Abstract
Standard-of-care multiparameter magnetic resonance imaging (MRI) scans of the brain were used to objectively subdivide glioblastoma multiforme (GBM) tumors into regions that correspond to variations in blood flow, interstitial edema, and cellular density. We hypothesized that the distribution of these distinct tumor ecological “habitats” at the time of presentation will impact the course of the disease. We retrospectively analyzed initial MRI scans in 2 groups of patients diagnosed with GBM, a long-term survival group comprising subjects who survived >36 month postdiagnosis, and a short-term survival group comprising subjects who survived ≤19 month postdiagnosis. The single-institution discovery cohort contained 22 subjects in each group, while the multi-institution validation cohort contained 15 subjects per group. MRI voxel intensities were calibrated, and tumor voxels clustered on contrast-enhanced T1-weighted and fluid-attenuated inversion-recovery (FLAIR) images into 6 distinct “habitats” based on low- to medium- to high-contrast enhancement and low–high signal on FLAIR scans. Habitat 6 (high signal on calibrated contrast-enhanced T1-weighted and FLAIR sequences) comprised a significantly higher volume fraction of tumors in the long-term survival group (discovery cohort, 35% ± 6.5%; validation cohort, 34% ± 4.8%) compared with tumors in the short-term survival group (discovery cohort, 17% ± 4.5%, P < .03; validation cohort, 16 ± 4.0%, P < .007). Of the 6 distinct MRI-defined habitats, the fractional tumor volume of habitat 6 at diagnosis was significantly predictive of long- or short-term survival. We discuss a possible mechanistic basis for this association and implications for habitat-driven adaptive therapy of GBM.
Keywords: glioblastoma, survival, MRI, habitats, cancer evolution
Introduction
Glioblastoma multiforme (GBM) typically exhibits substantial intratumoral heterogeneity at both microscopic and radiological spatial scales (1). Analysis of genomic patterns from The Cancer Genome Atlas (TCGA) database led to a general molecular model that identified 4 distinct “species” of GBM: proneural, neural, classical, and mesenchymal (2). However, more recent studies (3) found substantial spatial variations, so that, in some cases, all 4 species could be observed in different regions of the same tumor. Canoll et al. used RNA-sequencing and histological analysis of image-guided biopsies to show differences in cellular and molecular markers between tissue taken from the contrast-enhancing (CE) core and that from the nonenhancing (NE) margins of GBM tumors (4). Characteristic metabolic differences between the CE and NE regions in GBM have also been identified by 1H magnetic resonance spectroscopy (5). Machine learning on patterns in standard brain magnetic resonance imaging (MRI) images, and parameter maps from diffusion tensor imaging, and dynamic susceptibility contrast-enhanced (DSC)-MRI have been reported to correlate with molecular subtype and survival in newly diagnosed patients with GBM (6). Radiogenomic analysis informed by spatially localized biopsies has identified spatially complex distributions of molecularly distinct subpopulations in GBMs (7). Although such spatial variations in expression of molecular and pathologic markers, metabolism, and radiologic imaging patterns are known to exist in all solid tumors, the origin and the clinical significance of this heterogeneity remain subjects of investigation.
Heterogeneity within tumors may drive resistance to both untargeted and targeted therapies (8). Reliance on conventional maximum tolerated dose–based treatment regimens may accelerate the unopposed proliferation of resistant populations by eliminating the susceptible populations and the attendant competition for space and substrate. Enriquez-Navas et al. recently showed that an evolution-based adaptive therapeutic strategy that exploits such competition between subpopulations of tumor cells could prolong progression-free survival in preclinical models of breast cancer (9). An ongoing clinical trial in prostate cancer (10) has shown that evolutionary dynamics can be successfully integrated into clinical cancer treatment protocols, and it highlighted the unmet need for noninvasive metrics of intratumoral subpopulation changes during treatment.
In the present work, we build upon a conceptual model of GBMs as spatially heterogeneous complex adaptive systems in which tumor growth and response to therapy are governed by eco-evolutionary interactions between the tumor microenvironment and phenotypic properties of local cellular populations. This model posits an explicit and predictable link between macroscopic tumor features observed radiologically and the molecular-, cellular-, and tissue-scale properties of the underlying cancer cell populations. In this model, we hypothesize that radiologically apparent spatial heterogeneity within each GBM can be quantified by some combination of a small number of distinct eco-evolutionary “habitats,” each of which may have different patterns of growth and invasion and may respond differently to therapy (11). Our approach builds upon methods developed in landscape ecology to bridge spatial scales. For example, field biologists are often tasked with estimating species distribution within a large area such as a county or state. Methods developed in landscape ecology typically begin with an analysis of satellite imagery of the region. By combining image channels containing nonoverlapping information (RADAR, infrared and visible light, for example), the biologist can divide the whole region into a patchwork collection of distinct habitats. By sampling the species distribution within each distinct habitat, the geographic distribution of each species over the entire region can be estimated (12, 13).
Multispectral clustering on MRI images has been used before to quantify spatial variations within tumors. Vannier et al. recognized the analogy between multispectral remote-sensing satellite imagery and multiparametric MRI and showed that signatures for “scene components” in the radiologic images could be computed (14–16). This approach can be used to further objectively subdivide the tumor itself into spatially distinct subregions (“habitats”) that harbor distinct subpopulations of tumor cells (11, 17, 18). Spatial heterogeneity of GBMs at radiological scales presents as regional variations in contrast enhancement and edema, and we have used multispectral clustering to decompose each glioma into a small number of distinct “habitats” based on their intensity on different MRI sequences. Tumor voxels were clustered by the calibrated signal intensities on contrast-enhanced T1-weighted (T1W-CE) and fluid-attenuated inversion-recovery (FLAIR) sequences into 6 distinct “habitats” based on low- to medium- to high-contrast enhancement and low–high signal on FLAIR scans. The long-term survival (LTS) cohort (>36 months postdiagnosis) were found to have a significantly higher fraction of habitat 6 (high CE and high FLAIR signal intensity) compared with the short-term survival (STS) cohort (≤ 19 months postdiagnosis) in both the discovery and validation cohorts. We discuss a possible mechanistic basis for this association between habitat 6 and survival in GBM, and implications for habitats-driven adaptive therapy of GBM.
