Abstract
Transcription factors of the Nuclear Factor Y (NF-Y) family play essential functions in plant development and plasticity, including the formation of lateral root organs such as lateral root and symbiotic nodules. NF-Ys mediate transcriptional responses by acting as heterotrimers composed of three subunits, NF-YA, NF-YB, and NF-YC, which in plants are encoded by relatively large gene families. We have previously shown that, in the Phaseolus vulgaris × Rhizobium etli interaction, the PvNF-YC1 subunit is involved not only in the formation of symbiotic nodules, but also in the preference exhibited by the plant for rhizobial strains that are more efficient and competitive in nodule formation. PvNF-YC1 forms a heterotrimer with the PvNF-YA1 and PvNF-YB7 subunits. Here, we used promoter:reporter fusions to show that both PvNF-YA1 and PvNF-YB7 are expressed in symbiotic nodules. In addition, we report that knock-down of PvNF-YA1 and its close paralog PvNF-YA9 abolished nodule formation by either high or low efficient strains and arrested rhizobial infection. On the other hand, knock-down of PvNF-YB7 only affected the symbiotic outcome of the high efficient interaction, suggesting that other symbiotic NF-YB subunits might be involved in the more general mechanisms of nodule formation. More important, we present functional evidence supporting that both PvNF-YA1 and PvNF-YB7 are part of the mechanisms that allow P. vulgaris plants to discriminate and select those bacterial strains that perform better in nodule formation, most likely by acting in the same heterotrimeric complex that PvNF-YC1.
Keywords: nodulation, nitrogen fixation, Nuclear Factor Y, rhizobia, transcription factors
Introduction
Nitrogen (N) is an essential macronutrient for plant growth and development since it is part of many biological molecules such as nucleic acids, proteins, vitamins, and chlorophyll (Wang et al., 2012); however, its availability is frequently limited in soils of both natural and agronomical ecosystems. Most legume species overcome N limitation by establishing a symbiosis with N-fixing bacteria from different genera known as rhizobia. This interaction results in the formation of a new postembryonic root organ, the nodule, where bacteria allocate and convert atmospheric N to reduced forms that will be incorporated into the plant metabolism. The root nodule symbiosis (RNS) can be divided into three stages. In the pre-symbiotic stage, the two organisms recognize each other through an exchange of diffusible molecules. Under N limiting conditions, roots of legumes exudate flavonoids/isoflavonoids that are perceived by rhizobia (Weston and Mathesius, 2013), which activate the synthesis and secretion of key molecules called Nod factors (NFs; Denarie et al., 1996). NFs are perceived by the host plant inducing the first morphological response, the curling of the root hair around the bacterial microcolony, which results in the formation of an infection pocket (Lerouge et al., 1990). The second stage is the infection, which consists in the penetration of bacteria into host tissues through a tubular structure referred to as the infection thread (IT). Concomitantly with this infection process, cell divisions are initiated in the root cortex of the host to form the nodular primordium. The third and last stage consists in the development of the nodule and the release of bacteria from the ITs into the host cells to form organelle-like structures called symbiosomes (Popp and Ott, 2011), where biological N fixation will take place.
Morphological and developmental responses observed during RNS are initiated by the binding of NFs to LysM domain-containing receptors located at the plant plasma membrane, which results in the hierarchical activation of a set of transcriptions factors (TFs). Specific members of the Nuclear Factor Y (NF-Y) gene family of TFs have been implicated at different stages of the RNS, from epidermal infection to nodule development. NF-Ys are evolutionary conserved heterotrimeric TFs composed of three subunits (NF-YA, NF-YB, and NF-YC). Genes encoding NF-Y subunits have diversified in the plant lineage forming relatively large gene families with specific functions (Petroni et al., 2012; Laloum et al., 2013). In Medicago truncatula, two NF-YA subunits, MtNF-YA1 and MtNF-YA2, play important roles not only at early stages of rhizobial infection, but also at later stages of the RNS mediating both nodule organogenesis and the persistence of nodule meristems (Combier et al., 2006, 2008; Laloum et al., 2014; Laporte et al., 2014). On the other hand, M. truncatula MtNF-YC1 and MtNF-YC2 genes are required for nodule organogenesis, but not for intracellular infection by rhizobia (Baudin et al., 2015). Two NF-Y subunits of Lotus japonicus, LjNF-YA1 and LjNF-YB1, were identified as direct transcriptional targets of the master symbiotic regulator Nodule Inception (NIN). Knock-down of LjNF-YA1 by RNA interference (RNAi) arrested cell divisions associated with nodule formation, but did not affect epidermal infection. On the other hand, overexpression of LjNF-YA1 stimulates cell proliferation, a phenotype that was enhanced by co-expression of LjNF-YB1 (Soyano et al., 2013). In common bean (Phaseolus vulgaris), PvNF-YC1 was identified as a key TF required for both nodule organogenesis and infection by Rhizobium etli, the predominant species present in common bean nodules (Zanetti et al., 2010).
Phaseolus vulgaris originated in Mesoamerica and further expanded to South America, resulting into two gene pools at distinct centers of genetic diversification (CGDs): the Mesoamerican and the Southern Andes CGDs (Bitocchi et al., 2012). These gene pools have undergone parallel and independent domestication at each CGD, thus the characteristics of each gene pool are evident in both wild and domesticated accessions (Bitocchi et al., 2013). The abundance of R. etli strains in each CGD has been correlated with a polymorphism of the nodC gene of R. etli, which encodes an N-acetylglucosamine transferase involved in the first steps of NF synthesis. Strains bearing the nodC-α allele (hereafter nodC-α strains) are predominant in Mesoamerican soils, whereas those carrying the nodC-δ allele are highly represented in the Andean region (Aguilar et al., 2004). Wild and domesticated beans from each CGD are more efficiently nodulated by those strains that are more abundant in the soils of the cognate geographical region (Peltzer Meschini et al., 2008). More interesting, nodules of Mesoamerican beans co-inoculated with an equicellular mixture of both rhizobial strains were predominantly occupied by nodC-α strains (Aguilar et al., 2004), leading to the suggestion that Mesoamerican beans have developed molecular mechanisms that allow them to discriminate and select those strains that have coevolved in the same CGD. A key component of this mechanism is the above-mentioned PvNF-YC1 subunit of the NF-Y family of TFs (Zanetti et al., 2010). Overexpression of PvNF-YC1 in Mesoamerican beans was sufficient not only to improve the symbiotic outcome (i.e., nodule number and shoot dry weight) of the less efficient strains carrying the nodC-δ allele, but also to alter nodule occupancy by nodC-α and nodC-δ strains, exposing that competitions in the rhizosphere can be controlled by the plant (Zanetti et al., 2010). Since NF-Ys act as heterotrimers to promote transcriptional activation/repression, we sought to identify and characterize the NF-YA and NF-YB subunits that form the symbiotic functional heterocomplex. Given the symbiotic specific expression pattern exhibited by PvNF-YA1, PvNF-YA9, and PvNF-YB7 genes (Ripodas et al., 2015) and the physical interaction of PvNF-YC1 with PvNF-YA1 and PvNF-YB7 subunits (Baudin et al., 2015), we selected these members to conduct a functional characterization of their role in the RNS and in the strain preference observed in Mesoamerican beans. Here, we describe that simultaneous silencing of PvNF-YA1 and its closest homolog, PvNF-YA9, impaired nodule organogenesis triggered by either nodC-α or nodC-δ strains and reduced bacterial infection. Interestingly, overexpression of PvNF-YA1, but not PvNF-YA9, was sufficient to alter nodule occupancy in roots co-inoculated with nodC-α and nodC-δ strains. On the other hand, knock-down of PvFN-YB7 affected the number of nodules developed by a nodC-α, but not a nodC-δ strain, and altered nodule occupancy. All together, the results presented here highlight the functional implication of the heterotrimer formed by PvNF-YA1, PvNF-YB7 and PvNF-YC1 not only in the establishment of the RNS, but also in the mechanisms that determine strain specificity within the P. vulgaris ×R. etli interaction.
Materials and Methods
Biological Material and Generation of Composite Plants by Agrobacterium rhizogenes Transformation
Plant growth and transformation were performed essentially as previously described (Blanco et al., 2009; Zanetti et al., 2010). Briefly, P. vulgaris seeds were surface sterilized and germinated on 10% (w/v) agar-H2O for 2 days. Seedling were transferred to pots containing vermiculite and watered with Fahraeus media supplemented with 8 Mm KNO3. Five days after transplantation, P. vulgaris plants were inoculated in the stem with a saturated suspension of Agrobacterium rhizogenes strain K599 using a syringe. Approximately 10 days after transformation, when hairy roots have emerged from the inoculation sites, the main root system was removed by cutting the stem 1 cm below the site of inoculation. Composite plants consisting on a wild type aerial part and transgenic hairy roots were transferred to acrylic boxes containing agar-Fahraeus covered with paper. Alternatively, for co-inoculation experiments, composite plants were transferred to pots containing vermiculite and watered with Fahraeus media. R. etli strains SC15 (nodC-α) and 55N1 (nodC-δ) were previously reported (Aguilar et al., 2004). The R. etli strain CFNx5 (nodC-α) expressing the DsRed protein was previously generated and described (Battaglia et al., 2014).
