Abstract
We identified computational imaging features on 18F-fluorodeoxyglucose positron emission tomography (PET) that predict recurrence/progression in non–small cell lung cancer (NSCLC). We retrospectively identified 291 patients with NSCLC from 2 prospectively acquired cohorts (training, n = 145; validation, n = 146). We contoured the metabolic tumor volume (MTV) on all pretreatment PET images and added a 3-dimensional penumbra region that extended outward 1 cm from the tumor surface. We generated 512 radiomics features, selected 435 features based on robustness to contour variations, and then applied randomized sparse regression (LASSO) to identify features that predicted time to recurrence in the training cohort. We built Cox proportional hazards models in the training cohort and independently evaluated the models in the validation cohort. Two features including stage and a MTV plus penumbra texture feature were selected by LASSO. Both features were significant univariate predictors, with stage being the best predictor (hazard ratio [HR] = 2.15 [95% confidence interval (CI): 1.56–2.95], P < .001). However, adding the MTV plus penumbra texture feature to stage significantly improved prediction (P = .006). This multivariate model was a significant predictor of time to recurrence in the training cohort (concordance = 0.74 [95% CI: 0.66–0.81], P < .001) that was validated in a separate validation cohort (concordance = 0.74 [95% CI: 0.67–0.81], P < .001). A combined radiomics and clinical model improved NSCLC recurrence prediction. FDG PET radiomic features may be useful biomarkers for lung cancer prognosis and add clinical utility for risk stratification.
Keywords: radiomics, lung cancer, risk stratification, recurrence, PET
Introduction
Lung cancer remains the most common cause of cancer death worldwide, and the 5-year survival rates of non–small cell lung cancer (NSCLC) remain quite poor despite advances in diagnosis and treatment (1, 2). Further, many patients will develop recurrence or progression following primary treatment. The absolute risk of any recurrence at 5 years post-treatment ranges from 33% to 52%, with the majority occurring at a distant site (3, 4). Among prognostic factors for predicting outcomes in NSCLC, tumor stage based on the American Joint Committee on Cancer (AJCC) staging system is currently considered the best for predicting outcomes (5). More accurate clinical, imaging, and molecular biomarkers will be extremely useful for stratifying patients who are at a higher risk of recurrence and who might benefit from adjuvant or more aggressive treatment options (6).
Maximum standardized uptake value (SUVmax) on fluorine-18F fluoro-2-deoxy-D-glucose (FDG) positron emission tomography (PET) imaging has also been shown to predict recurrence or death in NSCLC (7). However, this is a single-voxel metric; we hypothesized that applying a radiomics approach to extract more complex information (eg, texture) from standard medical images could provide additional prognostic information (8, 9).
While recent work has evaluated the potential for radiomics features to augment traditional metrics of response (10–12), the majority of studies to date have focused on only the metabolic tumor volume (MTV) on PET and, to the best of our knowledge, no study has investigated the peritumoral region. Tumor invasion from the main mass can be defined by infiltration of stroma, blood vessels, or visceral pleura (13). Recent studies have also shown the potential for tumor cells to spread into air spaces in the lung tissue adjacent to the tumor volume (14). It is well known that these features may present as border spiculation, vascular convergence, or pleural attachment surrounding the tumor on anatomical imaging, and that they may result in subtle heterogeneous uptake on PET imaging (15).
We investigated the potential of FDG-PET radiomics to predict recurrence in NSCLC by (1) assessing the variability in radiomic feature extraction from PET images and (2) building and validating a radiomics model to predict time to recurrence. We hypothesize that computational imaging features in the tumor and surrounding area on FDG-PET can augment clinical features to improve recurrence prediction.
Methodology
Patient Selection
We retrospectively analyzed a total of 291 patients with NSCLC from 2 distinct cohorts of prospectively acquired patients (n = 145 and n = 146). The study was approved by our Institutional Review Board, and all subjects signed informed consent before participation. Our study was also compliant with the Health Insurance Portability and Accountability Act.
The training cohort consisted of subjects from a pool of patients with early-stage NSCLC referred for surgical treatment at 2 local medical centers between 2008 and 2012 with preoperative PET/computed tomography (CT) performed before surgery (n = 145). This data set is publicly available on The Cancer Imaging Archive (16, 17). We used a second cohort (n = 146) for model validation. This was a cohort from 3 local medical centers between 2010 and 2016. Subjects were selected from patients undergoing evaluation for lung cancer by PET/CT imaging before definitive treatment as part of an observational biomarker study. In both the training and validation cohorts, there were no patients that received neoadjuvant therapy.