Materials and Methods
Discovery Cohort
In this work, we have used the terms “discovery” (or training) and “validation” as they are understood in the field of machine learning, namely, to refer to the specific steps of training–validation–test in model development (19). Following IRB approval, patients with pathologically confirmed primary GBM and available preoperative T2-weighted (T2W), FLAIR, unenhanced T1W, and T1W-CE scans were identified retrospectively from a single participating institution. Median survival in glioblastoma is reported to be between 12 and 18 months postdiagnosis (20, 21). Recent estimates of 5-year survival rates for patients receiving maximal safe resection, concurrent radiotherapy and chemotherapy, and adjuvant chemotherapy are ∼10% (22). Our original intent was to investigate MRI habitats in high-grade gliomas from subjects who survived >5 years postdiagnosis. However, after application of the additional requirement that certain MRI scans be available at diagnosis, we had to downgrade this criterion to >3-year survival postdiagnosis of GBM so as to form cohorts with reasonable numbers of subjects. Thus, an LTS group comprising 22 subjects who survived >36 months postdiagnosis (median survival, 62.6 months; range, 36–107 months) was created. A control STS group of 22 subjects who survived <19 months postdiagnosis (median survival, 11.6 months; range, 2.5–19 month) was created to individually match to LTS subjects on age and calendar year of diagnosis.
Validation Cohort
Following IRB approval, patients with pathologically confirmed primary GBM and available preoperative T2W, FLAIR, T1W, and T1W-CE MRI scans were identified retrospectively from a multi-institutional database, matching on age and sex. The LTS group included 15 subjects who survived >36 months postdiagnosis (median survival, 86.6 months; range, 39–177 months), while the STS group included 15 subjects who survived ≤19 months postdiagnosis (median survival, 12.6 moths; range, 1.8–19 months).
Patient Population Statistics
Additional demographic and clinical covariates of relevance to this study are shown in Table 1.
Table 1.
Demographic and Clinical Characteristics of Patients in the Discovery and Validation Cohorts According to LTS and STS Status
| Characteristics | LTS | STS |
|---|---|---|
| Discovery Cohort | ||
| (N = 22) | (N = 22) | |
| Median Age (years) | 50.5 (range: 22–74) | 50.5 (range: 28–72) |
| Percent Male | 59.1 | 63.6 |
| Percent College Graduatea | 45.5 | 23.8 |
| Median KPS Scorea | 90% | 80% |
| Median Year Diagnosed | 2010 | 2011 |
| Percent Completed Stupp Protocolb | 37 | 0 |
| Median Survival (Months) | 67.7 (range: 36–126) | 11.5 (range: 2.5–19) |
| Validation Cohort | ||
| (N = 15) | (N = 15) | |
| Median Age (years) | 50 (range: 23–68) | 62 (range: 23–78) |
| Percent Male | 67 | 60 |
| Median Education (years) | Unknown | Unknown |
| Median KPS Score | 90c | 90d |
| Median Year Diagnosed | 2009 | 2009 |
| Percent Completed Stupp Protocol | 66.7 | 26.7 |
| Median Survival (months) | 86.6 (range: 39–177) | 12.6 (range: 1.8–19) |
a 1 STS missing education; 3 LTS and 6 STS missing KPS score.
b As defined in PubMed PMID: 15758009. Results based on 20 LTS and 16 STS patients with complete information on receipt of the chemoradiation protocol. A total of 7 patients underwent biopsy as the only form of surgery (1 LTS and 6 STS).
c 10 missing values.
d 11 missing values.
Image Registration
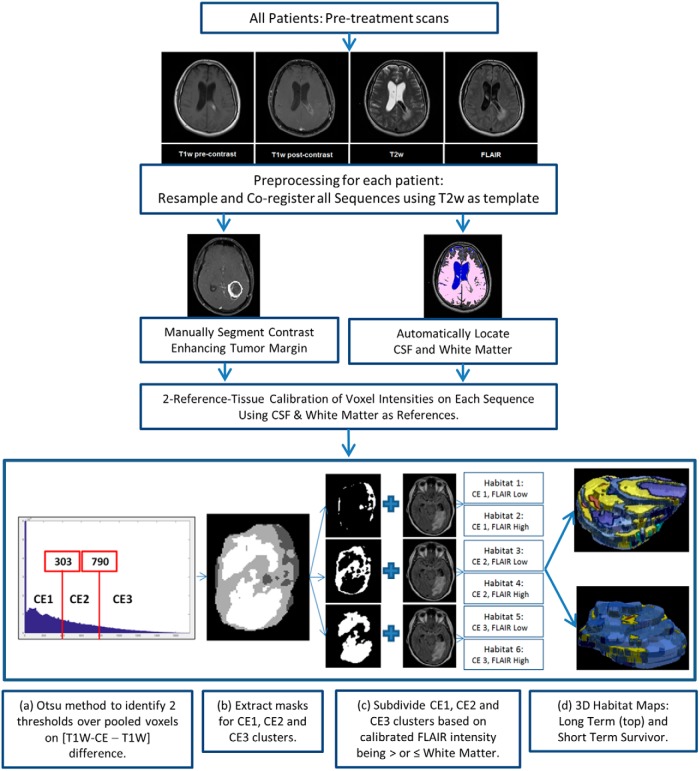
For each patient, the FLAIR, T1W, and T1W-CE images were coregistered with the T2W images using in-house MATLAB (MathWorks, Natick, MA) software (top panel in Figure 1). As part of this process, the FLAIR, T1W, and T1W-CE images were resampled to match pixel dimensions and slice thicknesses with the reference T2W images. Spatial alignment was performed using a combination of rigid and affine geometrical transformations.
Figure 1.
