Abstract
Cancers develop due to the accumulation of genetic and epigenetic alterations. Genetic alterations are induced by aging, mutagenic chemicals, ultraviolet light, and other factors; whereas, epigenetic alterations are mainly by aging and chronic inflammation. The accumulation and patterns of alterations in normal cells reflect our past exposure levels and life history. Most accumulated alterations are considered as passengers, but their accumulation is correlated with cancer drivers. This has been shown for aberrant DNA methylation but has only been speculated for genetic alterations. However, recent technological advancements have enabled measurement of rare point mutations, and studies have shown that their accumulation levels are indeed correlated with cancer risk. When the accumulation levels of aberrant DNA methylation and point mutations are combined, risk prediction becomes even more accurate. When high levels of alterations accumulate, the tissue has a high risk of developing cancer or even multiple cancers and is considered as a “cancerization field”, with or without expansion of physiological patches of clonal cells. In this review, we describe the formation of a cancerization field and how we can apply its detection in precision cancer risk diagnosis.
Introduction
Human cancers develop due to the accumulation of genetic and epigenetic alterations. Both alterations are now known to be present not only in cancer cells but also in normal cells long before cancer develops. Specific patterns of alterations are associated with exposure to environmental factors. The accumulation is associated with cancer risk and can be utilized for cancer risk diagnosis. Tissue with accumulated alterations is known as a “field for cancerization (cancerization field)”, with or without expansion of physiological patches of clonal cells.
Historically, the measurement of genetic alterations in normal cells has been difficult due to a technical limitation in measuring the extremely low frequency of genetic alterations. However, recent technological advancements have enabled their measurement, and cancer risk assessment using accumulated genetic alterations is now in sight. In this review, we describe the mechanisms by which genetic and epigenetic cancerization fields are induced, its characteristics, and how we can apply the field in precision cancer risk diagnosis.
Genetic and epigenetic alterations in normal cells reflect an individual’s life history and host responses
Inducers of genetic alterations
Genetic alterations are induced by aging, mutagenic chemicals, radiation, ultraviolet light (UV), oxygen radical, and other factors (Table 1). Aging is known to be the major cancer risk factor,1 and the total number of stem cell divisions, which depends on tissue types, is largely correlated with cancer risk.2 Actually, a substantial portion of somatic mutations accumulate early in an individual’s lifetime when stem cells show a high division rate.1 In addition to aging, exposure to mutagenic factors induces somatic mutations in human tissues that eventually lead to cancer.3 Exposure to UV light4 and radiation are also well-known inducers of somatic mutations. Furthermore, oxygen radicals,5 constantly produced even in physiological conditions and increased in inflammatory conditions, are also believed to induce somatic mutations.
Table 1.
Inducers of genetic and epigenetic alterations
| Type of alterations | Inducers | Reference |
|---|---|---|
| Genetic alterations | Aging | Rozhok et al.1 |
| Mutagenic chemicals | Cogliano et al.3 | |
| Acetaldehyde generated from ethanol | ||
| Aflatoxins from the fungus | ||
| Benzo(a)pyrene (B[a]P) in incomplete combustion of organic substances, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in cigarette smoke, and so on | ||
| Radiation | ||
| Ultraviolet light (UV) | Bykov et al.4 | |
| Oxygen radicals | Reardon et al.5 | |
| Epigenetic alterations | Aging | Issa et al.20 |
| Chronic inflammation | ||
| Ulcerative colitis | Issa et al.21 | |
| H. pylori infection-triggered gastritis | Maekita et al.22 Niwa et al.23 |
|
| HBV or HCV infection-triggered hepatitis | Nishida et al.24 | |
| Cigarette smoking | Liu et al.25 | |
| Estrogen (in vitro culture) | Cheng et al.27 |
