Abstract
The present study aimed to determine the expression profile and significance of microRNA-140-3p (miR-140-3p) in breast cancer (BC). miR-140-3p expression in BC tumor tissues and cell lines was measured by reverse transcription-quantitative polymerase chain reaction. Luciferase activity reporter assay and western blotting were used to assess the effect of miR-140-3p expression on tripartite motif 28 (TRIM28). Cell growth, migration, and invasion were assessed by Cell Counting Kit-8 assay, wound-healing assay and Transwell invasion assay, respectively. miR-140-3p expression was significantly reduced in BC tumor tissues compared with in adjacent normal tissues. Additionally, low miR-140-3p expression was found to predict poor prognosis of patients with BC. TRIM28 expression was significantly reduced by miR-140-3p overexpression in BC cell lines, and was inversely correlated with miR-140-3p in BC tissues. Overexpression of miR-140-3p also inhibited cell proliferation, migration and invasion compared with in the control group. In conclusion, the present study revealed that miR-140-3p inhibited the progression of BC partially by regulating TRIM28.
Keywords: microRNA-140-3p, tripartite motif 28, breast cancer, tumor suppressor
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer type and the second leading cause of tumor-associated mortality in women worldwide (1). In China, the top five commonly diagnosed types of cancer among women, in descending order, are: BC, lung and bronchial cancer, stomach cancer, colorectal cancer and esophageal cancer (2). Although significant progress regarding treatment methods for BC has been made, the prognosis of BC remains poor (3,4).
Extensive studies have revealed the critical role of microRNAs (miRNAs/miRs) in almost all cancer types (5,6). miRNAs are a class of non-coding RNAs that are widely involved in cell proliferation, invasion and apoptotic processes (7,8). miRNAs can be generally classified into two groups, namely oncogenic miRNAs and tumor suppressive miRNAs (5,6,8). The role of oncogenic miRNAs is to promote cancer development, while tumor suppressive miRNAs exert the opposite effect (5,6).
Numerous aberrantly expressed miRNAs have been identified in BC (5,6). Members of the miR-200 family have been reported to function as tumor suppressors in BC and prevent the epithelial-mesenchymal transition process (9,10). miR-206 was found to be downregulated in BC and is correlated with tumor progression through targeting cyclin D1 or notch3 (7,11,12). miR-140-3p downregulation results in lung cancer cell proliferation, invasion and migration stimulation by regulating ATPase H+ transporting accessory protein 2 (13). miR-140-3p expression is also elevated in chordoma and is correlated with worse recurrence-free survival (14). However, the role of miR-140-3p in BC remains unclear.
In the present study, miR-140-3p was revealed to be downregulated in BC tissues and cell lines. In vitro functional assays indicated that downregulation of miR-140-3p could promote BC cell proliferation and migration. The present study further revealed that tripartite motif 28 (TRIM28) was a direct target of miR-140-3p, which could help in understanding the regulatory mechanism of miR-140-3p in BC.
Materials and methods
Tissue samples and cell culture
BC and adjacent normal tissues were collected from 74 female patients (range, 43–68 years old; mean, 54.6 years old) between March 2010 and November 2012 at First Affiliated Hospital of Jiamusi University (Jiamusi, China). Tissues were immediately frozen in liquid nitrogen and stored at −80°C until further use. The study protocol was approved by the ethics committee of First Affiliated Hospital of Jiamusi University (Jiamusi, China). Written informed consent was obtained from all enrolled patients.
BC cell lines (MCF-7 and MDA-MB-453) and normal breast epithelial cells (MCF-10A) were purchased from American Type Culture Collection (Manassas, VA, USA). Cell lines were incubated in Dulbecco's modified Eagle's medium supplemented (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin in a 37°C humidified atmosphere containing 5% CO2.
