Abstract
Molecular testing is increasingly important in cancer diagnosis. Targeted next generation sequencing (NGS) is widely accepted method but structural variation (SV) detection by targeted NGS remains challenging. In the brain tumor, identification of molecular alterations, including 1p/19q co-deletion, is essential for accurate glial tumor classification. Hence, we used targeted NGS to detect 1p/19q co-deletion using a newly developed deep learning (DL) model in 61 tumors, including 19 oligodendroglial tumors. An ensemble 1-dimentional convolution neural network was developed and used to detect the 1p/19q co-deletion. External validation was performed using 427 low-grade glial tumors from The Cancer Genome Atlas (TCGA). Manual review of the copy number plot from the targeted NGS identified the 1p/19q co-deletion in all 19 oligodendroglial tumors. Our DL model also perfectly detected the 1p/19q co-deletion (area under the curve, AUC = 1) in the testing set, and yielded reproducible results (AUC = 0.9652) in the validation set (n = 427), although the validation data were generated on a completely different platform (SNP Array 6.0 platform). In conclusion, targeted NGS using a cancer gene panel is a promising approach for classifying glial tumors, and DL can be successfully integrated for the SV detection in NGS data.
Introduction
The paradigm of cancer diagnosis has recently changed from a histology-based approach to a strategy that combines histology and molecular features. In particular, molecular testing has become essential for diagnosis of tumors of the central nervous system1. For example, to diagnose oligodendroglial tumors, mutation in the IDH gene and co-deletion of 1p/19q must be detected. Currently, fluorescence in situ hybridization (FISH) is the gold standard method for detecting 1p/19q co-deletion2–6; however, this test can yield false positive results because FISH probes cannot cover the whole chromosome arm7; moreover, interpretation is time-consuming and labor-intensive.
Currently, next-generation sequencing (NGS) using a targeted cancer gene panel has become a popular diagnostic method, and has been adopted in many hospitals8. However, clinical NGS testing focuses mainly on detection of mutations such as single-nucleotide polymorphisms (SNPs) and small indels. Although copy number variation (CNV) can also be detected by targeted NGS, and a few software packages have been developed for this purpose9–11, CNV detection via NGS remains challenging, and manual reviews of CNV plots are required for accurate results. The 1p/19q co-deletion associated with oligodendroglial tumor is characterized by simultaneous whole-arm deletion of chromosome 1 and chromosome 19; therefore, arm-level evaluation is required. However, detection of such large deletion is not well supported by CNV detection programs for targeted NGS. In addition, using only tumor tissue without matched normal tissue makes it difficult to accurately detect CNV in clinical targeted NGS tests. Therefore, it is necessary to evaluate whether targeted NGS can detect 1p/19q co-deletion with sufficient accuracy for clinical use. If targeted NGS could be used to detect 1p/19q co-deletion as well as IDH mutation, it would have marked advantages over standard molecular tests such as FISH, immunohistochemistry, and Sanger sequencing.
Currently, many researchers are investigating the application of deep learning (DL) into various diagnostic areas. Many studies of DL have focused on diagnosis using histologic and radiologic images. Recently, however, DL has been applied to analysis of sequencing to improve the sensitivity and specificity of SNP and indel detection12. We speculated that CNV detection would also be a good candidate for application of DL. Hence, we developed a DL model for analyzing copy number variation in NGS data from a targeted cancer panel, with the goal of detecting 1p/19q co-deletion.