Fluid-attenuated inversion-recovery (FLAIR), T1-weighted (T1W), and contrast-enhanced T1-weighted (T1W-CE) images were coregistered with and resampled to match voxel dimensions in the reference T2W scans (top panel). A contour was manually drawn to circumscribe the CE tumor in all applicable slices on postregistration T1W-CE images (middle panel). Normal white matter and cerebral spinal fluid (CSF) were automatically segmented (middle panel, details in Figure 2). Voxel intensities were calibrated against white matter (WM) and CSF to permit cluster analysis of voxels pooled across patients on each type of magnetic resonance imaging (MRI) scan (middle panel). Pooled voxels from within the CE tumor mask were clustered into 6 habitats using the criteria listed in Table 1 (bottom panel). Also shown in the bottom panel is a 3D stack of maps of habitats 1–6 in an example tumor, for illustrative purposes.
Tumor Segmentation
In this work, we restricted our analysis of intratumoral “habitats” to the CE portion of the tumor volumes. For this purpose, a contour was manually drawn to circumscribe the CE tumor in all applicable slices on postregistration T1W-CE images (middle panel in Figure 1).
Intensity Calibration
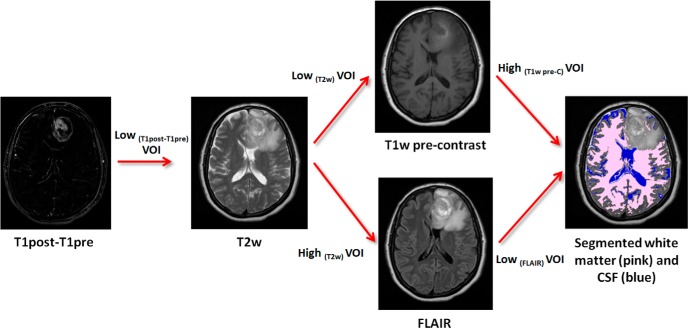
The next step in our image processing pipeline was intensity calibration (middle panel of Figure 1), the objective of which is to allow comparison of voxel intensities across patients on each given type of MRI scan. For this purpose, 2 reference normal tissue regions were automatically segmented as shown in Figure 2. In brief, intensities within the T1W-CE − T1W difference volume (ΔT1W) were clustered into low- and high-intensity classes using Otsu thresholding (23). Then, on T2W, voxels from the low-intensity class were subdivided further into low- and high-intensity clusters using Otsu thresholding. Voxels from the low cluster formed a volume of interest (VOI) that was applied to T1W, which was subdivided into low- and high-intensity clusters by Otsu thresholding, with the resulting voxels in the high-intensity class labeled as “normal white matter” (reference region 1). Voxels from the high T2W cluster formed a VOI mask that was applied to the FLAIR scan, and these were again subdivided into low- and high-intensity clusters using Otsu thresholding, and the low-intensity cluster was labeled as “CSF” (reference region 2). Voxel intensities on T2W, FLAIR, and precontrast T1W images were then linearly calibrated using “normal white matter” and “CSF” as reference tissues. The reference intensity values for these 2 tissues, respectively, were 81 and 183 on T2W, 587 and 464 on FLAIR, and 1099 and 748 on precontrast T1W, all in arbitrary units. These reference values were taken from the T2W, FLAIR, and precontrast T1W images of a patient chosen randomly from the discovery cohort, and do not carry any particular physiological meaning as such. Intensity calibration for T1W-CE was performed using the same linear transformation as computed for the associated precontrast T1W. Our input data comprise standard-of-care MRI images that were acquired with varying protocols per subject. Acquisition parameters such as the repetition time, echo time, and flip angle were not the same across all subjects for each scan type (T2W, FLAIR, T1W). Because MRI signal intensity is a nonlinear function of these acquisition parameters, linear calibration against 2 reference tissues may not necessarily be adequate for standardization of intensities per scan type. Fortunately, the range of excursions in these acquisition parameters across subjects was relatively small, and signal equation simulations indicated that calibration of raw signal intensity against 2 dissimilar reference tissues would provide satisfactory intensity calibration for other tissues with T1 and T2 values similar to or in-between those of the 2 reference tissues. The coefficient of variation of normal gray matter intensity across all patients was significantly smaller postcalibration as compared with precalibration on each of FLAIR, T1W, and T1W-CE images, and we took this to be evidence of successful intensity calibration (see online Supplemental Figure 1).
Figure 2.
Automatic segmentation procedure to locate WM and CSF volumes within the brain for use in intensity calibration. Intensities within the T1W-CE − T1W difference volume (ΔT1W) of a given subject were clustered into low- and high-intensity classes by Otsu thresholding. A mask of voxels in the low-intensity class was applied to the T2W image and further subdivided into low- and high-intensity clusters by Otsu thresholding. The resulting mask of voxels in the low-intensity cluster was applied to the T1W image, which was again subdivided into low- and high-intensity clusters with the high-intensity class labeled as “normal white matter” (reference region 1). The mask of high-intensity voxels from the T2W image was applied to the FLAIR image, and it was again subdivided into low- and high-intensity clusters with the resulting low-intensity cluster labeled as “CSF” (reference region 2).
Multispectral Clustering to Define Intratumoral Habitats
Calibration of intensities per MRI scan type allows us to pool voxels over multiple patients for combined cluster analysis. This series of steps is depicted in the bottom panel of Figure 1. In brief, the manually drawn CE tumor mask was applied to the calibrated ΔT1W difference volume of each patient in the discovery cohort, and the voxels within the mask were pooled over all subjects and clustered by Otsu thresholding into 3 levels of contrast enhancement: CE1 (low enhancement), CE2 (medium enhancement), and CE3 (high enhancement). The low-, medium- and high-contrast enhancement thresholds identified on the discovery cohort were refined on validation, specifically that the maximum value of ΔT1W difference intensity was capped at 5000 arbitrary units postcalibration before Otsu thresholding. This was done to manage the skewing of the clustering process by a long 1-sided tail on the ΔT1W difference intensity histogram in some patients. Each of these 3 clusters was further subclustered into 2 classes around a calibrated value of 600 on FLAIR, a threshold value that is similar to the mean intensity of normal white matter over all subjects after calibration. The final habitat definitions are listed in Table 2.
Table 2.
