Abstract
Quantitative mapping of hyperperfused and hypercellular regions of glioblastoma has been proposed to improve definition of tumor regions at risk for local recurrence following conventional radiation therapy. As the processing of the multiparametric dynamic contrast-enhanced (DCE-) and diffusion-weighted (DW-) magnetic resonance imaging (MRI) data for delineation of these subvolumes requires additional steps that go beyond the standard practices of target definition, we sought to devise a workflow to support the timely planning and treatment of patients. A phase II study implementing a multiparametric imaging biomarker for tumor hyperperfusion and hypercellularity consisting of DCE-MRI and high b-value DW-MRI to guide intensified (75 Gy/30 fractions) radiation therapy (RT) in patients with newly diagnosed glioblastoma was launched. In this report, the workflow and the initial imaging outcomes of the first 12 patients are described. Among all the first 12 patients, treatment was initiated within 6 weeks of surgery and within 2 weeks of simulation. On average, the combined hypercellular volume and high cerebral blood volume/tumor perfusion volume were 1.8 times smaller than the T1 gadolinium abnormality and 10 times smaller than the FLAIR abnormality. Hypercellular volume and high cerebral blood volume/tumor perfusion volume each identified largely distinct regions and showed 57% overlap with the enhancing abnormality, and minimal-to-no extension outside of the FLAIR. These results show the feasibility of implementing a workflow for multiparametric magnetic resonance-guided radiation therapy into clinical trials with a coordinated multidisciplinary team, and the unique and complementary tumor subregions identified by the combination of high b-value DW-MRI and DCE-MRI.
Keywords: pipeline, workflow, multiparametric, MRI, glioblastoma
Introduction
Conventional therapies for glioblastoma (GBM) continue to rely on anatomic imaging modalities for both surgery and radiation therapy (RT), including T1 gadolinium- (T1-Gd) enhanced and T2-weighted fluid attenuated inversion recovery (T2-FLAIR) sequences that do not provide biological information about the underlying disease. Multiple studies have shown the prognostic value of physiological magnetic resonance imaging (MRI) techniques such as proton spectroscopy and perfusion and diffusion MRI and measures such as progression-free survival (PFS) and overall survival (OS) in predicting treatment response in patients with GBM (1–9). These imaging techniques may show abnormal tumor infiltration beyond the contrast-enhanced or nonenhanced areas conventionally targeted by surgery and radiation, and these may potentially be used to guide radiation treatment, reduce tumor recurrence, and improve patient outcome (10).
Dynamic contrast-enhanced (DCE)-MRI assesses relative cerebral blood volume (rCBV), cerebral blood flow, and vascular permeability, which are associated with neovascularization and tumor growth and predict PFS and OS in patients with GBM (1, 2, 5, 11). While regions of elevated rCBV often overlap with regions of contrast enhancement, the nonenhancing, infiltrating tumor beyond this region may potentially be underestimated with perfusion MRI (12). In contrast, diffusion-weighted (DW) MRI may identify tumor phenotype by estimating water mobility in the tissue microenvironment as an indicator of tumor cellularity (13). Apparent diffusion coefficient (ADC) is inversely correlated with cellularity but it may be unreliable in regions of highly cellular tumor, normal brain tissue, edema, and micronecrosis, yielding elevated ADC compared with normal tissue using standard b-values of 0–1000 s/mm2 (10). At our center, we developed a novel DW-MRI technique using high b-value (b = 3000 s/mm2) to selectively isolate solid, often nonenhancing, tumor that is predictive of PFS and often extends beyond the high-dose radiation target (14). We have shown that a combination of these imaging techniques (DCE-MRI and high b-value DW-MRI) into a multiparametric imaging signature predicts PFS with spatial correspondence with patterns of failure, representing biologically high-risk tumor subvolumes identifiable before therapy (12).
Based on these findings, we wished to develop a phase II study to evaluate the feasibility and efficacy of using a multiparametric hypervascular/hypercellular MRI signature to identify areas at highest risk of failure before radiation treatment in patients with newly diagnosed GBM (NCT02805179). Building on a prior phase I/II study showing the safety and efficacy of radiation dose-escalation with concurrent temozolomide (15), this multiparametric advanced imaging technique was used to prospectively guide the boost volume for dose-intensified radiation. To conduct this trial, the development of a workflow was required to permit the integration of an advanced, multiparametric imaging technique into the radiation treatment planning process. Here, we report the workflow and imaging characteristics of the initial patients treated on this prospective clinical trial.
Methodology
Patient Population
Adult patients of ≥18 years of age with newly diagnosed, pathologically confirmed suptratentorial GBM following any extent of resection were enrolled on this University of Michigan IRB-approved clinical trial following study-specific informed consent. Research was conducted in compliance with the World Medical Association Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects. Eligibility included Karnofsky performance status ≥70, minimal life expectancy of 12 weeks, adequate organ function, and maximal contiguous volume of tumor based on advanced imaging-defined boost volume of <1/3 volume of brain. Patients unable to undergo MRI scans or with prior overlapping radiation therapy were excluded. All patients were treated with standard concurrent daily (75 mg/m2) and adjuvant monthly (150–200 mg/m2) temozolomide.
MRI and Computed Tomography Simulation
All patients underwent an MRI simulation and computed tomography (CT) simulation after surgery for radiation planning, within 14 days of commencing chemoradiation. Rigid alignment of the T1-weighted contrast-enhanced and T2 FLAIR MRI to the CT image volumes in the Eclipse image registration workspace was performed by the medical physicist and verified by the radiation oncologist.
