Abstract
Integumentary infections like pyoderma represent the main reason for antimicrobial prescription in dogs. Staphylococcus pseudintermedius and Pseudomonas aeruginosa are frequently identified in these infections, and both bacteria are challenging to combat due to resistance. To avoid use of important human antibiotics for treatment of animal infections there is a pressing need for novel narrow-spectrum antimicrobial agents in veterinary medicine. Herein, we characterize the in vitro activity of the novel peptide-peptoid hybrid B1 against canine isolates of S. pseudintermedius and P. aeruginosa. B1 showed potent minimum inhibitory concentrations (MICs) against canine S. pseudintermedius and P. aeruginosa isolates as well rapid killing kinetics. B1 was found to disrupt the membrane integrity and affect cell-wall synthesis in methicillin-resistant S. pseudintermedius (MRSP). We generated 28 analogues of B1, showing comparable haemolysis and MICs against MRSP and P. aeruginosa. The most active analogues (23, 26) and B1 were tested against a collection of clinical isolates from canine, of which only B1 showed potent activity. Our best compound 26, displayed activity against P. aeruginosa and S. pseudintermedius, but not the closely related S. aureus. This work shows that design of target-specific veterinary antimicrobial agents is possible, even species within a genus, and deserves further exploration.
Subject terms: Antibiotics, Bacterial infection
Introduction
Staphylococcus pseudintermedius is a commensal bacterium colonizing dog skin and mucosal sites1, and it is the predominant cause of canine pyoderma and otitis externa2. These common infections represent the main reason for antimicrobial prescription in dogs3. Over the last decade, methicillin-resistant S. pseudintermedius (MRSP) has been reported worldwide4, including sporadic infections in humans in contact with dogs5,6. Pseudomonas aeruginosa is another pathogen frequently involved in canine integumentary infections, in particular otitis externa7. P. aeruginosa is resistant to most antibiotics used in veterinary medicine. The presence of this pathogen and the increasing frequency of multidrug-resistant MRSP8, make treatment of dogs with integumentary infections difficult or even impossible in some cases9. In light of the few treatment options available against these pathogens, new therapeutic agents are needed, preferably drugs restricted to veterinary use and with a narrow spectrum10. This would limit their impact on the commensal microbiota11.
In recent years, antimicrobial peptides (AMPs) have attracted considerable interest as alternative anti-infectives12. AMPs are present in all multicellular organisms as part of their innate immune systems13. They show selective toxicity towards bacteria, rapid killing, broad-spectrum antimicrobial activity, and are active at micromolar concentrations or lower14. Furthermore, they possess immunomodulatory properties such as leukocyte recruitment and suppression of harmful inflammation15. Most AMPs exhibit their antimicrobial activity by disrupting the bacterial cell membrane; however, intracellular targets have also been reported16.
The main drawbacks of AMPs as therapeutics are toxicity and susceptibility to proteases17. Traditionally, these problems are overcome by chemical modification, such as cyclization or design of peptidomimetics, which are stable to proteases18. We and others have previously investigated antimicrobial N-alkylglycines (peptoids)19, β-peptoids (N-alkyl-β-alanine oligomers)20, β-peptides21, lysine-based α-peptide/β-peptoids22, and α/γ N-Acylated-N-aminoethylpeptides (AApeptides)23. Some studies have reported activity of peptides and peptidomimetics against veterinary pathogens24. However, only a few of them aimed at the design and optimization of AMPs with activity against S. pseudintermedius25–28 and canine strains of P. aeruginosa29.
The peptide-peptoid hybrid B1 (Fig. 1) has been previously identified and described as active against one clinical isolate of S. pseudintermedius and P. aeruginosa, as well as resistant to proteolytic degradation in conditions resembling in vivo metabolism30. The aim of the present study was to investigate the antimicrobial activity of B1 against a large collection of S. pseudintermedius and P. aeruginosa isolates from canine infections, determine time-kill kinetics and probe the mechanism of action against S. pseudintermedius. Furthermore, we designed and tested 28 analogues of B1 and selected the peptides 23 and 26 (Fig. 1) for their improved selectivity against S. pseudintermedius compared to B1 while retaining comparable activity against P. aeruginosa. Next, we aimed to get insight into the antibacterial activity of B1, 23 and 26 against other bacterial species causing infections in dogs. Finally, we closely investigated the selectivity of compounds 23 and 26 against S. pseudintermedius relative to S. aureus. This study demonstrates that design of peptide-based antimicrobials which target specific veterinary bacterial species is possible.
Figure 1.

Lead compound B1 and optimized structures 23 and 26.
Results
Antimicrobial activity and killing kinetics
Compound B1 was identified from a combinatorial library. Further evaluation of B1’s antimicrobial activity against a panel of 57S. pseudintermedius isolates from canine infections revealed consistently low MICs (2–4 µg/mL), irrespective of methicillin resistance (Fig. S5, Supplementary Information). Similar MICs (4–8 µg/mL) were observed for the three S. aureus isolates tested (data not shown). Furthermore, MICs of B1 against a panel of 50P. aeruginosa isolates ranged from 8–16 µg/mL.
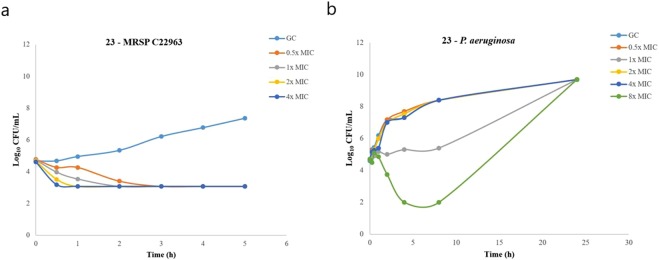
The time-kill kinetic assay showed complete killing of the tested MRSP strain E104 (MIC = 8–16 µg/mL) in 2 h at 2 × MIC, and in 1 h at 4 × MIC, revealing a rapid concentration-dependent effect (Fig. 2a). A similar but slightly inferior effect was detected against the clinical strain P. aeruginosa 26314, which was killed at 4x MIC in 2 h and at 2x MIC in 24 h (Fig. 2b). Lower concentrations (1x MIC) of B1 did not eliminate the Pseudomonas strain but resulted in delayed re-growth (Fig. 2b).
Figure 2.
Time kill kinetics for B1. Time kill kinetics for B1 against (a) MRSP E104 and (a) P. aeruginosa 26314. Time kill assays were performed in triplicate and presented as the average of three different samplings.
Mode of action of B1 against S. pseudintermedius E104
The mode of action of B1 was investigated by studying the effect of sub-inhibitory concentrations of B1 on membrane potential and macromolecule synthesis rate of the MRSP strain E104. The well-characterized antimicrobial nisin was used as a control. Growth curve analysis of B1 at 3 µg/ml showed cell lysis, as indicated by a decrease of OD over time (Fig. S6 Supplemental Material). However, at lower concentrations, B1 resulted in only minor growth inhibition. This is in contrast with nisin, which in addition to cell lysis at MIC concentration (5 µg/ml), significantly retarded growth, also at sub-inhibitory concentrations.
