Abstract
Paget's disease of bone (PDB) has a strong genetic component. Variants in SQSTM1 are found in up to 40% of patients with a family history of the disease, where a pattern of autosomal dominance with incomplete penetrance is apparent. By contrast, SQSTM1 variants are only found in up to 10% of patients with sporadic disease. It has been hypothesised that the remaining genetic susceptibility to PDB, particularly in familial cases, could be explained by rare genetic variants in loci previously identified by Genome Wide Association Studies. It is likely that polygenic factors are involved in many individuals. In this study we utilised whole exome sequencing to investigate predisposing genetic factors in an unsolved PDB kindred and identified a c.1189C > T p.L397F variant in DC-STAMP, also known as TM7SF4, that co-segregated with disease. DCSTAMP was identified as a gene of interest in PDB following Genome Wide Association Studies and has been previously shown to play critical roles in osteoclast fusion. The variant we identified has also been reported in association with PDB in a French-Canadian cohort however the significance of this variant was inconclusive. Targeted screening of DCSTAMP in our familial cohort of PDB patients revealed an additional 8 variants; however we did not find a significant association between any of these, including p.L397F, with PDB. Osteoclastogenesis assays from the affected proband and his unaffected brother demonstrated an increase in osteoclast number and nucleation, consistent with the pagetic phenotype. In converse to other established Paget's associated genetic variations such as SQSTM1, TNFRSF11A and OPTN, expression of the mutant DC-STAMP protein attenuated the activation of transcription factors NFκB and AP-1 when exogenously expressed. We found that the p.L397F variant did not influence the subcellular localization of the protein. Based on these findings we conclude that genetic variation in DCSTAMP is not a significant predisposing factor in our specific cohort of PDB patients and the p.L397F variant is unlikely to be a contributing factor in PDB pathogenesis.
Keywords: DC-STAMP, Paget's disease of bone, Signalling, Osteoclast
Highlights
-
•
Variants in DC-STAMP do not appear to be significantly associated with Paget’s disease of bone in our cohort.
-
•
The DC-STAMP p.L397F variant attenuates both NFkB and AP-1 signalling compared to the wild type protein.
-
•
No significant differences in cellular colocalisation were found between DC-STAMP wild type and p.L397F.
1. Introduction
Paget's disease of bone (PDB) is a chronic, progressive bone disease characterised by abnormalities in bone-resorbing osteoclasts which excessively resorb bone, coupled with subsequent disorganised bone formation leading to focal lesions of woven bone that are prone to fracture and deformity (Walsh, 2004). It can occur in families with an autosomal dominant monogenic mode of inheritance or sporadically, with no known familial involvement. In our centre 26% of patients report a family history of the disease (Britton et al., 2017). Variants in SQSTM1, encoding the protein p62, are at present the only consistent genetic risk factor for monogenic PDB. To date >20 variants in the gene are associated with PDB (Rea et al., 2013), and are found in up to 10% of patients with apparently sporadic disease and up to 40% of patients with familial disease (Ralston and Layfield, 2012). In families with SQSTM1 variants, the pattern of disease inheritance is autosomal dominant with high but incomplete penetrance (Morissette et al., 2006). In sporadic cases and non-SQSTM1 familial cases disease inheritance may be polygenic, in fact risk factors in 7 genes identified by Genome Wide Association Studies (GWAS) accounted for up to 13% of heritability (Albagha et al., 2011). Whilst the exact aetiology of PDB remains incompletely understood, both environmental and genetic factors are thought to contribute.
The role of p62 in PDB is thought to be via its regulation of osteoclast signalling pathways. It has been shown to interact directly with ubiquitinated tumour necrosis factor receptor associated factor 6 (TRAF6) via a TRAF6 interacting domain and atypical protein kinase C (aPKC), involved in the activation of NFκB, allowing the transcription factor to translocate to the nucleus and regulate gene transcription. Furthermore, p62 acts as a scaffold in the recruitment of the deubiquitinating enzyme cylindromatosis (CYLD) after sustained RANKL expression, resulting in deubiquitination of TRAF6 thereby preventing further expression of NFκB (Rea et al., 2013). Thus, p62 can positively or negatively regulate TRAF6-induced NFκB activity. Functional studies have shown that expression of most PDB-associated p62 variant proteins potentiate NFκB activity compared with expression of wild type p62 and may thereby stimulate osteoclastogenesis (Beyens et al., 2006; Rea et al., 2006; Najat et al., 2009; Rea et al., 2009); this may be due in part to disruption of CYLD recruitment (Rea et al., 2013). Variants in SQSTM1 are associated with increased disease severity, including a greater number of affected bones, younger age of onset, and an increased need for treatment compared with non-SQSTM1 mutation carriers (Visconti et al., 2010; Albagha et al., 2013). Thus an increased understanding of other underlying genetic factors associated with the disease may have important clinical implications.
Recent studies have focused on identifying additional genetic factors associated with PDB. GWAS using DNA from PDB patients that are negative for SQSTM1 variants identified an additional seven loci associated with disease predisposition (Albagha et al., 2011; Albagha et al., 2010; Chung et al., 2010). Furthermore, in line with the common disease rare variant theory, it has been predicted that much of the remaining genetic susceptibility to PDB results from rare variants, possibly within the loci identified in the GWAS (Beauregard et al., 2014). To date, all PDB associated variants in SQSTM1, including the highly recurrent p.P392L variant, have reported frequencies in the general population of <0.2% (Lek et al., 2016), thus supporting this claim. Targeted sequencing is a time consuming, costly and potentially inefficient method of identifying the remaining genetic factors, especially if they do not lie within these regions as predicted. Given that the protein coding regions of the genome have been predicted to harbour 85% of human disease causing variants, whole exome sequencing is likely to be a more efficient way of elucidating the remaining genetic variation in human disease (Lacey et al., 2014). A number of studies have utilised whole exome sequencing in the context of PDB, however these have benefited from either conducting the study in a region where the disease is exceptionally rare (Lu et al., 2017; Qi et al., 2017), or from a large kindred (Divisato et al., 2016). Targeted sequencing of genes close to or within a number of the loci identified in the GWAS has been performed, including TNFRSF11A (Gianfrancesco et al., 2012), OPTN (Obaid et al., 2015), RIN3 (Vallet et al., 2015), and DCSTAMP (Beauregard et al., 2014). In the latter study, the DCSTAMP c.1189C > T p.L397F variant was predicted to be damaging by in silico tools, however the association within that cohort was not significant (p = 0.09) (Beauregard et al., 2014).