The AJCC seventh edition system was used for staging. Pathological staging was used in the training cohort and a combination of clinical and pathological staging in the validation cohort. Demographic differences between the training and validation cohorts were assessed using the Wilcoxon rank-sum test for continuous variables and the χ2 test for categorical variables. All patients were followed per standard clinical protocol with clinical examination and imaging. We analyzed the combined endpoint of disease recurrence or progression. For stage I–IIIA subjects, we defined recurrence as either local, regional, or distant. For patients with stage IIIB–IV disease, we defined an event as any progression of disease. Time to event or last known follow-up was recorded from the date of pretreatment PET imaging.
Image Acquisition
Pretreatment FDG-PET/CT scans were acquired using a standard clinical protocol at 1 of 3 local medical centers. Images were acquired using either a GE Discovery VCT (GE Health care, Waukesha, WI), a GE Discovery LS PET/CT (GE Healthcare, Waukesha, WI), a Siemens Biograph mCT (Siemens Healthcare, Erlangen, Germany), or a Phillips Allegro/Gemini TF PET/CT (Phillips Healthcare, Cleveland, OH). Patients underwent scanning following fasting for a minimum of 6–8 h. A dose of 12–17 mCi of FDG was administered and patients underwent scanning from the skull base to mid-thigh using bed positions acquired every 2–5 minutes ∼45–60 minutes after injection. Manufacturer-specific CT-based attenuated correction was performed using ordered subset expectation maximization reconstruction.
Region of Interest Delineations
Pretreatment PET images were converted to SUV units normalized by body weight. Two research assistants (S.M. and S.B.) were trained by a board-certified physician in Nuclear Medicine (G.D.) in using MIM Version 6.6 (MIM Software Inc., Cleveland, OH) to contour tumor MTVs using the semiautomatic PET-edge gradient-based segmentation tool. Both observers contoured all images independently in the training cohort. A subset of 21 images considered difficult to contour were reviewed by the same physician and re-delineated if necessary. To assess intraobserver variability, observer 1 (S.M.) contoured all images a second time after a delay of 3 months. We calculated the Dice similarity coefficient (DSC), mean absolute distance (MAD) of the boundary, and absolute volume difference between each set of contours to assess inter- and intraobserver variability of the MTV regions in the training cohort. Observer 1 alone contoured all images in the validation cohort.
We then generated a 3-dimensional penumbra region extending outward 1 cm from the surface of the MTV to sample surrounding uptake by using a 3D distance transform with a threshold of 1 cm. This distance was intuitively chosen to sample enough surrounding tissue given the voxel sizes of the PET images, while avoiding oversampling normal tissue. In addition to the MTV alone, we also evaluated the following 2 additional regions: the MTV plus penumbra and the penumbra only (excluding the MTV).
Feature Extraction
We extracted radiomics features in the MTV, penumbra, and MTV plus penumbra regions in both cohorts using The Quantitative Image Feature Engine (18) implemented in MATLAB R2016B (The MathWorks, Natick, MA). In the MTV, features included size (n = 4), sphericity (n = 1), local volume-invariant integral (LVII) shape (n = 39), histogram intensity (n = 12), and gray-level co-occurrence matrix (GLCM) texture (n = 144) (19, 20), for a total of 200 features. Because the penumbra region was generated from the MTV, 44 size and shape measures were not calculated in the penumbra and MTV plus penumbra regions (because they would not be independent measurements), for a total of 156 features in each. This resulted in a total of 512 features for analysis as summarized in Table 1. We set a fixed intensity bin size of 0.2 SUV for texture feature calculation to allow a meaningful comparison between images on the same SUV scale. This discretization may also reduce the differences between multiple scanners used in this study (21).
Table 1.
Number of Extracted Features
| Region of Interest | Feature Type | Number of Features | Total Number of Features in ROI |
|---|---|---|---|
| Metabolic Tumor Volume (MTV) | Size | 4 | 200 |
| Sphericity | 1 | ||
| LVII shape | 39 | ||
| Intensity | 12 | ||
| GLCM texture | 144 | ||
| Penumbra | Intensity | 12 | 156 |
| GLCM texture | 144 | ||
| MTV + Penumbra | Intensity | 12 | 156 |
| GLCM texture | 144 | ||
| Total Number of Features | 512 | ||
We then calculated intraclass correlation coefficients (ICCs) across the 3 sets of outlines for each radiomic feature to assess inter- and intraobserver variability. Robust features, defined as those with ICCs >0.8 in the training cohort, were selected for further analysis (22, 23).
