Abstract
Long non-coding RNAs (lncRNAs) may serve an important role in cancer development and may also be suitable for use as prognostic biomarkers. At present, the role of lncRNAs in bladder cancer remains unclear. The present study examined the potential involvement of lncRNA LINC00460 in bladder urothelial carcinoma using data from The Caner Genome Atlas (TCGA) and cell line experiments. The results indicated that LINC00460 expression levels were increased in bladder urothelial carcinoma tissues and bladder cancer 5637 and T24 cell lines compared with corresponding normal controls (P<0.05). TCGA data indicated that LINC00460 expression was negatively correlated with a positive prognosis in patients with bladder urothelial carcinoma (P<0.05). Consistently, the downregulation of LINC00460 with short hairpin RNA significantly suppressed 5637 and T24 cell proliferation and migration. Therefore, it was suggested that strategies that target LINC00460 may be developed as novel therapeutic approaches for the treatment of bladder cancer. In addition, the expression level of androgen receptor (AR) was downregulated in bladder urothelial carcinoma tissues and exhibited a negative correlation with the expression level of LINC00460 (r=−0.43; P<0.0001), based on the data from TCGA. We hypothesized that LINC00460 may serve an oncogenic role by regulating the expression of AR.
Keywords: bladder cancer, long non-coding RNA, prognosis
Introduction
Bladder cancer is the seventh most common type of cancer in the male population worldwide, and eleventh including females (1). There are 2 major types of bladder cancer: Non-muscle invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC). Bladder cancer exhibits a high frequency of relapse and a poor clinical outcome once the tumors progress to muscle-invasive disease (2). Patients with MIBC have a poor prognosis due to the aggressive nature of the tumor and its resistance to chemo- and radiotherapy (3). Furthermore, for patients with MIBC, the risk of developing lymph node metastases is increased and chemotherapy is less effective in comparison with patients with NMIBC (4). Therapies and prognosis for this type of cancer depend on the clinical information and individual tumor pathology. However, in certain cases, even when tumors present with similar histology, they respond differently to the same treatment, resulting in different survival outcomes for the patients (5).
Protein-coding genes constitute only 2% of the total genome sequence; the remaining sequences produce various classes of functional non-coding RNAs (6). There are two major classes of non-coding RNAs, including small and long non-coding RNAs (lncRNAs). The difference between lncRNAs and small non-coding RNAs is primarily their size, with the length of lncRNAs defined as being >200 nucleotides. Similar to other non-coding RNAs, lncRNA transcripts are associated with a wide range of cancer types, including bladder cancer (7–10). Alterations in the expression levels of lncRNAs have been demonstrated to be associated with a number of important cellular functions, and may promote the migration and invasion of cancer cells (4,11). Non-coding RNAs have attracted increasing attention in previous years regarding their role in bladder cancer, with the number of studies on this topic increasing considerably. The combination of traditional methods of treatment with the use of non-coding RNAs may provide improved therapy for patients. The accumulation of data in this field may assist in elucidating the molecular profiles of patients with bladder cancer and contribute to the use of non-coding RNAs as tools for precision medicine to target critical genes in bladder cancer.
LINC00460 is a human lncRNA gene, transcribed from chromosome 13, measuring 935 bp; its function is poorly understood. Recent studies have demonstrated that LINC00460 is associated with kidney cancer, nasopharyngeal carcinoma and head and neck squamous cell carcinoma (12–14). A previous study demonstrated, using a bioinformatics approach, that LINC00460 may serve an important role in tumorigenesis and metastasis through the regulation of the cell cycle and cell death (15). During the analysis of the differential expression of lncRNAs in bladder cancer tissues in the present study, it was identified that LINC00460 was highly upregulated compared with the normal adjacent tissue. Furthermore, the upregulation of LINC00460 was demonstrated to be associated with the poor survival of patients. We hypothesized that this lncRNA may serve an important role in the regulation of biological processes in bladder urothelial carcinoma. However, the precise function and the underlying molecular mechanisms remain to be elucidated.