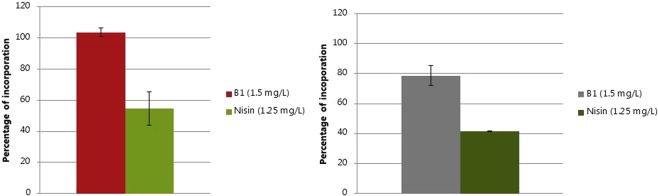
In order to determine if the lysis of MRSP E104 by B1 was due to the inhibition of cell wall biosynthesis, the synthesis rate of cell wall macromolecules was studied at sub-inhibitory concentrations (1.5 µg/mL). In addition, since cationic AMPs have been reported to interact with DNA due to their positive charge and hydrophobicity, the DNA replication was also measured. B1 resulted in a 20% reduction in cell wall synthesis without affecting DNA replication (Fig. 3). Exposure to nisin resulted in 50% inhibition of DNA synthesis (Fig. 3a) and 60% inhibition of cell wall synthesis (Fig. 3b)
Figure 3.
Effects of B1 and nisin on DNA (a) and cell wall (b) synthesis as measured by macromolecule biosynthesis analysis. Percentages of DNA and cell wall precursors incorporation with respect to unexposed control are presented as average values of two individual measurements.
To test the immediate effect of a sub-inhibitory concentration of B1 (1.5 µg/mL) on membrane potential, proton motive force (PMF) was measured using DiSC3(5) (3,3’-Dipropylthiadicarbocyanine Iodide), a fluorescent probe that concentrates in energized membranes and is released in the environment surrounding a cell upon membrane depolarization, thus increasing the intensity of its emission. As expected, no fluorescence increment was observed after nisin treatment (Fig. 4a,b). On the contrary, B1 caused significant membrane depolarization, as indicated by an increased DiSC3(5) fluorescence emission (Fig. 5a,b). Furthermore, the energy dissipation effect of B1 on the cell at the lysis concentration (1x MIC) was studied by flow cytometry analysis. Cell death (Fig. 5c, P2) and injury (P3) upon B1 exposure was indicated by the high ratio of propidium iodide (PI) staining relative to thiazole orange (TO) staining, whereas unexposed control cells were mainly stained with TO (Fig. 5b). Taken together, our studies indicated that B1 causes membrane depolarization and affects cell-wall synthesis but not DNA-synthesis.
Figure 4.
Effects of B1, Carbonyl cyanide m-chlorophenylhydrazone (CCCP) or nisin on DiSC3(5) (3,3′-Dipropylthiadicarbocyanine Iodide) fluorescence. Effects of B1, Carbonyl cyanide m-chlorophenylhydrazone (CCCP) or nisin on DiSC3(5) (3,3′-Dipropylthiadicarbocyanine Iodide) fluorescence plotted by emission spectra (a) and increment of fluorescence after exposure (b).
Figure 5.
Flow cytometry analysis of MRSP E104 exposed to B1. Four different conditions of cells are represented by boxes: P1 (unstained), P2 (dead cells), P3 (injured cells) and P4 (live cells). FL1 and FL3 axis represent green (thiazole orange, TO) and red fluorescence (propidium iodide, PI), respectively. (a) is a dotted plot of unstained cells, (b,c) are TO/PI stained unexposed and B1 exposed cells, respectively.
Analogues of B1
After characterizing B1 as a lead, we proceeded to generate an ensemble of 28 analogues (Table S1, Supplementary Information) in order to develop compounds with specific activity against S. pseudintermedius and P. aeruginosa, paired with low activity against methicillin-susceptible Staphylococcus aureus (MSSA). Compounds 2–29 are analogues of B1 (Fig. 1A). Compounds 2–5 (Table 1) contain L-Lys and peptoid residues. In compound 2, residue 6 (N-4-methylbenzylglycine) was replaced with N-benzylglycine resembling Phe; in compound 3, residue 4, N-butylglycine, was switched with residue 5, Lys, to generate a hydrophobic and cationic cluster. Compounds 4 and 5 are the reversed compounds of 2 and 3.
Table 1.
Sequence, Minimum Inhibitory Concentration in µg/mL and Haemolysis (µM) for compound B1 and 28 analogs.
| ID | Sequencea | Minimum Inhibitory Concentration (µg/ml) | Haemolysis (µM) | ||||
|---|---|---|---|---|---|---|---|
| MRSP C22963 | MSSA (ATCC 29213) | P. aeruginosa (26314) | EC10 | EC50 | %H 150(µM) | ||
| B1 | Lys-Lys-(NBu)Gly-Lys-(N1-Nal)Gly-(N4-MeBn)Gly-(N1-Nal)Gly | 2–4b | 8–16 | 8–16 | 64 | 230 | 32 |
| 2 | Lys-Lys-(NBu)Gly-Lys-(N1-Nal)Gly-(NPhe)Gly-(N1-Nal)Gly | 4 | 8–16 | 32 | — | — | <8 |
| 3 | Lys-Lys-Lys-(NBu)Gly-(N1-Nal)Gly-(NPhe)Gly-(N1-Nal)Gly | 4 | 16 | 32 | — | — | <8 |
| 4 | (N1-Nal)Gly-(NPhe)Gly-(N1-Nal)Gly-(NBu)Gly-Lys-Lys-Lys | 8 | 16 | 32 | 54 | — | 24 |
| 5 | (N1-Nal)Gly-(NPhe)Gly-(N1-Nal)Gly-Lys-(NBu)Gly-Lys-Lys | 8 | 16 | 32 | — | — | <8 |
| 6 | Lys-Lys-Leu-Lys-(1-Nal)Ala-Phe-(1-Nal)Ala | 8 | 32 | 8 | 5 | 58 | 96 |
| 7 | Lys-Lys-Lys-Leu-(1-Nal)Ala-Phe-(1-Nal)Ala | 8 | 16 | 16 | 8 | 140 | 55 |
| 8 | Lys-Lys-Leu-Lys-(2-Nal)Ala-Phe-(2-Nal)Ala | 8 | 16 | 32 | 56 | — | 34 |
| 9 | Lys-Lys-Lys-Leu-(2-Nal)Ala-Phe-(2-Nal)Ala | 8 | 16 | 16 | 46 | — | 39 |
| 10 | (1-Nal)Ala-Phe-(1-Nal)Ala-Lys-Leu-Lys-Lys | 8–16 | 16–32 | 64 | — | — | <8 |
| 11 | (2-Nal)Ala-Phe -(2-Nal)Ala-Lys-Leu-Lys-Lys | 4–8 | 16 | 64 | — | — | <8 |
| 12 | (1-Nal)Ala-Phe-(1-Nal)Ala-Leu-Lys-Lys-Lys | 16 | 32 | >64 | — | — | <8 |
| 13 | (2-Nal)Ala-Phe-(2-Nal)Ala-Leu-Lys-Lys-Lys | 8 | 32 | >64 | 110 | — | 15 |
| 14 | Lys-Lys-leu-Lys-(1-Nal)ala-phe-(1-Nal)ala | 4–8 | 16 | 16–32 | 28 | 128 | 58 |
| 15 | Lys-Lys-Lys-leu-(1-Nal)ala-phe-(1-Nal)ala | 2 | 8 | 8 | 8 | 40 | 88 |
| 16 | Lys-Lys-Lys-leu-(2-Nal)ala-phe-(2-Nal)ala | 2–4 | 8–16 | 32 | 18 | 118 | 60 |
| 17 | Lys-Lys-leu-Lys-(2-Nal)ala-phe-(2-Nal)ala | 8 | 16 | 32 | 8 | — | 40 |
| 18 | (2-Nal)ala-phe-(2-Nal)ala-Lys-leu-Lys-Lys | 16 | 32–64 | >64 | — | — | <8 |
| 19 | (2-Nal)ala-phe-(2-Nal)ala-leu-Lys-Lys-Lys | 16 | 32 | >64 | — | — | <8 |
| 20 | (1-Nal)ala-phe-(1-Nal)ala-Lys-leu-Lys-Lys | 16 | 32–64 | >64 | — | — | <8 |
| 21 | (1-Nal)ala-phe-(1-Nal)ala-leu-Lys-Lys-Lys | 16 | 32 | 32 | — | — | <8 |
| 22 | Lys-Lys-Lys-Leu-(2-Nal)Ala-Tyr-(2-Nal)Ala | 16 | >64 | >64 | 117 | — | 14 |
| 23 | Lys-Lys-Lys-Nle-(2-Nal)Ala-Phe-(2-Nal)Ala | 2–4 | 32–64 | 8 | 14 | 104 | 59 |
| 24 | Lys-Lys-Lys-Nle-(1-Nal)Ala-Phe-(1-Nal)Ala | 16–32 | >64 | 16 | 3 | 41 | 60 |
| 25 | Lys-Lys-Lys-leu-(1-Nal)ala-tyr-(1-Nal)ala | 2–4 | 32–64 | 8 | 38 | 138 | 53 |
| 26 | Lys-Lys-Lys-nle-(1-Nal)ala-phe-(1-Nal)ala | 4–8 | >64 | 8 | — | 63 | 56 |
| 27 | Lys-Lys-Lys-Nle-(2-Nal)Ala-Tyr-(2-Nal)Ala | 2–4 | 64 | 64 | 3 | 56 | 54 |
| 28 | Lys-Lys-Lys-leu-(2-Nal)ala-tyr-(2-Nal)ala | 2–4 | 32 | 32 | 6 | 50 | 64 |
| 29 | NLys-NLys-NLys-Leu-(2-Nal)Ala-Phe-(2-Nal)Ala | 4–8 | 16–32 | 32 | 9 | — | 46 |
aAll compounds were synthesized as peptide amides and isolated as TFA salts. bMRSP strain E104 (MIC = 8–16 µg/mL).
