Abstract
Inherent or acquired resistance to chemotherapeutic drugs is still an obstacle for the treatment of multiple myeloma (MM). MicroRNA dysregulation is related to the development of chemoresistance in cancers. However, its role in chemoresistance of MM is largely unknown. Here we demonstrated that miR-221/222 were upregulated in plasma cells from patients with MM, especially those with relapsed or refractory disease. Moreover, expression levels of miR-221/222 were inversely correlated with dexamethasone (Dex) sensitivity of human MM cell lines. Importantly, we found that Dex induced pro-death autophagy in MM cells and the inhibition of autophagy significantly decreased Dex-induced cell death. Mechanistically, autophagy-related gene 12 (ATG12) was identified as a novel target gene of miR-221/222, and miR-221/222 overexpression inhibited autophagy by directly targeting ATG12 and the p27kip (p27)-mammalian target of rapamycin (mTOR) pathway. Indeed, Dex treatment decreased the expression of miR-221/222, thereby activating the ATG12/p27-mTOR autophagy-regulatory axis and inducing cell death in Dex-sensitive MM cells. Furthermore, both in vitro and in vivo results showed that the inhibitions of miR-221/222 increased the expression of ATG12 and p27 and functionally induced extended autophagy and cell death of MM cells. In conclusion, our findings demonstrated the crucial role of the miR-221/222-ATG12/p27-mTOR autophagy-regulatory axis in Dex resistance of MM, and they suggest potential prediction and treatment strategies for glucocorticoid resistance.
Keywords: miR-221/222, autophagy, dexamethasone, drug resistance, multiple myeloma
The mechanisms of dexamethasone (Dex) resistance in multiple myeloma (MM) remain largely unknown. Hu and colleagues demonstrate that Dex induces pro-death autophagy in MM. Aberrant upregulation of miR-221/222 inhibits autophagy by directly targeting ATG12 and the p27-mTOR pathway, thereby promoting Dex resistance of MM cells.
Introduction
Multiple myeloma (MM) is the second most common hematological malignancy characterized by the neoplastic proliferation of plasma cells (PCs).1 It accounts for approximately 13% of all hematological malignancies, and it is still considered to be incurable.2 Although the introduction of novel drugs has significantly improved the outcome of MM patients, most patients eventually relapse or become refractory to current treatments.3, 4 Dexamethasone (Dex) is the most widely used chemotherapeutic drug in the combination regimens of MM treatment.1 Inherent or acquired resistance to Dex is broadly associated with poor prognosis in MM.5 Although mechanisms of Dex resistance, such as defective glucocorticoid receptor (GR) expression, deregulation of pro-apoptotic and anti-apoptotic proteins, and interactions with the kinome, have been proposed,6 the key determinants of Dex resistance remain largely unclear.
Nowadays, microRNAs are widely studied in MM for their important role in MM development, progression, and aggressiveness.7 microRNAs are a class of short non-coding RNAs that inhibit the expression of target genes through inducing either mRNA degradation or translational repression.8 They can function as either oncogenes or tumor suppressors depending on the genes they target.9 Many microRNAs are aberrantly expressed in MM,7, 10, 11 including miR-221/222, which have been reported to act as oncogenes in many cancer types.12, 13 Recently, accumulating evidence has shown that miR-221/222 are involved in the development of chemoresistance in a variety of cancers. Inhibition of miR-221/222 has a potential role in overcoming resistance of several chemotherapeutic drugs, including fulvestrant, tamoxifen, TRAIL, and melphalan.13 However, the role of miR-221/222 in the chemoresistance of MM is largely unknown.
Autophagy is an evolutionarily conserved mechanism by which protein aggregates and damaged organelles are eliminated through the lysosomal degradation pathway.14 This process regulates cellular homeostasis, recycles damaged organelles, overcomes nutrient deprivation, and relieves metabolic stress. However, excessive autophagy may lead to cell death.15 In MM, autophagy induced by chemotherapeutic drugs can either play a cytoprotective role, through relieving cellular stress caused by toxic insult, or play a cytotoxic role, through triggering autophagy-related cell death.16 Moreover, previous studies have shown that Dex-induced cell death in lymphoid leukemia is mediated through the initiation of autophagy. Induction of autophagy is required for acute lymphoblastic leukemia cells to overcome glucocorticoid resistance.17, 18, 19 Here we evaluated the role of miR-221/222 and autophagy in Dex resistance of MM, and we investigated the cross talk between miR-221/222 and autophagy. Our results demonstrated for the first time that miR-221/222 could inhibit Dex-induced cell death in MM through the inhibition of autophagy, by directly targeting autophagy-related gene 12 (ATG12) and the p27-mammalian target of rapamycin (mTOR) pathway.
Results
miR-221/222 Regulate the Sensitivity of MM Cells to Dex
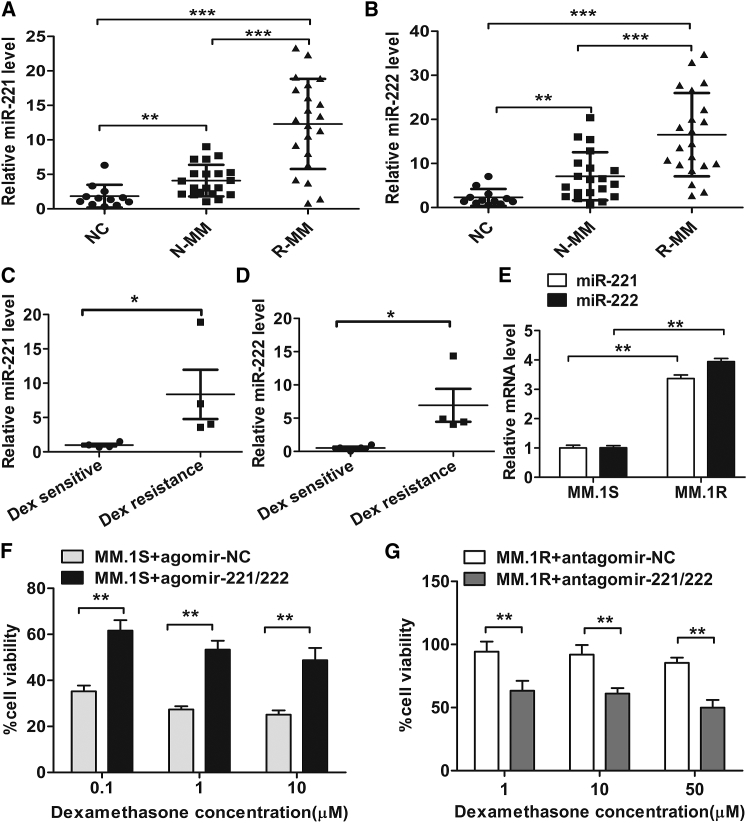
To investigate the role of miR-221 and miR-222 in the chemoresistance of MM cells, we first examined their expression levels in PCs from 12 healthy donors, 20 patients with newly diagnosed MM, and 21 patients with relapsed or refractory MM (Table S1). Compared with healthy controls, the expression levels of miR-221 and miR-222 were markedly upregulated in PCs from patients with newly diagnosed MM (p < 0.001), and they were further upregulated in PCs from patients with relapsed or refractory MM (p < 0.001) (Figures 1A and 1B). Next, we detected the expression of miR-221 and miR-222 in 8 human myeloma cell lines (HMCLs), and we evaluated the sensitivity of these cell lines to Dex. The results showed that MM.1S, U266, JJN3, and RPMI-8226 were Dex sensitive, while MM.1R, ARH-77, NCI-H929, and MR-20 were Dex resistant (Table S2). Compared with Dex-sensitive HMCLs, the expression levels of miR-221 and miR-222 were relatively higher in the Dex-resistant HMCLs (p < 0.05) (Figures 1C and 1D; Figures S1A and S1B).