Bladder cancer predominantly presents in men (16). Factors that are exclusive to males are likely to serve critical roles in the development of bladder cancer (17,18). An increasing amount of evidence has suggested the involvement of androgen receptor (AR) signaling in the development and progression of bladder cancer (19–21). The present study investigated the role of the lncRNA LINC00460 in bladder urothelial carcinoma using The Cancer Genome Atlas (TCGA) data and cell experiments. The results indicated that increased LINC00460 expression was a characteristic molecular change in bladder urothelial carcinoma tissues, and in 5637 and T24 cell lines. Therefore, the effects of aberrant LINC00460 expression on the biological behavior of 5637 and T24 cells were additionally investigated. The results provided novel insights into the function and mechanisms of LINC00460 in bladder urothelial carcinoma pathogenesis, and identified LINC0046 as a potential therapeutic target for cancer intervention.
Materials and methods
TCGA database
Gene expression data obtained by RNA sequencing and the corresponding clinical data for 412 patients (including 413 samples) with bladder urothelial carcinoma were downloaded from TCGA (https://cancergenome.nih.gov/). All RNA expression levels of the samples were normalized. The edgeR Bioconductor package was used to analyze P-values and fold-change (FC) using R (version 3.4.0) (22). A gene was defined as differentially expressed between cancerous and normal tissues when the false discovery rate-adjusted P<0.01 and the FC was ≥2-fold increased or decreased.
Cell culture and transfection
The expression of LINC00460 and the basic characterization of bladder urothelial carcinoma cell lines were initially investigated using the Expression Atlas database (23) and the American Type Culture Collection website (https://www.atcc.org/), respectively (Table I). Based on these results, the 5637 cell line was selected, as the expression of LINC00460 was the highest in these cells compared with the others included in the analysis. From the verification of LINC00460 expression using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in bladder cancer cell lines (T24, J82, TCCSUP and UM-UC-3), it was identified that LINC00460 was also upregulated in the T24 cell line compared with the normal bladder epithelial SV-HUC-1 cell line (data not show).
Table I.
Expression of LINC00460 and characterization of bladder cancer cell lines from the Expression Atlas database and the website of American Type Culture Collection cell lines.
| Cell line | Expression value, transcripts per million | Histological grade | Age | Sex |
|---|---|---|---|---|
| 5637 | 51 | Grade 2 | 68 | Male |
| U-BLC1 | 43 | Grade 3 | 84 | Female |
| HT-1197 | 11 | Grade 4 | 44 | Male |
| TCCSUP | 4 | Grade 4 | 67 | Female |
| J82 | 3 | Grade 3 | 58 | Male |
| RT-112 | 0.9 | Grade 2 | Not reported | Female |
| 253J | 0 | Grade 4 | 53 | Male |
| HT-1376 | 0 | Grade 3 | 58 | Female |
| RT4 | 0 | Grade 1 | 63 | Male |
| SW780 | 0 | Grade 1 | 80 | Female |
| T24 | 0 | Grade 3 | 81 | Female |
The 5637, T24, J82, TCCSUP, UM-UC-3 and SV-HUC-1 cells were purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). The 5637 and T24 cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the SV-HUC-1 cell line was cultured in F12 K medium (Gibco; Thermo Fisher Scientific, Inc.). J82, TCCSUP and UM-UC-3 cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.). These media were supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin, in a humidified atmosphere with 5% CO2 at 37°C.
A total of three short hairpin RNAs (shRNAs) targeting LINC00460 were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China) and transfected into 5637 and T24 cells with Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The three shRNA sequences specially targeting LINC00460 were designed and cloned into a pGU6/green fluorescent protein (GFP)/Neo-shRNA vector (Shanghai GenePharma Co., Ltd.). Cells (1×105 T24 cells/well; 3×105 5,637 cells/well) were seeded in six-well culture plates and transfected with the 2.5 µg sh-LINC00460 in each well. The most effective shRNA sequence (sh-3) in achieving knockdown of LINC00460 expression was selected for subsequent experiments 48 h after the transfection. The sh-LINC00460 sequences were as follows: sh-1, 5′-GCTAAGACCTAATAGCCAATA-3′; sh-2, 5′-GCCATCCACTTCAAAGTATTC-3′; and sh-3, 5′-ACCTTGGTCAAACGTTTAACC-3′. A scrambled shRNA was used as the negative control (sh-NC) in the experiments with the following sequence: 5′-GTTCTCCGAACGTGTCACGT-3′.
