Abstract
Actinobacteria able to produce varieties of bioactive natural products have been long appreciated by the field of drug discovery and development. Recently, a few of CRISPR/Cas9 systems bearing different types of replicons (pSG5 and pIJ101) were developed to efficiently edit their genomes. Despite wide application in gene editing, their utility in editing challenging DNA regions e.g. high sequence identity has not been compared. In this study, we confirmed that the widely used temperature-sensitive pSG5 replicon is indeed not suitable for editing modular polyketide synthase (PKS) genes due to causing unpredicted gene recombination. This problem can be addressed by replacing the pSG5 with the segregationally unstable pIJ101 replicon. By introducing a counter-selection marker CodA, convenient cloning sites in the single guide RNAs (sgRNAs) and homologous template scaffolds, we developed a new CRISPR-Cas9 system pMWCas9. This system was successfully used to delete/replace erythromycin PKS and other biosynthetic genes in Saccharopolyspora erythraea and Streptomyces sp. AL2110. By swapping the promoters of antB and antC with ermE and kasOp, we achieved a deacyl-antimycin hyper producer which produces a 9-fold higher yield than the original Streptomyces sp. AL2110 strain. Our results provide a robust and useful Cas9 tool for genetic studies in Actinobacteria.
Keywords: Biosynthesis, Polyketide synthase, CRISPR/Cas9, Antimycin, Actinobacteria
1. Introduction
Natural products (NPs) are critical for drug discovery and development [1]. Over the past decades, significant efforts have been devoted to understanding the logic of NP biosynthesis [2]. With this basis, biosynthetic pathways engineering or genome-based NP discovery have been successfully used to identify numerous novel bioactive compounds [3]. However, it was revealed that the majority of microbial metabolites repertoire indeed still have not yet been accessed. More than 90% of biosynthetic pathways are silenced under normal laboratory culturing conditions due to the absence of the essential regulatory signals to trigger biosynthesis [4]. To activate their biosynthesis, gene modification targeting different regulation stages is usually required [5,6].
Actinomycetes are among of the most prolific producers of diverse NPs, which contribute about 73% of total bacterial NPs and 45% of total microbial NPs [7]. The diverse genomic contents and relatively low growth rate make genetic manipulation of their genome much more changeling than other model organisms, such as Escherichia coli. The typical approach for genome editing is based on RecA mediated double-crossover [8]. However, the efficiency of inducing the second-crossover by this approach is low, which often takes weeks to accumulate a significant number of double-crossover mutants for PCR verification [8]. Although the introduction of a double strand break (DSB) at the genomic locus of interest can drastically increase the efficacy of double-crossover, previous approaches, such as using the meganuclease I-SceI, require prior integration of the endonuclease recognition site at the target locus [9]. The overall efficacy of this method is, thus, still very low.
Recently, the more efficient DSB mediated genome editing has been achieved by the type II clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) system [[10], [11], [12], [13]]. By co-expression with the customized single guide RNAs (sgRNAs), the Streptococcus pyogenes Cas9 nuclease can mediate a DSB in the sgRNA binding region; efficient double crossover can be achieved when the homologous templates are provided in the same plasmid. Currently, a few CRISPR systems have been developed and applied in editing actinomycetal genomes, among of which pCRISPR-Cas9, pCRISPomyces, pKCcas9dO and pWHU2653 are the four most widely used systems [[10], [11], [12], [13]]. These systems are mainly different in the replicon type as well as the way of expressing Cas9 nuclease. The pCRISPR-Cas9, pCRISPomyces and pKCcas9dO have a temperature sensitive pSG5 replicon, while pWHU2653 bears a segregationally unstable pIJ101 replicon; Cas9 protein in pCRISPR-Cas9 and pKCcas9dO is induced by thiostrepton (Tsr) while the nucleases in other two systems are constitutively expressed. Although all these systems have been successfully applied in genome editing, their efficiency in editing challenging DNA regions, e.g. with high sequence identity, has been not comparatively evaluated. Recently, the result shows that pKC1139 derived (with pSG5) plasmids can cause random recombination of the rapamycin and tylosin modular PKS genes when they were used to gene replacement [14]. It was assumed the pSG5 replicon might be the major reason for causing recombination. As three of the four widely used systems pCRISPR-Cas9, pCRISPomyces and pKCcas9dO are pSG5-based, thus evaluation their utility in editing modular PKS genes is necessary.
In this study, we confirmed that the pSG5 replicon is indeed the major reason for causing non-desired gene recombination, and the Cas9 system with pSG5 is not suitable to edit the modular PKS gene. Substitution the replicon by pIJ101 can overcome this problem. By integration of inducible expression of Cas9, convenient cloning sites in the sgRNAs region and homologous templates and a counter-selection marker codA, we further upgraded this Cas9 system. This new system was successfully used to efficiently edit the modular PKS and other genes in erythromycin and antimycin biosynthetic pathways. By engineering the promotor region in Streptomyces sp. AL2110, deacyl antimycins was dramatically improved to a 9-fold compared with the wild-type strain.
