from pseudoprogression in patients with a high-grade glioma. Am J Clin Oncol. 2014;0:1–7. [DOI] [PubMed] [Google Scholar]
- 3. Ellingson BM, Zaw T, Cloughesy TF, Naeini KM, Lalezari S, Mong S, Lai A, Nghiemphu PL, Pope WB. Comparison between intensity normalization techniques for dynamic susceptibility contrast (DSC)-MRI estimates of cerebral blood volume (CBV) in human gliomas. J Magn Reson Imaging. 2012;35:1472–1477. [DOI] [PubMed] [Google Scholar]
- 4. Chaskis C, Stadnik T, Michotte A, Van Rompaey K, D'Haens J. Prognostic value of perfusion-weighted imaging in brain glioma: a prospective study. Acta Neurochir (Wien). 2006;148:277–285. [DOI] [PubMed] [Google Scholar]
- 5. Maia ACM, Malheiros SMF, da Rocha AJ, Stávale JN, Guimarães IF, Borges LR, Santos AJ, da Silva CJ, de Melo JG, Lanzoni OP, Gabbai AA, Ferraz FA. Stereotactic biopsy guidance in adults with supratentorial nonenhancing gliomas: role of perfusion-weighted magnetic resonance imaging. J Neurosurg. 2004;101:970–976. [DOI] [PubMed] [Google Scholar]
- 6. Danchaivijitr N, Waldman AD, Tozer DJ, Benton CE, Brasil Caseiras G, Tofts PS, Rees JH, Jäger HR. Low-grade gliomas: do changes in rCBV measurements at longitudinal perfusion-weighted MR imaging predict malignant transformation? Radiology. 2008;247:170–178. [DOI] [PubMed] [Google Scholar]
- 7. Rossi Espagnet MC, Romano A, Mancuso V, Cicone F, Napolitano A, Scaringi C, Minniti G, Bozzao A. Multiparametric evaluation of low grade gliomas at follow-up: comparison between diffusion and perfusion MR with 18F-FDOPA PET. Br J Radiol. 2016;89:20160476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boxerman JL, Paulson ES, Prah MA, Schmainda KM. The effect of pulse sequence parameters and contrast agent dose on percentage signal recovery in DSC-MRI: implications for clinical applications. AJNR Am J Neuroradiol. 2013;34:1364–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu LS, Baxter LC, Smith KA, Feuerstein BG, Karis JP, Eschbacher JM, Coons SW, Nakaji P, Yeh RF, Debbins J, Heiserman JE. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. Am J Neuroradiol. 2009;30:552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barajas RF, Chang JS, Segal MR, Parsa AT, McDermott MW, Berger MS, Cha S. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology. 2009;253:486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galbán CJ, Lemasson B, Hoff BA, Johnson TD, Sundgren PC, Tsien C, Chenevert TL, Ross BD. Development of a multiparametric voxel-based magnetic resonance imaging biomarker for early cancer therapeutic response assessment. Tomography. 2015;1:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boxerman JL, Zhang Z, Safriel Y, Larvie M, Snyder BS, Jain R, Chi TL, Sorensen AG, Gilbert MR, Barboriak DP. Early post-bevacizumab progression on contrast-enhanced MRI as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 Central Reader Study. Neuro Oncol. 2013;15:945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Welker K, Boxerman J, Kalnin A, Kaufmann T, Shiroishi M, Wintermark M; American Society of Functional Neuroradiology MR Perfusion Standards and Practice Subcommittee of the ASFNR Clinical Practice Committee. ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. AJNR. Am J Neuroradiol. 2015;36:E41–E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Semmineh NB, Stokes AM, Bell LC, Boxerman JL, Quarles CC. A population-based digital reference object (DRO) for optimizing dynamic susceptibility contrast (DSC)-MRI methods for clinical trials. Tomography. 2017;3:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Semmineh N, Bell L, Stokes A, Hu L, Boxerman J, Quarles C. Optimization of acquisition and analysis methods for clinical dynamic susceptibility contrast (DSC) MRI using a population-based digital reference object. AJNR Am J Neuroradiol. 2018;39:1981–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27:859–867. [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer-Baese A, Lange O, Wismueller A, Hurdal MK. Analysis of dynamic susceptibility contrast MRI time series based on unsupervised clustering methods. IEEE Trans Inf Technol Biomed. 2007;11:563–573. [DOI] [PubMed] [Google Scholar]
- 18. Schmainda KM, Prah MA, Rand SD, Liu Y, Logan B, Muzi M, Rane SD, Da X, Yen YF, Kalpathy-Cramer J, Chenevert TL, Hoff B, Ross B, Cao Y, Aryal MP, Erickson B, Korfiatis P, Dondlinger T, Bell L, Hu L, Kinahan PE, Quarles CC. Multisite concordance of DSC-MRI analysis for brain tumors: results of a National Cancer Institute Quantitative Imaging Network Collaborative Project. AJNR Am J Neuroradiol. 2018;39:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelm ZS, Korfiatis PD, Lingineni RK, et al. . Variability and accuracy of different software packages for dynamic susceptibility contrast magnetic resonance imaging for distinguishing glioblastoma progression from pseudoprogression. J Med imaging (Bellingham). 2015;2:26001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DCE MRI Technical Committee. DCE MRI Quantification Profile, Quantitative Imaging Biomarkers Alliance. 2012; Version 1.0. Available from: http://rsna.org/QIBA_.aspx.