Nutrient availability is an important contributor to the ability of bacteria to establish successful infections in a host. Pseudomonas aeruginosa is an opportunistic pathogen in humans causing infections that are difficult to treat. In part, its success is attributable to a high degree of metabolic versatility. P. aeruginosa is able to sense and respond to varied and limited nutrient stress in the host environment. Two-component systems are important sensors-regulators of cellular responses to environmental stresses, such as those encountered in the host. This work demonstrates that the response by the two-component system TctD-TctE to the presence of citric acid has a role in biofilm formation, aminoglycoside susceptibility, and growth in P. aeruginosa.
KEYWORDS: Pseudomonas aeruginosa, biofilms, citric acid, two-component regulatory systems
ABSTRACT
The two-component system TctD-TctE is important for regulating the uptake of tricarboxylic acids in Pseudomonas aeruginosa. TctD-TctE accomplishes this through derepression of the gene opdH, which encodes a tricarboxylic acid-specific porin. Previous work from our lab revealed that TctD-TctE in P. aeruginosa also has a role in resistance to aminoglycoside antibiotics. The aim of this study was to further characterize the role of TctD-TctE in P. aeruginosa in the presence of citric acid. Here it was found that deletion of P. aeruginosa PA14 TctD-TctE (ΔtctED) resulted in a 4-fold decrease in the biofilm bactericidal concentrations of the aminoglycosides tobramycin and gentamicin when citric acid was present in nutrient media. Tobramycin accumulation assays demonstrated that deletion of TctD-TctE resulted in an increase in the amount of tobramycin retained in biofilm cells. The PA14 wild type responded to increasing concentrations of citric acid by producing less biofilm. In contrast, the amount of ΔtctED mutant biofilm formation remained constant or enhanced. Furthermore, the ΔtctED strain was incapable of growing on citric acid as a sole carbon source and was highly reduced in its ability to grow in the presence of citric acid even when an additional carbon source was available. Use of phenotypic and genetic microarrays found that this growth deficiency of the ΔtctED mutant is unique to citric acid and that multiple metabolic genes are dysregulated. This work demonstrates that TctD-TctE in P. aeruginosa has a role in biofilm development that is dependent on citric acid and that is separate from the previously characterized involvement in resistance to antibiotics.
IMPORTANCE Nutrient availability is an important contributor to the ability of bacteria to establish successful infections in a host. Pseudomonas aeruginosa is an opportunistic pathogen in humans causing infections that are difficult to treat. In part, its success is attributable to a high degree of metabolic versatility. P. aeruginosa is able to sense and respond to varied and limited nutrient stress in the host environment. Two-component systems are important sensors-regulators of cellular responses to environmental stresses, such as those encountered in the host. This work demonstrates that the response by the two-component system TctD-TctE to the presence of citric acid has a role in biofilm formation, aminoglycoside susceptibility, and growth in P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is a member of the gammaproteobacteria and an opportunistic pathogen in humans. It is a highly adaptable species and is readily able to sense and respond to its environment. In part, this is due to its genome encoding a large number of two-component systems (TCSs) that it uses to adapt to its surroundings (1, 2). This adaptability contributes to the ability of P. aeruginosa to colonize a variety of host environments (3, 4). Previous work has found that numerous TCSs in P. aeruginosa have a role in regulating virulence and the development of antibiotic resistance (5–9). TCSs are an extensively utilized means for bacteria to sense and respond to their environment (10–14). The basic makeup of a TCS consists of a membrane-bound sensor kinase and a cytoplasmic response regulator. Activation of the sensor kinase occurs through autophosphorylation of a histidine residue (14–16). The active kinase then phosphorylates its cognate response regulator, which activates it, and the active response regulator exerts its downstream transcriptional effects on gene expression (14–16). TCSs can potentially be involved in a multitude of regulatory pathways having diverse roles for cell responses (10, 17–20), including the development of antibiotic resistance (5, 8, 9, 21).
Salmonella enterica serovar Typhimurium, another pathogenic member of the gammaproteobacteria, contains an operon that encodes genes, named the tricarboxylic transporters (tct), which are responsible for the uptake of tricarboxylic carbon sources, such as citric acid (22). Within this operon are two genes for a TCS: tctD, which encodes a transcriptional regulator, and tctE, which encodes a histidine sensor kinase (22, 23). TctD-TctE senses tricarboxylic compounds in the environment and regulates the expression of the tct operon for uptake and metabolism of these compounds.
P. aeruginosa does not possess a fully conserved tct operon, but its genome does encode homologs of tctD and tctE (24). In P. aeruginosa, TctD-TctE acts on the expression of opdH through a mechanism of derepression in the presence of tricarboxylic acids. opdH encodes a porin (OpdH) required for the transport of tricarboxylic acids across the outer membrane permeability barrier of P. aeruginosa (24). Previous work from our lab found that a deletion of the P. aeruginosa operon tctED (previously described as PA0756-0757) in the PA14 wild-type strain resulted in a 4-fold increase in susceptibility to aminoglycoside antibiotics in biofilm cultures (25). Our current study questioned whether regulation of citrate metabolite (i.e., citric acid) uptake is involved in the aminoglycoside susceptibility phenotype observed in a tctED deletion (ΔtctED) strain.
In this study, we hypothesized that TctD-TctE is involved in biofilm formation as well as aminoglycoside resistance in P. aeruginosa. We observed that in biofilm cultures the ΔtctED mutant had a 4-fold increase in susceptibility to the aminoglycosides tobramycin and gentamicin when citric acid was present in the nutrient media. Furthermore, we found that there was a moderate but significant increase in the accumulation of tobramycin in ΔtctED mutant biofilms when citric acid was present in the growth medium. We further hypothesized that there are differences in biofilm formation in the ΔtctED mutant compared to PA14 in the presence of citric acid. We determined that in the presence of citric acid, the ΔtctED mutant showed no significant change in the amount of biofilm biomass, while the PA14 strain displayed reduced levels of biofilm biomass. It was also found that the ΔtctED mutant had a severely reduced level of growth when citric acid was present in the growth medium. The use of phenotypic microarrays determined that this phenotype was unique to citric acid as a carbon source. However, the use of a whole-genome expression microarray approach found that multiple metabolic genes not directly involved in citrate metabolism are dysregulated in the ΔtctED mutant relative to PA14. Here we demonstrate for the first time that TctD-TctE has a role in regulating biofilm formation in P. aeruginosa.
RESULTS
Loss of TctD-TctE leads to increased susceptibility to aminoglycosides in biofilm cultures.
Interest in studying TctD-TctE originated from previous findings from our lab demonstrating that the loss of expression of these proteins in the ΔtctED deletion strain (then designated the ΔPA0756-0757 strain) resulted in biofilms that were more susceptible to a subset of antibiotics, the aminoglycosides tobramycin and gentamicin (25). In this study, we wanted to explore whether inclusion of citric acid would have a further effect on the antibiotic susceptibility of the ΔtctED mutant. To explore this phenotype, we assayed for the minimal bactericidal concentrations (MBCs) of tobramycin and gentamicin with the addition of citric acid in the nutrient media (Table 1). We also included ciprofloxacin, a clinically relevant fluoroquinolone in the treatment of P. aeruginosa infections. Assays for determination of MBCs in planktonic cultures (MBC-P) containing 10 mM citric acid, in addition to antibiotic, found a 2-fold increase in susceptibility to both tobramycin and gentamicin in ΔtctED mutant cultures relative to PA14 cultures (Table 1). This increased susceptibility under planktonic conditions was not observed in our previous work, where citric acid was not included in the MBC assays (25). For assays for determination of MBCs in biofilms (MBC-B), a 4-fold increase in susceptibility to tobramycin and gentamicin was observed in the ΔtctED mutant compared to PA14 (Table 1). No difference in the MBCs between PA14 and the ΔtctED mutant was observed in either MBC-P or MBC-B assays with ciprofloxacin (Table 1). Additional antibiotics (including the antipseudomonal β-lactam ceftazidime, the fluoroquinolones levofloxacin and norfloxacin, chloramphenicol, and nalidixic acid) not previously tested (25) were assayed here. For all except one antibiotic, there were no differences in the MBC-Ps or MBC-Bs observed (see Table S1 in the supplemental material). A 2-fold increase in the MBC-P was observed when testing with the synthetic quinolone nalidixic acid.
