Summary
In traditional optical imaging, limited light penetration constrains high-resolution interrogation to tissue surfaces. Optoacoustic imaging combines the superb contrast of optical imaging with deep penetration of ultrasound, enabling a range of new applications. We used multispectral optoacoustic tomography (MSOT) for functional and structural neuroimaging in mice at resolution, depth, and specificity unattainable by other neuroimaging modalities. Based on multispectral readouts, we computed hemoglobin gradient and oxygen saturation changes related to processing of somatosensory signals in different structures along the entire subcortical-cortical axis. Using temporal correlation analysis and seed-based maps, we reveal the connectivity between cortical, thalamic, and sub-thalamic formations. With the same modality, high-resolution structural tomography of intact mouse brain was achieved based on endogenous contrasts, demonstrating near-perfect matches with anatomical features revealed by histology. These results extend the limits of noninvasive observations beyond the reach of standard high-resolution neuroimaging, verifying the suitability of MSOT for small-animal studies.
Keywords: photoacoustic imaging, hemodynamic response, whole-brain tomography, near-infrared neuroimaging, label-free interrogation, temporal coherence
Graphical Abstract
Highlights
-
•
Noninvasive functional and structural neuroimaging in mice using MSOT
-
•
Resting and activity-dependent response mapped on entire brain cross-sections
-
•
Connectivity between various structures revealed with coherence analysis
-
•
High-resolution structural tomography achieved based on endogenous contrasts
Olefir et al. apply multispectral optoacoustic (photoacoustic) tomography (MSOT) for noninvasive spatial and spectral mapping and decomposition of neural dynamics and organization of the intact mouse brain in vivo. The results extend the boundaries of noninvasive high-resolution observations beyond the reach of intravital optical neuroimaging in small-animal studies.
Introduction
Unveiling a structure-functional relationship in the nervous system is one of the key priorities of neuroimaging. In this pursuit, optical imaging has been the main workhorse, enabling over many decades the high-resolution structural brain imaging (Wilt et al., 2009). With superb contrast, resolution, and multiplexing capabilities, optical interrogation has also recently become an integral part of functional neuroimaging, empowering sensing of voltage and calcium dynamics of individual neurons and synaptic connections (Chen et al., 2011, Grienberger et al., 2014, Perron et al., 2009, Peterka et al., 2011, Stettler et al., 2006). Notwithstanding these advances, investigation of the brain using light has several fundamental limitations, due to diffraction and scattering, which constrain observations to the surface (Hillman, 2007, Hong et al., 2014, Ntziachristos, 2010). For microscopy, penetration of several hundred microns into brain has been achieved using two-photon excitation via cranial opening, which could be extended further by surgical removal of superficial tissue mass, with downsides of invasiveness and loss of circuit integrity (Dombeck et al., 2010, Helmchen and Denk, 2005, Kerr and Denk, 2008, Willem et al., 2015). Using three-photon GCaMP6 excitation, the activity of neurons in the dorsal hippocampus was recently recoded in vivo, albeit with major depth-dependent loss of spatial resolution (Ouzounov et al., 2017). For macroscopic brain imaging usage, the limits set by light scattering have been partly surmounted by the utilization of diffusive imaging methods such as diffuse optical tomography (DOT) and near-infrared spectroscopy (NIRS), which enabled measurements in thick samples, but with poor spatial resolution (Boas et al., 2004, Culver et al., 2003, Eggebrecht et al., 2014, Siegel et al., 1999).
Unlike optical imaging, high-resolution radiological methods such as fMRI and ultrasonography (US) enable excellent penetration with 3D isotropic visualization of intact biological samples. However, the principal downfall of fMRI is its low sensitivity, which limits spatial and especially temporal resolution (Denic et al., 2011, Hoyer et al., 2014, Jonckers et al., 2015, Logothetis, 2008), above all detrimental in small-animal studies, where miniature brain size and tiny structures downgrade the data quality (Farahani et al., 1990, Hoyer et al., 2014, Yang et al., 2006). Unlike fMRI, functional US (fUS) affords first-rate resolution in time and space, empowering exquisite connectivity analysis and hemodynamic readouts in murine and human applications (Errico et al., 2015, Gesnik et al., 2017, Macé et al., 2011, Osmanski et al., 2014). While suitable for mapping brain activation by sensory inputs, high-resolution fUS in adult murine models requires skull thinning or cranial opening (Macé et al., 2011, Osmanski et al., 2014). Using optimized US sequence with specialized probes, a recent report showed the feasibility of noninvasive transcranial fUS in young mice (< 1 month old [m.o.]), validating its utility for neuroimaging in behavioral studies (Tiran et al., 2017).
Optoacoustic (photoacoustic) imaging combines the superb contrast mechanisms of optical imaging with the high resolution and penetration depth of US (Beard, 2011, Ntziachristos et al., 2005, Taruttis and Ntziachristos, 2015). Despite unique capacities for multiscale and multispectral interrogation of thick biological samples, optoacoustic functional neuroimaging so far has remained largely confined to the cerebral cortex (Liao et al., 2012, Nasiriavanaki et al., 2014, Tang et al., 2015, Wang et al., 2003, Wang et al., 2013, Yao et al., 2015). The majority of functional neuroimaging reports have also neglected the multispectral assets of the methodology, using only a single illumination wavelength (Liao et al., 2012, Nasiriavanaki et al., 2014, Ovsepian et al., 2017, Wang et al., 2003, Yao and Wang, 2014). A few attempts at mapping functional dynamics of brain circuits and hemoglobin gradient changes in deeper brain compartments have been limited to studies of drug-induced seizure or solid tumor xenografts, with dubious physiological relevance (Burton et al., 2013, Gottschalk et al., 2017). Finally, reports using optoacoustic imaging for structural brain studies in vivo have succumbed to visualization of large superficial vessels and major anatomical references (Laufer et al., 2009, Lin et al., 2015, Razansky et al., 2011, Stein et al., 2009).
Thus, to date, the most valuable assets of optoacoustic interrogation neither have been properly validated nor utilized for brain imaging (Ovsepian et al., 2017). In this study, we set out to address some of the omission by testing the efficacy of multispectral optoacoustic tomography (MSOT) for high-resolution label-free functional and structural imaging with reconstruction of the entire mouse brain. We visualize, map, and decompose the spatial and spectral characteristics of a neurovascular response induced by whisker inputs on the entire forebrain cross-sections within intact mice. Based on the temporal coherence of hemodynamic signals and oxygen saturation gradients, we reconstruct the connectivity involved in processing and integration of somatosensory signals along the entire subcortical-cortical axis, which replicates closely the circuitry revealed by neurophysiological studies and tract tracing. Finally, we make original use of MSOT for label-free and contrast-based imaging of mouse brain anatomy and tract tracing ex vivo and in vivo, to relate volumetric data with results of light-microscopic observations.
Results
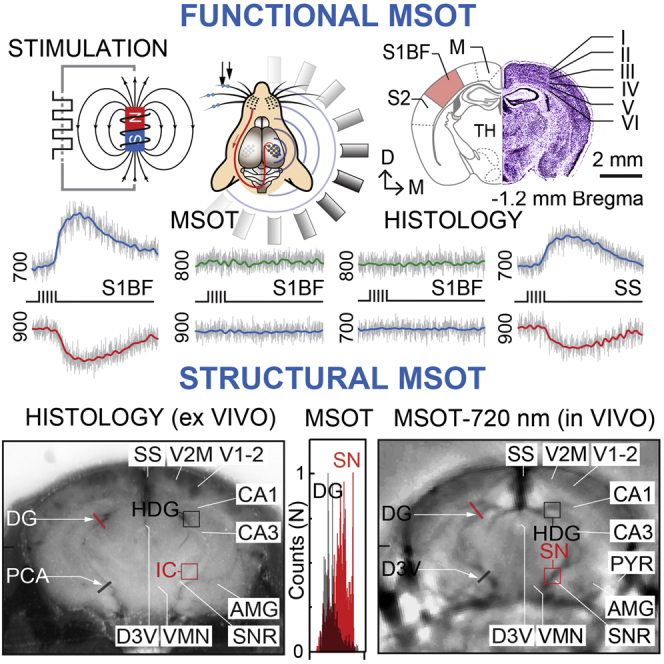
Despite outstanding capabilities for high-resolution interrogation and volumetric imaging of deep tissue, the vast majoroty of functional optoacoustic brain studies in small-animal models have so far been confined to the cortex. To date, activity-related changes in hemodynamic gradients within deep compartments of the brain have been unexplored. Taking advantage of the strong central representation and well-defined connections of somatosensory whisker projections of mice, we imaged the neurovascular responses related to activity and processing of whisker inputs on the entire forebrain cross-sections using MSOT (Figures S1–S3). We applied whisker stimulation (mechanical deflections) and monitored changes in deoxyhemoglobin (Hb) and oxyhemoglobin (HbO2) gradients in the brain, with computed tissue oxygenation (SO2) level.