Vector Construction
To generate localization and/or overexpression constructs, the open reading frame of PvNF-YA1 was amplified by PCR using primers PvNF-YA1 OE F, PvNF-YA1 OE R (Supplementary Table S1), and cloned into the pENTR/D-TOPO vector (Invitrogen), creating pENTR-NF-YA1. For overexpression, the pENTR-NF-YA1 was recombined into the destination vector p35S:HF-GATA (Mustroph et al., 2009). Later on, the FLAG-PvNF-YA1 fragment was amplified by PCR using specific primers and cloned into the pENTR/D-TOPO to create pENTR-FLAG-NF-YA1, which was subsequently recombined into the final destination vector pK7WG2D, which carries EgfpER as a screenable marker for early visualization and selection of the transgenic roots (Karimi et al., 2002). For subcellular localization, a translational fusion of PvNF-YA1 to the C-terminal end of GFP was generated by recombination of the pENTR-NF-YA1 with pMDC43 (Curtis and Grossniklaus, 2003). For the overexpression of PvNF-YA9, the ORF of this transcript was synthetized by Life Technologies to be used as level-0 module in GoldenGate cloning1. Assembled level-1 modules expressing the fusion FLAG-PvNF-YA9 under the control of the constitutive promoter CaMV35S and a pAtUbi:GFP fusion as a transgenic root selection marker were finally cloned in a level-2 binary vector backbone EC505051. For histochemical assays to measure glucuronidase activity, a 1.5 kb fragment of the promoter sequences of PvNF-YA1 and PvNF-YB7 was amplified with specific primers from common bean genomic DNA, cloned in the pENTR/D-TOPO vector, and finally introduced by recombination into the destination vector pKGWFS7, driving the expression of the fusion GFP-GUS. For silencing of PvNF-YA1/A9 by RNAi, a 217 bp fragment corresponding to the 3′ UTR of PvNF-YA1 was amplified by PCR, using PvNF-YA1/A9 RNAi F and PvNF-YA1/A9 RNAi R primers (Supplementary Table S1) and P. vulgaris cDNA as a template. Similarly, a fragment corresponding to the 3′ UTR was amplified using gene specific primers for knock-down of PvNF-YB7 (Supplementary Table S1). Each PCR product was cloned into the entry vector pENTR/D-TOPO and recombined into the destination vector pK7GWIWG2D (II) (Karimi et al., 2002) to finally produce PvNF-YA1/A9 RNAi and PvNF-YB7 RNAi constructs, respectively. All binary vectors were introduced into Agrobacterium tumefaciens GV3101 and/or A. rhizogenes K599 by electroporation and then used for agroinfiltration of Nicotiana benthamiana leaves or for the generation of transgenic hairy roots in P. vulgaris.
β-Glucuronidase Activity
Activity of the enzyme β-glucuronidase was determined in P. vulgaris roots and nodules formed at 7 and 14 dpi with strain CFNX5, a R. etli nodC-α strain that expresses the fluorescent protein Ds-Red. Roots were cut into 2 cm sections and then infiltrated with a solution of the dye reactive: 100 mM TRIS-HCl pH 7, 2 mM X-Gluc (5-bromo-4-cloro-3-indolil-β-D-glucoronic acid, 0.01% v/v Triton X-100, 50 mM NaCl, 2.0 mM potassium ferrocyanide). Tissue was incubated at 37°C for 1–10 h until color development. After incubation, roots were observed by bright field microscopy to visualize GUS staining in whole roots or, in some cases, selected roots and nodules were embedded in 4% (w/v) agarose and cut to thin sections (55 μm) using a Leica VT1000 S Vibrating blade microtome. Tissue sections were analyzed by bright field microscopy in an inverted microscope (OLYMPUS IX51).
Phenotypic Analysis
Composite plants were generated and inoculated as described (Peltzer Meschini et al., 2008; Blanco et al., 2009). Primary and lateral root length and density were measured as previously reported (Battaglia et al., 2014). Nodule quantification and dry weight determination were performed as previously described (Zanetti et al., 2010). Non-transgenic (not fluorescent roots) were excised from the root system before phenotypic analysis before inoculation with rhizobia, thus only nodules formed in transgenic roots were taken into account. IT quantification and classification was performed essentially as previously described by Battaglia et al. (2014). Briefly, composite plants were inoculated with a R. etli strain CFNx5 that constitutively expresses the fluorescent protein Ds-Red. ITs were visualized and quantified under UV light. For co-inoculation experiments, composite plants were transferred to pots containing vermiculite. Five days after transplantation, roots were inoculated with 10 ml of a mixture of R. etli strains SC15 and 55N1 (ratio 1:1) as previously described (Zanetti et al., 2010). Four weeks after co-inoculation, more than 100 nodules from 10 independent plants for each construct were excised, crushed, and plated in Congo Red-YEM agar plates. The color of the bacteria grown on this media was recorded. Bacterial DNA was extracted, subjected to PCR amplification of the nodC gene, and digested with HinfI to determine the restriction profile of the nodC gene as reported (Aguilar et al., 2004). All experiments were conducted in three independent biological replicates.
Subcellular Localization Assays
The PvNF-YA1-GFP construct was introduced in A. tumefaciens GV3101 and A. rhizogenes K599 by electroporation. A. tumefaciens carrying PvNF-YA1 was co-infiltrated into N. benthamiana leaves with the viral silencing inhibitor protein P19 (Voinnet et al., 2003) as described (Battaglia et al., 2014). The subcellular localization in common beans roots was done by generation of P. vulgaris composite plants expressing the PvNF-YA1-GFP fusion (Zanetti et al., 2010).
RNA Extraction and Quantitative RT-PCR
RNA extraction, cDNA synthesis, and RT-qPCR assays in common bean were performed as previously reported (Ripodas et al., 2013). Transcript levels for each of the target genes were normalized to the endogenous elongation factor 1 α (EF1α) transcript levels. Primer sequences for quantitative RT-PCR analyses are shown in Supplementary Table S1. Data shown are mean values obtained in three or four independent biological experiments with two or three technical repeats.
Microscopy and Imaging
Bright-field and epifluorescence imaging of ITs formation were performed as described (Battaglia et al., 2014). Confocal microscopy of P. vulgaris roots, N. benthamiana leaves, and ITs observation were made with a Leica confocal microscope (SP5) using 20X and 40X objectives. Samples were excited with argon laser and emission spectra used to detect the fluorescence were: GFP (498–550 nm) and DsRed (578–626 nm).
Western Blots
Proteins from transgenic root tissue of individual composite plants were extracted, separated into 12% SDS-PAGE, and subjected to immunoblot analysis using anti-FLAG antibody (1:500; Sigma–Aldrich) as previously described (Zanetti et al., 2010).
Results
PvNF-YA1 Is a Nuclear Localized TF Expressed in the Central Tissue of P. vulgaris Nodules
The current model for assembling of the heterotrimeric NF-Y complex proposes that the NF-YB interacts with NF-YC in the cytoplasm and the heterodimer translocates to the nucleus where it joins the NF-YA subunit. Consistent with this model, nuclear subcellular localization was described for several NF-YA proteins in different metazoan and plants species (Liu and Howell, 2010; Hackenberg et al., 2012; Baudin et al., 2015). In this study, nuclear subcellular localization of PvNF-YA1 was verified by expression of a translational fusion of PvNF-YA1 to the green fluorescent protein (GFP) into N. benthamiana leaf epidermal cells (Figure 1A). The GFP-PvNF-YA1 fusion protein was also detected in the nucleus of P. vulgaris root cells generated by A. rhizogenes–mediated transformation (Figure 1B), including epidermal root hairs (Figure 1C,D). As expected, free GFP was dispersed between the nucleus and the cytoplasm (Figure 1E).
FIGURE 1.
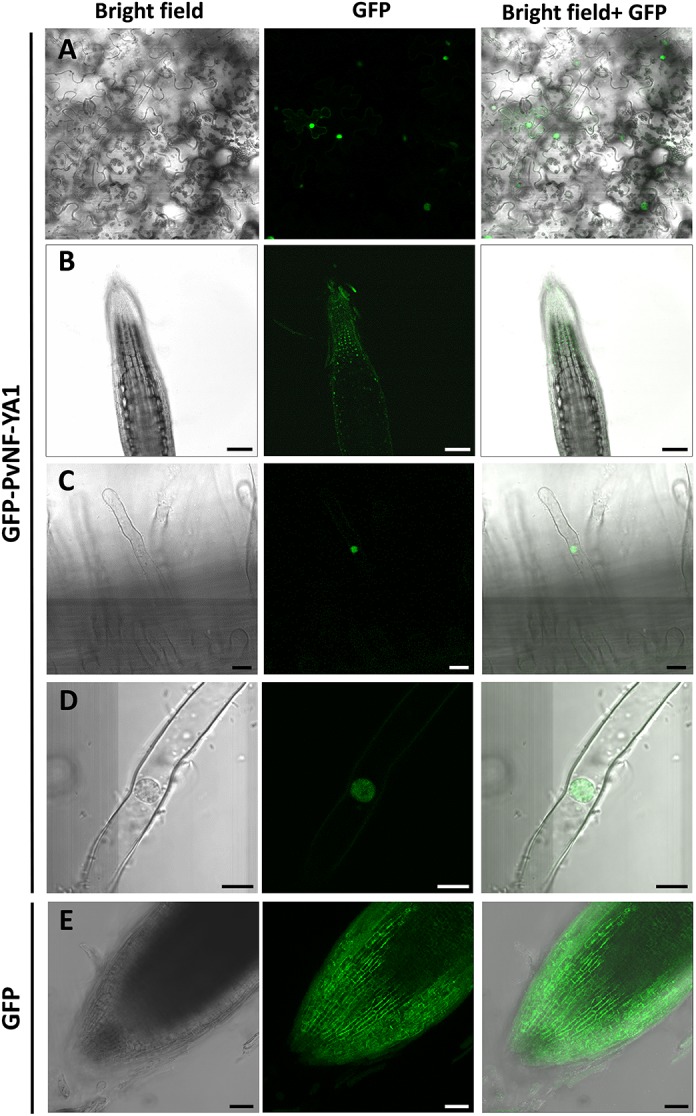
Subcellular localization of PvNF-YA1 in N. benthamiana leaves and P. vulgaris roots. (A) Subcellular localization of the translational fusion GFP-NF-YA1 in epidermal cells of leaves of N. benthamiana. Images obtained under white (bright field) and UV light (GFP) by confocal microscopy, as well as the merged image (bright field + GFP), are shown. Scale bar: 50 μm. P. vulgaris roots expressing the GFP-NF-YA1 fusion (B–D) or free GFP (E) were observed by confocal microscopy. Images show the tip of a lateral roots (B,E) or root hairs (C,D). Bright field, GFP channel, and merged (bright field + GFP) images are shown. The integration in Z-axis of 15 confocal sections is shown in E. Scale bars: 100 (B), 25 (C), 8 (D), and 50 μm (E).