Transfection of BC cell lines
miR-140-3p mimic (5′-UACCACAGGGUAGAACCACGG-3′), inhibitor (5′-CCGUGGUUCUACCCUGUGGUA-3′), and their corresponding negative controls (NC-mimic, 5′-GCAAGAGACAAGCGCUUAGCC-3′ and NC-inhibitor, 5′-GGUCCUGAUUCGUGCUACUCG-3′) were synthesized by Guangzhou Ribobio Co., Ltd. (Guangzhou, China). Small interfering RNA targeting TRIM28 (si-TRIM28, 5′-GACCAAACCTGTGCTTATGTT-3′) and NC (5′-GTTCTCCGAACGTGTCACGT-3′) was synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). A total of 2,000 cells (MCF-7 and MDA-MB-453) were seeded into 6-well plate and incubated until they reached 70–80% confluency. Transfection was conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 50 nM miRNA or 50 nM siRNA into MCF-7 and MDA-MB-453 cells, according to the manufacturer's protocols. After 48 h of transfection, cells were collected for following assays.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cell lines with the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. RNA was reverse transcribed to cDNA using PrimeScript™ RT reagent (Takara Biotechnology Co., Ltd., Dalian, China), following the manufacturer's protocols. miR-140-3p levels were quantified using SYBR Premix Ex Taq kit (Takara Biotechnology Co., Ltd.) on an ABI 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), with the following primers: miR-140-3p, forward 5′-ACACTCCAGCTGGGAGGCGGGGCGCCGCGGGA-3′, reverse 5′-CTCAACTGGTGTCGTGGA-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. The RT-qPCR condition was 95°C for 10 min followed by 40 cycles at 95°C for 15 sec, 60°C for 25 sec, and 72°C for 35 sec. Expression levels were measured using the 2−ΔΔCq method (15), with U6 small nuclear RNA used as an internal control.
Protein isolation and western blot analysis
Total protein was extracted using radioimmunoprecipitation assay buffer containing phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Haimen, China). The concentration of extracted samples was analyzed with Enhanced bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocols. A total of 50 µg extracted protein samples were separated via SDS-PAGE on a 10% gel and transferred to a polyvinylidene fluoride membrane. After blocking with 5% fat-free milk at 4°C for 4 h, the membranes were incubated with primary antibodies (TRIM28, cat. no. ab22553; dilution, 1:1,000, GAPDH, cat. no. ab9484; dilution, 1:1,000; both Abcam, Cambridge, MA, USA) at 4°C for overnight. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (cat. no. ab205719; dilution, 1:5,000; Abcam) at room temperature for 4 h. Band signals were detected using enhanced chemiluminescence (Beyotime Institute of Biotechnology) and analyzed with ImageJ v1.43 software (National Institutes of Health, Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded in 96-well plates at a density of 3,000 cells/well. Following transfection for 0, 24, 48 or 72 h, 10 µl CCK-8 reagent (Beyotime Institute of Biotechnology) was added to each well and incubated for another 2 h at 37°C. Optical density was measured at 450 nm using a microplate reader. All experiments were repeated at least three times.
Cell migration assay
Cells were seeded in 96-well plates at a density of 3×104 cells/well and cultured to ~90% confluence. A wound was created at the cell surface and washed twice with PBS. Images of the cells were captured after 0 and 24 h to record wound width. Cell migration rate was calculated by subtracting the value of the wound distance at 0 h from the value at 24 h after scratching.
Transwell invasion assay
For invasion assays, 4×104 cells in serum-free DMEM were placed into the upper chamber of an insert (8 µm, Corning Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences, San Jose, CA, USA). DMEM containing 10% FBS were added to the lower chamber. After 24 h of incubation, cells remaining on the upper membrane were removed with cotton wool, whereas migratory cells were stained with 10% Giemsa in methanol for 4 h at room temperature, imaged and counted using an inverted light microscope (magnification ×200; Canon, Inc., Tokyo, Japan).