TABLE 1.
MBCs for antibiotics in medium including 10 mM citric acid
| Culture and strain | MBC (μg/ml) |
||
|---|---|---|---|
| Tobramycin | Gentamicin | Ciprofloxacin | |
| Planktonic | |||
| PA14 | 64 | 128 | 4 |
| ΔtctED | 32 | 64 | 4 |
| Biofilm | |||
| PA14 | 400 | 800 | 40 |
| ΔtctED | 100 | 200 | 40 |
Minimal bactericidal concentrations (MBCs) for select antibiotics in media including 10 mM citric acid. Download Table S1, DOCX file, 0.02 MB (18.7KB, docx) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ΔtctED strain demonstrated increased accumulation of tobramycin in its biofilms.
To investigate this biofilm phenotype of tobramycin susceptibility, we questioned whether there was any greater accumulation of tobramycin in the ΔtctED mutant biofilms than in PA14 biofilms. We performed accumulation assays by growing biofilm cultures in the presence of citric acid (10 mM added to M63-arginine medium). The biofilms were then treated with a bactericidal concentration of tobramycin for the ΔtctED mutant (200 μg/ml) (Table 1) (26, 27). We included citric acid in this assay for the role that it plays in the activity of TctD-TctE (24) and also for its effects on cellular growth (28, 29) and biofilm formation (30). Biofilm cells were lysed and then assayed for the presence of tobramycin by measuring the diameter of the clearance zone on Escherichia coli lawns. M63 medium without added citric acid did not show any difference in tobramycin accumulation between the PA14 and ΔtctED strains (Fig. 1). However, when citric acid was present, there was a statistically significant (P < 0.05) increase in the zone of clearance observed, indicating a greater accumulation of tobramycin in ΔtctED mutant biofilms (Fig. 1). It is of note that a zone diameter change of ≥3 mm can clinically result in grouping of the bacteria to a susceptible or resistant category in antimicrobial disk susceptibility testing (31).
FIG 1.
Tobramycin accumulation assays of PA14 and the ΔtctED mutant under conditions where citric acid is present during growth of the biofilms. By the disk diffusion assay, disks were loaded with whole-biofilm lysate from cultures grown under conditions where citric acid was present. The results represented by the columns are averages from 3 biological repeats, and error bars are SEMs. The Bonferroni-Dunn method was used to determine significance, *, P < 0.05.
Efflux of antibiotics is a major mechanism of resistance for aminoglycosides, including tobramycin, in P. aeruginosa. Therefore, we were interested in testing whether a reduced level of efflux was contributing to the increased accumulation in the ΔtctED mutant by expressing the PA1875-1877 drug efflux system previously characterized by our lab (27) in both the PA14 and ΔtctED strains (producing the PA14efflux and ΔtctEDefflux strains). We reasoned that these strains would determine if heightened efflux could restore wild-type levels of tobramycin accumulation. We found that there was a moderate reduction of the clearance zone on E. coli lawns for both the PA14efflux and ΔtctEDefflux strains compared to that for the parental strains that did not express the efflux system. However, the trend of greater accumulation in the ΔtctEDefflux strain than in PA14efflux was still observed (Fig. 1).
Biofilm formation by the ΔtctED mutant is dysregulated in the presence of citric acid.
After observing that ΔtctED mutant biofilms have an increased accumulation of tobramycin in the presence of citric acid, we questioned whether there were differences in biofilm formation between the PA14 and ΔtctED strains using a crystal violet assay and microscopy.
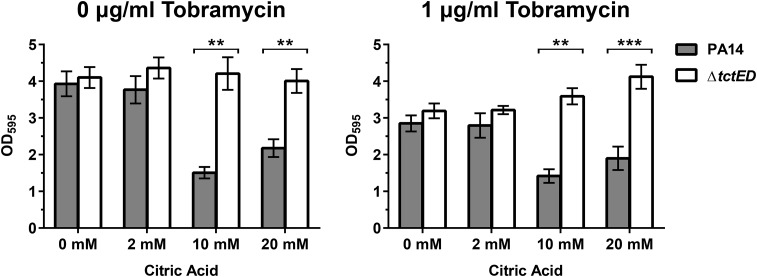
In M63 medium with 23 mM arginine as a carbon source and no added citric acid, the PA14 and ΔtctED strains displayed similar levels of biofilm formation, as determined by staining with crystal violet (Fig. 2). With the introduction of citric acid into the nutrient media, ΔtctED mutant biofilms maintained similar levels of staining regardless of the concentration of citric acid present in each growth medium (Fig. 2). Conversely, PA14 displayed an inverse relationship, where increasing concentrations of citric acid resulted in smaller amounts of biofilm formation. This observation was most apparent at the highest concentration of 20 mM citric acid. The decreased level of biofilm formation in PA14 suggests that biofilm formation is altered in the ΔtctED mutant. These experiments were repeated with subinhibitory levels of tobramycin to determine if there were any greater effects on the observed biofilm; however, the trends were the same as those found when no tobramycin was added (Fig. 2).
FIG 2.
Amount of biofilm formation compared between PA14 and the ΔtctED mutant in the presence of added citric acid. Conditions of no tobramycin and tobramycin at a subinhibitory concentration (1 μg/ml) were tested. The results represented by the columns represent the means from 3 biological replicates, and error bars are SEMs. The Bonferroni-Dunn method was used to determine significance. **, P < 0.01; ***, P < 0.001.
Flagella and type IV pili are important for early steps in biofilm formation in P. aeruginosa (32, 33). These are also crucial components for swimming as well as swarming motility. Since biofilm formation in the ΔtctED mutant strain was altered, we assayed the swarming and swimming motility phenotypes of the PA14 and ΔtctED strains. We observed no difference between the two strains, suggesting that the defect in biofilm formation occurs at later stages of biofilm formation (Fig. S1).
Swimming motility (A) and swarming motility (B) were no different in the ΔtctED strain relative to PA14. Plates were incubated for 18 h before images were captured. Images used are representative of those from 3 biological replicates. Swimming was performed on LB with 0.1% (wt/vol) agar. Swarming was performed as previously described by J. Overhage, M. Bains, M. D. Brazas, and R. E. W. Hancock (J Bacteriol 190:2671–2679, 2008). Download FIG S1, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further characterize the biofilm phenotype, we visualized PA14 and ΔtctED mutant biofilms by microscopy. Under conditions where no citric acid was added, PA14 biofilms were thick with macrocolonies present and covered the entire visual field. As increasing amounts of citric acid were added, the biofilms were flat and the surface was more sparsely populated in the presence of citric acid (Fig. 3A). To augment our analysis, we stained the biofilms that were formed in 12-well microtiter dishes with crystal violet. We observed that PA14 biofilm cultures decreased with increasing concentrations of citric acid (Fig. 3B). In contrast, the ΔtctED mutant maintained a high degree of biofilm formation in the presence of citric acid (Fig. 3A and B). In all concentrations of citric acid tested, the ΔtctED mutant maintained thick biofilms with visible dense macrocolonies (Fig. 3A). This biofilm growth appeared to increase with higher levels of citric acid, as observed by crystal violet staining (Fig. 3B). Typically, P. aeruginosa biofilms grow most densely at the air-liquid interface, observed as a line of crystal violet staining (Fig. 3B). This growth at the air-liquid interface was observed consistently in PA14. The ΔtctED mutant demonstrated higher levels of growth down beyond the air-liquid interface into the medium as the concentrations of citric acid increased (Fig. 3B). Taken together, these data suggest that the loss of tctED results in an inability to regulate biofilm formation in the presence of citric acid.