RT-qPCR assays
Total RNA was extracted from 5637, T24, J82, TCCSUP, UM-UC-3 and SV-HUC-1 cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Reverse transcription was conducted using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed to detect the expression of LINC00460 using SYBR® Premix Ex Taq™ II kit (Takara Biotechnology Co., Ltd.). RT-qPCR assays were performed using an Applied Biosystems QuantStudio 3 system (Applied Biosystems; Thermo Fisher Scientific, Inc.), with β-actin as an endogenous control. The primer sequences used were as follows: LINC00460 forward, 5′-CGAGAAGGCCACCTATGAGC-3′ and reverse, 5′-TGAAGTGGATGGCTCAGGAA-3′; β-actin forward, 5′-CCGTGAAAAGATGACCCAGATC-3′ and reverse, 5′-CACAGCCTGGATGGCTACGT-3′. A two-step qPCR was performed as follows: Initial denaturation in 95°C for 30 sec; 40 of cycles of denaturation at 95°C for 5 sec; and annealing and elongation at 60°C for 34 sec. The 2−ΔΔCq method was used to quantify LINC00460 (24).
Cell proliferation assay
Cells were seeded in 96-well plates at a density of 1.0×104 and 1.5×104 cells per well for T24 and 5637 cells, respectively. Following incubation at 37°C with 5% CO2 for 0, 24, 48 and 72 h, the absorbance of each sample was measured at 450 nm. Cell proliferation was evaluated using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Shanghai, China) according to the manufacturer's protocol.
Cell migration assay
The 5637 and T24 cells were seeded (1×106 cells/plate) in 6-well plates and incubated in serum-free RPMI-1640 medium. A wound was produced using a sterile 100 µl pipette tip when a confluent cell monolayer had formed. The size of the wound was measured using an inverted light microscope (magnification, ×200) and images were captured at 0 and 24 h.
Statistical analysis
SPSS 23.0 software (IBM Corp., Armonk, NY, USA) was used to perform statistical analysis. The differential expression levels of LINC00460 and AR between the cancerous and adjacent tissues were analyzed with t-tests. The differences in LINC00460 expression between bladder cancer 5637 and T24 cell lines and the normal bladder epithelial SV-HUC-1 cell line was assessed using a one-way analysis of variance with Tamhane's post-hoc test. The correlation between LINC00460 and AR was analyzed by Spearman's rank correlation analysis following logarithmic (log10) conversion of the original data. For the survival analysis, the LINC00460 expression data matrix and clinical data files were matched for each sample using the sample ID. The samples were divided into the LINC00460-low and LINC00460-high groups based on the cut-off of the median value, and according to sex. The LINC00460-high group contained the samples with an exact median value. A Kaplan-Meier plot was generated using the survival package in R (version 3.4.0) with log-rank tests. P<0.05 was considered to indicate a statistically significant difference.
Results
LINC00460 is upregulated in bladder urothelial carcinoma tissues and cell lines
The differences in LINC00460 expression between the bladder urothelial carcinoma tissues and the normal controls were analyzed using TCGA data. LINC00460 was significantly upregulated in bladder urothelial carcinoma tissues compared with the normal controls (P<0.0001; Fig. 1A). The expression level of LINC00460 was detected by RT-qPCR in bladder urothelial carcinoma and normal bladder epithelial cell lines. An increase in LINC00460 expression was observed in bladder cancer 5637 and T24 cell lines compared with the normal bladder epithelial SV-HUC-1 cell line (P<0.05; Fig. 1B).
Figure 1.