Figure 1.
miR-221/222 Regulate the Sensitivity of MM Cells to Dex
(A and B) Expression levels of miR-221 (A) and miR-222 (B) were determined by qRT-PCR in PCs from healthy individuals (NC, n = 12), patients with newly diagnosed MM (N-MM, n = 20), and patients with relapsed or refractory MM (R-MM, n = 21). p values were obtained using Mann-Whitney U rank-sum tests. **p < 0.01, ***p < 0.001. (C and D) qRT-PCR analysis of miR-221 (C) and miR-222 (D) expression levels in human MM cell lines (HMCLs). *p < 0.05. (E) qRT-PCR analysis of miR-221 and miR-222 expression levels in MM.1S and MM.1R cell lines. **p < 0.01. (F and G) MM.1S cells (F) transfected with agomir-221/222 or agomir-NC and MM.1R cells (G) transfected with antagomir-221/222 or antagomir-NC were treated with dexamethasone (Dex) at the indicated concentrations. After 48 h, cell viability was measured using the CCK-8 assay. **p < 0.01.
The MM.1R cell line is resistant to Dex and its parental sensitive counterpart is MM.1S. We found that the expression levels of miR-221 and miR-222 were significantly lower in MM.1S cells than in MM.1R cells (p < 0.01) (Figure 1E). To investigate the functional role of miR-221/222, MM.1S cells were transfected with agomir-221/222 or agomir-negative control (NC), and MM.1R cells were transfected with antagomir-221/222 or antagomir-NC (Figures S1C–S1E). As shown in Figures 1F and 1G, overexpressions of miR-221/222 dramatically reduced the sensitivity of MM.1S cells to Dex, while the inhibitions of miR-221/222 partially overcame the resistance of MM.1R cells to Dex. In line with these data, similar results were observed in Dex-sensitive U266 or JJN3 cells and Dex-resistant ARH-77 or NCI-H929 cells (Figures S1F–S1I). Collectively, these data indicated that the expression of miR-221/222 was dysregulated in MM cells, which may contribute to the Dex resistance of MM.
Induction of Autophagy Contributes to Dex-Induced MM Cell Death In Vitro
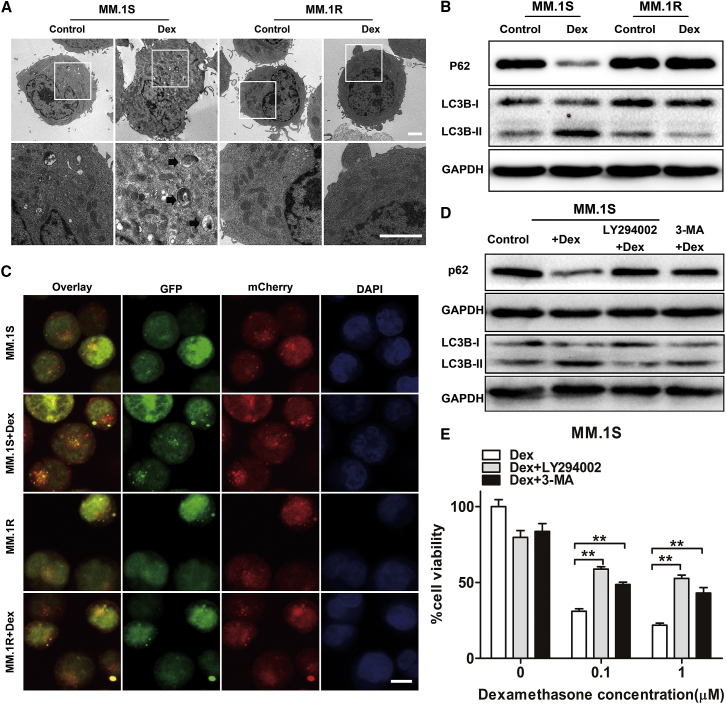
In response to chemotherapeutic drugs, autophagy can be induced and play either a pro-survival or a pro-death role, depending on the tumor type and treatment characteristics.20 Therefore, we next investigated whether autophagy was involved in Dex-induced cell death in MM cells. MM.1S or MM.1R cells were cultured with Dex for 24 h and then subjected to transmission electron microscopy. There were typical morphological features of autophagosomes and autolysosomes in Dex-treated MM.1S cells, but not in Dex-treated MM.1R cells (Figure 2A). When autophagy occurred, LC3B was transformed from a cytosolic form LC3B-I to a membrane form LC3B-II, and p62 was degraded. We found that Dex treatment increased the LCB-II:LC3B-I ratio and induced p62 degradation in Dex-sensitive MM.1S, U266, and JJN3 cells, but not in Dex-resistant MM.1R, ARH-77, and NCI-H929 cells (Figures 2B and S2A). Furthermore, we established MM cells expressing GFP-mCherry-LC3B fusion protein. Within the acidic lysosome, GFP fluorescence was quenched whereas mCherry fluorescence was stable. Hence, autophagosomes produced yellow dots (green plus red), whereas autolysosomes produced red dots. As shown in Figures 2C and S2B, MM.1S cells developed much more yellow and red dots than MM.1R cells after Dex treatment. Altogether, these results indicated that Dex induced the occurrence of autophagy in Dex-sensitive MM cells, but not in Dex-resistant MM cells.
Figure 2.
Role of Autophagy in Dex-Induced MM Cell Death
(A) Transmission electron microscopy was performed to detect morphological characteristics of MM.1S and MM.1R cells after treatment with Dex for 24 h. The arrows indicate autophagosomes or autolysosomes. Scale bar, 2 μm. (B) Western blot analysis of the expression levels of LC3B and p62 in MM.1S and MM.1R cells after treatment with Dex for 24 h. (C) Representative confocal images of MM cells expressing GFP-mCherry-LC3 fusion protein after Dex treatment for 24 h. Yellow (green plus red) dots indicate autophagosomes and free red dots indicate autolysosomes. Scale bar, 10 μm. (D) Western blot analysis of p62 and LC3B protein expression in MM.1S cells pretreated with autophagy inhibitor 3-MA or Ly294002 for 2 h followed by Dex (1 μM) for 24 h. Untreated MM.1S cells were used as the control. (E) MM.1S cells were pretreated with autophagy inhibitor 3-MA or Ly294002 for 2 h followed by Dex for 48 h. Cell viability was assessed using the CCK-8 assay. *p < 0.05, **p < 0.01.
Moreover, we showed that the addition of inhibitor of autophagy (3-methyladenine [3-MA] or LY294002) could inhibit Dex-induced autophagy in MM cells, as confirmed by reduced LC3B conversion and p62 degradation (Figure 2D), and significantly protect MM.1S, U266, and JJN3 cells from Dex-induced cell death (Figure 2E; Figures S2C and S2D). Therefore, we demonstrated that autophagy is critical for the induction of cell death following Dex treatment in MM.