2. Material and methods
2.1. Strains, chemicals, and general materials
Strains and plasmids used in this study are listed in Tables S1 and S2. Escherichia coli DH5α was used as cloning host. E. coli ET12567/pUZ8002 was used for intergeneric conjugation between E. coli and Sac. erythraea HL3168 E3 or S. sp. AL2110. Sac. erythraea HL3168 E3 was grown at 30 °C on corn steep liquor agar medium (10 g corn steep liquor, 10 g soluble starch, 3 g NaCl, 3 g (NH4)2SO4, 5 g CaCO3, 3 g yeast extract, 3 g tryptone, 20 g agar per liter, pH = 7.0) and S. sp. AL2110 on MS agar medium [8] (20 g soy flour, 20 g D-mannitol, 20 g agar per liter) for sporulation and at the same temperature. Both Sac. erythraea HL3168 E3 and S. sp. AL2110 were grown in tryptone soya broth (TSB) liquid medium [8] (30 g tryptone soya broth powder per liter) for the growth of mycelium and isolation of total DNA. Corn steep liquor or MS agar medium with the addition of 10 mM MgCl2 was used for intergeneric conjugation between E. coli and Sac. erythraea or Streptomyces, respectively. E. coli strains were maintained in LB medium (10 g tryptone, 5 g yeast extract, 10 g sodium chloride per liter) at 37 °C with the appropriate antibiotic selection (50 μg mL−1 apramycin, 50 μg mL−1 chloramphenicol, and 50 μg mL−1 kanamycin). For Sac. erythraea, trimethoprim (50 μg mL−1) and apramycin (50 μg mL−1) were used in corn steep liquor agar medium or TSB while nalidixic acid (30 μg mL−1) and apramycin (50 μg mL−1) were used in MS agar medium or TSB for Streptomyces. Soy bean oil medium (30 g soluble starch, 30 g soy flour, 40 g dextrin, 10 mL soybean oil, (NH4)2SO4 2 g, CaCO3 6 g per liter, pH = 7.0–7.2) was used for erythromycin production in Sac. erythraea HL3168 E3. M3-S (10 g glucose, 50 g soluble starch, 6 g (NH4)2SO4, 1.5 g CaCO3 per liter, pH = 7.0) was used for deacyl antimycins production in S. sp. AL2110.
Erythromycin and antimycin were purchased from Energy Chemical and Sigma Co. Ltd., respectively. Chromatography reagent ammonium acetate was purchased from Aladdin. Chromatography reagent acetonitrile and methanol were purchased from Sigma. All chemicals and reagents were purchased from Sinopharm Chemical Reagent Co., Ltd or Shanghai Sangon Biotech (China) Co., Ltd unless noted otherwise. HPLC analysis was carried out on a SHIMADZU LC-20A Prominence HPLC system. LC-MS analysis was carried out on a Thermo Instruments HPLC system connected to LCQ Fleet electrospray ionization (ESI) mass spectrometer (ThermoFisher Scientific Inc.). ESI-high resolution MS (ESI-HRMS) analysis was carried out on ESI-LTQ Orbitrap (ThermoFisher Scientific Inc.).
2.2. DNA manipulation
Primers used in this study are listed in Table S3. Restriction endonucleases and T4 DNA ligase were purchased from NEB. PrimeSTAR HS DNA Polymerase with GC Buffer and In-Fusion HD Cloning kit were purchased from Takara. Oligonucleotide primer synthesis was performed by Genwiz and DNA sequencing was performed by Biosune. Standard procedures were used for DNA purification, PCR, subcloning, and molecular analysis [15]. All kits and enzymes were used according to the manufacturers' recommendations.
2.3. Vector construction for eryAIII knock out in Sac. erythraea
A sgRNA scaffold containing NcoI-XbaI cloning site was introduced to pCRISPR-Cas9 to give pCAS9-1. The spacer inserts including gene specific 20 nt guide sequence were generated by annealing two 34 nt synthesized oligonucleotides, sgeryAIII-f and sgeryAIII-r. The annealed oligos were cloned into pCAS9-1 at NcoI-XbaI. The 1.5 kb UHA (upper homologous arm) and DHA (down homologous arm) sequences were then amplified by the primer pairs eryAIII-L-f/eryAIII-L-r and eryAIII-R-f/eryAIII-R-r from genomic DNA of Sac. erythraea HL3168 E3, spliced by overlap extension PCR and cloned into the above-mentioned plasmid at StuI by infusion cloning kit, generating plasmid pZW-1 for gene knock out. Primers am-ori-f/am-ori-r were used to amplify a fragment containing colE1 and aac(3)IV from pZW-1 and the fragment was cloned into pYH7 at XbaI-KpnI to give pZW-0, containing rep(pIJ101). Subsequently, an 8 kb fragment was released from plasmid pZW-1 by BsmI-StuI and cloned into pZW-0 to give pZW-2 (Fig. S1). pZW-1 and pZW-2 were then transformed into ET12567/pUZ8002 for subsequent introduction into Sac. erythraea by conjugation, respectively.