FIG 3.
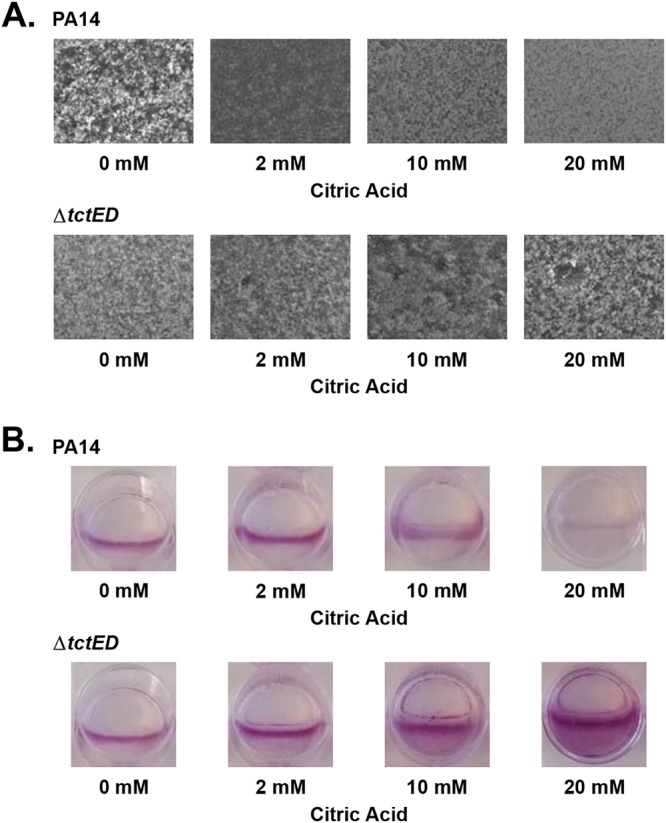
Phase-contrast microscopy (A) and 0.1% crystal violet staining (B) were used to assess PA14 and ΔtctED mutant biofilm cultures. Biofilms were grown statically in 12-well culture plates using the air-liquid interface assay described in Materials and Methods. Images are representative of those from 30 fields of view for each of 3 biological replicates. Crystal violet staining was performed on the same cultures used for microscopy after images were taken.
Addition of citric acid to our growth medium likely altered the pH. Therefore, we questioned whether the growth deficiency of the ΔtctED mutant was at all a result of changes in the pH of the medium. We measured the pH of M63 medium containing different concentrations of citric acid (0, 2, 10, and 20 mM), and the pH of M63 medium was adjusted accordingly (7.0, 6.8, 6.2, and 5.1, respectively). The growth of the ΔtctED strain showed that it displayed no difference from PA14 in its ability to grow in any of the pH-adjusted media (Fig. S2).
Growth assays confirmed that the ΔtctED strain (Δ) displayed no difference in ability to grow relative to PA14 (●) in varying pH. Cultures were grown for 16 h with shaking, taking measurements every 30 min. Data shown are the means for 3 biological replicates, and error bars represent the standard error of the mean (SEM). For points that have no error bar displayed, the SEM was too small to be visible on the graph. The absorbance at 600 nm (OD600) was measured and converted to the number of CFU per milliliter of volume based on measuring OD600 and plating cultures of PA14 and the ΔtctED mutant in a separate assay. Download FIG S2, EPS file, 0.2 MB (173.4KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ΔtctED strain has a growth deficiency in the presence of citric acid.
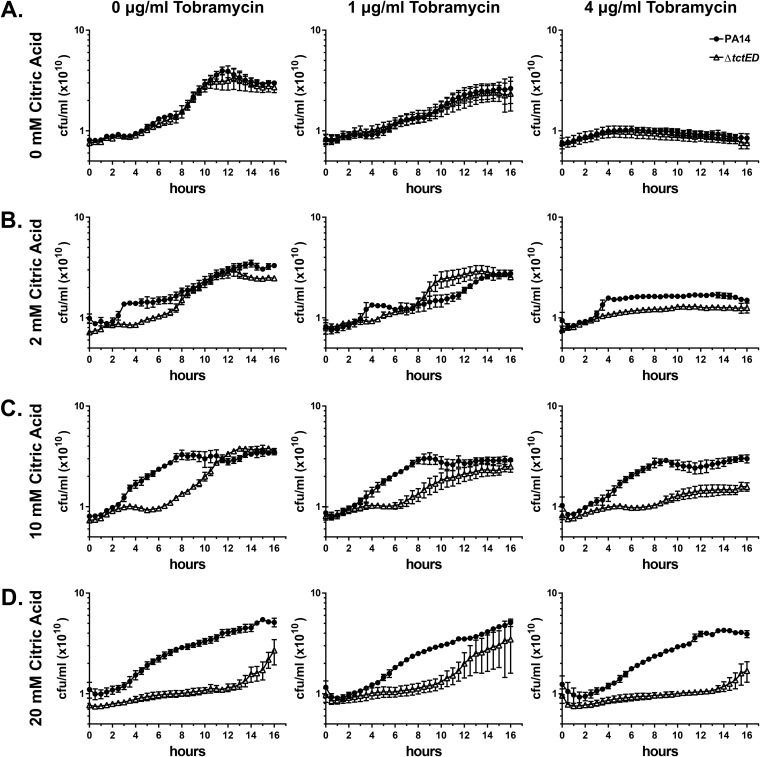
Since the ΔtctED strain displayed such a divergent biofilm phenotype in the presence of citric acid, we questioned whether there were any further growth phenotypes in citric acid. Previous research has found that high ratios of citrate metabolites relative to other carbon sources in nutrient media have an inhibitive effect on P. aeruginosa growth (28, 29). To test ΔtctED mutant growth in the presence of citric acid, we performed growth assays using M63 minimal medium containing arginine and various concentrations of both citric acid and tobramycin. When grown in higher concentrations of citric acid, reduced growth of the ΔtctED mutant relative to that of PA14 was observed (Fig. 4). In low and moderate concentrations (2 mM and 10 mM) of citric acid, there was a lag in growth of the ΔtctED strain, but it was able to achieve wild-type levels of culture densities by 16 h (Fig. 4B and C). The most extreme difference was observed at the highest concentration of citrate tested (20 mM), where the ΔtctED strain was severely reduced in its ability to grow compared to PA14 (Fig. 4D).
FIG 4.
The ΔtctED strain (Δ) exhibits a reduced ability to grow relative to PA14 (●) when citric acid is present in M63-arginine medium. Concentrations of 0 mM (A), 2 mM (B), 10 mM (C), and 20 mM (D) citric acid were tested with zero, subinhibitory (1 μg/ml), and inhibitory (4 μg/ml) concentrations of tobramycin. The data shown are the averages from 3 biological replicates, where error bars are the standard error of the mean (SEM). Where no error bars are visible, the SEM was less than what could be displayed on the graph.
Given that the ΔtctED strain displayed an aminoglycoside-specific susceptibility, it was of interest to determine if challenge of the cultures with tobramycin would exacerbate this ΔtctED mutant growth deficiency in the presence of citric acid. For growth assays with tobramycin, subinhibitory (1 μg/ml) and inhibitory (4 μg/ml) concentrations were used (25). Regardless of the concentrations of tobramycin present, the ΔtctED mutant had the same observable trend of reduced growth relative to that of PA14 (Fig. 4).