Previous expression analysis of NF-YA family members reveled that PvNF-YA1 transcripts accumulate at higher levels in nodules of 14 days post-inoculation than in roots or young nodules of 7 dpi (Ripodas et al., 2015). Here, we used a promoter:reporter fusion to investigate the activity of the PvNF-YA1 promoter in different tissues of roots and nodules. A construct comprising approximately 2 kb upstream of the translational initiation codon of PvNF-YA1 fused to the open reading frame of GUS reporter gene was introduced into P. vulgaris roots by A. rhizogenes-mediated transformation. Histological staining of non-inoculated ProPvNF-YA1:GUS hairy roots revealed GUS activity in the vascular tissue of primary roots, but not in lateral root primordia (Figure 2A). GUS staining was also observed in lateral root meristem (Figure 2B), as well as in the vascular tissue of non-inoculated lateral roots (Figure 2C,E). Upon inoculation with a R. etli strain SC15 (carrying the nodC-α allele), a strong and a more intense GUS staining was detected on curled root hairs and epidermal cells surrounding the infection foci of the rhizobia susceptible zone, which contains elongating root hairs, but not in fully elongated root hairs (Figure 2D,F). At later time points, GUS staining was detected in the dividing cells of nodule primordia (Figure 2G) as well as in the central tissue of nodules formed by a R. etli strain carrying the nodC-α allele (Figure 2H).
FIGURE 2.
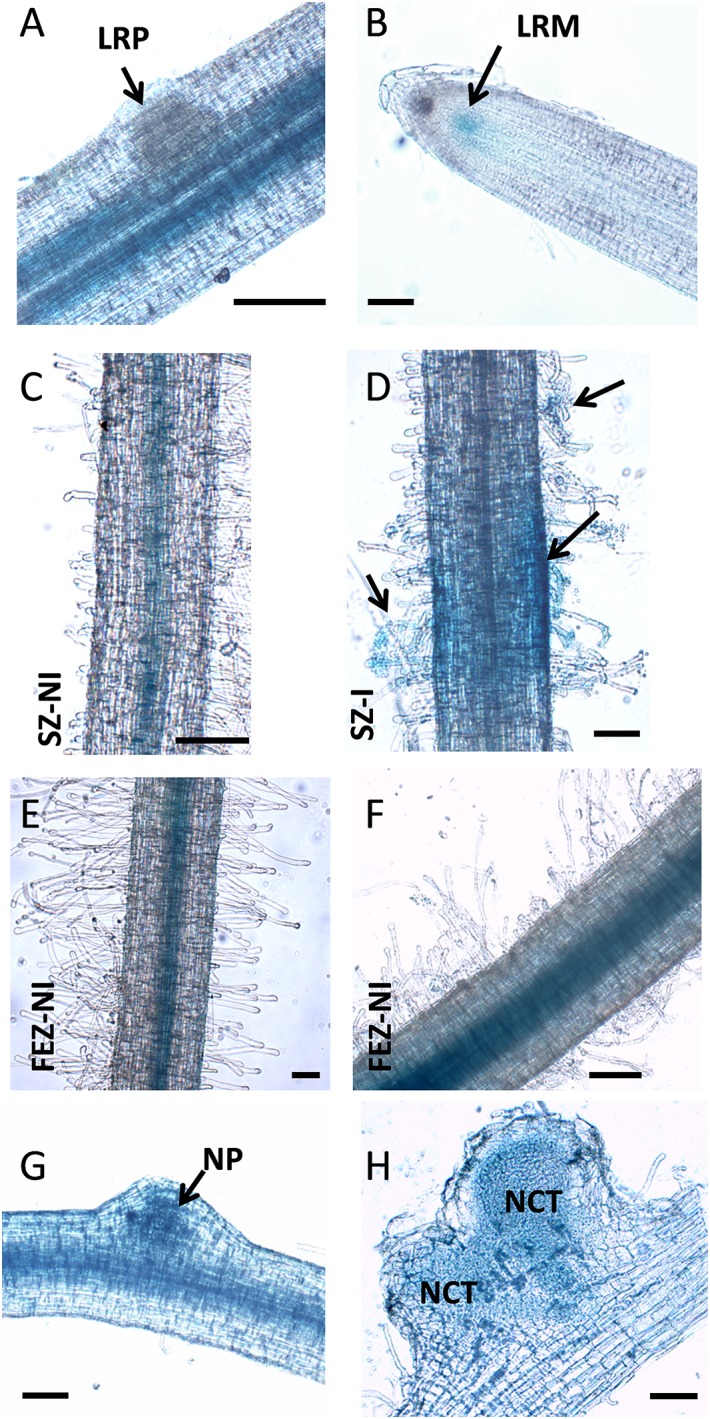
PvNF-YA1 promoter is active in roots and symbiotic nodules of P. vulgaris. Histochemical GUS staining of roots and nodule transformed with the ProPvNF-YA1:GFPGUS construct. (A) Non-inoculated (NI) primary root which containing a lateral root primordia (LRP). (B) Lateral root meristematic (LRM) zone of a NI lateral root. (C) Susceptible zone (SZ) of a NI lateral root. (D) SZ of lateral root inoculated (I) with R. etli strain SC15. Arrows point to infection sites. (E) Zone of fully elongated root hairs (FEZ) on a NI lateral root. (F) FEZ of a lateral root inoculated with R. etli strain SC15. (G) Nodule primordia (NP) or (H) nodules developed by R. etli strain SC15 at 6 or 14 dpi. NCT: nodule central tissue. Pictures are representative of images observed in more than three independent experiments. Bars: 200 μm.
Taken all together, the results presented in this section indicate that PvNF-YA1 is located in the nuclei of vascular tissues of primary and lateral roots, but also of rapidly dividing tissues such as lateral root meristems or nodule primordia. Importantly, PvNF-YA1 is responsive to rhizobia, being active at early stages of the RNS (root hair curling), as well as at later stages in N-fixing nodules.
PvNF-YA1 and PvNF-YA9 Are Symbiotic Subunits Required for Nodule Formation and Bacterial Infection
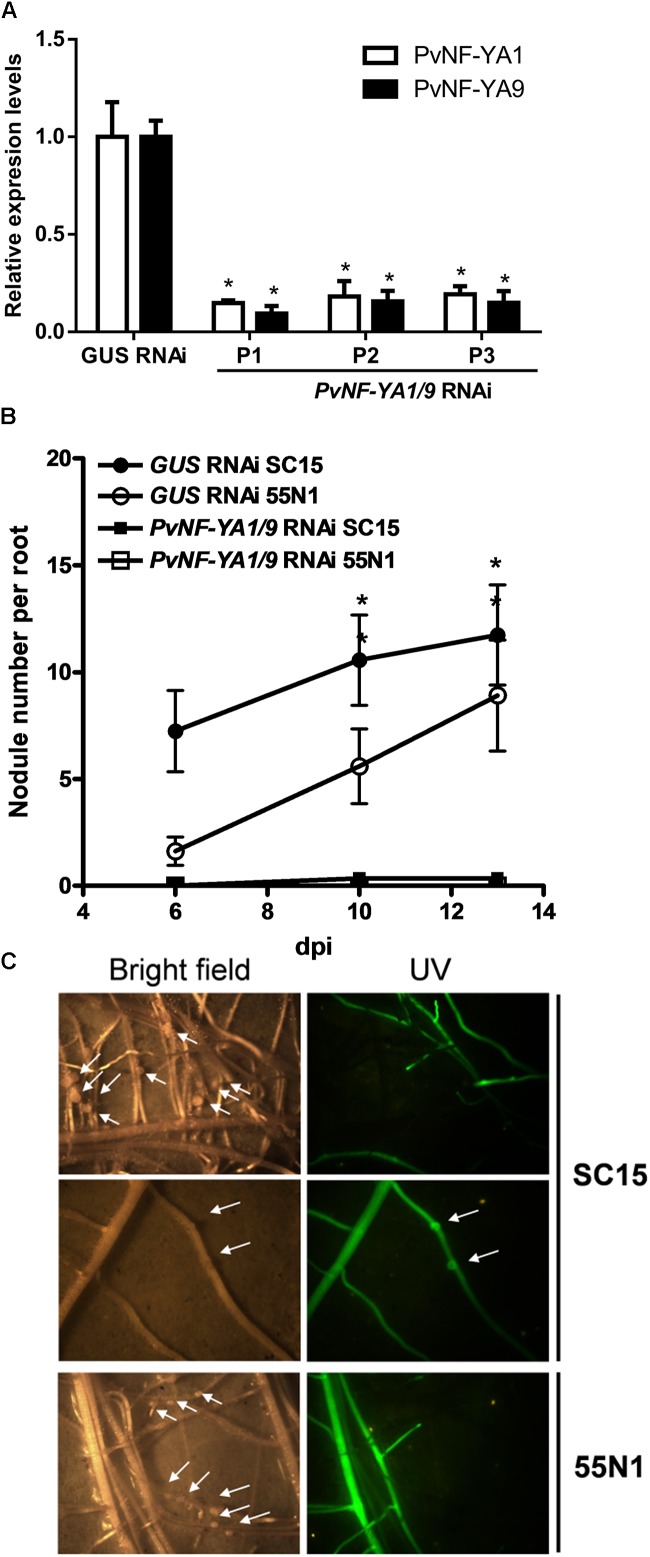
We have previously shown that both PvNF-YA1 and PvNF-YA9 mRNA levels increased in roots at early stages of the symbiotic interaction (i.e., 24 hpi) with R. etli. However, PvNF-YA1 transcript levels increased specifically upon inoculation with the more efficient nodC-α strain SC15, whereas PvNF-YA9 mRNAs accumulated at higher levels in response to both nodC-α and nocC-δ strains as compared with non-inoculated roots (Ripodas et al., 2015). Additionally, Laloum et al. (2014) showed that the orthologs of PvNF-YA1 and PvNF-YA9 in M. truncatula, MtNF-YA2 and MtNF-YA1, respectively, have partially redundant functions during RNS (Laloum et al., 2014). Thus, we questioned whether these two symbiotic NF-YA members play functions during the P. vulgaris ×R. etli interaction. Anticipating that PvNF-YA1 and PvNF-YA9 might display redundant functions, an RNAi construct based on a sequence of the 3′ untranslated region (UTR) of PvNF-YA1 mRNA that is highly similar to the 3′UTR of the PvNF-YA9 mRNA (Supplementary Figure S1) was designed to obtain P. vulgaris hairy roots with simultaneous reduction in PvNF-YA1 and PvNF-YA9 transcript levels. Transgenic roots were distinguished by visualization of the fluorescence emitted by the GFP protein, which is expressed under the control of the rolD promoter present in the T-DNA of the vector used for RNAi (see section “Materials and Methods”). Non-fluorescent roots were removed before collection of the tissue. RT-qPCR experiments verified that hairy roots from three independent PvNF-YA1/9 RNAi plants exhibit reduced PvNF-YA1 and PvNF-YA9 mRNA levels (∼80%), as compared with control roots transformed with a GUS RNAi construct (Figure 3A). The fragment used for RNAi includes the PvNF-YA1 binding sites for the microRNA miR169, a post-transcriptional mechanism that regulates most NF-YA family members (Combier et al., 2006; Ripodas et al., 2015). Notably, expression of this RNAi construct did not affect the expression of other members of NF-YA family (Supplementary Figure S2). Specific members of L. japonicus and Arabidopsis NF-YA families (e.g., LjNF-YA1, AtNF-YA2, and AtNF-YA10) have been implicated in root development (Soyano et al., 2013; Sorin et al., 2014), thus we performed a phenotypic analysis of the root system. Knock-down of PvNF-YA1/A9 RNAi did not alter the length of primary and lateral roots or the density of lateral roots (Table 1). Under symbiotic conditions, GFP-expressing transgenic PvNF-YA1/9 RNAi roots barely developed nodules either after inoculation with nodC-α (SC15) or nodC-δ (55N1) strains (Figure 3B,C). Only two bumps were detected in over 100 PvNF-YA1/9 RNAi roots examined in three independent experiments (Figure 2C). On the other hand, GUS RNAi roots or non-transgenic roots (non-fluorescent roots) that were not removed from the roots systems of PvNF-YA1/9 RNAi composite plants, developed a higher number of nodules when inoculated with the more efficient strain of R. etli SC15 as compared with the less efficient strain 55N1 (Figure 3B,C first and third panels), as previously described (Blanco et al., 2009; Zanetti et al., 2010). These results indicate that PvNF-YA1 and PvNF-YA9 play key functions in nodule formation.
