Abstract
The Quantitative Imaging Network of the National Cancer Institute is in its 10th year of operation, and research teams within the network are developing and validating clinical decision support software tools to measure or predict the response of cancers to various therapies. As projects progress from development activities to validation of quantitative imaging tools and methods, it is important to evaluate the performance and clinical readiness of the tools before committing them to prospective clinical trials. A variety of tests, including special challenges and tool benchmarking, have been instituted within the network to prepare the quantitative imaging tools for service in clinical trials. This article highlights the benchmarking process and provides a current evaluation of several tools in their transition from development to validation.
Keywords: cancer imaging, quantitative imaging, benchmarks, clinical translation, oncology
Introduction
A distinguishing advantage of any research network is the opportunity for the ensemble of member teams to collaborate in areas of shared interest, addressing common scientific or technological problems, to compare individual approaches, and ultimately to build consensus. As a result, the ensemble of teams in a research network is often greater than the sum of its parts. For the past 10 years, the National Cancer Institute (NCI) Quantitative Imaging Network (QIN) has provided a network environment where the development and validation of quantitative imaging (QI) analysis software tools designed to measure or predict response to cancer therapies in clinical trials have been pursued. The motivating hypothesis for the QIN has been that clinical trials in systemic or targeted chemo-, radiation-, or immunotherapies can benefit from the inclusion of QI methods in the treatment protocols. These methods involve the extraction of measurable information from medical images to assess the status or change of a disease.
To date, 36 multidisciplinary teams from academic institutions across the United States and Canada have participated in the NCI-funded research program. The current number of teams supported by the network is 20. These research teams discuss and resolve common challenges such as imaging informatics activities, clinical trial design and validation planning, and data acquisition and analysis issues, to name only a few. At the same time, each team is required to make technical and clinical progress on its individual research project.
The interest in QI as a method to gauge tumor progression or predict response to therapy predates the QIN. An early attempt at extracting numeric information from clinical images came in the form of RECIST (Response Evaluation Criteria in Solid Tumors) in 2000 (1, 2), based on earlier guidelines first published by the World Health Organization in 1981 (3). The RECIST criteria used a single straight line drawn across the widest dimension in a tumor image to provide a quantitative measure of tumor size. Size, suitability, and the number of lesions to be measured were stated in the original guidelines, and later revised in version 1.1 (4). Response criteria, measured by the change in linear dimension, were established to determine if the tumor was in complete response, partial response, or stable or progressive disease.
Although tumor shrinkage is an obvious desirable response to cancer therapy, it is not the only response that can occur, or in some cases, the response may be delayed in appearing (5). Furthermore, in a metastatic cancer setting a limited set of target lesions, as prescribed in RECIST 1.1, may not represent the overall tumor burden or response to therapy (6). These limitations restrict the usefulness of RECIST in some clinical trials. Often, immunotherapy trials, for example, show that complete response or stable response can occur after an initial increase in tumor burden (7, 8). Conventional RECIST criteria early in the therapy run the risk of labeling this initial increase as tumor progression, failing to account for the delayed onset of antitumor T cell response. Thus, a therapy under study in a clinical trial can be seen as failing. This has led to the development of iRECIST guidelines for response criteria in immunotherapy trials (9).
QI tools being developed and validated by QIN research teams measure far more than simple unidimensional tumor size, and the articles in this special issue of Tomography highlight a number of them. Physical attributes of tumors such as heterogeneity, diffusion and perfusion, and metabolic activity are being added to the more traditional size and shape measurements of QI to determine response to therapy. These attributes have been used in machine-based modeling studies driven by imaging data to characterize tumor growth (10–13). In addition, machine learning radiomics approaches for high-throughput extraction and analysis of quantitative image features are providing an even richer set of image parameters. These include intensity, texture, kurtosis, and skewness from which to extract measurement and prediction information on tumor progression (14–16).