Model Building and Validation
All radiomic features were normalized (Z-score transformation) before feature selection and model building. We further optimized the features through a generalized linear model via the least absolute shrinkage and selection operator (LASSO) (24) Cox regression using the glmnet package in R software version 3.4.3 (25). LASSO is a shrinkage and variable selection method for high-dimensional data, which was used to select top features to predict time to recurrence in the training cohort. The robust radiomic features and the 2 known clinical predictors (stage and SUVmax) were provided to LASSO. Alpha, the regularization parameter, was set to 1 (LASSO penalty) to minimize the number of selected features by shrinking most of the coefficients to zero and to minimize potential overfitting in the training cohort. In total, 100 randomizations of 4-fold cross-validation was used to reduce the effect of randomness in fold selection. The mean cross-validated error curves were averaged for each tuning parameter lambda value across all randomizations. The lambda and corresponding radiomic features associated with the minimum error were selected.
We built univariate and multivariate Cox proportional hazards models in the training cohort using the most frequently selected radiomic and/or clinical features. We evaluated the Akaike information criterion (AIC) to compare the quality of the different models, with lower AICs representing a higher quality model. We assessed the likelihood ratio P-value for the derived models to show recurrence prediction significance. HRs and 95% CIs were reported for individual variables. To evaluate nested models combining the clinical and/or radiomic features, the likelihood ratio test was used to compare the goodness of fit.
To verify prediction validity, we locked the coefficients of the variables in the top model generated from the training cohort and evaluated it in the validation cohort. The prognostic value was assessed using the concordance index with Noether's test to determine significance from random (0.5). We performed Kaplan–Meier analysis to separate high- and low-risk groups based on the median risk score in the training cohort. We performed a Student's t test for dependent samples to compare concordance indices between the models. All statistical analyses and model building were performed using R. Statistical significance was assessed at the P < .05 level.
Results
Patient Demographics
The training and validation cohorts were similarly matched with regard to median age (P = .057) and tumor location (P = .571) (Table 2). The training cohort had a higher proportion of males (P = .005) and adenocarcinoma histology (P = .035). There was a slightly higher proportion of stage IV patients in the validation cohort (P < .001), resulting in a larger percentage of patients who recurred/progressed (P = .038). The median time to recurrence was 14 months (range, 2–97) in the training cohort and 15 months (range, 1–59) in the validation cohort. The median follow-up time for censored patients without an event was 50 months (range, 1–115) in the training cohort and 32 months (range, 1–76) in the validation cohort.
Table 2.
Baseline Patient and Lesion Characteristics
| Training (n=145) | Validation (n=146) | P-value | ||
|---|---|---|---|---|
| Age, years | 69 (42–87) | 71 (41–96) | .057 | |
| Gender | Male | 109 (75%) | 87 (60%) | .005 |
| Tumor Location | Right upper lobe | 52 (36%) | 50 (34%) | .571 |
| Right middle lobe | 14 (10%) | 9 (6%) | ||
| Right lower lobe | 21 (14%) | 26 (18%) | ||
| Left upper lobe | 38 (26%) | 34 (23%) | ||
| Left lower lobe | 20 (14%) | 27 (19%) | ||
| Tumor Histology | Adenocarcinoma | 113 (78%) | 103 (71%) | .035 |
| Squamous cell | 29 (20%) | 30 (21%) | ||
| Non–small cell cancer not otherwise specified | 3 (2%) | 13 (9%) | ||
| Tumor Stage | 0a | 4 (3%) | 0 (0%) | <.001 |
| I | 89 (61%) | 100 (68%) | ||
| II | 28 (19%) | 13 (9%) | ||
| III | 21 (14%) | 17 (12%) | ||
| IV | 3 (2%) | 16 (11%) | ||
| Recurrence/Progression | Yes | 40 (28%) | 57 (39%) | .038 |
| No | 105 (72%) | 89 (61%) | ||
Variables shown as median (range) or number (%).
a Pathological stage 0 disease is defined as a carcinoma in situ (TisN0M0) as per the American Joint Committee on Cancer (AJCC) 7th edition staging system.
Segmentation Variability
Table 3 shows the Dice Similarity Coefficient (DSC), Mean Absolute Boundary Distance (MAD), and absolute volume difference between observers in the training cohort. Overall, semiautomatic segmentations were highly reproducible with an average DSC >0.9, MAD <1 mm, and volume differences <1 mL. When we inspected images with low DSC, high MAD, and/or high volume differences, we found that lesions that had the largest degree of variability tended to have a low uptake (eg, SUVmax <2), heterogeneous uptake, and/or were adjacent to structures with a similar metabolic uptake as the tumor (eg, the heart or mediastinum), making the precise boundary of the tumor difficult to determine. These features were evident in ∼20% of the cases.