HBV hepatitis B virus, HCV hepatitis C virus
Reflections of past exposures by mutation signatures in cancer tissues
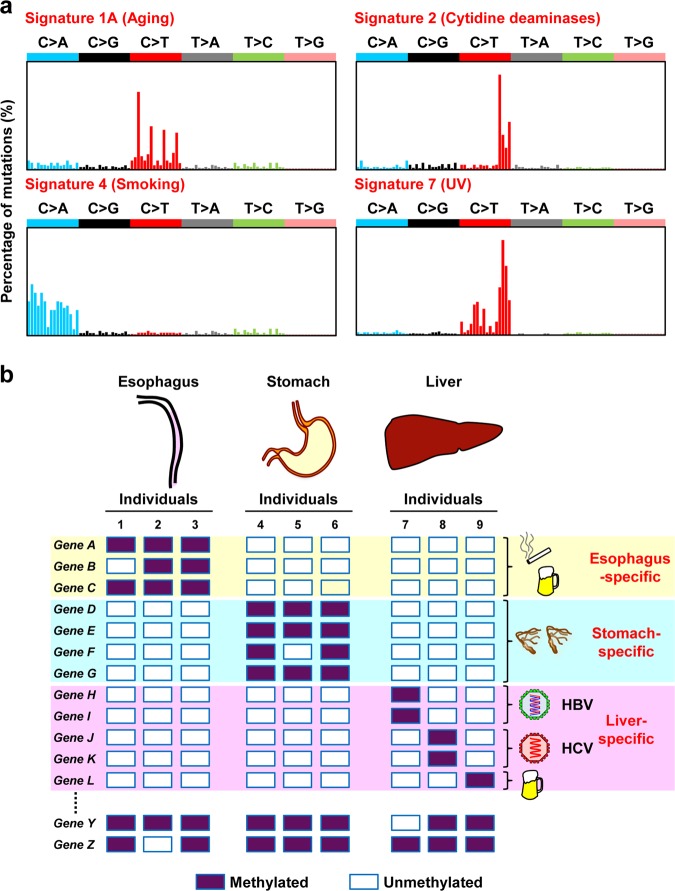
Exposure to a specific agent induces a specific combination of mutations, referred to as a “mutation signature” (Fig. 1a).6,7 Such mutation signatures have been mainly identified in cancer tissues, in which both driver and passenger mutations can be readily detected due to clonal expansion of cancer cells. Most recently, approximately 85,000,000 mutations in more than 23,829 cancers were classified into single base substitutions (SBS) in 96 trinucleotide contexts, doublet base substitutions (DBS), and small insertion and deletions (ID).6,7 Non-biased analysis was able to extract 67 characteristic patterns of SBS, and 49 of them were considered to be associated with exposure to specific carcinogens. Signature 1, characterized by C>T transitions at NpCpG trinucleotides, is associated with aging; signature 7, reflecting a large number of CC>TT dinucleotide mutations at dipyrimidines, with UV light; signature 4, characterized by C>A transversions with strand bias, with cigarette smoking; signature 2, characterized by C>T and C>G mutations at TpCpN trinucleotides, with excessive activity of cytidine deaminases (AID/APOBEC), which is observed in chronic inflammation. Mutation signatures in cancer tissues are composed of mutations accumulated not only before cancer development but also after development.
Fig. 1.
Reflection of past exposure to carcinogens in genetic and epigenetic alterations. a Reflection in the mutational signature. Exposure to a carcinogen induces a specific mutation signature (adapted from Alexandrov et al.7). b Reflection in methylated gene profiles. A different set of genes are aberrantly methylated depending on the tissue types and possibly on the inducers. Unexpressed genes are different among tissues, and specific genes are susceptible to methylation induction in individual tissues, such as Genes A, B, and C in the esophagus, Genes D, E, F, and G in the stomach, and Genes H, I, J, K, and L in the liver. Even among liver-specific susceptible genes, genes are methylated reflecting the cause of inflammation, such as Genes H and I by HBV infection, Genes J and K by HCV infection, and Gene L by alcohol
Specific patterns of copy number alterations can be also present in cancer tissues, referred to as a “copy number signature”. For example, copy number signatures in high-grade serous ovarian cancers can predict overall patient survival and the probability of platinum-resistant relapse.8 However, it is still unclear whether or not the copy number signature reflects past exposure to specific carcinogens.
Mutation signatures in normal tissues
Since a signature of somatic mutations in normal tissues is least influenced by the biological selection of specific mutations, it can provide vital information about past exposure to specific carcinogens. At the same time, a normal tissue is composed of a large number of clonal patches of cells,9 and a specific mutation, if ever, is present only in one of the many patches. DNA with the mutation constitutes only a very minor fraction in DNA from the bulk tissue, and it used to be very difficult to detect such somatic mutations.10 Thankfully, the development of new cutting-edge detection technologies11–14 has now enabled detection of rare mutations in a normal tissue exposed to a specific carcinogen,15 and mutation signatures can be now analyzed in normal tissues.
Signature 7 was detected in normal skin tissues, which are exposed to UV light.16 C>T transitions at GpCpN trinucleotides were detected in normal gastric mucosae exposed to gastritis triggered by Helicobacter pylori (H. pylori) infection, which induces up-regulation of AID.17 Signature 1 and signature 5, T>C transitions at ApTpN trinucleotides, were predominant in normal esophageal tissues,18,19 and C>A transversions were frequently detected in those of individuals with a severe smoking history.15 The signatures in these normal tissues were in good accordance with those in cancer tissues due to the same carcinogens. Therefore, mutation signatures reflect past exposure to carcinogens, namely an individual’s life history.