FIGURE 3.
Knock-down of PvNF-YA1/9 abolished nodule formation in P. vulgaris. (A) RT-qPCR analysis of the mRNA levels of PvNF-YA1 (white bars) and PvNF-YA9 (black bars) in GUS RNAi transgenic roots (control) and root systems collected from three independent PvNF-YA1/9 RNAi plants (P1, P2, and P3). Expression data were normalized to PveEF1α gene and are presented relative to the control roots. The error bars represent the SD of three technical replicates. Asterisks indicate statistical significant differences in a t-test with p < 0.05 comparing the values of each gene in each PvNF-YA1/9 RNAi plant with the control GUS RNAi. (B) Number of nodules formed per root in GUS RNAi (control) and PvNF-YA1/9 RNAi composite plants inoculated with the strains of R. etli SC15 and 55N1. Non-transgenic roots were removed before inoculation, thus only nodules formed in transgenic fluorescent hairy roots were registered. The error bars represent the SEM. Data are the average of three independent biological replicates with more than 60 transgenic roots for each condition. Asterisks indicate statistical significant differences in a t-test with p < 0.05 comparing. (C) Images illustrating the absence of nodules in fluorescent roots of plants transformed with the PvNF-YA1/9 RNAi construct and their presence in the non-fluorescent roots that were not excised from the root system. The arrows mark the presence of two nodule primordia or bumps. Pictures were taken at 10 dpi with strains SC15 (upper panels) or 55N1 (lower panels).
Table 1.
Phenotypic analysis of root architecture in GUS RNAi and PvNF-YA1/9 RNAi roots.
| Primary root | Lateral root | Lateral root | |
|---|---|---|---|
| lengtha (cm) | lengthb (cm) | densityc (n/cm ) | |
| GUS RNAi | 9.95 ± 0.40 | 2.20 ± 0.20 | 3.0 ± 0.20 |
| PvNF-YA1/9 RNAi | 9.15 ± 0.87 | 2.10 ± 0.15 | 2.46 ± 0.22 |
aNumber of primary roots analyzed > 50. bNumber of lateral roots analyzed > 100. cDensity is expressed as the number of lateral roots per cm of primary root. Number of primary roots analyzed > 30. Values between GUS RNAi and PvNF-YA1/9 RNAi roots were not significantly different in an unpaired t-test with P < 0.05.
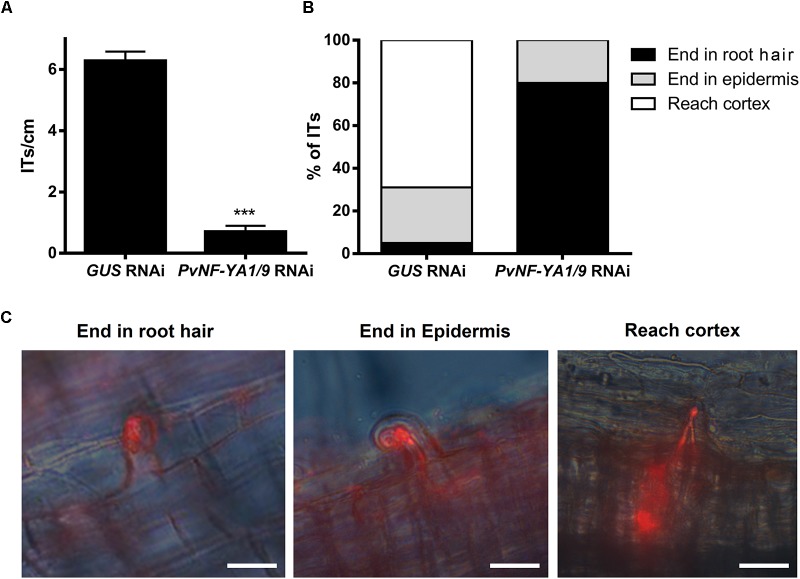
The availability of rhizobia strains expressing fluorescent markers allowed us to evaluate the number of infection events formed, as well as their progression toward cortical cells by fluorescence microscopy. Simultaneous knock-down of PvNF-YA1/A9 in P. vulgaris roots provoked an important reduction in the density of ITs as compared with control roots transformed with the GUS RNAi construct (Figure 4A). In addition, whereas in control plants 70% of ITs progressed and reached the cortical cell layer, nearly 80% of ITs formed in PvNF-YA1/9 RNAi roots aborted in the root hair, 20% of them ended at the base of the epidermal cells, but none progressed to the cortex (Figure 4B,C). These results suggested these two members of the NF-YA family of P. vulgaris would be required for both the initiation and progression of rhizobial infection events.
FIGURE 4.
Knock-down of PvNF-YA1/A9 impaired the initiation and progression of infection events. (A) Number of ITs formed per centimeter of root in GUS RNAi and PvNF-YA1/9 RNAi composite plants. ITs were quantified at 5 dpi with a R. etli strain CFNx5 (nodC-α) expressing the DsRed protein. Root segments from the susceptible zone were excised and visualized under fluorescent microscope. The number of roots segments analyzed was at least 50. Asterisks denote statistical significant differences in a t-test with p < 0.001. Data are the average of three independent biological replicates. (B) ITs formed were classified as events that reached the cortex (white bars), the epidermis (gray bars) or aborted in the root hair (black bars) and expressed as a percentage of the total of infection events. The number of ITs that abort in the root hairs or in the epidermis were significantly higher in the PvNF-YA1/9 RNAi plants than in the GUS RNAi in a t-test with p < 0.05, whereas no events were recorded that reach the cortex in PvNF-YA1/9 RNAi plants. Data are the average of three independent biological replicates. More than 50 root segments were analyzed in each biological replicate. (C) Images illustrating the classification shown in B as ITs that aborted in the root hair, in the epidermal cells, or that reached the cortical cells of GUS RNAi roots.
Knock-Down of PvNF-YA1/A9 Impairs Induction of Early Nodulins and Cell Cycle Genes
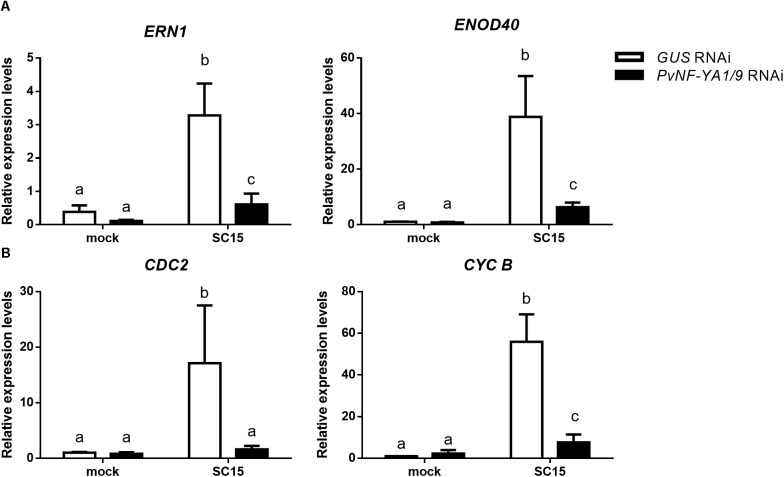
Considering the symbiotic phenotype observed in PvNF-YA1/9 RNAi roots and the well-known function of NF-Y heterotrimers as transcriptional modulators, we investigated whether knock-down of PvNF-YA1/9 affected the rhizobial-induced accumulation of nodulation marker transcripts. We selected ERN1 (ERF Required for Nodulation 1), a mRNA encoding a transcription factor of the ERF (Ethylene Response Factor) family required for nodulation and rhizobial infection (Middleton et al., 2007) and ENOD40 (Early Nodulin 40), a highly structured RNA required for cortical cell divisions that will form nodule primordia (Crespi et al., 1994). Previously, we have shown that these transcripts accumulated in P. vulgaris roots upon infection with R. etli strain SC15 and 55N1 (Zanetti et al., 2010; Mazziotta et al., 2013). RNA-sequencing data confirmed that these two nodulation marker transcripts accumulated to higher levels in wild type roots upon inoculation with either strain SC15 or 55N1 (Supplementary Figure S3; Dalla Via et al., 2015). Consistently with this observation, RT-qPCR experiments conducted in this study showed that both ERN1 and ENOD40 transcript levels increased more than 20 folds in control (GUS RNAi) hairy roots at 24 hpi as compared with non-inoculated GUS RNAi roots. However, PvNF-YA1/9 RNAi roots failed to increase ERN1 transcripts, and showed only a moderate increase (five folds) in ENOD40 transcript levels in response to rhizobial infection (Figure 5A and Supplementary Figure S4). These results suggest that PvNF-YA1/9 subunits might regulate directly or indirectly the expression of early nodulation genes in response to rhizobia.
FIGURE 5.