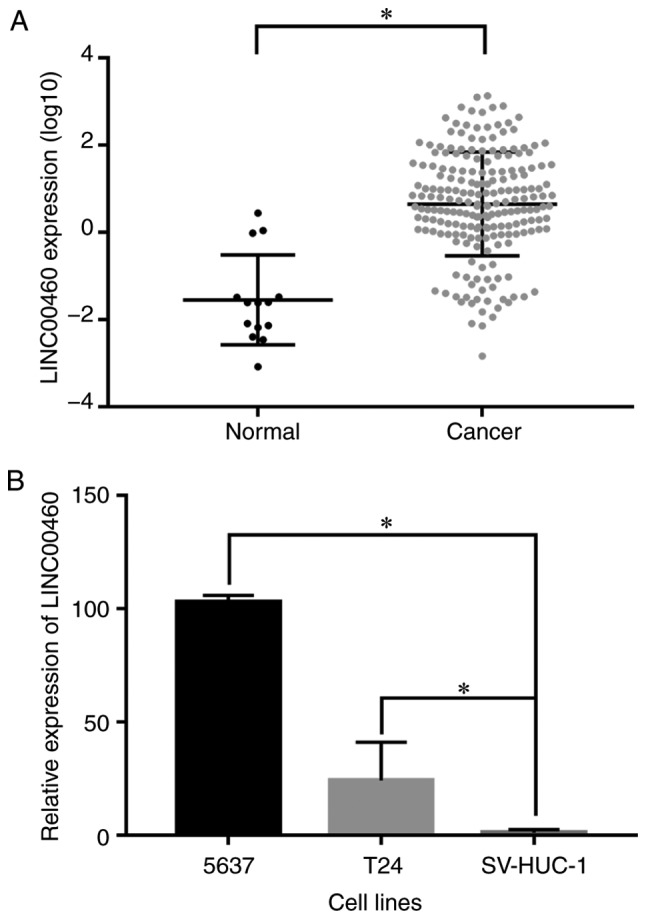
Expression of LINC00460 is upregulated in bladder cancer tissues and cell lines. (A) Relative expression of LINC00460 in bladder cancer tissues and normal controls. (B) Relative expression of LINC00460 in bladder cancer cell lines and a normal bladder epithelial cell line. *P<0.05.
Upregulation of LINC00460 is associated with poor survival
LINC00460 expression levels and the clinical data of 412 patients with bladder urothelial carcinoma were downloaded from TCGA. All patients, male and female, were divided into LINC00460-high and LINC00460-low groups using the mean expression value as the cut-off. The survival time was plotted using a Kaplan-Meier curve stratified by LINC00460-high and LINC00460-low groups. High levels of LINC00460 were significantly associated with a decreased survival time in male patients (P=0.03229; Fig. 2A). There was a similar association observed in the overall group (P=0.04678; Fig. 2B), but not among the female patients (P=0.42445; Fig. 2C).
Figure 2.
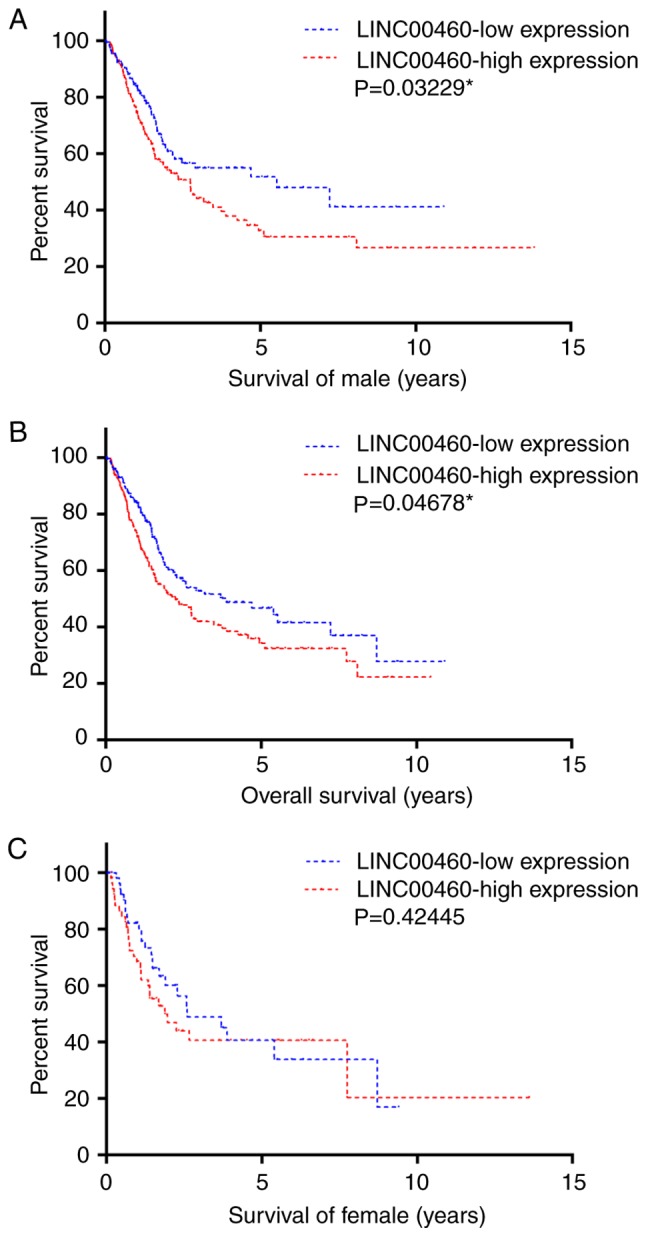
