Abstract
2,6-diaminopurine (DAP) is a nucleobase analog of adenine. When incorporated into double-stranded DNA (dsDNA), it forms three hydrogen bonds with thymine. Rare in nature, DAP substitution alters the physical characteristics of a DNA molecule without sacrificing sequence specificity. Here, we show that in addition to stabilizing double-strand hybridization, DAP substitution also changes the mechanical and conformational properties of dsDNA. Thermal melting experiments reveal that DAP substitution raises melting temperatures without diminishing sequence-dependent effects. Using a combination of atomic force microscopy (AFM), magnetic tweezer (MT) nanomechanical assays, and circular dichroism spectroscopy, we demonstrate that DAP substitution increases the flexural rigidity of dsDNA yet also facilitates conformational shifts, which manifest as changes in molecule length. DAP substitution increases both the static and dynamic persistence length of DNA (measured by AFM and MT, respectively). In the static case (AFM), in which tension is not applied to the molecule, the contour length of DAP-DNA appears shorter than wild-type (WT)-DNA; under tension (MT), they have similar dynamic contour lengths. At tensions above 60 pN, WT-DNA undergoes characteristic overstretching because of strand separation (tension-induced melting) and spontaneous adoption of a conformation termed S-DNA. Cyclic overstretching and relaxation of WT-DNA at near-zero loading rates typically yields hysteresis, indicative of tension-induced melting; conversely, cyclic stretching of DAP-DNA showed little or no hysteresis, consistent with the adoption of the S-form, similar to what has been reported for GC-rich sequences. However, DAP-DNA overstretching is distinct from GC-rich overstretching in that it happens at a significantly lower tension. In physiological salt conditions, evenly mixed AT/GC DNA typically overstretches around 60 pN. GC-rich sequences overstretch at similar if not slightly higher tensions. Here, we show that DAP-DNA overstretches at 52 pN. In summary, DAP substitution decreases the overall stability of the B-form double helix, biasing toward non-B-form DNA helix conformations at zero tension and facilitating the B-to-S transition at high tension.
Introduction
Compared to the canonical base adenine, 2,6-diaminopurine (DAP) (alternatively 2-aminoadenine) bears an additional amino group at position 2 of the purine molecule (Fig. 1). Despite this difference, the incorporation of DAP during PCR amplification yields no loss in sequence specificity, and in most instances, DAP-DNA is compatible with normal (A-T, G-C) DNA enzymology. DAP-DNA is interesting both biologically and structurally for nanoscale engineering (1, 2). Although nature has generally chosen to use the canonical bases, there are instances, such as in the genome of cyanophage S-2L, in which DAP substitution occurs (3, 4). The biological advantages (or disadvantages) of DAP substitution are not entirely understood. From a biophysics perspective, DAP substitution offers a way of manipulating the physical characteristics of a DNA molecule with applications, including the study of the interactions between DNA and proteins or drug candidates (5, 6, 7, 8, 9, 10), investigations of RNA-related mechanisms (11, 12), and even as novel dopants in DNA-based nanoelectronics (13). Characterizing how DAP substitution affects the physical properties of DNA yields insight into the relationship between the specific properties of individual bases and the biochemical characteristics of the whole double helix assembled with such bases.
Figure 1.
2,6-diaminopurine (bottom) is an analog of adenine with an additional amino group at position 2 of the purine molecule. When paired with thymine, it forms an additional hydrogen bond along the minor-groove side of the molecule. To see this figure in color, go online.
At the basepair level, the additional amino group is known to alter DNA conformation in two ways. First, in contrast to adenine, which forms two hydrogen bonds with thymine, DAP forms a third hydrogen bond with thymine (see Fig. 1). This additional bond strengthens the effective interaction between the paired backbones and, like sequences with high GC content, raises the melting temperature (1, 14). Second, the additional amino group extends into the minor groove of the B-form DNA helix (B-DNA), decreasing the compressibility of the groove through steric hindrance, reducing bending flexibility, and biasing the molecule to deviate from the typical B-form conformation at low tension (15, 16, 17, 18, 19). The decrease in bending flexibility results in DAP-containing molecules behaving as stiffer entropic springs, characterized by longer persistence lengths, and forming larger plectonemic loops upon supercoiling (6).
Above 60 pN, far beyond the entropic and elastic polymer-chain regimes, torsionally unconstrained DNA undergoes a conformational transition consisting of an abrupt 70% increase in molecule length at a characteristic tension threshold called the “overstretching” force (Fos) (20, 21, 22, 23). This transition involves both tension-induced melting (appearing as strand peeling or localized denaturation) and conversion to an unwound but still basepaired conformation called S-DNA (24, 25). The pathway by which DNA becomes overstretched is a function of local stability and is determined by buffer conditions and sequence (26, 27). Experiments using relatively short sequences have demonstrated that in physiological salt conditions, AT-rich regions tend to denature upon overstretching, whereas GC-rich regions remain hybridized (28, 29). Similarly, molecular dynamic simulations suggest that stronger base pairing in GC-rich regions maintains base registration and minimizes interstrand separation, producing a higher transition force (30). Above 150–200 mM monovalent salt levels, AT-rich regions also adopt the basepaired (S-DNA) conformation when overstretched (31, 32). In addition to base pairing, base stacking has been recognized as crucial to the conformational stability of DNA (33, 34). In fact, inter-base-stacking interactions contribute significantly to the free energy of double-helix formation (35, 36).
DAP-substituted DNA has not been studied in this high-tension regime, and the effects of its additional amine group are not intuitive. On the one hand, the third hydrogen bond with thymine should stabilize against tension-induced melting; hence, one might expect DAP-DNA to behave like GC-rich DNA and have a higher overstretching force. However, the importance of base stacking and other factors affecting conformational stability should not be overlooked. Stronger stacking interactions are thought to also play a role in stabilizing against overstretching (30, 37). Finally, the propensity of DAP-DNA to adopt conformations other than the canonical B-form in the low-tension regime suggests DAP may also facilitate the B-to-S transition, resulting in a lower overstretching force.
To further investigate the effect of DAP substitution upon DNA conformational stability in the low-tension (entropic) and high-tension (overstretching) regimes, we performed a series of atomic force microscopy (AFM) imaging, magnetic tweezer (MT)-based stretching, circular dichroism (CD) spectroscopy, and thermal melting experiments on equivalent WT and DAP-substituted DNA sequences. The AFM and MT experiments confirm the findings of Virstedt et al. and Peters et al. that under low tension, DAP-DNA is flexurally stiffer (14, 19). Interestingly, the AFM and MT assays yielded conflicting measurements of molecular contour length. In the AFM images (in which molecules were not subject to tension), DAP-DNA appeared shorter than wild-type (WT)-DNA; DAP-DNA had an average helical rise of 2.79 ± 0.12 Å/bp, compared to 3.11 ± 0.12 Å/bp for WT-DNA, indicating a possible difference in helical conformation. For comparison, B-form DNA has an approximate helical rise of 3.4 Å/bp, whereas A-form DNA has an approximate helical rise of 2.6 Å/bp (38, 39, 40). In contrast, when mild tension was applied in the MT assay, differences in the contour lengths of DAP-DNA and WT-DNA molecules were less pronounced (3.16 ± 0.27 and 3.25 ± 0.27 Å/bp, respectively). CD spectra confirmed that DAP substitution results in a conformational shift under tension-free conditions, showing an increase in left-versus-right (εL − εR) absorbance at 290 nm. However, the spectra of DAP molecules do not show the hallmark 270 nm peak and 240 nm shoulder of trifluoroethanol-induced A-form DNA (41, 42, 43). Although DAP-DNA has a shorter static contour length, similar to A-DNA, its helical characteristics, such as winding and propeller-twist angles, are likely distinct (43). Melting experiments also demonstrated that although DAP increases thermal stability, it does not increase melting temperatures to the degree of G-C basepairs. As a consequence, DAP-T stability is intermediate between AT pairs and GC pairs. Under high tension, DAP-DNA undergoes overstretching at a lower characteristic force, without denaturing. Together, these results reveal that DAP substitution decreases the overall stability of the B-form double helix at both ends of the tension spectrum, biasing toward non-B-DNA at zero tension and facilitating the B-to-S transition at high tension.