Background
If QI is to be useful in clinical trials as a method to measure or predict response to therapy, the methods must be developed on clinically available platforms such that the final validated tools would have value in multicenter clinical trials. To this end, the NCI QIN program was initiated in 2008. The support mechanism chosen for this effort was the cooperative agreement U01 mechanism. Here, successful applicants agree to collaborations and conditions established by NCI program staff. In the case of the QIN, these conditions include participation in a network of teams, joining in monthly teleconference meetings, and collaborating in several working groups.
Applications to the QIN are subject to the NIH peer-review process conducted 3 times each year. As a result, the network teams enter the program at different times and are thus at different stages in their tool development and validation at any given point in time. This creates a need to qualify the degree of development and validation each quantitative tool has attained. Accordingly, a system of benchmarking to assess tool maturity has been implemented.
Clinical Translation
The process of translating ideas and products from laboratory demonstration to clinical utility is the exercise of transferring stated features of the idea or product into realized benefits to the user. For example, the stated feature of improved sensitivity or specificity in an imaging protocol can translate into improved personalized care in the clinic. The tool developer must be aware of the nature of the clinical need for such a tool. Likewise, the clinical user must be realistic regarding the performance characteristics needed in a clinical decision support tool.
To ensure a strong connection between developer and clinical user, each QIN team is required to have a multidisciplinary composition that brings expertise in imaging physics and radiology along with informatics, oncology, statistics, and clinical requirements to the cancer problem being addressed. This gives each team multiple perspectives on the challenges of advancing decision support tools through the development and verification stages and on to the clinical validation stage.
Translation is not a simple move from bench to bedside. It requires a constant check on progress with a compass heading set by clinical need. There must be a set of guiding milestones to point the way through the translation landscape and to measure progress along the way. This, however, can be very difficult in a network of research teams, where each team is focused on a different imaging modality or approach and cancer problem.
A guiding pathway for QIN teams in this translation process continues to be the use of benchmarks for measuring progress toward clinical utility. Even though each team is working on a different application of QI for measurement or prediction of response to cancer therapy, they all share the challenges of bringing tools and methods into clinical utility. The benchmarks offer a ubiquitous pathway for all teams to move toward clinical workflow. As such, the benchmarks measure the tasks on the development side of the translation. There is no doubt that a set of benchmarks could be established for monitoring progress on the clinical side of the translation issue, but that is not a part of the QIN mission.
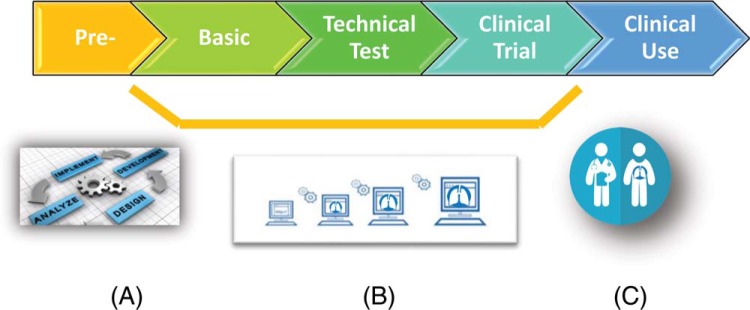
Figure 1 shows a schematic pathway from initial concept and development of tools and methods for clinical decision support all the way to final clinical use. The demarcations show that the benchmark grades represent milestones in the development toward the clinical use. The details of the benchmarks and the requirements to achieve each are given in the next section.
Figure 1.
Quantitative Imaging Network (QIN) benchmarks, described in the text and in Figure 2, designate key milestones toward the clinical translation of quantitative imaging (QI) tools from laboratory prototype (A) to scale up and optimization (B) to clinical use (C).
Benchmarking
For each team, the transition from the activities of tool development to clinical performance validation is a central part of the research, but this does not occur in a sudden step. There is a period where prototype tools are tested against retrospective image data from archives such as The Cancer Imaging Archive (TCIA) (http://www.cancerimagingarchive.net/) or other data sources to objectively assess tool performance. The benchmarking initiative allows investigators the opportunity to adjust their algorithms before committing to a specific prospective clinical trial.