Dendritic Cell Specific Transmembrane Protein (DCSTAMP), previously TM7SF4, is a seven transmembrane protein originally identified in dendritic cells (Hartgers et al., 2000) and later found to be abundantly expressed in osteoclasts with an apparent role in osteoclastogenesis (Rho et al., 2002). It has been shown to localise to the cell surface in both HEK293 cells (Hartgers et al., 2000) and osteoclasts (Kukita et al., 2004) as well as the endoplasmic reticulum in HEK293 and dendritic cells (Eleveld-Trancikova et al., 2005). Genetic ablation of DCSTAMP in mice impairs osteoclast fusion and thus attenuates bone resorption (Yagi et al., 2005). An Immunoreceptor Tyrosine-based Inhibitory motif has been identified on the cytoplasmic tail of DC-STAMP, suggestive of a signalling role, and deletion of this tail appears to prevent nuclear translocation of the critical osteoclast transcription factor NFATc1 (Chiu et al., 2017). In addition, NFATc1 expression is reduced in DCSTAMP knockout cells (Chiu et al., 2017). Both c-Fos (AP-1) and NFATc1 were required for DC-STAMP expression and the generation of multinucleated osteoclasts (Yagi et al., 2005).
The present study sought to identify predisposing genetic factors in an unsolved family with PDB. Here, we report that the DCSTAMP p.L397F variant, affecting a transmembrane loop, that co-segregated in a family with early onset PDB and was associated with hyper-activation of osteoclastogenesis. This effect was seemingly independent of NFκB and AP-1 transcriptional activity, which were reduced by exogenous expression of the variant. Mutational screening of our familial PDB cohort failed to yield any additional candidate variants within this gene with increased prevalence in cases, suggesting that DCSTAMP is unlikely to be a major predisposing factor of PDB in our current cohort.
2. Methods
2.1. Whole exome sequencing
In the course of recruitment to a study into the genetic basis of PDB (Rea et al., 2009), we identified a family with early onset, severe PDB in which affected individuals had no identified SQSTM1 variants based on Sanger sequencing of exons 7 and 8. The Sir Charles Gairdner Hospital Human Research Ethics Committee approved the study and subjects gave informed consent.
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Blood Midi Kit (QIAGEN) according to the manufacturer's instructions. 5 μg of genomic DNA from the proband and his affected son was sent to the Australian Genome Research Facility for whole exome sequencing using Agilent SureSelectXT Human All Exon V4 paired-end sequencing on the Illumina HiSeq2000. Sequence reads were aligned to the hg19 human reference genome and variants called according to the GATK best practices (McKenna et al., 2010). Based on the seemingly autosomal dominant inheritance seen in familial PDB cases with SQSTM1 variants, we only included variants that cosegregated with disease and were identified in both affected individuals. As all SQSTM1 variants associated with PDB to date have a MAF of <0.2% (Lek et al., 2016), we hypothesised that the causative variant in this family would also be relatively rare, so set a conservative MAF cut off within the ExAc database (Lek et al., 2016) of 1%. We removed synonymous and intronic variants, under the assumption that most disease-associated variants are exonic or impact protein coding regions (Choi et al., 2009), and due to ongoing difficulties with interpreting such variants. As the variant list remained extensive, variants within genes and loci previously published with an association to PDB, including from GWAS studies and recent next generation sequencing studies (Albagha et al., 2011; Albagha et al., 2010; Chung et al., 2010), as well as variants that had a consensus of a predicted deleterious or damaging phenotype by in silico prediction tools were prioritised.
2.2. Sanger sequencing
Sanger sequencing was used to confirm the variant identified in the whole exome sequencing study. Additionally, we performed a mutational screening analysis of DCSTAMP in our cohort of 63 familial PDB patients from 51 families who had previously been screened and shown to be negative for variants in exons 7 and 8 of SQSTM1. We also screened 24 healthy unrelated controls for variants in all 3 exons of DCSTAMP. An additional 28 SQSTM1 + ve familial patients from 17 families were screened for variants in exon 2 of DCSTAMP.
Genomic DNA was isolated using the High Pure PCR Template Preparation kit (Roche Applied Science) according to the manufacturer's protocol. The primers used for the amplification are as follows: exon 1A forward; 5’-GGCCTTTACGAAGGTCAGAGGAC, and reverse; 5’-TTGCCCTTAGGTTGCAAGTC, exon 1B forward; 5’-AGGCAGGAATGCTTTGATTG, and reverse; 5’-GATGTTTTCATACTTCCAACCAC, exon 1C forward; 5’-AAGCTACTTGCCTTTGCAGG, and reverse; 5’-CTTTTGCAAGACAGACCAGC, exon 2 forward; 5’-GCAGGGAAGTACTGAGAAAGAC, and reverse; 5’-CCCTTTCCCATTTCCCTCC, exon 3 forward; 5’-TTCCCAACATCTTTAACCCC and reverse; 5’-GAGTTTGAAGATAGTTTGCTGGC. 50 μL reactions were prepared containing 1× Colourless GoTaqⓇ Reaction Buffer, 2 mM MgCl2 (Exon reactions 1B and 1C) or 2.5 mM MgCl2 (Exons 1A, 2 and 3), 0.3 mM dNTPs, 0.6 μM each of forward and reverse primer, 20 ng Genomic DNA and 2 U GoTaqⓇ DNA Polymerase (Promega). Initial denaturation was performed at 94 °C for 5 min then cycled 32 times as follows: 94 °C for 45 s, 55 °C for 45 s (except for exon 2 at 68 °C) and 72 °C for 60s before a final extension step at 72 °C for 5 min. A 10 μL aliquot of each reaction was quality checked by electrophoresing through a 1% agarose gel and 20 μL of PCR product was sent to the Australian Genome Research Facility for dual directional sanger sequencing using the big Dye Terminator Version 3.1 ready reaction sequencing kit.