ATG12 and p27 Are Target Genes of miR-221/222 in MM
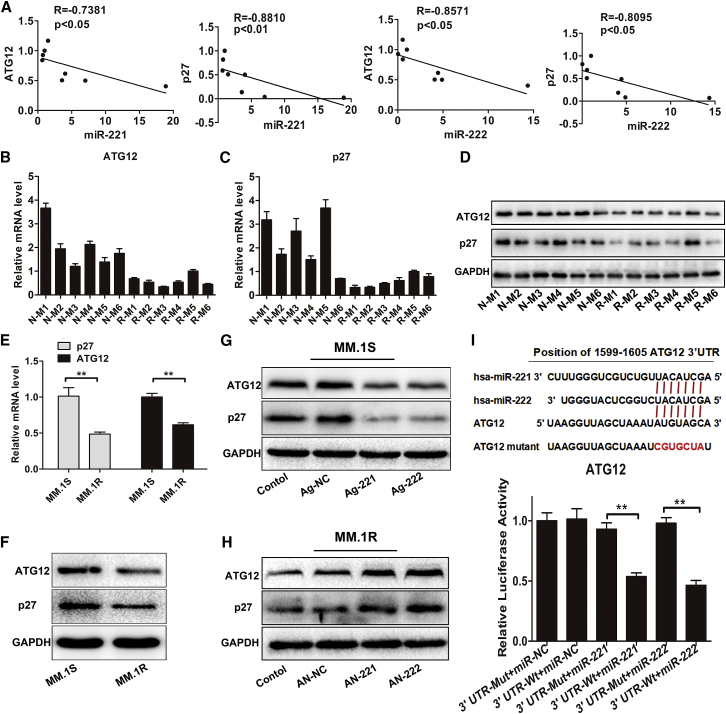
The functions of a microRNA are effectuated through its regulation of target genes. To predict the target genes of miR-221/222, we used bioinformatic microRNA target prediction software, including TargetScan, miRanda, and miRWalk. The results indicated that ATG12 and p27 may be potential target genes of miR-221/222 (data not shown). We next performed qRT-PCR, and we confirmed that miR-221/222 were inversely correlated with ATG12 and p27 mRNA expression in MM cell lines (Figure 3A; Figures S3A and S3B). Moreover, the protein levels of ATG12 and P27 were higher in miR-221/222 low-expressing MM cell lines than in miR-221/222 high-expressing MM cell lines (Figure S3C). Consistently, expression levels of miR-221/222 were lower (Figures S3D and S3E), while expression levels of p27 and ATG12 were higher (Figures 3B–3D) in PCs from newly diagnosed patients than in those from relapsed or refractory MM patients. Correlation analysis confirmed the inverse correlation between miR-221/222 and ATG12 or p27 mRNA expression in MM patients (Figure S3F).
Figure 3.
ATG12 and p27 Are the Direct Target Genes of miR-221 and miR-222
(A) Correlation of miR-221 and miR-222 expression levels with ATG12 and p27 mRNA levels in HMCLs was studied by way of the Spearman correlation test. (B–D) qRT-PCR (B and C) and western blot (D) analysis of ATG12 and p27 expression levels in PCs from patients with newly diagnosed MM (N-M) and patients with relapsed or refractory MM (R-M). (E and F) Expression levels of ATG12 and p27 were evaluated using qRT-PCR (E) and western blotting (F) in MM.1S and MM.1R cells. **p < 0.01. (G and H) MM.1S cells (G) were transfected with agomir-221 (Ag-221), agomir-222 (Ag-222), or agomir-NC (Ag-NC) and MM.1R cells (H) were transfected with antagomir-221 (AN-221), antagomir-222 (AN-221), or antagomir-NC (AN-NC), and then the expression levels of ATG12 and p27 were assessed using western blot analysis. Non-transfected MM cells were used as controls. (I) Dual-luciferase assays were performed in HEK293T cells after co-transfection with reporter constructs harboring wild-type (WT) or mutant (mut) ATG12 3′ UTR and plasmids expressing miR-221 or miR-222. **p < 0.01.
Furthermore, we found that expression levels of ATG12 and p27 were upregulated in MM.1S cells compared to MM.1R cells (p < 0.01) (Figures 3E and 3F). Transfection of agomir-221/222 inhibited ATG12 and p27 expression at both the mRNA and protein levels in MM.1S cells (Figure 3G; Figures S3G and S3H), whereas antagomir-221/222 led to the increased expression of ATG12 and p27 in MM.1R cells (Figure 3H; Figures S3I and S3J). Previous study has confirmed that p27 is a direct target of miR-221/222.21 To determine whether ATG12 was also a direct target of miR-221/222, the dual-luciferase reporter assay was performed. As shown in Figure 3I, overexpression of miR-221 or miR-222 significantly suppressed the relative luciferase activity of the reporter containing the wild-type 3′ UTR of ATG12, but not the mutant 3′ UTR (p < 0.01). Taken together, these results demonstrated that ATG12 and p27 were the targets of miR-221/222 and could be regulated by miR-221/222 in MM cells.
miR-221/222 Inhibit Autophagy and Trigger Dex Resistance of MM by Regulating ATG12 and p27 in MM
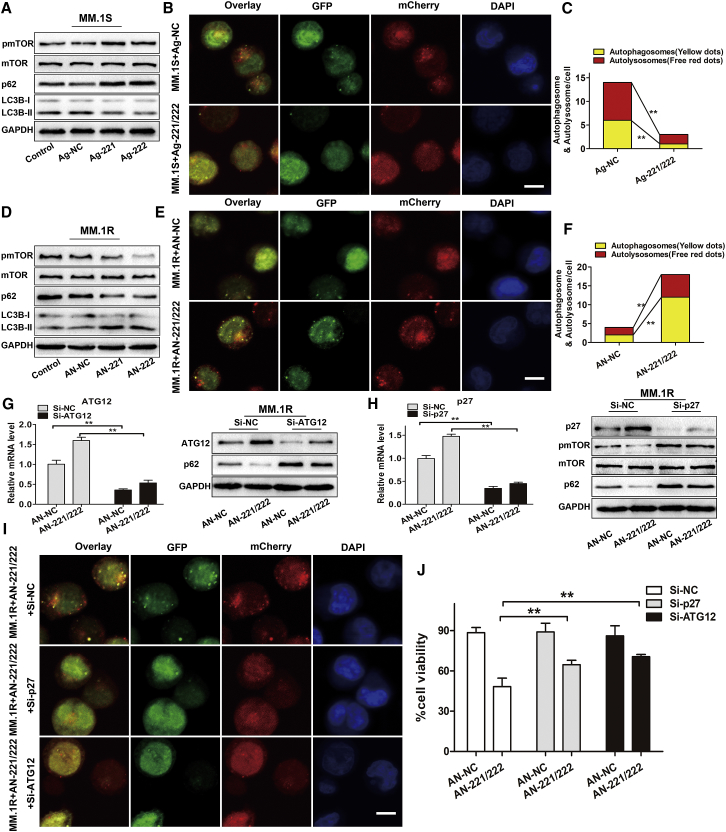
ATG12 is an important autophagy-related gene and is essential for autophagosome formation.22 p27 has been reported to be a positive regulator of autophagy by inhibiting the mTOR pathway.23 Therefore, we hypothesized that the upregulation of miR-221/222 may contribute to decreased autophagy through targeting ATG12 and p27. To verify this hypothesis, MM.1S or MM.1R cells were transfected with agomir-221/222 or antagomir-221/222, respectively. We observed that overexpressions of miR-221/222 reduced LC3B conversion and p62 degradation and increased mTOR phosphorylation in MM.1S cells (Figure 4A). Consistently, both the number of yellow and red dots was decreased in MM.1S cell transfected with agomir-221/222 (Figures 4B and 4C). In addition, downregulation of miR-221/222 led to the opposite effects in MM.1R cells (Figures 4D–4F).