Commissioning of Hardware and Software QA
All MRI scans were performed on a 3 T scanner (Skyra, Siemens Healthineers, Erlangen, Germany) in the Radiation Oncology Department. Routine quality assurance of this scanner consists of daily checks of intensity uniformity as well as weekly checks following the ACR phantom accreditation scanning protocol (16). T1 mapping is a critical element of DCE-MRI analysis. To assess the accuracy, repeatability, and interplatform reproducibility of T1 quantification from variable flip angles, we scanned a National Institute of Standards and Technology (NIST) T1 water phantom on our system, provided by our participation in an NCI Quantitative Imaging Network (QIN) multicenter collaborative project (17). We used the extended Tofts model to quantify DCE-MRI, which was implemented in an in-house functional image analysis tool (imFIAT) (18). The performance of our implementation of the extended Tofts model was evaluated using digital reference objects, that is, synthesized DCE phantoms with and without noise, which was fully reported previously (19). In addition, we participated in an NCI QIN multicenter arterial input function (AIF) challenge to validate and compare our AIF delineation procedure to others' (20). On the basis of these evaluations and validations, imFIAT has been granted a level-2 benchmark by the NCI QIN (21).
MRI Acquisition
All images were acquired on a 3 T scanner (Skyra) using a 20-channel head coil. Conventional images, such as 2-dimensional (2D) T2-FLAIR images, and 3-dimensional pre- and postcontrast T1-weighted images, were acquired. In addition, physiological image acquisitions are described in the following subsections.
Diffusion-Weighted Imaging.
DW images were acquired using a 2D RESOLVE pulse sequence with diffusion weighting in 3 orthogonal directions and b-values of 0 and 3000 s/mm2 (1 and 4, respectively) to reduce geometric distortion required for radiation treatment planning. RESOLVE is a multishot technique that uses 2D navigator correction with readout-segmented echo planar imaging (22). Thirty slices were acquired to cover the whole brain with echo time (TE)/repetition time (TR) = 81/7650 milliseconds, matrix size = 160 × 160, and slice thickness/gap = 4.0/1.2 mm for ∼4.23 minutes. DW images acquired with b = 3000 s/mm2 were used for target definition. In addition, DW images were acquired by a 2D echo planar spin echo pulse sequence with diffusion weighting in 3 orthogonal directions and 11 b-values from 0 to 2500 s/mm2 as a backup scan. Thirty slices were acquired with TE/TR = 93/8200 milliseconds, matrix size = 192 × 192, slice thickness/gap = 4.0/1.2 mm, parallel imaging factor of 4 and a single average for 5 minutes. Parallel imaging factor of 4 was used to reduce the echo training time and thereby reduce geometric distortion. A full characterization of geometric accuracy of DW images with these acquisition parameters was previously reported (14).
DCE Imaging.
DCE images were acquired by a 3D gradient echo pulse sequence, called TWIST, in the sagittal orientation to avoid the in-flow effect and ensure artery coverage for an input function delineation. To cover the whole brain, a field of view of 250 × 256 × 187 mm3 was used with a matrix of 128 × 128 × 104 to obtain an isotropic voxel size of ∼1.9, which allows reformatting of the images in an axial plane or other planes as desired. Other acquisition parameters included flip angle = ∼10°, TE/TR = ∼0.95/2.65 milliseconds, temporal resolution = ∼3 s, dynamic phase volumes = 60, and total acquisition time = 3 minutes. Contrast was injected after 5 dynamic image volumes to achieve sufficient baseline data points.
Acquisition for T1 Quantification.
3D gradient echo images with 4 flip angles (3°, 7°, 12°, and 16°), TE/TR = 2.27/5.34 milliseconds, a voxel size = ∼2 mm, and total acquisition time = 1:45 minutes before contrast injection were acquired to quantify native T1. Low spatial resolution B1 maps were acquired to correct flip angle errors in T1 quantification, with an acquisition time of ∼12 seconds.
Target Volume Definition and Data Transfer
Physician-defined volumes were delineated in the Eclipse treatment planning system (Varian Medical Systems) directly on the MRI scans. T2/FLAIR abnormality was defined on the FLAIR MRI (FLAIR∧GTV). The surgical cavity (Cavity∧GTV), residual contrast enhancement (Gd∧GTV), and combination of cavity and contrast enhancement (GTV_Low) were delineated on the T1-Gd MRI. Volumes were then exported from the treatment planning system to the image analysis software (functional image analysis tool or imFIAT), for creation of DCE-MRI tumor volumes (high CBV [hCBV]), and high b-value DW-MRI tumor volumes (hypercellular volume [HCV]).
Image Analysis
DCE Analysis.
Three-parameter Tofts model was used to quantify the fractional plasma volume (Vp), transfer constant of contrast (Ktrans), and the fractional volume of extravascular extracellular space (ve) (23). The model was programed using C++ with a GUI in a functional image analysis tool (imFIAT). A full characterization of performance of software using digital reference objects with a large range of physiological parameters, acquisition parameters, and added Gaussian noise has been previously published (19).
In brief, we used the general assumption that
| (1) |
where Ct is a contrast concentration in a voxel, and ΔR1 is a change in longitudinal relaxation rates after and before the contrast injection. If TR × R1 ≪1, which is generally satisfactory for brain normal tissue and tumors,
| (2) |
where ΔS is a change in gradient echo intensities after and before the contrast injection, Sbaseline is the averaged baseline gradient echo intensity before contrast injection, and R10 is the longitudinal relaxation rate before contrast injection. To obtain an AIF, 20 voxels with maximum intensity differences in a dynamic frame that was 1–2 time frames (∼4–7 seconds) before the enhancement peak were delineated. We participated in an NCI QIN challenge project of AIF delineation using our approach and our software (24). The parameters quantified from our AIF were well correlated with others (24).
T1 Calculation.
T1 maps were derived by fitting
| (3) |
where α is a flip angle, TR is repetition time, and S0 is margination amplitude, to the 4-flip angle T1-weighted images using Simplex algorithm.
Using equations (1) and (2), AIF, T10 and the 3-parameter Tofts model, Vp maps were calculated. Then, hematocrit of 0.45 was used to convert Vp to CBV as CBV = [Vp/(1 − 0.45) × 100 (ml/100 g)], where blood density (1 mL/g) was used.