2.4. Constructions of the CRISPR-Cas9 vectors
Primers cm-f/cm-r were used to amplify the chloramphenicol resistance gene (cat) from pACYCduet-1. Inducible promoter ptipA and thiostrepton resistance gene (tsr) were amplified from pCRISPR-Cas9 using two pairs of primers, tsr-f/tsr-r and ptipA-f/ptipA-r, respectively. Those three fragments were subsequently spliced together by overlap extension PCR, each end flanked by 39-bp homology sequence from pWHU2653. Then, the cat-ptipA-tsr fragment was inserted into pWHU2653 by PCR-targeting, replacing the constitutive promoter controlling the scas9 gene, the selecting marker cat was removed by PacI digestion and the rest of the vector was ligated by T4 ligase to give pMWcas9-1. To construct the double-enzyme digestion sgRNA cloning cassette, primers pmw-l-f/pmw-l-r were used to amplify the upstream fragment from pWHU2653 while another pair of primers, pmw-r-l/pmw-r-r were used for the downstream fragment amplifying. The two fragments were spliced together by overlap extension PCR, generates a 0.3 kb fragment. The 0.3 kb sgRNA cloning cassette was then inserted into pMWcas9-1 between XbaI-NheI using infusion cloning kit. Subsequently, the synthesized multiple cloning site (MCS), including HpaI, SpeI, StuI, was cloned into the HindIII site, giving plasmid pMWcas9 (Fig. S2).
2.5. Construction of genome editing vectors
The spacer inserts including gene specific 20 nt guide sequence were generated by annealing two 34 nt synthesized oligonucleotides, sgeryAIII2-f/sgeryAIII2-r for eryAIII knock out in Sac. erythraea HL3168 E3 and sgalpt-f/sgalpt-r for promoter antBp and antCp replacement in S. sp AL2110. The annealed oligos were cloned into pMWcas9 at EcoRI-XbaI, respectively. The 2.9 kb HA (homologous arm) was amplified from pZW-2 and cloned into the above-mentioned plasmid at HindIII by infusion cloning kit, generating plasmid pWHU2654 for gene knock out. The 1.5 kb UHA and DHA sequences were amplified from genomic DNA of the S. sp AL2110, using the primer pairs, alpt-l-f/alpt-l-r and alpt-r-f/alpt-r-r. The relevant UHA and DHA were then spliced together by overlap extension PCR, using primer pair alpt-l-f/alpt-r-r to generate approximately 3 kb HA fragments. These fragments were cloned into the above-mentioned plasmid at corresponding cloning site at MCS (shown in Table S3), generates vector pALPT. Promoter permE* and kasOp were amplified using primer pairs erme-f/erme-r and kasop-f/kasop-r from pWHU2653 and pkasop-T, respectively. The two promoters were then spliced together using primer pairs kasop-f/erme-r and introduced to pALPT at HpaI site via in vitro homologous recombination, generates pEKPT. Both pWHU2654 and pEKPT were subsequently used for genome editing.
For construction of the pWHU2653 based plasmid. A sgRNA scaffold including gene specific 20 nt guide sequence CTGCACATCACCCTCGACCA were amplified from pWHU2653 with primer pairs sg2653-eryaIII-a/sg2653-eryaIII-b and sg2653-eryaIII-c/sg2653-erya3-d, then spliced by overlap extension PCR and cloned into pWHU2653 at XbaI/NheI by infusion cloning kit. The 2.9 kb HA (homologous arm) was released from pWHU2654 with HindIII and cloned into the above-mentioned plasmid at HindIII by T4 ligase, generating plasmid pWHU2655 for gene knock out.
2.6. Transfer of the plasmids from E. coli to Sac. erythraea or S. sp. AL2110 by conjugation
The relevant plasmids first were transferred into E. coli ET12567/pUZ8002 cells by electroporation. Conjugation between E. coli ET12567/pUZ8002 and Sac. erythraea HL3168 E3 or S. sp. AL2110 was carried out as described previously [16,17]. The plates were incubated for 3–7 days at 30 °C, or until conjugates became visible.