Intratumoral Habitats' Definitions on Calibrated FLAIR and ΔT1W Intensities
| Calibrated FLAIR Image Intensity | Calibrated ΔT1W Difference Intensity | |
|---|---|---|
| Habitat 1 | ≤600 | ≤303 |
| Habitat 2 | >600 | ≤303 |
| Habitat 3 | ≤600 | 303 < ΔT1W ≤ 790 |
| Habitat 4 | >600 | 303 < ΔT1W ≤ 790 |
| Habitat 5 | ≤600 | >790 |
| Habitat 6 | >600 | >790 |
Statistics and Survival Analyses
Absolute tumor volume, habitat volumes, and habitat volume fractions for each habitat were computed. Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). Data normality was assessed using the D'Agostino–Pearson test, and significance of differences in habitat volumes between groups was assessed by 2-tailed unpaired t-tests. Survival analyses were performed using Kaplan–Meier survival curves, and statistical significance was computed using the log-rank test. For the Kaplan–Meier analysis, habitat volumes were dichotomized into 2 groups using the median score value.
Results
Mean tumor volumes at diagnosis were comparable between the LTS and STS groups in the discovery cohort (33 ± 6.6 cm3 vs. 37 ± 6.1 cm3, P = .62) (see online Supplemental Figure 2A). There was no statistically significant difference in mean tumor volumes at diagnosis between the LTS and STS groups in the validation cohort (33 ± 7.0 cm3 vs. 17 ± 4.8 cm3, P = .075), although there was a trend toward smaller tumor volumes in the STS group (see online Supplemental Figure 2B).
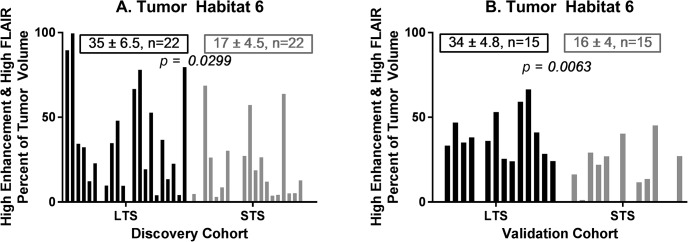
Figure 3 depicts differences in habitat 6 (high contrast enhancement and high FLAIR) content between a representative LTS subject (left; overall survival, 41+ months) and STS subject (right; overall survival, 3 months) at the time of tumor presentation before surgical intervention. In the discovery cohort habitat 6 comprised a significantly higher volume fraction (P < .03) of the tumor volume at diagnosis in long-term survivors (mean ± S.E.M. = 35% ± 6.5%; n = 22) compared with short-term survivors (mean ± S.E.M. = 17% ± 4.5%; n = 22) (Figure 4A). This finding was replicated in the validation cohort (P < .007), with habitat 6 comprising 34% ± 4.8% (n = 15) of the tumor volume in LTS subjects compared with 16% ± 4.0% (n = 15) of the tumor volume in STS subjects (Figure 4B).
Figure 3.
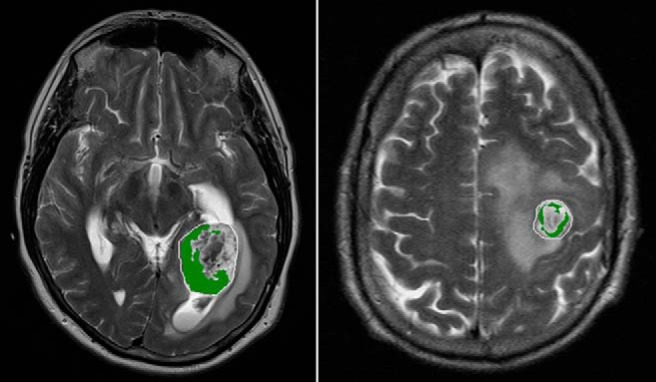
Habitat 6 (high enhancement and high FLAIR) on preoperative MRI comprises 23% of the tumor by volume in a long-term survivor (left, overall survival 41+ months) and 9% of the tumor by volume in a short-term survivor (right, overall survival 3 months).
Figure 4.
Habitat 6 (high enhancement and high FLAIR) was significantly higher in the LTS group relative to the STS group in both the (A) discovery and (B) validation cohorts.
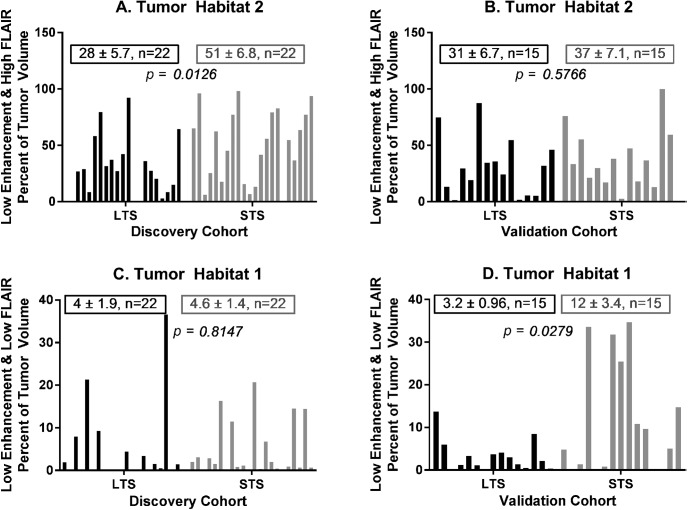
Habitat 2 (low enhancement and high FLAIR) comprised a significantly lower volume fraction (P = .0126) of the tumor at diagnosis in long-term survivors (mean ± S.E.M. = 28 ± 5.7; n = 22) relative to short-term survivors (mean ± S.E.M. = 51 ± 6.8; n = 22) in the discovery cohort (Figure 5A), but this was not replicated in the validation cohort (Figure 5B). In parallel, habitat 1 (low enhancement and low FLAIR) was not found to be significantly different between LTS and STS subjects in the discovery cohort (Figure 5C) but comprised a significantly lower volume fraction (P = .0279) of the tumor at diagnosis in long-term survivors (mean ± S.E.M. = 3.2 ± 0.96; n = 15) relative to short-term survivors (mean ± S.E.M. = 12 ± 3.4; n = 15) in the validation cohort (Figure 5D). Minor inconsistencies in FLAIR intensity calibration across the patients may be the root cause of this variable finding, given that Habitats 1 and 2 belong to the low and high FLAIR clusters, respectively.