Materials and Methods
DNA preparation
Normal and DAP-substituted DNA sequences were produced using the polymerase chain reaction (PCR). DAP-substituted DNA was produced by replacing dATP with DAP-5′-triphosphate (TriLink BioTechnologies, San Diego, CA) at the same concentration (200 μM) as the other three dNTPs (Fermentas, Waltham, MA).
Melting temperature characterization
The melting temperature of WT and DAP-substituted sequences was determined from fluorescently detected melting curves (44, 45). Three 155, 147, and 156 basepair-long portions of the pBR322 plasmid were selected containing 40, 54, and 65% GC content, respectively. The sequences were amplified using Taq DNA Polymerase (NEB, Ipswich, MA) in ThermoPol buffer. The primers used to produce 40, 55, and 65% amplicons are listed in Table S1. Thermocycling parameters are listed in Table S2. After PCR amplification, DNA samples were purified with a Qia Quick PCR cleanup kit (QIAGEN, Germantown, MD) and eluted in 10 mM Tris-HCl (pH 8.5). Amplicon lengths were verified by gel electrophoresis, and concentrations were measured by ultraviolet absorption using a Nanodrop Lite (Thermo Fisher Scientific, Waltham, MA). DNA was diluted to 10 ng/μL in 10 mM Tris-HCl (pH 7.4) with 15 mM KCl. Two intercalating dyes were used to assess melting: Syto-84 and Sybr-Green I (Thermo Fisher Scientific). Syto-84 was used at a concentration of 1 μM; at that concentration, the Syto family dyes have been shown to alter melting by no more than 0.6°C (Fig. S4) (46). For each sequence and type, at least six samples were measured and averaged. Sybr-Green I, which changes Tm by up to 10°C at the manufacturer’s suggested working concentration, was also titrated over a range of 0.0005–0.01% v/v 10,000×, assayed, and projected to yield the dye-free Tm (Fig. S5). Fluorescent intensity was recorded using a Bio-Rad C1000 quantitative PCR (qPCR) machine (Bio-Rad Laboratories, Hercules, CA) over a temperature range of 60–95°C in 0.5°C increments.
DNA for MT and AFM experiments
MT and AFM experiments measuring DNA mechanical parameters and overstretching were performed using 4642 bp-long (hereafter, 4.6 kb) DNA fragments. For the MT experiments, tethers were constructed from three components: a 4.6 kb-long core fragment containing either WT-DNA or DAP-DNA and two ∼1 kb flanking tails containing biotin or digoxigenin-11 dUTPs. The core fragment was prepared by PCR with Long Amp (NEB) using the pKLJ12wt plasmid (47) and primers 5′-AGCGTTGGCGCCGATTGCAGAATGAATTT and 5′- TGGGATCGGCCGAAAGGGCAGATTGATAGG, which contain KasI and EagI restriction sites (underlined), respectively. Thermocycle parameters are listed in Table S3. A single major amplicon around 4.6 kbp was produced (Fig. S6). The biotin- and digoxigenin-labeled tail fragments were also produced by PCR using Taq polymerase in standard buffer (New England BioLabs (NEB)). PCR solutions were supplemented with biotin-11 dUTP (Fermentas) and digoxigenin-11 dUTP (Roche, Indianapolis, IN) in a 1:9 ratio with respect to dTTP (6). The biotin-labeled fragment was amplified from pUC19 using the primer pair 5′-ATGATCCCCCATGTTGTGCA and 5′-TCAAGACGATAGTTACCGGATAAG to create a 1.8 kb biotin-labeled amplicon with a central KasI site. The digoxigenin-labeled fragment was amplified from pBluKSP using the primer pair 5′-TGGGTGAGCAAAAACAGGAAGGCA and 5′-GCGTAATCTGCTGCTTGCAA to create a 2 kb digoxigenin-labeled amplicon with a central EagI site. Thermocycle conditions are listed in Table S4.
After PCR amplification and column purification (Qia Quick PCR cleanup; QIAGEN), the core and tail fragments were digested with KasI and EagI-HF restriction enzymes (NEB) and purified again, and concentrations were measured by ultraviolet absorption. Restriction of the tails yields roughly 1 kb fragments with a single KasI or EagI sticky end. Restriction of the core fragment with KasI and EagI produces a 4.6 kb sequence with 5′GCGC and 5′GGCC overhangs at opposite ends. Note that this fragment sets the effective tether length in MT experiments because the randomly biotin- or digoxigenin-labeled handles will almost completely attach to the magnetic bead and the chamber surface, marginally contributing to the DNA extension. This is confirmed by MT measurements of DNA extension (see Fig. 3 C) and by Yan et al. (48). As a consequence, we expect that the MT measurements are mainly determined by the core fragments.
Figure 3.
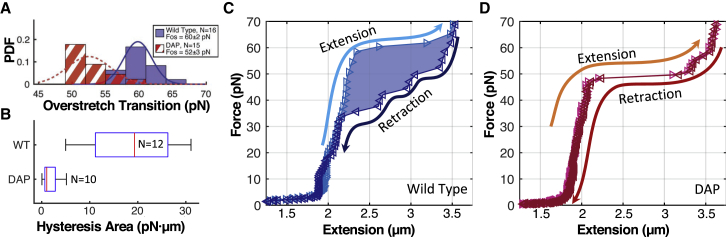
Worm-like chain fits of force versus length MT data. (A) Representative force versus length data for wild-type (blue) and DAP (red) DNA molecules with corresponding WLC fits (solid line for WT and dashed line for DAP) are shown. Fit errors correspond to 95% confidence limits. (B) Histogram of fit persistence length values is shown; DAP substitution increases Lp to 56 ± 5 from 45 ± 4 nm for WT (mean ± SD). Distributions are statistically distinct, p = 5.22 × 10−16. (C) Histogram of fit contour length values is shown. DAP substitution does not significantly decrease dynamic contour length, p = 0.103. N = 35 for WT and N = 48 for DAP in both histogram plots. To see this figure in color, go online.
To assemble the final tether, approximately 600 ng of the core fragment was mixed with 250 ng of each tail along with 30 U of T4 DNA ligase (NEB) and the recommended buffer to a total volume of 30 μL and incubated at 16°C for 4 h. After heat inactivation at 65°C for 20 min, the ligation mixture was used directly in the microchambers.