HCV Delineation
HCV was determined on DW images with b = 3000 s/mm2. A threshold was used from the normal tissue volume of interest (VOI) that was most contralateral to GBM. To obtain the normal brain VOI, an automated process was used to first extract the brain surface and find the middle line near the central fissure of the brain on T2-weighted images (b = 0). Then, the FLAIR abnormality volume was mirrored to the opposite side of the brain surface through the middle line. To remove CSF influence on the signals from the normal VOI, we remove all voxels with strong CSF signals by classifying CSF on T2-weighted images (b = 0) using fuzzy c-means. The VOI was eroded at least 5 mm from GTV∧FLAIR, and had ∼600 pixels per slice. Then, voxels within the GTV∧FLAIR on each slice were thresholded using mean + 2SD of the intensities in the VOI on the slice to account for DW intensity variations across slices. All these processes are fully automated. If a visual inspection of the normal brain VOI indicated the VOI inadequate, HCV could be recreated after adjusting the normal brain VOI by physician coauthors.
hCBV Delineation
hCBV was delineated on the CBV images with a threshold that was established in a previous study (12). Because normal white matter (WM) and gray matter (GM) have intrinsically different CBV values, the threshold value obtained from an uninvolved contralateral volume would vary depending on the ratio of GM to WM in that volume. We therefore segmented uninvolved contralateral GM in the frontal lobe (which has a higher CBV than uninvolved WM) and defined the hCBV tumor volume as the volume of tumor with CBV >1 SD above GM. This definition resulted in hCBV tumor volumes that predicted PFS and OS (12). Therefore, we used this definition in this pilot study of the clinical trial. This threshold was applied to GTV_Gd with 0–3 mm extension on CBV maps.
Volume Review
HCV and hCBV volumes were reviewed by the physician, neuroradiologist, and MRI physicist. Volumes were slightly edited during central review to remove components outside of the brain parenchyma for both HCV and hCBV, or overlap with blood vessels rather than parenchymal tumor volume for hCBV. Finalized tumor volumes were then imported from imFIAT back into the treatment planning system as binary image volumes associated with the HCV and hCBV image set. Images were automatically registered in the Eclipse Image Registration workspace to the original T1-post-Gd scan and checked by the clinical physicist. The clinical physicist then copied the HCV and hCBV volumes to the CT data set in the treatment planning system.
Image and Volume Registration and Delineation
The physician reviewed the HCV and hCBV volumes in the treatment planning system. CTV and PTV structures were then created as follows: CTV_Low was defined as a 1.7 cm expansion from GTV_Low delimited by normal anatomic boundaries. PTV_Low was defined 0.3 cm (0.2 cm positioning uncertainty with daily CBCT plus 0.1 cm MRI to CT registration uncertainty) as per institutional standard. For the advanced MRI (HCV and hCBV) boost target volumes, no CTV margin was used. PTV_High was defined as a 0.5 cm expansion from the HCV/hCBV volumes (0.2 cm positioning uncertainty + 0.2 cm RESOLVE DWI uncertainty + 0.1 cm MR to CT registration uncertainty).
PTV_Low was prescribed 60 Gy in 30 fractions and PTV_High was prescribed 75 Gy in 30 fractions using a simultaneous integrated boost technique. Volumetric modulated arc therapy using the Eclipse treatment planning system was used in all cases. The goal was to cover 100% of the target volumes with 95% of the prescribed dose, while maintaining conventional dose limits as utilized on cooperative group trials for high-grade glioma. This included maintaining optic chiasm and optic nerves ≤54 Gy, brainstem surface (ventral 3 mm of brainstem) ≤64 Gy, and brainstem core ≤55 Gy.
Results
Patient Characteristics and Imaging Subvolumes
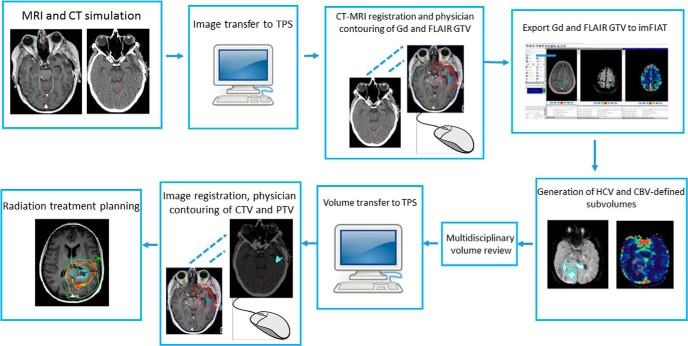
The initial 12 patients enrolled between September 2016 and June 2017 were included in this analysis. Baseline characteristics of patients are described in Table 1. All patients had IDH1 wild-type tumors by immunohistochemistry. Fifty percent of patients underwent gross total resection, 33% underwent subtotal resection, and the remainder underwent biopsy alone. The workflow for image acquisition, volume delineation and data transfer, and treatment planning is depicted in Figure 1. All patients initiated radiation within 6 weeks of surgery and within 2 weeks of simulation. Advanced volume processing was generally done within 24–36 hours of simulation.
Table 1.
Baseline Patient Characteristics
| Clinical Characteristic | No (%) |
|---|---|
| Median age (range) | 65–(51-77) |
| Male | 8 (67%) |
| Extent of resection | |
| Gross total resection | 6 (50%) |
| Subtotal resection | 4 (33%) |
| Biopsy | 2 (17%) |
| MGMT methylation status | |
| Positive | 3 (25%) |
| Negative | 9 (75%) |
| Tumor location | |
| Frontal lobe | 4 (33%) |
| Temporal lobe | 5 (42%) |
| Parietal lobe | 2 (17%) |
| Occipital lobe | 1 (8%) |
Figure 1.