Results
Summary of targeted NGS testing and diagnostic performance
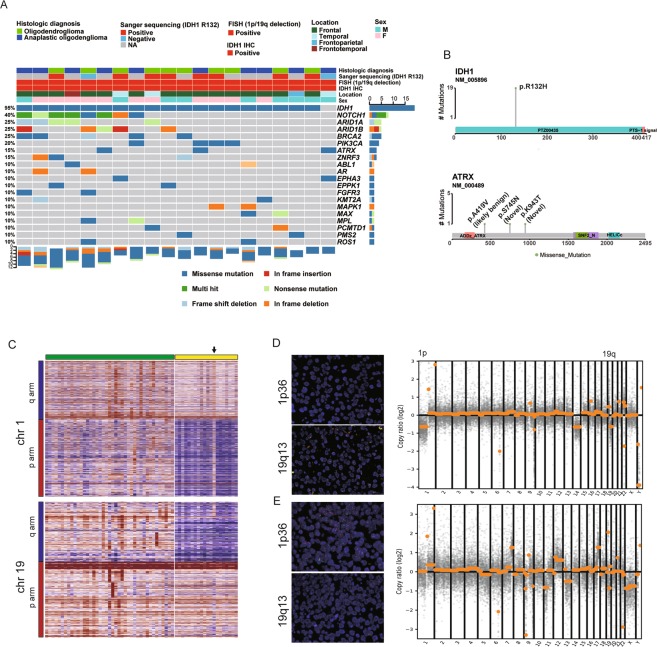
Single gene–based molecular test results for initial 20 cases, including evaluation of IDH1 mutation status by Sanger sequencing and immunohistochemistry and 1p/19q co-deletion status by FISH, are shown in Fig. 1A. Primary histologic diagnosis was based on histology and single gene–based molecular tests. Among the 20 cases, 2 were interpreted as negative for IDH1 mutation by Sanger sequencing, but targeted NGS testing revealed the R132H mutation in one of them (Fig. 1B). The other case that was negative in Sanger sequencing was also negative in targeted NGS testing. Thus, Sanger sequencing yielded a false negative result in one case. The other 18 cases were concordant between Sanger sequencing and targeted NGS testing for positive IDH mutation. ATRX mutations were found in three samples, but these mutations are not known to be of biological significance (Fig. 1B).
Figure 1.
Summary of detected variants using targeted next generation sequencing (NGS) in an initial set of 20 enrolled tumors with various clinicopathological factors (A). Mutations with protein annotation of IDH1 and ATRX (B). Copy number heatmap for 1p and 19q in 20 enrolled cases (red bar) and 41 controlled cases with no 1p/19q deletion (green bar) (C). Representative 1p/19q co-deletion in fluorescence in situ hybridization (FISH) and copy number variation (CNV) plot of targeted NGS (D), and one false-positive case (E).
Manual review of the copy number plot from the targeted NGS testing identified 1p/19q co-deletion in 19 of 20 cases (Fig. 1C, yellow bar, and Fig. 1D). One case harbored only partial deletion of 19q and no 1p deletion (Fig. 1C, arrow), and review of the FISH results by experts (S.J.N. and C.O.S.) suggested false positive (Fig. 1E). This case was also negative for IDH mutation in both Sanger sequencing and NGS testing. Therefore, this case was excluded for further analysis. None of the negative control samples harbored 1p/19q co-deletion (Fig. 1C, green bar). Overall, these findings demonstrate that targeted NGS testing can detect 1p/19q co-deletion and IDH gene mutation with high accuracy.
Mutations according to histologic grade of oligodendroglial tumors
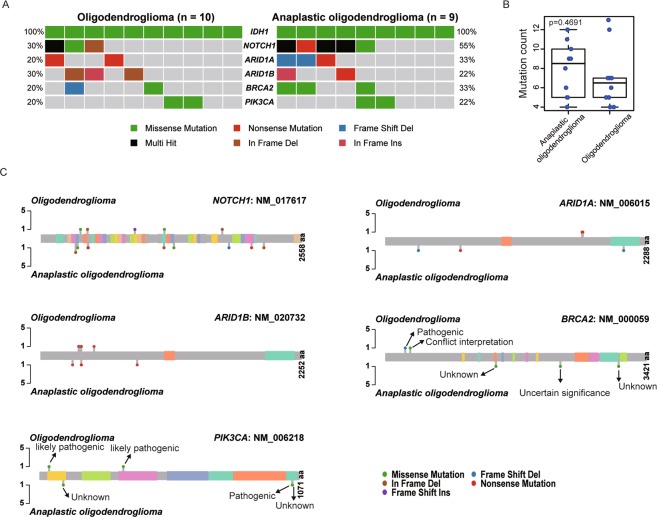
Oligodendroglioma (WHO grade II) and anaplastic oligodendroglioma (WHO grade III) have different histologic features and clinical behaviors1; therefore, it may be interesting to know how mutational features in this data set correlate with histologic grade. Thus, we additionally analyzed mutation features according to histologic grade. In addition to IDH1, frequently mutated genes included NOTCH1 (42.1%), ARID1A (26.3%), ARID1B (26.3%), BRCA2 (26.3%), and PIK3CA (21.0%), but mutation frequencies (Fig. 2A) and mutation number (Fig. 2B) did not differ significantly between oligodendroglioma and anaplastic oligodendroglioma. Details of the mutations in these five genes are shown in Fig. 2C. Interestingly, one pathogenic BRCA2 mutation (p.N257Kfs*17; rs80359671) was present, but the other BRCA2 mutations were of uncertain significance or their biological significance was previously unreported.