Luciferase activity assay
Using the online prediction algorithm TargetScan (http://www.targetscan.org), a total of 665 genes were identified to contain putative binding sites for miR-140-3p. From these predicted genes, TRIM28 was selected for further study as it was reported to be abnormally expressed in human cancers (16–18). The 3′untranslated region (3′-UTR) of TRIM28 obtained from genome was cloned into pMIRREPORT (Promega Corporation, Madison, WI, USA) and designated wild-type (Wt) 3′-UTR. A site-directed mutagenesis kit (Takara Biotechnology Co., Ltd.) was used to generate mutant (Mt) 3′-UTR. MCF-7 and MDA-MB-453 cells (2,000 cells/well) were co-transfected with 50 nM miR-140-3p mimic or NC, and 1 µg Wt or Mt 3′-UTR, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. Following transfection for 48 h, luciferase activities were measured with a dual-luciferase reporter assay system (Promega Corporation) following normalized to Renilla luciferase activity, according to the manufacturer's protocol.
Statistical analysis
Data were expressed as the means ± standard deviation and analyzed using GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). All experiments were repeated in triplicates. Students' t-test was used to compare two groups, and one-way analysis of variance and Tukey post-hoc test were used to make group comparisons. Pearson correlation coefficients were used for correlation analysis between miR-140-3p and TRIM28. The Kaplan-Meier method and log-rank test were used to estimate the effect of miR-140-3p on overall survival of patients with BC. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-140-3p and TRIM28 in BC tissues and cell lines
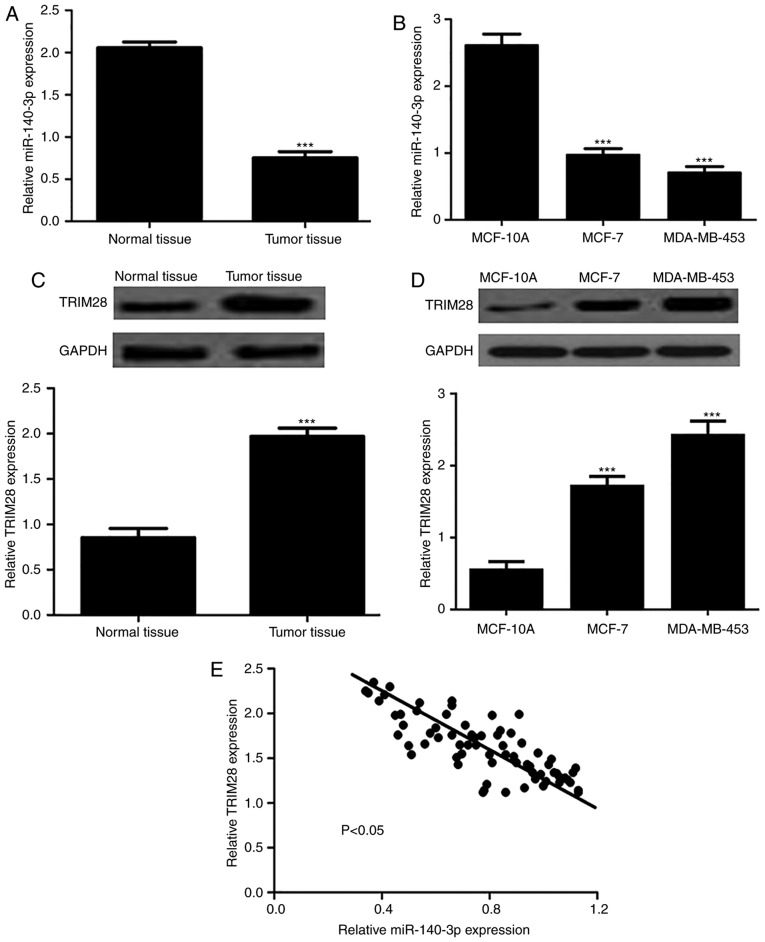
As shown in Fig. 1A and B, the expression levels of miR-140-3p were significantly reduced in BC tissues and cell lines compared with in normal tissues and cells. Conversely, TRIM28 expression was markedly elevated in BC tissues and cell lines (Fig. 1C and D). In addition, the correlation between miR-140-3p and TRIM28 in BC tissues was examined and an inverse correlation was identified (Fig. 1E). Collectively, these results demonstrated that in BC, miR-140-3p expression was reduced and TRIM28 was overexpressed.