Another initiative embraced by the QIN team members during their period of initial verification of tool performance has been team challenges. Here, several teams with sufficiently developed tools with similar quantitative measurement functions (segmentation, volume metrics, volume transfer constant, Ktrans, measurements, etc.) use a common data source, divided into training and test data sets, to determine and compare task-specific tool performance related to determining or predicting the therapeutic response. Within the QIN, these activities are referred to as challenges and collaborative projects (CCPs) (17) and have proven very useful in guiding the development of QI tools and analytic methods in preparation for more complete clinical validation studies. CCPs have been conducted at various points along the development pipeline, from basic concept to technical verification and preliminary clinical validation. Descriptions of CCP tasks, project design, and results have been disseminated through several peer-reviewed scientific publications (18–28).
The CCP activities highlighted the need to create a method for gauging the degree of development a tool had attained at any specific timepoint. This would help to evaluate challenge results when tools with widely different levels of development participated. To gauge the level of development for tools in the QIN, a benchmarking process was developed. A Task Force, comprising QIN members, was charged with the task of developing a system to stratify the level of progress made by teams in their efforts to develop QI tools for clinical workflow. In the context of QIN activities, a tool can be a software algorithm, a physical phantom, or a digital reference object used in the production or analysis of QI biomarkers for diagnosis and staging of cancer and for the prediction or measurement of response to therapy.
The Task Force developed QI Benchmarks as standard labels that signify the development, validation, and clinical translation of quantitative tools through a 5-tier benchmark system as shown in Figure 2 (29): pre-benchmark (level 1), basic benchmark (level 2), technical test benchmark (level 3), clinical trial benchmark (level 4), and clinical use benchmark (Level 5). In general, requirements for each benchmark designation require a peer-reviewed publication, where the scientific goals, methods, and results of the QI biomarker development or analysis are described. A benchmark is not automatically conferred on a QIN tool. The developer must make an application which includes the required information for that benchmark and conduct a discussion of the objective performance claim for the benchmark, best practices, and current limitations of the tool. In addition, it is important to note that candidates for each of the benchmarks must have fulfilled the requirements for the prior-level benchmark but not necessarily obtained it. The Coordinating Committee of QIN, consisting of the chairs of each of the Network Working Groups (30) and certain NCI program staff, reviews each benchmark application. If an application for a benchmark is rejected, the applicant will be allowed to address the concerns and resubmit the application.
Figure 2.
Five levels of QI benchmark for labeling of QI products. *In addition to the requirements listed for each level, candidates for benchmarks must have fulfilled the requirements for the prior-level benchmark, but not necessarily obtained that benchmark, to be considered for the current benchmark level.
The establishment of this benchmarking process will help to advance the field of QI in oncology by recognizing QI tools entering QIN (benchmark level 1), encouraging QIN investigators to participate in objective performance evaluation of their tools and methods (benchmark level 2), to streamline validation through dissemination of appropriately developed tools and methods to test sites (benchmark level 3), and to promote participation in oncology clinical trials (benchmark level 4) by providing objective evaluation of tool development to allow more accurate assessment or prediction of cancer therapies and eventual clinical use (benchmark level 5). It is anticipated that this initiative will help in proper placement of advanced tools and methods into prospective clinical trials and will streamline the process of translating such tools into the broader clinical community with adoption by industry.
Results
The current catalog of QIN tools contains 67 clinical decision support tools in various stages of development. Because of the staggered entrance of teams into the network, progress in development is not uniform across the network. This has created the need for benchmarking as a measurable way to evaluate tool development status. Of the tools listed in the catalog, there are ∼12 that are to the point of entering the clinical domain and qualifying for benchmark level 4 or 5.