Magnetic tweezing
The MT used in this work was built using an inverted, infinity-corrected microscope. A set of permanent magnets was mounted above the sample, between the stage and condenser optics. These magnets could be translated along and rotated around the optical axis to control the tension and supercoiling of the DNA tethers (49, 50). Similar designs and their application to DNA characterization have been thoroughly discussed by others (51, 52, 53, 54). The flow cell used for the MT experiments consists of a ∼50 μL capillary functionalized with antidigoxigenin antibody (Roche) (49). DNA tethers were formed by incubating the biotin and digoxigenin end-labeled fragments with streptavidin-coated superparamagnetic beads (Invitrogen, Carlsbad, CA) for ∼15 min. Conjugated tethers were isolated with a magnet and resuspended in 500 μL PTE buffer (phosphate-buffered saline (pH 7.4) with 1mM EDTA and 0.1% Tween). For low-force experiments, 1-μm diameter magnetic beads were used, whereas for overstretching measurements (in which forces in excess of 50 pN were needed), 4.5 μm beads (Invitrogen) were used. Finally, tether suspensions were incubated in the capillaries for ∼1 h before flushing out the excess, unbound tethers with 8–10 mL PTE buffer. All MT measurements were carried out in PTE buffer.
AFM-based imaging
Nanoscale images of WT-DNA and DAP-DNA deposited onto mica surfaces were acquired using a Nanowizard II (JPK Instruments, Berlin, Germany). WT and DAP molecules were diluted in 5 mM MgCl2 in DI-H2O to a final concentration between 0.2 and 0.3 ng/μL. 10 μL of each solution was deposited onto freshly cleaved mica substrates and incubated at room temperature for 5–10 min. The samples were then rinsed with 0.22 μm-filtered deionized ultrapure water and dried with a gentile nitrogen flow.
Images were acquired with the AFM operating in tapping mode, in air, using stiff silicon cantilevers (RTESO-Veeco, resonant frequencies of 300–350 kHz and spring constant ∼40 N/m; Veeco, Plainview, NY). 8 × 8 μm images were collected using a 0.5–1 Hz scan rate with 2048 × 2048 pixel resolution. Several hundred DNA molecules were recorded from several widely spaced areas of each sample and processed with a standard “flatten” filter in the JPK Data Processing software.
To morphologically characterize the two samples, we traced the DNA contours using a custom-tracing routine (55) and calculated the contour length, L0 (56). Only molecules in the range 1.1–1.8 μm surrounding the expected length were included. The average end-to-end distance of segments of the DNA molecules as a function of their separation along the contour was analyzed to estimate the persistence length (LP) according to the WLC model (57, 58, 59).
CD spectroscopy
WT-DNA and DAP-DNA molecules produced for the melting experiments and the 4.6 kb core fragments from the AFM/MT experiments were diluted to concentrations ranging from 10–30 ng/μL in 10 mM Tris-HCl (pH 7.4) with 150 mM KCl. The CD spectra of the respective samples were measured using a Jasco J-810 spectrometer (Jasco, Tokyo, Japan) in a quartz cuvette and blanked against the spectrum of Tris-KCl buffer without DNA. The spectrum of each sample was measured three times at wavelengths 210–310 nm (in 0.2 nm increments), averaged, and smoothed with a Savitzky-Golay filter of third order using a 31-step (6 nm) window width.
Results
To facilitate discussion of our findings, measured mechanical characteristics for WT and DAP-substituted DNA are summarized in Table 1. Unless otherwise noted, values reported in the text refer to mean ± SD. Confidence intervals, computed as 1.96× standard error, can be found in Table 1.
Table 1.
Summary of Results
| Parameter | Unit | Type | Mean | SD | N | 95% CI | p Value |
|---|---|---|---|---|---|---|---|
| L0 (MT) | μm | WT | 1.51 | 0.13 | 35 | ±0.04 | 0.103 |
| μm | DAP | 1.46 | 0.12 | 48 | ±0.03 | – | |
| L0 (AFM) | μm | WT | 1.45 | 0.12 | 22 | ±0.05 | 3.07 × 10−4 |
| μm | DAP | 1.30 | 0.12 | 20 | ±0.05 | – | |
| Axial Rise (MT) | Å/bp | WT | 3.25 | 0.27 | — | ±0.09 | — |
| Å/bp | DAP | 3.16 | 0.27 | — | ±0.07 | – | |
| Axial Rise (AFM) | Å/bp | WT | 3.11 | 0.12 | — | ±0.11 | — |
| Å/bp | DAP | 2.79 | 0.12 | — | ±0.11 | – | |
| Lp (MT) | nm | WT | 45 | 4 | 35 | ±1.2 | 5.22 × 10−16 |
| nm | DAP | 56 | 5 | 48 | ±1.5 | – | |
| Lp (AFM) | nm | WT | 56.2 | — | — | ±0.1 | — |
| nm | DAP | 79.9 | — | — | ±0.3 | – | |
| Fos | pN | WT | 60 | 2 | 16 | ±1.1 | 2.88 × 10−8 |
| pN | DAP | 52 | 3 | 15 | ±1.6 | – | |
| Hysteresis area | pN μm | WT | 19 | 9 | 12 | ±5.08 | 2.73 × 10−5 |
| pN μm | DAP | 2 | 1 | 10 | ±1.07 | – |
DAP substitution increases melting temperature but preserves nearest-neighbor effects
Under Watson-Crick base pairing, the two backbones of a DNA molecule are held together by hydrogen bonds formed between the basepairs. Consequently, to first order, one would naively expect the melting temperature to be proportional to the total number of basepair hydrogen bonds in a given sequence. However, the stacking interactions between A/T and G/C duplexes produce sequence-dependent effects (35, 60). Therefore, even if two relatively short sequences have the same length and GC fraction, they can have very different melting temperatures. Consequently, DAP substitution can be expected to affect melting temperatures in three ways: 1) the additional hydrogen bond stabilizes base pairing, 2) the stacking interactions of adenine and DAP are likely different, and 3) DAP’s extra amino group may alter conformational stability of B-DNA, which in turn affects hybridization.
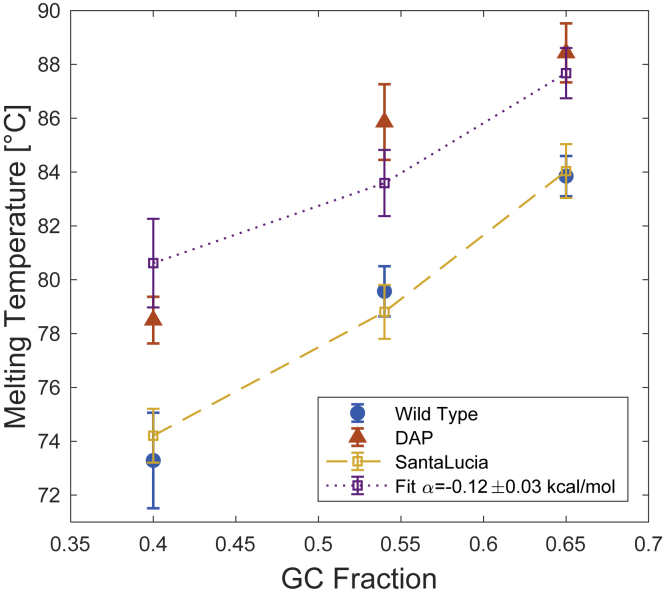
The melting temperature (Tm) of three sequences containing 40, 54, and 65% GC were compared, as shown in Fig. 2, in which measured (red and blue dots) and calculated (yellow and purple dots) values of Tm are presented as a function of GC percentage. The sequences were roughly the same length (155, 147, and 156 bp). The fraction of helical versus single-stranded DNA was measured by fluorescence in the presence of the intercalating dye Syto-84 using a Bio-Rad qPCR machine. The Syto-series dyes have been shown to shift melting temperatures by less than 0.6°C at 1 μM (Fig. S4) (46). Fluorescence curves were normalized to account for temperature dependence of the fluorescent dye (44, 45); details are included in the Supporting Materials and Methods. The Syto-84 findings were confirmed by roughly equivalent melting temperatures measured via Sybr-Green I (Fig. S5) using the titration protocol presented in Peters et al. (18).