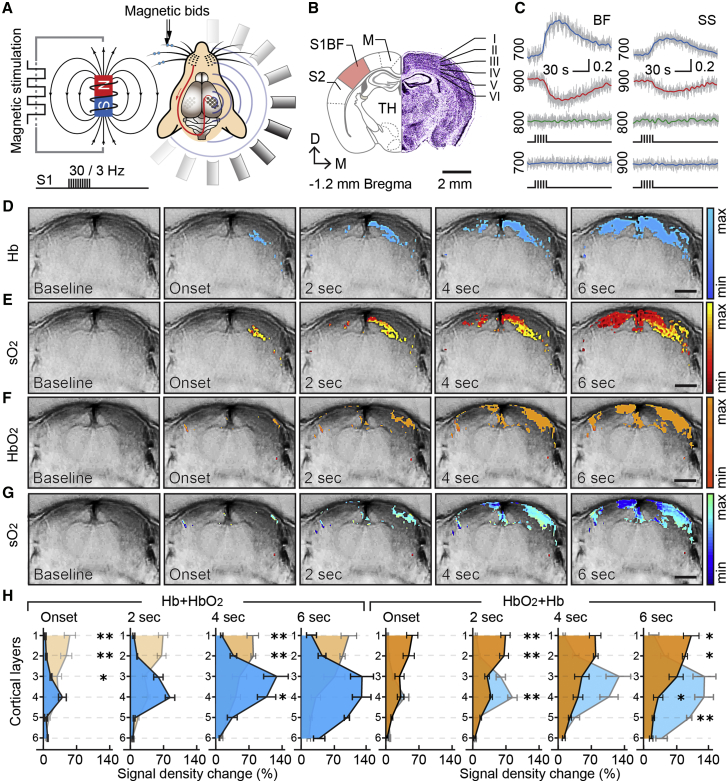
Figures 1A–1C show the experimental layout of functional MSOT studies and illustrates a mouse brain cross-section containing the somatosensory barrel cortex along with several major anatomical structures, together with traces of Hb, HbO2, and isosbestic point (700, 900, and 800 nm) signal changes as well as traces from negative control experiments collected from mice without whiskers stimulation. While in all experimental trials initial changes in Hb gradients and SO2 signals related to the activation of whisker inputs were most prominent in the somatosensory barrel field of the contralateral side, over a short period of time, the evoked responses spread over the adjacent somatosensory cortex hindlimb and primary motor areas, as well as ipsilateral somatosensory barrel and motor fields. Interestingly, under our experimental settings, the total blood volume (i.e., signal detected at 800 nm) in activated cortical regions remained largely unchanged, possibly due to weak intensity of whisker inputs. Figures 1D–1G illustrate a representative series of MSOT images overlaid with thresholded Hb, HbO2, and SO2 maps at consecutive time points from two experiments. Analysis of signal density changes across different layers of the somatosensory barrel field revealed a distinctly lamellar pattern of the hemodynamic response with significant differences evident in the signal onset and distribution across different cortical layers (Figure 1H). Unlike HbO2 changes concomitantly across multiple layers of the barrel cortex and reaching the highest intensity in the superficial compartments, the Hb response first appeared in L4–L5 and spread toward both the superficial and deeper cortical layers. Importantly, all alterations in hemodynamic signals related to the somatosensory whisker inputs were stimulus specific, as similar trails with non-magnetized whiskers showed no HbO2 or Hb changes (Figure 1C). Likewise, in similar experiments with two consecutive series of whisker stimulation, a strong activity-dependent depression in the response to the second stimulation series was observed, in line with history-dependent plasticity of the hemodynamic response (Figure S4).
Figure 1.
Functional MSOT of Whisker-Induced Response in the Somatosensory Barrel Cortex
(A) Schematic of the experimental design of whisker stimulation with pull-push magnet and brain imaging with MSOT.
(B) Representation of mouse brain cross-section containing anatomical references such as somatosensory barrel cortex (S1BF), S2 somatosensory field (S2), and motor cortex (M) (left), with Nissl-stained brain slice of corresponding plane showing cortical layers of the S1BF (right).
(C) Typical traces of HbO2 (900 nm), Hb (700 nm), and isosbestic (800 nm) signal from experimental group (top), and HbO2 and Hb signals from negative control group verifying that changes in Hb and HbO2 signals are specific to activation of whisker inputs.
(D–G) Consecutive time series of anatomical brain images overlaid with Hb (D), HbO2 (F), and SO2 (E and G) maps before (baseline) and after (2-s increments) whisker stimulation. Activation maps presented in different colors show pixel-wise changes in the intensity of MSOT signal related to the stimulation of whisker inputs, with signal intensity changes presented in arbitrary units.
(H) Distribution and dynamics of the whisker evoked Hb, HbO2, and SO2 signal density across different layers of S1BF of the somatosensory cortex. Graphs present means and SEM of signal density changes in different cortical layers from six independent trials with their comparison (∗p < 0.05; ∗∗p < 0.005; unpaired t test).
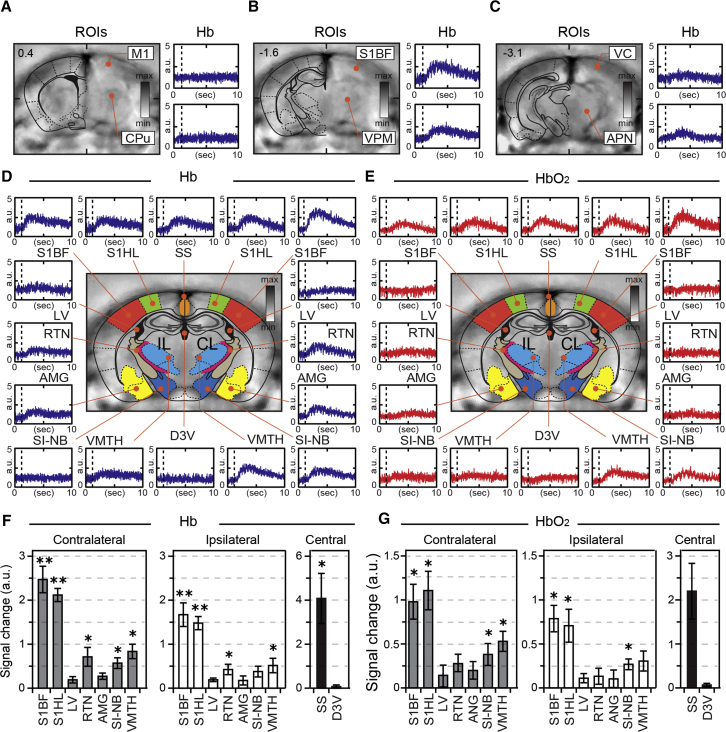
As somatosensory inputs to the barrel cortex are conveyed through the lemniscal route, via ventral and posterior thalamic relay nuclei, and through extrathalamic modulatory tracts, via basal forebrain projections (Brecht et al., 2004, Zaborszky et al., 2015), we set out to investigate whether neurovascular response changes related to whisker inputs can be detected in these and other functionally associated deep-brain structures. Figures 2A–2C show three consecutive MSOT cross-sections of a mouse brain with a series of Hb and HbO2 recordings related to the activation of whisker inputs. As can be readily seen, the amplitude of hemodynamic response strongly depends on the anatomical plane, with the strongest changes in Hb gradients confined to the cross-section containing the somatosensory barrel field and associated thalamic nuclei of the contralateral side. Extending the analysis of stimulation-evoked responses with SO2 measurements over extensive brain areas revealed activation of wider fields, involving ventral and reticular thalamic nuclei, basal forebrain, and amygdala, with however no consistent response detectable in areas corresponding to lateral ventricles (Figures 2D–2G).
Figure 2.
Location Specificity of Hemodynamic Changes across Mouse Brains Induced by Whisker Inputs
(A–C) Consecutive anatomical MSOT images of mouse brain overlaid with schematic maps of corresponding planes (A, B, and C; left part) with representative recordings of Hb signal from marked regions of interest (ROIs) (A, B, and C; right). Anatomical references: M1, motor cortex; CPu, caudate putamen; S1BF, somatosensory cortex barrel field; VPM, ventral posterior nucleus; VC, visual cortex; APN, anterior pretectal nucleus. Note that Hb changes are specific to the anatomical plane containing S1BF.
(D and E) Typical recordings of Hb (D) and HbO2 (E) signal from selected ROIs of the mouse brain. IL and CL, ipsilateral and contralateral to the stimulation side. RTN, reticular thalamic nucleus; SI-NB, substantia innominate nucleus basalis; S1HL, primary somatosensory, hindlimb; S1BF, primary somatosensory, barrel field; VMTH, ventromedial thalamic nucleus; LV, lateral ventricle; D3V, third dorsal ventricle; AMG, amygdala; SS, sagittal sinus; AMG, amygdala. For illustration purposes, all functional readouts have been inverted to represent more clearly the relative change.
(F and G) Summary graphs illustrating the mean values with SEM of Hb (F) and HbO2 (G) signal changes (i.e., peak amplitude) in different brain compartments pulled from six independent trials with their comparison (∗p < 0.05; ∗∗p < 0.005; unpaired t test).
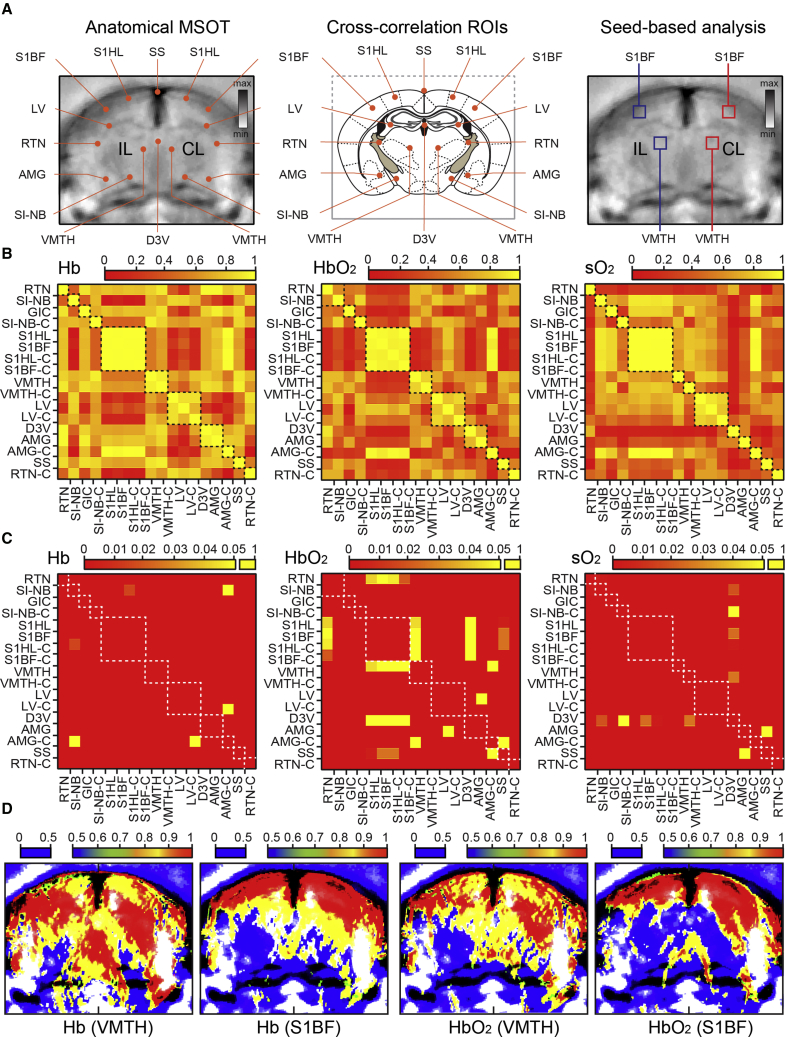
Next, the extent of the temporal coherence in the neurovascular response between multiple brain regions was analyzed, with results summarized in cross-correlation matrices of Hb, HbO2, and SO2 signals collected across entire brain cross-sections containing the somatosensory barrel field and related structures (Figures 3A–3D). Analogous measurements from negative control experiments, without activation of whisker inputs, are shown in Figures S5A and S5B. As can be readily appreciated, repetitive whisker stimulation activates rapid and temporally coherent hemodynamic response in the somatosensory barrel fields as well as in adjacent somatosensory hindlimb areas of both sides. In addition, coherent activation of a hemodynamic response was detectable in related reticular and ventral thalamic nuclei, as well as throughout the basal forebrain area, which correlated with the response of the barrel field in a statistically significant fashion (Figure 3C). Of note, the extent of functional coupling of evoked activity and correlation indexes were consistently higher in Hb and SO2 graphs compared to HbO2. Alterations in the hemodynamic signal induced by the activation of the somatosensory inputs were also readily detectable on the correlation seed maps of the entire brain cross-sections, when the contralateral ventromedial thalamic nucleus or the somatosensory barrel field were taken as the seed for such analysis (Figure 3D). Consistent with the results of cross-correlation maps, stimulation-related hemodynamic changes synchronized with the seed areas were detectable in reticular and ventral thalamic nuclei, as well as in the basal forebrain area.