2.3. Osteoclastogenesis assay
Peripheral blood samples were collected from the proband's son who was positive for the DCSTAMP p.L397F variant and his unaffected brother (negative for the variant). The affected brother was 65 years of age at the time of this study and his unaffected brother 67 years of age. Monocytes were isolated using Ficoll-Pacque™ Premium (GE Healthcare) according to manufacturer's instructions as described previously (Rea et al., 2006). Briefly, 20 mL of whole blood was mixed with 15 mL PBS, gently layered over 15 mL Ficoll-Pacque™ Premium in a 50 mL centrifuge tube and centrifuged at 1500 rpm in a Clements Orbital 900 centrifuge for 30 min at room temperature. The mononuclear cell interface layer/buffy coat was isolated and placed into a 50 mL centrifuge tube containing 12 mL PBS and centrifuged at 1500 rpm for 10 min at room temperature. The supernatant was discarded, and cells washed again with 12 mL PBS. The pellet was resuspended in 2 mL αMEM +10% FBS + 1× Glutamate +1× Pen/Strep supplemented with 25 ng/mL M-CSF. Cells were counted using a haemocytometer and seeded into a 96-well plate at a density of 2 × 105 cells per well. 48 h later media was replaced as above and further supplemented with 100 ng/mL GST-rRANKL. Media was changed by demi-depletion every 2–3 days. After 4 weeks, cells were fixed with 4% paraformaldahyde in PBS and stained for TRACP+. TRACP+ cells with 4 or more nuclei were considered osteoclasts.
2.4. Plasmids
The pEGFP N3 DC-STAMP wild type [WT] plasmid was generously provided by Professor Gosse Jansen (Jansen et al., 2009), with the DC-STAMP gene cloned into the vector prior to the N terminus of the EGFP sequence. The c.1189C > T p.L397F mutation was introduced into the wild-type sequence by site-directed mutagenesis (QuikChange kit; Stratagene) according to the manufacturer's protocol using primers designed in house as follows: Forward; 5’-TGGGACTGTTGTCCTCTATCTTTATGCAACTTAAAATCCTG, and reverse; 5’-CAGGATTTTAAGTTGCATAAAGATAGAGGACAAGAGTCCCA. Mutation status and sequence integrity were subsequently verified by sanger sequencing. The 3κB-luc-SV40, AP-1-luc and Renilla plasmids were obtained from Stratagene La Jolla, California. All plasmid DNA used for transfections was prepared using the EndoFree Plasmid Maxi Kit and eluted in endotoxin-free TE buffer (Qiagen).
2.5. Stable cell lines
HEK293 cells were seeded into 6-well plates at a density of 1 × 106 cells per well. Approximately 18 h later, cells were transfected using Lipofectamine 3000 (ThermoFisher Scientific) according to the manufacturer's protocol. 2.5 μg of either pN3 EGFP EV or pN3 EGFP DC-STAMP WT or pN3 EGFP DCSTAMP p.L397F that had been linearised with StuI was transfected into cells, with one well left as a non-transfected control. Approximately 48 h post transfection, each well was split across 6 wells in a 6-well plate. 4 h later, media was changed and supplemented with 0.5 mg/mL G418 to begin selection. Complete cell death in non-transfected cells was evident after 7 days and the cells discarded. Media was changed on the transfected cells every 2–3 days for 4 weeks to allow for substantial cell growth at which time cells were pooled and transferred to T25cm2 flasks. Expression of EGFP was confirmed periodically by fluorescence microscopy and cells were kept under G418 selection.
2.6. SDS page and western blot
Cells were lysed in RIPA buffer [150 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% w/v SDS, 0.5 mM EDTA, 0.5% w/v sodium deoxycholate, 1% v/v Triton-X100] with added complete proteinase inhibitor cocktail (Roche) and passed ten times through a 25 gauge needle, after which cell lysates were cleared by centrifugation for 30 mins at 14,000 rpm at 4 °C. Lysates were quantitated using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturers protocol for microplate assays. 30 μg protein lysate was incubated at 37 °C for 10 min then electrophoresed in 10% TGX Stain-Free Fast Cast Polyacrylamide gels (Bio-Rad). Gels were activated under UV for 5 min and then transferred overnight at 4 °C to a nitrocellulose membrane (Bio-Rad). Western blots were performed using A11122 anti-GFP primary antibody (Invitrogen by Thermo Fisher Scientific) followed by incubation with horse radish peroxidase conjugated rabbit secondary antibody (Promega). All incubations were performed at room temperature in TBS-T + 5% (w/v) skim milk powder. Membranes were washed three times with TBST (TBS containing 0.05% v/v Tween 20) prior to 1 h incubation with a species-appropriate HRP-conjugated secondary antibody in blocking solution. Membranes were washed three times with TBS-T prior to development using the Western Lightning Chemiluminescence reagent plus (PerkinElmer Life Sciences).
2.7. Reporter assays
HEK293 EGFP EV or DC-STAMP stable cells were seeded in clear 96-well plates at a density of 3 × 104 cells per well. Approximately 18 h later, cells were transfected with 40 ng 3κB-Luc-SV40-luciferase or pAP-1 luciferase and 10 ng of Renilla luciferase reporter per well with Lipofectamine 3000Ⓡ as per the manufacturer's protocol. 48 h post transfection the samples were processed using the Dual-GloⓇ Luciferase Assay System (Promega). Briefly, media was aspirated from each well and replaced with 75 μL of fresh DMEM +10% FBS + 1× Pen/Strep. 75 μL of Dual-GloⓇ Reagent was added to each well and the plate was placed on a rocker to mix for 10 min to allow lysis to occur. The entire 150 μL of lysate was then transferred to a white microplate, and firefly luminescence readings obtained with a BMG microplate reader. 75 μL of Dual-GloⓇ Stop & Glo was then added to each well, mixed gently and incubated at room temperature for 10 min before the Renilla luminescence was measured. The ratio of luminescence from experimental reporter:Renilla control reporter was calculated and then normalised to WT within the same experiment. Individual experiments were performed in quadruplicate. Data is expressed as the average +/− SEM of three independent experiments.
2.8. Confocal microscopy
HEK293 EGFP EV or DC-STAMP stable cells were seeded onto Poly-l-Lysine treated coverslips in 6-well plates at a density of 1 × 106 cells per well. 48 h later, cells were fixed with 4% paraformaldehyde in PBS and permeabilised in PBS supplemented with 0.1% Triton X for 5 min at room temperature. After washing, cells were blocked with PBS supplemented with 5% Donkey Serum for 30 min at room temperature. After removing the last wash solution, the following antibodies were used for staining; Rhodamine-conjugated Phalloidin (ThermoFisher Scientific R415) 2 h at room temperature; PDI (Abcam ab 2792–100) diluted 1:300 in PBS supplement with 1% 1 h at room temperature; GM130 (BD Biosciences 610822) diluted 1:300 in PBS supplement its 1% BSA 1 h at room temperature. After extensive washing, AlexaFlour 647 donkey anti-mouse (ThermoFisher Scientific A-31571) was added to the PDI and GM130 samples at a dilution of 1:500 for 1 h at room temperature. Again, after extensive washing, cells were stained with Hoechst 33342 (ThermoFisher Scientific H3570) dye diluted 1:10000 for 10 min at room temperature. Cells were extensively washed and the coverslips then mounted onto slides using Prolong Gold antifade mounting media (ThermoFisher Scientific P36930). All staining dilutions were in PBS supplemented with 1% BSA and all wash steps in PBS supplemented with 0.2% BSA.