Table 3.
Inter- and Intraobserver Variability in Metabolic Tumor Volume (MTV) PET-edge Segmentations
| Observera | Dice Similarity Coefficient (DSC) | Mean Absolute Boundary Distance (MAD, mm) | Absolute Volume Difference (mL)b |
|---|---|---|---|
| A vs a (Intra) | 0.916 (0.090) | 0.548 (0.544) | 0.71 (1.66) |
| A vs B (Inter) | 0.917 (0.087) | 0.559 (0.507) | 0.58 (0.92) |
| a vs B (Inter) | 0.904 (0.105) | 0.628 (0.631) | 0.79 (1.46) |
All values are the mean (standard deviation).
a Observer 1 contoured each tumor twice (A and a) and observer 2 contoured each lesion once (B).
b For reference, the average [range] volumes of all MTV contours by the three observers were 15.4 [0.4–297.8], 15.3 [0.4–296.9], and 15.3 [0.3–296.0] mL.
Feature Variability
Table 4 shows the ICCs of the 4 different classes of radiomic features in each of the 3 regions of interest. We found that a total of 435 of the 512 features (85%) had an ICC >0.8 (Table 5) and were considered robust to differences in the segmentations (22, 23).
Table 4.
Intraclass Correlation Coefficients for All FDG-PET Radiomic Features
| Feature Type | MTV | Penumbra | MTV + Penumbra | |||
|---|---|---|---|---|---|---|
| Inter- | Intra- | Inter- | Intra- | Inter- | Intra- | |
| Size | 0.996(0.99–1.00) | 0.994(0.99–1.00) | – | – | – | – |
| Intensity | 0.977(0.89–1.00) | 0.972(0.84–1.00) | 0.931(0.48–0.99) | 0.916(0.36–0.99) | 0.995(0.98–1.00) | 0.995(0.98–1.00) |
| Shape | 0.867(0.37–0.98) | 0.847(0.39–0.98) | – | – | – | – |
| Texture | 0.898(0.50–0.99) | 0.893(0.48–0.99) | 0.892(0.14–0.99) | 0.925(0.50–0.99) | 0.981(0.28–1.00) | 0.977(0.66–1.00) |
All values are shown as the mean (range).
Table 5.
Number (percent) of Robust FDG-PET Radiomic Features Selected in Each Category by Virtue of an ICC > 0.8
| Feature Type | MTV | Penumbra | MTV + Penumbra | |||
|---|---|---|---|---|---|---|
| Inter- | Intra- | Inter- | Intra- | Inter- | Intra- | |
| Size | 4 (100%) | 4 (100%) | – | – | – | – |
| Intensity | 12 (100%) | 12 (100%) | 11 (92%) | 11 (92%) | 12 (100%) | 12 (100%) |
| Shape | 27 (68%) | 30 (75%) | – | – | – | – |
| Texture | 115 (80%) | 115 (80%) | 118 (82%) | 131 (91%) | 144 (100%) | 142 (99%) |
Feature Selection and Model Training
Across the 100 randomizations, the average minimum cross-validation error was 10.5% at a lambda value of 0.1296 in the training cohort. This lambda generated 2 features with nonzero coefficients, stage, and 1 MTV plus penumbra GLCM texture feature (maximum probability). Although SUVmax has previously been shown to be associated with recurrence in NSCLC, it was not selected by LASSO as a top feature. However, it was found to be a significant univariate predictor in our cohort (Table 6), consistent with previous studies (7).
Table 6.