Figure 2.
Major variants detected in 19 oligodendroglial tumors, including 10 oligodendroglioma and 9 anaplastic oligodendroglioma (A). Total mutation counts in oligodendroglioma and anaplastic oligodendroglioma (B). Mutation locus of each variant between oligodendroglioma and anaplastic oligodendroglioma (C).
Development of ensemble one-dimensional convolution neural network for detection of 1p/19q co-deletion
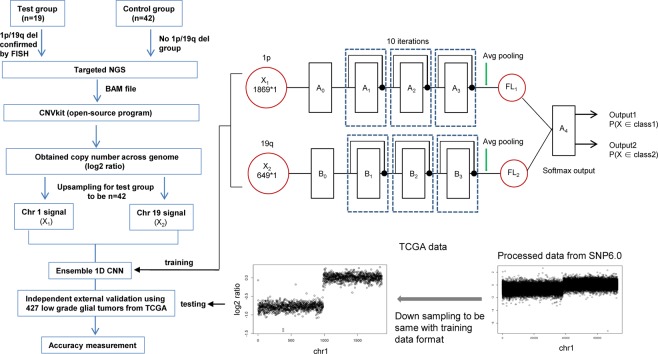
Finally, 19 cases with 1p/19q co-deletion and 42 control cases lacking such a deletion were used as a test group for model building. To address the group unbalance problem between test and control groups, the test group was up-sampled using SOMTE (Synthetic Minority Over-sampling Technique)13 to 42 to match the number in the control group. Log2 copy number ratios for chr1 and chr19 were extracted from whole chromosomes, and the signals were converted to 1869 × 1 (X1) and 649 × 1 (X2) vectors, respectively. These two data sets (chr1 andchr19) have completely different input variables. We used the ensemble method, a technique that creates multiple models and then combines them to obtain improved results, for the two data sets illustrated in Fig. 3. For the proposed ensemble model, we first applied a one-dimensional convolution neural network (1D-CNN) to the corresponding data set and obtained feature values for all cases after the uppermost pooling layer. We then concatenated these feature values and utilized them as input values of a fully connected neural network equipped with a softmax output layer for classification. External validation of this model was performed using 427 low-grade glial tumors from The Cancer Genome Atlas (TCGA) data. These data were generated using Affymetrix SNP array 6.0 platform. The log2 ratios were calculated from tumor and matched normal tissue, and then the log2 data were down-sampled to the same vector size used in the training model. The overall workflow is summarized in Fig. 3.
Figure 3.
Established pipeline for detection of 1p/19q co-deletion using ensemble 1D convolutional neural network (1D CNN) from targeted next generation sequencing (NGS) data and external validation using copy number data of 427 low grade glial tumors from SNP Array 6.0 platform downloaded from TCGA. A0 denotes the pretraining block, whose architecture is (170*1 convolution, 32)-(2*1 max pool)-(151*1 convolution, 64)-(2*1 max pool)-(51*1 convolution, 64)-(2*1 max pool), where (170*1 convolution, 32) indicates a convolution layer composed of 32 kernels with height = 170 and width = 1, and (2*1 max pool) indicates a max pooling layer of pooling windows with height = 2 and width = 1. B0 denotes the pretraining block, whose architecture is (150*1 convolution, 32)-(2*1 max pool)-(51*1 convolution, 64)-(2*1 max pool)-(51*1 convolution, 64)-(2*1 max pool). A1 denotes the first residual block, whose architecture is (1*1 convolution, 32)-(3*1 convolution, 32)-(1*1 convolution, 128); A2 the second residual block, whose architecture is (1*1 convolution, 64)-(3*1 convolution, 64)-(1*1 convolution, 256); and A3 the third residual block, whose architecture is (1*1 convolution, 128)-(3*1 convolution, 128)-(1*1 convolution, 512). The dotted rectangle implies that the procedures inside are executed over 10 iterations. FL1 and FL2 denote the 1024*1 flattened vectors. A4 denotes the 2048*2 softmax output layer. Output1 and Output2 indicate outputs from the softmax output layer, whose values are P(X ∈ class1) and P(X ∈ class2), respectively, where X = X1 ∪ X2. Classes 1 and 2 indicate 1p/19q co-deletion and no 1p/19q co-deletion, respectively.