Integrated workflow diagram for the implementation of an advanced dynamic contrast-enhanced (DCE)- and high b-value magnetic resonance (MR) imaging signature to guide dose-intensified radiotherapy. TPS = treatment planning system; Gd = T1-Gd-enhanced MRI; GTV = gross tumor volume; CTV = clinical target volume; PTV = planning target volume.
Characteristics and distributions of CBV in normal frontal GM, normal WM, and the hCBV tumor volumes are shown in Figure 2. Note that the CBV value + 2SD in the frontal WM was below the mean value in the normal frontal GM, and thus, it cannot be used to define the elevated CBV in the tumor volume.
Figure 2.
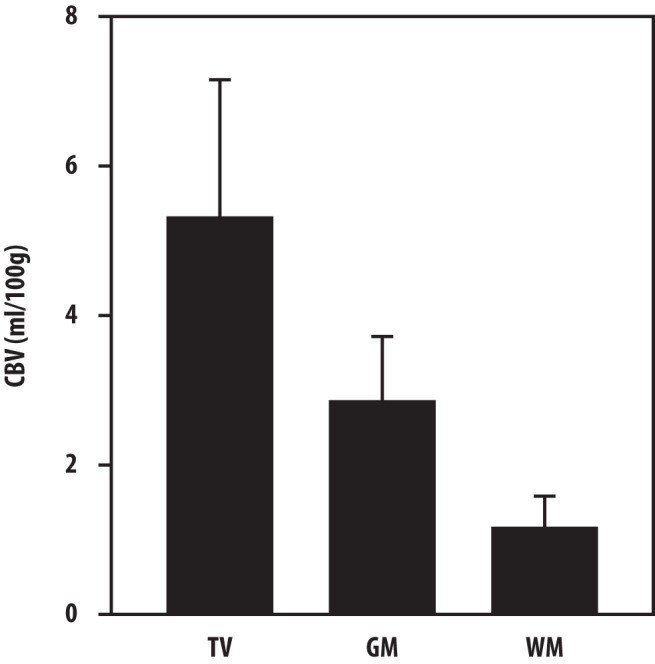
The averaged CBV values in the hCBV tumor volumes, normal frontal white matter (WM) and normal frontal gray matter (GM) in the 12 study patients. The error bars depict the averaged standard deviations of cerebral blood volume (CBV) in the 3 volumes of interest across the 12 patients. Note that the mean CBV value + 2SDs in the frontal WM is smaller than the mean value in the frontal GM, and thus, it cannot be used as a threshold value to define the elevated CBV in the tumor volume.
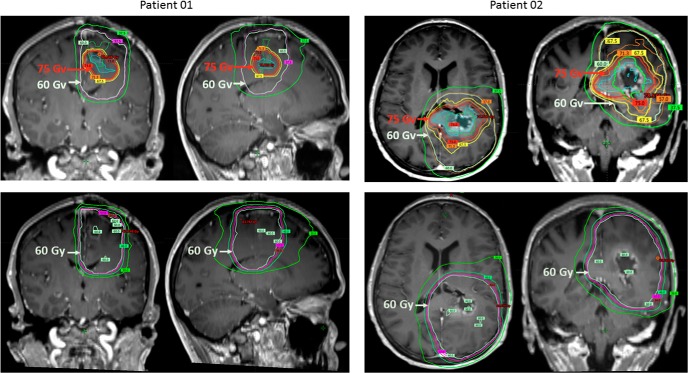
Characteristics of conventional and advanced MRI target volumes are listed in Table 2. On average, the Gd-enhanced volume was >3 times larger than those of either HCV or hCBV, and 1.8 times larger than the union (combination) of HCV and hCBV. HCV and hCBV identified largely distinct volumes with only ∼1 cc of overlap between the 2 (range, 0.002–6.8 cc). The enhancing component of the union of HCV and hCBV was only 57% (range, 0.3–0.9) of the volume, with an average of 4.1 cc (range, 1.4–7.9) extending outside of the enhancing region. Only 2 patients showed minimal extension of the union HCV and hCBV volume beyond FLAIR (0.06 cc and 0.04 cc, respectively), and FLAIR volumes were ∼10 times larger than the union of HCV and hCBV. An example of HCV and hCBV volumes that are largely nonoverlapping overlaid on the corresponding T1-Gd MRI is shown in Figure 3. Two examples of representative radiation plans for 2 different patients are shown in Figure 4. For comparison, example plans without the advanced MRI boost are also depicted. As showed, the advanced MRI boost volume was generally a smaller tumor subregion contained within the conventional target volume.
Table 2.
Volume and Overlap of Conventional and Advanced Imaging Subvolumes
| Target | Mean Volume (cc) | Range |
|---|---|---|
| GTV^Gd | 23.9 | 3.9–49.9 |
| GTV^FLAIR | 128.9 | 39.2–248.5 |
| GTV^HCV | 7.5 | 1.7–20.4 |
| GTV^hCBV | 6.6 | 0.5–18.2 |
| Union of HCV and hCBV | 13.1 | 2.3–31.8 |
| Overlap of HCV and hCBV | 0.9 | 0.002–6.8 |
| Overlap Gd and HCV | 5.3 | 0.4–17.4 |
| Overlap of Gd and hCBV | 4.5 | 0.3–15.0 |
| Overlap Gd and Union | 8.9 | 0.9–26.0 |
| Overlap FLAIR and HCV | 7.5 | 1.7–20.4 |
| Overlap FLAIR and hCBV | 6.5 | 0.5–18.2 |
| Overlap FLAIR and Union | 13.1 | 2.3–31.7 |
| HCV outside of Gd | 2.2 | 0.8–3.6 |
| hCBV outside of Gd | 2.0 | 0.0–6.1 |
| Union outside of Gd | 4.1 | 1.4–7.9 |
GTV˄Gd = Gadolinium enhanced target volume; GTV^FLAIR = FLAIR target volume; GTV˄HCV = Hypercellular high b-value DW-MRI target volume; GTV^hCBV = Hyperperfused DCE-MRI target volume.