Figure 1.
Expression of miR-140-3p in breast cancer tissues and cell lines. (A) miR-140-3p expression in breast cancer tissues and adjacent normal tissues. (B) miR-140-3p expression in breast cancer cell lines (MCF-7 and MDA-MB-453) and normal breast epithelial cells (MCF-10A). (C) TRIM28 expression in breast cancer tissues and adjacent normal tissues. (D) TRIM28 expression in breast cancer cell lines (MCF-7 and MDA-MB-453) and normal breast epithelial cells (MCF-10A). (E) Correlation of miR-140-3p and TRIM28 expression in breast cancer tissues. ***P<0.001. miR-140-3p, microRNA-140-3p; TRIM28, tripartite motif 28.
Clinical significance of miR-140-3p expression in BC
The median value of miR-140-3p expression was used to classify the enrolled patients with BC into high or low miR-140-3p expression groups. Kaplan-Meier curve revealed that low miR-140-3p expression predicted a worse 5-year overall survival for patients with BC compared with high miR-140-3p expression (Fig. 2).
Figure 2.
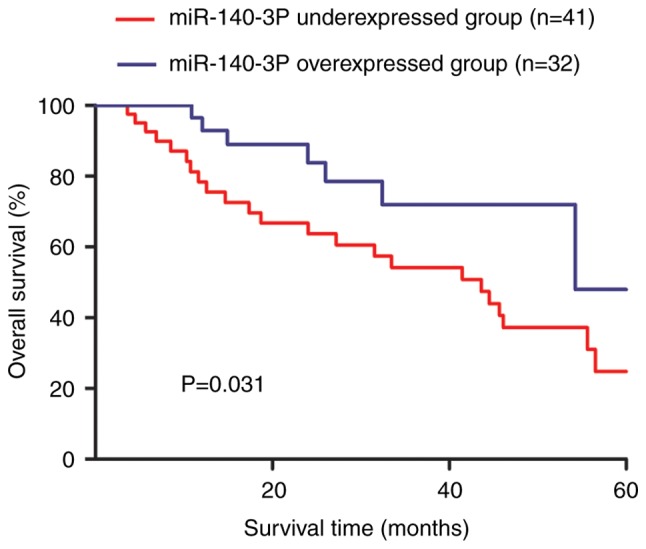
Kaplan-Meier curve and log-rank test were conducted to assess the effects of miR-140-3p expression on the 5-year overall survival of patients with breast cancer. miR-140-3p, microRNA-140-3p.