Image segmentation of tumor from surrounding tissue is an important tool function and serves as a first step in determining treatment planning regimens in oncology and many quantitative measurements of tumor status. Several QIN teams are developing segmentation tools for various applications. One such tool developed at Columbia University (New York, NY) performs image segmentation on solid tumors and has been shown in lung, liver, and lymph nodes as a semiautomatic software tool. The segmentation of magnetic resonance imaging (MRI) and/or computed tomography (CT) images across multiple slices yields quantitative information on tumor volume (31–33) and has been used in several clinical trials. This tool can be integrated into diagnostics, radiation-treatment planning, and tumor response assessment on commercial workstations.
Volumetric measurement of breast cancer tumors using dynamic contrast-enhanced MRI has been developed by the QIN team at the University of California at San Francisco (San Francisco, CA). The tool is an image processing and analysis package based on dynamic contrast-enhanced MRI contrast kinetics and has been approved on a commercial platform. It has proven useful in clinical trials performed by several groups in the NCI clinical trials network (34, 35). In addition to the analysis of algorithm performance, the validation of a breast phantom design has been reported (36). Features of the software package include image reconstruction, image registration, segmentation, and viewer/visualization. A commercial version is being used in ∼20 I-SPY clinical sites.
Auto-PERCIST (Positron Emission Tomography [PET] Response Criteria in Solid Tumors) is a software package for PET imaging and can provide clinical decision support through image segmentation, viewer/visualization, and response assessment. Similar to RECIST, the PERCIST package focuses on analyzing fludeoxyglucose-PET scans and evaluates if the study was performed properly from a technical standpoint. It establishes the appropriate threshold for the standardized uptake value corrected for lean body mass (SUL) evaluation of the lesion at baseline. Auto-PERCIST has been used to provide clinical assessment of therapy response in multicenter evaluations both here in the United States and in Korea, and a release of Auto-PERCIST for European oncology trials is planned. Although not completely developed under the QIN program, many of the features found in Auto-PERCIST were created and validated in the QIN program by teams originally at the Johns Hopkins University (Baltimore, MD) and currently at Washington University (St. Louis, MO). This tool has been used in several multicenter clinical trials, and details of its performance can be found in several publications (37–40).
Clinical support for evaluating tumor response can come in many forms. It be the algorithm, phantom, or digital reference object for direct analysis of images, and it can also be the workspace in which the software operates. Such is the case for ePAD, a Stanford University (Palo Alto, CA) web-based image viewing and annotation platform to enable deploying QI biomarkers into clinical trial workflow (41). It supports applications such as data collection, data mining, image annotation, image metadata archiving, and response assessment. This publicly available platform predates QIN, but many of the current quantitative functionalities of ePAD have been installed and validated under QIN support.
Conclusions
The list of benchmarked tools in QIN is growing. Constant updates are being made to the catalog as new QIN teams enter the network and existing teams progress in their development and validation of their QI tools in support of clinical trials (42, 43). This issue of Tomography highlights several QI tools and studies in the QIN. As the network moves forward, it has begun to focus on coordinated ways to approach clinical trial groups and interested commercial parties.
Acknowledgments
The QIN Challenge Task Force: Keyvan Farahani (NCI), Maryellen Giger (University of Chicago), Wei Huang (Oregon Health Sciences University), Jayashree Kalpathy-Cramer (Massachusetts General Hospital), Sandy Napel (Stanford University), Robert J. Nordstrom (NCI), Darrell Tata (NCI), and Richard Wahl (Washington University).