Figure 4.
miR-221/222 Inhibit Autophagy and Dex Sensitivity in MM Cells by Negatively Regulating ATG12 and p27
(A) Western blot analysis was used to evaluate the expression levels of pmTOR, mTOR, LC3B, and p62 in MM.1S cells transfected with agomir-221 (Ag-221), agomir-222 (Ag-222), or agomir-NC (Ag-NC) for 72 h. Non-transfected MM cells were used as controls. (B) Representative confocal images of MM.1S cells expressing GFP-mCherry-LC3 fusion protein and transfected with agomir-221/222 (Ag-221/222) or agomir-NC (Ag-NC) for 48 h. Scale bar, 10 μm. (C) The mean number of autophagosomes and autolysosomes per MM.1S cell is represented by yellow and red, respectively. **p < 0.01. (D) Western blot analysis was used to evaluate the expression levels of pmTOR, mTOR, LC3B, and p62 in MM.1R cells transfected with antagomir-221 (AN-221), antagomir-222 (AN-222), or antagomir-NC (AN-NC) for 72 h. Non-transfected MM cells were used as controls. (E) Representative confocal images of MM.1R cells expressing GFP-mCherry-LC3 fusion protein and transfected with antagomir-221/222 (AN-221/222) or antagomir-NC (AN-NC) for 48 h. Scale bar, 10 μm. (F) The mean number of autophagosomes and autolysosomes per MM.1R cell is represented by yellow and red, respectively. **p < 0.01. (G and H) MM.1R cells were co-transfected with antagomir-221/222 (AN-221/222) or antagomir-NC (AN-NC) and siRNA targeting ATG12 (G) or p27 (H) (Si-ATG12 or Si-p27) as indicated. Expression levels of ATG12 and p27 in MM.1R cells were evaluated by qRT-PCR and western blotting, and expression levels of pmTOR, mTOR, and p62 were detected by western blotting. **p < 0.01. (I) Representative confocal images of MM.1R cells expressing GFP-mCherry-LC3 fusion protein that were co-transfected with antagomir-221/222 and siRNAs for 48 h. Scale bar, 10 μm. (J) MM.1R cells co-transfected with antagomir-221/222 or antagomir-NC and siRNA targeting ATG12 or p27 were treated with Dex (50 μM) for 48 h, and cell viability was measured using the CCK-8 assay. **p < 0.01.
We next explored the function of ATG12 and p27 in autophagy and Dex-induced cell death in MM. MM.1R cells were co-transfected with antagomir-221/222 and small interfering RNAs (siRNAs) targeting ATG12 or p27. Results showed that the knockdown of ATG12 and p27 (Figures 4G and 4H) attenuated the effect of antagomir-221/222 on autophagy, as confirmed by reduced p62 degradation and a decrease of both yellow and red dots (Figures 4G–4I; Figure S4A), and significantly inhibited cell death of MM.1R induced by antagomir-221/222 plus Dex (Figure 4J). Although the combined treatment of antagomir-221/222 and Dex could also induce apoptotic cell death of MM.1R cells, as indicated by the increased cleavage of caspases-3 and PARP, the knockdown of p27 or ATG12 had no effect on caspases-3 and PARP (Figure S5A). What’s more, pre-treatment with pan-caspase inhibitor z-VAD-fmk completely blocked apoptosis (Figure S5B), but it only partially inhibited cell death of MM.1R induced by antagomir-221/222 plus Dex (Figure S5C). These results indicated that miR-221/222 could inhibit Dex-induced autophagic cell death, but not apoptosis, through the ATG12/p27-mTOR autophagy axis.
Consistently, the knockdown of ATG12 or p27 in MM.1S cells decreased the level of autophagy (Figures S4B–S4I) and inhibited Dex-induced cell death of MM.1S cells, without alteration of Dex-induced cleavage of caspases-3 and PARP (Figures S5D and S5E). Altogether, our data demonstrated that miR-221/222 regulated the autophagy and Dex sensitivity of MM by targeting ATG12 and p27.
Dex Decreases the Expression of miR-221/222 in MM Cells Both In Vitro and In Vivo
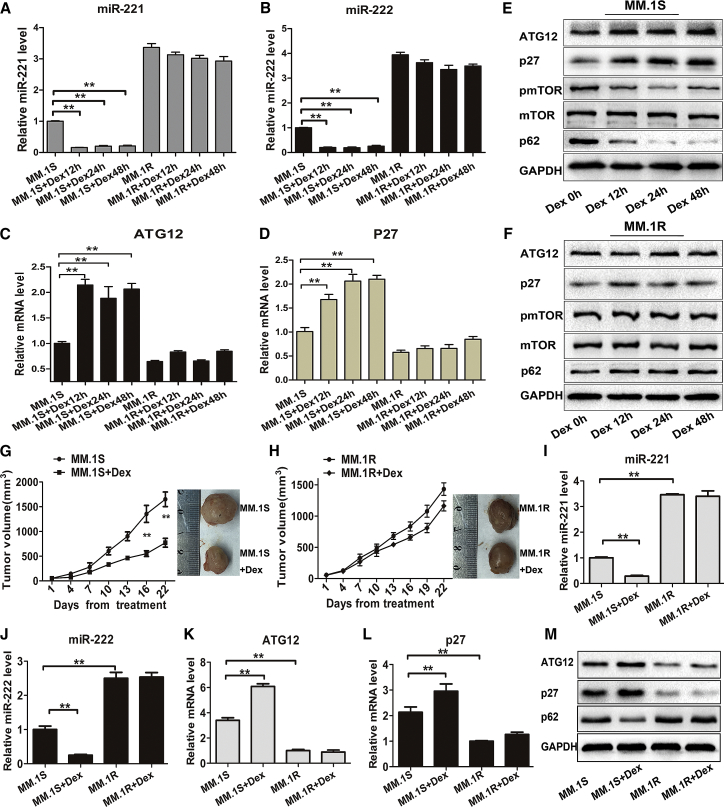
To further demonstrate the role of the miR-221/222-ATG12/p27-mTOR autophagy-regulatory axis in Dex-induced cell death, MM.1S and MM.1R cells were treated with Dex, and then the expression levels of miR-221/222, ATG12, p27, pmTOR, and p62 were evaluated. The results showed that miR-221/222 expressions in Dex-treated MM.1S cells were significantly decreased, whereas the expression of ATG12 or p27 was upregulated and that of pmTOR or p62 was decreased (Figures 5A–5E). In contrast, these changes were not observed in Dex-treated MM.1R cells (Figures 5A–5D and 5F).
Figure 5.
Dex Decreases the Expression of miR-221/222 in MM Cells Both In Vitro and In Vivo
(A–D) qRT-PCR analysis of miR-221 (A), miR-222 (B), ATG12 (C), and p27 (D) mRNA levels in MM.1S and MM.1R cells treated with Dex (1 μM) for different time points as indicated. **p < 0.01. (E and F) Western blot analysis of ATG12, p27, pmTOR, mTOR, and p62 protein levels in MM.1S (E) and MM.1R (F) cells in response to Dex (1 μM) treatment for different time points as indicated. (G and H) Growth curves and representative images of xenografted tumors. NOD/SCID mice were subcutaneously injected with MM.1S (G) or MM.1R (H) cells and then intraperitoneally treated with Dex (9 mg/kg) on days 1–4, 9–12, and 17–20. **p < 0.01. (I–L) qRT-PCR analysis of miR-221 (I), miR-222 (J), ATG12 (K), and p27 (L) mRNA levels in retrieved xenografted tumors. **p < 0.01. (M) Western blot analysis of ATG12, p27, and p62 protein levels in retrieved xenografted tumors.