2.7. Genome editing induced by CRISPR-Cas9
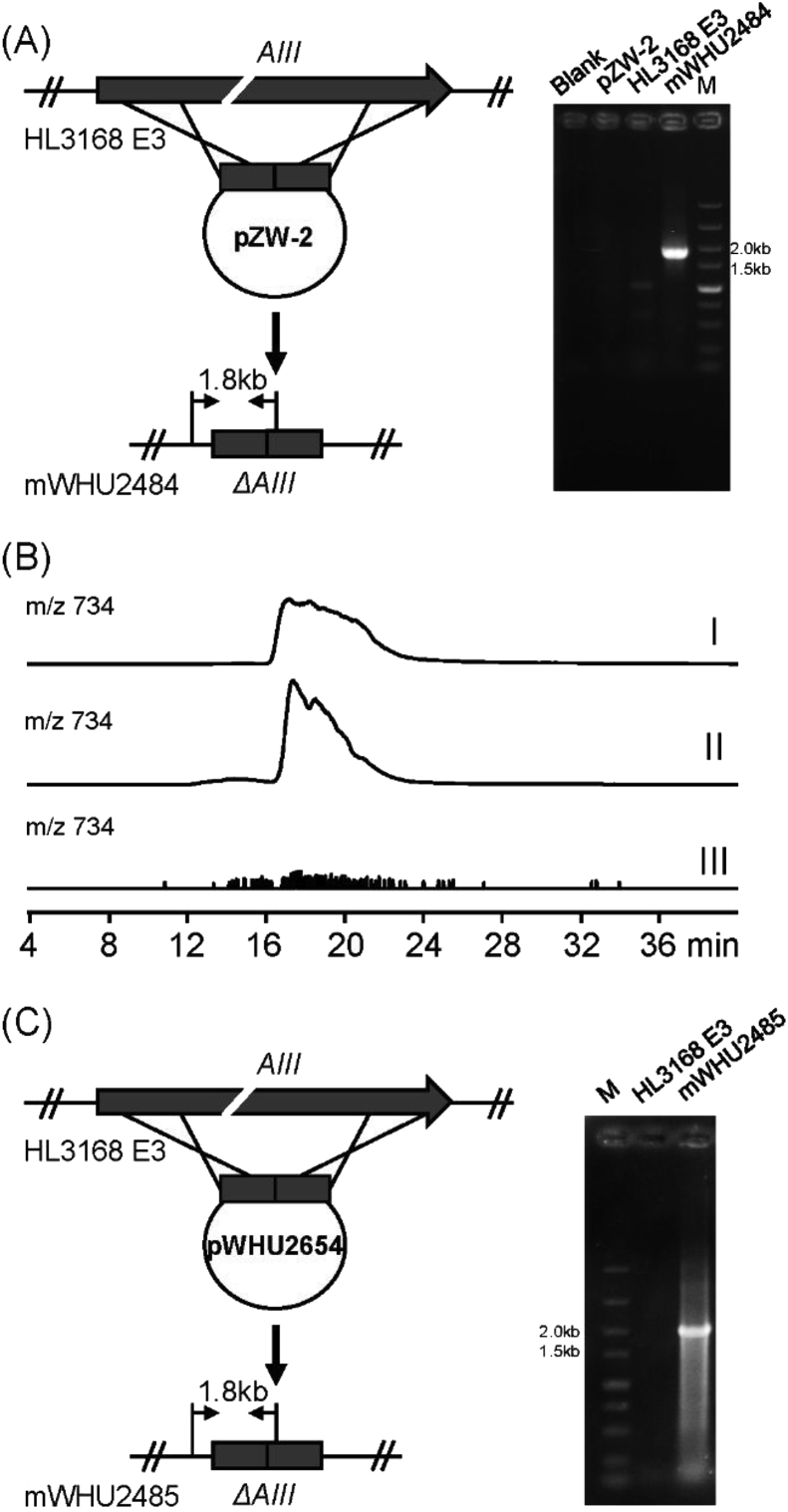
After conjugation, apramycin-resistant exconjugants were inoculated into 3 mL TSB medium again containing apramycin and trimethoprim and grown at 30 °C for 2 or 3 days. Subsequently, 2 μg mL−1 Tsr was added to induce the cleavage of their target DNA. The induced culture was then streaked onto MS agar plate to obtain single colonies. Each colony was inoculated into TSB liquid medium for growth of mycelium and genomic DNA was extracted and amplified by PCR using genotype confirmation primer pairs, ceryAIII-f/ceryAIII-r for eryAIII deletion and cekpt-f/cekpt-r for promoters' replacement (Fig. 1and Fig. S3), respectively. To obtain plasmid-free progeny, mycelium from the genotype confirmed culture was streaked on nonselective MS agar plate, followed by replica plating to MS agar plate with and without apramycin. For pZW-1, no Am-sensitive colonies were obtained after 15 rounds of culturing. The DNA of its single crossover mutant strain mWSW-1 was extracted and analyzed by PCR using primers ceryAIII-sc-f/ceryAIII-sc-r. The genomic DNA of the strain mWSW-2 (after 15 rounds of culturing) was extracted and analyzed by PCR using primer pairs of AT6-f/AT6-r and ceryAIII-f/ceryAIII-r. Genomic DNA of single apramycin-sensitive colonies was extracted and amplified by PCR using primers using primers pIJ101-f/pIJ101-r, giving eryAIII knocked out strain mWHU2484 (by pZW-2), mWHU2485 (by pWHU2654) and promoter antBp and antCp replaced strain mWHU2486.
Fig. 1.
Editing the erythromycin biosynthetic genes in Sac. erythraea by pIJ101-based Cas9 system. (A) Deleting the PKS gene eryAIII by pZW-2 and PCR evaluation of its deletion with the wild type HL3168 E3 and plasmid controls. The DSB region is indicated by a break. (B) Analysis of erythromycin production in the mWHU2484 and HL3168 E3 by LC-HRMS. Standard erythromycin (I), HL3168 E3 (II) and mWHU2484 (III). Erythromycin was indicated by selected ion chromatograms. (C) Deleting the PKS gene eryAIII by pWHU2654 (pMWCas9 derivative) and PCR evaluation of its deletion with the wild type HL3168 E3 control. The DSB region in eryAIII is indicated by a break.