Inducers of epigenetic alterations
Epigenetic alterations also appear to be induced by exposure to various environmental stimuli (Table 1). Aging was first shown to be correlated with increased levels of aberrant DNA methylation, possibly as a function of somatic cell replications.20 Subsequently, chronic inflammation triggered by various factors was also shown to be causally involved in inducing aberrant DNA methylation.21–24 Additionally, cigarette smoking was reported to induce aberrant DNA methylation in vitro,25 which was in line with the in vivo finding of high DNA methylation levels in normal esophageal tissues of individuals with a long smoking history.26 Estrogen treatment was also reported as an inducer of aberrant DNA methylation in cultured mammary epithelial cells.27
Reflection of past exposure and host responses in methylated gene profiles
A different set of genes are methylated depending on the tissue types (tumor types)28 and are likely methylated depending on the inducers (Fig. 1b). Mechanistically, genes that undergo DNA methylation are instructed by pre-existing epigenetic modifications.29 Namely, genes with high levels of trimethylation of histone H3 lysine 27 (H3K27me3) are susceptible to induction of aberrant DNA methylation,30–32 and genes lacking RNA polymerase II (pol II) binding are also likely to become methylated.33 Because the H3K27me3 status and distribution of pol II are different among the tissue types and are altered by exposure to different inducers, such as oxidative damage and colitis,34,35 exposure to a specific inducer likely leads to methylation of a specific set of genes according to the tissue type. Chemicals, such as cobalt compounds and cigarette smoke condensate, were also reported to induce alterations of the H3K27me3 status.25,36
Phenotypically, carcinogen-specific induction of aberrant DNA methylation has been demonstrated in hepatocellular carcinomas (HCCs) and cirrhotic liver tissues associated with HBV infection, HCV infection, or alcohol.37,38 HCV infection induced extensive methylation at more than 18,000 unique CpG sites, while HBV infection and alcohol-induced moderate methylation at 400–600 unique CpG sites, respectively.37 Once aberrant DNA methylation is induced at specific genes, the methylation profiles can persist throughout an individual’s lifetime, even after the inducer is no longer present.39
The accumulation level of epigenetic alterations reflects not only the type of inducer but also the duration of exposure to the inducer.39 Additionally, the accumulation level can reflect the differences in host responses to an environmental stimulus, which are determined by a single nucleotide polymorphism (SNP) of a specific gene. A well-established example is the influence of the IL1B genotype, which influences gastric cancer incidence in H. pylori-infected populations,40,41 on DNA methylation levels in normal gastric tissues.42 Individuals who have SNPs that secrete more IL-1β in response to H. pylori-triggered inflammation have higher levels of aberrant DNA methylation and higher risks of gastric cancer.
Taken together, analyzing genetic and epigenetic alterations accumulated in normal cells can provide information on an individual’s life-time exposure to environmental factors that induce genetic and epigenetic alterations that may eventually lead to cancer.
Formation of a field for cancerization by genetic and epigenetic alterations
What is the field for cancerization?
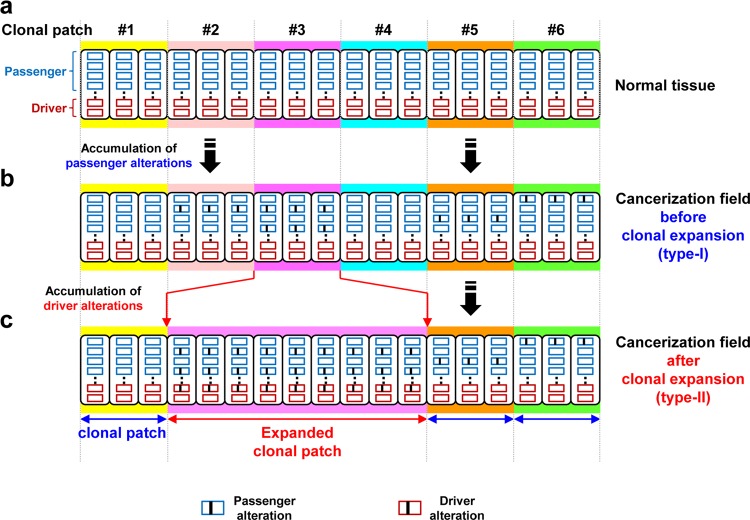
Some cancers, particularly those associated with chronic inflammation, often develop at multiple foci in a tissue. This phenomenon is known as the concept of “field cancerization” or “a field for cancerization”.43–45 The concept was first proposed by Slaughter et al. based upon the presence of multiple microscopic cancers in grossly normal mucosa of patients with oral squamous carcinomas.43 The finding was further advanced by detecting TP53 mutations in clonally expanded patches of cells, which may or may not be detected by routine microscopic examination, in multiple types of cancers (Fig. 2c).