Diagnostic accuracy of the DL model established using ensemble 1D-CNN
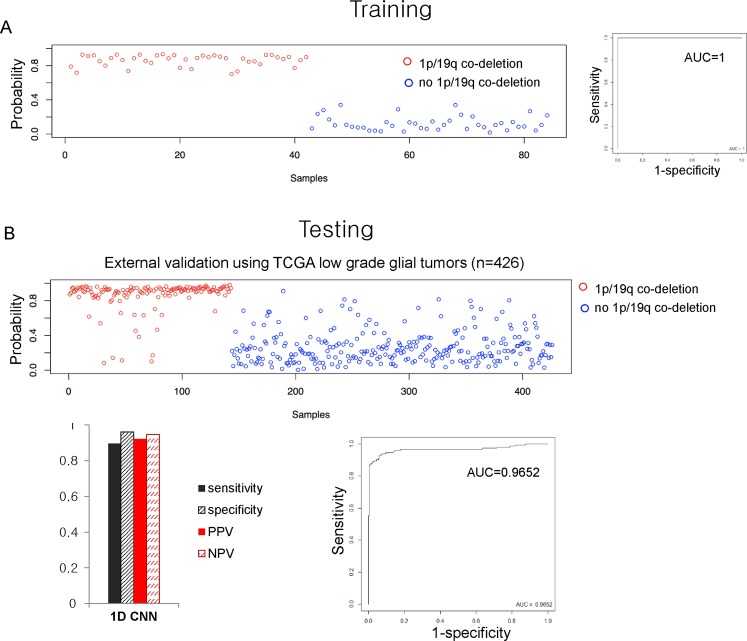
The build model was trained using the AMC (Asan Medical Center, Seoul, Korea) data set generated by targeted NGS. For detection of 1p/19q co-deletion, the area under the curve (AUC) of the trained DL model was 1 (Fig. 4A). The trained model was then tested using an independent data set consisting of 426 low-grade glial tumors from TCGA, processed as described above. The optimal cut-off point was determined by maximizing the Youden index (Sensitivity + Specificity − 1) based on the predicted probability of training data set, and then the optimal cut-off was used to calculate the sensitivity and specificity of the validation set (0.8951 and 0.9611, respectively; AUC, 0.9652) (Fig. 4B).
Figure 4.
Accuracy of deep learning model for detection of 1p/19q co-deletion. The accuracy of deep learning model in the training set to detect 1p/19q co-deletion from targeted next generation sequencing data was 100% (A). This model was externally validated using copy number data from the SNP Array 6.0 platform downloaded from TCGA and area under the curve (AUC) value was measured as 0.9652.
Discussion
In this study, we demonstrated that targeted NGS testing can accurately detect 1p/19q co-deletion. In addition, NGS testing has a comparative advantage in IDH gene detection: namely, NGS testing has greater sensitivity than Sanger sequencing and can cover the entire exome of the IDH1 and IDH2 genes. Recent studies also showed that targeted NGS testing is a promising diagnostic method for classifying glial tumors of the CNS14,15. However, the cancer gene panels used for targeted NGS are diverse, and different panels are used by various hospitals. To detect CNV by targeted NGS, a cancer panel must be designed to cover the whole chromosome, and internal controls to adjust for uneven amplification in NGS may be required. The cancer panel used in this study included hot spot mutations that occur frequently in the Korean population, but did not have an internal control to adjust CNV. Nonetheless, NGS testing using this cancer panel correctly detected 1p/19q co-deletion.
Recently, DL has been applied to diagnostics, focusing on imaging-based diagnoses such as mitotic count evaluation16,17 and detection of metastatic tumor in sentinel lymph nodes18. DL has the potential to be applied to all types of medical images, including radiologic imaging, EKG imaging, and photographs of skin lesions. However, the most important problem to overcome in this context is low accuracy in external validation. This accuracy issue can be attributed to many factors, including differences in background image color, inaccurate annotation of the target area, and heterogeneity of the target area. Therefore, images generated by computing are good candidates for application of DL. Recent studies reported major improvements in SNP/indel detection from NGS genomic data12. In this study, we showed that CNV from targeted NGS data can also be accurately detected using DL, even with a small training sample size. This was possible because all copy number data were generated in a regular format using a computational approach and the distinctive features of 1p/19q co-deletion in the CNV plot image. For external validation, we used CNV data from the SNP Array 6.0 platform. Even though completely different platforms were used to generate the training and validation sets, we achieved good accuracy in the validation set despite the small number of samples used for training.