2.8. Fermentation and metabolites analysis
For erythromycin, strains Sac. erythraea HL3168 E3 and its derived mutant mWHU2484 were inoculated into 50 mL TSB medium and grown at 30 °C for 3 or 4 days as the seed culture. The mycelium grown in the TSB medium was then fermented in 100 mL soy bean oil medium. (dextrin 4.0 g, soluble starch 3.0 g, soy flour 3.0 g, CaCO3 0.6 g, (NH4)2SO4 0.2 g, soy bean oil 1 mL, tap water to 100 mL, pH = 7.0–7.2) at 30 °C for 8 days. For deacyl antimycins, strains S. sp. AL2110 and its derived mutant mWHU2486 were grown on agar plates of the medium mannitol soya flour (MS) for sporulation. AL2110 and mWHU2486 maintained on MS media was chopped and inoculated into a 100 mL fermentation media M3-S (1.0 g glucose, 5 g soluble starch, 0.6 g (NH4)2SO4, 0.15 g CaCO3, tap water to 100 mL, pH = 7.0) and incubated at 30 °C for 5 days. After fermentation, each 100 mL culture broth was added with equal volume ethyl acetate and extracted under sonication for 15 min. The organic phase was transferred and dried by vacuum. Metabolites were subsequently re-dissolved by 1 mL methanol and filtrated by a 0.22 μm membrane to remove particles before HPLC analysis. The production yield of deacyl antimycins in AL2110 and mWHU2486 were deduced by fitting to the standard curve of antimycin sample (Sigma Co. Ltd.).
HPLC and LC-MS analysis of erythromycin were performed on a column of Venusil HLP C18 (2) (5 μm, 250 × 4.6 mm, Agela Technologies Inc.) at a flow rate of 1 mL min−1 and a UV detection at 210 nm over a 40 min gradient program: T = 0 min, 40% B; T = 10 min, 40% B; T = 11 min, 55% B; T = 36 min, 55% B; T = 37 min, 40% B; T = 40 min, 40% B. (A, H2O +10 mM CH3COONH4 pH = 9.7, and B, CH3CN). For deacyl antimycins, HPLC and LC-MS analysis were performed on a column of Inertsil ODS-3 (5 μm, 250 × 4.6 mm, GL Sciences B.V.) at a flow rate of 1 mL min−1 and a UV detection at 229 nm and 319 nm over a 30 min gradient program [18]: T = 0 min, 40% B; T = 20 min, 80% B; T = 21 min, 100% B; T = 25 min, 100% B; T = 26 min, 40% B; T = 30 min, 40% B. (A, H2O + 0.1% formic acid, and B, CH3CN + 0.1% formic acid).
3. Results and discussion
3.1. pCRISPR-cas9 causes unpredicted recombination of modular PKS genes
Constitutive expression of Cas9 will induce DSB immediately after the plasmid is transformed into the cell. As the rate of successful reparation of DSB by gene replacement is low, these kinds of Cas9 systems e.g. pWHU2653 often require a high plasmid transformation efficiency. This could be challenging for bacteria system recalcitrant to be transformed, for instance, many industrial strains. Thus, for getting higher transformation efficiency, the inducible expression is more advantageous over constitutive expression of the Cas9 nuclease. Based on this consideration, we chose pCRISPR-Cas9 for this study.
Erythromycin is an important family of antibiotics. Currently, its natural form and semi-synthetic derivatives, such as azithromycin, clarithromycin, dirithromycin, and roxithromycin have been widely utilized to treat Gram-positive pathogen infections [19]. Engineering its biosynthetic machinery, especially for the PKS genes, is promising to obtain novel bioactive analogues [20]. Recently, we confirmed that engineering the acyltransferase (EryAT6) domain can enormously broaden its substrate specificity to take highly diverse extender units [21]. Thus, to complement this engineered AT domain in vivo, we aim to delete the entire PKS gene eryAIII from the industrial strain Saccharopolyspora erythraea HL3168 E3 [16].
A sgRNA sequence targeted to the eryAIII and a ∼3 kb homologous template specific to the left and right boundaries of eryAIII were sequentially introduced into the pCRISPR-Cas9 to constitute the functional pZW-1 plasmid. This plasmid was transferred into the strain Sac. erythraea HL3168 E3 through E. coli conjugation with an efficiency of 4 cfu/mL broth. Efficient gene replacement was observed when the exconjugates were inoculated into the TSB media with 2 μg mL−1 Tsr. This strain was further inoculated onto the MS agar plate and grew in 39 °C for losing the delivery plasmid backbone. Isolated colonies (∼100) were picked for each time to testify the resistance of apramycin (Am). However, after fifteen rounds of culturing, we still failed to obtain any Am-sensitive colonies. PCR analysis showed that the Am resistant genes of the vector were indeed retained in the strains (Fig. S4). Isolation of plasmid from these strains was not successful, suggesting the plasmid indeed has been integrated into the genome. More so, the assumed to be eliminated eryAT6 (within the eryAIII) can be still detected by PCR in these colonies, though the expected eryAIII-deletion genotype was also observed through PCR analysis (Fig. S4). These results indicated that both gene deletion in the targeted region and unpredicted recombination event occurred.