Fig. 2.
Formation of a field for cancerization. a Normal tissues are assembled from clonal patches of normal cells. b Genetic and epigenetic alterations are potently induced by exposure to specific inducers, and passenger alterations are mainly accumulated in normal tissues without expansion. c Both passenger and driver alterations, which can induce monoclonal cell proliferation, are accumulated in normal tissues with expansion
In contrast, even without the expansion of clonal patches of cells, the cancerization field can be formed by accumulating mutations and/or aberrant DNA methylation. Especially, passenger alterations are far more frequent than driver alterations,15,46,47 and can be already accumulated in normal tissues without expansion of clonal patches (field type-I, Fig. 2b). The accumulation of passenger alterations is likely to be associated with the accumulation of driver alterations that have not yet caused the expansion of clonal patches. In contrast, driver alterations will eventually cause the expansion of clonal patches, and thus both passenger and driver alterations are accumulated in normal tissues with the expansion of clonal patches (field type-II, Fig. 2c). Such expansion of clonal patches is considered to be histologically normal initially, but can eventually lead to the formation of histologically distinguishable premalignant lesions.
Experimental demonstration of the presence of a cancerization field, however, has been difficult again because of the extremely low frequency of genetic alterations accumulated in normal cells. Historically, to overcome this issue, multiple transgenic animal models with marker genes were developed.48 A mutation in a marker gene can be selected by observing the altered function of the gene, such as plaque color and drug resistance, and rare mutations can be measured.49,50 Using these animal models, the frequency of somatic mutations in normal tissues was found to be in the levels of 10−6 to 10−4 per gene even after exposure to high levels of mutagenic chemicals. The animal models also showed increased mutation frequencies by exposure to various carcinogens.
Detection strategies for rare mutations in normal tissues
In human tissues, as repeatedly mentioned, the detection of rare mutations was previously very difficult. However, it was recently achieved by (1) using samples containing a limited number of clonal patches,16,18,19,51 and (2) enhancing the accuracy of next-generation sequencing.
The sampling strategy of only a limited number of clonal patches from a small piece of tissue was used to measure mutations accumulated in histologically normal human skin tissues16 and esophageal tissues.18,19 Epithelial organoids expanded from a single stem cell were used to measure mutations accumulated in normal adult tissue stem cells of the small intestine, colon, and liver.51 These studies revealed rare mutation characteristics of ultraviolet exposure or tissues of the origin. However, because of the high sequencing cost of analyzing a large number of human clinical samples, the association of these mutations with cancer risk could not be robustly established.
To measure rare mutations using next-generation sequencing, it is important to distinguish true mutations in a single or small number of DNA molecules from sequencing errors that arise in up to 1% of sequence reads covering a specific base position due to errors during PCR amplification and sequencing. This issue was overcome by multiple strategies for the preparation of sequencing libraries. By tagging individual DNA molecules with unique molecular barcodes, a true mutation is detected as a mutation in multiple sequence reads with the same barcode.10 Some of the molecular barcode-based methods, such as duplex sequencing,12 can distinguish a true mutation on both strands of a DNA duplex (fixed mutations) and a variation on either strand of a DNA duplex (derived from a pre-mutagenic lesion or a PCR error at early PCR cycles). However, these methods require a large number of sequencing reads, and it is very costly to analyze a large number of samples because of high sequencing cost.
To reduce sequencing cost, methods using a small number of template DNA molecules have been developed. Bottleneck sequencing improved the molecular barcoding system by adding a simple dilution step before amplification of the sequencing library.13 We also developed the 100 copy-based method, in which accurately quantified 100 copies of genomic DNA are used as a PCR template for amplicon sequencing and a true mutation (variant allele frequency, 1%) can be distinguished from sequencing errors (frequency, less than 1%).14 However, these methods cannot detect mutations at specific base positions, such as activating mutations of oncogenes. Bottleneck sequencing, which also adapts molecular barcodes, can distinguish a true mutation and a pre-mutagenic lesion.