Cox Proportional Hazards Model Statistics for Univariate Features in the Training Cohort
| Feature | Akaike Information Criterion | Likelihood Ratio | P-value | HR [95% CI] | Concordance [95% CI] |
|---|---|---|---|---|---|
| Stage | 341.7 | 19.98 | <.001 | 2.15[1.56–2.95] | 0.68[0.60–0.76] |
| Gray-level Cooccurrence Matrix Maximum Probability (MTV + Penumbra) | 347.5 | 14.18 | <.001 | 0.41[0.23–0.74] | 0.66[0.57–0.74] |
| SUVmax | 353.7 | 7.99 | .005 | 1.06[1.02–1.10] | 0.67[0.58–0.75] |
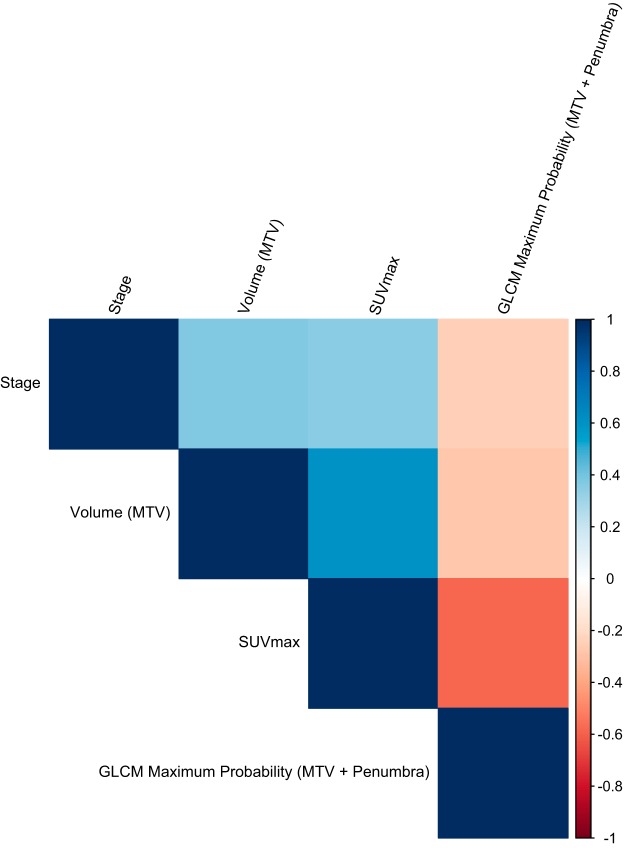
Figure 1 visualizes the Pearson correlation coefficients of the top features. For reference, correlation of the top features with MTV volume and SUVmax is also shown. All correlations were low and the radiomic feature showed no correlation with stage, volume, or SUVmax.
Figure 1.
Pearson correlation coefficient heatmap for the radiomic and standard clinical variables.
Univariate Cox regression model statistics, including the AIC, likelihood ratios, P-values, and HRs, are shown for the top features in Table 6. Both features were significant univariate predictors of time to recurrence. Overall, stage was the best univariate predictor.
Because stage was the best univariate predictor, the likelihood ratio test was performed to assess significant improvements to this well-established clinical model for recurrence prediction. Additional features were added to determine significant improvements to the model. Adding the MTV plus penumbra texture feature to stage significantly improved the model (P = .006). This multivariate model was a significant predictor of time to recurrence in the training cohort (likelihood ratio = 27.59, P < .001, concordance = 0.74 [95% CI: 0.66-0.81]). Both stage (HR = 1.92 [95% CI: 1.37–2.67], P < .001) and the radiomic texture feature (HR = 0.52 [95% CI: 0.30–0.91], P = .02) were significant covariates in the multivariate model. Adding SUVmax to stage did not significantly improve the clinical model performance (P = .22). It also did not significantly improve performance in the combined stage and radiomic model (P = .73).
Model Validation
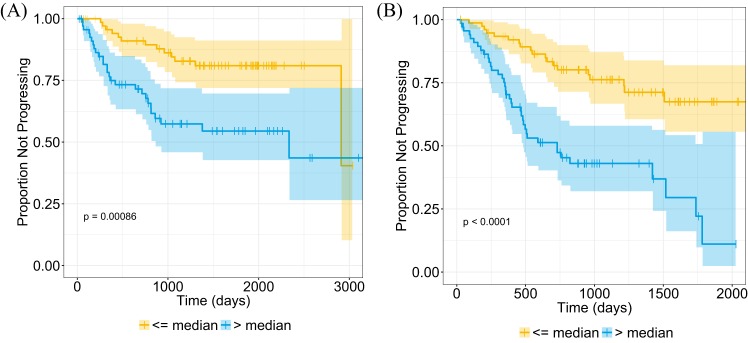
Univariate results were confirmed in the validation cohort (Table 7), with all features being significant predictors of time to recurrence. The locked multivariate model from the training cohort, which included stage and the radiomic texture feature, was a significant predictor in the validation cohort (concordance = 0.74 [95% CI: 0.67–0.81], Noether's P < .001). We separated the patients into high- and low-risk groups on the basis of the median risk score in the training cohort. Kaplan–Meier time-to-recurrence curves for the multivariate model in both cohorts are shown in Figure 2. Recurrence was lower in the group below the median model risk score.