To confirm that the citric acid growth deficiency observed in the ΔtctED mutant was due to the loss of TctD-TctE, a complementation strain (the tctED+ strain) was struck out on gradient agar plates containing medium that had a continuous gradient of increasing citric acid concentrations in one direction. We were able to determine that expression of TctD-TctE from a pJB866 plasmid restored the ability of the strain to grow on citric acid as a sole carbon source (Fig. 5). Additionally, complementation with tctED also restored wild-type levels of resistance to tobramycin (Fig. 5).
FIG 5.
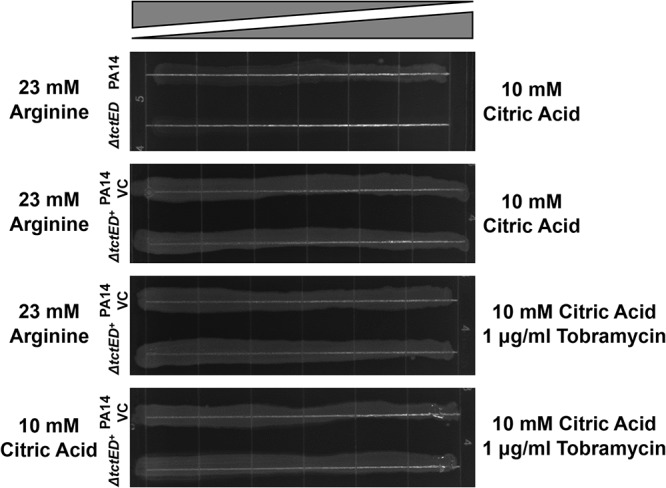
Complementation of the ΔtctED mutant with pJB866 harboring a wild-type copy of tctED (tctED+) restored growth on gradient agar plates. PA14 was transformed with pJB866 (PA14 VC [vector control]) to allow growth on selective medium.
The ΔtctED mutant growth deficiency is unique to citric acid as a carbon source.
To determine if the ΔtctED strain had a growth deficiency on different carbon nutrient sources, we utilized high-throughput phenotypic microarrays containing a variety of carbon sources (34). These arrays provide a measure of changes in absorbance from reduction of a dye occurring in the medium, which indirectly provides a measure for aerobic respiration and cell growth. These arrays can then provide insight into the ability of a bacterial strain to metabolize specific nutrients. The ΔtctED strain displayed a moderately reduced ability to grow on the carbon sources α-hydroxy-butyric acid (plate and well position, PM1 and E7, respectively) and d,l-carnitine (PM2A and H5) relative to PA14 (Table 2 and Fig. S3). Additionally, there was a moderate increase in the growth of the ΔtctED strain relative to that of PA14 on adenosine (PM1 and E12), α-keto-butyric acid (PM1 and D7), putrescine (PM2A and H8), and uridine (PM1 and D12) (Table 2 and Fig. S3). Most notably, the ΔtctED mutant was unable to grow on citric acid as the sole carbon source (PM1 and F2) (Fig. S3). This growth deficiency was confirmed with independent growth assays where either 2.2 mM glucose or 5 mM, 10 mM, or 20 mM citric acid was provided as the sole carbon source for the PA14 and ΔtctED strains. Under conditions where citric acid was the only carbon source, ΔtctED had no observable growth, while PA14 grew under all conditions (Fig. S4).
TABLE 2.
Assessment of ΔtctED mutant growth on various carbon sources through use of a phenotype microtiter assay
| Carbon source | ΔtctED mutant growth relative to PA14 |
|---|---|
| α-Hydroxy-butyric acid | − |
| α-Keto-butyric acid | + |
| Adenosine | + |
| Citric acid | − |
| d,l-Carnitine | − |
| Putrescine | + |
| Uridine | + |
Measures of oxidative phosphorylation performed on phenotypic microarray plates testing diverse carbon sources. PA14 (A) and ΔtctED (B) cells were grown for 24 h for these assays. Each curve represents increasing absorbance due to accumulation of the reduced form of a dye due to oxidative phosphorylation and is therefore an indirect measure of cell growth. In a single well of plates PM1 and PM2A, there is a unique carbon source available. Each plate is representative of 3 biological replicates. For each plate, well A1 contains no carbon source and is a negative control. Carbon sources present on the plate are available from the Biolog website (https://biolog.com/products-portfolio-overview/phenotype-microarrays-for-microbial-cells/). Download FIG S3, EPS file, 0.3 MB (342.6KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth assays measuring OD600 confirmed that the ΔtctED strain (Δ) could not use citric acid as a sole carbon source. The PA14 strain (●) was able to grow under all conditions tested. The carbon sources provided were 2.2 mM glucose (A) and 5 mM (B), 10 mM (C), and 20 mM (D) citric acid. Cultures were grown for 16 h with shaking, taking measurements every 30 min. The data shown are the means for 3 biological replicates, and error bars represent the standard error of the mean (SEM). For points that have no error bar displayed, the SEM was too small to be visible on the graph. Culture populations (number of CFU per milliliter) were calculated from the OD600 in a separate assay. Download FIG S4, EPS file, 0.1 MB (150.4KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Multiple genes that are involved in metabolism are dysregulated in the ΔtctED mutant.
DNA microarrays were used to better understand what pathways were dysregulated in the ΔtctED strain and possibly contributed to the observed growth deficiency and dysregulation of biofilm formation. It was found that under planktonic growth conditions 94 genes were dysregulated (>2-fold) and that under biofilm conditions 23 were significantly dysregulated (>2-fold) in the ΔtctED strain relative to the PA14 strain (Table 3). Of the 94 genes significantly dysregulated in planktonic cultures, 15 have roles in the metabolism in P. aeruginosa. In biofilm cultures, it was found that 8 of the 23 significantly dysregulated genes have a role in metabolic processes. Genes involved in metabolism that are significantly dysregulated are indicated by bold font in Table 3. Between planktonic and biofilm cultures, there was no overlap of the genes involved in metabolism, suggesting unique regulomes of TctD-TctE in each of these modes of growth. It was also notable that the transcriptional regulator genes cheY, pmrA, and dnr, as well as the two-component sensor gene phoQ, were significantly dysregulated in the ΔtctED strain between both the planktonic and biofilm conditions tested (Table 3).
TABLE 3.