Initially, we used a CNN with a whole CNV plot image to detect 1p/19q co-deletion, but this approach was associated with an unacceptably large computational workload. In addition, simultaneous detection of each 1p and 19q deletion is needed to detect the 1p/19q co-deletion, because this co-deletion is required for diagnosis of oligodendroglioma. In this study, we developed a new ensemble 1D-CNN method for detecting 1p/19q co-deletion from CNV data. The two input data sets for 1p and 19q have completely different input variables. For classification, we could combine the two data sets into a single data set and apply a classifier to these data, but to achieve better results, we used an ensemble method. Although the model was developed for the detection of the 1p/19q co-deletion, it could also be used to detect other chromosomal structural variations with similar patterns, such as arm level broad copy number amplification and/or deletion in other chromosomal loci, irrespective of tumor type. However, in this study, we did not test the detection ability of multiple forms of 1p/19q co-deletion, such as partial deletions, because our test group was small in number and did not include several multiple forms of partial deletions.
In this analysis, the relative order of bins on a chromosome was used to summarize the chromosome into a vector. The actual start location and length of each bin on chromosomes at 1 bp were omitted to reduce the length of the summarized vector. Consequently, the vector could not represent the actual scale and location of indel events. However, the adjusted log2 ratio in the CNR file from the CNVkit9 program was already adjusted to represent the length of the bin, and the indel events targeted in this study are of very large scale, occupying whole chromosome arms. Therefore, we determined that some distortion in the summarized vector is acceptable in exchange for accurate identification of arm deletion.
For the same reason, the TCGA data used as a validation set were preprocessed using the relative order of the bins and average log2 values. Moreover, if the summarized vector was made to represent actual scale and location, it would be ten millions of components in length for a single chromosome. Input of such a long vector would require too much computing power and data to train the CNN network.
As described above, we identified one false positive in the FISH test, suggesting that methods based on NGS could yield more accurate and objective results than conventional FISH.
In summary, this study showed that targeted NGS testing can accurately detect 1p/19q co-deletion, suggesting that this method represents a promising approach for classifying glial tumor. In addition, we applied DL to identify the co-deletion. This method yielded reproducible result in a validation set collected and processed at a different institution on a different platform. This result proves the robustness of the DL, and encourages application of DL to future NGS-based genomic analyses.
Materials and Methods
Samples
This was a retrospective study. The study initially enrolled 20 cases of oligodendroglial tumors (10 oligodendrogliomas and 10 anaplastic oligodendrogliomas), for which 1p/19q FISH results were available, between January 2015 and December 2016 at the Asan Medical Center, Seoul, Korea (Table 1). Oligodendroglial tumors were selected to detect the 1p/19q co-deletion because this tumor is known to harbor the 1p/19q co-deletion according to a recent up-dated classification1. Each deletion of 1p and 19q should occur at the chromosome arm level in a sample. The targeted NGS panel did not cover the whole chromosome; therefore, chromosome 1p deletion and 19q deletion were defined as complete segmental loss covered by NGS panel within chr1:1-125000000 and chr19:26500001-59128983, respectively, based on the hg19 human reference genome. Partial segmental loss within regions was classified as negative. Clinical information including age at diagnosis, tumor location, IDH1 Sanger sequencing results, and pathologic diagnosis was gathered from pathology reports. Negative control samples (n = 40) were selected randomly from 1500 targeted NGS data of Asan-CCGD (Asan Center for Cancer Genome Discovery), Asan Medical Center, Seoul, Korea. The study was approved by the institution review board of Asan Medical Center. The institution review board waived the requirement to obtain informed consent. All methods were performed in accordance with relevant guidelines and regulations.
Table 1.
