Abstract
GTP is an essential source of energy that supports a large array of cellular mechanochemical structures ranging from protein synthesis machinery to cytoskeletal apparatus for maintaining the cell cycle. However, GTP regulation during the cell cycle has been difficult to investigate because of heterogenous levels of GTP in asynchronous cell cycles and genetic redundancy of the GTP-generating enzymes. Here, in the unicellular red algae Cyanidioschyzon merolae, we demonstrated that the ATP–GTP-converting enzyme DYNAMO2 is an essential regulator of global GTP levels during the cell cycle. The cell cycle of C. merolae can be highly synchronized by light/dark stimulations to examine GTP levels at desired time points. Importantly, the genome of C. merolae encodes only two isoforms of the ATP–GTP-converting enzyme, namely DYNAMO1 and DYNAMO2. DYNAMO1 regulates organelle divisions, whereas DYNAMO2 is entirely localized in the cytoplasm. DYNAMO2 protein levels increase during the S-M phases, and changes in GTP levels are correlated with these DYNAMO2 protein levels. These results indicate that DYNAMO2 is a potential regulator of global GTP levels during the cell cycle.
Keywords: nucleoside diphosphate kinase, global GTP level, cell cycle, Cyanidioschyzon merolae
Introduction
The vital activities of eukaryotic cells are supported by various highly organized mechanochemical structures. For example, the ribosome complex synthesizes proteins by reading mRNA;1,2) the microtubule-based spindle separates chromosomes during mitosis;3) a part of the plasma membrane is utilized during endocytic activities to generate vesicles for cellular communications, such as hormone signaling and neurotransmission;4,5) and ring-shaped organelle division machineries cleave the mitochondria, peroxisomes, and chloroplasts at the middle to generate daughter organelles.6–8) All of these processes are mediated by GTPase proteins, including elongation factors,9) tubulin,10) and dynamin family members.11) These proteins undergo conformational changes and form highly ordered structures upon GTP hydrolysis. For example, the GTP-bound state of α- and β-tubulin form heterodimers that eventually organize into microtubules and are disassembled by the GTPase catalytic activity. Dynamin family members form a helical oligomer at the neck of a membrane constriction site, and their GTP hydrolysis activity generates elastic stress to pinch off the membrane. These findings indicate that sufficient GTP levels are needed to sustain the function of these GTPase-based machineries. However, it remains unclear how the GTP levels in eukaryotic cells are regulated, whether the global GTP level is temporally increased during specific stages of the cell cycle in which the machineries are working, and whether GTP is generated around the machinery when necessary.
In terms of dynamin-based machineries, recent studies have demonstrated the function of the nucleoside diphosphate kinase (NDPK) proteins in GTP synthesis.12–14) NDPK generates GTP by transferring one phosphate from ATP to GDP.15) The NDPK orthologs NM23-H1 and -H2 are involved in dynamin-mediated clathrin-dependent and -independent endocytosis in humans. Evidence from biochemical experiments strongly support that these NM23 proteins are the most likely ones to supply GTP to dynamin.13) Another NDPK homolog, namely NM23-H4, contains transit peptide that mediates the transport of NM23 proteins to the mitochondrial matrix. This protein provides a local GTP supply to the dynamin-like protein OPA113) at the inner mitochondrial membrane to maintain its membrane fusion function.16) Both in vivo and in vitro morphological and molecular genetic experiments have demonstrated that the NDPK-like protein DYNAMO1 is involved in the mitochondrial and peroxisomal division mediated by the dynamin-like protein Dnm1.14) DYNAMO1 contains a single NDPK domain, as identified by a proteomics study of a Dnm1-based organelle division machinery isolated from the unicellular red algae Cyanidioschyzon merolae.14) In that study, the overexpression of a mutant DYNAMO1 with a defective NDPK catalytic site showed a dominant negative mode and impaired constriction of mitochondrial and peroxisomal fission sites. Moreover, DYNAMO1 forms a ring-shaped complex with the GTPase Dnm1, resulting in a organelle division machinery that enhances the magnitude of the constriction force. Taken together, these findings indicate that GTP is generated by NDPK proteins around dynamin-mediated membrane fission rings. Interestingly, DYNAMO1 is localized at both the organelle division sites and in the cytosol, and its dysfunction does not affect the roles of the cytosolic GTP catalytic activity, such as that in protein translation,14) which indicates that this NDPK protein is not involved in the regulation of global GTP levels. Thus, which protein regulates the global GTP level during the cell cycle remains an important unanswered question because there are large quantities of cytosolic GTPase protein-based machineries, such as the ribosome complex and mitotic spindles, in the cytosol, both of which are regulated during cell cycle progression.17) To address this issue, investigating NDPK functions and GTP levels in the cytosol is essential. However, the genetic redundancy of NDPK makes it difficult to analyze the NDPK proteins in most eukaryotic cells.18) Moreover, cells divide randomly, which makes it difficult to determine the GTP levels in biochemical experiments. Notably, the genome of C. merolae contains only two isoforms of NDPK-like protein, namely DYNAMO1 and DYNAMO2.14,19,20) The cell cycle of this organism can be highly synchronized with the light/dark cycle, without the need of a pharmacological treatment. In this study, we demonstrated that DYNAMO2, a homolog of DYNAMO1, is entirely localized in the cytoplasm throughout the cell cycle progression and that its expression increases during the S-M phases. We analyzed the concentrations of nucleotides, including GTP, using liquid chromatography–electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) and showed that the GTP level increases from the S phase to the M phase in concert with the DYNAMO2 protein level. Because DYNAMO1 is specifically involved in organelle divisions in the M phase, DYNAMO2 is the more likely candidate to be involved in the regulation of the global GTP level in the cytosol.
Materials and methods
Phylogenetic analyses.