Figure 3.
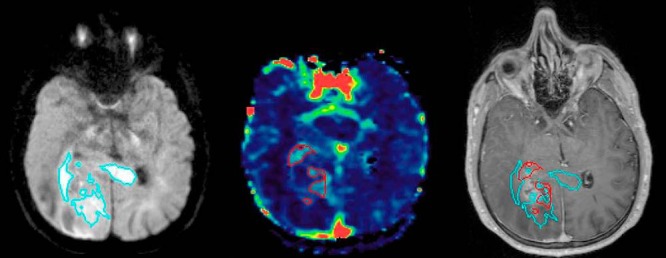
An example of a patient with largely nonoverlapping hypercellular tumor regions (TVHCV) identified by high b-value diffusion-weighted (DW)-magnetic resonance imaging (MRI) (cyan, left panel) and hyperperfused tumor regions (TVHCBV) identified by DCE-MRI (red, middle panel). Significant extension of TVHCV is showed beyond the T1 Gd-enhanced region (overlay on T1 Gd-enhanced image, right panel).
Figure 4.
Representative images from radiation plans from 2 different patients. The top row depicts images of radiation plans using advanced MRI to boost tumor subregions to 75 Gy. High-risk tumor targets are identified by advanced MRI (cyan) beyond the abnormal regions seen on T1 Gd-enhanced conventional MRI (green). The conformal 75-Gy isodose line targeting the advanced imaging tumor volume is depicted in red, and the larger 60-Gy isodose line targeting the anatomic T1-Gd-enhanced region with standard clinical margins is depicted in gray-white. The bottom row depicts images of comparison standard radiation plans for the same patients based on anatomic T1-Gd-enhanced MRI with standard clinical margins prescribed to 60 Gy. As showed, advanced MRI-identified boost regions prescribed to 75 Gy were often contained within standard anatomic MRI regions prescribed to 60 Gy.
Discussion
While the limitations of anatomic MRI for radiation therapy have been reinforced by multiple studies showing that tumor identified by advanced MRI techniques extending outside of conventionally defined volumes predicts patient prognosis independent of T1-Gd, T2-FLAIR, and other clinical factors, advanced imaging techniques have not been incorporated into routine radiation planning (10, 25). In this initial report of a prospective, single-arm phase II trial for patients with newly diagnosed GBM from a single institution, we describe the end-to-end process of delivery of dose-intensified RT to predicted, high-risk tumor subregions identified by multiparametric MRI. The advanced hypercellular and hyperperfusion tumor subvolumes were significantly smaller than the conventionally defined T1-Gd and T2/FLAIR abnormalities standardly targeted for radiation treatment planning, and identified distinct regions that were frequently nonenhancing and therefore excluded from standard radiation boost volume definition. Using physician-defined volumes on conventional T1-Gd and T2-FLAIR images, the semiautomated creation of advanced MR boost volumes was accomplished for real-time planning, yielding successful delivery of advanced imaging-defined dose-intensified RT in all patients beginning within 2 weeks of simulation.
Given the known limitations of conventional MRI for defining tumor extent and predicting outcome in patients with GBM, the use of advanced imaging including perfusion and diffusion-weighted MRI has been studied for more than a decade to assess physiologic phenotypes of prognostic significance in this disease. Dynamic susceptibility contrast (DSC) and DCE-MRI permit quantitative estimation of parameters reflective of tumor neovascularization that has been associated with tumor growth in GBM, including CBV, cerebral blood flow, and Ktrans (23, 26). Maximum CBV and pathologically verified tumor vascularity are correlated, and elevated mean relative CBV (rCBV) >1.75 in patients with low- and high-grade gliomas is associated with shorter time to progression (2). Additionally, these perfusion parameters are predictive of overall survival in patients with malignant gliomas (10, 11).
Given the well-known limitation in geometric accuracy of DSC images, we selected T1-weighted DCE images to estimate CBV for the purpose of radiation boost target definition. Several previous studies as well as ours have directly compared CBV values estimated by DSC and DCE images (27–31). An early study reported good correlation of median values in tumors between 2 CBV estimates (r = 0.67) and excellent pixel-by-pixel correlation between the 2 estimates in normal brain tissues (r = 0.96) in 9 patients with intraaxial cerebral tumors (27). Another study of 32 patients with high-grade glioma showed a weak-but-significant correlation between the 2 estimates in the enhancing tumor volumes in a pixel-by-pixel comparison (28). Another study including 17 healthy subjects and 9 patients with glioblastoma reported excellent correlations of the 2 CBV estimates in normal GM and WM (r = 0.9 and r = 0.89, respectively) and a good and significant correlation in the tumor (r = 0.67) (29). A recent study examined the diagnostic accuracy of glioma grades in 26 patients using the median tumor values of the 2 CBV estimates, which achieved a similar diagnostic accuracy (30). We have performed similar analysis in 20 patients with brain metastases, in which both DCE and DSC images were acquired in a single session with 2 contrast injections. We found good correlation between the 2 CBV estimates in both normal brain tissue and brain tumor volumes (r = 0.66–0.71) (unpublished data). These similarities and discrepancies depend upon several factors. The 2 imaging methods rely on considerably different contrast mechanisms and model theories, which can be affected by different physical and physiological conditions. Also, different acquisition parameters and modeling implementations, for example, correcting T1 and vascular leakage effects in DSC analysis, can affect the results.