Table 7.
Cox Proportional Hazards Model Statistics for Univariate Features in the Validation Cohort
| Feature | Akaike Information Criterion | Likelihood Ratio | P-value | HR [95% CI] | Concordance[95% CI] |
|---|---|---|---|---|---|
| Stage | 475.6 | 35.7 | <.001 | 2.13[1.69–2.68] | 0.69[0.63–0.76] |
| Gray-level Cooccurrence Matrix Maximum Probability (MTV + Penumbra) | 497.2 | 14.14 | <.001 | 0.50[0.33–0.76] | 0.66[0.60–0.72] |
| SUVmax | 506.1 | 5.24 | .02 | 1.03[1.01–1.05] | 0.67[0.61–0.73] |
Figure 2.
Kaplan–Meier curves for the multivariate stage and radiomic texture model risk scores in the training cohort (n = 145, P < .001) (A) and the validation cohort (n = 146, P < .001) (B). Patients have been stratified on the basis of median risk value in the training cohort. The shaded regions represent the 95% confidence intervals (CI) and “+” indicates censored data.
The multivariate model including stage and the radiomic feature significantly outperformed the best performing clinical model of stage in the training (P = .036) and validation (P = .033) cohorts. The combined model also outperformed the radiomic feature alone in both the training cohort (P = .019) and the validation cohort (P < .001).
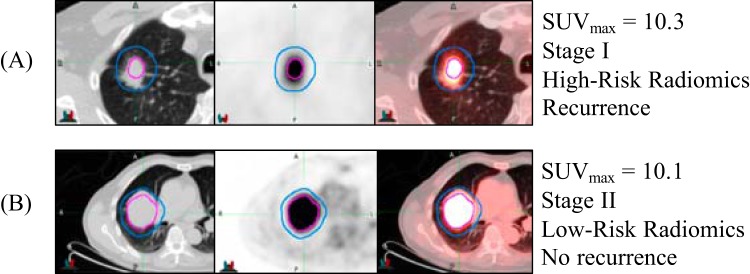
Figure 3 exemplifies 2 patients with similar SUVmax that would typically be considered to be at a high risk of recurrence. Yet, the combined model including radiomics correctly predicted the recurrence status of each patient on the basis of the median risk value. Based on qualitative inspection, the high-risk patient had more heterogeneous uptake in the penumbra region compared with the low-risk patient.
Figure 3.
Example computed tomography (CT) image (left), corresponding positron emission tomography (PET) image (middle), and fused PET/CT images (right) for 2 patients, where the metabolic tumor volume (MTV) is encircled in magenta and the penumbra in between the magenta and blue outlines. Patients (A) and (B) had relatively high SUVmax values, but the radiomics model distinguished the high-risk patient (A) who recurred at 16-month follow-up and the low-risk patient (B) who had not recurred at just under 5 years of follow-up.
Discussion
We show here evidence that texture in the MTV and nearby surrounding region can predict recurrence in NSCLC. Furthermore, augmenting this radiomic feature with stage significantly improved performance over stage alone, which was validated in an independent data set. This model also showed potential value in risk-stratifying patients with NSCLC who are at high versus low risk of recurrence or progression. A general rule in modeling studies is that 10 patients are needed for every feature selected in the model (8). To minimize overfitting, our final model consisted of only 2 features. However further studies on larger sample sizes with additional features may improve prognostic performance and applicability to other cohorts.
The radiomic feature selected was a GLCM texture feature in the combined MTV plus penumbra volume. This feature, which describes local texture variations, suggests that patients whose PET images show a more heterogeneous texture, specifically in the penumbra region surrounding the MTV, are more likely to recur. This suggests the importance of image data in the surrounding region for recurrence prediction. This region may contain uptake not measured in the MTV (and not by the SUVmax) and could indicate areas of disease adjacent to the primary mass. The texture being detected in this region may be indicative of an invasive component of the tumor, for example, spiculations or tumor spread through blood vessels, but this requires further investigation (15).
Notably, size or shape features, including the commonly used metrics of maximum axial diameter and 3D volume, were not selected as predictive features. SUVmax was also not selected, and adding it to clinical or combined models did not significantly improve performance. This suggests that texture features may provide more useful information than traditional metrics for predicting recurrence/progression.