Figure 5.
Habitat 2 (low enhancement and high FLAIR) was significantly lower in the LTS group relative to the STS group in the discovery cohort (panel A), but this difference was not recapitulated in the validation cohort (panel B). Habitat 1 (low enhancement and low FLAIR) was not significantly different between the LTS and STS groups in the discovery cohort (panel C), but was significantly lower in the LTS group in the validation cohort (panel D).
Habitat 3 (medium enhancement and low FLAIR), habitat 4 (medium enhancement and high FLAIR), and habitat 5 (high enhancement and low FLAIR) were not significantly different between the LTS and STS groups in either the discovery or validation cohorts (see online Supplemental Figure 3).
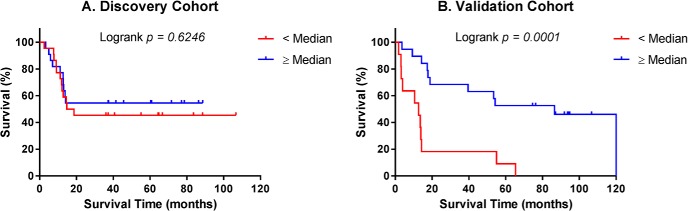
Median percent of tumor volume occupied by habitat 6 in the discovery cohort (5.77%) was used as a cutpoint to dichotomize patients into high and low habitat 6 fraction groups. Kaplan–Meier survival analyses were then carried out separately in the discovery and validation cohorts using the prespecified cutpoint (5.77%) established for the discovery cohort, as a stringent test of reproducibility. Based on the median cutpoint, low and high fractions of habitat 6 were not associated with overall survival in the discovery cohort (Figure 6A; P = .62), but were statistically significant with respect to overall survival in the validation cohort (Figure 6B; P = .0001). In the discovery cohort, Kaplan–Meier 3-year survival rates were 45% and 55% in the < median versus ≥ median subgroups, respectively. In the validation cohort corresponding 3-year survival rates were 18% and 68%, respectively.
Figure 6.
Kaplan–Meier plots of overall survival in the discovery cohort (A). Survival of patients with habitat 6 volume fraction ≥ median (5.77%, n = 22) and < median (n = 22). Kaplan–Meier plots of overall survival in the validation cohort (B). Survival of patients with habitat 6 volume fraction ≥ median from the discovery cohort (5.77%, n = 19) and < median (n = 11).
Discussion
The overall goal of our work is to develop noninvasive imaging biomarkers that can be used to drive evolution-based adaptive therapeutic strategies for GBM. For any biomarker to be clinically useful, it must be computable reliably and reproducibly (24). MRI parameters such as ADC, T1, and T2, and with some limitations, also model-dependent parameters such as relative cerebral blood volume (rCBV), relative cerebral blood flow, and Ktrans, are comparable between data sets when standardized protocols are utilized (25–35). Parameter maps are therefore attractive for computing tumor habitats consistently across patients and scan dates, but these maps are not routinely collected as part of standard-of-care imaging. The subjects in our study received their initial diagnostic scans at a variety of institutions including at community radiology facilities, as a result of which there was great variability in the type and quality of scans that were available for retrospective analysis. In particular, we were unable to curate sufficient numbers of LTS subjects with available ADC maps at diagnosis. We therefore sought to compute intratumoral habitats using FLAIR, T1W, and T1W-CE scans after calibrating raw MRI pixel intensities against 2 reference tissues.
High signal on ΔT1W is indicative of either good perfusion or high microvascular leakiness. High intensity on FLAIR images in glioma represents a mixture of vasogenic edema, which arises from leakage of plasma into regions with low cell density, and tumor cell infiltration along long white matter tracts (36). Our retrospective study shows, in both a discovery cohort and a validation cohort, that tumors in LTS subjects have a significantly higher fraction of habitat 6 (high contrast enhancement and high FLAIR signal intensity) than STS. Particularly striking is the similarity in habitat 6 content of LTS tumors between the discovery and validation cohorts (35% and 34%, respectively) and of STS tumors between the discovery and validation cohorts (17% and 16%, respectively). We divided tumor regions with high signal intensity on ΔT1W calibrated difference images into 2 distinct habitats with either high or low FLAIR signal. Low FLAIR signal would be expected in regions with high contrast enhancement stemming from good perfusion, which would be conducive to high cellular density, although not necessarily where the enhancement arises from microvascular leakiness. Our results demonstrate the high contrast enhancement and high FLAIR signal habitat is strongly associated with patient survival.
In a preliminary study of pretreatment MRI examinations from 32 patients with GBM enrolled in the TCGA, Gatenby et al. showed that GBM tumor habitats defined on FLAIR and T1W-CE images could be used to differentiate patients who survived <400 days from patients who survived ≥400 days postdiagnosis (37). A follow-up study indicated that incorporating information from 3 MRI sequences, namely, T2W, FLAIR, and T1W-CE, improved prediction of survival time in patients with GBM (38). LaViolette et al. similarly clustered voxels into low, medium, and high classes on T1W, T1W-CE, FLAIR and apparent diffusion coefficient of water (ADC) maps to divide GBM tumors into 81 habitats, and identified 5 specific habitats that when present at higher volumes correlated with poorer prognosis (39). Recently, Juan-Albarracín et al. analyzed preoperative DSC-MRI and FLAIR scans of 50 patients with GBM to compute tumor habitats on the basis of rCBV, relative cerebral blood flow, and edema, and they found a surprising correlation between longer survival times and lower indices of perfusion (40). Boonzaier et al. report that tumor habitats reflecting low ADC values intersecting with high rCBV values demonstrate a significantly elevated choline-to-N-acetylaspartate ratio on 1H magnetic resonance spectroscopy, and that a higher proportion of this habitat within the NE region of GBM is associated with poor overall survival (41). Interpatient diversity in overall imaging patterns of growth and invasion has been associated with tumor aggressiveness and clinical outcomes across patients (42–45). Our investigation leverages unique resources of data including patients with exceptionally long follow-up for prognosis in glioblastoma.