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- NCI
- National Cancer Institute
- QIN
- Quantitative Imaging Network
- CCPs
- challenges and collaborative projects
- MRI
- magnetic resonance imaging
- PET
- positron emission tomography
References
- 1. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubenstein L, Verweij J, Glabbeke MV, van Oosterom AT, Christian MC, Gwyther SG. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–216. [DOI] [PubMed] [Google Scholar]
- 2. Padhani AR, Ollivier L. The RECIST (Response Evaluation Criteria in Solid Tumors) criteria: implications for diagnostic radiologists. Br J Radiol. 2001;74:983–986. [DOI] [PubMed] [Google Scholar]
- 3. Park JO, Lee SI, Song SY, Kim K, Kim WS, Jung CW, Park YS, Im YH, Kang WK, Lee MH, Lee KS, Park K. Measuring response in solid tumors: comparison of RECIST and WHO response criteria. Jpn J Clin Oncol. 2003. October;33:533–537. [DOI] [PubMed] [Google Scholar]
- 4. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubenstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- 5. Huang B. Some statistical considerations in the clinical development of cancer immunotherapies. Pharm Stat. 2018;17:49–60. [DOI] [PubMed] [Google Scholar]
- 6. Khul CK, Alparslan Y, Schmoee J, Sequeria B, Keulers A, Brummendorf TH, Keil S. Validity of RECIST version 1.1 for response assessment in metastatic cancer: a prospective multireader study. Radiology. 2018. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7. Evangelista L, de Jong M, Del Vecchio S, Cai W. The new era of cancer immunotherapy: what can molecular imaging do to help? Clin Transl Imaging. 2017;5:299–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carter BW, Bhosale PR, Yang WT. Immunotherapy and the role of imaging, Cancer. 2018;124:2906–2922. [DOI] [PubMed] [Google Scholar]
- 9. Seymur L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, Lin NU, Litiere S, Dancey J, Chen A, Hodi FS, Therasse P, Hoekstra OS, Shankar LK, Wolchok JD, Ballinger M, Caramella C, de Vries EG; RECIST Work Group. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18:e143–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hogea C, Davatzikos C, Biros G. An image-driven parameter estimation problem for a reaction-diffusion glioma growth model with mass effects. J Math Biol. 2008;56:793–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yankeelov TE, Atuegwu N, Hormuth D, Weis JA, Barnes Sl, Miga MI, Rericha EC, Quanranta V. Clinically relevant modeling of tumor growth and treatment response. Sci Trans Med. 2013;5:51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weis JA, Miga MI, Arlinghaus LR, Li X, Abramson V, Chakravarthy AB, Pendyala P, Yankeelov TE. Predicting the response of breast cancer to neoadjuvant therapy using a mechanically coupled reaction-diffusion model. Cancer Res. 2015;75:4697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roque T, Risser L, Kersemans V, Smart S, Allen D, Kinchesh P, Gilchrist S, Gomes AL, Schnabel JA, Chappell MA. A DECR-MRI driven 3-D reaction-diffusion model of solid tumor growth. IEEE Trans Med Imaging. 2018;37:724–732. [DOI] [PubMed] [Google Scholar]
- 14. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout G, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aerts HJWL, Rios Velazques E, Leijennar RTH, Parmar C, Grossmann P, Cavalho S, Bussink J, Monshouwer R, Haibe-Kains Rietveld D, Hoeders F, Rietbergen MM, Leemans R, Dekker A, Quackenbush J, Gilles RJ, Lambin P. Decoding tumor phenotype by noninvasive imaging using quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L, Lian Z, Liu J, Luo X, Pei S, Mo Z, Huang W, Ouyang F, Guo B, Liang L, Chen W, Liang C, Zhang S. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23:4259–4269. [DOI] [PubMed] [Google Scholar]