Compounds 6–13 contain L-lysine and L-amino acids (Leu, Phe, 1-Nal, 2-Nal) instead of the peptoid residues N-butylglycine, N-1-naphthylmethylglycine and N-4-methylbenzylglycine, respectively. In compound 6, peptoid residues have been replaced with the corresponding amino acids and in 7, residue Leu4 and residue Lys5 have been switched to obtain a hydrophobic and cationic cluster. Compounds 8 and 9 differ from 6 and 7 for the presence of 2- instead of 1-Nal. Compounds 10–13 are the reverse of 6–9. Compounds 14–21 are analogues of 6–13 in which L-Lys residues have been retained and Leu, 1-Nal, 2-Nal and Phe have been replaced by the corresponding D-stereoisomers.
Based on the data for 1–21, we synthesized a second set of compounds (22–29) maintaining three L-Lys at the N-terminus except for 29. Compounds 22–23 are analogues of 9 in which Phe6 and Leu4 were replaced by Nle and Tyr, respectively. Compound 23 is an analogue of 7 in which Leu4 was replaced with Nle. Compounds 25 and 26 are derived from 15, in which D-Phe6 and D-Leu4 have been replaced by D-Nle and D-Tyr, respectively. Compounds 27 and 28 contain combinations of substitutions introduced in compounds 22–26. Finally, compound 29 is an analogue of 9 in which Lys residues have been replaced by N-(4-aminobutyl)glycine.
MICs of the analogues were determined against S. pseudintermedius C22963, MSSA (ATCC 29213), and P. aeruginosa 26314. The sequence of each analog, the MICs and hemolysis data against red blood cells are reported in Table 1.
Compounds 2–21 were generally less active than B1 with MICs of 2–16 µg/mL against S. pseudintermedius C22963, 8–64 µg/mL against S. aureus, and 8- >64 µg/mL against P. aeruginosa 26314. Hemolysis at 150 µM for compounds 2–17 ranged from <8–96% while the reverse sequences 18–21 were not hemolytic.
To further improve selectivity and hemolysis, we synthesized a second set of compounds (22–29) maintaining three L-Lys at the N-terminus except for 29. Five of the compounds featuring three Lys (or Lys-like residues, Fig. 1) at the N-terminus (23, 25, 26, 27 and 28) showed 16-fold better activity against the MRSP strain (MIC 2–4 µg/mL) than against the MSSA strain (MIC 32- >64 µg/mL). Furthermore, they showed moderate activity against MRSA USA300 strain FPR3757 (32- >64 µg/mL) (data not shown). In addition, compound 23, 25 and 26 retained the activity level of B1 against P. aeruginosa (8 µg/mL). The haemolytic values of compounds 22–29 ranged from 46 to 64% at 150 µM except for compound 22 (14%). The two most promising analogues (23 and 26) were selected for further studies due to (i) the low MICs observed in MRSP and P. aeruginosa, (ii) the relatively higher antimicrobial effect against S. pseudintermedius compared to S. aureus, and (iii) a moderate to low-level of hemolysis EC50 of 104 µM and 63 µM, respectively (Table 1).
Time-kill experiments of 23 and 26
Time-kill experiments showed that 26 (Fig. 6). was superior to both B1 and 23 (Fig. 7a) and displayed complete killing of MRSP C22963 within 1 h at only 0.5x MIC Similarly to B1, these two analogues exhibited complete and concentration-dependent killing of the MRSP strain within 2 h at all tested concentrations at or above the MIC. Surprisingly, 23 was not able to completely kill P. aeruginosa, even at the highest tested concentration (8x MIC) (Fig. 7b). We also intended to do time-kill curves for 26 against P. aeruginosa (MIC = 16 µg/mL). To our disappointment, 26 precipitated after 15 min in the media, and it was not possible to get reproducible results.
Figure 6.

Time kill curve of compound 26 against MRSP C22963. Time kill curve of compound 26 against MRSP C22963 at four different concentrations. Time kill assays were performed in triplicate and presented as the average of three different samplings.
Figure 7.
Time kill kinetics for 23. Time kill kinetics for 23 against (a) MRSP C22963 and (a) P. aeruginosa 26314. Time kill assays were performed in triplicate and presented as the average of three different samplings.
Antimicrobial activity of B1, 23 and 26 against a selection of canine pathogens
In order to get further insight into the antimicrobial spectrum of B1, 23 and 26, we tested MICs against a collection of clinical isolates representing other canine pathogens (Corynebacterium auriscanis, Enterococcus faecalis, Enterococcus faecium, Streptococcus canis, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumonia, Pasteurella canis, Proteus mirabilis) (Table 2). All three compounds showed activity against Gram-positive bacteria with MICs ranging between 2–16 µg/mL, except for Enterococcus (>64 mg/mL). However, B1, 23 and 26 were substantially less active against the Gram-negative bacteria with most of the MICs in the range of 32- >64 µg/mL, P. aeruginosa and Pasteurella canis being the only exceptions. For the latter species, the effect varied consistently between B1, 23 and 26 (8 µg/mL, >64 µg/mL and >64 µg/mL, respectively).
Table 2.