miR-140-3p inhibits cell proliferation, migration and invasion of BC cell lines in vitro
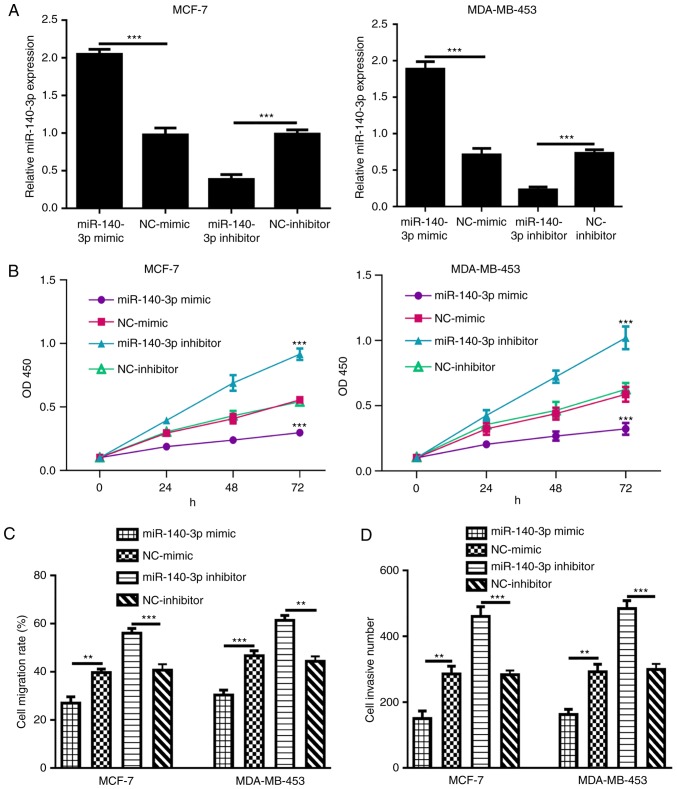
Proliferation and migration are indicators of the malignancy of tumor cells (19). RT-qPCR results demonstrated that the expression levels of miR-140-3p were significantly elevated following miR-140-3p mimic transfection, but were reduced by miR-140-3p inhibitor transfection (Fig. 3A). Subsequently, a CCK-8 assay demonstrated that cell proliferation in the miR-140-3p mimic group was significantly lower than in the NC-mimic group (Fig. 3B). Conversely, miR-140-3p inhibitor significantly elevated cell proliferation compared with NC-inhibitor (Fig. 3B). As shown in Fig. 4C, cell migration in the miR-140-3p mimic-transfected group was significantly decreased compared with that in the NC-mimic group, whereas migration in the miR-140-3p inhibitor group was markedly increased compared with that in the NC-inhibitor group. Transwell invasion assays indicated that cell invasion was enhanced by miR-140-3p inhibitor but decreased by miR-140-3p mimic (Fig. 3D).
Figure 3.
miR-140-3p overexpression inhibits breast cancer cell proliferation, migration and invasion. (A) miR-140-3p expression, (B) cell proliferation, ***P<0.001 miR-140-3p mimic vs. NC-mimic, or miR-140-3p inhibitor vs. NC-inhibitor (C) cell migration and (D) cell invasion in breast cancer cell lines (MCF-7 and MDA-MB-453) with synthetic RNA transfection. ***P<0.001, **P<0.01. miR-140-3p, microRNA-140-3p; NC, negative control; OD, optical density.
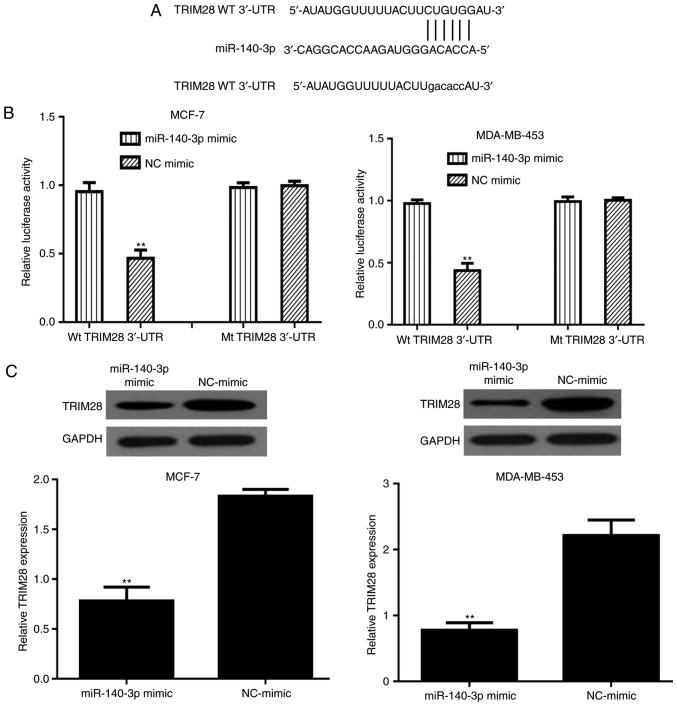
Figure 4.