A maximum-likelihood tree was constructed with the PHYLogeny Inference Package (PHYLIP) version 3.69521) using an alignment of the amino acid sequences of the following 56 NDPK domain-containing proteins: C. m., C. merolae (DYNAMO1_CML110c, DYNAMO2_CMK060c); T. p., Thalassiosira pseudonana (TpNDPK1_XP_002295246.1, TpNDPK2_XP0022911211, TpNDPK3_XP0022867331); O. t., Ostreococcus tauri (OtNDPK1_XP_022841083.1, OtNDPK2_XP_022840003.1); D. d., Dictyostelium discoideum (DdNDPK-A_XP_644519.1, DdNDPK-B_XP_641417.1); S. p., Schizosaccharomyces pombe (SpNDPK_P49740.1); S. c., Saccharomyces cerevisiae (SsNDPK_P36010.); P. p., Physcomitrella patens (PpNDPK1_XP_024368299.1, PpNDPK3_XP_024398552.1, PpLOC112289340_XP_024390257.1, PpLOC112277920_XP_024366539.1); C. r., Chlamydomonas reinhardtii (CrNDPK1_XP_001698246.1, CrNDPK2_XP_001702884.1); A. t., Arabidopsis thaliana (AtNDPK1_NP_567346.2, AtNDPK3_NP_192839.1, AtNDPK4_NP_567690.1, AtNDPK2_NP_568970.2, AtNDPK5_NP_173184.2); O. s., Oryza sativa spp. japonica (OsNPDK1-A_XP_015614147.1, OsNDPK1-B_XP_015647142.1, OsNDPK3_XP_015639333.1, OsNDPK4_XP_015618263.1, OsNDPK5_XP_015623738.1); C. e., Caenorhabditis elegans (CeNDPK-A_NP_492761.1, CeY48G8AL.15_NP_001021779.1); D. m., Drosophila melanogaster (DmAwdC_NP_476761.3, DmAwdE_NP_001287624., DmNmdyn-D6_NP_572965.1); D. r., Danio rerio (DrNDPK-b_NP_571001.2, DrNDPK-A_XP_021326629.1, DrNDPK3_NP_001349197.1, DrNDPK-B_NP_571002.1, DrNDPK4_NP_957489.1, DrNDPK5_NP_001002516.1, DrNDPK6_NP_571672.2); X. l., Xenopus laevis (XlNDPK-A_P70010.1, XlNDPK3_NP_001087358.1, XlNDPK4_NP_001084697.1, XlNDPK5L_NP_001087794.1, XlNDPK6S_001089757.1); M. m., Mus musculus (MmNM23-M1_P15532.1, MmNM23-M2_Q01768.1, MmNM23-M3_Q9WV85.3, MmNM23-M4_Q9WV84.1, MmNM23-M5_Q99MH5.2, MmNM23-M6_O88425.1); and H. s., Homo sapiens (HsNM23-H1_P15531.1, HsNM23-H2_P22392.1, HsNM23-H3_Q13232.2, HsNM23-H4_O00746.1, HsNM23-H5_P56597.1, HsNM23-H6_O75414.3). The sequences were collected by BLAST searches of the National Center for Biotechnology Information databases of the respective species using DYNAMO1 of the red alga C. merolae as the query. Sequences of the NDPK domains were automatically aligned using CLUSTAL X, version 2.0.9.22) For phylogenetic analyses, ambiguously aligned regions were manually arranged or deleted using BioEdit Sequence Alignment Editor, version 4.8.10 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), resulting in 130 amino acids (including inserted gaps) that were subsequently used. The local bootstrap probabilities were calculated using the CONSENSE program from the PHYLIP package.
Antibodies used for immunoblotting analysis and immunofluorescence microscopy.
To generate anti-DYNAMO2 antisera in rabbit, the open reading frame of the CMK060C protein from C. merolae was amplified by PCR using the following primers: 5′-ACCATCAC atgttcgttccttctttaggtttctc-3′ and 5′-AGCTAATT ttcataaacccaacgagcaacc-3′ (InFusion sticking regions are capitalized). The amplified DNA fragment was InFusion-cloned into the amplified PQE vector using the following primers: 5′-TTATGAA aattagctgagcttggactcctg-3′ and 5′-CGAACAT gtgatggtgatggtgatgcg-3′ (InFusion sticking regions are capitalized). XL1-Blue strain cells were transformed with this plasmid, cultured at 37 ℃ for 12 h in 100-ml Luria–Bertani (LB) medium, scaled up to 1-l LB medium, and incubated further at 37 ℃ for 2 h and then at 18 ℃ for 1 h. Isopropyl β-D-1 thiogalactopyranoside was added at a final concentration of 0.1 mM, and after a further 12 h of incubation at 18 ℃, cells were harvested by centrifugation at 1,000 ×g for 10 min. Cell pellets were resuspended in 200-ml HEPES buffer (HB250) containing 250 mM NaCl, 20 mM HEPES-KOH, pH 7.5, 2 mM EGTA, 1 mM MgCl2, 1 mM dithiothreitol, and a complete protease inhibitor cocktail (Roche, Basel, Switzerland). After homogenizing cells by sonication for 10 min, recombinant DYNAMO2 was purified using a His-Trap column (GE Healthcare, Chicago, IL, USA) and subcutaneously injected into a rabbit for immunization (T.K. Craft Corp., Gunma, Japan). The other antibodies used in this study were a rabbit anti-α-tubulin antibody23) and a rabbit anti-Dnm1 antibody.24)
Phase contrast and immunofluorescence microscopy.
C. merolae cells were fixed and blocked as described previously.23) Phase-contrast and immunofluorescence images were captured using a fluorescence microscope (BX51; Olympus, Tokyo, Japan). Immunofluorescence profiles were acquired using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
LC-ESI-MS/MS analysis of nucleotides during the cell cycle.