Antimicrobial activity of B1, 23 and 26 against a collection of clinical isolates from canine (µg/mL).
| Bacteria | B1 | 23 | 26 |
|---|---|---|---|
| Acinetobacter baumannii, 27065, 16 D1, dog, wound, 2010 | 64 | 64 | >64 |
| Corynebacterium auriscanis, 31551, 54 C6, dog, ear, 2013 | 4 | 8 | 32 |
| E. coli, 30235, 23 A6, dog, wound, 2012 | 64 | 32 | 32 |
| Enterococcus faecalis, 27404, 17 C7, dog, wound, 2011 | 64 | >64 | 64 |
| Enterococcus faecium, 30951, 24 C1, dog, ear, 2013 | 16 | 64 | >64 |
| Klebsiella pneumoniae, 26233, 11 H5, dog, wound, 2010 | >64 | >64 | >64 |
| Pasteurella canis, 31096, 24 C8, dog, skin, 2013 | 8 | >64 | >64 |
| Proteus mirabilis, 25178, 9 A4, dog, ear, 2009 | >64 | >64 | >64 |
| Pseudomonas aeruginosa (26314, 12 C5, dog, urine, 2010). | 16 | 64 | >64 |
| Streptococcus canis, 26740-1, 14 H1, dog, ear, 2010 | 4 | 16 | >64 |
| Staphylococcus aureus, 27266, 16 G9, dog, skin, 2010 | 8 | 32 | 64 |
| Staphylococcus pseudintermedius (22963, 3 B9, dog, 2007). | 2 | 4 | 4 |
Selectivity of 23 and 26 between S. pseudintermedius and S. aureus
Finally, we tested the selectivity of 23 and 26 against clinical isolates of S. pseudintermedius (n = 10) vs S. aureus (n = 10) (Table S7, supplemental material). We found that the two compounds were generally more active against S. pseudintermedius (2–16 µg/mL) than S. aureus (32- >64 µg/mL).
Discussion
The aims of this study were to (i) characterize the antimicrobial activity of the novel peptide-peptoid hybrid B1 against a large collection of two common dog integumentary pathogens S. pseudintermedius and P. aeruginosa; (ii) probe the mode of action against S. pseudintermedius E104; (iii) conduct a structure-activity study of B1 involving 28 analogues and identify compounds with potent activity against representative strains of S. pseudintermedius and P. aeruginosa, paired with weak activity against S.aureus; (iv) test the most promising of these analogues (23 and 26) as well as B1 against a broad panel of other canine pathogens; (v) test for selectivity within the Staphylococcus genus between S. pseudintermedius and S. aureus.
B1 showed low MICs against 50 canine P. aeruginosa (8–16 µg/mL), 57S. pseudintermedius (2–4 µg/mL). Killing kinetics showed that B1 kills MRSP and P. aeruginosa in less than 30 min at 8 x MIC. Our MIC and time-kill data for MRSP and P. aeruginosa are comparable with previous literature reports. Mohamed et al. designed and tested synthetic peptides (8 to 16 amino acids) against MSSP and MRSP. The most effective peptides displayed a MIC50 and MIC90 of 1 and 2 µM, respectively25. Molchanova et al.22 reported 22 different α-peptide/β-peptoid hybrids containing cationic and hydrophobic residues in a 1:1 ratio that were active against MRSP (2–8 µg/mL) as well as other relevant Gram-positive and Gram-negative bacteria. The same authors also identified fluorinated antimicrobial lysine-based peptidomimetics with activity against methicillin-resistant S. pseudintermedius26. Finally, Cabassi and coworkers identified a peptide (AMP2041) with activity against human and animal multidrug resistant P. aeruginosa isolates31, including strains of canine origin. In a parallel study to the present, we have characterized the in vitro pharmacokinetic properties of B1, including hemolytic activity and stability to proteases. The measured hemolytic activity of B1 was 32% at 150 µM and the compound showed only 38% degradation after 24 hours exposure to the mix of protease of bacterial origin Pronase. Furthermore, B1 was suitable for topical delivery from cream formulation and showed no skin penetration after administration30. These data, combined with our results from the present study, suggest that B1 may be suitable as antimicrobial for topical treatment of canine superficial pyoderma.
To investigate the mode of action of B1, we used the well-characterized clinical MRSP ST71 strain E104 (MIC = 8–16 µg/mL), which is resistant to β-lactams, ciprofloxacin, clindamycin, doxycycline, and trimethoprim/sulfamethoxazole8. We found that the primary mode of action of B1 is on the bacterial membrane and secondarily on cell wall synthesis. However, B1 has no effect on DNA replication. Membrane activity was supported by a reduction of the initial OD in the growth curve study at 3 µg/mL (Fig. S6, Supplementary information), indicating cell lysis. Furthermore, B1 caused significant membrane depolarization as seen by an increased DiSC3(5) fluorescence emission (Fig. 4a,b). We obtained further support for the membrane permeabilization by the rapid cell death observed in the flow cytometry assay (Fig. 5).
The primary mode of action of B1 is in agreement with the classical membrane-targeting mechanism reported for a number of antimicrobial peptides, e.g. magainin II32 and Cecropin B33. Besides membrane disruption, AMPs may have intracellular targets as reviewed recently34. These include protein35, DNA36, and cell-wall synthesis37. Here, B1 reduced cell wall synthesis by 20% (Fig. 3b). A few antimicrobial peptides such as plectasin have been reported to inhibit cell wall synthesis in Gram-positive bacteria. Plectasin targets the bacterial cell wall precursor lipid II as determined by advanced NMR38.
Our finding that B1 does not affect DNA synthesis (Fig. 3a), is in agreement with literature reports that most AMPs do not have this target. However, some exceptions are known, e.g. indolicidin39. Furthermore, LP5, a compound similar to B1, has been reported to inhibit DNA replication and induce SOS response in S. aureus40. This may be due to a difference in net charge +4 (B1) and +6 (LP5), respectively.
In the mechanism of action study of B1, we used the well-characterized antimicrobial agent nisin as control. This antimicrobial compound used in food presevation interacts with the peptidoglycan precursor molecule lipid II, which leads to membrane depolarization and bacterial cell death41. The mechanism of nisin has been studied using a plethora of different techniques42,43.In our study, nisin had an inhibitory effect on MRSP growth at sub-lysis concentrations (Fig. S6 Supplementary Information). Simultaneous cell lysis and growth inhibition effects by nisin may be linked to the dual mode of action of this AMP, which encompasses both membrane pore formation and cell wall synthesis inhibition. Exposure to nisin resulted in 60% inhibition of cell wall synthesis, and 50% inhibition of DNA synthesis (Fig. 3a,b). We did not investigate the mechanism of B1 on P. aeruginosa, but a few previous reports on the AMP killing mode of action of P. aeruginosa exist: using fluorescence microscopy and field emission scanning electron microscopy, Memariani et al. found that the 14-mer AMP PV3, kills bacteria by disrupting the cell membrane44. Furthermore, Scocchi and coworkers investigated the mechanism of killing against P. aeruginosa strain PAO1 and additional three isolates and observed that Bac7 (1–35) inactivated the target cells by disrupting their cellular membranes45.
We synthesized 28 analogues of B1 by (i) altering the peptoid residues position in the sequence; (ii) substituting peptoid residues for L-amino acids; (iii) modifying the chirality of the amino acid components and/or altering their position in the sequence. Like B1 and most AMPs, these analogues are cationic and hydrophobic. The rationale for their design is further discussed in results section. Besides S. pseudintermedius and P. aeruginosa, all the analogues were also tested against MSSA (ATCC 29212). Our two best compounds, 23 and 26, showed slightly higher MICs (Table 1) against S. pseudintermedius (2–4 µg/mL and 4–8 µg/mL respectively) and P. aeruginosa (8 µg/mL) compared to B1 and considerably worse activity against MSSA (32–64 µg/mL, >64 µg/mL), respectively).