Previous work in the field of radiomics has evaluated FDG-PET features for outcome prediction in lung cancer. Jansen et al. found the GLCM energy texture feature was a significant predictor of overall survival in oligometastatic NSCLC (26). Others have shown that texture features may be beneficial for predicting local control, distant metastasis, and disease-free survival in lung cancer (10–12). However, the majority of studies to date have focused on only the MTV. To the best of our knowledge, ours is the first study that evaluates the lung tumor penumbral region of PET images for recurrence prediction. Future work integrating CT imaging features or molecular data may improve prognostic performance.
Our study investigated PET/CT images from multiple scanners and institutions, potentially introducing variability in image data and quality and therefore the construction of a predictive model. We used a standard acquisition protocol across all institutions to minimize this variability (27, 28). This may still result in signal variations in the tumor and penumbra regions; therefore, further studies investigating single scanners are warranted and may improve model performance.
Previous work has also shown that PET radiomic features are dependent more on delineation variability than on reconstruction algorithm (29) and that texture features are less affected by difference in scanners (30). Many radiomic features also show high test–retest stability with repeat PET imaging (31). The PET-edge segmentation tool we used for tumor segmentation showed high reproducibility with associated radiomic feature robustness. Segmentations were performed with commercially available software (MIM Software, Inc.), making it an easily deployed and integrated system.
Our work is also applicable in a “real world,” nonresearch setting, where different scanners and images of variable quality are routinely used for clinical assessment. However, additional external validation of this radiomics model is warranted to determine the impact of different scanners and acquisition protocols on model predictions.
Our study has several limitations. The primary limitation is that the penumbra region was not restricted to the lung volume, that is, it may at times have included the adjacent chest wall, major blood vessels, and/or mediastinum. However, as features were selected from within this region, it is providing relevant information for the prediction of recurrence. The effect of this and the efforts to minimize it remain the subject of further investigation. Owing to differences in breathing between the PET and CT images, accurate registration of the lung boundary is challenging. We also investigated only a single distance of 1 cm for the penumbra region; it is possible that larger or smaller distances could improve or degrade performance. Another limitation is the inherent low resolution of the PET images, limiting the amount of information we can analyze for each tumor owing to lower voxel quantities for smaller tumors. Finally, the sample sizes analyzed were relatively small, and validation of this model in larger data sets is warranted.
In conclusion, a PET texture feature in the metabolic tumor volume and surrounding region augmented staging for NSCLC recurrence prediction. This model may be useful in identifying patients who are at a higher risk of recurrence or progression and may assist physicians in determining what patients may benefit from adjuvant or personalized treatment options at the time of diagnosis.
Acknowledgments
Equal contribution: “S.N and V.S.N contributed equally to this work.”
The authors would like to acknowledge MIM Software Inc. for their assistance with segmentation software, Jalen Benson and Weiruo Zhang for their assistance with clinical data curation, and the Stanford Data Studio for statistical consulting. The authors would like to acknowledge funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellowship and the National Cancer Institute (NCI) R01 CA160251, U01 CA187947, and U01 CA196405.
Disclosures: Dr. Sandy Napel is a Consultant for Carestream Health Inc., on the Medical Advisory Board for Fovia Inc., a Scientific Advisor for EchoPixel Inc., and a Scientific Advisor for RADLogics Inc. However, he is not an employee of any of these companies and none of these conflicts are related to the data used and research completed in this manuscript.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- NSCLC
- Non–small cell lung cancer
- MTV
- metabolic tumor volume
- FDG
- fluoro-2-deoxy-D-glucose
- PET
- positron emission tomography
- CT
- computed tomography
- DSC
- Dice similarity coefficient
- MAD
- mean absolute distance
- GLCM
- gray-level co-occurrence matrix
- LASSO
- least absolute shrinkage and selection operator
- AIC
- Akaike information criterion
- HR
- hazard ratio
- CI
- confidence interval
- ICCs
- intraclass correlation coefficients
References