Association between the expression levels of LINC00460 and the overall survival of patients with bladder cancer. A high expression of LINC00460 was associated with poor survival in (A) male patients and (B) the whole patient cohort, but not (C) female patients.
Downregulation of LINC00460 inhibits the proliferation and migration of bladder urothelial carcinoma cells
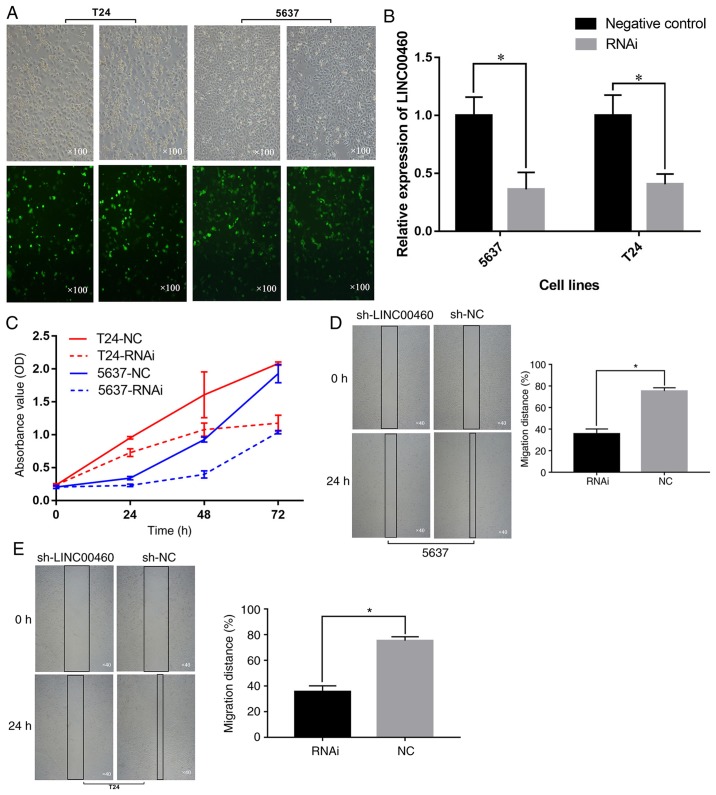
To explore the function of LINC00460, an shRNA targeting LINC00460 was transfected into 5637 and T24 cells. GFP visualization indicated that the shRNA was efficiently transfected into cells (Fig. 3A). RT-qPCR indicated that LINC00460 was significantly downregulated at 48 h after transfection of shRNA in the 5637 and T24 cell lines compared with the control group (P<0.05; Fig. 3B). A CCK-8 assay demonstrated that the downregulation of LINC00460 inhibited the proliferation of 5637 and T24 cells in vitro (P<0.05; Fig. 3C). In addition, the effect of LINC00460 on the migration capacity of 5637 and T24 cells was observed via a wound-healing assay. The wound-healing assay revealed that the knockdown of LINC00460 decreased the migration distance of cells (Fig. 3D and E). Overall, the results demonstrated that the silencing of LINC00460 may inhibit the proliferation and migration abilities of 5637 and T24 cells.