C. merolae 10D cell cultures were sub-cultured at 1 × 107 cells/ml as described previously.25) Cells were harvested every 2 h from 0 h to 24 h after the onset of 12-h light/12-h dark cycle. For the 0-h sample, cells were collected immediately after the light was turned on. For the 12-h sample, cells were collected immediately after the light was turned off. For the 24-h sample, cells were harvested immediately after the light was turned on next. The number of cells in each sample was adjusted to 5 × 108 cells. Cells in the samples were centrifuged, and cell pellets were quickly frozen in liquid nitrogen. Frozen samples were dissolved in 500-µl chilled methanol at −30 ℃ and 1.25× volumes of chloroform. Subsequently, 2 mM raffinose, as an internal standard used for normalization between samples, was added, and all samples were lyophilized and dissolved in methanol:water (1:1 v/v). After centrifugation at 10,000 ×g for 5 min, the supernatant was evaporated at room temperature under nitrogen for 6 h, dissolved in water containing 20-µg/ml ethylenediaminetetraacetic acid, and then subjected to LC-ESI-MS/MS analysis. LC-ESI-MS/MS was performed using a 4000 QTRAP quadrupole linear ion-trap hybrid mass spectrometer (AB Sciex) with an ACQUITY UPLC System (Waters, Milford, MA, USA). Samples were injected into an ACQUITY UPLC BEH C18 column (1 × 150 mm; Waters, Milford, MA, USA) and then directly subjected to ESI-MS/MS analysis. A 10-µl aliquot was separated by step-gradient elution with mobile phases A (water, 0.1% formic acid, and 0.028% ammonia) and B (20% acetonitrile, 0.1% formic acid, and 0.028% ammonia) at the following ratios: 95:5 (for 0–5 min), 0:100 (5–25 min), 0:100 (25–30 min), and 95:5 (30–40 min). The flow rate was 50 µl/min at 30 ℃. The source temperature was 400 ℃, the declustering potential was −85, and the collision cell exit potential was −11. The ion transitions at m/z 506 > 79, 426 > 79, 522 > 79, 442 > 79, and 503 > 179 for ATP, ADP, GTP, GDP, and raffinose, respectively, were used in the multiple reaction monitoring mode. Data were analyzed and quantified using Analyst software (AB Sciex, Tokyo, Japan). Nucleotides were purchased from Sigma-Aldrich. The degradation rate of nucleotides during sample preparation was measured using commercial nucleotides (Table 1). Thus, the true value of nucleotide concentrations N(t) was corrected from the measured value Nm(t) using the following formula:
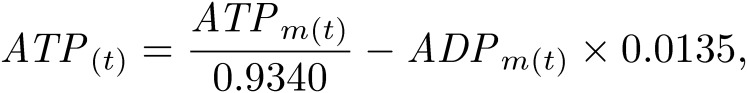 |
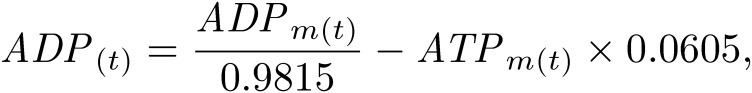 |
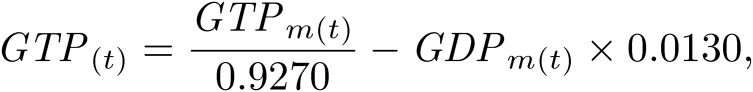 |
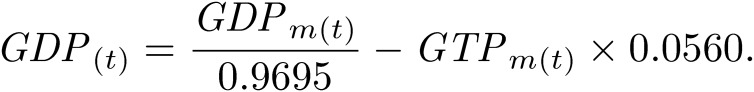 |
The changes in nucleotide concentrations ΔN/N0 were calculated using the following formula:
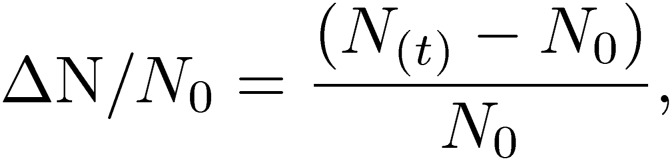 |
where N0 is the nucleotide concentration at 0 h.
Table 1.
Degradation of nucleotides during sample preparation assessed by LC-ESI-MS/MS. Each parameter is represented as percentages of total adenosine nucleotides (left) and guanosine nucleotides (right) remaining after the analysis. n = 3, mean ± SD.
| ATP addition | ADP addition | GTP addition | GDP addition | ||
|---|---|---|---|---|---|
| ATP | 93.400 ± 0.849 | 1.350 ± 0.495 | GTP | 92.700 ± 3.394 | 1.300 ± 0.424 |
| ADP | 6.050 ± 1.845 | 98.150 ± 0.636 | GDP | 5.600 ± 3.536 | 96.950 ± 1.061 |
| AMP | 0.550 ± 0.636 | 0.350 ± 0.354 | GMP | 1.650 ± 0.212 | 1.750 ± 0.636 |
Quantification and statistical analysis.
Experiments were performed using at least two independent cultures unless specified otherwise. The immunofluorescence microscopy profiles of DYNAMO2 protein levels during the cell cycle were chosen randomly to sample unbiased populations. The cytosol was defined as the region excluding the chloroplast. The Mann–Whitney test was used to determine the Gaussian distribution of the data, and Pearson’s skewness test was used to determine the skewness of the data set using GraphPad Prism 7.4. Confidence levels are shown in each graph.