Hemolysis at 150 µM is a commonly used parameter in the literature for comparison of antimicrobial peptides or peptidomimetics46. The hemolytic activity of 23 and 26 was 59% and 56% at 150 µM, respectively. A variation of the therapeutic index, selectivity index, SI, is often used in the field and is defined as the ratio between the concentration leading to 50% lysis of human erythrocytes and the minimum concentration inhibiting bacterial growth SI = (HC50/MIC) for the bacterium being considered47. The selectivity indices for MRSP C22963 are: B1 (82), 23 (37), 26 (11). Typically, selectivity indices are below 100, although higher values have been reported48.
The SI values are not a major issue for drugs intended for topical use, since a highly hemolytic compound like the steroid antibiotic fusidic acid is being used for against human skin disorders and canine pathogenic staphylococci49. Compound 23 showed slower killing kinetics against MRSP C22963 and P. aeruginosa than B1, and was not able to eliminate P. aeruginosa but resulted in delayed re-growth. Notably, compound 26 was able to kill MRSP C22963 within 1 h at only 0.5x MIC, which was faster than both B1 and 23 and fully comparable with literature reports25.
We tested the activity of B1 and the analogues 23 and 26 against a broader range of canine pathogens. They displayed antimicrobial activity (Table 2) against a collection of Gram-positive clinical isolates (2- >64 µg/mL) and less against the Gram-negative isolates (8- >64 µg/mL). Generally, B1 proved a lot more active than 23 and especially 26 (Table 2). In addition to S. pseudintermedius, S. aureus and P. aeruginosa, we observed increased activity for B1 (4–16 µg/mL) against other bacteria such as: Corynebacterium auriscanis associated with canine otitis externa50, Streptococcus canis which causes respiratory, cutaneous, genital and urinary infections in various animal species51, Pasteurella canis which is a well-known major pathogen of infections caused by dog bites52, and Enterococcus faecium which is an important nosocomial pathogen53. However, we observed no significant activity against bacterial isolates belonging to the species Acinetobacter baumannii, Escherichia coli, Enterococcus faecilis and Klebsiella pneumoniae.
Having established the antimicrobial spectrum of 23 and 26, we tested the selectivity of 23 and 26 against clinical isolates of S. pseudintermedius (n = 10) vs S. aureus (n = 10) (Table S7, supplementary information). We found that the two compounds were generally more active against S. pseudintermedius (2- >16 µg/ml) than S. aureus (32- >64 µg/mL), especially compound 26. AMPs with enhanced selectivity against a target bacterial genus have been previously reported54 and selectivity within a genus has been observed in few other studies55. For example, Guo et al. published a peptide, C16G2, which is able of killing S. mutans selectively but not closely related streptococcal species. Our finding is significant, as we demonstrated that short amphipathic peptidomimetics can maintain activity against S. pseudintermedius, even when their efficacy against S. aureus decreases multiple folds. This suggests that the former might be more susceptible to membrane-active agents, or susceptible to a wider range of agents. These observations should aid the design of novel therapeutics for the treatment of S. pseudintermedius infections in animals, for which B1, 23 and 26 pose as promising lead candidates. The underlying reason for the higher activity of compounds 23 and 26 against S. pseudintermedius over S. aureus is unknown, but might be related to peptides mechanism of interactions with the bacterial cell envelope. The spatial orientation of amino acid side-chains upon interacting with the bacterial membrane is often fundamental for the activity of AMPs56. The compounds B1, 23 and 26 all contain seven residues, which is too short to display α-helical structure57. Therefore, they most likely form random coils. Furthermore, 26 contains D-amino acids, which are known to disrupt α-helix structure58. Finally, the presence of even a single peptoid residue in an α-helix has been reported to result in a significant reduction of the helical content56.
Similarities and differences among bacterial membranes of different species are crucial in determining the spectrum of activity of amphipathic AMPs59. Since the structure of B1, 23 and 26 are closely related, we speculate that the mechanism of action of 23 and 26 is similar to that of B1. Compound B1 is an L-peptide/peptoid hybrid, 23 is a full L-peptide containing non-canonical amino acids, whilst 26 is an L/D-peptide hybrid (Fig. 1). The affinity of antimicrobial peptides for bacterial cells is due to their amphiphilic properties (hydrophobic and positively charged). The interaction with the bacterial membrane is regulated, for both peptides and peptoids, by their hydrophobicity. Also, the presence of peptoid residues in the backbone of B1 may correspond to a higher structural flexibility and a more cell-penetrating action of B1. On the other hand, the peptides 23 and 26 may have a more pronounced membrane-disrupting activity, which may correlate with the faster killing effect observed against S. pseudintermedius.
Veterinary medicine needs antimicrobials which are tuned to a veterinary spectrum and are not shared with human medicine. These considerations are in line with the “One Health” view of infectious diseases, which acknowledges that humans and animals share the same pool of bacterial pathogens. Moreover, MRSP has been isolated in humans, highlighting its zoonotic potential and therapeutic challenge60. Therefore, it is in the interest of veterinary medicine and public health that novel antimicrobial agents target P. aeruginosa and S. pseudintermedius in companion animals selectively and not closely related species within a genus, including S. aureus.
In this perspective, we have characterized a novel peptide-peptoid hybrid B1 with antimicrobial activity against both S. pseudintermedius and P. aeruginosa, the main cause of pyoderma in dogs. A structure-activity study identified two compounds, 23 and 26, with potent activity against the aforementioned species, paired with poor activity against the closely related S. aureus. Our results represent a first step towards the design of peptide-based antimicrobials with a pathogen-targeted spectrum, even within a genus. Such investigation deserves further exploration towards the rationale design of drugs selective for veterinary pathogens.
Materials and Methods
Synthesis of peptides and peptidomimetics
The synthesis of the peptides and peptidomimetics was performed as described by Oddo et al.61. Briefly, peptides were synthesized by Fmoc solid phase peptide synthesis (SPPS). Peptidomimetics were prepared by a combination of the above and sub-monomer peptoid synthesis. Following TFA-cleavage, precipitation in ether and lyophilisation, the compounds were purified (>95%) by preparative HPLC and the purity was determined through analytical HPLC. The identity of each compound was verified by MALDI-TOF-MS. The compounds used in this study are shown in Supplementary Information (Table S1).
Antimicrobial susceptibility testing
Activity of B1 was tested on 50P. aeruginosa isolates and 57S. pseudintermedius isolates (including 7 MRSP) that had been isolated in the diagnostic laboratory Sund Vet Diagnostik (University of Copenhagen) from various infections in dogs between 2009 and 2011. Representative MRSP (C22963), MSSA, and P. aeruginosa (26314) were used for testing the first set of B1 analogues.
For antibacterial spectrum the following strains were used: Corynebacterium auriscanis, 31551, (54 C6, dog, ear, 2013); Enterococcus faecalis, 27404, (17 C7, dog, wound, 2011); Enterococcus faecium, 30951, (24 C1, dog, ear, 2013); Streptococcus canis, 26740-1, (14 H1, dog, ear, 2010); Staphylococcus aureus, 27266, (16 G9, dog, skin, 2010); Staphylococcus aureus, 27266, (16 G9, dog, skin, 2010); Staphylococcus pseudintermedius 22963, (3 B9, dog, 2007). Acinetobacter baumannii, 27065, (16 D1, dog, wound, 2010); E. coli, 30235, 23 A6, dog, wound, 2012; Klebsiella pneumoniae, 26233, (11 H5, dog, wound, 2010); Pasteurella canis, 31096, (24 C8, dog, skin, 2013); Proteus mirabilis, 25178, (9 A4, dog, ear, 2009); Pseudomonas aeruginosa 26314, 12 C5, dog, urine, 2010).