Genes with significant changes in expression (>2-fold) in the ΔtctED mutant relative to PA14 grown in planktonic and biofilm cultures
| Culture and gene locus | Gene namea | Gene function | Fold change in expressionb |
|---|---|---|---|
| Planktonic | |||
| PA14_01310 | 4.80 | ||
| PA14_70010 | 4.73 | ||
| PA14_01320c | coIII | Cytochrome c oxidase, subunit III | 4.51 |
| PA14_60190 | clpB | ClpB protein | 3.81 |
| PA14_07660 | 3.77 | ||
| PA14_49130 | dctA | C4-Dicarboxylate transport protein | 3.63 |
| PA14_70040 | dadA | d-Amino acid dehydrogenase, small subunit | 3.06 |
| PA14_38840 | 2.88 | ||
| PA14_17550 | 2.82 | ||
| PA14_07680 | 2.78 | ||
| PA14_68840 | 2.75 | ||
| PA14_58690 | 2.68 | ||
| PA14_61600 | 2.68 | ||
| PA14_61610 | 2.60 | ||
| PA14_44480 | 2.57 | ||
| PA14_02520 | 2.54 | ||
| PA14_45620 | cheY | Two-component response regulator CheY | 2.48 |
| PA14_17700 | 2.48 | ||
| PA14_10730 | 2.44 | ||
| PA14_24770 | 2.38 | ||
| PA14_68260 | 2.36 | ||
| PA14_45610 | cheZ | Chemotaxis protein CheZ | 2.29 |
| PA14_16260 | 2.29 | ||
| PA14_15030 | leuA | 2-Isopropylmalate synthase | 2.28 |
| PA14_61840 | 2.28 | ||
| PA14_60520 | 2.26 | ||
| PA14_16680 | 2.19 | ||
| PA14_30830 | 2.17 | ||
| PA14_44860 | 2.15 | ||
| PA14_71720 | 2.14 | ||
| PA14_19870 | ldh | Leucine dehydrogenase | 2.08 |
| PA14_05060 | 2.06 | ||
| PA14_24760 | 2.05 | ||
| PA14_26590 | 2.04 | ||
| PA14_51850 | 2.01 | ||
| PA14_27210 | efp | Translation elongation factor P | −2.01 |
| PA14_66140 | −2.01 | ||
| PA14_17080 | pyrH | Uridylate kinase | −2.02 |
| PA14_64560 | −2.02 | ||
| PA14_65605 | parC | Topoisomerase IV subunit A | −2.03 |
| PA14_54370 | lepA | GTP-binding protein LepA | −2.04 |
| PA14_25490 | −2.04 | ||
| PA14_18350 | −2.04 | ||
| PA14_61770 | prs | Ribose-phosphate pyrophosphokinase | −2.06 |
| PA14_61780 | −2.06 | ||
| PA14_17060 | rpsB | 30S ribosomal protein S2 | −2.09 |
| PA14_25740 | tmk | Thymidylate kinase | −2.10 |
| PA14_51840 | −2.12 | ||
| PA14_62710 | pnp | Polyribonucleotide nucleotidyltransferase | −2.12 |
| PA14_21760 | capB | Cold acclimation protein B | −2.13 |
| PA14_65160 | −2.14 | ||
| PA14_14440 | valS | Valyl-tRNA synthetase | −2.15 |
| PA14_09030 | rpmD | 50S ribosomal protein L30 | −2.15 |
| PA14_66230 | waaG | UDP-glucose:(heptosyl) LPS alpha 1,3-glucosyltransferase | −2.15 |
| PA14_17675 | dgkA | Diacylglycerol kinase | −2.16 |
| PA14_09050 | secY | Secretion protein SecY | −2.18 |
| PA14_70640 | rubA1 | Rubredoxin 1 | −2.20 |
| PA14_73420 | rnpA | Ribonuclease P protein component | −2.20 |
| PA14_25760 | holB | DNA polymerase III, delta prime subunit | −2.21 |
| PA14_57580 | rpsI | 30S ribosomal protein S9 | −2.22 |
| PA14_09020 | rpsE | 30S ribosomal protein S5 | −2.23 |
| PA14_63120 | −2.24 | ||
| PA14_25730 | −2.25 | ||
| PA14_65170 | rpsR | 30S ribosomal protein S18 | −2.25 |
| PA14_58130 | mreC | Rod shape-determining protein MreC | −2.26 |
| PA14_41250 | tig | Trigger factor | −2.27 |
| PA14_08860 | rplD | 50S ribosomal protein L4 | −2.30 |
| PA14_12550 | −2.33 | ||
| PA14_09000 | rplF | 50S ribosomal protein L6 | −2.36 |
| PA14_17220 | lpxB | Lipid A-disaccharide synthase | −2.40 |
| PA14_25630 | rpmF | 50S ribosomal protein L32 | −2.41 |
| PA14_09010 | rplR | 50S ribosomal protein L18 | −2.44 |
| PA14_00580 | −2.48 | ||
| PA14_07560 | rpsU | 30S ribosomal protein S21 | −2.48 |
| PA14_63110 | −2.48 | ||
| PA14_14610 | −2.49 | ||
| PA14_63150 | pmrA | Two-component regulator system response regulator | −2.49 |
| PA14_62830 | tpiA | Triosephosphate isomerase | −2.53 |
| PA14_61820 | −2.55 | ||
| PA14_09040 | rplO | 50S ribosomal protein L15 | −2.58 |
| PA14_65180 | rpsF | 30S ribosomal protein S6 | −2.58 |
| PA14_15980 | rimM | 16S rRNA processing protein | −2.59 |
| PA14_73410 | −2.62 | ||
| PA14_58570 | −2.68 | ||
| PA14_25270 | aroP1 | Aromatic amino acid transport protein AroP1 | −2.76 |
| PA14_44060 | sdhC | Succinate dehydrogenase (C subunit) | −2.80 |
| PA14_58120 | mreD | Rod shape-determining protein MreD | −2.91 |
| PA14_67560 | typA | Regulatory protein TypA | −2.93 |
| PA14_52340 | −3.04 | ||
| PA14_27370 | −3.52 | ||
| PA14_15970 | rpsP | 30S ribosomal protein S16 | −3.55 |
| PA14_70180 | rpmG | 50S ribosomal protein L33 | −3.59 |
| PA14_15990 | trmD | tRNA (guanine-N1)-methyltransferase | −3.60 |
| PA14_39060 | −4.14 | ||
| Biofilm | |||
| PA14_72260 | 5.54 | ||
| PA14_02520 | 5.26 | ||
| PA14_42860 | 3.51 | ||
| PA14_46900 | 3.41 | ||
| PA14_56540 | 2.94 | ||
| PA14_55750 | 2.92 | ||
| PA14_22350 | 2.52 | ||
| PA14_56690 | 2.47 | ||
| PA14_44470 | hemN | Oxygen-independent coproporphyrinogen III oxidase | 2.46 |
| PA14_60700 | ccpR | Cytochrome c551 peroxidase precursor | 2.44 |
| PA14_49170 | phoQ | Two-component sensor PhoQ | 2.11 |
| PA14_52800 | acsA | Acetyl coenzyme A synthetase | 2.08 |
| PA14_06870 | dnr | Transcriptional regulator Dnr | 2.06 |
| PA14_20890 | rfaD | ADP-l-glycero-d-mannoheptose 6-epimerase | 2.02 |
| PA14_18360 | 2.02 | ||
| PA14_09980 | dkgB | 2,5-Diketo-d-gluconate reductase B | −2.06 |
| PA14_47420 | −2.13 | ||
| PA14_55590 | −2.19 | ||
| PA14_57960 | ptsN | Nitrogen regulatory IIA protein | −2.21 |
| PA14_00990 | −2.22 | ||
| PA14_56980 | −2.23 | ||
| PA14_52070 | −2.24 | ||
| PA14_10170 | fepB | Ferrienterobactin-binding periplasmic protein precursor | −2.70 |
Where no gene name is provided, it is an uncharacterized gene with a conserved hypothetical gene product.
Values are the means for 2 biological replicates.
Boldface describes genes involved in metabolism.
DISCUSSION
The aim of this study was to further characterize the role of the TCS TctD-TctE in P. aeruginosa. We were able to build on previous findings and further characterize the aminoglycoside susceptibility of the ΔtctED deletion strain in the presence of citric acid. In this work, we also made the novel finding of a growth deficiency and biofilm dysregulation in the presence of citric acid for the ΔtctED strain.
First, to further characterize previous observations of an increased susceptibility to the aminoglycosides tobramycin and gentamicin in the ΔtctED mutant, we performed assays for MBC-P and MBC-B with citric acid included in the nutrient media. We found that there was a 4-fold increase in ΔtctED mutant susceptibility to tobramycin and gentamicin in MBC-B assays, similar to what was observed previously (Table 1). Interestingly, there was also a 2-fold increase in ΔtctED mutant susceptibility to these antibiotics in MBC-P assays that included 10 mM citric acid. Previous findings revealed no difference in MBCs between PA14 and the ΔtctED mutant in the absence of citric acid (25). It is likely that this small increase in planktonic susceptibility observed in the ΔtctED mutant was due to the added stress of a growth deficiency in the presence of citric acid that we observed here (Fig. 4) and that is discussed below. It was also noted here that there was a 2-fold greater MBC-P of the quinolone antibiotic nalidixic acid (see Table S1 in the supplemental material). The ΔtctED mutant had an MBC-P of 1,024 μg/ml, whereas an MBC-P of 512 μg/ml was observed in PA14. These MBC-Ps are higher than those of other antibiotics, such as tobramycin, gentamicin, and ciprofloxacin (32, 64, and 4 μg/ml in the ΔtctED mutant, respectively). It is possible that the differences seen with nalidixic acid are attributable to the fact that there is already a high intrinsic resistance to nalidixic acid in P. aeruginosa.