Figure 2.
Melting curves of three sequences of WT (blue, solid circle) and DAP-DNA (red, solid triangles) with GC fractions of 0.4, 0.54, and 0.65 were measured using the fluorescence of Syto-84 (1 μM); error bars indicate SD. WT results closely match Tm predicted by the SantaLucia nearest-neighbor model (yellow); error bars indicate model accuracy (61). The DAP melting temperatures were fit with a modified SantaLucia model (purple). A least-squares fit yields α = −0.12 ± 0.03 kcal/mol. To see this figure in color, go online.
For short sequences, melting approximates a binary process wherein the entire sequence separates in unison. With that approximation, melting temperatures can be estimated by
| (1) |
where Tm is the temperature at which 50% of double-stranded DNA (dsDNA) molecules will be melted, ΔH is the sequence-dependent enthalpy, ΔS is the sequence-dependent entropy, R is the gas constant, and [dsDNA] is the concentration of dsDNA in solution.
Under the SantaLucia nearest-neighbor model, ΔH and ΔS are equal to the sum of empirically determined, nearest-neighbor terms and are corrected to account for salt-dependent effects (61, 62). Although the SantaLucia model is intended for short sequences, we find that it yields predictions within ±0.8°C of our measured melting temperatures (Fig. 2, blue and yellow dots), well within the predicted accuracy of the model (61).
DAP substitution increases melting temperatures. However, it does not eliminate sequence-dependent effects as illustrated by the fact that Tm of each sequence is increased by 6–8°C. If sequence did not matter (i.e., melting was determined primarily by the number of H-bonds), one would expect each of the DAP-DNA sequences to melt at approximately the same temperature.
To underscore this point, we compare our data to a modified version of the SantaLucia model. Under this modified model, the enthalpy of each A/T pair is supplemented with an additional factor (α), accounting for both the aggregate effect of the additional hydrogen bond, changes in stacking energy, and perturbations to conformational stability. For example, under the modified scheme a sequence of 5′-GATCG-3′ would have a total enthalpy
| (2) |
A least-squares fit to the DAP melting data yields α = −0.12 ± 0.03 kcal/mol. Although the modified model does not perfectly capture the Tm for each sequence (notably the 54% sequence see Fig. 2 purple and red symbols), it does preserve the general trend.
Zero-to-low force regime: DAP substitution increases persistence length, decreases static but not dynamic contour length
We mechanically characterized how DAP substitution alters the dynamic conformational properties of dsDNA using MT-based force spectroscopy. 4.6 kbp DNA tethers were subjected to tensions ranging over 0.001–5 pN, and their time-averaged, end-to-end lengths were measured. Only molecules in the range 1.1–1.8 μm, surrounding the expected 1.5 μm length, were included in the analysis. Below 10 pN, DNA acts as an entropic spring and is well characterized by a worm-like chain (WLC) polymer model. The force-length dependence for a WLC is approximately
| (3) |
where Le is the end-to-end distance, L0 is the molecule’s contour length, and Lp is the persistence length, which characterizes the stiffness (63). For each molecule, force versus length measurements were fit to Eq. 3, yielding estimates of L0 and Lp. Representative measurements and accompanying fits for a WT and DAP molecule are included in Fig. 3. DAP substitution increased persistence length from 45 ± 4 nm for WT to 56 ± 5 nm for DAP (p = 5.22 × 10−16). However, the dynamic contour length only decreased from 1.51 ± 0.13 μm to 1.46 ± 0.12 μm, a change that is not statistically distinct (p = 0.103, see Fig. 3, B and C).
To extend these findings, we also measured static contour and persistence lengths via AFM imaging. WT and DAP-DNA molecules were deposited onto freshly cleaved mica surfaces and imaged in tapping mode. Representative images of both WT and DAP samples are shown in Fig. 4. Contour lengths were measured by tracing each DNA filament using an image recognition algorithm and computing the integrated length (55). Molecules that were significantly longer or shorter than the expected 1.1–1.8 μm range were discarded from the data set. A histogram of contour lengths for WT and DAP molecules is included in Fig. 5 A. As indicated, DAP substitution decreased the average contour length from 1.45 ± 0.12 to 1.30 ± 0.12 μm (p = 3.07 × 10−4), corresponding to axial rises per basepair of 3.11 and 2.79 Å/bp, respectively, consistent with previous measurements (6, 14).
Figure 4.
Representative images of (A) wild-type and (B) DAP-DNA molecules captured via AFM. To see this figure in color, go online.
Figure 5.
Contour and persistence length measured via analysis of AFM images. (A) Contour length was calculated by tracing individual molecules in each AFM image. DAP substitution reduced the length (mean ± SD). The distributions are statistically different in a two-tailed t-test (p = 3.07 × 10−4). (B) Mean-squared distance versus separation along each molecular contour is shown. Data points (blue circles and red squares) correspond to the average square distance, aggregated for all molecules of a given type (WT and DAP, respectively). Data was fit to Eq. 4, yielding estimates of the average persistence length; ± error indicates 95% confidence interval. To see this figure in color, go online.
Persistence length was estimated from the measured mean-square displacement between points along the contours of molecules in two dimensions (55). For a WLC confined to two dimensions, the average squared distance between two points along the chain is given by
| (4) |
where L is the curvilinear distance along the chain between the points, and Lp is the molecule’s persistence length (57, 59). Using the x-y coordinates generated by the tracing routine, it is straightforward to compute the distance between points separated by increasing contour lengths. The data for molecules from either WT or DAP samples were pooled, and the means of the squared end-to-end distances were computed as a function of separation along the contour and fit to Eq. 4 (results shown in Fig. 5 B). The excluded volume effects were negligible given the low DNA concentration and the limiting of contour length separations to less than 200 nm when fitting Eq. 4 (58, 64). WT-DNA yielded Lp = 56.2 ± 0.1 nm, whereas DAP substitution increased the persistence length to Lp = 79.9 ± 0.3 nm (95% confidence interval). Analyzing overlapping or nonoverlapping segment data produced similar estimates (Fig. S7; Table S5).
Both MT and AFM reveal a significant increase in persistence length because of DAP substitution, indicating that DAP molecules are flexurally stiffer (i.e., harder to bend). This result is consistent with what has been reported previously for experiments using AFM and optical tweezing (14, 19). Similarly, previously reported MT experiments measuring DNA under supercoiling found that DAP substitution increased the size of plectonemic writhes, a finding that is consistent with DAP-DNA being harder to bend (6).