3.2. pSG5 causes random recombination of PKS gene which can be solved by using pIJ101 replicon
Recently, a similar result was also observed in editing the rapamycin and tylosin PKS genes by the pKC1139-based plasmid [14]. It was assumed that the integrated plasmid (via single crossover) can constitute a replication fork when the culturing temperature was lowered for inducing the second recombination event to lose the plasmid. This pSG5-mediated replication fork collides with the natural fork leading to the temporary arrest and subsequent genome instability and rearrangements. To repair this catastrophic event, flanking PKS genes are forced to exert homologous recombination for removing the pSG5 encoding region, which leads to many unpredicted recombination events. However, pCRISPR-Cas9 system is different from pKC1139 integrated into genome, which exerts double crossover in plasmid form. Thus, the mechanism for causing random recombination should be different as assumed before.
To verify the essential role of pSG5 in this event, the pSG5 replicon was replaced by the segregational instability pIJ101 replicon together with a Tsr resistance gene from the plasmid pYH7 [22]. The generated plasmid pZW-2 (Fig. S1) was introduced into Sac. erythraea HL3168 E3 through the same procedure. Identical to pZW-1, efficient gene replacement was observed when 2 μg mL−1 Tsr was added into the culture (Fig. 1A). This strain was inoculated onto the MS agar plate to lose the delivery plasmid. Unlike the aforementioned pSG5-derived Cas9 system, all the tested colonies (80) from five-round culture have successfully lost the delivery plasmid. The deletion of eryAIII was further verified by both PCR and fermentation analysis (Fig. 1B), which confirms the pIJ101 is effective for editing modular PKS gene. The mechanism for pSG5-induced recombination is currently not clear. We assumed that the elevated temperature (39 °C) to prevent the replication of pSG5 may cause the emergency reaction of bacteria. As such reactions, for instance, SOS response can extensively induce DNA repair systems; and genes with high sequence identity can probably undergo homogeneous recombination. Taken together these results confirmed that the pSG5 replicon indeed is the major factor for causing the recombination even and using pIJ101 replicon can overcome this problem.
3.3. Construction of a new Cas9 system and used to edit biosynthetic genes of erythromycin in Sac. erythraea
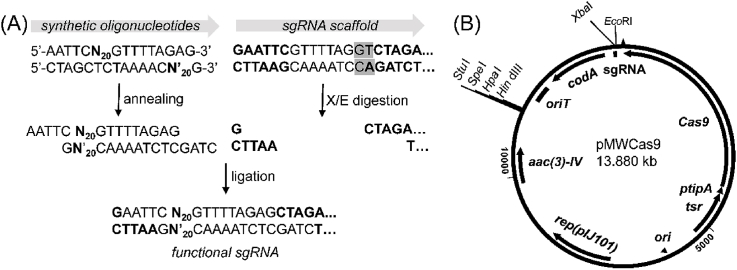
As the pZW-2 is not convenient for gene cloning, we further set out to construct a new pIJ101-based Cas9 vector (Fig. 2). The tsr and tipA cassette from pCRISPR-CAS was introduced into the pWHU2653 via PCR-Targeting to replace the original constitutive promoter. The resulted plasmid was then infused with a sgRNA cassette containing a EcoRI-XbaI cloning site via in vitro recombination. In this cassette, two nucleotides A and G in the original sgRNA scaffold were mutated into G and T for introducing a unique XbaI site. For installation of the target sequence, two short primers (each is 34 nt in a form of 5′-AATTCN20GTTTTAGAG-3′ and 5′-CTAGCTCTAAAACN′20G-3’ (N20 is the target sequence and N′20 is its complementary sequence) were mixed and annealed to form a double stranded DNA. This DNA fragment contains the correct nucleotides AG and has terminal cohesive ends complementary to the NcoI and XbaI sticky ends. Through ligation, both target sequences and correct nucleotides AG can be efficiently introduced into sgRNA scaffold to constitute the functional sgRNA (Fig. 2A). Finally, a 44 bp synthetic oligonucleotide was inserted into the restriction site of HindIII for introducing a multi-cloning site to insert homologous template. This new plasmid pMWCas9 has a pIJ101 replicon, a cloning site in the sgRNA region, a multi-cloning site for the homologous template and a codA counter-selectable marker, thus could be more robust and convenient than its parent Cas9 systems.
Fig. 2.
The Cas9 vector pMWCas9 (GenBank accession number: MH683611). (A) Cloning strategy of sgRNA. The original two nucleotides AG (shaded) of the sgRNA scaffold were changed into GT in order to introduce a unique XbaI site. The sticky ends generated by EcoRI and XbaI digestion (X/E) can be ligated with the annealed double stranded synthetic oligonucleotides to form the functional sgRNA. N20 is the target sequence should be 3 bp upstream of a PAM sequence. (B) Map of pMWCas9, the backbone is a segregationally unstable pIJ101 replicon, cas9 is controlled by the thiostrepton inducible tipA promoter, the sgRNA cassette is under control of the permE* promoter, apramycin (aac(3)-IV) and codA serve as the selection and counter-selection markers, respectively. The unique cloning sites StuI, SpeI, HpaI and HindIII are for insertion of the homologous template sequence. This plasmid can shuttle between E. coli and Actinomycetes.