Rhizobial induction of early nodulins and cell cycle genes is compromised by silencing of PvNF-YA1/A9. Roots expressing GUS (white bars) or PvNF-YA1/9 RNAi (black bars) constructs were inoculated with the R. etli strain SC15 or mock inoculated with the media used to growth the rhizobia (mock). At 24 hpi, transcript levels of (A) nodulin and (B) cell cycle genes were determined by RT-qPCR. Values were normalized to PvEF1α and presented relative to the values of the mock-inoculated GUS RNAi roots. Error bars represent the SEM and different letters indicate that values are significantly different in a t-test with p < 0.05. Data are the average of three independent biological replicates, each performed with at least five independent plants.
In the context of nodulation, it has been shown that genes involved in cell cycle progression are activated at early time points after inoculation with rhizobia (Soyano et al., 2013; Breakspear et al., 2014). Transcripts of two cell cycle genes, cyclin B (CYCB) and CDC2, accumulated to higher levels in P. vulgaris roots in response to strain SC15, but not in response to strain 55N1 (Zanetti et al., 2010; Supplementary Figure S3). Moreover, we have shown that PvNF-YC1 is required for accumulation of CYCB and CDC2 mRNAs during the symbiosis of P. vulgaris with R. etli (Zanetti et al., 2010). Thus, we assessed whether silencing of PvNF-YA1/A9 affected rhizobial-induced transcript accumulation of these cell cycle genes. As shown in Figure 5B, PvNF-YA1/9 RNAi roots failed to accumulated higher transcript levels of CDC2 and CYCB upon rhizobial infection in contrast with that observed in GUS RNAi roots. These results were consistently observed in three independent biological replicates (Supplementary Figure S4), suggesting a possible role of these NF-YA members in the re-activation of mitotic divisions of those cells committed for symbiosis, which is required for initiation of nodule organogenesis.
Overexpression of PvNF-YA1, but Not PvNF-YA9, Enhances the Symbiotic Outcome of a Less Efficient and Competitive Rhizobial Strain
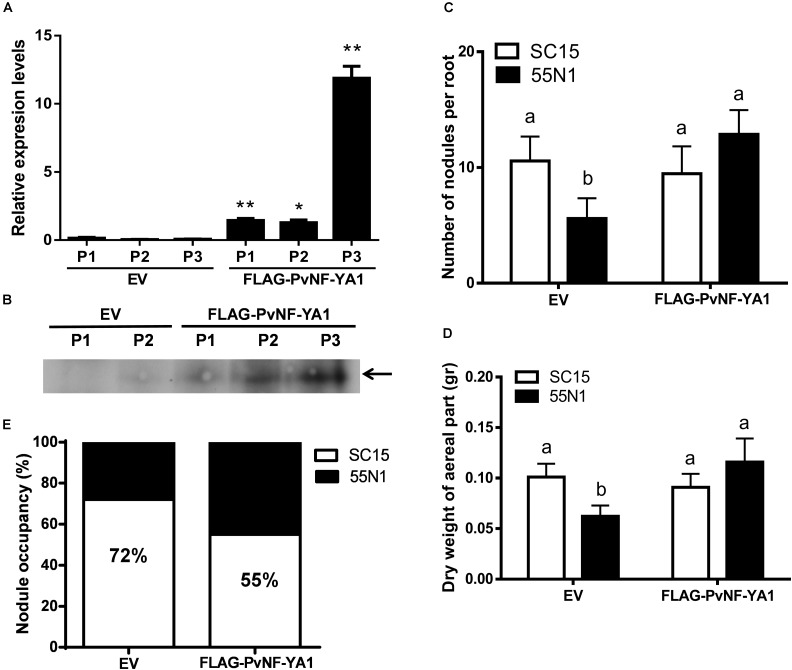
The function of PvNF-YA1 and PvNF-YA9 in the establishment of the interaction between P. vulgaris and different rhizobia strains was investigated using transgenic roots that express a FLAG-tagged version of these subunits under the control of the nearly constitutive promoter Cauliflower Mosaic Virus 35S (CaMV35S), designated as FLAG-PvNF-YA1 and FLAG-PvNF-YA9. Roots transformed with the FLAG-PvNF-YA1 construct exhibited 2–10 fold times higher levels of PvNF-YA1 mRNAs than control roots transformed with the empty vector (EV) as revealed by RT-qPCR in three different composite plant (Figure 6A). Western blot experiments using anti-FLAG antibodies verified the accumulation of the FLAG-PvNF-YA1 proteins and showed a relatively good correlation with PvNF-YA1 transcript levels in the three independent composite plants analyzed here (Figure 6B). Phenotypic analysis of FLAG-NF-YA1 composite plants reveals that overexpression of PvNF-YA1 had no effect on the length of the main and lateral roots (Table 2). FLAG-PvNF-YA1 roots showed a slight increase in the number of lateral roots developed by cm of root (density of lateral roots) compared to control plants transformed with the EV; however, this difference was not statistically significant (Table 2).
FIGURE 6.
Overexpression of PvNF-YA1 enhances nodulation of the less efficient R. etli strain 55N1 and alters nodule occupancy in P. vulgaris. (A) PvNF-YA1 mRNA levels measured by RT-qPCR in control plants [empty vector (EV)] or independent roots of FLAG-PvNF-YA1. Error bars represent the SD of two technical replicates. Expression data were normalized to PveEF1α and presented as relative to EV. P1, P2, and P3 indicate the plant number. Asterisks indicate statistical significant differences in a t-test with P < 0.05. (B) Western blot analysis with α-FLAG antibody on root extracts from EV and FLAG-PvNF-YA1 plants. The band observed in the last three lines corresponds to the FLAG-PvNF-YA1 fusion of 38 kDa (arrow). P1, P2, and P3 correspond to the pool of transgenic roots of three different plants. (C) Number of nodules per root developed in control roots (EV) or FLAG-PvNF-YA1 at 10 dpi with strains of R. etli SC15 (white bars) or 55N1 (black bars). The error bars represent the SEM of the number of nodules formed per root present in more than 50 independent transgenic roots. Different letters indicate significantly different values in a t-test with p < 0.05. Data are the average of three independent biological experiments. (D) Dry weight of the aerial tissue of EV or FLAG-PvNF-YA1 composite plants at 21 dpi with strains of R. etli SC15 or 55N1. Error bars represent the SEM of at least 20 independent plants. Different letters indicate significantly different values in a t-test with p < 0.05. Data are the average of three independent biological experiments. (E) Percentage of nodule occupancy by strains SC15 (white) and 55N1 (black) in co-inoculation experiments. Nodule occupancy was determined at 21 dpi by examination of nodC polymorphic profiles of bacteria isolated from more than 100 individual nodules collected from a minimum of 10 co-inoculated plants for each construction. The results are the average of three independent biological experiments.
Table 2.
Phenotypic analysis of root architecture in EV and FLAG-PvNF-YA1 roots.
| Primary root | Lateral root | Lateral root | |
|---|---|---|---|
| lengtha (cm) | lengthb (cm) | densityc (n/cm) | |
| EV | 11.21 ± 0.45 | 1.18 ± 0.09 | 4.7 ± 0.5 |
| FLAG-PvNF-YA1 | 10.15 ± 0.71 | 1.24 ± 0.12 | 5.8 ± 0.8 |
aNumber of primary roots analyzed > 50. bNumber of lateral roots analyzed > 50. cDensity is expressed as the number of lateral roots per cm of primary root. Number of primary roots analyzed > 25. Values between EV and FLAG-PvNF-YA1 roots were not significantly different in an unpaired t-test with P < 0.05.
To evaluate whether overexpression of PvNF-YA1 affects the symbiotic outcome of P. vulgaris with different R. etli strains, FLAG-PvNF-YA1 and EV roots were inoculated with either the nodC-α strain SC15 or the nodC-δ strain 55N1 of R. etli. Overexpression of NF-YA1 did not produce a significant difference in the number of nodules per root formed by strain SC15 or in the dry weight of the aerial part (Figure 6C,D, white bars). However, upon inoculation with strain 55N1, FLAG-PvNF-YA1 roots formed a significantly greater number of nodules per root than control EV roots (Figure 6C, black bars). This increase in the number of nodules was correlated with an increase in the dry weight of the aerial part of the plant, which could be associated with increased efficiency of N fixation in the FLAG-PvNF-YA1 plants inoculated with 55N1 (Figure 6D, black bars). Notoriously, both parameters—nodule number and dry weight—of FLAG-PvNF-YA1 plants inoculated with 55N1 reached values comparable to those measured when plants were challenged with SC15. All these results show the same trend previously reported in plants overexpressing PvNF-YC1 (Zanetti et al., 2010). On the other hand, constitutive expression of FLAG-PvNF-YA9 in P. vulgaris roots did not affect the number of nodules formed by neither SC15 nor 55N1 strains of R. etli (Supplementary Figures S5A,B).
Previous studies have shown that Mesoamerican accession of P. vulgaris are preferentially nodulated by strains of rhizobia carrying the nodC-α allele when co-inoculated with strains with de nodC-δ allele (Aguilar et al., 2004). Moreover, overexpression of the PvNF-YC1 subunit was sufficient to alter nodule occupancy by these strains (Zanetti et al., 2010). Based on these previous observations and having found that the overexpression of PvNF-YA1 affects the interaction with the less efficient and competitive strain 55N1, we proceeded to assess the role of PvNF-YA1 and PvNF-YA9 in the strain preference observed in Mesoamerican beans. Roots of composite plants were co-inoculated with an equicellular mixture of R. etli strains SC15 and 55N1. Nodules were collected at 21 dpi and the identity of the strain contained within individual nodules was evaluated by analyzing the phenotype of the strains growing in Congo red-YEM (Yeast Extract Mannitol) agar plates or the genotype by detection of the polymorphism of the nodC gene as previously described (Zanetti et al., 2010; Supplementary Figure S6). In control roots, 72% of the nodules were occupied by strain SC15 and only 28% by strain 55N1. Interestingly, FLAG-PvNF-YA1 roots showed an increase of 17% in the nodule occupancy by the strain 55N1 as compared to the control plants (Figure 6E), indicating that constitutive and ectopic expression of PvNF-YA1 is sufficient to alter nodule occupancy in Mesoamerican beans. On the contrary, overexpression of FLAG-PvNF-YA9 did not alter nodule occupancy by SC15 and 55N1 strains (Supplementary Figure S5C). Thus, these results suggest that PvNF-YA1, but not PvNF-YA9, might play functions in the strain preference observed in Mesoamerican beans by the strains carrying the nodC-α allele.