High-force regime: DAP substitution decreases the tension at which overstretching occurs
DNA overstretching is characterized by a sudden, roughly 1.7× increase in tether length when tension exceeds a critical value. Under physiological buffer conditions, overstretching typically occurs around 60–70 pN. The transition exhibits both force-induced melting, wherein the phosphate backbones separate, enabling each strand to stretch independently, as well as the formation of a double-stranded structure called S-DNA. The propensity of a DNA molecule to form S-DNA rather than undergo force-induced melting is sequence dependent. Simulations have shown that whereas G-C pairs persist through the transition, A-T pairs often form melted bubbles (30, 37). Similarly, various pulling experiments and theoretical models have also demonstrated that at low-to-moderate monovalent salt conditions (<150 mM), AT-rich sequences tend to melt more easily than GC-rich sequences (24, 28, 29, 65, 66, 67, 68, 69), whereas under high-salt conditions interstrand charge repulsion is reduced, and AT-rich sequences also form S-DNA (31, 32). Moreover, cyclic pulling experiments demonstrate that force-induced melting is associated with hysteresis, such that once melted, the complementary strands reanneal slowly (21, 28, 32, 70).
To investigate how DAP affects overstretching, we analyzed the force-extension behavior of several molecules in the high-force regime. Un-nicked molecules (verified by a twist versus extension curve) were excluded from the analysis because torsional constraint fixes the helical linking number, forcing hyperextended molecules to adopt a P-DNA configuration (25, 71, 72, 73, 74). Nicks occur randomly during the preparation of the DNA tether. However, un-nicked molecules, for both WT and DAP-DNA, were ∼40% of the total number of sample molecules, and assuming random nicking along the chain, a simple calculation (Poisson distribution) was used to estimate that the probability of nicking adjacent to a basepair was 0.02% (Fig. S8). Using this probability in simulations, we can estimate that less than 26% of the molecules analyzed had more than one nick (Fig. S9). Thus, nicks did not strongly bias the overstretching measurements nor the observed hysteresis. Remarkably, DAP-DNA overstretched at a significantly lower transition force than WT-DNA. The overstretching transition force (Fos) was characterized by calculating the midpoint of the transition plateau. As shown in Fig. 6 A, DAP substitution lowered the transition force to Fos = 52 ± 3 pN compared to 60 ± 2 pN for WT-DNA (p = 2.88 × 10−8); data were collected in 150 mM phosphate-buffered saline.
Figure 6.
Overstretching under high tension. (A) Histogram of overstretching transition force is shown. Distributions are statistically distinct, p = 2.88 × 10−8. (B) Box and whisker plot of area between extension and relaxation curves are shown. Box edges indicate the 25th and 75th percentiles. Whiskers indicate the extent of data. Red line indicates the median. Nearly all WT molecules exhibited significant hysteresis H = 19 ± 9 pN μm; DAP molecules showed little hysteresis H = 2 ± 1 pN μm. (C) Example force-extension cycle for WT-DNA is shown. As tension is increased (light blue trace), the molecule undergoes force-induced melting. Upon relaxation (dark blue), molecule length decreases with a different characteristic curve owing to the presence of single-stranded DNA. (D) Example force-extension cycle for DAP-DNA is shown. Little hysteresis is observed between extension and relaxation, indicating basepair persistence and the adoption of the S-form. To see this figure in color, go online.
Cyclic extension and relaxation also revealed that DAP-DNA exhibits very little hysteresis through the overstretching transition. The lack of hysteresis in the DAP curves indicates that peeling does not occur even in the presence of nicks, in support of DAP bases favoring S-DNA formation rather than melting. By comparison, nearly all WT molecules show significant hysteresis, indicating that force-induced melting constituted the bulk of extensional increase. To quantify the difference in DAP and WT behavior, we calculated the area between the extension and relaxation traces for each molecule (see Fig. 6, C and D for illustration). The reannealing process can be influenced by several factors, notably ionic strength of the solution and loading rate (24). Here, both DAP and WT hysteresis was measured in 150 mM phosphate-buffered saline. Additionally, MTs essentially operate as force clamps. In these experiments, each data point is recorded over a period of 45–60 s, with at least 1 s between successive measurements. The entire hysteresis measurement requires at least 20 min. Consequently, the molecules are stretched at an almost zero loading rate, and the DNA remains overstretched for several minutes. Results are shown in Fig. 6 B. The smaller secondary plateau in Fig. 6 D at ∼56 pN appeared in some but not all measurements; therefore, we do not speculate as to its origin. In summary, WT-DNA had an average hysteresis area of 19 ± 9 pN μm, whereas DAP-DNA averaged 2 ± 1 pN μm (p = 2.73 × 10−5).
CD spectroscopy confirms DAP substitution induces conformational shift
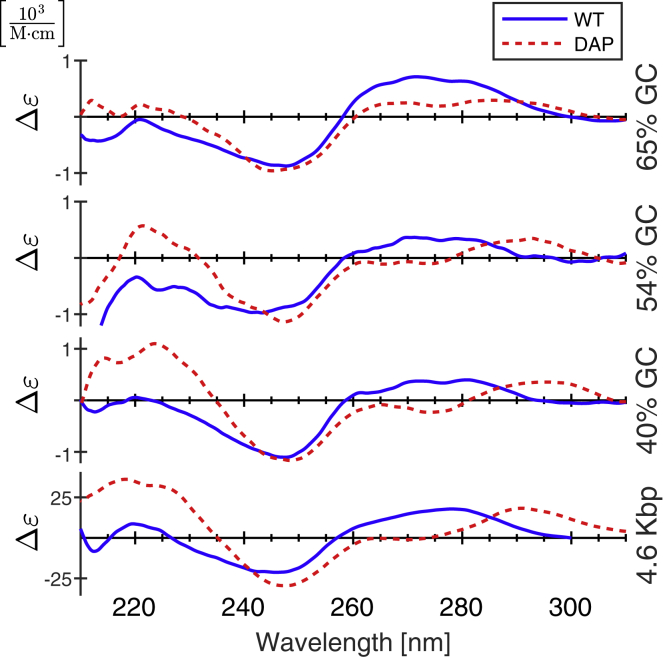
The CD spectra of WT and DAP versions of the 40, 54, and 65% GC and 4.6 kb core fragments were measured over the range 210–310 nm. Results are shown in Fig. 7. The spectra of all four WT molecules show the expected behavior of B-DNA, specifically a local maximum in the 270–280 nm range and a local minimum around 250 nm (41, 42). The spectrum of 4.6 kb DAP molecules is very similar to the spectra reported by Peters et al. and Virstedt et al. for DAP-substituted molecules without extensive AT or GC repeats, namely the 4.6kb DAP spectra shows a peak near 290 nm, a shoulder between 260 and 270 nm, and a minimum near 250 nm (14, 19). Interestingly, the 40, 54, and 65% GC sequences show a progressive shift from non-WT behavior toward a WT-like spectrum. 40% molecules, which have the most DAP, show a peak at 295 nm, a minimum at 248 nm, and a modest local minimum at 274 nm. As DAP content decreases (and GC content increases), the minimum at 274 nm fades.
Figure 7.
CD spectra of WT (blue, solid lines) and DAP-substituted molecules (red, dashed lines). To see this figure in color, go online.
Discussion
DAP substitution stabilizes interstrand pairing but does not eliminate sequence effects
The melting experiments clearly demonstrate that DAP substitution increases the thermal stability of double-stranded molecules but does not raise melting temperatures to the expected level of equivalent length poly-GC fragments. The melting temperature of each roughly 150 bp sequence was increased by ∼5–10°C (between 80 and 90°C).