Pathologic and demographic features of 20 cases.
| No | Diagnosis | FISH (1p36/19q13) | IDH1 by Sanger sequencing | Location | Sex | Age (yrs) |
|---|---|---|---|---|---|---|
| 1 | oligodendroglioma | positive | R132H | frontal | M | 39 |
| 2 | oligodendroglioma | positive | R132H | temporal | M | 56 |
| 3 | oligodendroglioma | positive | R132H | frontal | F | 62 |
| 4 | oligodendroglioma | positive | Negative | frontal | F | 42 |
| 5 | oligodendroglioma | positive | R132H | frontal | M | 34 |
| 6 | oligodendroglioma | positive | R132H | frontal | F | 55 |
| 7 | oligodendroglioma | positive | R132H | frontal | M | 40 |
| 8 | oligodendroglioma | positive | R132H | temporal | F | 63 |
| 9 | oligodendroglioma | positive | NA | frontal | M | 41 |
| 10 | oligodendroglioma | positive | NA | frontal | F | 29 |
| 11 | anaplastic oligodendroglioma | positive | NA | frontal | F | 45 |
| 12 | anaplastic oligodendroglioma | positive | NA | frontal | M | 65 |
| 13 | anaplastic oligodendroglioma | positive | R132H | frontal | F | 70 |
| 14 | anaplastic oligodendroglioma | positive | Negative | temporal | M | 63 |
| 15 | anaplastic oligodendroglioma | positive | NA | frontoparietal | M | 58 |
| 16 | anaplastic oligodendroglioma | positive | NA | frontal | F | 52 |
| 17 | anaplastic oligodendroglioma | positive | NA | frontal | M | 54 |
| 18 | anaplastic oligodendroglioma | positive | NA | frontal | F | 66 |
| 19 | anaplastic oligodendroglioma | positive | NA | frontotemporal | F | 51 |
| 20 | anaplastic oligodendroglioma | positive | NA | frontal | F | 48 |
FISH, in situ hybridization; NA, not available; M, male; F, female.
DNA extraction
Matched slides stained with hematoxylin and eosin (H&E) from each formalin-fixed and paraffin-embedded (FFPE) tissue sample were reviewed by two pathologists (H.S.H and S.J.N). The tumor area was marked to determine tumor purity and to guide tumor DNA extraction. Genomic DNA (gDNA) was extracted according to our previous report19. Briefly, 2 ~ 5 sections (6 μm thick) from the indicated area in each FFPE tissue were obtained. After de-paraffinization with xylene and ethanol, gDNA was isolated using the NEXprep FFPE Tissue Kit (#NexK-9000; Geneslabs, Seongnam, Korea). Quantification was performed using the Qubit™ dsDNA HS Assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
DNA library construction and high-throughput sequencing
Targeted NGS was performed using MiSeq (Illumina, Inc., San Diego, CA, USA) with OncoPanel AMCv3 (OP-AMCv3, developed in-house by Asan-CCGD) to include the exons of 199 genes (575,147 bp) and partial introns from 8 genes often rearranged in cancer (209,397 bp) to detect fusion genes and additional small (10,534 bp) specific single nucleotide polymorphism loci for CNV analysis. Overall, the panel covered 823,971 bp. A DNA library was prepared as described in our previous report using the S1 method20; briefly, gDNA shearing with S1 enzyme, end repair, A-tailing, and ligation with the TruSeq adaptor using the SureSelect XT reagent kit (Agilent Technologies, Santa Clara, CA, USA). Each library was constructed with sample-specific barcodes 6 bp in size and quantified using the Qubit kit. Eight libraries were pooled (yielding a total of 720 ng) for hybrid capture using the Agilent SureSelect XT custom kit (OP-AMCv3 RNA bait, 1.2 Mb; Agilent Technologies). The concentration of the enriched target was measured using quantitative polymerase chain reaction (qPCR; Kapa Biosystems, Inc., Woburn, MA, USA). DNA libraries that passed quality checks were sequenced using MiSeq.
Genomic data analysis
Sequenced reads were mapped to the human reference genome (NCBI build 37) using the Burrows-Wheeler Aligner (version 0.5.9)21 with default options. PCR-duplicates were removed using the Picard tool. Then de-duplicated reads were realigned at known indel positions using GATK IndelRealigner, and base quality was recalibrated with GATK TableRecalibration22–24. Somatic mutations of single nucleotide variants and short indels were called in tumor tissue with matched normal tissue using MuTect (1.1.7)25 and SomaticIndelocator in GATK, respectively. Germline variants from somatic variant candidates were filtered out using the common dbsnp database (build 141; found in ≥1% of samples) and a panel of normal samples. Filtered somatic variants were annotated with the Variant Effect Predictor (v79), and then converted to MAF file using vcf2maf (v1.6.12).