TRIM28 is a direct target of miR-140-3p. (A) miR-140-3p binding site within the 3′-UTR of TRIM28 was predicted by TargetScan. (B) miR-140-3p mimic suppressed the luciferase activity of cells containing the Wt TRIM28 3′-UTR but not those containing Mt TRIM28 3′-UTR. **P<0.01 miR-140-3p vs. NC-mimic. (C) TRIM28 expression in breast cancer cell lines with miR-140-3p mimic or NC-mimic transfection. **P<0.01. miR-140-3p, microRNA-140-3p; Mt, mutant; NC, negative control; TRIM28, tripartite motif 28; UTR, untranslated region; Wt, wild-type.
TRIM28 is a direct target of miR-140-3p
The binding site between miR-140-3p and the 3′-UTR of TRIM28 is presented in Fig. 4A. The mutant sequence at the binding site between the 3′-UTR of TRIM28 and miR-140-3p is also shown in Fig. 4A. To confirm this prediction, a luciferase reporter system with TRIM28 Wt 3′-UTR or Mt 3′-UTR and miR-140-3p mimic or NC-mimic co-transfection was constructed. As shown in Fig. 4B, the luciferase activity of cells transfected with Wt 3′-UTR was suppressed by the miR-140-3p mimic. However, the miR-140-3p mimic did not alter the luciferase activity in cells transfected with Mt 3′-UTR (Fig. 4B). To validate whether TRIM28 can be regulated by miR-140-3p, the levels of TRIM28 in miR-140-3p mimic- or NC-mimic-transfected cells were measured. As expected, the levels of TRIM28 were downregulated by miR-140-3p mimic in BC cell lines (Fig. 4C).
Effects of miR-140-3p on cell proliferation, migration and invasion are partially reversed by TRIM28
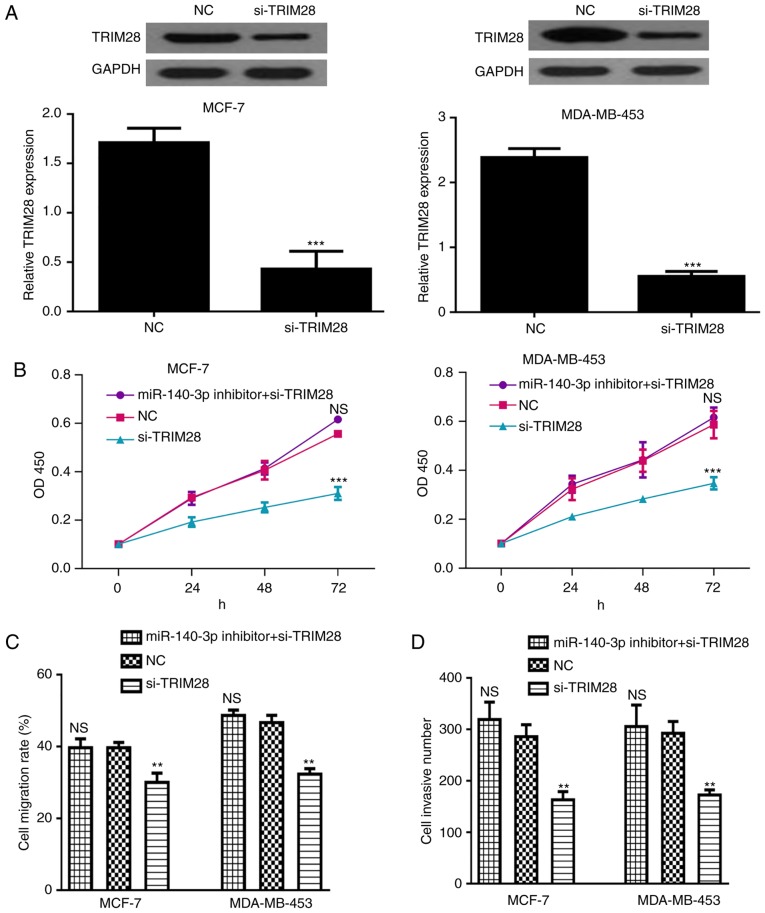
To determine whether TRIM28 can reverse the inhibitory effects of miR-140-3p, si-TRIM28 and miR-140-3p inhibitor were co-transfected into BC cells. Transfection with si-TRIM28 could downregulate the expression of TRIM28 (Fig. 5A). In addition, the inhibitory effects of si-TRIM28 on cell proliferation, migration and invasion were reversed by miR-140-3p inhibitor, suggesting that TRIM28 may be a functional target of miR-140-3p (Fig. 5B-D).