Result and discussion
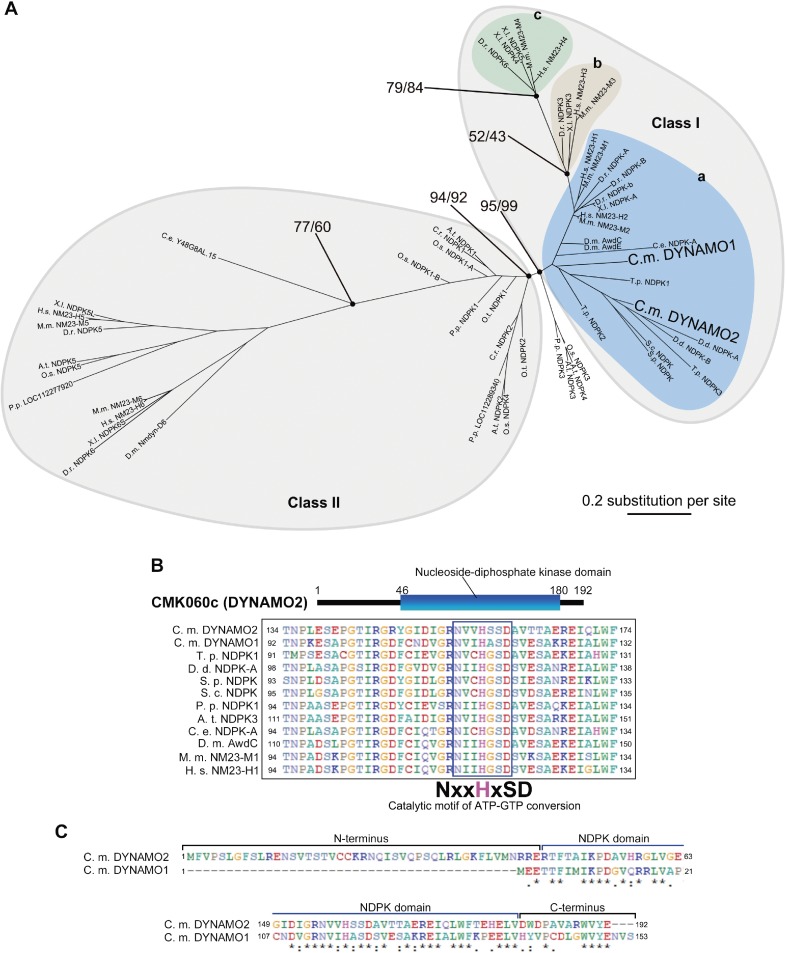
The genome of C. merolae encodes two isoforms of NDPK-like proteins, namely DYNAMO1 (CML110C) and DYNAMO2 (CMK060C).19,20) The phylogenetic analysis of NDPK-containing proteins from four kingdoms, namely Plantae, Opisthokonta, Amoebozoa, and Chromista, revealed two large classes of NDPKs: Classes I and II (Fig. 1A). Class I NDPKs are well conserved among red algae, diatoms, amoebae, fungi, and animals and are further classified into three subgroups, Groups I-a, I-b, and I-c. Group I-a forms the base of Class I tree and includes DYNAMO1, an NDPK involved in mitochondrial and peroxisomal division,14) and Awd and NM23-H1, which are involved in endocytosis in flies and humans, respectively.12,13) This suggests that the basic function of Class I NDPKs originated from the dynamin family member-dependent membrane fission event. Groups I-b and I-c branched from Group I-a, and NM23-H4 in Group I-c plays a role in OPA1-mediated mitochondrial inner membrane fusion.13,16) The membrane fusion dynamin OPA1 likely initially acquired from holozoa and fungi,26) suggesting that branched phylogeny within Group I likely reflects acquired events of dynamin-related family members. These results demonstrate that the uncharacterized NDPK-like protein DYNAMO2 belongs to an evolutionarily basic compartment of the class I NDPK family. Similar to other NDPK family member proteins, DYNAMO2 contains an NDPK domain and a catalytic motif for ATP–GTP conversion including the histidine phosphorylation site (Fig. 1B). Thus, DYNAMO2 is the functional NDPK protein most likely to be conserved among eukaryotes except in Plantae. Both DYNAMO2 and DYNAMO1 have NDPK domains, but DYNAMO2 has an N-terminal extension and the first half of its C-terminal tail is different from that of DYNAMO1 (Fig. 1C). Therefore, we expected that these two isoforms have different subcellular functions.
Figure 1.
Classification of DYNAMO2. (A) The maximum-likelihood phylogenetic tree of NDPK proteins. The numbers at the nodes are local bootstrap values. Branch lengths are proportional to the number of amino acid substitutions, which are indicated by the scale bar below the tree. (B) The predicted domain architecture of CMK060C/DYNAMO2 is aligned and compared with those of its homologous proteins. C. m., Cyanidioschyzon merolae; T. p., Thalassiosira pseudonana; D. d., Dictyostelium discoideum; S. p., Schizosaccharomyces pombe; S. c., Saccharomyces cerevisiae; P. p., Physcomitrella patens; A. t., Arabidopsis thaliana; C. e., Caenorhabditis elegans; D. m., Drosophila melanogaster; M. m., Mus musculus; H. s., Homo sapiens. (C) Amino acid sequence alignment between DYNAMO2 and DYNAMO1. Upper and lower panels show N- and C-terminal regions, respectively.