2.9. Statistical analysis
Association analysis for variants identified in the screening study was performed using Fisher's exact test for allelic tests due to the small sample size. Luciferase assay data was analysed by ANOVA using the car package in R Studio, followed by the Multiple Comparison of Means with Tukey's Contrasts in the multicomp package in R Studio.
3. Results
3.1. Patient details
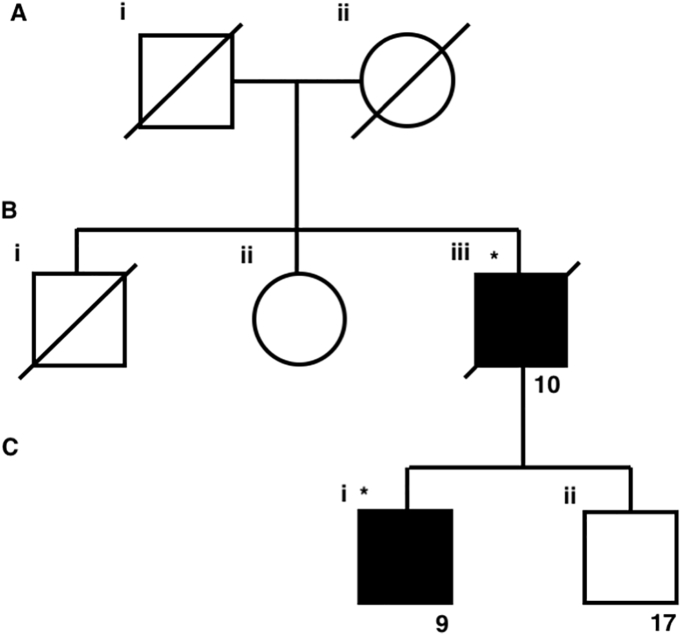
Within this kindred are two affected individuals: the proband and one of his two sons. The proband had a severe pagetic phenotype with early age of onset with diagnosis at 40 years of age, markedly elevated pre-treatment plasma alkaline phosphatase (ALP) activity of 1650 IU/L (reference range < 135 IU/L), and polyostic disease involving the L5 vertebra, right hemipelvis, sacrum, right femur, right fibula and right tibia. His affected son was diagnosed with PDB at 42 years of age with a pre-treatment ALP of 511 IU/L and involvement of the left hemipelvis. A second son, currently aged 69 years, had no sign of disease at the time of the study with an ALP of 61 IU/L and is thus categorised as unaffected. However, as this individual was not subjected to x-ray screening we cannot definitively rule out the presence of asymptomatic disease.
3.2. Genetic screening
Using our initial broad filtering method, we identified >300 uncommon, possibly pathogenic exonic variants that were present in both affected individuals, including a variant in DCSTAMP, c.1189C > T p.L397F which was immediately of interest. DCSTAMP is within a region on chromosome 8 found to be associated with PDB in GWAS studies (Albagha et al., 2011; Chung et al., 2010), and this variant was identified in a cohort of 267 French-Canadian PDB patients, however the association with PDB did not reach significance (p = 0.09) (Beauregard et al., 2014). In our study, this variant was the only plausible pathogenic exonic variant in the vicinity of the 7 GWAS loci, as it had a population frequency reported in gnomAD of <1% and was predicted to be pathogenic. As all reported variants in SQSTM1 associated with PDB to date have population frequencies of <0.2% (Lek et al., 2016), we predicted that in line with the common disease rare variant theory, that the genetic susceptibility to PDB in this kindred may result from rare variants with a population frequency of <1% (Beauregard et al., 2014). The p.L397F variant was confirmed in the affected individuals via Sanger sequencing, and was not present in the unaffected son, showing co-segregation with disease within the limitations of this small kindred.
3.3. Cohort screening
Initial screening of DC-STAMP, which included the 63 familial SQSTM1 negative PDB patients and 24 healthy, unrelated controls, revealed a total of 9 variants; 4 synonymous, 3 missense, 1 intronic, and 1 in the 3’ UTR (Table 2). All variants had previously been reported in variant databases. With the exception of the p.L397F variant, all of the missense variants were common with a minor allele frequency (MAF) of >5% in the gnomAD Non-Finnish European population (Table 2). The p.L397F variant was not identified in our controls, but additional screening of exon 2 of DC-STAMP in our SQSTM1 positive subgroup of familial patients identified a further two individuals with this variant (Table 1). None of the DCSTAMP variants, in either the SQSTM1 positive or negative subgroups, were statistically more highly represented in cases compared to the group of 24 healthy, unrelated controls (Table 2).
Fig. 1.
Family pedigree of a p.L397F carrier family. Individuals affected by PDB are blocked in black. Diagonal crosses indicate individual is deceased. Asterisks indicate that the individual is a carrier of the p.L397F variant. Individuals assigned with a number have DNA available. Numbers indicate laboratory reference for the individual.
Table 2.
Mutational Screening of DCSTAMP. SQSTM1 mutation positive and negative individuals with PDB were screened for single nucleotide polymorphisms (SNP) in DCSTAMP. The Gnomad non-Finnish European (NFE) minor allele frequencies (MAF) for the variants identified are included for comparison. p values indicate significant difference between variant presence in patients versus controls. SIFT scores are used as an indicator of pathogenicity.