Figure 5.
miR-140-3p regulates cell proliferation, migration and invasion through targeting TRIM28. (A) TRIM28 expression in breast cancer cell lines (MCF-7 and MDA-MB-453) following si-TRIM28 or NC transfection. (B) Cell proliferation, (C) cell migration and (D) cell invasion in breast cancer cell lines (MCF-7 and MDA-MB-453) following miR-140-3p inhibitor or si-TRIM28 transfection. ***P<0.001 or **P<0.01 si-TRIM28 vs. NC. miR-140-3p, microRNA-140-3p; NC, negative control; NS, not significant; OD, optical density; si-TRIM28, TRIM28 small interfering RNA; TRIM28, tripartite motif 28.
Discussion
It has been demonstrated that an improved understanding of tumor progression mechanisms could greatly improve the survival of cancer patients (5–8). In recent decades, the importance of miRNAs in tumors has attracted more attention (5–7). miR-140 is located at the intron of the WW domain containing E3 ubiquitin protein ligase 2 gene and produces two mature microRNAs: miR-140-5p and miR-140-3p (20). miR-140-5p has been reported to act as a tumor suppressor in several cancer types, including gastric cancer, BC and ovarian cancer (21–23). miR-140-3p has also been revealed to be abnormally expressed in several types of cancer, including lung cancer and spinal chordoma (13,14). However, whether miR-140-3p serves a role in BC remains largely unknown.
In the present study, miR-140-3p expression was downregulated in BC tissues and cell lines compared with in adjacent normal tissues and a normal cell line. Furthermore, low miR-140-3p expression predicted a poor prognosis of patients with BC. Functional assays revealed that miR-140-3p overexpression inhibited BC cell proliferation, migration and invasion. These results collectively suggested that miR-140-3p may function as a tumor suppressor in BC. Using a miRNA target prediction algorithm, it was suggested that TRIM28 may contain a putative binding site for miR-140-3p in its 3′-UTR. Additionally, TRIM28 expression was upregulated in BC tissues and cell lines.
TRIM28 belongs to the TRIM protein family and serves a critical role in a wide range of biological activities (24,25). Hao et al revealed that high TRIM28 expression is a predictor for poor prognosis in patients with BC (16). Wei et al reported that TRIM28 enhances BC metastasis through direct interaction with twist-related protein 1 (TWIST1), to protect TWIST1 from degradation, suggesting that TRIM28 could be a target for BC treatment (17). These studies highlight the importance of TRIM28 in the progression of BC. A previous study demonstrated that TRIM28 expression could be regulated by miR-491 in glioma (18). Therefore, this study investigated whether TRIM28 is also a downstream target of miR-140-3p. A luciferase activity reporter assay and western blot analysis were performed to validate TRIM28 as a direct target of miR-140-3p. Additionally, an inverse correlation between miR-140-3p and TRIM28 expression was detected in BC tissues. Furthermore, BC cells were co-transfected with si-TRIM28 and miR-140-3p inhibitor, revealing that the inhibitory effects of si-TRIM28 on BC cell behaviors may be reversed by miR-140-3p inhibitor. These results demonstrated that TRIM28 was an effector for the role of miR-140-3p on BC cell behaviors.