For testing the selectivity of 23 and 26 against clinical isolates of S. pseudintermedius vs S. aureus the following strains were used.
S. aureus strains
25054, (8 G6, dog, wound, 2009); 27266, (16 G9, dog, skin, 2010 also used in exp above); 28264,(20 B1, dog, wound, 2011); 30935, (24 B9, dog, joint, 2013); 36968, (61 A9, dog, wound, 2016); 37595,(65 D2, dog, joint, 2016); 37708-2, (66 C6, dog, skin, 2016); 38200, (68 E9, dog, skin, 2016); 38565-1, (70 A5, dog, skin, 2017); 38841, (70 G5, dog, urine, 2017).
S. pseudintermedius strains
26071, (11 E5, dog, skin, 2009); 26092-2, (11 E4, dog, skin, 2009); 26959, (15 F8, dog, wound, 2010); 27364, (17 A7, dog, wound, 2011); 27382, (17 B8, dog, ear, 2011); 27382, (17 B8, dog, ear, 2011); 31524, (54 C3, dog, ear, 2013); 33228, (55 E2, dog, skin, 2014); 35890, (59 B8, dog, ear, 2015); 37526-1, (65 B4, dog, skin, 2016); 37535-1, (65 B6, dog, skin, 2016); 37535-1, (65 B6, dog, skin, 2016); 38637, (70 C3, dog, wound 2017); 38820, (70 F9, dog, urine 2017).
MIC determination was performed by broth microdilution according to the Clinical and Laboratory Standard Institute (CLSI, M31-A3, 2008)62. In brief, each bacterial strain was diluted to concentration of 5 × 105 CFU/mL in Mueller Hinton broth II (MHB II) media (Oxoid) and added to a two-fold serial dilution of peptides and peptide-peptoid hybrids concentrations ranging from 1 to 64 µg/ml in 96-well plates (Nunc Internationals, Rochester, NY). The MICs were determined as a lowest concentration showing no visible growth after incubation for 18 hours at 37 °C. Experiments were performed in triplicates on two different days.
Time kill curves
Time kill assays were performed in triplicate, meaning that every value is the average of three different samplings. Time–kill kinetic assays were performed using P. aeruginosa (26314), MRSP E104 and/or MRSP C22963 to determine the cell killing activity of B1 and two of its most promising analogues (compounds 23 and 26) based on prior antimicrobial susceptibility testing. The method reported by Blondeau et al.63 was followed with minor modifications. Briefly, the assay was performed in MHB II with concentrations corresponding to 0.5, 1, 2 and 4 times the MIC of the strain. After 0, 15, 30, 60, 120, 180 and 300 min of growth, 100 μL aliquots were collected and 10-fold serially diluted. Twenty μL of cell suspension from each dilution were spotted in triplicate on blood agar plates followed by 16–18 h incubation at 37 °C and determination of colony forming units (CFU).
Haemolytic activity
The EC10 (10% maximal effective concentration), EC50 (half maximal effective concentration) values and the percentage of haemolysis at 150 μM were determined for all compounds as previously described64. Briefly, two-fold serial dilutions (2.35 to 150 μM) of compounds in phosphate buffer saline (PBS) were mixed with equal volume of 0.5% v/v suspension of fresh human red blood cells (RBC) in the same buffer. After 1 h incubation at 37 °C, plates were centrifuged and aliquots of the supernatants were transferred to clear 96-well plates. Absorbance at 414 nm was measured and normalized using a negative (PBS, 0%) and a positive (melittin, 100%) control. The EC10 and EC50 are concentrations at which 10% and 50% of RBC were lysed, respectively, as interpolated graphically by the y-axis intersection of the plotted data.
Macromolecule biosynthesis rate
Macromolecule biosynthesis rate was measured in MRSP E104 following a protocol adapted from Ling et al.65. Briefly, E104 overnight culture was sub-cultured 1:100 in MHB II and grown up to OD 0.2 at 600 nm. Cells were pelleted down by centrifugation and resuspended in fresh medium followed by incubation for 20 min with B1 or nisin at 1.5 µg/ml and 1.25 µg/ml, respectively, and radiolabeled precursor: (50µCi) 3H-Thymidine (PerkinElmer) and (5µCi) 3H-glucosamine hydrochloride per ml for DNA and cell wall, respectively. A positive control without antimicrobial was maintained. After incubation, samples were precipitated with equal volume of cold 30% TCA (Sigma) on ice. Precipitates were filtered on a membrane filter and subsequently subjected to two washes of cold 15% TCA and two washes of cold water using vacuum manifold. Subsequently, filters were air dried overnight and then transferred to 10 ml scintillation vials. Finally, scintillation fluid (3 ml) was added to each vial and 3H count was taken in Beckman Coulter LS6500 liquid scintillation counter for one minute. The radioactive counts of the control samples were considered to have 100% precursor incorporation and macromolecule synthesis. The percentage rates of the antimicrobial-exposed samples were calculated accordingly. Each experiment was performed with replicates and the average rates of incorporation were plotted.
DiSC3(5) fluorescence-based membrane potential study
Freshly sub-cultured E104 cells were labelled with 1 µM 3,3-Dipropylthiadicarbocyanine iodide [DiSC3(5)] (Sigma) in MHB II. The fluorescence spectra of labelled cells were plotted in the LS50B luminescence spectrometer (PerkinElmer) at excitation/emission wavelengths 546 nm/573 nm using time drive application of FLWINLAB software. After reading an initial stable emission spectra of DiSC3(5), labelled cells were treated with B1 or nisin or protonophore CCCP at 1, 1.5 and 1.25 µg/ml, respectively and the change of fluorescence over time was recorded. The increment of DiSC3(5) fluorescence upon addition of antimicrobials or CCCP (FUafter-treatment – FUbefore-treatment) was plotted. This experiment was performed twice and each time with two technical replicates.
Flow cytometry analysis of antimicrobial-exposed cells
Freshly grown cultures (OD 0.2 at 600 nm) of MRSP E104 were exposed to 3 µg/ml of B1 or 2.5 µg/ml of nisin for 1 h and diluted 1:10 in flow cytometry analysis buffer (PBS supplemented with 1 mM EDTA, 0.01% Tween 20 and filtered by 0.22 µm membrane). Diluted cells were stained with 420 nM thiazole orange (TO, Sigma) and 48 µM propidium iodide (PI, Sigma) and analysed using BD ACCURI C6 flow cytometer (BD Biosciences). Unstained cells were used as negative control. Finally, the differentially labelled bacterial populations in antimicrobial-exposed cultures were plotted under FL3(red)/FL1(green) axis using the instrument’s software and categorised into four different conditions: P1 (unstained), P2 (dead cells), P3 (injured cells) and P4 (live cells).
Supplementary information
Acknowledgements
Birgitte Simonsen is thanked for excellent technical help. This work was supported by Marie Curie Actions under the Seventh Frame Programme (Initial Training Network Grant TRAIN-ASAP, contract number 289285), the University of Copenhagen Centre for Control of Antibiotic Resistance (UC-Care) and the Augustinus Foundation, Bdr. Hartmanns Foundation and The Hørslev Foundation (PRH).