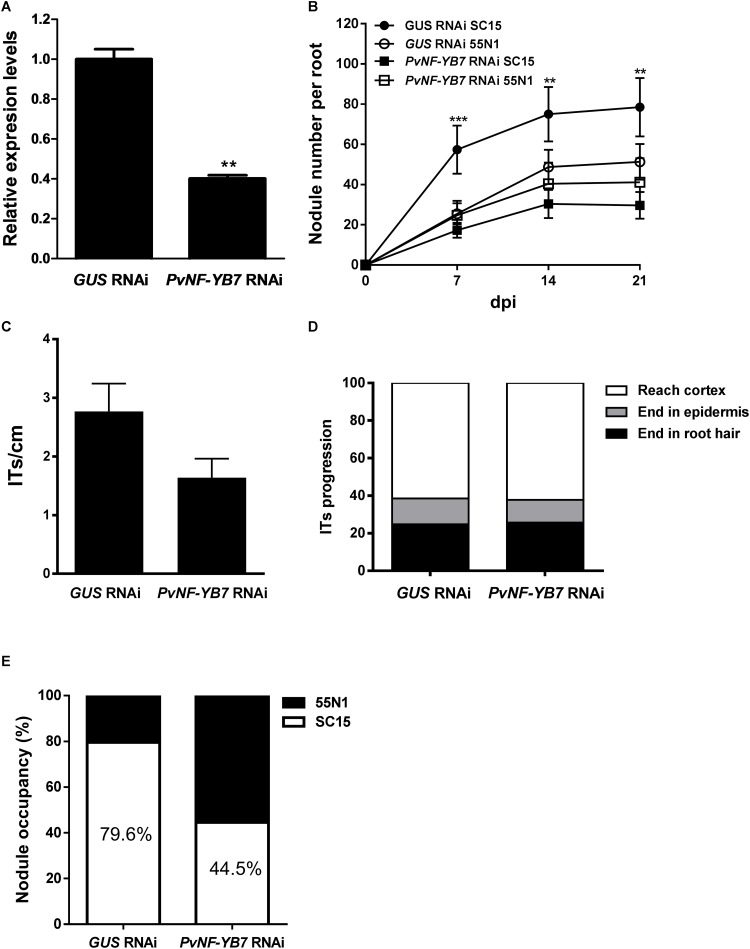
PvNF-YB7 Is Expressed in the Nodule Central Tissue and Required for the Selection of the Highly Efficient Nodulation Strain of R. etli
As already mentioned, the PvNF-YB7 subunit forms a heterotrimer in planta with PvNF-YA1 and PvNF-YC1 subunits, and PvNF-YB7 mRNAs accumulate at higher levels in symbiotic nodules upon inoculation with R. etli. Thus, we focused our analysis in this specific member of the PvNF-YB family. A spatial expression analysis using a promoter:GUS construct was performed in transgenic hairy roots of P. vulgaris. Roots transformed with the PromPvNF-YB7:GUS construct exhibited GUS staining in the vascular tissue of the elongation zone of primary roots, as well as in the lateral root primordia (Figure 7A). GUS staining was also detected in the vasculature of emerged lateral roots (Figure 7B). In already developed lateral roots, expression of PvNF-YB7 was observed in a small number of cells of the lateral root meristem as well as in the vasculature of the susceptible zone (Figure 7C,D). Under symbiotic conditions, GUS staining was observed in the curled root hairs of the susceptible zone (Figure 7E), in the dividing tissue of nodule primordia (Figure 7F), as well as in the base of developing nodule (Figure 7G) and in the central tissue of nodules developed by the R. etli nodC-α strain SC15 (Figure 7H). These results indicate that the PvNF-YB7 promoter is active during nodule formation and support the notion that this subunit plays a relevant function in the establishment of symbiotic nodules. Then, we investigated the functional relevance of PvNF-YB7 in the establishment of an efficient symbiotic interaction and in the strain preference in Mesomearican beans using RNAi-mediated gene silencing. Expression of an RNAi construct specifically designed to silence PvNF-YB7 in P. vulgaris roots resulted in a significant reduction (>40%) of PvNF-YB7 mRNA levels (Figure 8A). Interestingly, PvNF-YB7 RNAi roots inoculated with the nodC-α strain SC15 developed nearly half of the nodules formed in control GUS RNAi roots; however, when inoculated with strain 55N1, PvNF-YB7 RNAi and GUS RNAi roots developed a similar number of nodules (Figure 8B), indicating that PvNF-YB7 subunit might be one of the molecular components of P. vulgaris that are required for the establishment of an interaction with high efficient strains of R. etli. Rhizobial infection was slightly, but not significantly reduced by knock-down of PvNF-YB7 as determined by the frequency of ITs formed by an RFP labeled R. etli strain carrying the nodC-α alelle (Figure 8C). In addition, progression of the infection events was not altered by the reduction in PvNF-YB7 levels (Figure 8D), suggesting that this member of the NF-YB family is not strictly required for the initiation and elongation of ITs. Since silencing of PvNF-YB7 affected nodule formation with SC15 but not with 55N1, we questioned whether this silencing could affect the strain selectivity observed in Mesoamerican beans. Notably, upon co-inoculation with a mixture of strains SC15 and 55N1, the occupancy of the nodules by strain SC15 was reduced in more than 35% in PvNF-YB7 RNAi roots as compared with GUS RNAi roots (Figure 8E). All together, the results presented here support a role for PvNF-YB7 in nodule organogenesis triggered specifically by the high efficient strain SC15.
FIGURE 7.
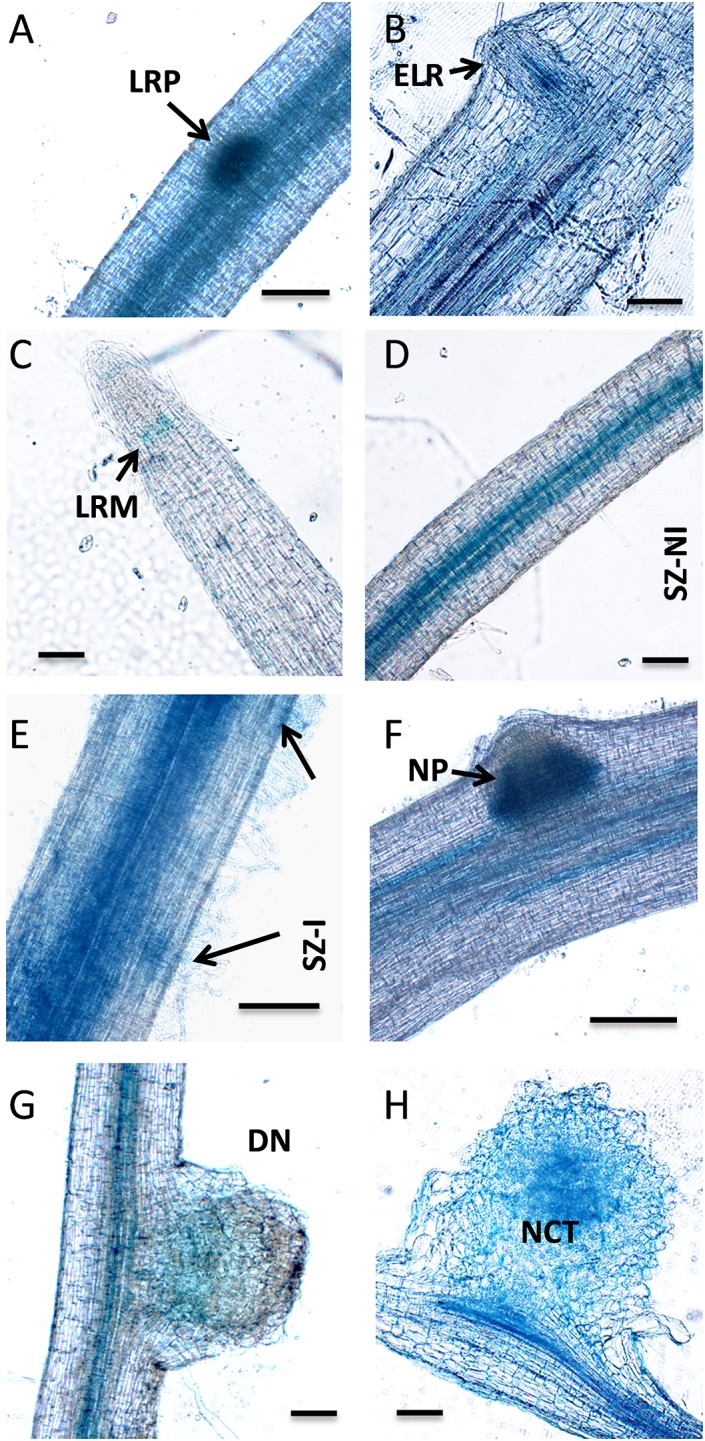
Tissue-specific expression analysis of PvNF-YB7 in roots and nodules of P. vulgaris. Histochemical GUS staining of roots and nodules transformed with the ProPvNF-YB7:GUS construct. (A) Non-inoculated (NI) primary root which containing a lateral root primordia (LRP). (B) Thin section of a primary root containing an emerged lateral root (ELR). (C) Lateral root meristem (LRM) of a NI lateral root. (D) Susceptible zone (SZ) of a NI lateral root. (E) SZ of lateral root inoculated (I) with R. etli strain SC15. Arrows point to infection sites. (F) Nodule primordia (NP) of 6 dpi, (G) developing nodule (DN) of 14 dpi, and (H) mature nodule of 21 dpi formed by strain SC15. NCT: nodule central tissue. Pictures are representative of images observed in more than three independent experiments. Bars: 200 μm.
FIGURE 8.