Figure 3.
Temporal Coherence of Hemodynamic Changes Induced by Stimulation of Whisker Inputs in Mouse Brain
(A) Structural MSOT images (left and right panels) with marked ROIs used in cross-correlation analysis (left) and in seed-based correlation mapping the functional connectome (right), along with schematized map of corresponding brain plane with anatomical references (middle). For abbreviations, see Figure 2 legend.
(B and C) Correlation matrix of Hb, HbO2, and SO2 illustrating the degree of temporal coherence of hemodynamic response induced by whisker inputs (B) and corresponding graph of the distribution of p values of same ROIs (C).
(D) Seed-based correlation maps of the same brain illustrating areas with temporally coherent changes in Hb and HbO2 signals (i.e., co-activation) in response to whisker inputs. Whisker input driven changes of the hemodynamic signals in the contralateral somatosensory cortex barrel field (S1BF) and contralateral ventro-medial thalamic nucleus (VMTH) have been used as seeds for current coherence maps, with the degree of correlation presented in the color bars.
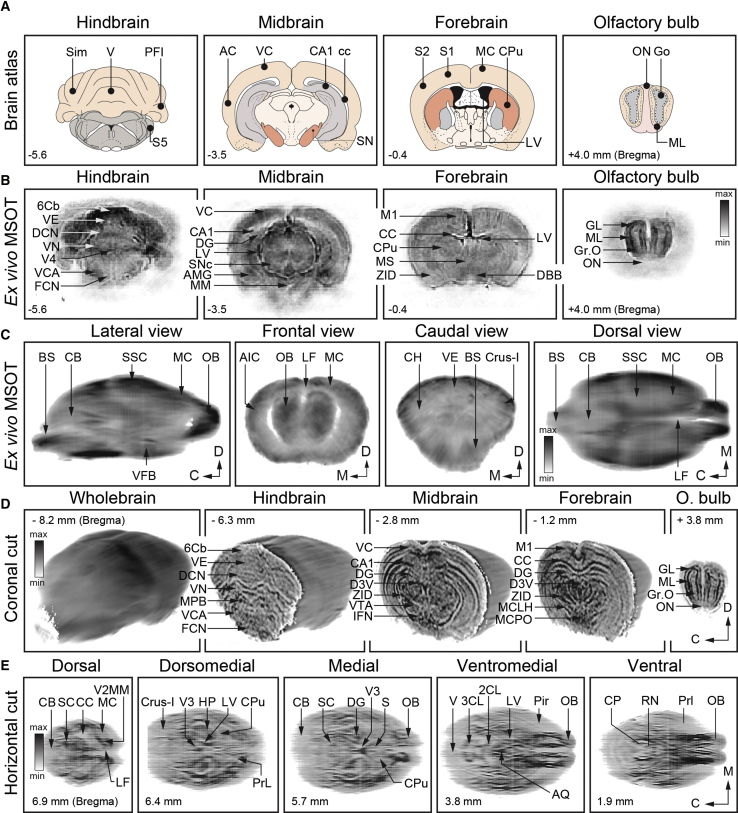
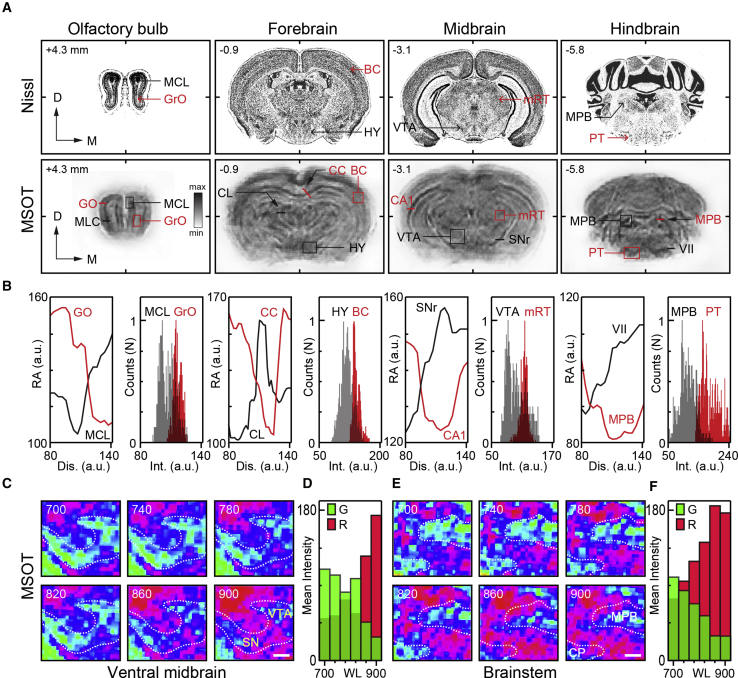
Having verified the in vivo functional neuroimaging capacities of MSOT across the entire cross-section of the mouse brain, we set to explore and characterize its capabilities as a high-resolution anatomical imaging tool, to verify the accurate assignment of functional readouts to various brain structures. Serial cross-sectional anatomical images of the mouse brain were acquired along the rostro-caudal axis at 0.1 mm steps in vivo followed by ex vivo imaging without perfusion or after perfusion. Figures 4A–4E present consecutive coronal and horizontal anatomical scans of an ex vivo mouse brain at 740-nm wavelength and relate them with anatomical references from the mouse brain atlas. As is clearly visible, at the specified wavelength as well as throughout almost the entire range of the NIR window (700–900 nm), the endogenous absorbers of perfused (bloodless) brains strongly contribute to the formation of anatomical MSOT images (Figures 4C–4E and S6). Indeed, the distinctly stratified and compartmented appearance of brains in coronal cross-sections with a lamellar outlook of olfactory bulbs, along with numerous structural details within the inner forebrain, midbrain, and hindbrain, including cerebellum and brainstem, recapitulate with high fidelity the schematized features of the corresponding anatomical planes from the brain atlas (Figures 4A–4E; Videos S4 and S5). To verify more rigorously the relationship of the structural features as revealed by ex vivo MSOT with major anatomical formations of the mouse brain, we compared coronal MSOT scans with structural details on matching stereotactic matching cross-sections enhanced with Nissl staining, captured at low magnification light microscopy (Figures 5A and 5B). As can be readily seen, there is a near-perfect correspondence between the coarse and fine features of the two series of images, with structures revealed by histology also clearly visible and quantifiable on MSOT cross-sections. Importantly, spectral decomposition of images revealed highly localized and specific traits across various brain regions, with melanin-rich midbrain dopaminergic nuclei as well as myelinated bundles of the midbrain, hypothalamus, and medulla readily discernable. In the ventral midbrain and dorsal medulla, for instance, neuro-melanin-rich substantia nigra pars compacta (SNc) and adrenergic pontine-reticular nuclei can be readily delineated on the MSOT scans (Figures 5C–5F).
Figure 4.
Structural Brain Imaging with MSOT Ex Vivo
(A and B) Schematized anatomy of a mouse brain at four different coronal planes (Bregma coordinates underneath) (A) with corresponding MSOT cross-sections of non-perfused ex vivo brain (B). Note exquisite structural details revealed at all planes and depths throughout entire brain cross-sections. ON, olfactory nerve; Go, granule cells of olfactory bulb; ML, mitral cell layer; S1 and S2, somatosensory cortex 1 and 2; MC, motor cortex; CPu, caudate putamen; LV, lateral ventricle; AC and VC, auditory and visual cortices; CA1, hippocampal CA1 area; cc, corpus callosum; SN, substantia nigra; Sim, simple lobule; V, vermis; PFI, para-floccules; S5, trigeminal nucleus; Gr.O, granule cells of olfactory bulb; GL, granule cell layer; MS, medal septum; DBB, diagonal band Broca; DG, dentate gyrus; LV, lateral ventricle; SNc, substantia nigra pars compacta; AMG amygdala; MM, medial mammillary nucleus; 6Cb, cerebellar lobule 6; VE, vermis; DCN, deep cerebellar nucleus; VN, vestibular nucleus; V4, fourth ventricle; VCN, ventral cochlear nucleus; FNC, facial nucleus.
(C) Lateral, frontal, caudal, and dorsal views of reconstructed mouse brain, from left to right. Maximum intensity projections. C, caudal; D, dorsal; M, medial; BS, brainstem; CB, cerebellum; SSC, somatosensory cortex; MC, motor cortex; OB, olfactory bulb; AIC, agranular insular cortex; LF, longitudinal fissure; CH, cerebellar hemisphere; VE, vermis; Crus-1, cerebellar crus 1.
(D) MSOT of the whole brain and series of cross-sections at four different planes. 6Cb, sixth cerebellar lobule; VE, vermis; DCN, deep cerebellar nucleus; VN, vestibular nuclear complex; MPB, medial parabrachial nucleus; VCA, ventral cochlear nucleus; FCN, facial nucleus; VC, visual cortex; CA1, CA1 area; DG, dentate gyrus; D3V, dorsal 3 ventricle; ZID, zona incerta dorsalis; VTA, ventral tegmental area; IFN, interfascicular nucleus; M1, motor cortex; CC, corpus callosum; MCLH, magnocellular lateral hypothalamus; MCPO, magnocellular preoptic nucleus; GL, glomerular layer; ML, mitral cell layer; Gr.O, granule cell layer; ON, olfactory nerve.