- 1. Lang-Lazdunski L. Surgery for nonsmall cell lung cancer. Eur Respir Rev. 2013;22:382–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 3. Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res. 2014;3:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Consonni D, Pierobon M, Gail MH, Rubagotti M, Rotunno M, Goldstein A, Goldin L, Lubin J, Wacholder S, Caporaso NE, Bertazzi PA, Tucker MA, Pesatori AC, Landi MT. Lung cancer prognosis before and after recurrence in a population-based setting. J Natl Cancer Inst. 2015;107:djv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non–small cell lung cancer, Version 6.2015. J Natl Compr Canc Netw. 2015;13:515–524. [DOI] [PubMed] [Google Scholar]
- 6. Pignon J-P, Tribodet H, Scagliotti GV, Douillard J-Y, Shepherd FA, Stephens RJ, Le Chevalier T. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. [DOI] [PubMed] [Google Scholar]
- 7. Liu J, Dong M, Sun X, Li W, Xing L, Yu J. Prognostic value of 18F-FDG PET/CT in surgical non-small cell lung cancer: a meta-analysis. PLoS One. 2016;11:e0146195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeda K, Takanami K, Shirata Y, Yamamoto T, Takahashi N, Ito K, Takase K2, Jingu K. Clinical utility of texture analysis of 18F-FDG PET/CT in patients with Stage I lung cancer treated with stereotactic body radiotherapy. J Radiat Res. 2017;58:862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, Rossi A, Chiti A, Sollini M. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018:45:207–217. [DOI] [PubMed] [Google Scholar]
- 12. Wu J, Aguilera T, Shultz D, Gudur M, Rubin DL, Loo Jr BW, et al. Early-stage non–small cell lung cancer: quantitative imaging characteristics of 18F fluorodeoxyglucose PET/CT allow prediction of distant metastasis. Radiology. 2016;281:270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol. 2011;6:244–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kadota K, Nitadori J-i, Sima CS, Ujiie H, Rizk NP, Jones DR, Adusumilli PS, Travis WD. Tumor spread through air spaces is an important pattern of invasion and impacts the frequency and location of recurrences after limited resection for small stage I lung adenocarcinomas. J Thorac Oncol. 2015;10:806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren J, Zhou J, Ding W, Zhong B. Clinicopathological characteristics and imaging features of pulmonary adenocarcinoma with micropapillary pattern. Zhonghua Zhong Liu Za Zhi. 2014;36:282–286. [Article in Chinese] [PubMed] [Google Scholar]
- 16. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bakr S, Gevaert O, Echegaray S, Ayers K, Zhou M, Shafiq M, Zheng H, Benson JA, Zhang W, Leung ANC, Kadoch M, D Hoang C, Shrager J, Quon A, Rubin DL, Plevritis SK, Napel S. A radiogenomic dataset of non-small cell lung cancer. Sci Data. 2018;5:180202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an open-source, modular engine for 3D quantitative feature extraction from volumetric medical images. J Digit Imaging. 2018;31:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haralick RM. Statistical and structural approaches to texture. Proceedings of the IEEE. 1979;67:786–804. [Google Scholar]
- 20. Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Cybern. 1973:610–621. [Google Scholar]
- 21. Leijenaar RT, Nalbantov G, Carvalho S, Van Elmpt WJ, Troost EG, Boellaard R, Aerts HJ, Gillies RJ, Lambin P. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9:e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pavic M, Bogowicz M, Würms X, Glatz S, Finazzi T, Riesterer O, Roesch J, Rudofsky L, Friess M, Veit-Haibach P, Huellner M, Opitz I, Weder W, Frauenfelder T, Guckenberger M, Tanadini-Lang S. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018;57:1070–1074. [DOI] [PubMed] [Google Scholar]
- 24. Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological). 1996:267–88. [Google Scholar]
- 25. Team RCR. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2013. [Google Scholar]
- 26. Jensen GL, Yost CM, Mackin DS, Fried DV, Zhou S, Gomez DR. Prognostic value of combining a quantitative image feature from positron emission tomography with clinical factors in oligometastatic non-small cell lung cancer. Radiother Oncol. 2018;126:362–367. [DOI] [PubMed] [Google Scholar]
- 27. Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging (Bellingham). 2015;2:041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beichel RR, Smith BJ, Bauer C, Ulrich EJ, Ahmadvand P, Budzevich MM, Gillies RJ, Goldgof D, Grkovski M, Hamarneh G, Huang Q, Kinahan PE, Laymon CM, Mountz JM, Muzi JP, Muzi M, Nehmeh S, Oborski MJ, Tan Y, Zhao B, Sunderland JJ, Buatti JM. Multi-site quality and variability analysis of 3D FDG PET segmentations based on phantom and clinical image data. Med Phys. 2017;44:479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, Hoekstra OS, Smit EF, Boellaard R. Repeatability of radiomic features in non-small-cell lung cancer [18F] FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsujikawa T, Tsuyoshi H, Kanno M, Yamada S, Kobayashi M, Narita N, Kimura H, Fujieda S, Yoshida Y, Okazawa H. Selected PET radiomic features remain the same. Oncotarget. 2018;9:20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leijenaar RT, Carvalho S, Velazquez ER, Van Elmpt WJ, Parmar C, Hoekstra OS, Boellaard R, Dekker AL, Gillies RJ, Aerts HJ, Lambin P. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]