Knock-down of PvNF-YB7 reduces nodule formation and occupancy by the high efficient R. etli strain SC15. (A) RT-qPCR analysis of the mRNA levels of PvNF-YB7 in GUS RNAi transgenic roots (control) and in roots from three independent PvNF-YB7 RNAi plants. Expression data were normalized to PveEF1α gene and are presented relative to the control roots. Results are the average of two or three technical replicates. The error bars represent the SD. Asterisks indicate statistical significant differences in a t-test with p < 0.01. The results are representative of three independent biological experiments. (B) Number of nodules per root formed in GUS (control) and PvNF-YB7 RNAi composite plants inoculated with the strains of R. etli SC15 and 55N1. The error bars represent the SEM. Data are the average of three independent biological replicates (n > 50 for each condition). Two or three asterisks indicate statistical significant differences in a t-test with p < 0.01 or p < 0.001. (C) Number of ITs formed per centimeter of root in GUS and PvNF-YB7 RNAi composite plants. ITs were quantified at 5 dpi with a R. etli strain CFNx5 (nodC-α) expressing the DsRed protein. Data are the average of three independent biological replicates each with more than 50 roots segments from the susceptible zone. No statistical significant differences were found in a t-test with p < 0.05. Data are the average of three independent biological replicates. (D) ITs were classified as events that reached the cortex (white bars), the epidermis (gray bars), or aborted in the root hair (black bars) and expressed as a percentage of the total infection events. Data are the average of three independent biological replicates each with more than 50 independent roots segments. (E) Percentage of nodule occupancy by strains SC15 (white) and 55N1 (black) in co-inoculation experiments in GUS and PvNF-YB7 RNAi roots. Nodule occupancy was determined at 21 dpi by examination of nodC polymorphic profiles of bacteria isolated from more than 100 individual nodules obtained from at least 10 composite plants. Results are the average of three independent biological replicates.
Discussion
Nuclear Factor Y TFs act as heterotrimers to activate or repress expression of their target genes. Since the individual subunits of the complex are encoded by relatively large gene families in plant genomes, it is crucial to elucidate the composition of the heterotrimers that would be acting in specific tissues or during the activation of morphogenetic programs, such as rhizobial infection and nodule organogenesis, which will largely rely on the tissue-specific expression pattern of individual members of these gene families. Our previous studies have shown that PvNF-YA1, PvNF-YA9, PvNF-YB7 and PvNF-YC1 are expressed in nodules (Ripodas et al., 2015). Here, the use of promoter:GFP-GUS constructs revealed that the promoters of both PvNF-YA1 and Pv-NF-YB7 are active in infected root hairs, as well as in the central tissue of N-fixing nodules developed by the strain of R. etli carrying the nodC-α allele. The expression pattern of these two subunits is reminiscent of that described for LjNF-YA1 and LjNF-YB1 in the legume L. japonicus, in which LjNF-YA1 plays a crucial role in nodule formation (Soyano et al., 2013). In addition, it is consistent with that reported for MtNF-YA1 promoter at early stages of symbiosis between M. truncatula and Sinorhizobium meliloti, in which expression was detected in infected root hairs of the susceptible zone and in the central region of developing nodules (Laporte et al., 2014). The expression pattern of PvNF-YA1 and PvNF-YB7 partially overlap in roots under non-symbiotic conditions. Expression of PvNF-YB7, but not PvNF-YA1, was detected in lateral root primordia, suggesting that they might be acting in different heterotrimeric NF-Y complex at initial steps of lateral root formation, prior emergence. In already emerged lateral roots, expression of PvNF-YA1 and PvNF-YB7 overlap in vascular and meristematic tissue, suggesting that both subunits might be part of a heterotrimer involved in post-emergence lateral root growth. This speculation is consistent with the role assigned for LjNF-YA1 and LjNF-YB1 subunits in lateral root growth (Soyano et al., 2013). Since PvNF-YA1 and PvNF-YB7, together with PvNF-YC1, have been shown to be part of the same hetertorimeric complex in planta and their expression overlap in developing nodules formed by the more efficient strain of R. etli, it is possible to speculate that these two subunits might act in concert to promote nodule formation and/or development during the high efficient interaction established between Mesoamerican beans and strains of R. etli carrying the nodC-α allele. This speculation is supported by the phenotype observed in plants with altered levels of these NF-Y subunits. The results presented here revealed that both PvNF-YA1/A9 and PvNF-YB7 RNAi roots showed a significant reduction in the number of nodules formed by the strain SC15. A previous study indicated that silencing of PvNF-YC1 also prevented nodule formation by this R. etli strain (Zanetti et al., 2010), indicating that the three subunits might act in concert in nodule formation and development triggered by the more efficient strain SC15. On the other hand, when plants were inoculated with the less efficient strain 55N1, nodule formation was also diminished in PvNF-YA1/9 RNAi and PvNF-YC1 RNAi roots, but not in PvNF-YB7 RNAi roots. These results indicate that other NF-YB subunits, e.g., PvNF-YB10 or PvNF-YB12, which are also expressed in nodules (Ripodas et al., 2015), might fulfill this function when plants are challenged by a strain that is less efficient in nodule formation, presumably by heterotrimerization with PvNF-YC1 and PvNF-YA1 or PvNF-YA9. This was not unexpected since expression of a RNAi designed to specifically silence MtNF-YB16 or a less specific RNAi that simultaneous knock-down four NF-YB gene family members (MtNF-YB16/B18/B6/B11) in M. truncatula did not revealed any noticeable symbiotic phenotype upon inoculation with S. meliloti (Baudin et al., 2015). In addition, two independent studies based on yeast-two hybrid assays performed with Arabidopsis NF-Y subunits revealed that NF-YB subunits tend to be more promiscuous in heterotrimer formation than NF-YA and NF-YC subunits (Calvenzani et al., 2012; Hackenberg et al., 2012). The fact that PvNF-YB7 RNAi plants form nodules with both R. etli strains points toward a low degree of specificity in the NF-YB association for nodule formation, as previously proposed for the M. truncatula ×S. meliloti interaction (Baudin et al., 2015). However, the observation that PvNF-YB7 seems to be required for a high efficient nodule formation leads to speculate that the symbiotic outcome, at least in the case of P. vulgaris ×R. etli interaction, might depend on strict NF-Y heterotrimer formation.
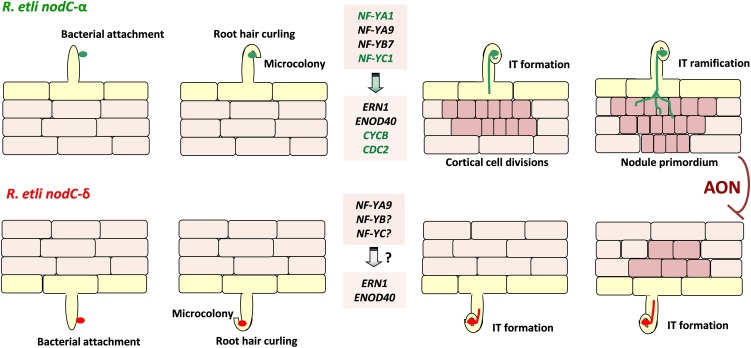
An interesting finding of this study is the observation that overexpression of PvNF-YA1, but not of PvNF-YA9, increased the efficiency of nodule formation when plants were inoculated with the less efficient strain 55N1; moreover, it was sufficient to increase the proportion of nodules occupied by 55N1 when co-inoculated with the higher competitive strain SC15. It must also be noted that PvNF-YA1 proved to be responsive only to strain SC15, whereas PvNF-YA9 expression is activated by both strains (Ripodas et al., 2015). Thus, PvNF-YA9 appears to be part of the more general pathway triggered by high or low efficient strains, which might explain why overexpression of this subunit did not altered nodule number or their occupancy. On the other hand, PvNF-YA1 seems to be part of the signaling pathway activated only by the high efficient strain SC15. The phenotype observed in PvNF-YA1 overexpressing roots is reminiscent of that previously described in plants overexpressing PvNF-YC1 (Zanetti et al., 2010). Considering the fact that PvNF-YA1 and PvNF-YC1 are able to interact in the same NF-Y trimeric complex (Baudin et al., 2015), we suggest that increased levels of both PvNF-YA1 and PvNF-YC1 could help to improve the symbiotic capacity of less efficient rhizobia. Moreover, these two NF-Y subunits are part of a symbiotic heterotrimeric TF that might work in the activation of the genetic program that allows Mesoamerican plants to discriminate between strains that are more or less efficient in nodule formation. A striking result regarding nodule occupancy by SC15 and 55N1 strains was obtained in PvNF-YB7 RNAi plants. In these plants, nodules occupied by SC15 decreased by about 35% compared to controls. This might be most likely a consequence of the reduced capacity of these plants to form nodules upon inoculation with strain SC15. Thus, we concluded that PvNF-YB7 is required for high efficient nodule organogenesis in Mesoamerican P. vulgaris roots. This observation also reinforces the hypothesis that this specific NF-YB subunit might be an additional component of the genetic programs that determine the selectivity displayed by P. vulgaris plants for those strains that perform better in nodule formation. Based on this and previous studies (Zanetti et al., 2010; Ripodas et al., 2015), we propose a model that attempt to explain how PvNF-YA1, PVNF-YB7, and PvNF-YC1 function to control the strain preference observed in Mesoamerican accessions of P. vulgaris (Figure 9). In this model, specific activation of PvNF-YA1 and PvNF-YC1 in response to nodC-α strains, such as SC15, lead to a rapid activation of the morphological responses, such as cortical cell divisions (possibly due the specific and rapid activation of cell cycle genes) and progression of IT toward nodule primordia. These responses result in the formation of nodules occupied by nodC-α strains. On the other hand, nodC-δ strains fail to rapidly induce PvNF-YA1 and PvNF-YC1, thus these morphological responses occurred slower in the presence of strains such as 55N1; as a consequence, nodules formed by these strains will be delayed. During symbiosis, existing N-fixing nodules inhibit the formation of new nodules by a mechanism referred to as autoregulation of nodulation (AON), which involves long distance signaling from root to shoot and back again (Ferguson et al., 2014). Within this context, the earliest formation of nodules occupied by a nodC-α strain will inhibit the formation of new nodules that might be colonized by a nodC-δ strain, explaining why in co-inoculation experiments or in the field, the majority of the nodules formed in Measomerican beans are occupied by nodC-α strains.
FIGURE 9.