- 17. Farahani K, Kalpathy-Cramer J, Chenevert TL, Rubin DL, Sunderland JJ, Nordstrom RJ, Buatti J, Hylton N. Computational challenges and collaborative projects in the NCI quantitative imaging network. Tomography. 2016;2:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang W, Li X, Chen Y, Li X, Chang MC, Oborski MJ, Malyarenko DI, Muzi M, Jajamovich GH, Fedorov A, Tudorice A, Gupta SN, Laymon CM, Marro KI, Dyvorne HA, Miller JV, Barboriak DP, Chenevert TL, Yankeelov TE, Mountz JM, Kinahan PE, Kikinis R, Taouli B, Fenessay F, Kalpathy-Cramer J. Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Transl Oncol. 2014;7:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, Aryal MP, LaViolette PS, Oborski MJ, O'Sullivan F, Abramson RG, Jafari-Khouzani K, Afzal A, Tudorica A, Moloney B, Gupta SN, Besa C, Kalpathy-Cramer J, Mountz JM, Laymon CM, Muzi M, Schmainda K, Cao Y, Chenevert TL, Taouli B, Yankeelov TE, Fennessy F, Li X. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge. Tomography. 2016;2:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chenevert TL, Malyarenko DI, Newitt D, Jayatilake M, Tudorica A, Fedorov A, Kikinis R, Liu TT, Muzi M, Oborski MJ, Laymon CM, Li X, Yankeelov TE, Kalpathy-Cramer J, Mountz JM, Kinahan PE, Rubin DL, Fennessy F, Huang W, Hylton N, Ross BD. Errors in quantitative image analysis due to platform-dependent image-scaling. Transl Oncol. 2014;7:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalpathy-Cramer J, Zhao B, Goldgof D, Gu Y, Wang X, Yang H, Tan Y, Gillies R, Napel S. A comparison of lung nodule segmentation algorithms: methods and results from a multi-institutional study. J Digit Imaging. 2016;29:476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalpathy-Cramer J, Mammonov A, Zhao B, Lu L, Cherezov D, Napel S, Echegaray S, Rubin D, McNitt-Gray M, Lo P, Sieren JC, Uthoff J, Dilger SKN, Driscoll B, Yeung I, Hadjiski L, Cha K, Balagurunathan Y, Gillies R, Goldgof D. Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography. 2016;2:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmainda KM, Prah MA, Rand SD, Liu Y, Logan B, Muzi M, Rane SD, Da X, Yen YF, Kalpathy-Cramer J, Chenevert TL, Hoff B, Ross B, Cao Y, Aryal MP, Erickson B, Korfiatis P, Dondlinger T, Bell L, Hu L, Kinahan PE, Quarles CC. Multisite concordance of DSC-MRI analysis for brain tumors: results of a national cancer institute quantitative imaging network collaborative project. AJNR Am J Neuroradiol. 2018;39:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malyarenko DI, Wilmes LJ, Arlinghuas LR, Jacobs MA, Juang W, Helmer KG, Taouli B, Yankeelov TE, Newitt D, Chenevert TL. QIN DAWG validation of gradient nonlinearity bias correction workflow for quantitative diffusion-weighted imaging in multicenter trials. Tomography. 2016;2:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malyarenko D, Fedorov A, Bell L, Prah M, hectors S, Arlinghaus L, Muzi M, Solaiyappan M, Jacobs M, Fung M, Shukla-Dave A, McManus K, Boss M, Taouli B, Yankeelov TE, Quarles CC, Schmainda K, Chenevert TL, Newitt DC. Toward uniform implementation of parametric map Digital Imaging and Communication in Medicine standard in multisite quantitative diffusion imaging studies. J Med Imaging. 2018; 5:011006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newitt DC, Malyarenko D, Chenevert TL, Quarles CC, Bell L, Fedorov A, Fennessy F, Jacobs MA, Solaiyappan M, Hectors S, Taouli B, Muzi M, Kinahan PE, Schmainda KM, Prah MA, Taber EN, Kroenke C, Huang W, Arlinghaus LR, Yankeelov TE, Cao Y, Aryal M, Yen YF, Kalpathy-Cramer J, Shukla-Dave A, Fung M, Liang J, Boss M, Hylton N. Multisite concordance of apparent diffusion coefficient measurements across the NCI Quantitative Imaging Network. J Med Imaging. 2018;5:011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beichel RR, Smith BJ, Bauer C, Ulrich EJ, Ahmadvand P, Budzevich MM, Gillies RJ, Goldgof D, Grkovski M, Hamarneh G, Huang Q, Kinahan PE, Laymon M, Mountz JP, Nehmeh S, Oborski MJ, Tan Y, Zhao B, Sunderlnd JJ, Buatti JM. Multi-site quality and variability analysis of 3D FDG PET segmentations based on phantom and clinical image data. Med Phys. 2017;44:479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balagurunathan Y, Beers A, Kalpathy-Cramer J, McNitt-Gray M, Hadjiski L, Zhao B, Zhu J, Yang H, Yip SSF, Aerts HJWL, Napel S, Cherezov D, Cha K, Chan HP, Flores C, Garcia A, Gillies R, Goldgof D. Semi-automated pulmonary nodule interval segmentation using NLST data. Med Phys. 2018;45:1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. https://imaging.cancer.gov/programs_resources/specialized_initiatives/qin/about/default.htm (accessed Nov 28, 2018).