To further verify these findings in vivo, MM.1S-xenografted or MM.1R-xenografted mice were intraperitoneally treated with Dex. Compared with PBS controls, Dex treatment significantly decreased tumor volumes of MM.1S-xenografted mice (p < 0.01) (Figure 5G), but not MM.1R-xenografted mice (Figure 5H). Consistent with our in vitro findings, Dex markedly decreased the expressions of miR-221/222 and p62, and it increased the expressions of ATG12 and p27 in MM.1S-xenografted mice, but not in MM.1R-xenografted mice (Figures 5I–5M). Based on these findings, we concluded that Dex-induced miR-221/222 reduction may contribute to the occurrence of pro-death autophagy in MM.
The Inhibition of miR-221/222 Improves the Autophagy and Dex Sensitivity of MM Cells In Vivo
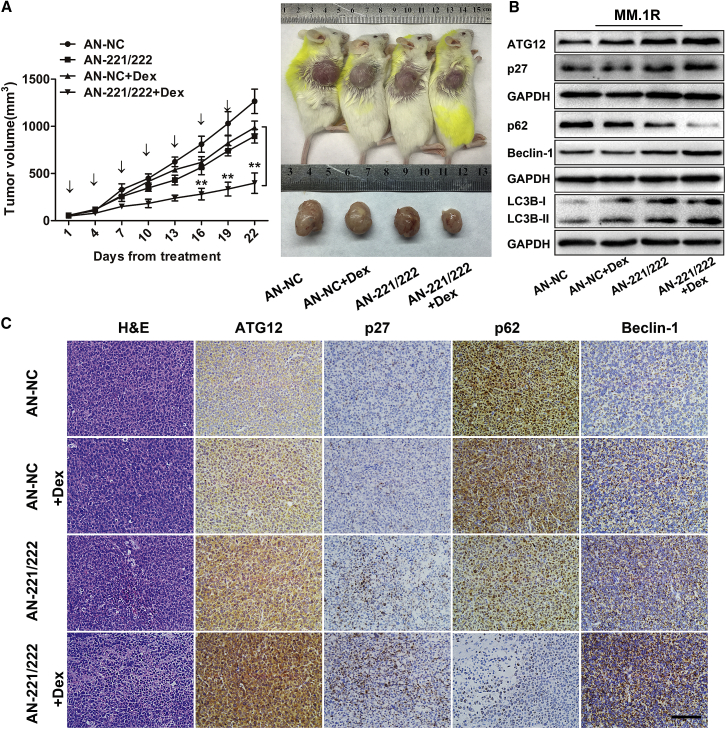
Finally, we evaluated whether miR-221/222 could regulate autophagy, thereby modulating Dex sensitivity of MM cells in vivo. MM.1R-xenografted mice were randomized to receive 4 different treatments, as indicated in Figure 6A. The results showed that treatment with antagomir-221/222 decreased the expression levels of miR-221/222 in excised tumors (Figure S6). Compared with control groups, combined treatment with antagomir-221/222 and Dex overcame Dex resistance, as evidenced by a remarkable tumor growth inhibition (p < 0.01) (Figure 6A). We also found that treatment with antagomir-221/222 could inhibit tumor growth when used alone, which is in accordance with a previous report.24 Consistent with our in vitro findings, we observed increased expression of both ATG12 and p27, as well as increased LC3B-II, decreased p62, and increased Beclin-1 in excised tumors treated with antagomir-221/222 (Figure 6B). Moreover, combination treatment with antagomir-221/222 plus Dex induced further upregulation of both ATG12 and p27, as well as extended autophagy in tumor tissues (Figures 6B and 6C). Taken together, these in vivo data further indicated that miR-221/222 could inhibit the autophagy pathway in MM cells and targeting miR-221/222 could sensitize MM cells to Dex treatment.
Figure 6.
Inhibition of miR-221/222 Promotes Autophagy and Restores Dex Sensitivity of MM Cells In Vivo
(A) Growth curves and representative images of tumors from MM.1R-xenografted mice. Antagomir-NC (AN-NC) or antagomir-221/222 (AN-221/222) was injected intratumorally every 3 days for seven times, and Dex (9 mg/kg) was injected intraperitoneally on days 1–4, 9–12, and 17–20. **p < 0.01. (B and C) Western blot (B) and immunohistochemical analyses (C) of p27, ATG12, LC3B, p62, and Beclin-1 protein levels in tumor tissues from MM.1R-xenografted mice treated with antagomir-NC, antagomir-221/222, antagomir-NC plus Dex, or antagomir-221/222 plus Dex. Scale bar, 50 μm.
Discussion
Glucocorticoids are fundamental drugs used in the treatment of MM, and its resistance is closely correlated with a poor prognosis in MM.5 Recently, increasing evidence has demonstrated that microRNAs are involved in drug resistance in hematological malignancies through modulating drug transporter-related proteins, cell cycle-related proteins, drug targets, autophagy, tumor microenvironment, cell survival signaling, and apoptosis pathways.25 However, there are relatively few studies on the role of microRNAs in the chemoresistance of MM. Here we investigated the roles of miR-221/222 in modulating Dex resistance in MM cells, and we reported for the first time that, by targeting ATG12 and p27, miR-221/222 mediated the Dex sensitivity of MM cells through regulating autophagy.
In this study, we confirmed that miR-221 and miR-222 were upregulated in PCs from MM patients, especially those with relapsed or refractory disease. Importantly, expressions of miR-221/222 were inversely correlated with Dex sensitivity of HMCLs. Recent studies have shown that miR-221 and miR-222 were widely upregulated and considered to be crucial oncogenes in a variety of solid and hematologic malignancies, including glioblastoma, breast cancer, hepatocellular carcinomas, prostate cancer, pancreatic cancer, papillary thyroid cancer, chronic lymphocytic leukemia, and acute and chronic myeloid leukemia.12, 13 By inhibiting tumor suppressor proteins, such as p27Kip1 (p27), PUMA, and Bim, miR-221/222 promoted resistance to fulvestrant, tamoxifen, TRAIL, gefitinib, melphalan, and many other chemotherapeutic drugs.13 Therefore, whether miR-221/222 were involved in the drug resistance of other chemotherapeutic drugs in MM needs further exploration.
Next, we identified that Dex induced autophagy in Dex-sensitive MM cell lines, but not in Dex-resistant MM cell lines. Importantly, the inhibition of autophagy significantly decreased Dex-induced cell death in Dex-sensitive MM cell lines. It is universally accepted that basal levels of autophagy are essential for MM cells to degrade misfolded proteins and maintain energy balance.26 In response to some anti-MM drugs, such as doxorubicin and carfilzomib, autophagy was induced to play a cytoprotective role and contributed to attenuate cell death.27, 28 However, excessive autophagy might lead to MM cell death.29, 30 Growing evidence has demonstrated that anti-MM drugs, such as FK866 and iron chelators, facilitated MM cell death through inducing autophagic cell death.31, 32 In addition, autophagy can be induced by FTY720 and clioquinol to promote apoptosis.33, 34 Therefore, whether autophagy plays a pro-survival or a pro-death role in MM may depend on treatment characteristics. Better understanding the exact role of autophagy in MM induced by different anti-MM drugs is essential for improving MM therapy. Our findings that Dex promoted cell death of MM cells by inducing pro-death autophagy may provide new insights into the mechanism of Dex resistance in MM.