Quality checks for fastq files were performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Quality checks for analysis-ready BAM files, including total read number, % PF reads, % selected bases, mean target depth, % target not covered, % target bases covered 30×, and % duplication for cleaned BAM files, were performed using the calculateHSMetrics and MarkDuplicates modules of the Picard package. Integrative Genomics Viewer (v2.4) was used to view the BAM file26.
Copy number analysis
Copy number ratio (log2) was obtained using the CNVkit9. BAM files generated by the pipeline described were used as input files for CNVkit using default parameters without normal tissue. The copy number ratios were obtained and used for downstream analysis. Each bin’s adjusted log2 ratio, chromosomal number, and relative order in that chromosome were extracted from the CNR file. By arranging the adjusted log2 ratio value according to the bin’s relative order, each chromosome was summarized as a vector. The vectors summarizing chromosomes 1 and 19 were 1869 and 649 dimensions in length, respectively. For each case, the CNV plots for chromosomes 1 and 19 were plotted from these vectors. All CNV plots obtained using the log2 ratio were reviewed manually by two pathologists (H.S.P. and C.O.S.) to determine the presence of the 1p/19q co-deletion.
Fluorescence in situ hybridization
FISH was performed on FFPE blocks using probes for 1p36 and 19q13 (Vysis, Downers Grove, IL, USA). Signals were evaluated for each locus-specific FISH probe on an Olympus BX51TRF microscope (Olympus, Tokyo, Japan) equipped with a triple-pass filter (DAPI/Green/Orange; Vysis), and at least 50–100 non-overlapping nuclei containing a minimum of two control signals with intact morphology were evaluated. No target signal or a single signal was interpreted as a deletion, and losses with any disproportion of centromere and target signals in a nucleus with more than one 1p36 or 19q13 signal (ratio 3/2, 4/3, 4/2, 5/3, etc.) were scored FISH imbalances. Because FISH imbalances often, but not always, correspond to LOH, combined criteria were used, defining 1p and 19q deletion as a combined target-to-control signal ratio <0.75 or cut-off of a nucleus with a 1 or 0 target signal >50%.
IDH1 sequencing
The genomic region spanning codon 132 of IDH1 was amplified by PCR with primers 5′-TGAGAAGAGGGTTGAGGAGTTC-3′ (forward) and 5′-CACATACAAGTTGGAAATTTCTGG-3′ (reverse). The amplicon was sequenced with the forward primer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster, CA, USA).
External validation using The Cancer Genome Atlas (TCGA) data set
Low-grade glial tumors (n = 426) were used for independent external validation of the DL model for detection of 1p/19q co-deletion. Copy number (CN) log2 data generated using Affymetrix SNP Array 6.0 platform were downloaded from The Cancer Genome Atlas (TCGA). CN probes were extracted, and the CN log2 ratio for tumor-matched normal tissue was calculated for each sample. The processed log2 CN data were reformatted to correspond to the targeted NGS data, i.e., chromosome 1 log2 CN data were reformatted as 1896 × 1 vectors, and chromosome 19 log2 CN data were reformatted as 649 × 1 vectors. Bins in chromosome 1 and 19 were gathered into 1896 and 649 groups, respectively, according to their relative order on the chromosome, and reformatting was performed by averaging the corresponding groups. The representative log2 ratio value for each group was calculated by the averaging log2 value of the bins corresponding to that group. Vectors from reformatted CN data were plotted in the same manner as CNV plots made from targeted NGS data. All CNV plots were reviewed manually by two pathologists (H.S.P. and C.O.S.) to determine the presence of the 1p/19q co-deletion.
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRFK) (NRF-2017M3A9G5061671 and NRF-2017M3A9G5061673), the Basic Science Research Program through NRFK (NRF-2017R1C1B2009899 and NRF-2018R1D1A1B07042349), and the Technology Innovation Program for Fostering New Post-Genome Industry funded by the Ministry of Trade, Industry & Energy (MOTIE) (#10067796 and #10067407) funded by the Korean government.