Figure 3.
Downregulation of LINC00460 inhibits the proliferation and migration of bladder cancer cells. (A) The green fluorescent protein images indicate the efficiency of transfection. Magnification, ×100. (B) Reverse transcription quantitative polymerase chain reaction assays demonstrated that LINC00460 was significantly downregulated following short hairpin RNA transfection. (C) The downregulation of LINC00460 inhibited the proliferation of bladder cancer cells in vitro. The knockdown of LINC00460 decreased the distance of migration in bladder cancer (D) 5637 and (E) T24 cells. Magnification, ×40. The experiments were performed in triplicate and data are expressed as the mean ± standard deviation. *P<0.05. OD, optical density; NC, negative control; RNAi, RNA interference; sh, short hairpin.
AR is downregulated in bladder urothelial carcinoma tissues and is negatively correlated with LINC00460 expression
AR expression was analyzed in bladder urothelial carcinoma and adjacent tissues, and its correlation with LINC00460 expression, using TCGA data. AR was significantly downregulated in bladder urothelial carcinoma tissues compared with the normal tissues (P<0.05; Fig. 4A) and was negatively correlated with the expression of LINC00460 (P<0.0001; r=−0.4715; Fig. 4B).
Figure 4.
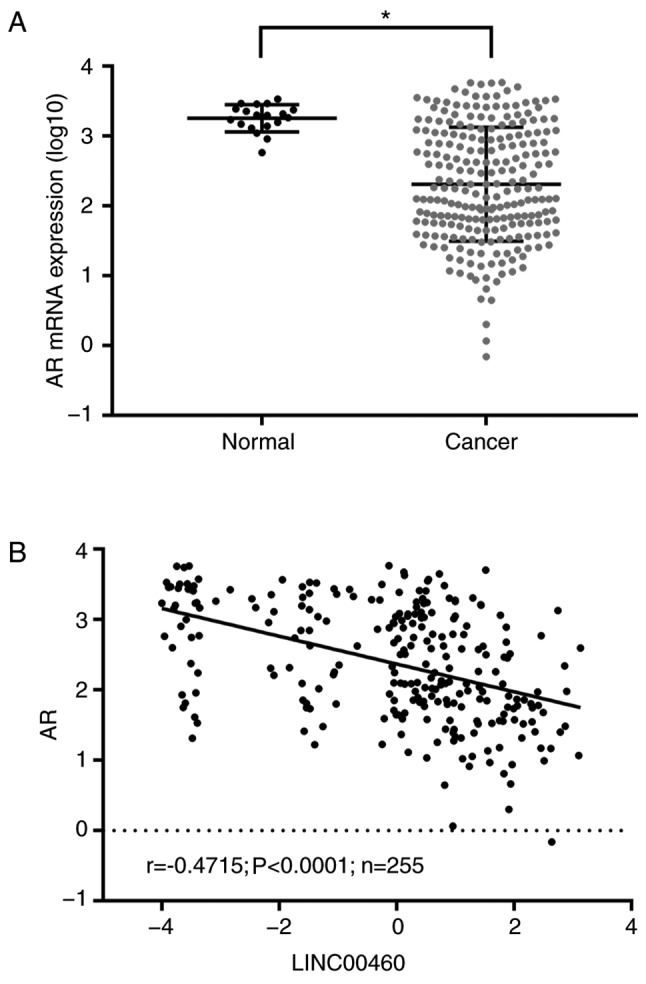
AR is downregulated in bladder urothelial carcinoma tissues and its expression is negatively correlated with the expression of LINC00460. (A) AR mRNA expression was significantly downregulated in bladder urothelial carcinoma tissues. *P<0.05. (B) AR mRNA expression was negatively correlated with the expression of LINC00460 (r=−0.43; P<0.0001; n=255). AR, androgen receptor.