Increasing evidence has indicated an important role of microRNAs in regulating autophagy. For example, miR-100 and miR-155 promoted autophagy by inhibiting the mTOR pathway,35, 36 while miR-23b and miR-200c inhibited autophagy through targeting ATG12 or Foxp3.37, 38 Using gain- or loss-of-function studies, we found that miR-221/222 could negatively regulate autophagy in MM, and ATG12, an important autophagy-related protein,22 was identified as a new target of miR-221/222. As with ATG12, miR-221/222 also regulated p27, which has been reported to show potent anti-tumor effect by inducing autophagic cell death.39 Furthermore, our results showed that knockdown of ATG12 or p27 antagonized the autophagy-inducing and Dex-sensitizing activity of miR-221/222 antagomirs in Dex-resistant MM cells. Of note, silencing ATG12 or p27 in Dex-sensitive MM cells significantly inhibited autophagy, accompanied by reduced sensitivity to Dex. Previous studies have found that miR-221/222 could inhibit apoptosis in MM cells.40, 41 Although our data indeed showed that targeting miR-221/222 could increase the apoptosis of MM cells induced by Dex, we proved that cell death mediated through the miR-221/222-ATG12/p27-mTOR autophagy axis was autophagic cell death, but not apoptosis. Thus, these results suggest that ATG12 and p27 are two key players in miR-221/222-mediated autophagy inhibition and Dex resistance in MM.
Finally, we showed that, compared with Dex-resistant MM cells, the basal expression levels of miR-221/222 were lower in Dex-sensitive MM cells. In addition, Dex treatment could further decrease miR-221/222 expression in Dex-sensitive MM cells, accompanied by the upregulated expressions of ATG12 and p27, as well as downregulated pmTOR and p62. These data indicated that the miR-221/222-ATG12/p27-mTOR autophagy-regulatory axis was directly involved in the Dex-induced occurrence of excessive autophagy and cell death in MM cells. Previous studies have reported that transcription factors play an important role in regulating microRNA expression. For example, c-Jun and nuclear factor κB (NF-κB) were found to induce the expression of miR-221/222 in prostate carcinoma and glioblastoma,42 while the promyelocytic leukemia zinc finger (PLZF) was reported as a negative regulator of miR-221/miR-222 expression in melanoma.43 In ongoing studies, we are investigating the potential upstream regulators of miR-221/222 in MM.
Collectively, our data reveal that overexpressions of miR-221/222 promote Dex resistance of MM cells through the inhibition of autophagy by targeting ATG12 and p27. The findings of the novel miR-221/222-ATG12/p27-mTOR autophagy-regulatory axis provide new insights into the underlying mechanisms of Dex resistance, and they suggest that the inhibitions of miR-221/222 and induction of pro-death autophagy may be promising therapeutic strategies for the treatment of Dex-resistant MM.
Materials and Methods
MM Cell Lines and Patient Samples
Human MM cell lines MM.1S, MM.1R, RPMI-8226, U266, NCI-H929, and ARH-77 were obtained from the American Type Culture Collection (Manassas, VA, USA). MR20 was kindly provided by Dr. Ruben Carrasco (Harvard University, USA). All cell lines were cultured in RPMI-1640 (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco). JJN3 was obtained from DSMZ (Germany), and it was cultured in isometric DMEM (Gibco) and Iscove’s MDM (IMDM; Gibco) with 20% FBS.
Primary PCs from 12 healthy donors, 20 patients with newly diagnosed MM, and 21 patients with relapsed or refractory MM were obtained from bone marrow aspirates using CD138+ magnetic beads selection (Miltenyi Biotec, Auburn, CA, USA), according to the recommended protocols. All MM patients were diagnosed according to the International Myeloma Working Group diagnostic criteria for MM (Table S1). These studies were conducted in accordance with the Declaration of Helsinki, and studies were approved by the institutional review board of Huazhong University of Science and Technology.
Quantitative Real-Time Amplification of MicroRNAs and mRNAs
Total RNA was isolated using TRIzol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). qRT-PCR for miR-221, miR-222, and U6 small nuclear RNA (snRNA) was performed using TaqMan microRNA reverse transcription kit, TaqMan Universal Master Mix II, and TaqMan microRNA assay kits for miR-221 (000524), miR-222 (000525), and U6 snRNA (001973) (Applied Biosystems, Foster City, CA, USA). U6 snRNA was used as an internal control. For mRNA studies, cDNA was synthesized with PrimeScript RT reagent Kit (Takara, Dalian, China), and then qRT-PCR was performed using SYBR Green RT-PCR Kit (Takara). The primers for ATG12, p27, and GAPDH are shown in Table S3. GAPDH was used as an internal control. All PCR reactions were carried out in triplicate on ABI 7500 FAST Real Time PCR System (Applied Biosystems), and relative expression was evaluated by the comparative 2−ΔΔCT method.
Western Blotting
A total of 20–40 μg cell lysates was subjected to electrophoresis on 8%–15% SDS-polyacrylamide gels and transferred to polyvinyldene fluoride membranes. Primary antibodies included anti-phospho(p)-mTOR, anti-mTOR, anti-PARP (Cell Signaling Technology, Danvers, MA, USA), anti-LC3B, anti-SQSTM1/p62, anti-p27, anti-GAPDH, anti-ATG12, and anti-cleaved Caspase-3 (Abcam, Cambridge, UK).
Cell Transfection
Chemically modified hsa-miR-221/222 mimics (agomir-221/222) or inhibitors (antagomir-221/222) were either used to increase or inhibit miR-221 and miR-222 expressions. MM cells were transiently transfected with agomir-221/222 or agomir-NC (GenePharma, Shanghai, China) at a final concentration of 100 nM and antagomir-221/222 or antagomir-NC (Ribobio, Guangzhou, China) at a final concentration of 200 nM, using HiPerFect Transfection Reagent (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. Similar conditions were applied for transfection with 50 nM Silencer Select siRNA for ATG12 (ATG12 siRNA) or p27 (p27 siRNA) (Ribobio). The sequences of siRNAs are shown in Table S4.
In Vitro Luciferase Reporter Assay
HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with 2 μg plasmids expressing wild-type Luc-ATG12 or mutant Luc-ATG12 (GeneChem, Shanghai, China); 0.4 μg empty plasmids; plasmids expressing miR-221, miR-222, or miR-NC; and 0.02 μg Renilla construct in 24-well plates. At 48 h after transfection, cell extracts were prepared, and luciferase reporter assays were performed using the Dual-Luciferase Assay Kit (Promega, Madison, WI, USA). Firefly Luciferase activities were normalized to parallel Renilla activities.
Cell Viability Assay
Cell Counting Kit-8 (CCK-8) assay was performed to evaluate cell viability, according to the manufacturer’s instructions (Dojindo Laboratories, Kumamoto, Japan). For combination experiments with microRNAs, 8 × 105 MM.1S, U266, or JJN3 cells were transfected with agomir-221/222 or agomir-NC (NC) and MM.1R, ARH-77, or NCI-H929 cells were transfected with antagomir-221/222 or antagomir-NC (NC) in 6-well plates. After 24 h, MM cells were re-seeded in 96-well plates (3 × 104 cells/well) and treated with Dex (Sigma-Aldrich, St. Louis, MO, USA) at the indicated concentrations for 48 h. For the combination of autophagy inhibitors with Dex experiments, MM cells were seeded in 96-well plates (3 × 104 cells/well), pretreated with autophagy inhibitor 3-MA (500 μM, Sigma-Aldrich) or Ly294002 (2.5 μM, Sigma-Aldrich) for 2 h, followed by Dex (1 μM) for 48 h, and then subjected to CCK-8 assay. Similar conditions were performed for combination experiments with siRNAs.