(E) Horizontal MSOT sections of the mouse brain at five (D, dorsal; DM, dorso-medial; M, medial; MV, medio-ventral; and V, ventral) planes with reference to interaural plane (distance in millimeters). CB, cerebellum; SC, superior colliculus; CC, corpus callosum; MC, motor cortex; V2MM, secondary visual cortex; LF, longitudinal fissure; V3, ventricle 3; HP, hippocampus; LV, lateral ventricle; CPu, caudate putamen; PrL, pre-limbic cortex; DG, dentate gyrus; S, septum; OB, olfactory bulb; V, vermis; 3CL, 3 cerebellar lobule; 2CL, 2 cerebellar lobule; AQ, aqueduct cerebral; Pir, piriform cortex; CP, cerebellar peduncle; RN, reticular nucleus; Prl, prelimbic cortex.
Figure 5.
Morphometry and Correlation of Anatomical Features Revealed by MSOT with Those of Histochemistry
(A) Typical Nissl-stained mouse brain sections at consecutive planes (top) with corresponding MSOT images (bottom) (millimeters from bregma) (modified with permission from http://atlas.brain-map.org/atlas). Note close resemblance between major features of two sets of images. MCL, mitral cell layer; GrO, granule cell layer; BC, barrel cortex; HY, hypothalamus; VTA, ventral tegmental area; mRT, mesencephalic reticular thalamic nucleus; MPB, medial parabrachial nucleus; PT, pyramidal tract; CL, central thalamic nucleus; CC, corpus callosum; SN, substantia nigra; CA1, hippocampal CA1 region; VII, facial nucleus.
(B) Intensity profile graphs and absorption density distribution histograms of selected structures marked in (A) verifying the feasibility of semiquantitative morphometry using MSOT scans of ex vivo mouse brain.
(C and D) MSOT images of the VTA and SNc area of the midbrain taken at different wavelengths (C) with representation of spectral changes attributable to melanin-rich structures (blue) in the region (D). (C) Scale bar: 100 μm.
(E and F) Spectral map of the MPB and cerebellar peduncle (CP) region of medulla (E) with representation of spectral changes (F) presumably due to presence of melanin in the region (between 700 and 900 nm). Scale bar: 100 μm. Note wavelength-dependent changes in the spectral content of MSOT images over the analyzed range (ratio of red and green pixels in C and E plotted in D and F).
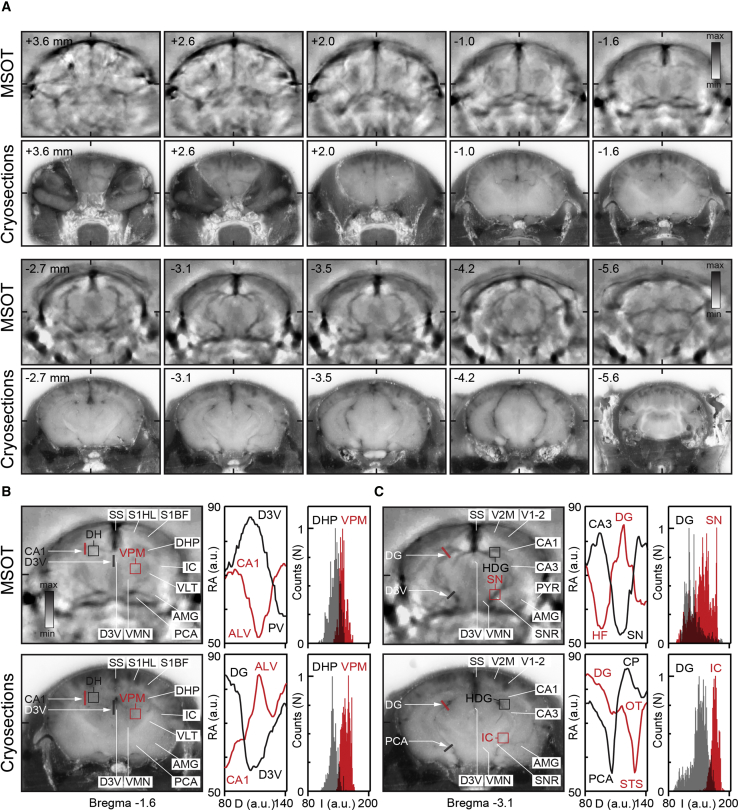
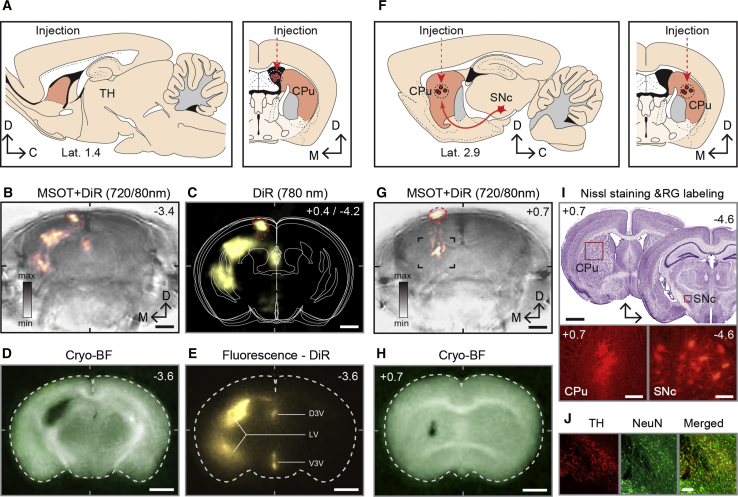
Because hemoglobin is the principal endogenous chromophore and the main source of optoacoustic signal in vivo, its fairly uniform presence throughout the brain degrades structural features formed by other endogenous absorbers. To more rigorously define the anatomical brain imaging capabilities of MSOT in vivo, we compared the major structural features exposed by MSOT in the living animal with those captured with camera on the cryosliced frozen mouse head. As illustrated on a series of images (Figure 6A; Video S6), numerous major anatomical features visible on coronal cryosections were also readily discernable on MSOT cross-sections acquired at 720 nm, showing a near-perfect cross-correspondence. Similar to the ex vivo MSOT brain images, the structures revealed by anatomical MSOT in vivo could be quantified using morphometric tools and methods (Figures 6B and 6C), although the contrast and prominence of superficial and deep structural details are more distinguishable in ex vivo images. To find out whether the deep neuroimaging imaging capabilities of MSOT in vivo can be further enhanced using exogenous contrast agents, we applied Di-R for visualizing the ventricular system of the mouse brain and for tracing nigrostriatal dopaminergic projections. The high extinction coefficient of Di-R in aqueous medium makes it ideal for imaging the ventricular system as well as tracing water-rich axonal bundles. Two days after tracer injection in the dorsal chamber of the lateral ventricle, or in the caudate nucleus, in vivo MSOT scans of the entire brain were acquired, followed by perfusion of mice and histological verification of injection sites and visualization of the labeling of neuronal tracts, using light microscopy (Figures 7A–7J). In mice injected with Di-R in the dorsal cistern of the lateral ventricle, 3D reconstruction and spectral unmixing showed strong labeling of the ventricular system of the injected side, with dorsal, lateral, and ventral chambers readily visible (Figures 7A–7C). These anatomical features revealed with MSOT were further verified by low-power fluorescence imaging of corresponding brain cross-sections (Figures 7D and 7E). Similarly, in mice receiving Di-R injection in the dorsal striatum, the site of tracer infusion could be visualized on in vivo MSOT cross-sections, as also confirmed by ex vivo cryo-slicing and histology (Figures 7F–7H). We could not, however, visualize the nigro-striatal dopaminergic projections or SNc nucleus on whole-brain MSOT scans, even though the presence of Di-R dopaminergic axons and neurons were detectable with immunostaining and light microscopy (Figures 7I and 7J).
Figure 6.
Label-Free Anatomical MSOT of Intact Mouse Brain In Vivo
(A) Series of consecutive MSOT cross-sections acquired in vivo (MSOT, average of 10 frames) and corresponding low-power images of the same frozen mouse brain captured on the cryo-slicer (cryosections). Note numerous exquisite structural details revealed by MSOT at all anatomical planes and depths throughout the entire brain cross-sections, with their nearly perfect correspondence with those captured using a high-resolution digital camera.
(B and C) Anatomical images and intensity profile graphs with absorption density distribution analysis of selected anatomical structures (bregma –1.6 mm, B, and bregma –3.1 mm, C) (red and black lines and boxed areas) demonstrating the utility of label-free MSOT for semiquantitative morphometry and neuroanatomical measurements in vivo in intact mice. SS, sagittal sinus; S1HL, somatosensory cortex hindlimb area; S1BF, somatosensory cortex barrel field; DHP, dorsal hippocampus; CA1, hippocampal CA1 area; VPM, ventral posterior-medial thalamic nucleus; IC, internal capsule; VLT, ventrolateral thalamic nucleus; VMN, ventromedial thalamic nucleus; AMG, amygdala; D3V, third ventricle; PCA, posterior central artery; PV, periventricular nucleus; ALV, stratum alveus; SN, substantia nigra; CP, cerebral peduncle; OT, optical tract; HF, hippocampal fissure; STS, sagittal transverse sinus; DG, dentate gyrus. Scale bar: 100 μm.
Figure 7.
Enhancing Structural Brain Imaging with MSOT Using Exogenous Contrasts
(A–E) Visualizing mouse ventricular system with MSOT using NIR Di-R tracer.
(A) A schematic of the sagittal and coronal brain sections showing the site of intraventricular injection of a tracer. Anatomical references: CPu, caudate putamen; TH, thalamus.
(B and C) MSOT cross-section of the mouse brain overlaid with unmixed and reconstructed images of the cerebral ventricles (B) with corresponding simplified schematics of brain cross-sections between −2.6 and −4.0 mm of bregma (C).
(D and E) Histological verification of the Di-R injection site (D) and ventricular labeling using cryo-slicing and fluorescence light microscopy (E). Anatomical references: D3V, dorsal 3 ventricle; LV, lateral ventricle; V3V ventral 3 ventricle.