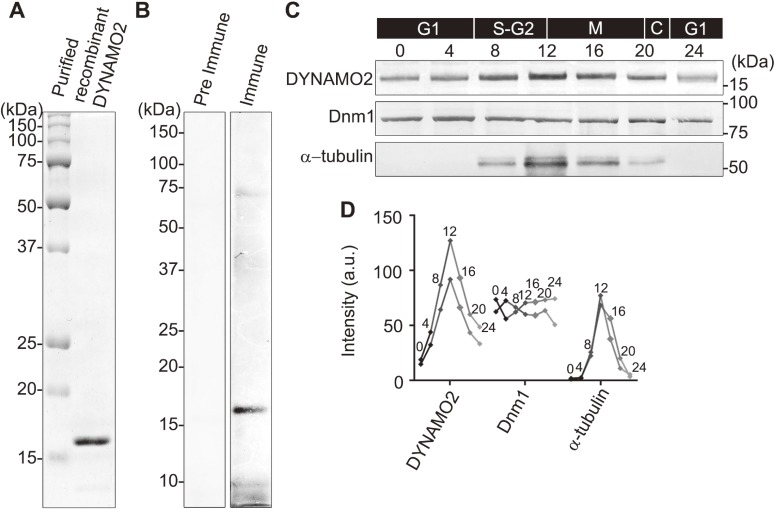
To determine the function of DYNAMO2, a rabbit anti-DYNAMO2 antibody was prepared (Fig. 2A and B). The protein expression profile of DYNAMO2 was characterized using a synchronized C. merolae culture (Fig. 2C and D). DYNAMO2 was found to be expressed throughout the cell cycle with an increase at 8–20 h, i.e., after the onset of light/dark stimulation, corresponding to the S-M phase.25,27) The protein level peaked at 12 h at around the G2-M phase. Further, a comparison of the expression level of two major GTPases, α-tubulin and the dynamin-like GTPase Dnm1, with that of DYNAMO2 revealed that Dnm1 was expressed throughout the cell cycle as its levels were found to be equivalent between the cytosolic pool and division sites of mitochondria and peroxisomes, consistent with previously reported results.23,28) The expression of α-tubulin was limited to the S-M phase and peaked at 12 h after the onset of the light/dark cycle, similar to as the expression of DYNAMO2. As C. merolae lacks a typical cytoskeleton, the tubulin function in this organism is mostly limited to the formation of mitotic spindles.24,29) Collectively, these results suggest that the dynamic changes in DYNAMO2 expression are similar to those in α-tubulin expression rather than Dnm1 expression, which mediates mitochondrial and peroxisomal fission supported by DYNAMO1 activity.
Figure 2.
Protein expression profile of DYNAMO2. (A) An SDS-PAGE gel showing the recombinant DYNAMO2 used for antibody generation. (B) Anti-DYNAMO2 antibody was characterized by immunoblotting of the whole cell lysates of C. merolae compared with the pre-immunization serum. (C) Protein expression throughout the cell cycle was assessed by immunoblotting for DYNAMO2, Dnm1, and α-tubulin using the whole cells lysates of C. merolae. Samples were collected at 0, 4, 8, 12, 16, 20, and 24 h after the onset of the light/dark cycle. (D) Protein levels of DYNAMO2, Dnm1, and α-tubulin were quantified from the immunoblots using ImageJ. Inset numbers indicate the time after the onset of the light/dark cycle. Line graphs of two independent experiments are shown.
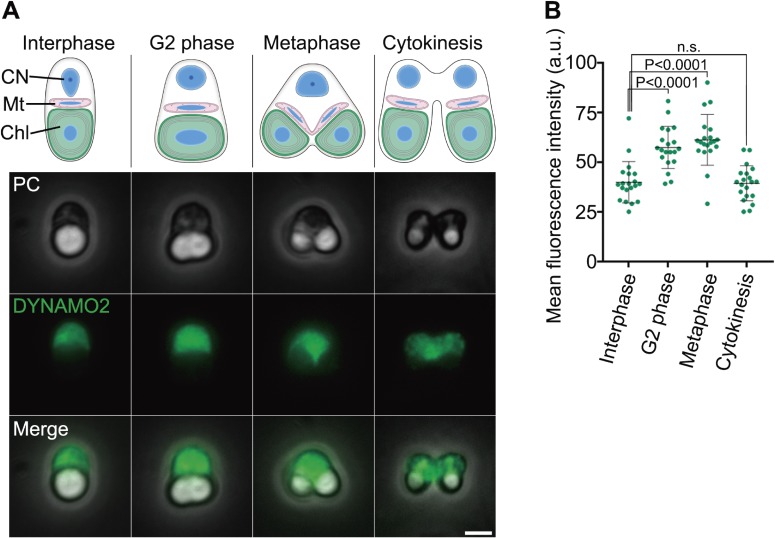
Next, we determined the subcellular localization of DYNAMO2 by phase-contrast immunofluorescence microscopy (Fig. 3). After methanol fixation of C. merolae, its cytosol and chloroplast were observed as a dark region surrounded by the edge of the cell and a bright white structure, respectively (Fig. 3A). In C. merolae, the mitotic cycle progression is correlated with chloroplast division30,31) (Fig. 3A, upper panel). The chloroplast has a spherical structure during the interphase and elongates to divide during the G2-M phase transition. During the metaphase, the chloroplast divides by binary division and segregates into daughter cells during cytokinesis. Thus, the cell cycle stages are identified based on the chloroplast morphology. Throughout the cell cycle, DYNAMO2 signals were detected in the entire cytoplasm (Fig. 3A, lower panel). Moreover, the signal intensities in the cytosol were higher in the cells during G2 phase and mitosis than in the cells during the interphase and cytokinesis (Fig. 3B). Immunostaining did not reveal dots of intense DYNAMO2 signal around the division sites of mitochondria and peroxisomes, in contrast to the results recently reported for DYNAMO1.14) These findings suggest that DYNAMO2 is not involved in mitochondrial and peroxisomal division, although it belongs to class I NDPKs that have functional relationship with dynamin. This result is supported by a previous genetic study on mammalian cells that reported non-membranous functions of the NDPK family members, such as NM23-H1, involving interactions with microtubules and regulation of intracellular signaling mediated by small GTPases.32,33) We assumed that class I-a NDPKs possess at least two functions of NDPKs: one in dynamin-related membrane fission machinery and another in the regulation of GTP levels in the cytoplasm.
Figure 3.
Localization of DYNAMO2. (A) A schema of the C. merolae cell structures at each cell cycle phase is shown (upper panel), and DYNAMO2 localization in typical cells at each cell cycle phase (middle and lower panels) was assessed by immunofluorescence microscopy (lower panel). CN; cell nucleus, Mt; mitochondrion, Chl; chloroplast, PC; phase-contrast, DYNAMO2; anti-DYNAMO2 antibody. Scale bar, 2 µm. (B) Immunofluorescence signal of DYNAMO2 was quantified using ImageJ. n.s., not significant; P, p-value (Mann–Whitney test).