Model illustrating the proposed function of the PvNF-YA1/-YB7/-YC1 heterotrimer in the strain preference observed in the P. vulgaris × R. etli interaction. Perception of R. etli strains nodC-α (represented in green, upper panels) by Mesoamerican P. vulgaris plants results in the root hair curling that entrap bacteria to form a microcolony. This perception also rapidly (within 24 hpi) and locally induces expression of genes encoding the PvNF-YA1 (and PvNF-YA9) and PvNF-YC1 subunits, which form a hetrotrimeric transcription factor with PvNF-YB7. Heterotrimer formation activates the expression of nodulation (ERN1 and ENOD40) and the cell cycle (CYCB and CDC2) genes, promoting infection thread (IT) formation and cortical cell divisions. The IT progresses, ramifies, and reaches the nodule primordium to produce a functional nodule occupied by the nodC-α strain. On the other hand, perception of nodC-δ strains (represented in red, lower panels) leads to root hair curling, microcolony formation, and induction of PvNF-YA9, ERN1, and ENOD40 in a similar way that nodC-α strains. However, induction of PvNF-YA1 and PvNF-YC1, as well as CYCB and CDC2, is not observed at 24 hpi, delaying cortical cell divisions and the progression of ITs that contain the nodC-δ strain. As a consequence of these local responses, when both strains are present (e.g., in coinoculation experiments or in the field when both bacteria are present in the soil with even abundance) nodules are mainly occupied by the nodC-α strains. Since nodulation is systemically repressed by a systemic mechanisms referred as autoregulation of nodulation (AON), new nodule formation triggered by the nodC-δ strain is arrested once the appropriate number of nodules is reached. In this model, genes specifically induced at 24 hpi with the nodC-α strain are colored in green. PvNF-YA9 and other NF-YB and NF-YC subunits, as well as ERN1 and ENOD40, are activated also by the nodC-δ strain.
Infection events and their progression to the cortical cells were drastically reduced by introduction of the PvNF-YA1/9 RNAi construct in P. vulgaris roots. Moreover, no ITs reaching the cortex were detected in PvNF-YA1/9 RNAi roots, at least at the time points analyzed here, indicating that they might have aborted either in the root hair or the epidermis. Thus, these two NF-YA subunits play not only an important role in nodule organogenesis and development, but also in the epidermal and/or cortical responses that lead to a successful infection of the nodule. This phenotype resembles that previously reported by Laloum et al. (2014) in M. truncatula plants with reduced levels of MtNF-YA1 and MtNF-YA2 (the orthologs of PvNF-YA9 and PvNF-YA1, respectively), where both frequency and progression of ITs were impaired. This phenotype is more pronounced than that observed in M. truncatula that are null mutants for the MtNF-YA1 allele, which exhibited an increased number of ITs, but the morphology of ITs was abnormal and IT growth was arrested (Laporte et al., 2014). In contrast, L. japonicus plants with reduced levels of a single NF-YA subunit, LjNF-YA1 (the ortholog of PvNF-YA9 and MtNF-YA1 of P. vulgaris and M. truncatula, respectively), produced a normal amount of ITs without evidenced of IT arresting (Soyano et al., 2013), suggesting some functional redundancy with other NF-YA subunits in this legume species. Based on our results and those described by others, it is possible to conclude that two symbiotic NF-YA subunits play partially redundant and essential functions in the sophisticated intracellular infection by rhizobia observed in many legume species. On the other hand, expression of the PvNF-YB7 RNAi construct did not produce any strong phenotype in the infection by rhizobia; except by a mild, but not significant reduction in the frequency of ITs. This indicates that other NF-YB subunits of P. vulgaris might exert redundant functions in the genetic program that leads to this intracellular mechanism of infection.
Nuclear Factor Y subunits have been involved in the control of cell division activities during the development of lateral root organs, either lateral roots or symbiotic nodules (Zanetti et al., 2010; Soyano et al., 2013; Sorin et al., 2014). Here, we found that lateral root growth or density was not significantly affected by knock-down of PvNF-YA1 and PvNF-YA9, indicating that these subunits do not seem to be required or play redundant functions with other NF-YA members during the activation of cell divisions related to the initiation or growth of lateral roots. However, rhizobial induction of cell cycle genes such as CDC2 and CYCB was almost completely abolished in PvNF-YA1/9 RNAi roots, which is well correlated with the absence of nodule formation observed in these roots. In addition, induction of ENOD40, which is also required for cortical cell division activity during formation of nodule primordia, is reduced in PvNF-YA1/9 RNAi roots. These results indicate that PvNF-YA1 and PvNF-YA9 participate not only in the transcriptional activation of early markers of infection such as ERN1, a direct target of the symbiotic NF-Y complex (Laloum et al., 2014; Baudin et al., 2015), but also in the control of cell division activities during nodule formation. A previous study revealed that the PvNF-YC1 subunit is also involved in the control of CDC2 and CYCB during nodule formation (Zanetti et al., 2010). Future experiments will help to elucidate whether these and other cell cycle related genes are direct transcriptional targets of the symbiotic complex formed by the PvNF-YA1/9, PvNF-YB7 and PvNF-YC1 in P. vulgaris during the interaction with high or low efficient strains of R. etli.
Author Contributions
MZ, FB, and CR conceived the research and designed the experiments. CR, MC, and JC performed the experiments with help from JV. MZ, CR, and JC analyzed the experimental results. CR and MZ wrote the manuscript. All the authors discussed the results and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) PICT2013-0384, PICT2014-0321, and PICT 2016-0582.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00221/full#supplementary-material
References
- Aguilar O. M., Riva O., Peltzer E. (2004). Analysis of Rhizobium etli and of its symbiosis with wild Phaseolus vulgaris supports coevolution in centers of host diversification. Proc. Natl. Acad. Sci. U.S.A. 101 13548–13553. 10.1073/pnas.0405321101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Rípodas C., Clúa J., Baudin M., Aguilar O. M., Niebel A., et al. (2014). A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol. 164 1430–1442. 10.1104/pp.113.230896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M., Laloum T., Lepage A., Rípodas C., Ariel F., Frances L., et al. (2015). A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 169 2761–2773. 10.1104/pp.15.01144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitocchi E., Bellucci E., Giardini A., Rau D., Rodriguez M., Biagetti E., et al. (2013). Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 197 300–313. 10.1111/j.1469-8137.2012.04377.x [DOI] [PubMed] [Google Scholar]
- Bitocchi E., Nanni L., Bellucci E., Rossi M., Giardini A., Zeuli P. S., et al. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. U.S.A. 109 E788–E796. 10.1073/pnas.1108973109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F. A., Meschini E. P., Zanetti M. E., Aguilar O. M. (2009). A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21 2797–2810. 10.1105/tpc.108.063420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26 4680–4701. 10.1105/tpc.114.133496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., et al. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7:e42902. 10.1371/journal.pone.0042902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J. P., de Billy F., Gamas P., Niebel A., Rivas S. (2008). Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 22 1549–1559. 10.1101/gad.461808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J. P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., et al. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20 3084–3088. 10.1101/gad.402806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M. D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E., et al. (1994). enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13 5099–5112. 10.1002/j.1460-2075.1994.tb06839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Via V., Narduzzi C., Aguilar O. M., Zanetti M. E., Blanco F. A. (2015). Changes in the common bean transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol. 169 1356–1370. 10.1104/pp.15.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarie J., Debelle F., Prome J. C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65 503–535. 10.1146/annurev.bi.65.070196.002443 [DOI] [PubMed] [Google Scholar]
- Ferguson B. J., Li D., Hastwell A. H., Reid D. E., Li Y., Jackson S. A., et al. (2014). The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J. 12 1085–1097. 10.1111/pbi.12216 [DOI] [PubMed] [Google Scholar]
- Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. (2012). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5 876–888. 10.1093/mp/ssr107 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Laloum T., Baudin M., Frances L., Lepage A., Billault-Penneteau B., Cerri M. R., et al. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79 757–768. 10.1111/tpj.12587 [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18 157–166. 10.1016/j.tplants.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M. F., et al. (2014). The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 65 481–494. 10.1093/jxb/ert392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344 781–784. 10.1038/344781a0 [DOI] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 782–796. 10.1105/tpc.109.072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta L., Reyonoso M. A., Aguilar O. M., Blanco F. A., Zanetti M. E. (2013). Transcriptional and functional variation of NF-YC1 in genetically diverse accessions of Phaseolus vulgaris during the symbiotic association with Rhizobium etli. Plant Biol. 15 808–818. 10.1111/j.1438-8677.2012.00683.x [DOI] [PubMed] [Google Scholar]
- Meschini E. P., Blanco F. A., Zanetti M. E., Beker M. P., Küster H., Pühler A., et al. (2008). Host genes involved in nodulation preference in common bean (Phaseolus vulgaris)-Rhizobium etli symbiosis revealed by suppressive subtractive hybridization. Mol. Plant Microbe Interact. 21 459–468. 10.1094/MPMI-21-4-0459 [DOI] [PubMed] [Google Scholar]
- Middleton P. H., Jakab J., Penmetsa R. V., Starker C. G., Doll J., Kaló P., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234. 10.1105/tpc.106.048264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M. E., Jang C. J., Holtan H. E., Repetti P. P., Galbraith D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 18843–18848. 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R. W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24 4777–4792. 10.1105/tpc.112.105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C., Ott T. (2011). Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr. Opin. Plant Biol. 14 458–467. 10.1016/j.pbi.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Ripodas C., Castaingts M., Clua J., Blanco F., Zanetti M. E. (2015). Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris). Front. Plant Sci. 5:761. 10.3389/fpls.2014.00761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripodas C., Via V. D., Aguilar O. M., Zanetti M. E., Blanco F. A. (2013). Knock-down of a member of the isoflavone reductase gene family impairs plant growth and nodulation in Phaseolus vulgaris. Plant Physiol. Biochem. 68 81–89. 10.1016/j.plaphy.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Sorin C., Declerck M., Christ A., Blein T., Ma L., Lelandais-Brière C., et al. (2014). A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 202 1197–1211. 10.1111/nph.12735 [DOI] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9:e1003352. 10.1371/journal.pgen.1003352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. Mar. 33 949–956. 10.1046/j.1365-313X.2003.01676.x [DOI] [PubMed] [Google Scholar]
- Wang D., Maughan M. W., Sun J., Feng X., Miguez F., Lee D., et al. (2012). Impact of nitrogen allocation on growth and photosynthesis of miscanthus (Miscanthus × giganteus). Glob. Change Biol. Bioenergy 4 688–697. 10.1111/j.1757-1707.2012.01167.x [DOI] [Google Scholar]
- Weston L. A., Mathesius U. (2013) Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 39 283–297. 10.1007/s10886-013-0248-5 [DOI] [PubMed] [Google Scholar]
- Zanetti M. E., Blanco F. A., Beker M. P., Battaglia M., Aguilar O. M. (2010). A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22 4142–4157. 10.1105/tpc.110.079137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.