Furthermore, the enhancement of the accuracy of next-generation sequencing has been also achieved by developing computational and statistical strategies to exclude sequences of low confidence, such as sequencing errors.16,18,52 Sequencing errors are excluded by comparing the frequencies of detected mutations with those of the background sequencing errors. In addition, the detection methods of somatic mutations at a single-cell level are now being attempted.53 However, current methods suffer from amplification bias and the high cost of library preparation, and multiple methods to reduce the bias and cost are being developed.52,54
Involvement of genetic alterations in a field
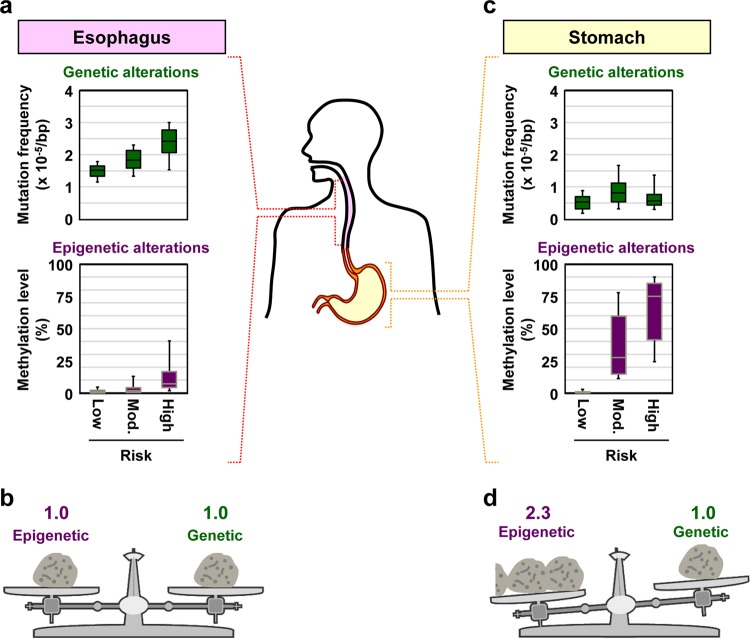
The impact of accumulation of point mutations in normal tissues on cancer risk was demonstrated using the 100 copy-based method. Frequencies of point mutations were measured in esophageal tissues with a low, intermediate, or high risk of esophageal squamous cell carcinoma (ESCC) (Fig. 3a). The mutation frequency in esophageal tissues (non-cancerous tissues) of ESCC patients (high-risk group) was 1.4-fold higher than that in esophageal tissues of healthy people without any exposure to ESCC risk factors (low-risk group). Even among people exposed to the risk factors, the mutation frequency in the high-risk group was higher than that in the intermediate-risk group (healthy people with exposure to the risk factors). The 100 copies of genomic DNA were obtained from a piece of biopsy (about 0.75 mm3), corresponding to about 750,000 cells, and were likely to contain a large number of patches. Therefore, the mutations detected were likely to be derived from independent patches, and to be involved in cancerization field type-I. In contrast, in normal gastric tissues, a risk level-dependent increase in the mutation frequency was not observed (Fig. 3c). Namely, the mutation frequency in non-cancerous gastric tissues of gastric cancer patients (high-risk group) was similar to that in healthy people with H. pylori infection.
Fig. 3.
Different impacts of genetic and epigenetic alterations on cancer risk. Both genetic and epigenetic alterations are involved in forming a field for cancerization, but their relative contributions differ depending on the cancer types. a, b The impacts of genetic and epigenetic alterations are similar for ESCCs. Box plots represent median (center line), upper, and lower quartiles (box limits), and 91 and 9 percentiles (whiskers). The original data were obtained from our previous publication.15 c, d The impact of epigenetic alterations is higher than that of genetic alterations for gastric cancer
Most accumulated genetic alterations are considered as passengers, but their accumulation is correlated with cancer drivers. The functions of mutations accumulated in normal tissues can be assessed by the ratio of non-synonymous mutations/synonymous mutations. In normal skin tissues, NOTCH1 and NOTCH2 mutations showed a significant excess of non-synonymous mutations, and clustered in the extracellular epidermal growth factor-like domain.16 In normal esophageal tissues, NOTCH1 mutations were present in 25–42% of the cells, and were considered to be associated with clonal expansion.18,19 In benign lesions of melanoma patients, which are clearly with clonal expansion, BRAF V600E mutation was detected.55 In normal blood cells, specific chromosomal alterations were detected in expanded clonal cells, and were associated with increased risk of hematological malignancies.56 These mutations are considered to be involved in the formation of cancerization field type-II, which is associated with the expansion of clonal patches.