| SNP | Controls |
SQSTM1-ve |
SQSTM1 + ve |
PDB all |
Gnomad NFE MAF | SIFT | Polyphen | |||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF (n = 24) | MAF (n = 63) | Unadjusted p | MAF (n = 28) | Unadjusted p | MAF (n = 91) | Unadjusted p | ||||
| c.315C > T p.I105I | 6 (12.00%) | 16 (12.70%) | 1.00 | 13.11% | ||||||
| c.574 G > C p.E192Q | 6 (12.00%) | 16 (12.70%) | 1.00 | 13.12% | 0.41 T | 0.051 B | ||||
| c.1030–9 T > C Intronic |
35 (72.92%) | 103 (81.75%) | 0.30 | 35 (62.50%) | 0.30 | 138 (75.82%) | 0.71 | 72.14% | ||
| c.1046 A > G p.D349G | 5 (10.42%) | 17 (13.49%) | 0.80 | 6 (10.71%) | 1.00 | 23 (12.64%) | 0.81 | 13.57% | 0.69 T | 0.00 B |
| c.1189C > T p.L397F | 0 (0.00%) | 2 (1.59%) | 1.00 | 2 (3.57%) | 0.50 | 4 (2.20%) | 0.58 | 1.39% | 0.78 T | 0.779 P |
| c.1233C > T p.S411S | 0 (0.00%) | 1 (0.79%) | 1.00 | 0 (0.00%) | 1.00 | 1 (0.55%) | 1.00 | 0.02% | ||
| c.1341 T > C p.I447I | 0 (0.00%) | 1 (0.79%) | 1.00 | 0.16% | ||||||
| c.1395 A > G p.A465A | 0 (0.00%) | 4 (3.17%) | 0.58 | 5.02% | ||||||
| c.*259 G > C 3’UTR | 6 (12.00%) | 17 (13.49%) | 0.80 | 12.66% | ||||||
Table 1.
Phenotype and SQSTM1 genotype information for individuals identified as carriers of the p.L397F DCSTAMP variant.
| ID | Age at diagnosis | Pre-treatment ALP (IU/L)* | # Bones Affected | SQSTM1 mutation |
|---|---|---|---|---|
| 9 | 42 | 511 | 1 | n.p. |
| 10 | 40 | 1650 | 5 | n.p. |
| 78 | 36 | 3070 | 19 | p.K378X |
| 276 | 42 | 360 | 5 | p.P392L |
• ID = laboratory reference for individual.
• *Reference range for alkaline phosphatase (ALP) <135 IU/L.
• n.p. = not present.
3.4. DC-STAMP p.L397F + monocytes have increased osteoclastogenic potential
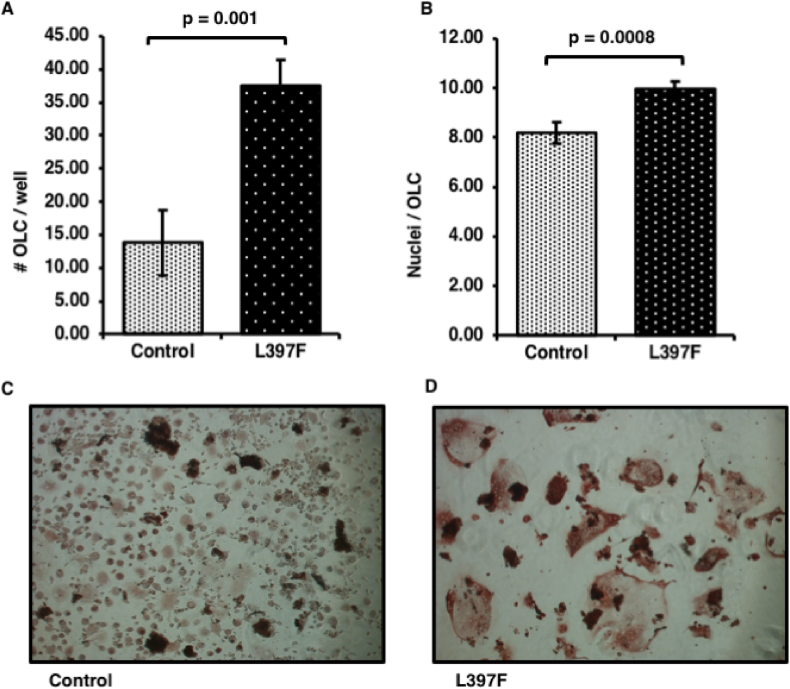
We obtained peripheral blood samples from the affected and unaffected sons of the proband (who is deceased), isolated monocytes and performed in vitro osteoclastogenesis assays. The affected son was 65 years of age at the time of this study and the unaffected son 67 years of age. Consistent with a pagetic phenotype, we found that the osteoclast like cells (OLC) from the affected individual were more numerous than his affected brother (Fig. 2A), with 37.50 ± 3.95 cells per well for the affected vs 13.83 ± 4.93 for the unaffected (p = 0.001). In addition, the OLC were hyper-nucleated (Fig. 2B) with 9.98 ± 0.28 nuclei per cell for the affected vs 8.20 ± 0.42 for the unaffected, (p = 0.0008).
Fig. 2.
Osteoclastogenesis of human monocytes from p.L397F carrier patient and unaffected control. Human monocytes were isolated from whole blood of individuals 9, who is affected by PDB and is a carrier of the p.L397F variant, and his unaffected brother, 17, who does not carry the variant (Fig. 1). Cells were seeded into 96-well plates and cultured in the presence of RANKL and M-CSF for 4 weeks before fixing and staining for TRACP. (A) Average number of OLC's formed per well. (B) Average number of nuclei per OLC. (C) Representative image of control OLCs from individual 17 taken at 10× magnification. (D) Representative image of p.L397F affected OLCs taken at 10× magnification.
3.5. Osteoclast signalling pathways
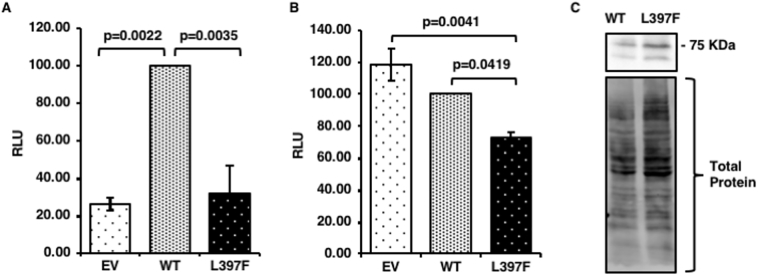
As osteoclastogenesis can be enhanced in PDB due to increased signalling, we performed luciferase assays to determine whether the variant had an effect on the activity of the NFκB and AP-1 transcription factors, both of which have critical roles in osteoclast survival and bone resorption. Due to low levels of expression in transient transfections, we created HEK293T cells stably expressing either EGFP or EGFP DC-STAMP (wild type, [WT] or mutant [p.L397F]). Previous studies have shown that expression of PDB-associated SQSTM1 mutant proteins activate NFκB and AP-1 compared with the wild type counterpart (Rea et al., 2006; Najat et al., 2009; Rea et al., 2009), and components of both of these transcription factors also play critical roles in the activation of NFATc1; DC-STAMP facilitates NFATc1 transcription (Chiu et al., 2017; Kim and N, 2014). Compared to control cells expressing GFP, expression of wild type DC-STAMP increased NFκB activity (p = 0.0022), but had no effect on AP-1 activation (Fig. 3A and B, respectively). The NFκB activation of the p.L397F mutant DC-STAMP expressing cells was indifferent to GFP expressing controls, whereas AP1 signalling was significantly impaired by the mutant (p = 0.0037). We found that compared to wild type DC-STAMP, the p.L397F mutant had an inhibitory effect on both NFκB signalling (p = 0.0035) and AP-1 signalling (p = 0.0419). Both the EGFP DC-STAMP WT and mutant constructs expressed 2 bands, representing glycosylated and unglycosylated forms of the DC-STAMP protein as previously described (Eleveld-Trancikova et al., 2005), and were found to express at similar levels after correction for total protein loading (Fig. 3C).