Additional physiologic properties of malignant gliomas may be assessed with DW-MRI, which has been used to assess the mobility of water molecules in the tissue microenvironment as a surrogate for tumor cellularity, and is a method for therapeutic response assessment as first shown in patients with glioma (6, 7, 13). An inverse correlation is observed between ADC and brain tumor cellularity in preclinical studies (13). However, known limitations of this approach for isolating tumor cellularity from normal brain tissue, edema, and micronecrosis in the heterogeneous GBM microenvironment may lead to unpredictably elevated ADC compared with normal tissue. This limitation may be mitigated through the use of high b-value DW-MRI (3000–4000 s/mm2) versus 0 and 800–1000 s/mm2 to attenuate signals due to edema (14, 32–34). We investigated the prognostic value of this approach, showing that the hypercellular tumor region (HCV) identified before RT using high b-value DW-MRI correlates with worse PFS in patients with newly diagnosed GBM treated with standard chemoradiation (14). We determined that in contrast to DCE-MRI alone, the combined use of high b-value MRI with DCE-MRI identifies largely spatially distinct regions with mean overall of only 21% (12). Moreover, the combination of these modalities correlated with patterns of failure and progression, and therefore, these are rationally targeted for intensified radiation treatment (12).
A limited number of studies are prospectively evaluating the incorporation of advanced imaging for the radiation treatment of patients with GBM. These include ongoing studies using proton MR spectroscopic imaging and amino acid positron emission tomography to guide radiation treatment in patients with GBM. Proton MR spectroscopic imaging detects chemical compounds reflective of cellular turnover and proliferation and correlates with histologic tumor cell density and survival in patients with GBM (35–38), although its use for radiation treatment has been limited to select centers with imaging expertise. Amino acid positron emission tomography including 11C-Methionine and 18F-radiolabeled 3,4-dihydroxy-6-[18F]fluoro-L-phenylalanine [18F]F-DOPA tracers is also under evaluation for targeting of potentially aggressive tumor regions beyond conventional MRI in ongoing trials in the United States and Europe. Studies have shown significant correlation in the standard uptake values of 11C-MET and 18F-FDOPA with nearly identical patterns of spatial uptake (39); both have been shown to be prognostic for survival and recurrence and potentially complementary to MRI, although not widely adopted and limited to research centers with expertise in complex radiotracer synthesis or on-site cyclotrons (40–42).
Our study represents the first report of the prospective implementation of a multiparametric imaging signature that is integrated in the RT workflow to guide intensified RT against distinct, poor prognosis phenotypes in patients with GBM. Initial implementation of the real-time use of a multiparametric MR signature for radiation planning involved QA and commissioning of DW- and DCE-MRI for clinical usage before clinical implementation. Implementation of an advanced MR biomarker for radiation treatment requires close coordination between the radiation oncologist, imaging physics, and clinical physics teams to delineate tumor volumes, process and transfer data between treatment planning and advanced imaging software, and ensure timely initiation of treatment. Limitations of this approach include the phenotypic and biologic diversity of GBM, and whether a multiparametric signature is sufficient to characterize this heterogeneity and guide treatment in this disease. To address this, we are acquiring and correlating other physiological imaging modalities with advanced MRI, as well as acquiring longitudinal imaging to evaluate whether temporal changes in advanced imaging features may be used to predict outcome and further tailor therapy.
In this report, we show the feasibility of the real-time use of a multiparametric MR signature to guide radiation treatment against prognostic, unique tumor subregions that substantially differ from the T1-Gd-enhancing high-risk boost volumes standardly defined by conventional MRI. Survival outcomes are awaited from this study, and future directions include translation of this workflow to a second site to validate the generalizability of this novel radiotherapeutic approach for patients with GBM.
Acknowledgments
This study was funded in part by NIH/NCI grant U01CA183848.
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- DCE
- Dynamic contrast-enhanced
- DW
- diffusion-weighted
- MRI
- magnetic resonance imaging
- GBM
- glioblastoma
- T1-Gd
- T1 gadolinium
- T2-FLAIR
- T2-weighted fluid attenuated inversion recovery
- MRI
- magnetic resonance imaging
- rCBV
- relative cerebral blood volume
- ADC
- apparent diffusion coefficient
- CT
- computed tomography
- imFIAT
- in-house functional image analysis tool
- AIF
- arterial input function
- 2D
- 2-dimensional
- TE
- echo time
- TR
- repetition time
- HCV
- hypercellular volume
- VOI
- volume of interest
- hCBV
- high CBV
- WM
- white matter
- GM
- gray matter
- DSC
- dynamic susceptibility contrast
References
- 1. Cao Y, Tsien CI, Nagesh V, Junck L, Ten Haken R, Ross BD, Chenevert TL, Lawrence TS. Survival prediction in high-grade gliomas by MRI perfusion before and during early stage of RT [corrected]. Int J Radiat Oncol Biol Phys. 2006;64:876–885. [DOI] [PubMed] [Google Scholar]