(F) A schematic of the sagittal and coronal brain sections with the site of NIR Di-R infusion revealed in dorsal striatum for retrograde labeling of SN dopaminergic projection neurons.
(G and H) Anatomical MSOT cross-section of the mouse brain overlaid with unmixed and reconstructed images of tracer injection plane (G) with histological verification (H).
(I) Validation of the injection sites and the location of SN shown in Nissl-stained brain sections and in fluorescence microscopic images of the injections site in CPu and retrogradely labeled neurons in the SNc.
(J) Immunofluorescence verification of dopaminergic neurons using anti-tyrosinase antibody and neuron-specific marker NeuN.
Discussion
High-resolution imaging of large-scale neural dynamics and the organization of the brain in intact animal models are of major relevance to basic and translational neuroscience, relating specific functional processes to various structures without disruption of tissue homeostasis and integrity. Despite the major advances in neuroimaging, there is pressing need for technologies and methods enabling noninvasive scalable observations of static and dynamic processes in the brain. This is particularly pertinent to small-animal neuroimaging applications, where miniature scales and weak signals impose major challenges. As a highly promising interrogation method, optoacoustic imaging has shown great potential for a range of applications in murine neuroimaging. However, up until now, all of the major assets of optoacoustic imaging have been underrated or overlooked (Ovsepian et al., 2017). Much emphasis has been placed on resolving the vasculature on the brain surface and analysis of the neurovascular response in the cortex using an open-skull approach (Liao et al., 2012, Nasiriavanaki et al., 2014, Tang et al., 2015, Wang et al., 2003, Wang et al., 2013, Yao et al., 2015), with little progress made on systematic studies of processes and functions unfolding in the deeper compartments within the intact brain. Recently, an attempt has been made to visualize the functional connectome across an entire brain cross-section in craniotomized rats; however, images were acquired showing grossly distorted vascularization, possibly due to data overprocessing (Li et al., 2017).
In this study, we applied MSOT for high-resolution mapping of large-scale neurovascular dynamics in cross-sections of the mouse brain in response to somatosensory whisker inputs. We visualized and spectrally decomposed hemodynamic changes related to processing and allocation of whisker signals along the entire subcortical-cortical axis of the forebrain, to map Hb and HbO2 gradients and tissue oxygenation level changes in multiple anatomical planes and brain compartments. Using the same modality, we also obtained scans of mouse brains with significant structural detail, based on intrinsic chromophores and exogenous contrast agents. These unveiled a close-to-perfect match between key anatomical features as revealed by MSOT with those discovered using customary histology and microscopy. The strong central representation along with the well-defined connectivity and discreet response to whisker inputs (Brecht and Sakmann, 2002, Feldmeyer et al., 2013, Petersen, 2007) render the whisker somatosensory system an ideal experimental model, enabling access to fundamental neurobiological processes using neuroimaging and electrophysiological methods (Berger et al., 2007, Grinvald et al., 1986, Petersen et al., 2001, Yang et al., 1996). In agreement with results of optical imaging and neurophysiological reports, our MSOT scans showed strong activation of the barrel cortex by repetitive whisker inputs. The stimulation and type of anesthesia used in this study were based on careful consideration of numerous reports that investigated functional dynamics within this sensory system (Berger et al., 2007, Jonckers et al., 2015, Petersen et al., 2003, Yang et al., 1997). Unlike focusing on cellular-resolution microscopic readouts of functional changes in small neuronal groups within the superficial cortical layers, or low-resolution surrogate blood-oxygen-level-dependent (BOLD) signal of fMRI, we mapped and spectrally decomposed the activation and processing of somatosensory inputs by the brain at high spatial and temporal resolution. Although our observations are in general agreement with results of earlier reports showing whisker-induced hemodynamic changes in the contralateral barrel field (Feldmeyer et al., 1999, Petersen, 2007, Schubert et al., 2003), we were also able to detect expanded HbO2 and Hb alterations involving the ipsilateral barrel cortex as well as throughout thalamic nuclei, caudal basal forebrain nuclei, and other subcortical structures. These findings are in line with neuroanatomical data on ascending lemniscal projections, which relay whisker somatosensory inputs via ventral and posterior thalamic nuclei and through extra-thalamic circuits, activating a range of subcortical structures (Brecht et al., 2004, Zaborszky et al., 2015). In this context, it is important to note that the intensity and pattern of brain responses to somatosensory inputs from whiskers can be affected by multiple factors, including the type of anesthesia, stimulus intensity, and number of activated whiskers, with prolonged strong inputs capable of activating BOLD signal over wider brain areas, including the ipsilateral to the stimulation side cortical and subcortical structures, via long-range associative connections (Alonso et al., 2008, Logothetis, 2008, Mirabella et al., 2001).
Closer analysis of the initiation and spread of the hemodynamic signals showed that Hb changes in the cortex can faithfully replicate the order of processing and integration of whisker inputs given that thalamo-cortical projections target and synchronously activate the L4 neurons, with spread of the activity from therein across entire cortical columns (Bruno and Sakmann, 2006, Petersen, 2007, Yu et al., 2014). Indeed, such activation patterns caused a faster response onset and more robust Hb signal in deep cortical layers. In contrast, HbO2 signal changes appeared more diffuse, with higher signal intensity predominantly in the superficial layers. Although we do not have a ready explanation for such dissociation between Hb and HbO2 signals, the compensatory nature of HbO2 response, which in the cerebral cortex is enabled largely by activation of the superficial vascular bed via penetrating arterioles feeding deeper tissue, could account for the stronger HbO2 signal in the superficial layers (Nishimura et al., 2007, Schaffer et al., 2006, Yu et al., 2014). We would like to stress, however, that at present the coincidence and extent of coupling between the neuronal activity and BOLD responses remain open for debate and require further in-depth studies. Nevertheless, the specificity of activation of whisker inputs as demonstrated herein was verified by the absence of Hb and HbO2 signals in anatomical planes lacking representations of somatosensory whisker inputs (Figures 2A–2C).
With respect to anatomical MSOT imaging, it is critical to note that, within the NIR range (700–900 nm), in addition to Hb, other endogenous absorbers can contribute constructively toward the formation of structural images, revealing exquisite anatomical details across entire brain cross-sections. In intact animals, as we have shown, these features are strongly masked by an overwhelming hemoglobin signal but can be partly recovered using special imaging conditions and data processing. While the contribution of individual absorbers in the formation of anatomical images remains to be characterized, water, cytochromes, lipid, and melanin are expected to play a major role (Jacques, 2013, Ovsepian et al., 2017, Weber et al., 2016, Yao and Wang, 2014). As shown, structures such as the cerebral cortex, hippocampus, corpus callosum, basal ganglia, thalamic nuclei, ventricular system, olfactory bulb, cerebellum, together with a range of brainstem formations could be readily distinguished and quantified on MSOT scans by simple semi-quantitative analytical tools and spectral decomposition methods. It is reasonable to assume that, with its relatively uniform distribution in the brain, water absorption, like that of hemoglobin, also contributes toward the background signal, while the different density of proteins and lipids are expected to play key roles toward formation of structural features in the neuron-rich cerebral cortex and hippocampus, as well as in deep nuclei. Likewise, fat is expected to contribute and shape myelinated bundles and white-matter-rich hubs (Ovsepian et al., 2017). Clearcut appearance of neuro-melanin-rich midbrain dopaminergic and adrenergic nuclei (Barden and Levine, 1983, Saper and Petito, 1982) as revealed with MSOT are also of considerable interest as this could potentially serve as a biomarker for studies of neurodegenerative disease affecting these major supra-spinal hubs. As shown here, anatomical MSOT neuroimaging can be further enhanced using exogenous contrast agents. However, at this point, mapping the connectome and major neural projections with tracers seems to be over-ambitious due to the inherent low sensitivity of MSOT, urging further research and optimization of hardware and experimental procedures.
In summary, our results show the powerful capabilities of MSOT for functional and structural neuroimaging, and highlight some limitations and areas for future optimization. Given additional research and refinement, MSOT is expected to advance noninvasive neuroimaging of small-animal models onto a new level, offering an impressive means for addressing a wide range of outstanding questions, with implications for basic neurobiology and translational neuroscience.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken, polyclonal anti-TH | Abcam | CN76422 |
| Rabbit, polyclonal anti-NeuN | Abcam | CN104225 |
| AlexaFluor594, goat anti-chicken TH | Abcam | N150176 |
| AlexaFluor488 goat anti-rabbit NeuN | Life Technologies | A-11008 |
| Experimental Models: Organisms/Strains | ||
| Nude-FOXN1 Nu/Nu mice | Jackson Laboratories | 000819-B6.Cg-Foxn1 < nu > J |
| CD1 mice | Charles River | Crl:CD1(ICR)-022 |
| Software and Algorithms | ||
| IgorPro | Wavemetr, Oregon | Version 6.1 |
| A model-based reconstruction algorithm | IBMI GmbH | Dean-Ben al., 2012 |
| Other | ||
| NIR Di-R | Thermo-Fisher, DE | D12731 |
| MSOT256-TF | iThera Med. GmbH | 256-TF |
| Nd-YAG laser pumped OPO | InnoLas Las. GmbH | Model SL450 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be provided by the Lead Contact, Vasilis Ntziachristos (v.ntziachristos@helmholtz-muenchen.de).
Experimental Model and Subject Details
Young adult Hsd:Athymic Nude-FOXN1 Nu/Nu and CD1 mice used in all experiments described in the study (Table S1) were housed in a special housing facility at 21°C and 36% ± 2% humidity. The light-dark cycles were maintained at 12 h, with food and water provided ad libitum. All experiments involving living animals were conducted in accordance with the institutional guidelines and procedures approved by the government of Upper Bavaria, in compliance with German Federal and European Union law. Necessary steps have been taken to reduce animal use and suffering.