Further investigation of aminoglycoside susceptibility in the ΔtctED mutant led us to determine that there was a moderate increase in the accumulation of tobramycin in ΔtctED mutant biofilms grown in the presence of 10 mM citric acid (Fig. 1). A major mechanism of P. aeruginosa resistance to aminoglycosides is through efflux of the compound (35–37). We expressed an efflux system that contributes to biofilm-specific aminoglycoside resistance (27) in the PA14 and ΔtctED strains. Expression of this efflux system reduced the zones of clearance in E. coli and thereby indicated reduced tobramycin accumulation in biofilms of all P. aeruginosa strains (Fig. 1). However, there was still a moderately greater accumulation in the ΔtcED mutant than in PA14 (Fig. 1). This result demonstrates that efflux does not contribute to the tobramycin susceptibility observed in ΔtctED mutant biofilms.
Because the aminoglycoside susceptibility in the ΔtctED mutant was biofilm specific, we decided to further investigate biofilm formation in the presence of citric acid. Our analysis indicated that the ΔtctED mutant is unable to respond appropriately to the presence of citric acid. When biofilm cultures were grown, PA14 responded to increasing concentrations of citric acid with a reduction in biofilm formation, whereas the ΔtctED strain continued to form thick biofilms regardless of the increasing concentrations of citric acid (Fig. 2 and 3).
As stated above, ΔtctED displayed a consistently high biofilm mass for all concentrations of citric acid present in the medium tested. However, there were decreases in planktonic ΔtctED mutant culture growth (Fig. 4) with increases in the concentration of citric acid in the medium. It was also observed that the ΔtctED mutant could not grow if citric acid was provided as the only carbon source (Fig. S3 and S4). Through the use of phenotypic assays which measure oxidation levels in the medium by oxidative phosphorylation, therefore indicating the metabolic activity of the cultures, it was found that the ΔtctED mutant was uniquely inactive in citric acid-containing medium and that no other carbon source gave such low levels of activity relative to that of PA14 (Table 2 and Fig. S3). Taken together, these data suggest that the loss of TctD-TctE in the ΔtctED mutant has an effect on planktonic and biofilm growth due to an inability to properly sense and respond to citric acid present in the environment. Further support for this comes from the observation that complementing the ΔtctED mutant with a plasmid expressing TctD-TctE restored the ability to grow on citric acid as the sole carbon source, as seen in gradient plants (Fig. 5). It is known that high relative concentrations of citrate metabolites can be inhibitive of growth in P. aeruginosa (28, 29). It is therefore likely that the loss of tctED results in the dysregulation of sensation and the response to citric acid in the medium, leading to an inhibition of growth.
Other modes of growth were otherwise unaffected in the ΔtctED mutant when grown on regular M63 medium lacking citric acid. Through the use of swarming and swimming assays, we determined that there was no difference in motility between the PA14 and ΔtctED strains (Fig. S1). Both flagella and pili, which are important for motility, also have roles in forming biofilm structures in P. aeruginosa (32, 33). The lack of any motility defect in the ΔtctED mutant adds support to the idea that the loss of the TCS results in a dysregulation of signaling in biofilm formation and growth.
The heightened accumulation of tobramycin in the ΔtctED mutant may also be explained by a lack of response to citric acid in the medium. With the ΔtctED mutant maintaining higher levels of adherent biofilms than PA14, the accumulation of tobramycin may be at least in part due to a larger amount of biofilm cells and extracellular matrix in the ΔtctED mutant. With the greater accumulation of tobramycin in the matrix, the ΔtctED mutant would then acquire increasing localized concentrations of tobramycin and increased killing. It has been found previously that small molecules, such as aminoglycosides, readily permeate into the extracellular matrix of biofilms (38–40). It is possible that the accumulation observed is at least in part due to more tobramycin being retained in the matrix and biofilm cells.
While the ΔtctED mutant biofilm cultures displayed higher MBC-Bs (Table 1) and greater accumulation of tobramycin in medium containing citric acid (Fig. 1), it should be noted that when planktonic cultures of the ΔtctED mutant were challenged with tobramycin, there was only a slight change in the susceptibility phenotype (Fig. 4). This fits with previous data (25) and findings obtained in this study (Table 1) that there are minimal differences in ΔtctED planktonic cultures and that the greatest increases in susceptibility relative to that of PA14 are observed in biofilm cultures. This suggests that TctD-TctE activity has greater involvement in P. aeruginosa when it is developing as a biofilm than when it is developing as a planktonic culture. In biofilms, there are subpopulations of cells with various levels of metabolic activity (3, 39, 41). Because of the localized higher density of cells, there can be limitations of nutrients in those locations. It is reasonable that systems such as TctD-TctE would be more heavily relied upon in biofilms.
It is likely that other factors contribute to the role that TctD-TctE has in aminoglycoside susceptibility, considering that it is a TCS ultimately regulating transcriptional responses. This was evident based on genome-wide expression data from this study, which found that a wide range of genes were dysregulated in the ΔtctED mutant. Additionally, phenotypic microarray data suggested that the role that TctD-TctE has in P. aeruginosa is complex, affecting multiple different regulatory pathways between citrate uptake. A large portion of genes (46 of 94 in planktonic cultures and 15 of 23 in biofilms) that were dysregulated in the ΔtctED mutant are of unknown function in P. aeruginosa (Table 3). Of the genes that do have characterized functional roles, a high number of genes that are involved in metabolism through electron transport, catabolism, anabolism, or transport were represented (15 of 48 in planktonic cultures and all those in biofilms). Additionally, other members of two-component systems were represented, such as cheY, phoQ, and pmrA (Table 3). Many genes for ribosomal subunits, DNA replication, and cell shape were represented, but these might have been dysregulated indirectly as a result of reduced growth in the ΔtctED mutant.
Phenotypic microarray analysis found that the ΔtctED mutant was completely defective in its ability to grow with citric acid as a sole carbon source. Interestingly, the ΔtctED mutant had moderately increased growth relative to PA14 in medium with carbon sources of α-keto-butyric acid, adenosine, putrescine, and uridine (Table 2) and moderately decreased growth in α-hydroxy-butyric acid and d,l-carnitine. It has recently been shown that the metabolic activity of bacteria alone can have a significant effect on susceptibility to antibiotic treatment (42, 43). It is possible that metabolic dysregulation in general also has a contribution to the aminoglycoside susceptibility observed in the ΔtctED mutant.
In conclusion, this study has further resolved some contributors to the role that TctD-TctE has in resistance and tolerance to aminoglycosides in P. aeruginosa through finding that the loss of TctD-TctE results in increased aminoglycoside susceptibility in the biofilm when citric acid is present in the environment. Furthermore, we were able to characterize the biofilm phenotype of consistent biofilm mass as well as a growth deficiency due to the loss of TctD-TctE in P. aeruginosa when citric acid is present. This work emphasizes the importance of how TCSs responsible for sensing and responding to environmental cues, even those for metabolites, can have significant impacts on biofilm development and when cells encounter dynamic and changing environmental conditions, which is important for developing improved methods of treating infections caused by bacterial pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed Table 4. All cultures except those specified were grown overnight at 37°C with constant shaking in LB from single colonies grown on LB agar plates. For the experiments, overnight cultures were subcultured into M63 minimal medium [22 mM KH2PO4, 40 mM K2HPO4, 15 mM (NH4)2SO4], using 0, 2, 10, and 20 mM citric acid and 23 mM arginine as carbon sources. For strains carrying pJB866 constructs, the medium was supplemented with 2 mM m-toluic acid for promoter induction and 10 μg/ml tetracycline for maintenance of plasmids. The only exception for the growth medium under experimental conditions was for growth assays conducted using Biolog plates on an OmniLog system, where the buffer and nutrient sources provided by the manufacturer of PM1 and PM2A plates were used (https://biolog.com/products-portfolio-overview/phenotype-microarrays-for-microbial-cells/).