- 2. Law M, Young RJ, Babb JS, Peccerelli N, Chheang S, Gruber ML, Miller DC, Golfinos JG, Zagzag D, Johnson G. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology. 2008;247:490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X, Jin H, Lu Y, OH J, Chang S, Nelson SJ. Identification of MRI and 1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR Biomed. 2004;17:10–20. [DOI] [PubMed] [Google Scholar]
- 4. Li Y, Lupo JM, Parvataneni R, Lamborn KR, Cha S, Chang SM, Nelson SJ. Survival analysis in patients with newly diagnosed glioblastoma using pre- and postradiotherapy MR spectroscopic imaging. Neuro Oncol. 2013;15:607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirai T, Murakami R, Nakamura H, Kitajima M, Fukuoka H, Sasao A, Akter M, Hayashida Y, Toya R, Oya N, Awai K, Iyama K, Kuratsu JI, Yamashita Y. Prognostic value of perfusion MR imaging of high-grade astrocytomas: long-term follow-up study. AJNR Am J Neuroradiol. 2008;29:1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galban CJ, Hoff BA, Chenevert TL, Ross BD. Diffusion MRI in early cancer therapeutic response assessment. NMR Biomed. 2017;30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chenevert TL, Stegman LD, Taylor JM, Robertson PL, Greenberg HS, Rehemtulla A, Ross BD. Diffusion magnetic resonance imaging: an early surrogate marker of therapeutic efficacy in brain tumors. J Natl Cancer Inst. 2000;92:2029–2036. [DOI] [PubMed] [Google Scholar]
- 8. Moffat BA, Chenevert TL, Lawrence TS, Meyer CR, Johnson TD, Dong Q, Tsien C, Mukherji S, Quint DJ, Gebarski SS, Robertson PL, Junck LR, Rehemtulla A, Ross BD. Functional diffusion map: a noninvasive MRI biomarker for early stratification of clinical brain tumor response. Proc Natl Acad Sci U S A. 2005;102:5524–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galbán CJ, Chenevert TL, Meyer CR, Tsien C, Lawrence TS, Hamstra DA, Junck L, Sundgren PC, Johnson TD, Ross DJ, Rehemtulla A, Ross BD. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat Med. 2009;15:572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao Y, Tseng CL, Balter JM, Teng F, Parmar HA, Sahgal A. MR-guided radiation therapy: transformative technology and its role in the central nervous system. Neuro Oncol. 2017;19(Suppl_2):ii16–ii29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao Y, Nagesh V, Hamstra D, Tsien CI, Ross BD, Chenevert TL, Junck L, Lawrence TS. The extent and severity of vascular leakage as evidence of tumor aggressiveness in high-grade gliomas. Cancer Res. 2006;66:8912–8917. [DOI] [PubMed] [Google Scholar]
- 12. Wahl DR, Kim MM, Aryal MP, Hartman H, Lawrence TS, Schipper MJ, et al. Combining perfusion and high B-value diffusion MRI to inform prognosis and predict failure patterns in glioblastoma. Int J Radiat Oncol Biol Phys. 2018;102:757–764. [DOI] [PubMed] [Google Scholar]
- 13. Sugahara T, Korogi Y, Kochi M, Ikushima I, Shigematu Y, Hirai T, Okuda T, Liang L, Ge Y, Komohara Y, Ushio Y, Takahashi M. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging. 1999;9:53–60. [DOI] [PubMed] [Google Scholar]
- 14. Pramanik PP, Parmar HA, Mammoser AG, Junck LR, Kim MM, Tsien CI, et al. Hypercellularity components of glioblastoma identified by high b-value diffusion-weighted imaging. Int J Radiat Oncol Biol Phys. 2015. 15;92:811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsien CI, Brown D, Normolle D, Schipper M, Piert M, Junck L, Heth J, Gomez-Hassan D, Ten Haken RK, Chenevert T, Cao Y, Lawrence T. Concurrent temozolomide and dose-escalated intensity-modulated radiation therapy in newly diagnosed glioblastoma. Clin Cancer Res. 2012;18:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phantom test guidance for the ACR MRI accreditation program. American College of Radiology; Reston, VA: 1998. [Google Scholar]
- 17. Bane O, Hectors SJ, Wagner M, Arlinghaus LL, Aryal MP, Cao Y, Chenevert TL, Fennessy F, Huang W, Hylton NM, Kalpathy-Cramer J, Keenan KE, Malyarenko D, Mulkern RV, Newitt DC, Russek SE, Stupic KF, Tudorica A, Wilmes LJ, Yankeelov TE, Yen YF, Boss MA, Taouli B. Accuracy, repeatability, and interplatform reproducibility of T1 quantification methods used for DCE-MRI: Results from a multicenter phantom study. Magn Reson Med. 2018;79:2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao Y. WE-D-T-6C-03: Development of image software tools for radiation therapy assessment. Med Phys. 2005;32:2136–2136. [Google Scholar]
- 19. Cao Y, Li D, Shen Z, Normolle D. Sensitivity of quantitative metrics derived from DCE MRI and a pharmacokinetic model to image quality and acquisition parameters. Acad Radiol. 2010;17:468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, Aryal MP, LaViolette PS, Oborski MJ, O'Sullivan F, Abramson RG, Jafari-Khouzani K, Afzal A, Tudorica A, Moloney B, Gupta SN, Besa C, Kalpathy-Cramer J, Mountz JM, Laymon CM, Muzi M, Schmainda K, Cao Y, Chenevert TL, Taouli B, Yankeelov TE, Fennessy F, Li X. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge. Tomography. 2016;2:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farahani K, Kalpathy-Cramer J, Chenevert TL, Rubin DL, Sunderland JJ, Nordstrom RJ, Buatti J, Hylton N. Computational challenges and collaborative projects in the NCI Quantitative Imaging Network. Tomography. 2016;2:242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Porter DA, Heidemann RM. High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magn Reson Med. 2009;62:468–475. [DOI] [PubMed] [Google Scholar]
- 23. Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, Port RE, Taylor J, Weisskoff RM. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. [DOI] [PubMed] [Google Scholar]