Transmission Electron Microscopy
Cells were seeded and subjected to Dex treatment in 6-well plates. After 24 h, cells were collected and washed twice with PBS. Then, cell pellets were fixed with 2.5% phosphate-buffered glutaraldehyde and stored at 4°C before embedding. After washing with PBS, the cells were postfixed with 1% OsO4 (Servicebio, Wuhan, China), dehydrated with an increasing gradient of ethanol and acetone, and then embedded in Spurr’s resin. Ultrathin sections (50–70 nm) were obtained on an electron microscope (EM) UC6 ultramicrotome (Leica Microsystems, Wetzlar, Germany) and adhered to uncoated copper grids. The sections were then stained with 4% uranyl acetate and lead citrate prior to viewing on a Tecnai G2 12 transmission electron microscope (FEI, Hillsboro, OR, USA).
GFP-mCherry-LC3B Transfection and Confocal Microscopy
MM cells were infected with adenovirus harboring vector expressing GFP-mcherry-LC3B fusion protein, according to the manufacturer’s instructions (Vigenebio, Jinan, China). After the induction of autophagy, MM cells were collected and seeded on glass slides coated with polylysine (Servicebio) for 30 min at room temperature. Cells then were fixed with 4% paraformaldehyde for 30 min and washed 3 times with PBS for 5 min/wash. The fixed cells were counterstained with DAPI (Antgene, Wuhan, China) for nuclear staining for 15 min, washed three times with PBS, and examined using a Nikon Eclipse Ti laser-scanning confocal microscope (Nikon, Japan). The number of autophagosomes and autolysosomes per cell was counted by yellow and free red dots in the overlay images, respectively. At least 15 cells of each group were counted. The data represent the means from at least three independent experiments (**p < 0.01).
Animal Model and Treatment
Female NOD/SCID mice (4–6 weeks, Beijing Vital River Laboratory Animal Technology, China) were subcutaneously injected with 1 × 107 MM.1S cells or MM.1R cells in 200 μL serum-free RPMI-1640 medium to establish human Dex-sensitive or Dex-resistant MM xenografts. The treatment began from when the tumors were palpable (about 3 weeks after injection). In the Dex treatment experiment, MM.1S-xenografted or MM.1R-xenografted mice were randomly divided into two groups and then intraperitoneally injected with 9 mg/kg Dex or PBS on days 1–4, 9–12, and 17–20, respectively. Each group contained six mice.
In the antagomir-221/222 combined with Dex treatment experiment, mice were randomized to receive 4 different treatments. The groups were antagomir-NC plus PBS (n = 6), antagomir-221/222 plus PBS (n = 6), antagomir-NC plus Dex (n = 6), and antagomir-221/222 plus Dex (n = 6). Antagomir-NC or antagomir-221/222 (20 nmol) was dissolved with 50 μL PBS and injected intratumorally every 3 days for seven times, and Dex (9 mg/kg) was injected intraperitoneally on days 1–4, 9–12, and 17–20. Tumor volume (V) was monitored every 3 days, and it was calculated using the following formula: V = 0.5 × a × b2, where a and b represent the longest and shortest diameters, respectively. All mice were sacrificed at day 23. All experimental procedures and protocols had been approved by the Committee on Animals Handling of Huazhong University of Science and Technology.
Immunohistochemistry
Paraffin-embedded sections of tumor tissues from xenograft mice were incubated with antibodies directed against p27, ATG12, p62, and Beclin-1 (Abcam) at 4°C overnight. Subsequently, the sections were treated with polymer, developed with DAB-Chromogen, and counterstained with hematoxylin.
Statistical Analysis
All values are presented as the mean ± SD. Mann-Whitney U rank-sum test was used to evaluate miR-221/222 differences between independent groups of MM samples and MM cell lines. The correlations between miR-221/222 and p27 or ATG12 in MM samples and MM cell lines were studied by way of Spearman correlation test. Other comparisons were determined by the unpaired t test. All analyses were performed with GraphPad Prism 5 and p < 0.05 was considered statistically significant.
Author Contributions
Conceptualization, C.S., Y.H., and J.X.; Methodology, J.X., Y.H., C.S., and Y.S.; Investigation, J.X. and Y.S.; Formal Analysis, J.X., F.F., Y.S., Z.C., H.H., J.Z., J.D., and L.A.; Writing – Original Draft, J.X., Y.H., C.S., and F.F.; Writing – Review & Editing, Y.H., C.S., J.X., and F.F.; Resources, A.X., S.M., L.C., and B.Z.; Project Administration, Y.H. and C.S. All of the authors agreed to submit the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81670197 to C.S.), the Clinical Research Physician Program of Tongji Medical College, HUST (to C.S.), and the Key Project of Hubei Provincial Natural Science Foundation (2014CFA064 to Guohui Cui). The authors thank Pei Zhang and An-Na Du from The Core Facility and Technical Support, Wuhan Institute of Virology, for their help with producing EM micrographs.
Footnotes
Supplemental Information includes six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2019.01.012.
Contributor Information
Chunyan Sun, Email: suncy0618@163.com.
Yu Hu, Email: dr_huyu@126.com.