Fig. 3.
DC-STAMP p.L397F variant attenuates NFκB and AP-1 activity. HEK293 cells stably expressing pEGFP N3 Empty Vector (EV), or pEGFP N3 DC-STAMP WT or p.L397F were co-transfected with a Renilla internal control expression plasmid and either (A) an NFκB luciferase reporter plasmid or (B) a AP-1 luciferase reporter plasmid. After 48 h, luciferase activities were read and firefly luciferase values were first normalised to Renilla, and then compared directly to the wildtype level set at 100%. (C) Expression levels of pEGFP N3 DC-STAMP WT or p.L397F were determined by western blot against GFP with total protein used as a loading control. Individual assays were performed in quadruplicate, and data is the average relative luciferase units (RLU) of 3 independent experiments ± Standard Error. p values indicate significant differences compared to cells expressing wild type DC-STAMP.
3.6. Cellular localisation of DC-STAMP
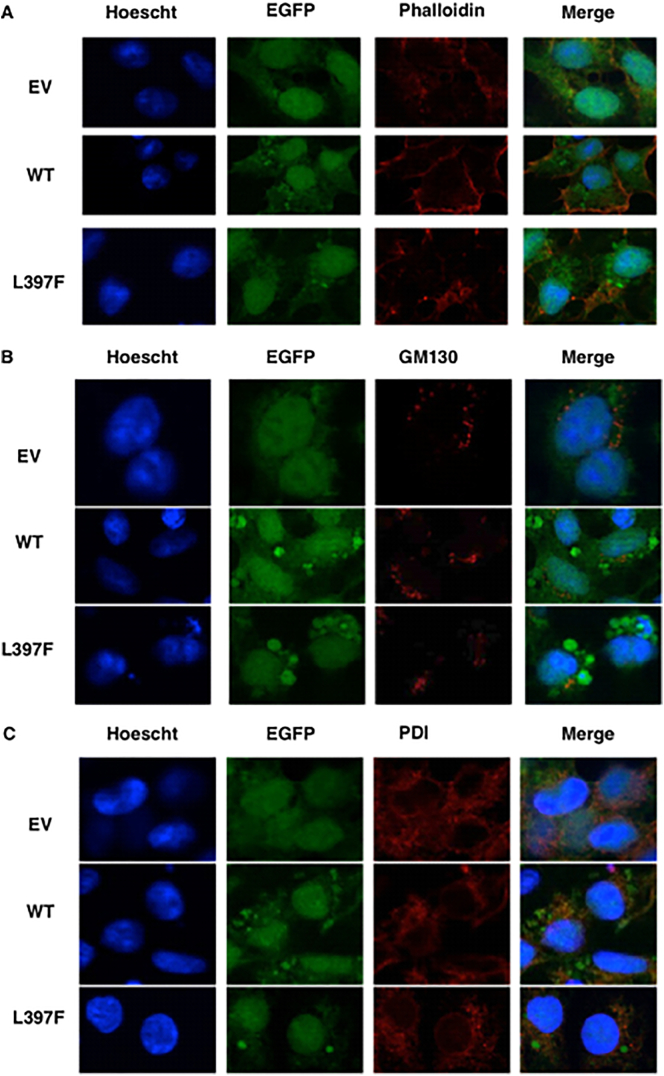
We hypothesised that as the p.L397P substitution occurs within a transmembrane domain of the receptor that the mutant protein may mislocalise within the cell. To test this, we used confocal microscopy to image cells stably expressing EGFP DC-STAMP (WT or p.L397F) or EGFP alone. To assist us in determining the localisation of the receptor, we used a Rhodamine-conjugated phalloidin probe to label F-actin (Fig. 4A), PDI antibody to visualise the ER (Fig. 4B) or a GM1130 antibody to localise the cis-Golgi (Fig. 4C), plus the nuclear stain Hoescht 33,342. EGFP was expressed in the nucleus and cytoplasm in a diffuse manner. Both WT and p.L397F mutant DC-STAMP showed nuclear staining in addition to cytoplasmic punctate staining. Partial colocalisation with the ER marker was apparent for both DC-STAMP WT and p.L397F (Pearson's coefficient = 0.46 and 0.51, respectively) and were not significantly different. Due to its diffuse expression, EGFP was also partly co-localised with the ER (Pearson's co-efficient = 0.41). Neither WT nor p.L397F DC-STAMP colocalised to the cis-Golgi or actin markers.
Fig. 4.
p.L397F variant does not affect cellular localisation of DC-STAMP. HEK293 cells stably expressing EGFP or EGFP DC-STAMP (wild type, [WT] or mutant [p.L397F]) were seeded onto cover slips. The following day cells were fixed and stained with antibodies or probes for sub-cellular localisation a) Phalloidin to F-actin b) PDI an ER marker or c) GM130 a cis-Golgi marker and the nuclear stain Hoechst. Images are representative of the majority of cells from independent replicate experiments.
4. Discussion
In this study we identified a variant in DCSTAMP (p.L397F) that co-segregated in a family with unusually young onset of PDB and a proband that had presented with a severe phenotype. DCSTAMP falls within a locus highlighted by GWAS in patients with PDB (Albagha et al., 2011; Chung et al., 2010). An additional study reported the same variant as potentially associated with PDB and used in silico prediction to suggest the variant may be pathogenic (Beauregard et al., 2014). While our method of variant prioritization certainly has the potential to have excluded other potential causal variants, we were limited by an extensive list of variants due to a small kindred and the likely autosomal dominant pattern of inheritance. Based on previous studies highlighting DCSTAMP as a gene of interest in PDB (Albagha et al., 2011; Chung et al., 2010), and the association of this particular p.L397F variant with PDB (Beauregard et al., 2014), it was decided that it warranted further investigation.