Method Details
Multispectral optoacoustic tomography (MSOT)
The MSOT (MSOT256-TF, iThera Medical GmbH, Munich, Germany) used for all imaging experiments has a cylindrically curved detector array (R = 40 mm), providing 270° angular coverage of the imaging specimen (Olefir et al., 2016). Figure S1A shows a schematic representation of MSOT. The detector containing 256 elements manufactured from a piezo-composite material covers up to 7.5 MHz, with 5 MHz ± 10% (−6 dB) center frequency. All channels are digitized in parallel at 10 Hz repetition and sampling rate of 40 MHz, using a custom designed data acquisition (DAQ) card. Illumination within the near infra-red (NIR, 680-940 nm) is provided by a tunable Nd-YAG laser pumped optical parametric oscillator (OPO, InnoLas Laser GmBH, Krailling, Germany) with 9 ns pulse duration, 90 mJ peak pulse energy at 750 nm wavelength with a repetition rate of 10 Hz. The laser beam multiplexed to five fiber bundles is arranged over the detector array to provide circular illumination of the imaging plane, which is fixed or moved stepwise in a rostrocaudal direction with 0.1 mm increment.
Brain imaging with MSOT
Male mice (4-8 weeks old) were used for all experiments, which were conducted under general isoflurane anesthesia. To avoid direct contact with the US coupling water medium in the imaging chamber, animals were wrapped in optically and acoustically transparent foil prior to their placement in the holder in a supine position relative to the MSOT detector array. After in vivo imaging sessions, mice were injected with a lethal dose of ketamine (180 mg/kg) and processed for ex vivo brain imaging experiments, cryo-slicing and immunohistochemistry. For ex vivo MSOT, brains were excised without or after perfusion (4% PFA in 0.1 M PBS) followed by 24 h post-fixation in 4% PFA. Wavelength selection was based on experimental requirements as well as consideration of light fluence maps, and absorption spectra of endogenous chromophores of the brain (Figures S1B–S1D).
For all functional brain imaging studies, mice were anesthetized, with customized magnetic beads attached to 6-10 large whiskers of rows B, C and D of the left side, which was used as the actuator of whisker stimulation. Anesthetized mice (1.2 - 1.8% vaporized isoflurane in air at 0.8 L/min) were placed in the MSOT, with the level of isoflurane subsequently reduced (0.7 - 1.0%) to achieve stable and shallow breathing, predictive of a highly responsive brain state. A custom-made rostral mask was utilized to ensure sufficient space for free whisker movement upon application of the magnetic force, while seamless contact was maintained between the mouse head and the coupling water medium (Figure S2A; Video S1. Testing of the Custom-Made Device for Mechanical Stimulation (Deflection) of Whiskers Using Magnetic Force, Related to Figures 1 and 2, Video S2. Testing of the Custom-Made Device for Mechanical Stimulation (Deflection) of Whiskers Using Magnetic Force, Related to Figures 1 and 2, Video S3. Testing of the Custom-Made Device for Mechanical Stimulation (Deflection) of Whiskers Using Magnetic Force, Related to Figures 1 and 2). An ‘in-house’ designed electromagnetic unit containing an induction coil with a metallic rod in the middle was placed in the imaging chamber in close proximity to the mouse rostrum, to generate sufficient magnetic force for whisker deflections. The latter was controlled by a customized AC amplifier and an impulse generator unit (Figure S2B) to deliver rectangular current pulses (100 ms) at 3-5 Hz frequency.
An “in-house” designed electromagnetic unit was used to generate sufficient magnetic force for whisker deflections at varying frequencies.
The selection of imaging planes was based on the coordinates described in the mouse brain atlas (Franklin and Paxinos, 2008), as well as on anatomical references visualized by the MSOT in vivo, to ensure that selected cross-sections contain the somatosensory cortex barrel field and connected thalamic nuclei. As an additional reference, we used in vivo MSOT brain images co-registered with ex vivo scans, with the latter highlighting exquisite structural details. A series of functional images from adjacent planes (1.5 mm rostral or caudal from the central plane) were also acquired to ensure brain structures displaying the most pronounced signals had been captured in MSOT studies. To safeguard fast acquisition of multispectral data, measurements of stimulation induced activities were conducted at three wavelengths: 700 nm, 800 nm, 900 nm, which correspond to the absorption optima of deoxy- and oxy- Hb (Hb = 700 nm and HbO2 = 900 nm) and isosbestic point, a measure of the total blood volume (805 nm) (Figures S1D, S3A, and S3B). For both, structural and functional imaging studies, raw data were collected without averaging at ∼3.3 Hz per each wavelength. Based on selected wavelengths, changes in Hb gradients and blood volume could be tracked over time and projected on anatomical MSOT cross-sections. After 60 s baseline imaging, whisker were stimulated over 10-30 s without interruption of the data collection, which was followed by 60 s post-stimulation recordings. For control experiments, the same protocols were followed with whiskers being not decorated with magnetic beads.
For ex vivo structural MSOT studies, non-perfused or perfused brains fixed with PFA were attached to a custom-made transparent rod placed in the center of the MSOT detector array, and imaged within 680-940 nm range, with 100 nm increment and 200 μm stepwise movements of the sample in the rostrocaudal direction. For in vivo structural MSOT, experimental procedures were similar to that of functional imaging studies except measurements were acquired along the entire rostro-caudal axis of the brain at 200 μm steps. Throughout these experiments, mice were supplied with air or transiently (30-45 s) challenged with 10% CO2, to achieve a better contrast and visibility of deep structures. All structural imaging experiments were followed by euthanizing the mice with ketamine overdose (180 mg/kg), deep freezing and slicing (100 μm) of mice brains for correlative imaging using a Leica Cryostat CM1950.
Di-R injection into the brain and immunohistochemistry
Under deep anesthesia (ketamine 80 mg/kg and xylazine 10 mg/kg) the head of the mouse was fixed in a stereotactic frame with the skull surgically opened under local anesthesia (bupivacaine, 0.5%). Using a dental drill, small holes were bored over the dorsal cistern of the lateral ventricle (−0.34 mm bregma, 1.4 mm lateral, 1.5 mm deep) or over the caudate nucleus (+0.5 mm bregma, 1.5 mm lateral, 2.7 mm deep) (Franklin and Paxinos, 2008). NIR Di-R (Thermo-Fisher, Germany) was slowly pressure-injected (1.0 μL over 5-6 min) into the lateral ventricle or the dorsal striatum with a Hamilton syringe (22-gauge needle) followed by careful removal of the injector and wound stitching. In control experiments, similar procedures were followed except mice were injected with an equivalent volume of DMSO only. After surgery, animals were placed back into their home cage. Following 4 days recovery period, animals were used for in vivo and ex vivo MSOT brain imaging at peak Di-R absorption wavelength (Figure S1D) followed by histology and immunostaining. For the latter, coronal PFA fixed sections containing the substantia nigra were dried on a heating plate and treated with 4% PFA for 7 min, followed by rinsing with TBST (0.1 mM). The slices were blocked with 10% goat serum in TBS and Triton (0.1 mM and 0. 3%, respectively) for 1 hour at room temperature. After washes of blocking solution, tissue was incubated with primary antibodies (chicken, polyclonal anti-tyrosine hydroxylase, 1:50 and rabbit, polyclonal anti-NeuN, 1:50) (Abcam, CN76422, and CN104225, respectively) diluted in TBS- Triton 0.3% in a moisture chamber at 4°C overnight. On the following day the secondary antibodies (AlexaFluor594, goat anti-chicken 1:200 for tyrosine hydroxylase and AlexaFluor488 goat anti-rabbit 1:200 for NeuN) (Abcam N150176 and Life Technologies A-11008) were added for 1 h in a moisture chamber at room temperature before and after rinsing with TBS-Triton 0.3% for 3 times over 10 min. Tissue sections were subsequently mounted with Prolong Gold anti-fade mounting media containing DAPI and viewed in a Zeiss Axio-Imager M2 Microscope. For negative control sections, all procedures were kept the same except that goat serum in TBS was used instead of primary antibodies. For ex vivo validation of the injection site and structural MSOT results, we performed immunohistochemistry on 10 μm thick frozen sections cut by the Leica Cryostat CM1950.
Quantification and Statistical Analysis
MSOT images were generated using a model-based reconstruction algorithm (Dean-Ben et al., 2012) with Tikhonov regularization. All acquired frames were co-registered to suppress the motion artifacts with rigid registration of the algorithm based on mutual information, which was supplied in MATLAB. Every frame was filtered using a Gaussian filter, with standard deviation (SD) of 2 pixels (200 μm). Similar to functional MRI data (Jonckers et al., 2015), in the resulting time-series, every pixel in the frame has an associated time trace representing the change of image intensity at the corresponding location. As functional imaging studies were performed at 3 wavelengths, all acquired data consisted of 3 time series. For every pixel, the corresponding intensity and time profile were filtered in the Fourier domain, with a low-pass filter in the range of up to 0.1 Hz, to remove high-frequency noise and any irrelevant signal fluctuations. A time trace of a hemodynamic response was generated based on the canonical Hemodynamic Response Function (HRF) convolved with a ramp function representing the stimulation pattern ( if the stimulation is on at time and otherwise), resulting in the expected response function, : .
For temporal coherence analysis and determining the extent of the correlation of the activity between various brain subfields, intensity traces were correlated with . The absolute correlation values obtained for each pixel were plotted to generate a spatial correlation map. The correlation map threshold was tuned to discard pixels that do not correlate with the expected response strongly (correlation value below 0.5) or significantly (, where is the total number of traces considered for a specific wavelength). For every pixel , the corresponding baseline intensity variability range was computed as , where and are the mean and standard deviation of the pixel intensity during the baseline measurement. For illustration purposes of brain activation, thresholded pixels altered by activation were overlaid in color over the anatomical images. A pixel shown as active at time has a corresponding correlation value higher than 0.5 and the corresponding intensity value at time exceeding the baseline variability range. Pixel color is coded to represent the deviation of its value from . Pixels with the corresponding time traces having negative values were discarded as such values have no physical meaning. This, however, did not mean that the corresponding time trace did not bear information on activation. For every obtained multispectral frame, sO2 was computed using a linear unmixing algorithm. To achieve better accuracy of sO2 estimation, all frames were calibrated for 4 cm water absorption prior to spectral decomposition. For illustration purposes of thresholded pixels, the corresponding computed sO2 values were overlaid in color on anatomical images, with pixels labeled as activated at either 700 nm or 900 nm illumination wavelengths.
For brain circuit analysis, 17 regions of interest (ROIs) were identified in MSOT brain cross-sections. Time traces corresponding to pixels in a particular region were averaged to improve the SNR. The resulting time series were compared pairwise. Correlation values as well as the corresponding P values were computed and presented. Furthermore, for each of the investigated time series, the activation amplitude was computed as , where is the mean value of during the baseline measurement. For seed-based correlation analysis, several seed regions have been identified. For every region, the corresponding traces have been averaged and the resulting time series was correlated to every trace outside of the seed region. The resulting correlation maps were thresholded as described above.