Author Contributions
I.G., A.E., B.J. N.M. and P.D. performed the experiments. I.G., A.E., B.J. N.M. A.O., P.D., L.G. and PRH conceived and designed the study. I.G., B.J. N.M., P.D. and P.R.H. wrote the manuscript. All authors contributed to the final version of the manuscript and approved it.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/4/2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39042-3.
References
- 1.Peacock, S. J. & Paterson, G. K. In Annual Review of Biochemistry, Vol 84 Vol. 84 Annual Review of Biochemistry (ed R. D. Kornberg) 577–601 (2015). [DOI] [PubMed]
- 2.Morell EA, Balkin DM. Methicillin-Resistant Staphylococcus Aureus: A Pervasive Pathogen Highlights the Need for New Antimicrobial Development. The Yale Journal of Biology and Medicine. 2010;83:223–233. [PMC free article] [PubMed] [Google Scholar]
- 3.Weese JS, Van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Veterinary Microbiology. 2010;140:418–429. doi: 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Harrison, E. M. et al. A Shared Population of Epidemic Methicillin-Resistant Staphylococcus aureus 15 Circulates in Humans and Companion Animals. mBio5, 10.1128/mBio.00985-13 (2014). [DOI] [PMC free article] [PubMed]
- 5.Gomez-Sanz E, Torres C, Lozano C, Zarazaga M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comparative Immunology Microbiology and Infectious Diseases. 2013;36:83–94. doi: 10.1016/j.cimid.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Stegmann R, Burnens A, Maranta CA, Perreten V. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. Journal of Antimicrobial Chemotherapy. 2010;65:2047–2048. doi: 10.1093/jac/dkq241. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin JL, McCamley RN. Otitis-Externa Associated With Pseudomonas-Aeruginosa Infection. Veterinary Record. 1978;103:343–343. doi: 10.1136/vr.103.15.343-a. [DOI] [PubMed] [Google Scholar]
- 8.Brochmann RP, Helmfrid A, Jana B, Magnowska Z, Guardabassi L. Antimicrobial synergy between carprofen and doxycycline against methicillin-resistant Staphylococcus pseudintermedius ST71. BMC Veterinary Research. 2016;12:126. doi: 10.1186/s12917-016-0751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. International Journal of Medical Microbiology. 2011;301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Paul NC, Bärgman SC, Moodley A, Nielsen SS, Guardabassi L. Staphylococcus pseudintermedius colonization patterns and strain diversity in healthy dogs: A cross-sectional and longitudinal study. Veterinary Microbiology. 2012;160:420–427. doi: 10.1016/j.vetmic.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Collignon P. Antibiotic resistance: are we all doomed? Internal Medicine Journal. 2015;45:1109–1115. doi: 10.1111/imj.12902. [DOI] [PubMed] [Google Scholar]
- 12.Mercer DK, O’Neil DA. Peptides as the next generation of anti-infectives. Future Medicinal Chemistry. 2013;5:315–337. doi: 10.4155/fmc.12.213. [DOI] [PubMed] [Google Scholar]
- 13.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 14.Hancock R. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 15.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Critical Reviews in Biotechnology. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 16.Haney, E. F., Mansour, S. C. & Hancock, R. E. W. In Antimicrobial Peptides: Methods and Protocols (ed Paul R. Hansen) 3–22 (Springer New York, 2017). [DOI] [PubMed]
- 17.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discovery Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Molchanova, N., Hansen, P. R. & Franzyk, H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules22, 10.3390/molecules22091430 (2017). [DOI] [PMC free article] [PubMed]
- 19.Lee J, et al. Effect of side chain hydrophobicity and cationic charge on antimicrobial activity and cytotoxicity of helical peptoids. Bioorganic & Medicinal Chemistry Letters. 2018;28:170–173. doi: 10.1016/j.bmcl.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 20.Hamper BC, Kolodziej SA, Scatez AM, Smith RG, Cortez E. Solid phase synthesis of b-peptiods: N-substituted b-aminopropionic Acid Oligomers. J. Org. Chem. 1998;63:708–718. doi: 10.1021/jo971675w. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, DeGrado W. De novo design, synthesis, and characterization of antimicrobial beta-peptides. J Am Chem Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 22.Molchanova N, Hansen PR, Damborg P, Nielsen HM, Franzyk H. Lysine-Based α-peptide/β-peptoid peptidomimetics: Influence of hydrophobicity, fluorination and distribution of cationic charge on antimicrobial activity and cytotoxicity. ChemMedChem. 2017;20:312–318. doi: 10.1002/cmdc.201600553. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, et al. Short Antimicrobial Lipo-α/γ-AA Hybrid Peptides. ChemBioChem. 2014;15:2275–2280. doi: 10.1002/cbic.201402264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoro D, Maddox CW. Canine antimicrobial peptides are effective against resistant bacteria and yeasts. Veterinary Dermatology. 2014;25:35–e12. doi: 10.1111/vde.12091. [DOI] [PubMed] [Google Scholar]
- 25.Mohamed MF, Hammac GK, Guptill L, Seleem MN. Antibacterial Activity of Novel Cationic Peptides against Clinical Isolates of Multi-Drug Resistant Staphylococcus pseudintermedius from Infected Dogs. PLoS One. 2014;9:e116259. doi: 10.1371/journal.pone.0116259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molchanova N, Hansen P, Damborg P, Franzyk H. Fluorinated antimicrobial lysine-based peptidomimetics with activity against methicillin-resistant Staphylococcus pseudintermedius. J. Peptide Sci. 2018 doi: 10.1002/psc.3098.. [DOI] [PubMed] [Google Scholar]
- 27.Fazakerley, J., Crossley, J., McEwan, N., Carter, S. & Nuttall, T. In vitro antimicrobial efficacy of β‐defensin 3 against Staphylococcus pseudintermedius isolates from healthy and atopic canine skin. Vet Dermatol. 21(5), 463–468 (2010). [DOI] [PubMed]
- 28.Itoh H, et al. Total Synthesis and Biological Mode of Action of WAP-8294A2: A Menaquinone-Targeting Antibiotic. The Journal of Organic Chemistry. 2017;83:6924–6935. doi: 10.1021/acs.joc.7b02318. [DOI] [PubMed] [Google Scholar]
- 29.Cabassi CS, et al. Activity of AMP2041 against human and animal multidrug resistant Pseudomonas aeruginosa clinical isolates. Annals of Clinical Microbiology and Antimicrobials. 2017;16:17. doi: 10.1186/s12941-017-0193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco I, et al. In Vitro ADME Properties of Two Novel Antimicrobial Peptoid-Based Compounds as Potential Agents against Canine Pyoderma. Molecules. 2018;23:630. doi: 10.3390/molecules23030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabassi CS, et al. Broad-spectrum activity of a novel antibiotic peptide against multidrug-resistant veterinary isolates. The Veterinary Journal. 2013 doi: 10.1016/j.tvjl.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Smith DK, Moulding K, Chen HM. The Dependence of Membrane Permeability by the Antibacterial Peptide Cecropin B and Its Analogs, CB-1 and CB-3, on Liposomes of Different Composition. J. Biol. Chem. 1998;273:27438–27448. doi: 10.1074/jbc.273.42.27438. [DOI] [PubMed] [Google Scholar]
- 34.Le, C. F., Fang, C. M. & Sekaran, S. D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrobial Agents and Chemotherapy61, 10.1128/aac.02340-16 (2017). [DOI] [PMC free article] [PubMed]