TABLE 4.
Strains and plasmids used in this study
| Strain | Description | Reference or source |
|---|---|---|
| Strains | ||
| PA14 | P. aeruginosa wild-type strain | 25 |
| ΔtctED | PA14 containing a chromosomal deletion of tctED | 25 |
| PA14 VC | PA14 complemented with pJB866 | 25 |
| tctED+ | ΔtctED mutant complemented with pJB866::tctED | 25 |
| PA14efflux | PA14 complemented with pJB866::PA1875-1877 | This study |
| ΔtctEDefflux | ΔtctED mutant complemented with pJB866::PA1875-1877 | This study |
| Plasmids | ||
| pJB866 | Pm m-toluic acid-inducible promoter; tetracycline resistant | 44 |
| pJB866::tctED | pJB866 carrying a Pm-regulated insertion of tctED | 25 |
| pJB866::PA1875-1877 | pJB866 carrying PA1875-1877 drug efflux system genes | 27 |
Biofilms grown for microscopy were cultured using an air-liquid interface assay described previously (25). Overnight cultures were subcultured 1/100 into fresh medium in 12-well plates. Cultures were incubated statically for 24 h at 37°C with the plates propped up at an angle of ∼30 to 45°. Before microscopy, the media with planktonic cultures were removed and the wells were washed once with sterile M63 buffer. A volume of 200 μl of fresh M63 buffer was added to the wells to prevent the biofilms from drying out while performing microscopy, and the culture plate was put directly on the microscope stage for analysis.
The absorbance, or the optical density at a 600-nm wavelength (OD600), was measured to determine culture growth. The number of CFU per milliliter of volume for growth curves was calculated by measuring the OD600 of separate PA14 and ΔtctED mutant cultures, plating out dilutions, and performing colony counts to determine the number of CFU per milliliter. The PA14 and ΔtctED strains displayed no difference in the relationship of the OD600 measurements to the plated colony counts. Based on the medium and conditions used in this study, an OD600 of 1.0 represents a culture population of 6 × 1010 CFU/ml for P. aeruginosa. This value was used to calculate all values of the number of CFU per milliliter from OD600 measurements.
MBC assays.
Assays for determination of minimal bactericidal concentrations (MBCs) in biofilms (MBC-Bs) and in planktonic cultures (MBC-Ps) were utilized to assess susceptibility to antibiotics (25–27). An aliquot of overnight cultures was diluted (1/50) into fresh M63 medium with arginine as described above and then used for final inoculation of antibiotic-containing medium. For the MBC assays, a final concentration of 10 mM citric acid was added to the medium. To prepare MBC-B assay mixtures, diluted cultures were grown statically for 24 h at 37°C to form mature biofilms. After 24 h, the planktonic cultures and medium were removed and fresh medium containing antibiotics was added. Twofold serial dilutions of antibiotics ranged from 2.5 to 160 μg/ml for ciprofloxacin, 12.5 to 800 μg/ml for gentamicin, and 6.25 to 400 μg/ml for tobramycin. The preformed biofilms were incubated in the presence of antibiotics for 24 h at 37°C. For the MBC-P assays, diluted cultures were treated with antibiotics and incubated for 24 h at 37°C. Antibiotic concentration ranges were 0.5 to 32 μg/ml for ciprofloxacin, 2 to 128 μg/ml for gentamicin, and 1 to 64 μg/ml for tobramycin. After incubation, the cultures used for both MBC-B and MBC-P determination were spotted onto LB agar plates and grown overnight at 37°C. The concentration in the culture from which spots that displayed no growth upon subculture were sampled was taken as the MBC.
Tobramycin accumulation assays.
Tobramycin accumulation assays were performed as previously described (27). Briefly, static biofilms of the PA14 and ΔtctED strains were grown in 12-well microtiter plates for 24 h with or without 10 mM citric acid supplemented in the medium. Biofilms were treated with 200 μg/ml tobramycin for 8 h and subsequently rinsed with M63 buffer. The biofilms were treated with 0.1 M glycine (pH 3) at 37°C to lyse the biofilm cells. After 16 h, the mixture of lysed cells and glycine buffer was evaporated to dryness, resuspended in sterile water, and absorbed into sterile Whatman paper disks. The disks were then placed on agar plates spread with E. coli DH5α. The plates were incubated at 37°C for 16 h, and the zone of clearing around the disk was used as an indication of the amount of tobramycin in the biofilm lysate.
Crystal violet staining.
Assessment of biofilm mass was done by staining with 0.1% (wt/vol) crystal violet. Static biofilms in the respective media were grown for 24 h in 96-well plates. The cell population was first taken by reading the OD600 and converting the value to the number of CFU per milliliter. Planktonic cultures were then removed, and 100 μl of 0.1% crystal violet were added to the wells and allowed to stain for 20 min. The stain was then removed, the wells were washed, and the crystal violet absorbed by the biofilms was solubilized with 70% ethanol. The OD595 were then read as an assessment of the amount of biofilm mass adherent within the wells.
Microscopy.
A Leica DMI6000 B inverted microscope with the companion Leica Application Suite software provided (v1.5.1, build 869) was used. Images are representative of those from 3 biological replicates, for each of which 30 fields of view were assessed.
Phenotype microarrays testing various carbon sources.
Phenotypic assessment of growth for a collection of carbon sources was performed using Biolog MicroPlates. Overnight cultures were diluted to a McFarland standard of 0.25 in buffer with dye added; both the buffer and the dye were provided by Biolog, Inc. Plates PM1 and PM2A were utilized to assess the growth on various carbon sources. Assays were performed on a GEN III OmniLog ID system, where the plates were incubated for 24 h and readings were taken every 15 min. Three biological replicates were performed.
Gradient agar plates.
Agar plates containing continual transitions of one nutrient medium to another were prepared as previously described (38). Culture plates were set at an angle as one nutrient medium was poured, allowed to set, and laid flat before the other nutrient medium was poured on top, generating a gradient of one medium to another. Overnight cultures of P. aeruginosa strains were then dragged along the gradient using an inoculating loop.
RNA isolation.
RNA was isolated by lysis of whole cells grown under planktonic and biofilm conditions. Planktonic cultures were grown by subculturing overnight cultures into fresh M63 medium containing arginine as the sole carbon source. The cultures were then grown at 37°C with constant shaking for 4 to 5 h until reaching an OD600 approaching 0.5, before pelleting and lysing the cells. Biofilm cultures were grown by spotting 48 5-μl aliquots of an overnight culture onto M63-arginine agar plates. The plates were first grown for 24 h at 37°C and then grown for another 24 h at room temperature before harvesting the biofilm cultures. RNA was isolated from the cultures by first lysing the pelleted cultures using the TRIzol reagent. RNA was purified using a PureLink RNA minikit according to the instructions of Thermo Fisher Scientific, Inc. RNA samples were cleared of DNA through the use of DNase digestion, and samples were checked for DNA contamination by PCR.
Microarray analysis.
Microarrays were performed using an Affymetrix GeneChip system for the P. aeruginosa PAO1 annotated genome. Samples were sent to the Genome Québec and Innovation Centre at McGill University, Montréal, QC, Canada, for quality control testing and performing the microarrays. Raw data were received as .cel files and analyzed in-house. Expression Console software, build 1.3.1.187, and the annotated P. aeruginosa PAO1 library Pae_G1a were acquired from the Affymetrix website. While PA14 background strains were used in this study, the close genetic homology of the core genome between P. aeruginosa strains made it sufficient to use PAO1 DNA microarray chips and the Pae_G1a library. Data normalization was performed using the robust multiarray average method, and the housekeeping gene rpoD was selected for expression normalization. Gene expression changes are represented as the mean fold change in expression in the ΔtctED mutant relative to that in PA14 under planktonic or biofilm conditions for 2 biological replicates of each condition.