We went on to perform genetic screening of DCSTAMP in our familial cohort. We identified the p.L397F variant in an additional two affected individuals, both of whom are also carriers of SQSTM1 variants (Rea et al., 2006), further suggesting that this variant may be involved in disease pathogenesis. Thus, our study sought to investigate the potential pathogenicity of this variant through a combination of in vitro functional studies that utilised both stable cell lines and patient derived osteoclast-like cells.
DC-STAMP has been previously reported to play a critical role in osteoclast fusion. It is a large transmembrane receptor protein that localises to the cell surface of osteoclasts (Kukita et al., 2004). Knockout in mice has been shown to disrupt osteoclast fusion and thus attenuates, but does not completely abolish, bone resorption (Yagi et al., 2005). It has been hypothesised that genetic variation in DCSTAMP would contribute to the hyper-activation of osteoclastogenesis in PDB by stimulating cell fusion specifically (Albagha et al., 2011). Accordingly, we found that the number of osteoclasts and the number of nuclei per osteoclast were significantly increased in the affected son of the proband who is a carrier of the variant, compared with the non-affected, non-mutation carrying son. It is possible that the presence of the mutation led to increased fusion of monocytic cells leading to more multinucleated osteoclast like cells in the patient cultures. It is also possible that the presence of the mutation, or other unknown genetic factors, increased survival of the patient OCLs. It is also possible, given our osteoclastogenesis assay could only be performed once due to the overseas location of one of the brothers, that the differences are due to chance. However, as osteoclastogenesis and osteoclast survival are regulated in part via Receptor Activator of NFκB Ligand-induced signalling pathways, thus we investigated the effects of the p.L397F variant on transcription factor activation.
Unlike other PDB-associated variants in SQSTM1, TNFRSF11A and OPTN, that potentiate osteoclastogenesis via elevated NFκB activity (Beyens et al., 2006; Rea et al., 2006; Najat et al., 2009; Rea et al., 2009; Gianfrancesco et al., 2012; Obaid et al., 2015), we found that expression of DC-STAMP p.L397F had no effect on NFκB activity compared with cells expressing GFP alone. However, when compared with wild type DC-STAMP expressing cells NFκB activity was attenuated. This finding was unexpected, as the enhanced osteoclast formation seen in patient-derived cells would suggest that signalling could be increased and it is contrary to the assumption that different genetic factors contribute to the pagetic phenotype through similar mechanisms. We found that compared with cells expressing either GFP or wild type DC-STAMP, cells expressing the p.L397F variant had transcriptional suppression of AP-1. It has been reported that DC-STAMP influences both the expression and nuclear translocation of the critical transcription factor NFATc1, an effect that arises from the cytoplasmic tail of the protein (Chiu et al., 2017). The AP-1 component c-Fos is critical in the induction of NFATc1 (Kim and N, 2014), although it is again unclear how suppression of AP-1 by DC-STAMP would contribute to a pagetic phenotype. As the p.L397F variant lies outside of this region, instead resulting in a missense change within the seventh transmembrane domain of the protein, it is unlikely to directly influence NFATc1. This suggests that our observations may be a general phenomenon of dampened transcription with expression of DC-STAMP p.L397F. To further investigate the potential effects of this variant on DC-STAMP function, we determined whether it influenced the subcellular targeting of DC-STAMP. We found that the proteins were localised similarly in our stable cell lines, with both partially colocalising to the ER as has been described for DC-STAMP previously. Thus, alternate mechanisms may exist that account for the dampening effects on NFκB/AP-1 signalling in stable cells and the hyper-activation of osteoclastogenesis observed in the affected son harboring the p.L397F variant. We note that our study is limited in that our signalling assays were performed in HEK293 cells, as we were unable to express the DC-STAMP protein in osteoclasts. It is therefore important to interpret the results with caution. It is possible that the increased osteoclastogenesis we observed with patient-derived cells may be due to an as yet unidentified genetic factor. Alternatively, it may reflect unregulated fusion of osteoclast progenitor cells expressing the p.L397F mutant which would be in keeping with the established role of DC-STAMP in osteoclast fusion. However, the mechanisms underlying this effect on fusion remain largely unknown and are therefore difficult to assess. These possibilities form the basis of our on-going investigations.
Overall, our study highlights the need for more sophisticated methods of gene discovery in complex diseases such as PDB, which is further hindered by a lack of availability of DNA from affected family members due to the late onset of symptoms. Whole exome sequencing can be useful, particularly where GWAS studies have highlighted loci of interest, however data analysis tools beyond co-segregation analysis and in silico filtering are required to reduce the size of candidate gene lists. The DCSTAMP p.L397F variant was identified as the most plausible variant in our initial list of 300 possible candidates, subsequent mutational screening of our familial cohort of PDB patients failed to identify any significant association between variants in this gene and PDB. Our functional studies indicated no changes to localization, and a lack of hyperactivation of key osteoclastogenic pathways, these findings, combined with a previous report of showing only marginal association to PDB, we conclude that DCSTAMP is unlikely to be a significant driver of PDB in our familial cohort.
Acknowledgements
This work was supported by a Sir Charles Gairdner Hospital Research Advisory Committee project grant to JPW and SLR.