To verify the function of this plasmid, it was then subjected to deleting the PKS gene eryAIII. As similar to the above mentioned, sgRNA and homolouges template were individually cloned into the EcoRI-XbaI site and HindIII site. Identical sgRNA and homologous template were also introduced into the pWHU2653. Both constructed plasmids (pWHU2654 and pWHU2655) were then introduced into the Sac. erythraea HL3168 E3. Like assumed, the transformation of the pWHU2653 system (pWHU2655) was not successful, while pMWCas9 system (pWHU2654) can be efficiently introduced into Sac. erythraea with the efficiency of 21–83 cfu/ML (Fig. S5), confirming inducible expression is more advantageous over constitutive expression of the Cas9 nuclease. Like the pZW-2, both of efficient double crossover and loss of the delivery plasmid were achieved by the pMWCas9 system, which resulted in rapid deletion of eryAIII (Fig. 1C). This result confirms the robustness and usefulness of the new pIJ101-based Cas9 system for editing highly repetitive modular PKS genes.
3.4. Editing the promotor region in S. sp. AL2110 to increase the production yield of deacyl-antimycin
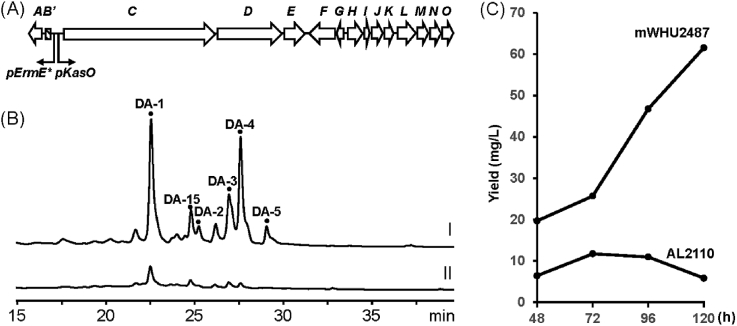
Finally, to demonstrate broad utility of this Cas9 system in other Actinomycetes species, it was used to edit the regulatory regions between antB and antC in the antimycin gene clusters from S. sp. NRRL AL2110 [17]. Antimycin is a group of industrial useful NRPS-PKS bydrid metabolites. Their biosynthesis has been elucidated by us and other groups [[23], [24], [25], [26], [27]]. It was revealed that binding of FscRI in the DNA region between antB and antC is essential for transcription of antimycin biosynthetic genes [27]. In order to remove the regulatory effect and boost the transcription of the ant genes, the promotor regions between antB and antC were replaced by a cassette containing two constitutive strong promotors permE* and kasOp (Fig. 3A) [8,28] through Cas9 mediated double crossover. Like in Sac. erythraea, both efficient double crossover and loss of the delivery plasmid was achieved. This recombinant strain was further sent for fermentation analysis. To our gratification, the production yield of deacyl-antimycins is significantly improved in this strain (62 mg/L at D5) which is 9-fold higher than the wild type strain AL2110 (6.4 mg/L at D5) (Fig. 3B and C). This strain provides an excellent basis for further engineering the biosynthesis of antimycin to produce novel bioactive structural varieties. Taken together, these results demonstrated that this new cas9 system is very robust and useful to effectively edit Actinomycetal genomes.
Fig. 3.
Engineering the promotors of antB and antC to increase the production of deacyl antimycins in S. sp. AL2110. (A) Schematic illustration of promoters engineering in the antimycin gene cluster. Constitutive promoters permE* and kasOp were introduced by pMWCas9 to replace the promoters of antB and antC which are responsible for the transcription of antA (antB has been deleted in AL2110) and antC to antE respectively. (B) Deacyl antimycins (DA-1-5 and DA-10, see Fig. S6) productions in the engineered strain mWHU2487 (I) and wild type AL2110 (II). (C) The production yield of deacyl antimycins in pmWHU2478 and AL2110 during the fermentation period (D2-D5). At the D5, mWHU2487 produces deacyl antimycins (62 mg/L) ∼9 fold higher than AL2110 (6.4 mg/L).
4. Conclusion
In summary, we confirmed that the pSG5 replicon is the major reason for causing unpredicted recombination and not suitable for editing DNA region with high sequence identity e.g. modular PKS genes. This problem can be overcome using the pIJ101 replicon. Our newly developed Cas9 system with pIJ101 replicon, inducible Cas9 cassette, codA counter selection marker and convenient cloning sites in the sgRNA scaffolds and homologous template can largely facilitate plasmid construction, and offer robust and efficient genome editing, which is highly important for study Actinobacteria. Finally, both eryAIII deleted Sac. erythraea and ANTs yield-boosted Streptomyces.sp. AL2110 strains provide a convenient platform for further engineering their structural diversity through pathway engineering.
Author contributions
JM, SW, WZ and CL conducted the experiments. XQ, LZ and ZD planned and supervised the study. XQ and JM wrote the paper. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no competing financial interest(s).