In conclusion, these findings clearly demonstrated that miR-140-3p may act as a tumor suppressor and that its effects were exerted by regulating TRIM28 in BC. Additionally, restoration of miR-140-3p expression may be a novel strategy for BC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YZ, BW and HW conceived and designed the study. YZ, BW, YW, GC, QL and HW performed the experiments. YZ, BW, YW, GC, QL and HW wrote the paper. YZ, BW, YW, GC, QL and HW reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
The collection and the use of all tissue samples were approved by the Ethics Committee of First Affiliated Hospital of Jiamusi University (Jiamusi, China). Written informed consent was obtained from all participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Tzanninis IG, Kotteas EA, Ntanasis-Stathopoulos I, Kontogianni P, Fotopoulos G. Management and outcomes in metaplastic breast cancer. Clin Breast Cancer. 2016;16:437–443. doi: 10.1016/j.clbc.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenblick A, Pondé N, Piccart M. Metastatic breast cancer: The Odyssey of personalization. Mol Oncol. 2016;10:1147–1159. doi: 10.1016/j.molonc.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M. The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomed Pharmacother. 2016;78:129–139. doi: 10.1016/j.biopha.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Asghari F, Haghnavaz N, Baradaran B, Hemmatzadeh M, Kazemi T. Tumor suppressor microRNAs: Targeted molecules and signaling pathways in breast cancer. Biomed Pharmacother. 2016;81:305–317. doi: 10.1016/j.biopha.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Gravgaard KH, Lyng MB, Laenkholm AV, Søkilde R, Nielsen BS, Litman T, Ditzel HJ. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134:207–217. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 10.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliman SJ, Howley BV, Mehta DS, Fearnhead HO, Kemp DM, Barkley LR. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis. 2014;3:e113. doi: 10.1038/oncsis.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong XM, Zhang GH, Huo YK, Zhao XH, Cao DW, Guo SF, Li AM, Zhang XR. MicroRNA-140-3p inhibits proliferation, migration and invasion of lung cancer cells by targeting ATP6AP2. Int J Clin Exp Pathol. 2015;8:12845–12852. [PMC free article] [PubMed] [Google Scholar]
- 14.Zou MX, Huang W, Wang XB, Lv GH, Li J, Deng YW. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol. 2014;7:4877–4885. [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Hao L, Leng J, Xiao R, Kingsley T, Li X, Tu Z, Yang X, Deng X, Xiong M, Xiong J, Zhang Q. Bioinformatics analysis of the prognostic value of Tripartite Motif 28 in breast cancer. Oncol Lett. 2017;13:2670–2678. doi: 10.3892/ol.2017.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei C, Cheng J, Zhou B, Zhu L, Khan MA, He T, Zhou S, He J, Lu X, Chen H, et al. Tripartite motif containing 28 (TRIM28) promotes breast cancer metastasis by stabilizing TWIST1 protein. Sci Rep. 2016;6:29822. doi: 10.1038/srep29822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi Z, Cai S, Cai J, Chen L, Yao Y, Chen L, Mao Y. miR-491 regulates glioma cells proliferation by targeting TRIM28 in vitro. BMC Neurol. 2016;16:248. doi: 10.1186/s12883-016-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi: 10.1186/s12943-017-0708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang Z, Mao J. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24:386–392. doi: 10.1038/cgt.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. 2015;75:117–122. doi: 10.1016/j.biopha.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 24.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Chen DT, Kurtyka C, Rawal B, Fulp WJ, Haura EB, Cress WD. Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem. 2012;287:40106–40118. doi: 10.1074/jbc.M112.380865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.