Experimentally, the possibility of contamination of an extremely small number of cancer cells in non-cancerous tissues cannot be completely excluded, as HER2-positive cancer cells were detected in adjacent normal breast tissues.57
Involvement of epigenetic alterations in a field
The impact of accumulation of aberrant DNA methylation on cancer risk was shown much more easily because its accumulation levels in normal tissues are high and can be readily measured. In the stomach, DNA methylation levels in non-cancerous gastric tissues increased in the order of healthy people without H. pylori infection (low risk), healthy people with past H. pylori infection (intermediate risk), patients with a single gastric cancer, and patients with multiple gastric cancers (high risk).22,58 In the mammary glands, the DNA methylation levels of RASSF1A in non-cancerous tissues of breast cancer patients were higher than those in reduction mammoplasty (normal) tissues.59 Additionally, the presence of an epigenetic field for cancerization was shown for colon cancers,60 esophageal adenocarcinomas,61,62 liver cancers,24,63 urothelial cancers,64 and cervical cancers.65
Most epigenetic alterations accumulated in normal tissues are considered to be passengers, which are involved in cancerization field type-I. However, their accumulation levels are correlated with those of drivers of cancer. Passenger genes tend to be methylated in a larger fraction of normal cells compared to driver genes. Therefore, measuring DNA methylation levels of passenger genes is technically easy and thus accurate, and is considered to be suitable for cancer risk diagnosis. As for the epigenetic alterations of driver genes, methylation-silencing of several tumor-suppressor genes, such as CDKN2A (p16), CDH1 (E-cadherin), and SMARCA166 has been reported. Alterations in these driver genes accumulate only at low levels but are considered to be causally involved in the formation of a field. Taken all together, accumulation of genetic and epigenetic alterations in normal tissues can be associated with cancer risk.
Different impacts of genetic and epigenetic alterations and their combination
Both genetic and epigenetic alterations are involved in forming a field for cancerization, but their contributions differ depending on the cancer type.15 In normal esophageal tissues, both the mutation frequency and DNA methylation levels increased according to the risk level of ESCCs (Fig. 3a), indicating that the impact of genetic and epigenetic alterations on cancer risk is similar for ESCCs (Fig. 3b). In normal gastric tissues, DNA methylation levels increased in a risk level-dependent manner, but the mutation frequency did not (Fig. 3c). The impact of epigenetic alterations was 2.3-fold higher than that of genetic alterations in the gastric tissues (Fig. 3d).
The different impacts of genetic and epigenetic alterations on cancer risk are considered to depend on the major carcinogens involved and their carcinogenic mechanisms. In the esophagus, cigarette smoking and alcohol intake, which induce both genetic and epigenetic alterations, are major risk factors of ESCCs. Therefore, the impact of genetic and epigenetic alterations is considered to be similar in the esophagus. In the stomach, chronic inflammation triggered by H. pylori infection, a strong inducer of aberrant DNA methylation, is nearly an exclusive risk factor for gastric cancers. Therefore, the impact of epigenetic alterations is considered stronger in the stomach.
The involvement of both genetic and epigenetic alterations in ESCC risk led to the idea that combining both alterations can achieve a more precise risk diagnosis.15 Traditional risk factors only (aging, cigarette smoking, alcohol drinking, and betel quid chewing) showed a reasonable predictive power (c-index; range 0–1) of 0.851. The addition of the mutation frequency to the traditional risk factors improved the power to 0.894, and further addition of the methylation level improved the prediction power even more to 0.926. This showed that the combined measurement of both genetic and epigenetic alterations can achieve precise cancer risk estimation, and provide cancer risk information that cannot be obtained from life history of exposure to traditional risk factors. Additionally, how genetic and epigenetic alterations are combined should be optimized depending on the cancer types due to their different impacts.
Clinical implementation of precision cancer risk diagnosis
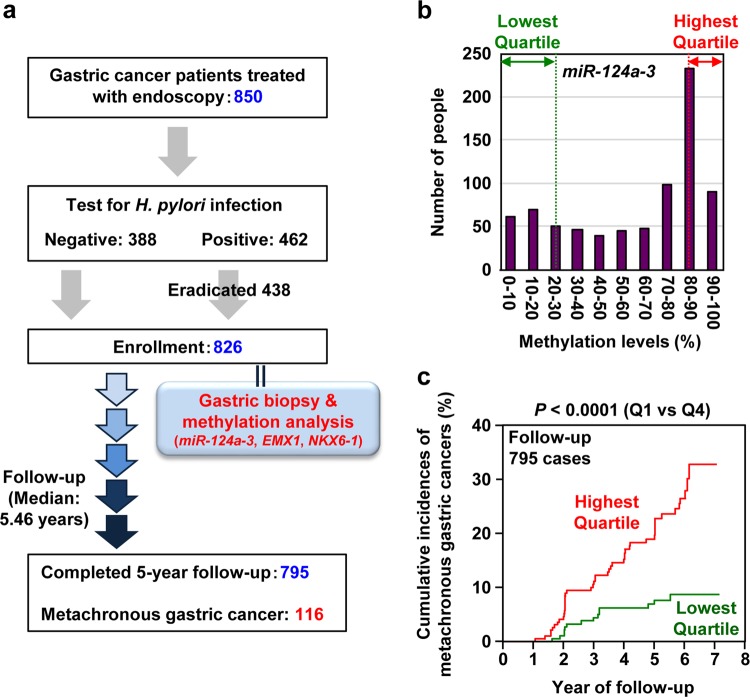
Even a prospective clinical study has demonstrated the usefulness of measuring the accumulation of alterations in normal tissues for cancer risk diagnosis. It has been considered and is now shown that aberrant DNA methylation (methylation burden) has a much larger impact on gastric cancer risk than mutations. Therefore, in this prospective study,67,68 only the methylation burden in a gastric mucosa was measured for 826 gastric cancer patients treated by endoscopic submucosal dissection (Fig. 4a). Only three pre-selected marker genes were measured to avoid multiple testing. After a median follow-up period of 5.46 years, patients who had the highest methylation burden (highest quartile) had a 3-fold higher risk of developing a metachronous gastric cancer than those who had the lowest burden (Fig. 4b, c).