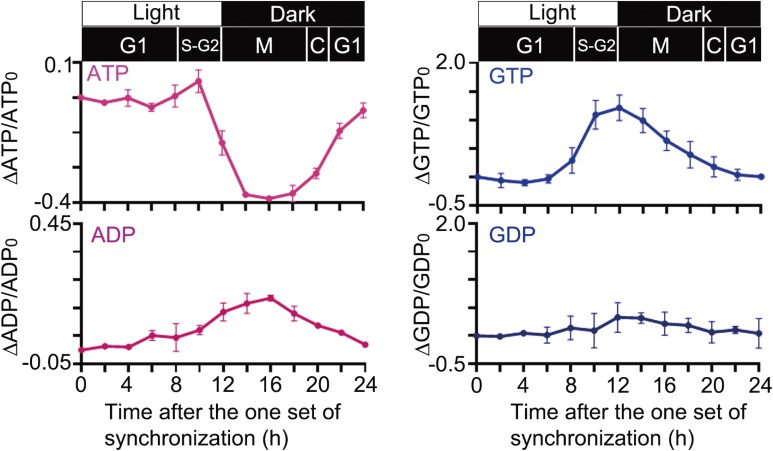
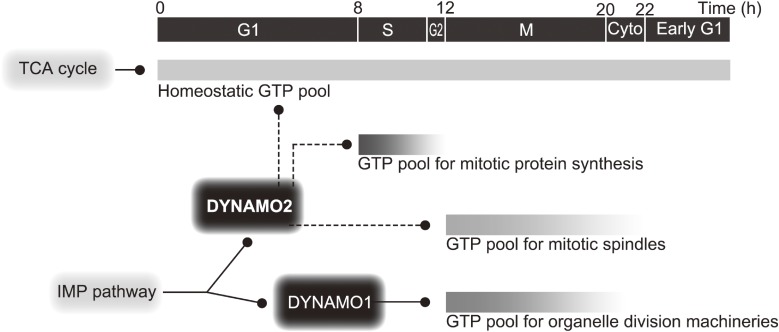
C. merolae contains only two isoforms of NDPKs, DYNAMO1 and DYNAMO2, of which DYNAMO1 is specifically involved in mitochondrial and peroxisomal division.14) Moreover, DYNAMO2 expression was found to be increased during the M phase, localized entirely in the cytoplasm. This finding suggests that DYNAMO2 is a global GTP regulator during the cell cycle. To examine whether the global GTP level changes during the cell cycle in concert with the DYNAMO2 protein level, we measured the global levels of nucleotides, including GTP, throughout the cell cycle by LC-ESI-MS/MS (Fig. 4). The ATP level was slightly increased at 8–10 h after the onset of synchronization corresponding to the S phase and rapidly decreased from 12 h onward. The ATP level was lower in the mitotic phase than in the interphase, suggesting that ATP production via oxidative phosphorylation or glycolytic reaction was low or that ATP production could not compensate for the ATP consumption during mitosis, as proposed previously.34) Both mitochondria and plastids are involved in ATP production, which is driven by light stimulation.35) In fact, our data showed a rapid decrease in the ATP level from 12 h onward, i.e., during transition to the dark period; thus, light-driven ATP synthesis may be attributable to the low ATP level observed during the M phase. On the other hand, the GTP level was elevated during the mitotic S phase. An increase in the GTP level started at 8 h, peaked at 10–12 h corresponding to the S-G2 phases, and decreased almost to the basal level at 22 h. These profiles were well correlated with the DYNAMO2 expression profile, suggesting that this NDPK-like protein is intimately involved in the changes in the GTP level during the cell cycle. However, with regard to the GTP supply, the possibility of the involvement of other mechanisms of GTP synthesis, such as the succinyl-CoA synthase pathway in the TCA cycle, cannot be excluded.36) Mitochondrial respiration is downregulated during mitochondrial fission in mammals;37) it has higher activity during mitochondrial fusion state rather than fission in mammalian culture cells and plant cells.38,39) However, because mitochondrial division only occurs during the M phase in C. merolae23,40) and the ATP level decreases during the M phase (Fig. 4), the contribution of the succinyl-CoA synthase pathway is less likely.
Figure 4.
Changes in the relative amount of each nucleotide as measured by LC-ESI-MS/MS. Cells were harvested every 2 h after the onset of the light/dark cycle. n = 2, mean ± SD.
Interestingly, the GDP level was constant during the cell cycle, although there was burst of GTP concentration during the S-M phases (Fig. 4). This result suggests that a balance between GDP synthesis and consumption is maintained during the cell cycle. GDP synthesis is mediated by guanylate kinase using GMP as a substrate.41) The initial metabolite of GMP is inosine monophosphate or guanosine,42,43) suggesting that the metabolic pathways involving these two metabolites are involved in the regulation of GTP and GDP levels during the S-M phases or that GDP is generated from a preexisting GMP pool during the S-M phases. Our results indicate that the NDPK-like protein DYNAMO2 is a potential regulator of the global GTP level during the S-M phase, but more detailed investigations in C. merolae using molecular genetic approaches are warranted.
Here, we demonstrated that one of the two isoforms of NDPK-like protein, i.e., DYNAMO2, is localized in the cytosol. Its expression is elevated during the S-M phase and well correlated with the dynamics of the global GTP level that is temporally elevated during the S-M phase. Based on our results, we hypothesize two functions of DYNAMO2 during the cell cycle (Fig. 5). First, DYNAMO2 regulates the global GTP level throughout the cell cycle progression. Particularly, DYNAMO2 expression and the global GTP levels peak at the S phase, strongly suggesting the importance of GTP during this period. In general, protein translation peaks during the G1-S and G2-M transition states because cellular energy is mostly invested in ensuring accurate cell division.44–46) We assume that the rapid increases in the DYNAMO2 and GTP levels around the S-G2 phases contribute to the synthesize of proteins required for the cell division process. In fact, most of the division-related proteins in C. merolae, such as α-tubulin, centromere histone H3, Mda1, and plastid-dividing ring 1, are newly synthesized around the S phase.29,40,47,48) Second, DYNAMO2 regulates the M phase-specific subcellular structures, such as the microtubules. Increase in DYNAMO2 levels is consistent with the increase in α-tubulin expression. Because the function of microtubules is involved only in spindle apparatus formation in C. merolae,29) their GTP requirement is limited to that during the M phase. Interestingly, α-tubulin molecules are dispersed in the cytoplasm during the S phase and are organized into microtubules during the G2/M phase transition.31) Because the global GTP level peaked at the S-G2 phase, one of the functions of DYNAMO2 is likely to mediate this microtubule transition or to maintain the spindle apparatus during mitosis.