Standard-of-care therapy in newly diagnosed GBM is maximal safe surgical resection followed by concomitant radiation therapy and temozolomide for 6 weeks, followed by adjuvant temozolomide for 6 monthly cycles (46). Thereafter, subjects in our retrospective study would each also have received a variety of investigational and/or palliative treatments, including extended cycles of temozolomide. Our findings suggest that one or more characteristics of the radiologically visible initial tumor mass define an intrinsic prognostically relevant tumor feature that continues to influence patient outcome months, and even years, after diagnosis. It is possible that the radiologic appearance of habitat 6 is a shared feature of disparate favorable markers in GBM, such as Isocitrate Dehydrogenase (IDH) mutation status (47), mesenchymal subtype (48) or lymphocyte cytokines such as CXCR4 (49). Alternately, one can hypothesize that components of the immune system in the LTS subjects retain the ability to recognize tumor antigens present in the original mass that are retained in the recurrent mass. Immune infiltrates in the tumor would be consistent with the MRI characteristics of habitat 6, namely, high contrast-enhancement and high tumor-associated edema. Pathological studies have shown that increased CD8+ T cell infiltrates in newly diagnosed GBM is associated with long-term survival (50), and we hypothesize that increased FLAIR signal in well-perfused—and presumably cellular—regions may be indicative of interstitial edema related to inflammatory changes caused by an immune response. A definitive biological interpretation of our finding requires further investigation.
Known weaknesses in our study include that the numbers in each survival group stratum were small and statistical power correspondingly limited to detect all but strong associations in the data. Specifically, while our analysis detected a significant difference between the LTS and STS groups in both the discovery and validation cohorts (Figure 4), on an individual patient basis, we could observe survival differences by a binary analysis around the median habitat 6 content in only the validation cohort (Figure 6). The need to improve calibration of raw MRI image intensities is revealed in the inconsistent significances of Habitats 1 and 2 in the discovery and validation cohorts (Figure 5). Additional covariates may also impinge upon our analysis. For example, in the discovery cohort, LTS and STS subjects were matched for parameters such as patient age and year of diagnosis, but LTS patients were nonetheless more educated and more likely to survive the completion of standard treatment. In the validation cohort, the LTS and STS groups were not matched for patient age and treatment regimens. It is unclear how these group differences might explain the present findings.
Only about 5% of patients with GBM undergoing standard of care survive ≥5 years postdiagnosis (46). Investigation of a cohort of rare long-term survivors identifies a “habitat” on initial multiparametric MRI scans that is significantly different than in a control cohort. Our working hypothesis is that habitat 6 corresponds to a microenvironment that selects for glioma cells that are either innately less aggressive or are more amenable to control by tumor-infiltrating leukocytes. Habitat imaging has the potential to provide noninvasive longitudinal biomarkers of intratumoral evolutionary and ecological dynamics for the informed application of adaptive therapy to manage GBM.
Supplemental Materials
Acknowledgments
We wish to thank the following people for their contributions to this work: Joo Kim, MD, for guidance on whole brain segmentation; Lila Kis and Doniya Milani for assistance with local data curation. We also wish to acknowledge research support from the National Institutes of Health (U54 CA193489; P30 CA076292 (IRAT Core); R01 CA116174; R01 NS060752, R01 CA164371, U54 CA210180, U54 CA143970, U54 CA193489, U01 CA220378), the James S. McDonnell Foundation (grant no. 220020400), the Ivy Foundation, and a 2017 Moffitt Team Science Award (Drs. Egan and Raghunand).
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- MRI
- Magnetic resonance imaging
- GBM
- glioblastoma multiforme
- FLAIR
- fluid-attenuated inversion-recovery
- CE
- contrast-enhancing
- NE
- nonenhancing
- DSC
- dynamic susceptibility contrast-enhanced
- T1W
- unenhanced T1-weighted
- T1W-CE
- contrast-enhanced T1-weighted
- LTS
- long-term survival
- STS
- short-term survival
- T2W
- T2-weighted
- VOI
- volume of interest
- rCBV
- relative cerebral blood volume
- ΔT1W
- T1W-CE − T1W difference volume
References
- 1. Hu LS, Ning S, Eschbacher JM, Gaw N, Dueck AC, Smith KA, Nakaji P, Plasencia J, Ranjbar S, Price SJ, Tran N, Loftus J, Jenkins R, O'Neill BP, Elmquist W, Baxter LC, Gao F, Frakes D, Karis JP, Zwart C, Swanson KR, Sarkaria J, Wu T, Mitchell JR, Li J. Multi-parametric MRI and texture analysis to visualize spatial histologic heterogeneity and tumor extent in glioblastoma. PLoS One. 2015;10:e0141506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gill BJ, Pisapia DJ, Malone HR, Goldstein H, Lei L, Sonabend A, Yun J, Samanamud J, Sims JS, Banu M, Dovas A, Teich AF, Sheth SA, McKhann GM, Sisti MB, Bruce JN, Sims PA, Canoll P. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci U S A. 2014;111:12550–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Autry A, Phillips JJ, Maleschlijski S, Roy R, Molinaro AM, Chang SM, Cha S, Lupo JM, Nelson SJ. Characterization of metabolic, diffusion, and perfusion properties in GBM: contrast-enhancing versus non-enhancing tumor. Transl Oncol. 2017;10:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, Bi Y, Pal S, Davuluri RV, Roccograndi L, Dahmane N, Martinez-Lage M, Biros G, Wolf RL, Bilello M, O'Rourke DM, Davatzikos C. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 2016;18:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, Plasencia J, Dueck AC, Peng S, Smith KA, Nakaji P, Karis JP, Quarles CC, Wu T, Loftus JC, Jenkins RB, Sicotte H, Kollmeyer TM, O'Neill BP, Elmquist W, Hoxworth JM, Frakes D, Sarkaria J, Swanson KR, Tran NL, Li J, Mitchell JR. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017;19:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol. 2015;12:69–70. [DOI] [PubMed] [Google Scholar]