Accession number(s).
The data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE114431.
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Sciences and Engineering Research Council and Cystic Fibrosis Canada held by T.-F.M. P.K.T. was supported by an Ontario Queen Elizabeth II Graduate Scholarship in Science and Technology and a University of Ottawa Excellence Scholarship.
We are grateful to Karine Hébert and Franco Pagotto from the Bureau of Microbial Hazards of the Food Directorate at Health Canada for use of and assistance with the OmniLog apparatus. We also thank Stephanie Halmhofer for her comments and editing, Sandeep Tamber for providing insight on the manuscript, and Clayton Hall for assistance with the isolation of RNA samples.
REFERENCES
- 1.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 2.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. 2011. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J 5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberhardt MA, Puchałka J, Fryer KE, Martins dos Santos VAP, Papin JA. 2008. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol 190:2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock REW. 2012. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother 56:6212–6222. doi: 10.1128/AAC.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gooderham WJ, Hancock REW. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 9.Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol 178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alm E, Huang K, Arkin A. 2006. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S, Hirakawa H, Muta S, Kuhara S. 2010. Identification and classification of a two-component system based on domain structures in bacteria and differences in domain structure between Gram-positive and Gram-negative bacteria. Biosci Biotechnol Biochem 74:716–720. doi: 10.1271/bbb.90746. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigue A, Quentin Y, Lazdunski A, Méjean V, Foglino M. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol 8:498–504. doi: 10.1016/S0966-842X(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 13.Salazar ME, Laub MT. 2015. Temporal and evolutionary dynamics of two-component signaling pathways. Curr Opin Microbiol 24:7–14. doi: 10.1016/j.mib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 15.Jacob-Dubuisson F, Mechaly A, Betton JM, Antoine R. 2018. Structural insights into the signalling mechanisms of two-component systems. Nat Rev Microbiol 16:585–593. doi: 10.1038/s41579-018-0055-7. [DOI] [PubMed] [Google Scholar]
- 16.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Heeb S, Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol Plant Microbe Interact 14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Lu CD. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol 189:5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tatke G, Kumari H, Silva-Herzog E, Ramirez L, Mathee K. 2015. Pseudomonas aeruginosa MifS-MifR two-component system is specific for α-ketoglutarate utilization. PLoS One 10:e0129629. doi: 10.1371/journal.pone.0129629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamorano L, Moyà B, Juan C, Mulet X, Blázquez J, Oliver A. 2014. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactams, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother 58:5084–5095. doi: 10.1128/AAC.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somers JM, Kay WW. 1983. Genetic fine structure of the tricarboxylate transport (tct) locus of Salmonella typhimurium. Mol Gen Genet 190:20–26. doi: 10.1007/BF00330319. [DOI] [PubMed] [Google Scholar]
- 23.Widenhorn KA, Somers JM, Kay WW. 1989. Genetic regulation of the tricarboxylate transport operon (tctI) of Salmonella typhimurium. J Bacteriol 171:4436–4441. doi: 10.1128/jb.171.8.4436-4441.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamber S, Maier E, Benz R, Hancock REW. 2007. Characterization of OpdH, a Pseudomonas aeruginosa porin involved in the uptake of tricarboxylates. J Bacteriol 189:929–939. doi: 10.1128/JB.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Fritsch M, Hammond L, Landreville R, Slatculescu C, Colavita A, Mah TF. 2013. Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One 8:e61625. doi: 10.1371/journal.pone.0061625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Mah TF. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng FM, Dawes EA. 1973. Chemostat studies on the regulation of glucose metabolism in Pseudomonas aeruginosa by citrate. Biochem J 132:129–140. doi: 10.1042/bj1320129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiting PH, Midgley M, Dawes EA. 1976. The regulation of transport of glucose, gluconate and 2-oxogluconate and of glucose catabolism in Pseudomonas aeruginosa. Biochem J 154:659–668. doi: 10.1042/bj1540659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, supplement M100, 28th ed, p 35 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 33.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 34.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hocquet D, Vogne C, El Garch F, Vejux A, Gotoh N, Lee A, Lomovskaya O, Plésiat P. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 47:1371–1375. doi: 10.1128/AAC.47.4.1371-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin Microbiol Infect 10:12–26. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 37.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart PS. 2003. Diffusion in biofilms. J Bacteriol 185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 40.Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, Andrews-Polymenis HL. 2014. Identification of novel factors involved in modulating motility of Salmonella enterica serotype Typhimurium. PLoS One 9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. 2012. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol 194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meylan S, Porter CBM, Yang JH, Belenky P, Gutierrez A, Lobritz MA, Park J, Kim SH, Moskowitz SM, Collins JJ. 2017. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol 24:195–206. doi: 10.1016/j.chembiol.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng B, Su Y, Li H, Han Y, Guo C, Tian Y, Peng X. 2015. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab 21:249–261. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S. 1997. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid 38:35–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimal bactericidal concentrations (MBCs) for select antibiotics in media including 10 mM citric acid. Download Table S1, DOCX file, 0.02 MB (18.7KB, docx) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Swimming motility (A) and swarming motility (B) were no different in the ΔtctED strain relative to PA14. Plates were incubated for 18 h before images were captured. Images used are representative of those from 3 biological replicates. Swimming was performed on LB with 0.1% (wt/vol) agar. Swarming was performed as previously described by J. Overhage, M. Bains, M. D. Brazas, and R. E. W. Hancock (J Bacteriol 190:2671–2679, 2008). Download FIG S1, EPS file, 1.4 MB (1.4MB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth assays confirmed that the ΔtctED strain (Δ) displayed no difference in ability to grow relative to PA14 (●) in varying pH. Cultures were grown for 16 h with shaking, taking measurements every 30 min. Data shown are the means for 3 biological replicates, and error bars represent the standard error of the mean (SEM). For points that have no error bar displayed, the SEM was too small to be visible on the graph. The absorbance at 600 nm (OD600) was measured and converted to the number of CFU per milliliter of volume based on measuring OD600 and plating cultures of PA14 and the ΔtctED mutant in a separate assay. Download FIG S2, EPS file, 0.2 MB (173.4KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Measures of oxidative phosphorylation performed on phenotypic microarray plates testing diverse carbon sources. PA14 (A) and ΔtctED (B) cells were grown for 24 h for these assays. Each curve represents increasing absorbance due to accumulation of the reduced form of a dye due to oxidative phosphorylation and is therefore an indirect measure of cell growth. In a single well of plates PM1 and PM2A, there is a unique carbon source available. Each plate is representative of 3 biological replicates. For each plate, well A1 contains no carbon source and is a negative control. Carbon sources present on the plate are available from the Biolog website (https://biolog.com/products-portfolio-overview/phenotype-microarrays-for-microbial-cells/). Download FIG S3, EPS file, 0.3 MB (342.6KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth assays measuring OD600 confirmed that the ΔtctED strain (Δ) could not use citric acid as a sole carbon source. The PA14 strain (●) was able to grow under all conditions tested. The carbon sources provided were 2.2 mM glucose (A) and 5 mM (B), 10 mM (C), and 20 mM (D) citric acid. Cultures were grown for 16 h with shaking, taking measurements every 30 min. The data shown are the means for 3 biological replicates, and error bars represent the standard error of the mean (SEM). For points that have no error bar displayed, the SEM was too small to be visible on the graph. Culture populations (number of CFU per milliliter) were calculated from the OD600 in a separate assay. Download FIG S4, EPS file, 0.1 MB (150.4KB, eps) .
Copyright © 2019 Taylor et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.