Supplemental Information
References
- 1.Kumar S.K., Callander N.S., Alsina M., Atanackovic D., Biermann J.S., Chandler J.C., Costello C., Faiman M., Fung H.C., Gasparetto C. Multiple Myeloma, Version 3.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2017;15:230–269. doi: 10.6004/jnccn.2017.0023. [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A., Anderson K. Multiple myeloma. N. Engl. J. Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S.K., Rajkumar S.V., Dispenzieri A., Lacy M.Q., Hayman S.R., Buadi F.K., Zeldenrust S.R., Dingli D., Russell S.J., Lust J.A. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S.K., Lee J.H., Lahuerta J.J., Morgan G., Richardson P.G., Crowley J., Haessler J., Feather J., Hoering A., Moreau P., International Myeloma Working Group Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study. Leukemia. 2012;26:149–157. doi: 10.1038/leu.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson P.G., Sonneveld P., Schuster M.W., Irwin D., Stadtmauer E.A., Facon T., Harousseau J.L., Ben-Yehuda D., Lonial S., Goldschmidt H., Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N. Engl. J. Med. 2005;352:2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 6.Smith L.K., Cidlowski J.A. Glucocorticoid-induced apoptosis of healthy and malignant lymphocytes. Prog. Brain Res. 2010;182:1–30. doi: 10.1016/S0079-6123(10)82001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi C., Chng W.J. MicroRNA: important player in the pathobiology of multiple myeloma. BioMed Res. Int. 2014;2014:521586. doi: 10.1155/2014/521586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong Y.W., Ferland-McCollough D., Jackson T.J., Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 10.Pichiorri F., Suh S.S., Ladetto M., Kuehl M., Palumbo T., Drandi D., Taccioli C., Zanesi N., Alder H., Hagan J.P. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., Chen L., Barlogie B., Stephens O., Wu X., Williams D.R., Cartron M.A., van Rhee F., Nair B., Waheed S. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. USA. 2010;107:7904–7909. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Martino M.T., Rossi M., Caracciolo D., Gullà A., Tagliaferri P., Tassone P. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin. Ther. Targets. 2016;20:1099–1108. doi: 10.1517/14728222.2016.1164693. [DOI] [PubMed] [Google Scholar]
- 13.Song J., Ouyang Y., Che J., Li X., Zhao Y., Yang K., Zhao X., Chen Y., Fan C., Yuan W. Potential Value of miR-221/222 as Diagnostic, Prognostic, and Therapeutic Biomarkers for Diseases. Front. Immunol. 2017;8:56. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galluzzi L., Vitale I., Abrams J.M., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., Dawson T.M., Dawson V.L., El-Deiry W.S., Fulda S. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun Z., Zhichao J., Hao Y., Ou J., Ran Y., Wen D., Qun S. Targeting autophagy in multiple myeloma. Leuk. Res. 2017;59:97–104. doi: 10.1016/j.leukres.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Laane E., Tamm K.P., Buentke E., Ito K., Kharaziha P., Oscarsson J., Corcoran M., Björklund A.C., Hultenby K., Lundin J. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009;16:1018–1029. doi: 10.1038/cdd.2009.46. [DOI] [PubMed] [Google Scholar]
- 18.Bonapace L., Bornhauser B.C., Schmitz M., Cario G., Ziegler U., Niggli F.K., Schäfer B.W., Schrappe M., Stanulla M., Bourquin J.P. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J. Clin. Invest. 2010;120:1310–1323. doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidari N., Hicks M.A., Harada H. GX15-070 (obatoclax) overcomes glucocorticoid resistance in acute lymphoblastic leukemia through induction of apoptosis and autophagy. Cell Death Dis. 2010;1:e76. doi: 10.1038/cddis.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulda S. Autophagy in Cancer Therapy. Front. Oncol. 2017;7:128. doi: 10.3389/fonc.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafrè S.A. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otomo C., Metlagel Z., Takaesu G., Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su M., Wang J., Wang C., Wang X., Dong W., Qiu W., Wang Y., Zhao X., Zou Y., Song L. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 2015;22:986–999. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Martino M.T., Gullà A., Cantafio M.E., Lionetti M., Leone E., Amodio N., Guzzi P.H., Foresta U., Conforti F., Cannataro M. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget. 2013;4:242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie L., Jing R., Qi J., Lin Z., Ju S. Drug resistance-related microRNAs in hematological malignancies: translating basic evidence into therapeutic strategies. Blood Rev. 2015;29:33–44. doi: 10.1016/j.blre.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Hoang B., Benavides A., Shi Y., Frost P., Lichtenstein A. Effect of autophagy on multiple myeloma cell viability. Mol. Cancer Ther. 2009;8:1974–1984. doi: 10.1158/1535-7163.MCT-08-1177. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y., Gao Y., Chen L., Gao G., Dong H., Yang Y., Dong B., Chen X. Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin. Cancer Res. 2011;17:3248–3258. doi: 10.1158/1078-0432.CCR-10-0890. [DOI] [PubMed] [Google Scholar]
- 28.Jarauta V., Jaime P., Gonzalo O., de Miguel D., Ramírez-Labrada A., Martínez-Lostao L., Anel A., Pardo J., Marzo I., Naval J. Inhibition of autophagy with chloroquine potentiates carfilzomib-induced apoptosis in myeloma cells in vitro and in vivo. Cancer Lett. 2016;382:1–10. doi: 10.1016/j.canlet.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Lamy L., Ngo V.N., Emre N.C., Shaffer A.L., 3rd, Yang Y., Tian E., Nair V., Kruhlak M.J., Zingone A., Landgren O., Staudt L.M. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435–449. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zismanov V., Lishner M., Tartakover-Matalon S., Radnay J., Shapiro H., Drucker L. Tetraspanin-induced death of myeloma cell lines is autophagic and involves increased UPR signalling. Br. J. Cancer. 2009;101:1402–1409. doi: 10.1038/sj.bjc.6605291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cea M., Cagnetta A., Fulciniti M., Tai Y.T., Hideshima T., Chauhan D., Roccaro A., Sacco A., Calimeri T., Cottini F. Targeting NAD+ salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood. 2012;120:3519–3529. doi: 10.1182/blood-2012-03-416776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pullarkat V., Meng Z., Donohue C., Yamamoto V.N., Tomassetti S., Bhatia R., Krishnan A., Forman S.J., Synold T.W. Iron chelators induce autophagic cell death in multiple myeloma cells. Leuk. Res. 2014;38:988–996. doi: 10.1016/j.leukres.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Liao A., Hu R., Zhao Q., Li J., Li Y., Yao K., Zhang R., Wang H., Yang W., Liu Z. Autophagy induced by FTY720 promotes apoptosis in U266 cells. Eur. J. Pharm. Sci. 2012;45:600–605. doi: 10.1016/j.ejps.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Cao B., Li J., Zhou X., Juan J., Han K., Zhang Z., Kong Y., Wang J., Mao X. Clioquinol induces pro-death autophagy in leukemia and myeloma cells by disrupting the mTOR signaling pathway. Sci. Rep. 2014;4:5749. doi: 10.1038/srep05749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge Y.Y., Shi Q., Zheng Z.Y., Gong J., Zeng C., Yang J., Zhuang S.M. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget. 2014;5:6218–6228. doi: 10.18632/oncotarget.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan G., Xie W., Liu Z., Xu W., Lao Y., Huang N., Cui K., Liao M., He J., Jiang Y. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70–79. doi: 10.4161/auto.26534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P., Zhang J., Zhang L., Zhu Z., Fan J., Chen L., Zhuang L., Luo J., Chen H., Liu L. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143.e12. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 38.Wang S., Wu J., Ren J., Vlantis A.C., Li M.Y., Liu S.Y.W., Ng E.K.W., Chan A.B.W., Luo D.C., Liu Z. MicroRNA-125b Interacts with Foxp3 to Induce Autophagy in Thyroid Cancer. Mol. Ther. 2018;26:2295–2303. doi: 10.1016/j.ymthe.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q., Xie W., Kuhn D.J., Voorhees P.M., Lopez-Girona A., Mendy D., Corral L.G., Krenitsky V.P., Xu W., Moutouh-de Parseval L. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gullà A., Di Martino M.T., Gallo Cantafio M.E., Morelli E., Amodio N., Botta C., Pitari M.R., Lio S.G., Britti D., Stamato M.A. A 13 mer LNA-i-miR-221 Inhibitor Restores Drug Sensitivity in Melphalan-Refractory Multiple Myeloma Cells. Clin. Cancer Res. 2016;22:1222–1233. doi: 10.1158/1078-0432.CCR-15-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J.J., Chu Z.B., Hu Y., Lin J., Wang Z., Jiang M., Chen M., Wang X., Kang Y., Zhou Y. Targeting the miR-221-222/PUMA/BAK/BAX Pathway Abrogates Dexamethasone Resistance in Multiple Myeloma. Cancer Res. 2015;75:4384–4397. doi: 10.1158/0008-5472.CAN-15-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galardi S., Mercatelli N., Farace M.G., Ciafrè S.A. NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39:3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felicetti F., Errico M.C., Bottero L., Segnalini P., Stoppacciaro A., Biffoni M., Felli N., Mattia G., Petrini M., Colombo M.P. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.