- 9. Enriquez-Navas PM, Kam Y, Das T, Hassan S, Silva A, Foroutan P, Ruiz E, Martinez G, Minton S, Gillies RJ, Gatenby RA. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Transl Med. 2016;8:327ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J, Cunningham JJ, Brown JS, Gatenby RA. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat Commun. 2017;8:1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- 13. Turner MG. Landscape ecology: what is the state of the science? Annu Rev Ecol Evol Syst. 2005;36:319–344. [Google Scholar]
- 14. Vannier MW, Butterfield RL, Jordan D, Murphy WA, Levitt RG, Gado M. Multispectral analysis of magnetic resonance images. Radiology. 1985;154:221–224. [DOI] [PubMed] [Google Scholar]
- 15. Vannier MW, Butterfield RL, Rickman DL, Jordan DM, Murphy WA, Biondetti PR. Multispectral magnetic resonance image analysis. Crit Rev Biomed Eng. 1987;15:117–144. [PubMed] [Google Scholar]
- 16. Gohagan JK, Spitznagel EL, Murphy WA, Vannier MW, Dixon WT, Gersell DJ, Rossnick SL, Totty WG, Destouet JM, Rickman DL, et al. . Multispectral analysis of MR images of the breast. Radiology. 1987;163:703–707. [DOI] [PubMed] [Google Scholar]
- 17. Carano RA, Ross AL, Ross J, Williams SP, Koeppen H, Schwall RH, Van Bruggen N. Quantification of tumor tissue populations by multispectral analysis. Mag Reson Med. 2004;51:542–551. [DOI] [PubMed] [Google Scholar]
- 18. Barck KH, Willis B, Ross J, French DM, Filvaroff EH, Carano RA. Viable tumor tissue detection in murine metastatic breast cancer by whole-body MRI and multispectral analysis. Magn Reson Med. 2009;62:1423–1430. [DOI] [PubMed] [Google Scholar]
- 19. Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. [DOI] [PubMed] [Google Scholar]
- 20. Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. [DOI] [PubMed] [Google Scholar]
- 21. Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ Jr., Mehta MP. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. [DOI] [PubMed] [Google Scholar]
- 23. Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. [Google Scholar]
- 24. Hayes DF. Biomarker validation and testing. Mol Oncol. 2015;9:960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leach MO, Morgan B, Tofts PS, Buckley DL, Huang W, Horsfield MA, Chenevert TL, Collins DJ, Jackson A, Lomas D, Whitcher B, Clarke L, Plummer R, Judson I, Jones R, Alonzi R, Brunner T, Koh DM, Murphy P, Waterton JC, Parker G, Graves MJ, Scheenen TW, Redpath TW, Orton M, Karczmar G, Huisman H, Barentsz J, Padhani A. Imaging vascular function for early stage clinical trials using dynamic contrast-enhanced magnetic resonance imaging. Eur Radiol. 2012;22:1451–1464. [DOI] [PubMed] [Google Scholar]
- 26. Malyarenko D, Galban CJ, Londy FJ, Meyer CR, Johnson TD, Rehemtulla A, Ross BD, Chenevert TL. Multi-system repeatability and reproducibility of apparent diffusion coefficient measurement using an ice-water phantom. J Magn Reson Imaging. 2013;37:1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ellingson BM, Bendszus M, Boxerman J, Barboriak D, Erickson BJ, Smits M, Nelson SJ, Gerstner E, Alexander B, Goldmacher G, Wick W, Vogelbaum M, Weller M, Galanis E, Kalpathy-Cramer J, Shankar L, Jacobs P, Pope WB, Yang D, Chung C, Knopp MV, Cha S, van den Bent MJ, Chang S, Yung WK, Cloughesy TF, Wen PY, Gilbert MR. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol. 2015;17:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taouli B, Beer AJ, Chenevert T, Collins D, Lehman C, Matos C, Padhani AR, Rosenkrantz AB, Shukla-Dave A, Sigmund E, Tanenbaum L, Thoeny H, Thomassin-Naggara I, Barbieri S, Corcuera-Solano I, Orton M, Partridge SC, Koh DM. Diffusion-weighted imaging outside the brain: consensus statement from an ISMRM-sponsored workshop. J Magn Reson Imaging. 2016;44:521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, Aryal MP, LaViolette PS, Oborski MJ, O'Sullivan F, Abramson RG, Jafari-Khouzani K, Afzal A, Tudorica A, Moloney B, Gupta SN, Besa C, Kalpathy-Cramer J, Mountz JM, Laymon CM, Muzi M, Schmainda K, Cao Y, Chenevert TL, Taouli B, Yankeelov TE, Fennessy F, Li X. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge. Tomography. 2016;2:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, Boellaard R, Bohndiek SE, Brady M, Brown G, Buckley DL, Chenevert TL, Clarke LP, Collette S, Cook GJ, deSouza NM, Dickson JC, Dive C, Evelhoch JL, Faivre-Finn C, Gallagher FA, Gilbert FJ, Gillies RJ, Goh V, Griffiths JR, Groves AM, Halligan S, Harris AL, Hawkes DJ, Hoekstra OS, Huang EP, Hutton BF, Jackson EF, Jayson GC, Jones A, Koh DM, Lacombe D, Lambin P, Lassau N, Leach MO, Lee TY, Leen EL, Lewis JS, Liu Y, Lythgoe MF, Manoharan P, Maxwell RJ, Miles KA, Morgan B, Morris S, Ng T, Padhani AR, Parker GJ, Partridge M, Pathak AP, Peet AC, Punwani S, Reynolds AR, Robinson SP, Shankar LK, Sharma RA, Soloviev D, Stroobants S, Sullivan DC, Taylor SA, Tofts PS, Tozer GM, van Herk M, Walker-Samuel S, Wason J, Williams KJ, Workman P, Yankeelov TE, Brindle KM, McShane LM, Jackson A, Waterton JC. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klaassen R, Gurney-Champion OJ, Wilmink JW, Besselink MG, Engelbrecht MRW, Stoker J, Nederveen AJ, van Laarhoven HWM. Repeatability and correlations of dynamic contrast enhanced and T2* MRI in patients with advanced pancreatic ductal adenocarcinoma. Magn Reson Imaging. 2018;50:1–9. [DOI] [PubMed] [Google Scholar]