Anatomical MSOT images were generated through averaging 10 frames of the same anatomical planes acquired at a single specified excitation wavelength. For assessment and presentation of the Di-R data, reconstructed MSOT images were analyzed with a QL-shrinkage detection algorithm (Tzoumas et al., 2016) that estimates the spatial distribution of a contrast agent in the recorded multispectral dataset based on absorption spectrum.
In whole brain imaging studies with tracer injection, the resulting images and spatial distribution of Di-R were rendered in 3D and overlaid with anatomical images using Amira (ThermoFisher). All color maps are generated with MATLAB, while IgorPro was used for plotting graphs. Final figures were prepared using IgorPro (6.1, Wavemetrics, Oregon) and Adobe Illustrator (CS6 package). Where applicable, the mean values and standard error of the means (SEM) have been presented in graphs, to illustrate the distribution of the parameters of interest. Unpaired and paired Students t tests and one-way analysis of variance (ANOVA) have been used for variance analysis and comparison, with p values less than 0.05 defining differences as statistically significant.
Acknowledgments
The research leading to these results has received funding from the Deutsche Forschungsgemeinschaft (DFG), Germany (Reinhard Koselleck award “High resolution near-field thermoacoustic sensing and imaging”, NT 3/9-1; Gottfried Wilhelm Leibniz Prize 2013, NT 3/10-1); EIT Health (FiBrUS—Mammography matters: Fighting breast cancer with workflow optimized ultrasound screening; 18202); the European Union project CosmoPHOS (FP7-NMP-2012-LARGE-6, Contract 310337); and the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 761214. The materials presented and views expressed here are the responsibility of the author(s) only. The EU Commission takes no responsibility for any use made of the information set out. S.O.O. was also supported by the Project LO1611 from the MEYS under the NPU I program.
Author Contributions
S.V.O. and I.O. designed the study. I.O., A.G., H.Y., J.M.-N., and S.V.O. performed the experiments. A.G. and G.S. developed the stimulation system. S.G. provided technical support. I.O., H.Y., J.M.-N., P.S., and S.V.O. conducted data analysis and interpretation. S.V.O., I.O., V.B.O., and V.N. wrote the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: March 5, 2019
Footnotes
Supplemental Information includes six figures, one table, and six videos and can be found with this article online at https://doi.org/10.1016/j.celrep.2019.02.020.
Contributor Information
Vasilis Ntziachristos, Email: v.ntziachristos@helmholtz-muenchen.de.
Saak V. Ovsepian, Email: saak.ovsepian@gmail.com.
Supplemental Information
References
- Alonso B.d.C., Lowe A.S., Dear J.P., Lee K.C., Williams S.C., Finnerty G.T. Sensory inputs from whisking movements modify cortical whisker maps visualized with functional magnetic resonance imaging. Cereb. Cortex. 2008;18:1314–1325. doi: 10.1093/cercor/bhm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barden H., Levine S. Histochemical observations on rodent brain melanin. Brain Res. Bull. 1983;10:847–851. doi: 10.1016/0361-9230(83)90218-6. [DOI] [PubMed] [Google Scholar]
- Beard P. Biomedical photoacoustic imaging. Interface Focus. 2011;1:602–631. doi: 10.1098/rsfs.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T., Borgdorff A., Crochet S., Neubauer F.B., Lefort S., Fauvet B., Ferezou I., Carleton A., Lüscher H.R., Petersen C.C. Combined voltage and calcium epifluorescence imaging in vitro and in vivo reveals subthreshold and suprathreshold dynamics of mouse barrel cortex. J. Neurophysiol. 2007;97:3751–3762. doi: 10.1152/jn.01178.2006. [DOI] [PubMed] [Google Scholar]
- Boas D.A., Dale A.M., Franceschini M.A. Diffuse optical imaging of brain activation: approaches to optimizing image sensitivity, resolution, and accuracy. Neuroimage. 2004;23(Suppl 1):S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Brecht M., Sakmann B. Whisker maps of neuronal subclasses of the rat ventral posterior medial thalamus, identified by whole-cell voltage recording and morphological reconstruction. J. Physiol. 2002;538:495–515. doi: 10.1113/jphysiol.2001.012334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M., Krauss A., Muhammad S., Sinai-Esfahani L., Bellanca S., Margrie T.W. Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J. Comp. Neurol. 2004;479:360–373. doi: 10.1002/cne.20306. [DOI] [PubMed] [Google Scholar]
- Bruno R.M., Sakmann B. Cortex is driven by weak but synchronously active thalamocortical synapses. Science. 2006;312:1622–1627. doi: 10.1126/science.1124593. [DOI] [PubMed] [Google Scholar]
- Burton N.C., Patel M., Morscher S., Driessen W.H., Claussen J., Beziere N., Jetzfellner T., Taruttis A., Razansky D., Bednar B., Ntziachristos V. Multispectral opto-acoustic tomography (MSOT) of the brain and glioblastoma characterization. Neuroimage. 2013;65:522–528. doi: 10.1016/j.neuroimage.2012.09.053. [DOI] [PubMed] [Google Scholar]
- Chen X., Leischner U., Rochefort N.L., Nelken I., Konnerth A. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–505. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- Culver J.P., Durduran T., Furuya D., Cheung C., Greenberg J.H., Yodh A.G. Diffuse optical tomography of cerebral blood flow, oxygenation, and metabolism in rat during focal ischemia. J. Cereb. Blood Flow Metab. 2003;23:911–924. doi: 10.1097/01.WCB.0000076703.71231.BB. [DOI] [PubMed] [Google Scholar]
- Dean-Ben X.L., Ntziachristos V., Razansky D. Acceleration of optoacoustic model-based reconstruction using angular image discretization. IEEE Trans. Med. Imaging. 2012;31:1154–1162. doi: 10.1109/TMI.2012.2187460. [DOI] [PubMed] [Google Scholar]
- Denic A., Macura S.I., Mishra P., Gamez J.D., Rodriguez M., Pirko I. MRI in rodent models of brain disorders. Neurotherapeutics. 2011;8:3–18. doi: 10.1007/s13311-010-0002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck D.A., Harvey C.D., Tian L., Looger L.L., Tank D.W. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggebrecht A.T., Ferradal S.L., Robichaux-Viehoever A., Hassanpour M.S., Dehghani H., Snyder A.Z., Hershey T., Culver J.P. Mapping distributed brain function and networks with diffuse optical tomography. Nat. Photonics. 2014;8:448–454. doi: 10.1038/nphoton.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico C., Pierre J., Pezet S., Desailly Y., Lenkei Z., Couture O., Tanter M. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature. 2015;527:499–502. doi: 10.1038/nature16066. [DOI] [PubMed] [Google Scholar]
- Farahani K., Sinha U., Sinha S., Chiu L.C., Lufkin R.B. Effect of field strength on susceptibility artifacts in magnetic resonance imaging. Comput. Med. Imaging Graph. 1990;14:409–413. doi: 10.1016/0895-6111(90)90040-i. [DOI] [PubMed] [Google Scholar]
- Feldmeyer D., Egger V., Lubke J., Sakmann B. Reliable synaptic connections between pairs of excitatory layer 4 neurones within a single “barrel” of developing rat somatosensory cortex. J. Physiol. 1999;521:169–190. doi: 10.1111/j.1469-7793.1999.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D., Brecht M., Helmchen F., Petersen C.C., Poulet J.F., Staiger J.F., Luhmann H.J., Schwarz C. Barrel cortex function. Prog. Neurobiol. 2013;103:3–27. doi: 10.1016/j.pneurobio.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Franklin K.B.J., Paxinos G. Elsevier; 2008. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- Gesnik M., Blaize K., Deffieux T., Gennisson J.L., Sahel J.A., Fink M., Picaud S., Tanter M. 3D functional ultrasound imaging of the cerebral visual system in rodents. Neuroimage. 2017;149:267–274. doi: 10.1016/j.neuroimage.2017.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk S., Fehm T.F., Deán-Ben X.L., Tsytsarev V., Razansky D. Correlation between volumetric oxygenation responses and electrophysiology identifies deep thalamocortical activity during epileptic seizures. Neurophotonics. 2017;4:011007. doi: 10.1117/1.NPh.4.1.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C., Chen X., Konnerth A. NMDA receptor-dependent multidendrite Ca2+ spikes required for hippocampal burst firing in vivo. Neuron. 2014;81:1274–1281. doi: 10.1016/j.neuron.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Lieke E., Frostig R.D., Gilbert C.D., Wiesel T.N. Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature. 1986;324:361–364. doi: 10.1038/324361a0. [DOI] [PubMed] [Google Scholar]
- Helmchen F., Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hillman E.M. Optical brain imaging in vivo: techniques and applications from animal to man. J. Biomed. Opt. 2007;12:051402. doi: 10.1117/1.2789693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G., Diao S., Chang J., Antaris A.L., Chen C., Zhang B., Zhao S., Atochin D.N., Huang P.L., Andreasson K.I. Through-skull fluorescence imaging of the brain in a new near-infrared window. Nat. Photonics. 2014;8:723–730. doi: 10.1038/nphoton.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer C., Gass N., Weber-Fahr W., Sartorius A. Advantages and challenges of small animal magnetic resonance imaging as a translational tool. Neuropsychobiology. 2014;69:187–201. doi: 10.1159/000360859. [DOI] [PubMed] [Google Scholar]
- Jacques S.L. Optical properties of biological tissues: a review. Phys. Med. Biol. 2013;58:R37–R61. doi: 10.1088/0031-9155/58/11/R37. [DOI] [PubMed] [Google Scholar]
- Jonckers E., Shah D., Hamaide J., Verhoye M., Van der Linden A. The power of using functional fMRI on small rodents to study brain pharmacology and disease. Front. Pharmacol. 2015;6:231. doi: 10.3389/fphar.2015.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J.N., Denk W. Imaging in vivo: watching the brain in action. Nat. Rev. Neurosci. 2008;9:195–205. doi: 10.1038/nrn2338. [DOI] [PubMed] [Google Scholar]