Author Contributions
C.O.S., I.S. and C.H. planned and designed the study. H.P., S.M.C., J.S., C.H., I.S. and C.O.S. wrote the manuscript. S.M.C., J.H.O., E.J.C., J.Y.L., D.H.K., S.J.J. and C.O.S. generated sequence data and analyzed the data. H.S.H. and S.J.N. collected clinical data and reviewed histology. J.S., H.P., C.H., I.S. and C.O.S. developed the deep learning model. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hosub Park and Sung-Min Chun contributed equally.
Contributor Information
Changha Hwang, Email: chwang@dankook.ac.kr.
Insuk Sohn, Email: insuks@gmail.com.
Chang Ohk Sung, Email: co.sung@amc.seoul.kr.
References
- 1.Louis DN, et al. The World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Fallon KB, et al. Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. Journal of neuropathology and experimental neurology. 2004;63:314–322. doi: 10.1093/jnen/63.4.314. [DOI] [PubMed] [Google Scholar]
- 3.Fuller CE, Perry A. Fluorescence in situ hybridization (FISH) in diagnostic and investigative neuropathology. Brain pathology (Zurich, Switzerland) 2002;12:67–86. doi: 10.1111/j.1750-3639.2002.tb00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzales M, Dale S, Susman M, Mills J. Quantitation of chromosome 1p and 19q deletions in glial tumours by interphase FISH on formalin-fixed paraffin-embedded tissue. Journal of clinical neuroscience: official journal of the Neurosurgical Society of Australasia. 2006;13:96–101. doi: 10.1016/j.jocn.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Jeon YK, et al. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization. Neuropathology: official journal of the Japanese Society of Neuropathology. 2007;27:10–20. doi: 10.1111/j.1440-1789.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- 6.Reddy KS. Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer genetics and cytogenetics. 2008;184:77–86. doi: 10.1016/j.cancergencyto.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Sim J, et al. Comparison of 1p and 19q status of glioblastoma by whole exome sequencing, array-comparative genomic hybridization, and fluorescence in situ hybridization. Medical oncology (Northwood, London, England) 2018;35:60. doi: 10.1007/s12032-018-1119-2. [DOI] [PubMed] [Google Scholar]
- 8.Jennings LJ, et al. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. The Journal of molecular diagnostics: JMD. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS computational biology. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome biology. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Povysil G, et al. panelcn.MOPS: Copy-number detection in targeted NGS panel data for clinical diagnostics. Human mutation. 2017;38:889–897. doi: 10.1002/humu.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poplin, R. et al. Creating a universal SNP and small indel variant caller with deep neural networks. bioRxiv, 10.1101/092890 (2016). [DOI] [PubMed]
- 13.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 14.Carter JH, et al. Targeted Next-Generation Sequencing in Molecular Subtyping of Lower-Grade Diffuse Gliomas: Application of the World Health Organization’s2016 Revised Criteria for Central Nervous System Tumors. The Journal of molecular diagnostics: JMD. 2017;19:328–337. doi: 10.1016/j.jmoldx.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Zacher A, et al. Molecular Diagnostics of Gliomas Using Next Generation Sequencing of a Glioma-Tailored Gene Panel. Brain pathology (Zurich, Switzerland) 2017;27:146–159. doi: 10.1111/bpa.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha M, Chakraborty C, Racoceanu D. Efficient deep learning model for mitosis detection using breast histopathology images. Computerized medical imaging and graphics: the official journal of the Computerized Medical Imaging Society. 2018;64:29–40. doi: 10.1016/j.compmedimag.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Medical image computing and computer-assisted intervention: MICCAI… International Conference on Medical Image Computing and Computer-Assisted Intervention. 2013;16:411–418. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 18.Ehteshami Bejnordi B, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. Jama. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon MJ, et al. Genomic Alterations of Anaplastic Thyroid Carcinoma Detected by Targeted Massive Parallel Sequencing in a BRAF(V600E) Mutation-Prevalent Area. Thyroid: official journal of the American Thyroid Association. 2016;26:683–690. doi: 10.1089/thy.2015.0506. [DOI] [PubMed] [Google Scholar]
- 20.Chun SM, et al. Next-Generation Sequencing Using S1 Nuclease for Poor-Quality Formalin-Fixed, Paraffin-Embedded Tumor Specimens. The Journal of molecular diagnostics: JMD. 2018;20:802–811. doi: 10.1016/j.jmoldx.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current protocols in bioinformatics. 2013;43:11.10.11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]