- 35.Graf M, et al. Proline-rich antimicrobial peptides targeting protein synthesis. Natural Product Reports. 2017;34:702–711. doi: 10.1039/C7NP00020K. [DOI] [PubMed] [Google Scholar]
- 36.Amiche M, Seon AA, Pierre TN, Nicolas P. The dermaseptin precursors: a protein family with a common preproregion and a variable C-terminal antimicrobial domain. FEBS Letters. 1999;456:352–356. doi: 10.1016/S0014-5793(99)00964-3. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and Mode of Action of Lantibiotics. Chemical Reviews. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 38.Schneider T, et al. Plectasin, a Fungal Defensin, Targets the Bacterial Cell Wall Precursor Lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 39.Subbalakshmi C, Sitaram N. Mechanism of antimicrobial action of indolicidin. Fems Microbiology Letters. 1998;160:91–96. doi: 10.1111/j.1574-6968.1998.tb12896.x. [DOI] [PubMed] [Google Scholar]
- 40.Gottschalk S, et al. The antimicrobial lysine-peptoid hybrid LP5 inhibits DNA replication and induces the SOS response in Staphylococcus aureus. BMC microbiology. 2013;13:192. doi: 10.1186/1471-2180-13-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.‘t Hart P, Oppedijk SF, Breukink E, Martin NI. New Insights into Nisin’s Antibacterial Mechanism Revealed by Binding Studies with Synthetic Lipid II Analogues. Biochemistry. 2016;55:232–237. doi: 10.1021/acs.biochem.5b01173. [DOI] [PubMed] [Google Scholar]
- 42.Hsu, S.-T. D. et al. The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nature Structural &Amp; Molecular Biology 11, 963, 10.1038/nsmb830, https://www.nature.com/articles/nsmb830#supplementary-information (2004). [DOI] [PubMed]
- 43.Breukink E, et al. Use of the Cell Wall Precursor Lipid II by a Pore-Forming Peptide Antibiotic. Science. 1999;286:2361. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 44.Memariani H, et al. Mechanism of action and in vitro activity of short hybrid antimicrobial peptide PV3 against Pseudomonas aeruginosa. Biochemical and Biophysical Research Communications. 2016;479:103–108. doi: 10.1016/j.bbrc.2016.09.045. [DOI] [PubMed] [Google Scholar]
- 45.Runti, G. et al. The Mechanism of Killing by the Proline-Rich Peptide Bac7(1-35) against Clinical Strains of Pseudomonas aeruginosa Differs from That against Other Gram-Negative Bacteria. Antimicrobial Agents and Chemotherapy61, 10.1128/aac.01660-16 (2017). [DOI] [PMC free article] [PubMed]
- 46.Soler M, et al. Identification of BP16 as a non-toxic cell-penetrating peptide with highly efficient drug delivery properties. Organic & Biomolecular Chemistry. 2014;12:1652–1663. doi: 10.1039/C3OB42422G. [DOI] [PubMed] [Google Scholar]
- 47.Ilić N, et al. Selective antimicrobial activity and mode of action of adepantins, glycine-rich peptide antibiotics based on anuran antimicrobial peptide sequences. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1828;1004-1012:2013. doi: 10.1016/j.bbamem.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Jahnsen RD, et al. Tailoring Cytotoxicity of Antimicrobial Peptidomimetics with High Activity against Multidrug-Resistant Escherichia coli. Journal of Medicinal Chemistry. 2014;57:2864–2873. doi: 10.1021/jm401335p. [DOI] [PubMed] [Google Scholar]
- 49.Frosini, S. M., Bond, R., Loeffler, A. & Larner, J. Opportunities for topical antimicrobial therapy: permeation of canine skin by fusidic acid. Bmc Veterinary Research13, 10.1186/s12917-017-1270-6 (2017). [DOI] [PMC free article] [PubMed]
- 50.Aalbaek B, et al. Coryneform bacteria associated with canine otitis externa. Vet Microbiol. 2010;145:292–298. doi: 10.1016/j.vetmic.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 51.Galperine T, et al. Streptococcus canis infections in humans: Retrospective study of 54 patients. Journal of Infection. 2007;55:23–26. doi: 10.1016/j.jinf.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Kim B, Pai H, Lee KH, Lee Y. Identification of Pasteurella canis in a Soft Tissue Infection Caused by a Dog Bite: The First Report in Korea. Annals of Laboratory Medicine. 2016;36:617–619. doi: 10.3343/alm.2016.36.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tremblay, C. L., Charlebois, A., Masson, L. & Archambault, M. Characterization of hospital-associated lineages of ampicillin-resistant Enterococcus faecium from clinical cases in dogs and humans. Frontiers in Microbiology4, 10.3389/fmicb.2013.00245 (2013). [DOI] [PMC free article] [PubMed]
- 54.Eckert R, et al. Adding selectivity to antimicrobial peptides: Rational design of a multidomain peptide against Pseudomonas spp. Antimicrobial Agents and Chemotherapy. 2006;50:1480–1488. doi: 10.1128/aac.50.4.1480-1488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo LH, et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7569–7574. doi: 10.1073/pnas.1506207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uggerhøj LE, Munk JK, Hansen PR, Güntert P, Wimmer R. Structural features of peptoid–peptide hybrids in lipid–water interfaces. FEBS Letters. 2014;588:3291–3297. doi: 10.1016/j.febslet.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Manning MC, Illangasekare M, Woody W. R. Circular dichroism studies of distorted α-helices, twisted β-sheets, and β-turns. Biophysical Chemistry. 1988;31:77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 58.Grieco P, et al. The effect of d-amino acid substitution on the selectivity of temporin L towards target cells: Identification of a potent anti-Candida peptide. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828:652–660. doi: 10.1016/j.bbamem.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 59.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self defence? Biochem Soc. Trans. 2001;29:598–601. doi: 10.1042/bst0290598. [DOI] [PubMed] [Google Scholar]
- 60.Pomba C, et al. Public health risk of antimicrobial resistance transfer from companion animals. Journal of Antimicrobial Chemotherapy. 2017;72:957–968. doi: 10.1093/jac/dkw481. [DOI] [PubMed] [Google Scholar]
- 61.Oddo A, et al. An all-D amphipathic undecapeptide shows promising activity against colistin-resistant strains of Acinetobacter baumannii and a dual mode of action. Antimicrobial Agents and Chemotherapy. 2016;60:592–599. doi: 10.1128/aac.01966-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Standards, N. C. f. C. L. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved standard (2012).
- 63.Blondeau JM, Borsos S, Blondeau LD, Blondeau BJ. In vitro killing of Escherichia coli, Staphylococcus pseudintermedius and Pseudomonas aeruginosa by enrofloxacin in combination with its active metabolite ciprofloxacin using clinically relevant drug concentrations in the dog and cat. Veterinary Microbiology. 2012;155:284–290. doi: 10.1016/j.vetmic.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 64.Munk JK, Ritz C, Fliedner FP, Frimodt-Moller N, Hansen PR. Novel method to identify the optimal antimicrobial peptide in a combination matrix using anoplin as an example. Antimicrob Agents Chemother. 2014;58:1063–1070. doi: 10.1128/aac.02369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature517, 455–459, 10.1038/nature14098, http://www.nature.com/nature/journal/v517/n7535/abs/nature14098.html#supplementary-information (2015). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.