Fig. 4.
A multicenter prospective clinical study to predict cancer risk by measuring accumulated alterations. a Study design of the prospective study. Among the 826 patients enrolled, 116 patients developed metachronous gastric cancer after 1 year of enrollment with a median follow-up period of 5.46 years. b Distribution of the DNA methylation level of one of the pre-selected marker genes, miR-124a-3, in non-cancerous gastric tissues. Patients in the highest quartile had DNA methylation levels of 88.1–91.8%, while those in the lowest quartile had those of 8.3–23.0%. c The impact of methylation burden on the risk of metachronous gastric cancer. The highest quartile had a 3-fold higher risk of developing a metachronous gastric cancer than the lowest quartile. Risk prediction among patients who have already been treated for the first cancer is generally very difficult, but was achieved by measuring the methylation burden. This figure was modified from our previous study68
Perspective
Determining an individual’s future cancer risk by measuring genetic and epigenetic alterations is a promising approach for precision cancer risk diagnosis. One potential limitation is that the target tissue needs to be collected for this strategy. However, the recent clinical practice involves routine biopsies of a variety of organs, such as gastrointestinal tract, liver, skin, prostate, and breast. Additionally, the causal involvement of epigenetic alterations is now suggested for various human chronic disorders, such as neurodegenerative and metabolic disorders. Therefore, risk diagnosis by measuring epigenetic alterations has the potential to be expanded to various human disorders other than cancer.
Acknowledgements
The authors are grateful to Dr. H. Yamada for her excellent assistance in designing the figures and to Dr. N. Hattori for her careful reading of the manuscript. The original work described here was supported by the National Cancer Center Research and Development Fund, and The Japan Agency for Medical Research and Development.
Author contributions
H.T. and T.U. collected relevant information and wrote the manuscript.
Competing interests
T.U. is a recipient of funding from Sysmex and Miraca Inc., and has filed a patent for detecting rare point mutations using a small number of sequencing templates.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rozhok AI, DeGregori J. The evolution of lifespan and age-dependent cancer risk. Trends Cancer. 2016;2:552–560. doi: 10.1016/j.trecan.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cogliano VJ, et al. Preventable exposures associated with human cancers. J. Natl. Cancer Inst. 2011;103:1827–1839. doi: 10.1093/jnci/djr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bykov VJ, Hemminki K. Assay of different photoproducts after UVA, B and C irradiation of DNA and human skin explants. Carcinogenesis. 1996;17:1949–1955. doi: 10.1093/carcin/17.9.1949. [DOI] [PubMed] [Google Scholar]
- 5.Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov, L. et al. The repertoire of mutational signatures in human cancer. bioRxiv (2018).