Fitting the melting data against our modified SantaLucia model shows that, relative to adenine, DAP substitution stabilizes the per-base hybridization energy (α in Fig. 2) by ΔΔE = −0.12 ± 0.03 kcal/mol. Interestingly, this is smaller than one might expect, given reported DAP-stacking energies measured by the “dangling-bond” assay (18, 19, 36). Peters et al. report that stacking interactions of a single DAP base with a neighboring GC basepair contributes approximately −5 kcal/mol to the enthalpy of hybridization, compared with −4 kcal/mol for adenine (18, 19). In other words, combining the effect of DAP’s extra amino group with the improved stacking, one would expect the total-per-base change in energy to decrease by at least ΔΔE ≤ −1 kcal/mol. Instead, our findings suggest that factors, such as conformational instabilities resulting from steric clashes in the minor groove, partially offset the stabilizing effects of the extra hydrogen bond and stronger base stacking.
DAP-DNA resists force-induced melting upon overstretching
In addition to having increased thermal stability, DAP-DNA also resists force-induced melting. Whereas WT molecules exhibited significant hysteresis when relaxing from the overstretching transition, DAP molecules show little to no hysteresis. This suggests that a DAP-DNA molecule’s two polynucleotide backbones remain hybridized during overstretching, whereas the length of WT molecules fluctuates until the tension drops enough to allow reannealing (67).
DAP substitution flexurally stiffens DNA yet lowers the barrier to adopt non-B-form conformations
Several studies have shown that DAP substitution stiffens the molecule against lateral bends (6, 14, 19). The low-tension AFM and MT experiments presented here confirm that finding. However, the results at higher tension also reveal that DAP substitution lowers the energetic barrier of conformation changes, lowering its resistance to overstretching.
In line with the previous reports (14, 19), our low-tension MT and AFM experiments demonstrate that DAP substitution increases persistence length. It has been argued that DAP’s extra amino group, which protrudes into the minor groove (Fig. 1), alters the helical conformation, and resists flexural bends that would compress the minor groove (14, 38). Indeed, DAP substitution would seem to add steric clashes between the exocyclic amino group and the nitrogen at position 3 of adjacent purines as observed by Calladine (75, 76). Increased helical and decreased propeller twist might minimize steric clashes between adjacent purines in the minor groove of DAP-substituted DNA.
The CD spectra of WT and DAP molecules shown in Fig. 7 confirm that DAP substitution alters the helical conformation, although the spectra of DAP-DNA are not indicative of A-DNA. In line with what has been reported by others (14, 18, 19), we see that DAP-DNA has a peak near 290 nm, a local minimum or shoulder near 275 nm, and a minimum at 250 nm. This is distinct from B-DNA, which is characterized by a peak between 270 and 280 nm and a minimum near 245 nm, but also distinct from the spectra of A-DNA, formed by titration with trifluoroethanol, which show a maximum at 270 nm, a minimum at 210 nm, a shoulder near 240 nm, and a zero at 300 nm and beyond (41, 42, 43). Most telling is the decrease in ellipticity at 275 nm in DAP-substituted versus WT-DNA molecules. Such a decrease correlates with some combination of increased helical and decreased propeller twist accompanied by decreased helical rise (77). These changes ameliorate the steric repulsions between 3′-pyrimidine-5′-purine basepairs created by adding exocyclic amines in the minor groove (75) to shift the molecule to a helical configuration with a flattened CD spectrum from 260 to 270 nm that resembles that of C-DNA (78). X-ray diffraction experiments revealed that C-form DNA produced in lithium chloride precipitates had a helical twist of 38.6° (79), slightly increased over the 36° of B-form (43).
Interestingly, whereas DAP substitution resulted in a significant reduction in contour length as measured by AFM (from 3.11 down to 2.79 Å/bp, a 10% decrease), it only had a marginal effect when measured with MT (3.25 to 3.16 Å/bp, a 3% decrease). Although the zero-tension AFM results suggest DAP decreases the helix pitch, the change does not persist once low-to-moderate tension is applied. Considering the CD spectroscopy results, one explanation for the discrepancy is that an unperturbed DAP molecule adopts a non-canonical conformation but quickly reverts back to B-DNA when mildly stretched.
If DAP-DNA more readily adopts noncanonical forms at zero tension to minimize steric clashes between adjacent purines, one might expect those steric clashes to also facilitate conformational changes at high tension. This is vividly displayed in overstretching experiments on DAP-substituted DNA. For WT-DNA in physiological salt conditions, the characteristic overstretching transition around 60 pN is caused by a combination of factors, including force-induced melting (through strand peeling and melting bubble formation) and adoption of the S-DNA conformation (24, 32, 65, 70, 80). Several studies have shown that force-induced melting tends to originate at AT-rich regions (28, 29, 30, 37). In particular, a detailed study conducted by Bosaeus et al. comparing the overstretching behavior of short 70% AT and 60% GC oligonucleotides (oligos) found that whereas AT sequences melt and GC sequences form S-DNA, the transition threshold for GC oligos was similar if not slightly higher than AT oligos: 63.3 ± 1.1 pN for GC versus 61.5 ± 2.6 pN for AT (28, 29). Before the Bosaeus et al. study, other authors had similarly argued that a S-DNA formation likely required more energy than AT separation but less than GC separation (80, 81, 82). Our melting and hysteresis experiments demonstrate that DAP-DNA base pairing is significantly more stable than WT-DNA. Therefore, naively, one might expect DAP-DNA to behave like GC-rich sequences, with an overstretching transition equal to or slightly greater than WT molecules. Instead we find that DAP-DNA has a significantly lower transition tension (52 ± 3 vs 60 ± 2 pN for WT-DNA). This indicates that DAP substitution not only disrupts B-DNA at low tension but also lowers the energetic barrier separating B- and S-form DNA.
Conclusions
Diaminopurine is a nucleobase analog of adenosine, bearing an additional amino group on its 2′-carbon, which participates in a third hydrogen bond when base paired with thymine. Because the additional amine resides on the minor-groove side of the DAP-T basepair, it not only stabilizes DAP-T pairs but significantly changes dsDNA conformation and mechanics. Through AFM imaging, mechanical characterization using a MT, and CD spectroscopy, we show that DAP substitution biases DNA molecules to adopt a non-canonical helix at zero applied tension and resist flexural bending at low-to-moderate tension (<10 pN) yet also decreases the overstretching threshold from roughly 60 to 52 pN. From an energetics perspective, the steric clashes of DAP’s extra amino group penalize entropic bends (increasing the persistence length) but also raise the conformational free energy of the B-form helix, facilitating axial extension and initiating the B-to-S transition at a lower tension.
Author Contributions
D.K., M.C., D.D., and F.M. wrote and edited the manuscript. D.K. and D.D. produced the DAP-DNA constructs and oversaw the melting and CD experiments. M.C. and D.S. conducted the MT experiments. V.C. and R.C. performed the AFM experiments.
Acknowledgments
We thank Y. Lee and G. Malla for conducting DNA melting experiments, D. Katz for the use of the qPCR thermocycler, V. Conticello and S. Hughes for use and training on the CD spectrometer, and L. Bonazzi, E. Chiodi, L. Finzi, C. Marrano, N. Missana, L. Nardo, and R. Ziano for useful discussion, help in technical assembly of the MT apparatus, DNA sample preparation, and critical reading of the manuscript.
This work was supported by the National Institutes of Health grant R01 GM084070. Funding for the open access charge came from University of Milano-Bicocca, Italy.
Editor: Keir Neuman.
Footnotes
Matteo Cristofalo and Daniel Kovari contributed equally to this work.