Acknowledgements
The authors are grateful to Prof. Wen Liu (Shanghai Institute of Organic Chemistry) for gifting the strain of Sac. erythraea HL3168 E3, Dr. Ming Jiang (Shanghai Jiao Tong University) for helping to develop the HPLC condition of erythromycin and Prof. Yuhui Sun (Wuhan University) for kindly providing the pWHU2653 and pYH7. This work was supported by the National Nature Science Foundation of China Grants (Nos. 31570057, 31430002, 31320103911 and 31770063), Taishan Scholarship and “the Fundamental Research Funds for the Central Universities 22221818014.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2019.02.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C.T., Tang Y. Royal Society of Chemistry; 2017. Natural product biosynthesis: chemical logic and enzymatic machinery. [Google Scholar]
- 3.Katz L., Baltz R.H. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 4.Nett M., Ikeda H., Moore B.S. Genomic basis for natural product biosynthetic diversity in the Actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B., Tian W., Wang S., Yan X., Jia X. Activation of natural products biosynthetic pathways via a protein modification level regulation. ACS Chem Biol. 2017;12:1732–1736. doi: 10.1021/acschembio.7b00225. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood D.A. Oxford University Press, Inc.; 2007. Streptomyces in nature and medicine: the antibiotic makers. [Google Scholar]
- 8.Kieser T., Bibb M., Butter M., Chater K.F., Hopwood D.A. The John Innes Foundation; 2001. Practical streptomyces genetics. [Google Scholar]
- 9.Fernández-Martínez L.T., Bibb M.J. Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in Actinomycetes. Sci Rep. 2014;4:7100. doi: 10.1038/srep07100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Y., Charusanti P., Zhang L., Weber T., Lee S.Y. CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 11.Cobb R.E., Wang Y., Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng H., Wen S., Xu W., He Z., Zhai G. Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Appl Microbiol Biotechnol. 2015;99:10575–10585. doi: 10.1007/s00253-015-6931-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin. 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 14.Wlodek A., Kendrew S.G., Coates N.J., Hold A., Pogwizd J. Diversity oriented biosynthesis via accelerated evolution of modular gene clusters. Nat Commun. 2017;8:1206. doi: 10.1038/s41467-017-01344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- 16.Wang Y., Wang Y., Chu J., Zhuang Y., Zhang L. Improved production of erythromycin A by expression of a heterologous gene encoding S-adenosylmethionine synthetase. Appl Microbiol Biotechnol. 2007;75:837–842. doi: 10.1007/s00253-007-0894-z. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y., Chen J., Zhang L., Zheng Q., Han Y. Multiplexing of combinatorial chemistry in antimycin biosynthesis: expansion of molecular diversity and utility. Angew Chem Int Ed. 2013;52:12308–12312. doi: 10.1002/anie.201305569. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L., Mori T., Zheng Q., Awakawa T., Yan Y. Rational control of polyketide extender units by structure-based engineering of a crotonyl-CoA carboxylase/reductase in antimycin biosynthesis. Angew Chem Int Ed Engl. 2015;54:13462–13465. doi: 10.1002/anie.201506899. [DOI] [PubMed] [Google Scholar]
- 19.Katz L., Ashley G.W. Translation and protein synthesis: Macrolides. Chem Rev. 2005;105:499–528. doi: 10.1021/cr030107f. [DOI] [PubMed] [Google Scholar]
- 20.Dunn B.J., Khosla C. Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface. 2013;10 doi: 10.1098/rsif.2013.0297. 20130297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Zhang W., Zhang H., Tian W., Wu L. Structural basis of a broadly selective acyltransferase from the polyketide synthase of splenocin. Angew Chem Int Ed. 2018;57:5823–5827. doi: 10.1002/anie.201802805. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y., He X., Liang J., Zhou X., Deng Z. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl Microbiol Biotechnol. 2009;82:303–310. doi: 10.1007/s00253-008-1793-7. [DOI] [PubMed] [Google Scholar]
- 23.Yan Y., Zhang L., Ito T., Qu X., Asakawa Y. Biosynthetic pathway for high structural diversity of a common dilactone core in antimycin production. Org Lett. 2012;14:4142–4145. doi: 10.1021/ol301785x. [DOI] [PubMed] [Google Scholar]
- 24.Sandy M., Rui Z., Gallagher J., Zhang W. Enzymatic synthesis of dilactone scaffold of antimycins. ACS Chem Biol. 2012;7:1956–1961. doi: 10.1021/cb300416w. [DOI] [PubMed] [Google Scholar]
- 25.Ilka S., Christian P., DJ S., Bertrand A., Pierre L. An unprecedented 1,2-shift in the biosynthesis of the 3-aminosalicylate moiety of antimycins. Chembiochem. 2012;13:769–773. doi: 10.1002/cbic.201200033. [DOI] [PubMed] [Google Scholar]
- 26.Chang C., Huang R., Yan Y., Ma H., Dai Z. Uncovering the formation and selection of benzylmalonyl-CoA from the biosynthesis of splenocin and enterocin reveals a versatile way to introduce amino acids into polyketide carbon scaffolds. J Am Chem Soc. 2015;137:4183–4190. doi: 10.1021/jacs.5b00728. [DOI] [PubMed] [Google Scholar]
- 27.McLean T.C., Hoskisson P.A., Seipke R.F. Coordinate regulation of antimycin and candicidin biosynthesis. mSphere. 2016;1:e00305–e00316. doi: 10.1128/mSphere.00305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Li X., Wang J., Xiang S., Feng X. An engineered strong promoter for Streptomycetes. Appl Environ Microbiol. 2013;79:4484–4492. doi: 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.