References
- Albagha O.M. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget's disease of bone. Nat. Genet. 2010;42(6):520–524. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albagha O.M.E. Genome-wide association identifies three new susceptibility loci for Paget's disease of bone. Nat. Genet. 2011;43(7):685–U104. doi: 10.1038/ng.845. [DOI] [PubMed] [Google Scholar]
- Albagha O.M. Common susceptibility alleles and SQSTM1 mutations predict disease extent and severity in a multinational study of patients with Paget's disease. J. Bone Miner. Res. 2013;28(11):2338–2346. doi: 10.1002/jbmr.1975. [DOI] [PubMed] [Google Scholar]
- Beauregard M. Identification of rare genetic variants in novel loci associated with Paget's disease of bone. Hum. Genet. 2014;133(6):755–768. doi: 10.1007/s00439-013-1409-x. [DOI] [PubMed] [Google Scholar]
- Beyens G. Identification and molecular characterization of a novel splice-site mutation (G1205C) in the SQSTM1 gene causing Paget's disease of bone in an extended American family. Calcif. Tissue Int. 2006;79(5):281–288. doi: 10.1007/s00223-006-0122-3. [DOI] [PubMed] [Google Scholar]
- Britton C. The changing presentation of Paget's disease of bone in Australia, a high prevalence region. Calcif. Tissue Int. 2017;101(6):564–649. doi: 10.1007/s00223-017-0312-1. [DOI] [PubMed] [Google Scholar]
- Chiu Y.H. Dendritic cell-specific transmembrane protein (DC-STAMP) regulates osteoclast differentiation via the Ca2+/NFATc1 Axis. J. Cell. Physiol. 2017;232(9):2538–2549. doi: 10.1002/jcp.25638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P.Y.J. The majority of the genetic risk for Paget's disease of bone is explained by genetic variants close to the CSF1, OPTN, TM7SF4, and TNFRSF11A genes. Hum. Genet. 2010;128(6):615–626. doi: 10.1007/s00439-010-0888-2. [DOI] [PubMed] [Google Scholar]
- Divisato G. ZNF687 mutations in severe Paget disease of bone associated with giant cell tumor. Am. J. Hum. Genet. 2016;98(2):275–286. doi: 10.1016/j.ajhg.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleveld-Trancikova D. The dendritic cell-derived protein DC-STAMP is highly conserved and localizes to the endoplasmic reticulum. J. Leukoc. Biol. 2005;77(3):337–343. doi: 10.1189/jlb.0804441. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F. A nonsynonymous TNFRSF11A variation increases NFkappaB activity and the severity of Paget's disease. J. Bone Miner. Res. 2012;27(2):443–452. doi: 10.1002/jbmr.542. [DOI] [PubMed] [Google Scholar]
- Hartgers F.C. DC-STAMP, a novel multimembrane-spanning molecule preferentially expressed by dendritic cells. Eur. J. Immunol. 2000;30:3585–3590. doi: 10.1002/1521-4141(200012)30:12<3585::AID-IMMU3585>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Jansen B.J. OS9 interacts with DC-STAMP and modulates its intracellular localization in response to TLR ligation. Mol. Immunol. 2009;46(4):505–515. doi: 10.1016/j.molimm.2008.06.032. [DOI] [PubMed] [Google Scholar]
- Kim J.H., N K. Regulation of NFATc1 in osteoclast differentiation. J. Bone Metab. 2014;21:233–241. doi: 10.11005/jbm.2014.21.4.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukita T. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J. Exp. Med. 2004;200(7):941–946. doi: 10.1084/jem.20040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey S., Chung J.Y., Lin H. A comparison of whole genome sequencing with exome sequencing for family-based association studies. BMC Proc. 2014;8(Suppl 1 Genetic Analysis Workshop 18Vanessa Olmo):S38. doi: 10.1186/1753-6561-8-S1-S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. A FKBP5 mutation is associated with Paget's disease of bone and enhances osteoclastogenesis. Exp. Mol. Med. 2017;49(5):e336. doi: 10.1038/emm.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette J., Laurin N., Brown J.P. Sequestosome 1: mutation frequencies, haplotypes, and phenotypes in familial Paget's disease of bone. J. Bone Miner. Res. 2006;21(Suppl. 2):P38–P44. doi: 10.1359/jbmr.06s207. [DOI] [PubMed] [Google Scholar]
- Najat D. Characterization of a non-UBA domain missense mutation of sequestosome 1 (SQSTM1) in Paget's disease of bone. J. Bone Miner. Res. 2009;24(4):632–642. doi: 10.1359/jbmr.081204. [DOI] [PubMed] [Google Scholar]
- Obaid R. Optineurin negatively regulates osteoclast differentiation by modulating NF-kappaB and interferon signaling: implications for Paget's disease. Cell Rep. 2015;13(6):1096–1102. doi: 10.1016/j.celrep.2015.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X. Familial early-onset Paget's disease of bone associated with a novel hnRNPA2B1 mutation. Calcif. Tissue Int. 2017;101(2):159–169. doi: 10.1007/s00223-017-0269-0. [DOI] [PubMed] [Google Scholar]
- Ralston S.H., Layfield R. Pathogenesis of Paget disease of bone. Calcif. Tissue Int. 2012;91(2):97–113. doi: 10.1007/s00223-012-9599-0. [DOI] [PubMed] [Google Scholar]
- Rea S.L. A novel mutation (K378X) in the sequestosome 1 gene associated with increased NF-kappaB signaling and Paget's disease of bone with a severe phenotype. J. Bone Miner. Res. 2006;21(7):1136–1145. doi: 10.1359/jbmr.060405. [DOI] [PubMed] [Google Scholar]
- Rea S.L. Sequestosome 1 mutations in Paget's disease of bone in Australia: prevalence, genotype/phenotype correlation, and a novel non-UBA domain mutation (P364S) associated with increased NF-kappaB signaling without loss of ubiquitin binding. J. Bone Miner. Res. 2009;24(7):1216–1223. doi: 10.1359/jbmr.090214. [DOI] [PubMed] [Google Scholar]
- Rea S.L. New insights into the role of sequestosome 1/p62 mutant proteins in the pathogenesis of Paget's disease of bone. Endocr. Rev. 2013;34(4):501–524. doi: 10.1210/er.2012-1034. [DOI] [PubMed] [Google Scholar]
- Rho J. Gene expression profiling of osteoclast differentiation by combined suppression subtractive hybridization (SSH) and cDNA microarray analysis. DNA Cell Biol. 2002;21(8):541–549. doi: 10.1089/104454902320308915. [DOI] [PubMed] [Google Scholar]
- Vallet M. Targeted sequencing of the Paget's disease associated 14q32 locus identifies several missense coding variants in RIN3 that predispose to Paget's disease of bone. Hum. Mol. Genet. 2015;24(11):3286–3295. doi: 10.1093/hmg/ddv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti M.R. Mutations of SQSTM1 are associated with severity and clinical outcome in Paget disease of bone. J. Bone Miner. Res. 2010;25(11):2368–2373. doi: 10.1002/jbmr.132. [DOI] [PubMed] [Google Scholar]
- Walsh J.P. Paget's disease of bone. Med. J. Aust. 2004;181(5):262–265. doi: 10.5694/j.1326-5377.2004.tb06265.x. [DOI] [PubMed] [Google Scholar]
- Yagi M. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 2005;202(3):345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]