Supporting Materials and Methods, nine figures, and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30063-3.
Contributor Information
Domenico Salerno, Email: domenico.salerno@unimib.it.
David Dunlap, Email: ddunlap@emory.edu.
Supporting Material
References
- 1.Bailly C., Waring M.J. The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Res. 1998;26:4309–4314. doi: 10.1093/nar/26.19.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrum J.P., Ricardo A., Szostak J.W. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′,3′-dideoxyribonucleoside-5′-phosphorimidazolide monomers. J. Am. Chem. Soc. 2009;131:14560–14570. doi: 10.1021/ja906557v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirnos M.D., Khudyakov I.Y., Vanyushin B.F. 2-aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977;270:369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- 4.Khudyakov I.Y., Kirnos M.D., Vanyushin B.F. Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine. Virology. 1978;88:8–18. doi: 10.1016/0042-6822(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 5.Cerami A., Reich E., Goldberg I.H. The interaction of actinomycin with DNA: requirement for the 2-amino group of purines. Proc. Natl. Acad. Sci. USA. 1967;57:1036–1042. doi: 10.1073/pnas.57.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández-Sierra M., Shao Q., Dunlap D. E. coli gyrase fails to negatively supercoil diaminopurine-substituted DNA. J. Mol. Biol. 2015;427:2305–2318. doi: 10.1016/j.jmb.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailly C., Payet D., Waring M.J. PCR-based development of DNA substrates containing modified bases: an efficient system for investigating the role of the exocyclic groups in chemical and structural recognition by minor groove binding drugs and proteins. Proc. Natl. Acad. Sci. 1996;93:13623–13628. doi: 10.1073/pnas.93.24.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomasz M., Das A., Waring M.J. The purine 2-amino group as the critical recognition element for sequence-specific alkylation and cross-linking of DNA by mitomycin C. J. Am. Chem. Soc. 1998;120:11581–11593. [Google Scholar]
- 9.Marco E., Negri A., Gago F. Role of stacking interactions in the binding sequence preferences of DNA bis-intercalators: insight from thermodynamic integration free energy simulations. Nucleic Acids Res. 2005;33:6214–6224. doi: 10.1093/nar/gki916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng Y.D., Ge H., Henderson R.M. Atomic force microscopy study of the structural effects induced by echinomycin binding to DNA. J. Mol. Biol. 2005;345:745–758. doi: 10.1016/j.jmb.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 11.Véliz E.A., Easterwood L.M., Beal P.A. Substrate analogues for an RNA-editing adenosine deaminase: mechanistic investigation and inhibitor design. J. Am. Chem. Soc. 2003;125:10867–10876. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert S.D., Mediatore S.J., Batey R.T. Modified pyrimidines specifically bind the purine riboswitch. J. Am. Chem. Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 13.Kawai K., Kodera H., Majima T. Long-range charge transfer through DNA by replacing adenine with diaminopurine. J. Am. Chem. Soc. 2010;132:627–630. doi: 10.1021/ja907409z. [DOI] [PubMed] [Google Scholar]
- 14.Virstedt J., Berge T., Travers A.A. The influence of DNA stiffness upon nucleosome formation. J. Struct. Biol. 2004;148:66–85. doi: 10.1016/j.jsb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Howard F.B., Miles H.T. 2NH2A X T helices in the ribo- and deoxypolynucleotide series. Structural and energetic consequences of 2NH2A substitution. Biochemistry. 1984;23:6723–6732. doi: 10.1021/bi00321a068. [DOI] [PubMed] [Google Scholar]
- 16.Lankaš F., Cheatham T.E., III, Šponer J. Critical effect of the N2 amino group on structure, dynamics, and elasticity of DNA polypurine tracts. Biophys. J. 2002;82:2592–2609. doi: 10.1016/s0006-3495(02)75601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travers A.A., Thompson J.M. An introduction to the mechanics of DNA. Philos. Trans. A Math. Phys. Eng. Sci. 2004;362:1265–1279. doi: 10.1098/rsta.2004.1392. [DOI] [PubMed] [Google Scholar]
- 18.Peters J.P., Yelgaonkar S.P., James Maher L., III Mechanical properties of DNA-like polymers. Nucleic Acids Res. 2013;41:10593–10604. doi: 10.1093/nar/gkt808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters J.P., Mogil L.S., Maher L.J., III Mechanical properties of base-modified DNA are not strictly determined by base stacking or electrostatic interactions. Biophys. J. 2014;107:448–459. doi: 10.1016/j.bpj.2014.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cluzel P., Lebrun A., Caron F. o. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 21.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 22.Marko J.F. Stretching must twist DNA. Europhys. Lett. 1997;38:183–188. [Google Scholar]
- 23.Marko J.F. DNA under high tension: overstretching, undertwisting, and relaxation dynamics. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1998;57:2134–2149. [Google Scholar]
- 24.Zhang X., Chen H., Yan J. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc. Natl. Acad. Sci. USA. 2013;110:3865–3870. doi: 10.1073/pnas.1213740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Léger J.F., Romano G., Marko J.F. Structural transitions of a twisted and stretched DNA molecule. Phys. Rev. Lett. 1999;83:1066–1069. [Google Scholar]
- 26.Zhang X., Chen H., Yan J. Two distinct overstretched DNA structures revealed by single-molecule thermodynamics measurements. Proc. Natl. Acad. Sci. USA. 2012;109:8103–8108. doi: 10.1073/pnas.1109824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenner J.R., Williams M.C., Bloomfield V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosaeus N., El-Sagheer A.H., Nordén B. Tension induces a base-paired overstretched DNA conformation. Proc. Natl. Acad. Sci. USA. 2012;109:15179–15184. doi: 10.1073/pnas.1213172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosaeus N., El-Sagheer A.H., Nordén B. Force-induced melting of DNA--evidence for peeling and internal melting from force spectra on short synthetic duplex sequences. Nucleic Acids Res. 2014;42:8083–8091. doi: 10.1093/nar/gku441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bongini L., Lombardi V., Bianco P. The transition mechanism of DNA overstretching: a microscopic view using molecular dynamics. J. R. Soc. Interface. 2014;11:20140399. doi: 10.1098/rsif.2014.0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King G.A., Gross P., Peterman E.J. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 2013;110:3859–3864. doi: 10.1073/pnas.1213676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Qu Y., Yan J. Interconversion between three overstretched DNA structures. J. Am. Chem. Soc. 2014;136:16073–16080. doi: 10.1021/ja5090805. [DOI] [PubMed] [Google Scholar]
- 33.Yakovchuk P., Protozanova E., Frank-Kamenetskii M.D. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vologodskii A., Frank-Kamenetskii M.D. DNA melting and energetics of the double helix. Phys. Life Rev. 2018;25:1–21. doi: 10.1016/j.plrev.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Sen A., Nielsen P.E. Hydrogen bonding versus stacking stabilization by modified nucleobases incorporated in PNA.DNA duplexes. Biophys. Chem. 2009;141:29–33. doi: 10.1016/j.bpc.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Guckian K.M., Schweitzer B.A., Kool E.T. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bongini L., Pongor C., Bianco P. An AT-barrier mechanically controls DNA reannealing under tension. Nucleic Acids Res. 2016;44:7954–7962. doi: 10.1093/nar/gkw604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Travers A.A. The structural basis of DNA flexibility. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2004;362:1423–1438. doi: 10.1098/rsta.2004.1390. [DOI] [PubMed] [Google Scholar]