- 30. The Working Groups of the Quantitative Imaging Network are: (1) Clinical Trials Design and Development, (2) Bioinformatics IT and Data Sharing, (3) Image Analysis and Performance Metrics, (4) MRI, and (5) PET-CT.
- 31. Tan Y, Lu L, Bonde A, Wang D, Qi J, Schwartz HL, Zhao B. Lymph node segmentation by dynamic programming and active contours. Med Phys. 2018;45:2054–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan J, Schwartz LH, Zhao B. Semi-automatic segmentation of liver metastases on volumetric CT images. Med Phys. 2015;42:6283–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tan Y, Schwartz LH, Zhao B. Segmentation of lung tumors on CT scans using watershed and active contours. Med Phys. 2013;40:043502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hylton NM. Vascularity assessment of breast lesions with gadolinium-enhanced MR imaging. Magn Reson Imaging Clin N Am. 1999;7:411–420. [PubMed] [Google Scholar]
- 35. Partridge SC, Heumann EJ, Hylton NM. Semi-automated analysis for MRI of breast tumors. Stud Health Technol Inform. 1999;62:259–260. [PubMed] [Google Scholar]
- 36. Keenan KE, Wilmes LJ, Aliu SO, Newitt DC, Jones EF, Boss MA, Stupic KF, Russek SE, Hylton NM, Design of a breast phantom for quantitative MRI. J Magn Reson Imaging. 2016;44:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O JH, Lodge MA, Wahl RL. Practical PERCIST: a simplified guide to PET response criteria in solid tumors 1.0. Radiology. 2016;280:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hyun OJ, Luber BS, Leal JP, Wang H, Bolejack V, Schuetze SM, Schwartz LH, Helman LJ, Reinke D, Baker LH, Wahl RL. Response to early treatment evaluated with 18F-FDG PET and PERCIST 1.0 predicts survival in patients with Ewing sarcoma family of tumors treated with a monoclonal antibody to the insulinlike growth factor 1 receptor. J Nucl Med. 2016;57:735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O JH, Wahl RL. PERCIST in perspective. Nucl Med Mol Imaging. 2018;52:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao XR, Zhang Y, Yu YH. Use of 18F-FDG PET/CT to predict short-term outcomes early in the course of chemoradiotherapy in stage III adenocarcinoma of the lung. Oncol Lett. 2018;16:1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rubin DL, Willrett D, O'Connor MJ, Hage C, Kurtz C, Moreira DA. Automated tracking of quantitative assessments of tumor burden in clinical trials. Trans Oncol. 2014;7:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yankeelov TE, Mankoff DA, Schwartz LH, Lieberman FS, Buatti JM, Mountz JM, Erickson BJ, Fennessy FM, Huang W, Kalpathy-Cramer J, Wahl RL, Linden HM, Kinahan PE, Zhao B, Hylton NM, Gillies RJ, Clarke L, Nordstrom R, Rubin DL. Quantitative imaging in cancer clinical trials. Clin Cancer Res. 2016;22:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mountz JM, Yankeelov TE, Rubin DL, Buatti JM, Erikson BJ, Fennessy FM, Gillies RJ, Huang W, Jacobs MA, Kinahan PE, Laymon CM, Linden HM, Mankoff DA, Schwartz LH, Shim H, Wahl RL. Letter to cancer center directors: progress in quantitative imaging as a means to predict and/or measure tumor response in cancer therapy trials. J Clin Oncol. 2014;32:2115–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]