- 24. Huang W, Chen Y, Fedorov A, Li X, Jajamovich GH, Malyarenko DI, Aryal MP, LaViolette PS, Oborski MJ, O'Sullivan F, Abramson RG, Jafari-Khouzani K, Afzal A, Tudorica A, Moloney B, Gupta SN, Besa C, Kalpathy-Cramer J, Mountz JM, Laymon CM, Muzi M, Schmainda K, Cao Y, Chenevert TL, Taouli B, Yankeelov TE, Fennessy F, Li X. The impact of arterial input function determination variations on prostate dynamic contrast-enhanced magnetic resonance imaging pharmacokinetic modeling: a multicenter data analysis challenge. Tomography. 2016;2:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao Y, Sundgren PC, Tsien CI, Chenevert TT, Junck L. Physiologic and metabolic magnetic resonance imaging in gliomas. J Clin Oncol. 2006;24:1228–1235. [DOI] [PubMed] [Google Scholar]
- 26. Rosen BR, Belliveau JW, Aronen HJ, Kennedy D, Buchbinder BR, Fischman A, Gruber M, Glas J, Weisskoff RM, Cohen MS, et al. Susceptibility contrast imaging of cerebral blood volume: human experience. Magn Reson Med. 1991;22:293–299; discussion 300–303. [DOI] [PubMed] [Google Scholar]
- 27. Haroon H, Patankar T, Zhu X, Li K, Thacker N, Scott M, Jackson A. Comparison of cerebral blood volume maps generated from T2* and T1 weighted MRI data in intra-axial cerebral tumours. Br J Radiol. 2014;80:161–168. [DOI] [PubMed] [Google Scholar]
- 28. Alcaide-Leon P, Pareto D, Martinez-Saez E, Auger C, Bharatha A, Rovira A. Pixel-by-pixel comparison of volume transfer constant and estimates of cerebral blood volume from dynamic contrast-enhanced and dynamic susceptibility contrast-enhanced MR imaging in high-grade gliomas. AJNR Am J Neuroradiol. 2015;36:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Artzi M, Liberman G, Nadav G, Vitinshtein F, Blumenthal DT, Bokstein F, Aizenstein O, Ben Bashat D. Human cerebral blood volume measurements using dynamic contrast enhancement in comparison to dynamic susceptibility contrast MRI. Neuroradiology. 2015;57:671–678. [DOI] [PubMed] [Google Scholar]
- 30. Santarosa C, Castellano A, Conte GM, Cadioli M, Iadanza A, Terreni MR, Franzin A, Bello L, Caulo M, Falini A, Anzalone N. Dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging for glioma grading: preliminary comparison of vessel compartment and permeability parameters using hotspot and histogram analysis. Eur J Radiol. 2016;85:1147–1156. [DOI] [PubMed] [Google Scholar]
- 31. Bazyar S, Ramalho J, Eldeniz C, An H, Lee YZ. Comparison of cerebral blood volume and plasma volume in untreated intracranial tumors. PloS One. 2016;11:e0161807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mardor Y, Roth Y, Ochershvilli A, Spiegelmann R, Tichler T, Daniels D, Maier SE, Nissim O, Ram Z, Baram J, Orenstein A, Pfeffer R. Pretreatment prediction of brain tumors' response to radiation therapy using high b-value diffusion-weighted MRI. Neoplasia. 2004;6:136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mardor Y, Pfeffer R, Spiegelmann R, Roth Y, Maier SE, Nissim O, Berger R, Glicksman A, Baram J, Orenstein A, Cohen JS, Tichler T. Early detection of response to radiation therapy in patients with brain malignancies using conventional and high b-value diffusion-weighted magnetic resonance imaging. J Clin Oncol. 2003;21:1094–1100. [DOI] [PubMed] [Google Scholar]
- 34. Chu HH, Choi SH, Ryoo I, Kim SC, Yeom JA, Shin H, Jung SC, Lee AL, Yoon TJ, Kim TM, Lee SH, Park CK, Kim JH, Sohn CH, Park SH, Kim IH. Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology. 2013;269:831–840. [DOI] [PubMed] [Google Scholar]
- 35. McKnight TR, von dem Bussche MH, Vigneron DB, Lu Y, Berger MS, McDermott MW, Dillon WP, Graves EE, Pirzkall A, Nelson SJ. Histopathological validation of a three-dimensional magnetic resonance spectroscopy index as a predictor of tumor presence. J Neurosurg. 2002;97:794–802. [DOI] [PubMed] [Google Scholar]
- 36. Vigneron D, Bollen A, McDermott M, Wald L, Day M, Moyher-Noworolski S, Henry R, Chang S, Berger M, Dillon W, Nelson S. Three-dimensional magnetic resonance spectroscopic imaging of histologically confirmed brain tumors. Magn Reson Imaging. 2001;19:89–101. [DOI] [PubMed] [Google Scholar]
- 37. Dowling C, Bollen AW, Noworolski SM, McDermott MW, Barbaro NM, Day MR, Henry RG, Chang SM, Dillon WP, Nelson SJ, Vigneron DB. Preoperative proton MR spectroscopic imaging of brain tumors: correlation with histopathologic analysis of resection specimens. AJNR Am J Neuroradiol. 2001;22:604–612. [PMC free article] [PubMed] [Google Scholar]
- 38. Croteau D, Scarpace L, Hearshen D, Gutierrez J, Fisher JL, Rock JP, Mikkelsen T. Correlation between magnetic resonance spectroscopy imaging and image-guided biopsies: semiquantitative and qualitative histopathological analyses of patients with untreated glioma. Neurosurgery. 2001;49:823–829. [DOI] [PubMed] [Google Scholar]
- 39. Becherer A, Karanikas G, Szabo M, Zettinig G, Asenbaum S, Marosi C, Henk C, Wunderbaldinger P, Czech T, Wadsak W, Kletter K. Brain tumour imaging with PET: a comparison between [18F]fluorodopa and [11C]methionine. Eur J Nucl Med Mol Imaging. 2003;30:1561–1567. [DOI] [PubMed] [Google Scholar]
- 40. Bell C, Dowson N, Puttick S, Gal Y, Thomas P, Fay M, Smith J, Rose S. Increasing feasibility and utility of (18)F-FDOPA PET for the management of glioma. Nucl Med Biol. 2015;42:788–795. [DOI] [PubMed] [Google Scholar]
- 41. Lee IH, Piert M, Gomez-Hassan D, Junck L, Rogers L, Hayman J, Ten Haken RK, Lawrence TS, Cao Y, Tsien C. Association of 11C-methionine PET uptake with site of failure after concurrent temozolomide and radiation for primary glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2009;73:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Albert NL, Weller M, Suchorska B, Galldiks N, Soffietti R, Kim MM, et al. Response assessment in neuro-oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18:1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]