- 39.Dickerson R.E. DNA structure from A to Z. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- 40.Kulkarni M., Mukherjee A. Understanding B-DNA to A-DNA transition in the right-handed DNA helix: perspective from a local to global transition. Prog. Biophys. Mol. Biol. 2017;128:63–73. doi: 10.1016/j.pbiomolbio.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Kypr J., Kejnovská I., Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranjbar B., Gill P. Circular dichroism techniques: biomolecular and nanostructural analyses- a review. Chem. Biol. Drug Des. 2009;74:101–120. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 43.Bloomfield V.A., Crothers D.M., Tinoco I. University Science Books; Sausalito, CA: 2000. Nucleic Acids: Structures, Properties and Functions. [Google Scholar]
- 44.Smith E., Jones M.E., Drew P.A. Quantitation of DNA methylation by melt curve analysis. BMC Cancer. 2009;9:123. doi: 10.1186/1471-2407-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanford L.N., Kent J.O., Wittwer C.T. Quantum method for fluorescence background removal in DNA melting analysis. Anal. Chem. 2013;85:9907–9915. doi: 10.1021/ac4024928. [DOI] [PubMed] [Google Scholar]
- 46.Gudnason H., Dufva M., Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson L.M., Yang H. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc. Natl. Acad. Sci. USA. 2008;105:5827–5832. doi: 10.1073/pnas.0705570105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan Y., Leng F., Dunlap D. Protein-mediated looping of DNA under tension requires supercoiling. Nucleic Acids Res. 2018;46:2370–2379. doi: 10.1093/nar/gky021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salerno D., Brogioli D., Mantegazza F. Magnetic tweezers measurements of the nanomechanical properties of DNA in the presence of drugs. Nucleic Acids Res. 2010;38:7089–7099. doi: 10.1093/nar/gkq597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tempestini A., Cassina V., Mantegazza F. Magnetic tweezers measurements of the nanomechanical stability of DNA against denaturation at various conditions of pH and ionic strength. Nucleic Acids Res. 2013;41:2009–2019. doi: 10.1093/nar/gks1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strick T.R., Allemand J.F., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 52.Seol Y., Neuman K.C. Magnetic tweezers for single-molecule manipulation. Methods Mol. Biol. 2011;783:265–293. doi: 10.1007/978-1-61779-282-3_15. [DOI] [PubMed] [Google Scholar]
- 53.De Vlaminck I., Dekker C. Recent advances in magnetic tweezers. Annu. Rev. Biophys. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- 54.Kriegel F., Ermann N., Lipfert J. Probing the mechanical properties, conformational changes, and interactions of nucleic acids with magnetic tweezers. J. Struct. Biol. 2017;197:26–36. doi: 10.1016/j.jsb.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 55.Cassina V., Manghi M., Mantegazza F. Effects of cytosine methylation on DNA morphology: an atomic force microscopy study. Biochim. Biophys. Acta. 2016;1860:1–7. doi: 10.1016/j.bbagen.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Cassina V., Seruggia D., Mantegazza F. Atomic force microscopy study of DNA conformation in the presence of drugs. Eur. Biophys. J. 2011;40:59–68. doi: 10.1007/s00249-010-0627-6. [DOI] [PubMed] [Google Scholar]
- 57.Podestà A., Indrieri M., Dunlap D. Positively charged surfaces increase the flexibility of DNA. Biophys. J. 2005;89:2558–2563. doi: 10.1529/biophysj.105.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 59.Wiggins P.A., van der Heijden T., Nelson P.C. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat. Nanotechnol. 2006;1:137–141. doi: 10.1038/nnano.2006.63. [DOI] [PubMed] [Google Scholar]
- 60.Šulc P., Romano F., Louis A.A. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 2012;137:135101. doi: 10.1063/1.4754132. [DOI] [PubMed] [Google Scholar]
- 61.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.SantaLucia J., Jr., Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 63.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 64.Faas F.G., Rieger B., Cherny D.I. DNA deformations near charged surfaces: electron and atomic force microscopy views. Biophys. J. 2009;97:1148–1157. doi: 10.1016/j.bpj.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu H., Chen H., Yan J. Two distinct overstretched DNA states. Nucleic Acids Res. 2010;38:5594–5600. doi: 10.1093/nar/gkq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rouzina I., Bloomfield V.A. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys. J. 2001;80:882–893. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitelam S., Pronk S., Geissler P.L. There and (slowly) back again: entropy-driven hysteresis in a model of DNA overstretching. Biophys. J. 2008;94:2452–2469. doi: 10.1529/biophysj.107.117036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams M.C., Wenner J.R., Bloomfield V.A. Entropy and heat capacity of DNA melting from temperature dependence of single molecule stretching. Biophys. J. 2001;80:1932–1939. doi: 10.1016/S0006-3495(01)76163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romano F., Chakraborty D., Louis A.A. Coarse-grained simulations of DNA overstretching. J. Chem. Phys. 2013;138:085101. doi: 10.1063/1.4792252. [DOI] [PubMed] [Google Scholar]
- 70.Fu H., Chen H., Yan J. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res. 2011;39:3473–3481. doi: 10.1093/nar/gkq1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allemand J.F., Bensimon D., Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc. Natl. Acad. Sci. USA. 1998;95:14152–14157. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bryant Z., Stone M.D., Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 73.King G.A., Peterman E.J., Wuite G.J. Unravelling the structural plasticity of stretched DNA under torsional constraint. Nat. Commun. 2016;7:11810. doi: 10.1038/ncomms11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh J., Purohit P.K. Structural transitions in torsionally constrained DNA and their dependence on solution electrostatics. Acta Biomater. 2017;55:214–225. doi: 10.1016/j.actbio.2017.03.052. [DOI] [PubMed] [Google Scholar]
- 75.Calladine C.R. Mechanics of sequence-dependent stacking of bases in B-DNA. J. Mol. Biol. 1982;161:343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- 76.Saenger W. Springer; New York: 1984. Principles of Nucleic Acid Structure, Springer Advanced Texts in Chemistry. [Google Scholar]
- 77.Johnson B.B., Dahl K.S., Zhurkin V.B. Correlations between deoxyribonucleic acid structural parameters and calculated circular dichroism spectra. Biochemistry. 1981;20:73–78. doi: 10.1021/bi00504a013. [DOI] [PubMed] [Google Scholar]
- 78.Sprecher C.A., Baase W.A., Johnson W.C. Conformation and circular dichroism of DNA. Biopolymers. 1979;18:1009–1019. doi: 10.1002/bip.1979.360180418. [DOI] [PubMed] [Google Scholar]
- 79.Marvin D.A., Spencer M., Hamilton L.D. The molecular configuration of deoxyribonucleic acid. III. X-ray diffraction study of the C form of the lithium salt. J. Mol. Biol. 1961;3:547–565. doi: 10.1016/s0022-2836(61)80021-1. [DOI] [PubMed] [Google Scholar]
- 80.Cocco S., Yan J., Marko J.F. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E Stat. Nonlin. Soft. Matter. Phys. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 81.Rief M., Clausen-Schaumann H., Gaub H.E. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 82.Clausen-Schaumann H., Rief M., Gaub H.E. Mechanical stability of single DNA molecules. Biophys. J. 2000;78:1997–2007. doi: 10.1016/S0006-3495(00)76747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.