- Laufer J., Zhang E., Raivich G., Beard P. Three-dimensional noninvasive imaging of the vasculature in the mouse brain using a high resolution photoacoustic scanner. Appl. Opt. 2009;48:D299–D306. doi: 10.1364/ao.48.00d299. [DOI] [PubMed] [Google Scholar]
- Li L., Zhu L., Ma C., Lin L., Yao J., Wang L., Maslov K., Zhang R., Chen W., Shi J., Wang L.V. Single-impulse panoramic photoacoustic computed tomography of small-animal whole-body dynamics at high spatiotemporal resolution. Nat. Biomed. Eng. 2017;1:0071. doi: 10.1038/s41551-017-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L.D., Lin C.T., Shih Y.Y., Duong T.Q., Lai H.Y., Wang P.H., Wu R., Tsang S., Chang J.Y., Li M.L., Chen Y.Y. Transcranial imaging of functional cerebral hemodynamic changes in single blood vessels using in vivo photoacoustic microscopy. J. Cereb. Blood Flow Metab. 2012;32:938–951. doi: 10.1038/jcbfm.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Xia J., Wong T.T., Li L., Wang L.V. In vivo deep brain imaging of rats using oral-cavity illuminated photoacoustic computed tomography. J. Biomed. Opt. 2015;20:016019. doi: 10.1117/1.JBO.20.1.016019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis N.K. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- Macé E., Montaldo G., Cohen I., Baulac M., Fink M., Tanter M. Functional ultrasound imaging of the brain. Nat. Methods. 2011;8:662–664. doi: 10.1038/nmeth.1641. [DOI] [PubMed] [Google Scholar]
- Mirabella G., Battiston S., Diamond M.E. Integration of multiple-whisker inputs in rat somatosensory cortex. Cereb. Cortex. 2001;11:164–170. doi: 10.1093/cercor/11.2.164. [DOI] [PubMed] [Google Scholar]
- Nasiriavanaki M., Xia J., Wan H., Bauer A.Q., Culver J.P., Wang L.V. High-resolution photoacoustic tomography of resting-state functional connectivity in the mouse brain. Proc. Natl. Acad. Sci. USA. 2014;111:21–26. doi: 10.1073/pnas.1311868111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Schaffer C.B., Friedman B., Lyden P.D., Kleinfeld D. Penetrating arterioles are a bottleneck in the perfusion of neocortex. Proc. Natl. Acad. Sci. USA. 2007;104:365–370. doi: 10.1073/pnas.0609551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat. Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V., Ripoll J., Wang L.V., Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat. Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- Olefir I., Mercep E., Burton N.C., Ovsepian S.V., Ntziachristos V. Hybrid multispectral optoacoustic and ultrasound tomography for morphological and physiological brain imaging. J. Biomed. Opt. 2016;21:86005. doi: 10.1117/1.JBO.21.8.086005. [DOI] [PubMed] [Google Scholar]
- Osmanski B.F., Pezet S., Ricobaraza A., Lenkei Z., Tanter M. Functional ultrasound imaging of intrinsic connectivity in the living rat brain with high spatiotemporal resolution. Nat. Commun. 2014;5:5023. doi: 10.1038/ncomms6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounov D.G., Wang T., Wang M., Feng D.D., Horton N.G., Cruz-Hernández J.C., Cheng Y.T., Reimer J., Tolias A.S., Nishimura N., Xu C. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat. Methods. 2017;14:388–390. doi: 10.1038/nmeth.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsepian S.V., Olefir I., Westmeyer G., Razansky D., Ntziachristos V. Pushing the boundaries of neuroimaging with optoacoustics. Neuron. 2017;96:966–988. doi: 10.1016/j.neuron.2017.10.022. [DOI] [PubMed] [Google Scholar]
- Perron A., Mutoh H., Launey T., Knöpfel T. Red-shifted voltage-sensitive fluorescent proteins. Chem. Biol. 2009;16:1268–1277. doi: 10.1016/j.chembiol.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka D.S., Takahashi H., Yuste R. Imaging voltage in neurons. Neuron. 2011;69:9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C.C. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Petersen R.S., Panzeri S., Diamond M.E. Population coding of stimulus location in rat somatosensory cortex. Neuron. 2001;32:503–514. doi: 10.1016/s0896-6273(01)00481-0. [DOI] [PubMed] [Google Scholar]
- Petersen C.C., Grinvald A., Sakmann B. Spatiotemporal dynamics of sensory responses in layer 2/3 of rat barrel cortex measured in vivo by voltage-sensitive dye imaging combined with whole-cell voltage recordings and neuron reconstructions. J. Neurosci. 2003;23:1298–1309. doi: 10.1523/JNEUROSCI.23-04-01298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razansky D., Buehler A., Ntziachristos V. Volumetric real-time multispectral optoacoustic tomography of biomarkers. Nat. Protoc. 2011;6:1121–1129. doi: 10.1038/nprot.2011.351. [DOI] [PubMed] [Google Scholar]
- Saper C.B., Petito C.K. Correspondence of melanin-pigmented neurons in human brain with A1-A14 catecholamine cell groups. Brain. 1982;105:87–101. doi: 10.1093/brain/105.1.87. [DOI] [PubMed] [Google Scholar]
- Schaffer C.B., Friedman B., Nishimura N., Schroeder L.F., Tsai P.S., Ebner F.F., Lyden P.D., Kleinfeld D. Two-photon imaging of cortical surface microvessels reveals a robust redistribution in blood flow after vascular occlusion. PLoS Biol. 2006;4:e22. doi: 10.1371/journal.pbio.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Kötter R., Zilles K., Luhmann H.J., Staiger J.F. Cell type-specific circuits of cortical layer IV spiny neurons. J. Neurosci. 2003;23:2961–2970. doi: 10.1523/JNEUROSCI.23-07-02961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel A., Marota J.J., Boas D. Design and evaluation of a continuous-wave diffuse optical tomography system. Opt. Express. 1999;4:287–298. doi: 10.1364/oe.4.000287. [DOI] [PubMed] [Google Scholar]
- Stein E.W., Maslov K., Wang L.V. Noninvasive, in vivo imaging of blood-oxygenation dynamics within the mouse brain using photoacoustic microscopy. J. Biomed. Opt. 2009;14:020502. doi: 10.1117/1.3095799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler D.D., Yamahachi H., Li W., Denk W., Gilbert C.D. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron. 2006;49:877–887. doi: 10.1016/j.neuron.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Tang J., Xi L., Zhou J., Huang H., Zhang T., Carney P.R., Jiang H. Noninvasive high-speed photoacoustic tomography of cerebral hemodynamics in awake-moving rats. J. Cereb. Blood Flow Metab. 2015;35:1224–1232. doi: 10.1038/jcbfm.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taruttis A., Ntziachristos V. Advances in real-time multispectral optoacoustic imaging and its applications. Nat. Photonics. 2015;9:219–227. [Google Scholar]
- Tiran E., Ferrier J., Deffieux T., Gennisson J.L., Pezet S., Lenkei Z., Tanter M. Transcranial functional ultrasound imaging in freely moving awake mice and anesthetized young rats without contrast agent. Ultrasound Med. Biol. 2017;43:1679–1689. doi: 10.1016/j.ultrasmedbio.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoumas S., Kravtsiv A., Gao Y., Buehler A., Ntziachristos V. Statistical molecular target detection framework for multispectral optoacoustic tomography. IEEE Trans. Med. Imaging. 2016;35:2534–2545. doi: 10.1109/TMI.2016.2583791. [DOI] [PubMed] [Google Scholar]
- Wang X., Pang Y., Ku G., Xie X., Stoica G., Wang L.V. Noninvasive laser-induced photoacoustic tomography for structural and functional in vivo imaging of the brain. Nat. Biotechnol. 2003;21:803–806. doi: 10.1038/nbt839. [DOI] [PubMed] [Google Scholar]
- Wang L., Maslov K., Wang L.V. Single-cell label-free photoacoustic flowoxigraphy in vivo. Proc. Natl. Acad. Sci. USA. 2013;110:5759–5764. doi: 10.1073/pnas.1215578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Beard P.C., Bohndiek S.E. Contrast agents for molecular photoacoustic imaging. Nat. Methods. 2016;13:639–650. doi: 10.1038/nmeth.3929. [DOI] [PubMed] [Google Scholar]
- Willem M., Tahirovic S., Busche M.A., Ovsepian S.V., Chafai M., Kootar S., Hornburg D., Evans L.D., Moore S., Daria A. η-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature. 2015;526:443–447. doi: 10.1038/nature14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt B.A., Burns L.D., Wei Ho E.T., Ghosh K.K., Mukamel E.A., Schnitzer M.J. Advances in light microscopy for neuroscience. Annu. Rev. Neurosci. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.X., Smith M.B., Wang J. Vol. 26. Springer; 2006. Magnetic Susceptibility Effects in High Field MRI. [Google Scholar]
- Yang X., Hyder F., Shulman R.G. Activation of single whisker barrel in rat brain localized by functional magnetic resonance imaging. Proc. Natl. Acad. Sci. USA. 1996;93:475–478. doi: 10.1073/pnas.93.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Hyder F., Shulman R.G. Functional MRI BOLD signal coincides with electrical activity in the rat whisker barrels. Magn. Reson. Med. 1997;38:874–877. doi: 10.1002/mrm.1910380604. [DOI] [PubMed] [Google Scholar]
- Yao J., Wang L.V. Photoacoustic brain imaging: from microscopic to macroscopic scales. Neurophotonics. 2014;1:011003. doi: 10.1117/1.NPh.1.1.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Wang L., Yang J.M., Maslov K.I., Wong T.T., Li L., Huang C.H., Zou J., Wang L.V. High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat. Methods. 2015;12:407–410. doi: 10.1038/nmeth.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Qian C., Chen D.Y., Dodd S.J., Koretsky A.P. Deciphering laminar-specific neural inputs with line-scanning fMRI. Nat. Methods. 2014;11:55–58. doi: 10.1038/nmeth.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborszky L., Csordas A., Mosca K., Kim J., Gielow M.R., Vadasz C., Nadasdy Z. Neurons in the basal forebrain project to the cortex in a complex topographic organization that reflects corticocortical connectivity patterns: an experimental study based on retrograde tracing and 3D reconstruction. Cereb. Cortex. 2015;25:118–137. doi: 10.1093/cercor/bht210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An “in-house” designed electromagnetic unit was used to generate sufficient magnetic force for whisker deflections at varying frequencies.