- 7.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macintyre G, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018;50:1262–1270. doi: 10.1038/s41588-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanpain C, Simons BD. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 2013;14:489–502. doi: 10.1038/nrm3625. [DOI] [PubMed] [Google Scholar]
- 10.Sloan DB, Broz AK, Sharbrough J, Wu Z. Detecting rare mutations and DNA damage with sequencing-based methods. Trends Biotechnol. 2018;36:729–740. doi: 10.1016/j.tibtech.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang ML, et al. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2016;113:9846–9851. doi: 10.1073/pnas.1607794113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita S, et al. A novel method to quantify base substitution mutations at the 10(-6) per bp level in DNA samples. Cancer Lett. 2017;403:152–158. doi: 10.1016/j.canlet.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita S, et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc. Natl. Acad. Sci. USA. 2018;115:1328–1333. doi: 10.1073/pnas.1717340115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martincorena I, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu T, et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology. 2014;147:407–417. doi: 10.1053/j.gastro.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Martincorena I, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoyama A, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 20.Issa JP, et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat. Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 21.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 22.Maekita T, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin. Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 23.Niwa T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 24.Nishida N, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene. 2010;29:3650–3664. doi: 10.1038/onc.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oka D, et al. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426. doi: 10.1002/cncr.24394. [DOI] [PubMed] [Google Scholar]
- 27.Cheng AS, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 29.Takeshima H, Ushijima T. Methylation destiny: Moira takes account of histones and RNA polymerase II. Epigenetics. 2010;5:89–95. doi: 10.4161/epi.5.2.10774. [DOI] [PubMed] [Google Scholar]
- 30.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 32.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 33.Takeshima H, Yamashita S, Shimazu T, Niwa T, Ushijima T. The presence of RNA polymerase II, active or stalled, predicts epigenetic fate of promoter CpG islands. Genome Res. 2009;19:1974–1982. doi: 10.1101/gr.093310.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Hagan HM, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshima H, et al. Induction of aberrant trimethylation of histone H3 lysine 27 by inflammation in mouse colonic epithelial cells. Carcinogenesis. 2012;33:2384–2390. doi: 10.1093/carcin/bgs294. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Ke Q, Costa M. Alterations of histone modifications by cobalt compounds. Carcinogenesis. 2009;30:1243–1251. doi: 10.1093/carcin/bgp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hlady RA, et al. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget. 2014;5:9425–9443. doi: 10.18632/oncotarget.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert MP, et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J. Hepatol. 2011;54:705–715. doi: 10.1016/j.jhep.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 39.Takeshima H, et al. Degree of methylation burden is determined by the exposure period to carcinogenic factors. Cancer Sci. 2017;108:316–321. doi: 10.1111/cas.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Omar EM, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 41.Loh M, et al. Meta-analysis of genetic polymorphisms and gastric cancer risk: variability in associations according to race. Eur. J. Cancer. 2009;45:2562–2568. doi: 10.1016/j.ejca.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 42.Yoo EJ, et al. Influence of IL1B polymorphism on CpG island hypermethylation in Helicobacter pylori-infected gastric cancer. Virchows Arch. 2010;456:647–652. doi: 10.1007/s00428-010-0918-4. [DOI] [PubMed] [Google Scholar]
- 43.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 44.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 45.Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat. Rev. Cancer. 2018;18:19–32. doi: 10.1038/nrc.2017.102. [DOI] [PubMed] [Google Scholar]
- 46.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nohmi T, Suzuki T, Masumura K. Recent advances in the protocols of transgenic mouse mutation assays. Mutat. Res. 2000;455:191–215. doi: 10.1016/S0027-5107(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 49.Gossen JA, et al. Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proc. Natl. Acad. Sci. USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohler SW, et al. The use of transgenic mice for short-term, in vivo mutagenicity testing. Genet. Anal. Tech. Appl. 1990;7:212–218. doi: 10.1016/0735-0651(90)90003-X. [DOI] [PubMed] [Google Scholar]
- 51.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018;19:269–285. doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linnarsson S, Teichmann SA. Single-cell genomics: coming of age. Genome Biol. 2016;17:97. doi: 10.1186/s13059-016-0960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shain AH, et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med. 2015;373:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 56.Loh PR, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559:350–355. doi: 10.1038/s41586-018-0321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sadanandam A, et al. Genomic aberrations in normal tissue adjacent to HER2-amplified breast cancers: field cancerization or contaminating tumor cells? Breast Cancer Res. Treat. 2012;136:693–703. doi: 10.1007/s10549-012-2290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakajima T, et al. Metachronous gastric cancers after endoscopic resection: how effective is annual endoscopic surveillance? Gastric Cancer. 2006;9:93–98. doi: 10.1007/s10120-006-0372-9. [DOI] [PubMed] [Google Scholar]
- 59.Yan PS, et al. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin. Cancer Res. 2006;12:6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 60.Shen L, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J. Natl. Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 61.Jin Z, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112–4115. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee YC, et al. Revisit of field cancerization in squamous cell carcinoma of upper aerodigestive tract: better risk assessment with epigenetic markers. Cancer Prev. Res. 2011;4:1982–1992. doi: 10.1158/1940-6207.CAPR-11-0096. [DOI] [PubMed] [Google Scholar]
- 63.Kondo Y, et al. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis—a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 64.Nishiyama N, et al. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–240. doi: 10.1111/j.1349-7006.2009.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verhoef VM, et al. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): a randomised controlled non-inferiority trial. Lancet Oncol. 2014;15:315–322. doi: 10.1016/S1470-2045(14)70019-1. [DOI] [PubMed] [Google Scholar]
- 66.Takeshima H, et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015;357:328–338. doi: 10.1016/j.canlet.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 67.Asada K, et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut. 2015;64:388–396. doi: 10.1136/gutjnl-2014-307094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maeda M, et al. High impact of methylation accumulation on metachronous gastric cancer: 5-year follow-up of a multicentre prospective cohort study. Gut. 2017;66:1721–1723. doi: 10.1136/gutjnl-2016-313387. [DOI] [PMC free article] [PubMed] [Google Scholar]