Figure 5.
A schema of DYNAMO2 function. DYNAMO2 is important for cytosolic GTP generation during the cell cycle rather than for membrane remodeling, which is mediated by its homolog DYNAMO1. Time indicates time after the onset of the light/dark synchronization.
GTP is an essential energy source for GTPases, including tubulin, dynamin, and elongation factors. Many mitotic processes, such as structural rearrangements for chromosome segregation, organelle division, and RNA translation, are highly GTP-demanding during specific cell cycle phases. The molecular function of DYNAMO2 remains obscure, but our finding about the dynamics of DYNAMO2 and the global GTP level during the cell cycle elucidates the basic mechanism underlying the energy demand of the GTPase-based molecular machineries.
Acknowledgements
This work was supported in part by grants from the Japan Society for the Promotion of Science Fellowships (no. 14J04556 to Y.I.); MEXT-Supported Program for the Strategic Research Foundation at Private Universities (no. JWU2014-1018 to T.K.); Core Research for Evolutional Science and Technology Program of Japan Science and Technology Agency (to T.K.); Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid for Scientific Research (no. JP16H04813 to T.K; nos. JP24247038, JP25112518, JP25116717, JP26116007, JP15K14511, and JP15K21743 to Y.F.); and the Takeda Science Foundation (to Y.F.), Naito Foundation (to Y.F.), and Japan Foundation for Applied Enzymology and Novartis Foundation (Japan) for the Promotion of Science (to Y.F.). We thank O. Misumi (Yamaguchi University) and F. Yagisawa (Ryukyu University) for providing C. merolae 10D and K. Itoh (Johns Hopkins University) for study discussion and manuscript preparation.
References
- 1).Palade G.E. (1955) A small particulate component of the cytoplasm. J. Biophys. Biochem. Cytol. 1, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Roberts, R.B. (1958) Microsomal Particles and Protein Synthesis. Pergamon, London. [Google Scholar]
- 3).Van Beneden E. (1876) Recherches sur les Dicyemides, survivants actuels d’un embranchement des Mésozoaires. Bull. Acad. R. Belg. 41, 1160–1205. [Google Scholar]
- 4).Anderson R.G., Brown M.S., Goldstein J.L. (1977) Role of the coated endocytic vesicle in the uptake of receptor-bound low density lipoprotein in human fibroblasts. Cell 10, 351–364. [DOI] [PubMed] [Google Scholar]
- 5).Miller T.M., Heuser J.E. (1984) Endocytosis of synaptic vesicle membrane at the frog neuromuscular junction. J. Cell Biol. 98, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Kuroiwa T., Kawano S., Hizume M. (1977) Studies on mitochondrial structure and function in Physarum polycephalum. V. Behaviour of mitochondrial nucleoids throughout mitochondrial division cycle. J. Cell Biol. 72, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Mita T., Kanbe T., Tanaka K., Kuroiwa T. (1986) A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130, 211–213. [Google Scholar]
- 8).Imoto Y., Kuroiwa H., Ohnuma M., Kawano S., Kuroiwa T. (2012) Identification of peroxisome-dividing ring in Cyanidioschyzon merolae based on organelle partner hypothesis. Cytologia 77, 515–522. [Google Scholar]
- 9).Moller W., Schipper A., Amons R. (1987) A conserved amino acid sequence around Arg-68 of Artemia elongation factor 1 alpha is involved in the binding of guanine nucleotides and aminoacyl transfer RNAs. Biochimie 69, 983–989. [DOI] [PubMed] [Google Scholar]
- 10).Mitchison T., Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- 11).Hinshaw J.E., Schmid S.L. (1995) Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. [DOI] [PubMed] [Google Scholar]
- 12).Krishnan K., Richa R., Sujata R., Madhuri S., Michael M., Radhakrishnan N., et al. (2001) Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron 30, 197–210. [DOI] [PubMed] [Google Scholar]
- 13).Boissan M., Guillaume M., Qinfang S., Lorena G., Jérôme G., Maryse R., et al. (2014) Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 344, 1510–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Imoto Y., Abe Y., Honsho M., Okumoto K., Ohnuma M., Kuroiwa H., et al. (2018) Onsite GTP fuelling via DYNAMO1 drives division of mitochondria and peroxisomes. Nat. Commun. 9, 4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Norman A.W., Wedding R.T., Black M.K. (1965) Detection of phosphohistidine in nucleoside diphosphokinase isolated from Jerusalem artichoke mitochondria. Biochem. Biophys. Res. Commun. 20, 703–709. [DOI] [PubMed] [Google Scholar]
- 16).Schlattner U., Tokarska-Schlattner M., Ramirez S., Yulia Y.T., Andrew A.A., Dariush M., et al. (2013) Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: A cardiolipin-dependent switch. J. Biol. Chem. 288, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Byers B., Goetsch L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Lacombe M.L.L., Munier A., Mehus J.G., Lambeth D.O. (2000) The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32, 247–258. [DOI] [PubMed] [Google Scholar]
- 19).Matsuzaki M., Misumi O., Shin-i T., Maruyama S., Takahara M., Miyagishima S., et al. (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. [DOI] [PubMed] [Google Scholar]