- 32. Sorace AG, Wu C, Barnes SL, Jarrett AM, Avery S, Patt D, Goodgame B, Luci JJ, Kang H, Abramson RG, Yankeelov TE, Virostko J. Repeatability, reproducibility, and accuracy of quantitative MRI of the breast in the community radiology setting. J Magn Reson Imaging. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malyarenko D, Fedorov A, Bell L, Prah M, Hectors S, Arlinghaus L, Muzi M, Solaiyappan M, Jacobs M, Fung M, Shukla-Dave A, McManus K, Boss M, Taouli B, Yankeelov TE, Quarles CC, Schmainda K, Chenevert TL, Newitt DC. Toward uniform implementation of parametric map Digital Imaging and Communication in Medicine standard in multisite quantitative diffusion imaging studies. J Med Imaging (Bellingham). 2018;5:011006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newitt DC, Malyarenko D, Chenevert TL, Quarles CC, Bell L, Fedorov A, Fennessy F, Jacobs MA, Solaiyappan M, Hectors S, Taouli B, Muzi M, Kinahan PE, Schmainda KM, Prah MA, Taber EN, Kroenke C, Huang W, Arlinghaus LR, Yankeelov TE, Cao Y, Aryal M, Yen YF, Kalpathy-Cramer J, Shukla-Dave A, Fung M, Liang J, Boss M, Hylton N. Multisite concordance of apparent diffusion coefficient measurements across the NCI Quantitative Imaging Network. J Med Imaging (Bellingham). 2018;5:011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bane O, Hectors SJ, Wagner M, Arlinghaus LL, Aryal MP, Cao Y, Chenevert TL, Fennessy F, Huang W, Hylton NM, Kalpathy-Cramer J, Keenan KE, Malyarenko DI, Mulkern RV, Newitt DC, Russek SE, Stupic KF, Tudorica A, Wilmes LJ, Yankeelov TE, Yen YF, Boss MA, Taouli B. Accuracy, repeatability, and interplatform reproducibility of T1 quantification methods used for DCE-MRI: results from a multicenter phantom study. Magn Reson Med. 2018;79:2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Villanueva-Meyer JE, Mabray MC, Cha S. Current clinical brain tumor imaging. Neurosurgery. 2017;81:397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou M, Hall L, Goldgof D, Russo R, Balagurunathan Y, Gillies R, Gatenby R. Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results. Transl Oncol. 2014;7:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA. Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J Magn Reson Imaging. 2017;46:115–123. [DOI] [PubMed] [Google Scholar]
- 39. McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, Schmainda KM, LaViolette PS. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Juan-Albarracín J, Fuster-Garcia E, Pérez-Girbés A, Aparici-Robles F, Alberich-Bayarri Á, Revert-Ventura A, Martí-Bonmatí L, García-Gómez JM. Glioblastoma: vascular habitats detected at preoperative dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging predict survival. Radiology. 2018;287:944–954. [DOI] [PubMed] [Google Scholar]
- 41. Boonzaier NR, Larkin TJ, Matys T, van der Hoorn A, Yan JL, Price SJ. Multiparametric MR imaging of diffusion and perfusion in contrast-enhancing and nonenhancing components in patients with glioblastoma. Radiology. 2017;284:180–190. [DOI] [PubMed] [Google Scholar]
- 42. Baldock AL, Ahn S, Rockne R, Johnston S, Neal M, Corwin D, Clark-Swanson K, Sterin G, Trister AD, Malone H, Ebiana V, Sonabend AM, Mrugala M, Rockhill JK, Silbergeld DL, Lai A, Cloughesy T, McKhann GM 2nd, Bruce JN, Rostomily RC, Canoll P, Swanson KR. Patient-specific metrics of invasiveness reveal significant prognostic benefit of resection in a predictable subset of gliomas. PLoS One. 2014;9:e99057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swanson KR, Rostomily RC, Alvord EC. A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: a proof of principle. Br J Cancer. 2008;98:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Szeto MD, Chakraborty G, Hadley J, Rockne R, Muzi M, Alvord EC Jr., Krohn KA, Spence AM, Swanson KR. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 2009;69:4502–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baldock AL, Yagle K, Born DE, Ahn S, Trister AD, Neal M, Johnston SK, Bridge CA, Basanta D, Scott J, Malone H, Sonabend AM, Canoll P, Mrugala MM, Rockhill JK, Rockne RC, Swanson KR. Invasion and proliferation kinetics in enhancing gliomas predict IDH1 mutation status. Neuro Oncol. 2014;16:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. [DOI] [PubMed] [Google Scholar]
- 47. Price SJ, Allinson K, Liu H, Boonzaier NR, Yan JL, Lupson VC, Larkin TJ. Less invasive phenotype found in isocitrate dehydrogenase-mutated glioblastomas than in isocitrate dehydrogenase wild-type glioblastomas: a diffusion-tensor imaging study. Radiology. 2017;283:215–221. [DOI] [PubMed] [Google Scholar]
- 48. Naeini KM, Pope WB, Cloughesy TF, Harris RJ, Lai A, Eskin A, Chowdhury R, Phillips HS, Nghiemphu PL, Behbahanian Y, Ellingson BM. Identifying the mesenchymal molecular subtype of glioblastoma using quantitative volumetric analysis of anatomic magnetic resonance images. Neuro Oncol. 2013;15:626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma X, Shang F, Zhu W, Lin Q. CXCR4 expression varies significantly among different subtypes of glioblastoma multiforme (GBM) and its low expression or hypermethylation might predict favorable overall survival. Expert Rev Neurother. 2017;17:941–946. [DOI] [PubMed] [Google Scholar]
- 50. Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, Parsa AT. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.