- 20).Nozaki H., Takano H., Misumi O., Terasawa K., Matsuzaki M., Maruyama S., et al. (2007) A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Felsenstein J. (1981) Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 22).Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- 23).Nishida K., Takahara M., Miyagishima S., Kuroiwa H., Matsuzaki M., Kuroiwa T. (2003) Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl. Acad. Sci. U.S.A. 100, 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Fujiwara T., Yoshida Y., Kuroiwa T. (2009) Synchronization of cell nuclear, mitochondrial and chloroplast divisions in the unicellular red alga Cyanidioschyzon merolae. Cytologia (Tokyo) 74, 1. [Google Scholar]
- 25).Suzuki K., Ehara T., Osafune T., Kuroiwa H., Kawano S., Kuroiwa T. (1994) Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 63, 280–288. [PubMed] [Google Scholar]
- 26).Muñoz-Gómez S.A., Slamovits C.H., Dacks J.B., Baier K.A., Spencer K.D., Wideman J.G. (2015) Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr. Biol. 25, 1489–1495. [DOI] [PubMed] [Google Scholar]
- 27).Fujiwara T., Misumi O., Tashiro K., Yoshida Y., Nishida K., Yagisawa F., et al. (2009) Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 16, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Imoto Y., Abe Y., Okumoto K., Honsho M., Kuroiwa H., Kuroiwa T., et al. (2017) Defining the dynamin-based ring organizing center on the peroxisome-dividing machinery isolated from Cyanidioschyzon merolae. J. Cell Sci. 130, 853–867. [DOI] [PubMed] [Google Scholar]
- 29).Nishida K., Yagisawa F., Kuroiwa H., Nagata T., Kuroiwa T. (2005) Cell cycle-regulated, microtubule-independent organelle division in Cyanidioschyzon merolae. Mol. Biol. Cell 16, 2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Miyagishima S., Itoh R., Toda K., Kuroiwa H., Kuroiwa T. (1999) Real-time analyses of chloroplast and mitochondrial division and differences in the behavior of their dividing rings during contraction. Planta 207, 343–353. [Google Scholar]
- 31).Imoto Y., Fujiwara T., Yoshida Y., Kuroiwa H., Maruyama S., Kuroiwa T. (2010) Division of cell nuclei, mitochondria, plastids, and microbodies mediated by mitotic spindle poles in the primitive red alga Cyanidioschyzon merolae. Protoplasma 241, 63–74. [DOI] [PubMed] [Google Scholar]
- 32).Gallagher B.C., Parrott K.A., Szabo G., de Otero S. (2003) A receptor activation regulates cortical, but not vesicular localization of NDP kinase. J. Cell Sci. 116, 3239–3250. [DOI] [PubMed] [Google Scholar]
- 33).Hartsough M.T., Morrison D., Salerno M., Palmieri D., Ouatas T., Patrick J., et al. (2002) Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J. Biol. Chem. 277, 32389–32399. [DOI] [PubMed] [Google Scholar]
- 34).Maeshima K., Matsuda T., Shindo Y., Imamura H., Tamura S., Imai R., et al. (2018) A transient rise in free Mg2+ ions released from ATP-Mg hydrolysis contributes to mitotic chromosome condensation. Curr. Biol. 28, 444–451. [DOI] [PubMed] [Google Scholar]
- 35).Mills, J.D. (1996) The regulation of the chloroplast ATP synthase, CFo–CF1. In Oxygenic Photosynthesis: The Light Reactions (eds. Ort, D.R. and Yocum, C.F.). Kluwer Academic Publishers, Dordrecht, pp. 469–485. [Google Scholar]
- 36).Berg, J.M., Tymoczko, J.L. and Stryer, L. (2002) Biochemistry, 5th ed. W. H. Freeman, New York. [Google Scholar]
- 37).Schmitt K., Grimm A., Dallmann R., Oettinghaus B., Restelli L.M., Witzig M., et al. (2018) Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 27, 657–666. [DOI] [PubMed] [Google Scholar]
- 38).Mishra P., Carelli V., Manfredi G., Chan D.C. (2014) Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19, 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Jaipargas E.A., Barton K.A., Mathur N., Mathur J. (2015) Mitochondrial pleomorphy in plant cells is driven by contiguous ER dynamics. Front. Plant Sci. 6, 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Nishida K., Yagisawa F., Kuroiwa H., Yoshida Y., Kuroiwa T. (2007) WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. U.S.A. 104, 4736–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Hall S.W., Kühn H. (1986) Purification and properties of guanylate kinase from bovine retinas and rod outer segments. Eur. J. Biochem. 161, 551–556. [DOI] [PubMed] [Google Scholar]
- 42).Lagerkvist U. (1958) Biosynthesis of Guanosine 5′-Phosphate II. Amination of xanthosine 5′-phosphate by purified enzyme from pigeon liver. J. Biol. Chem. 233, 143–149. [PubMed] [Google Scholar]
- 43).Woods R.A., Roberts D.G., Friedman T., Jolly D., Filpula D. (1983) Hypoxanthine: Guanine phosphoribosyltransferase mutants in Saccharomyces cerevisiae. Mol. Gen. Genet. 191, 407–412. [DOI] [PubMed] [Google Scholar]
- 44).Pyronnet S., Pradayrol L., Sonenberg N. (2000) A cell cycle-dependent internal ribosome entry site. Mol. Cell 5, 607–616. [DOI] [PubMed] [Google Scholar]
- 45).Pyronnet S., Sonenberg N. (2001) Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11, 13–18. [DOI] [PubMed] [Google Scholar]
- 46).Kronja I., Orr-Weaver T.L. (2011) Translational regulation of the cell cycle: When, where, how and why? Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 3638–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Maruyama S., Kuroiwa H., Miyagishima S.Y., Tanaka K., Kuroiwa T. (2007) Centromere dynamics in the primitive red alga Cyanidioschyzon merolae. Plant J. 49, 1122–1129. [DOI] [PubMed] [Google Scholar]
- 48).Yoshida Y., Kuroiwa H., Misumi O., Yoshida M., Ohnuma M., Fujiwara T., et al. (2010) Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 329, 949–953. [DOI] [PubMed] [Google Scholar]