Discussion
The majority of cases of bladder cancer are non-muscle invasive at the time of initial diagnosis, and the initial treatment options generally focus on tumor resection, and the prevention of recurrence or progression to muscle-invasive disease (25). The 5-year recurrence-free survival rate ranges from 62–89% for localized muscle invasive bladder cancer following a radical cystectomy (26). However, for patients with metastatic disease, the 5-year survival is decreased, at ~5% (27). The standard drugs used in chemotherapy for the perioperative therapy for MIBC and metastatic disease are cisplatin-based. The median overall survival time with chemotherapy treatment is slightly >1 year, with an objective response rate of 40–60% (28–30). In May 2016, atezolizumab was approved as a second-line therapy for patients with locally advanced or metastatic urothelial carcinoma that had progressed during or following platinum-containing chemotherapy; it became the first available immunotherapeutic antibody to target programmed death-ligand 1. Although targeted therapies have become standard for numerous other malignancies, the number of approved targeted agents in bladder cancer is limited. lncRNAs modulate the expression of genes that are pivotal in the pathways associated with bladder cancer development and progression; for example, the HOX transcript antisense RNA/zinc finger E-box binding homeobox 1 (ZEB1) interaction affects epithelial-mesenchymal transition in bladder cancer cell lines (31), and metastasis associated lung adenocarcinoma transcript 1/Epithelial cadherin (32) and urothelial cancer associated 1/ZEB1/zinc finger E-box binding homeobox 2 (33) interact to affect the invasion and metastasis of bladder cancer cells. Data from the Cancer Research Network (CRN) database (34) indicated that LINC00460 is upregulated in 14 types of cancer, suggesting that LINC00460 may be a regulator in cancer cell development (35). The role of LINC00460 in bladder cancer was selected for analysis in the present study, as the expression fold change in bladder urothelial carcinoma was the highest among all the types of cancer included in the CRN database.
The present study explored the role of LINC00460 in bladder urothelial carcinoma using TCGA data and cell experiments. The results demonstrated that LINC00460 was upregulated in bladder cancer tissues compared with the corresponding normal controls, while the effect of LINC00460 on the prognosis for bladder urothelial carcinoma was only observed in male patients. Concomitantly, the results also indicated that AR was downregulated in bladder urothelial carcinoma tissues, which was negatively correlated with LINC00460 expression. Epidemiological and clinical data have suggested that males are 3–4 times more likely to develop bladder urothelial carcinoma compared with females (16,36). Emerging preclinical evidence has indicated the involvement of AR signaling in the development and progression of bladder urothelial carcinoma; androgens, β-catenin, cluster of differentiation 24, cyclins, epidermal growth factor receptor/receptor tyrosine-protein kinase erbB-2, ETS domain-containing protein Elk-1, several AR coregulators and orphan receptors have been demonstrated to directly or indirectly modulate several molecules/pathways involved in bladder urothelial carcinoma cell proliferation (19,20,37–40). As a result, we hypothesized that LINC00460 may serve its oncogenic role by regulating the expression of AR.
The effects of LINC00460 on the proliferation and migration of 5637 and T24 cells were determined using gain- and loss-of-function approaches. The data revealed that the downregulation of LINC00460 inhibited the proliferation and migration of 5637 and T24 cells. Zhang et al (15) performed a regulatory network analysis of LINC00460, and the results indicated that LINC00460 was associated with various biological processes, consistent with the results from the present study.
However, the function of LINC00460 was only investigated in bladder urothelial carcinoma cell lines. An in vivo model is required to confirm the results. In addition, the mechanisms underlying the effect of LINC00460 on 5637 and T24 cells are not yet fully characterized. There was a recurring node, double-strand-break repair protein rad21 homolog, between mRNAs and transcription factors associated with LINC00460, as identified through bioinformatics methods in a study conducted by Zhang et al (15), which was previously demonstrated to affect cell growth in breast cancer (35). The results from the present study suggested that the expression level of AR mRNA was downregulated in bladder urothelial carcinoma tissues and was negatively correlated with LINC00460. LINC00460 functions as a competing endogenous RNA to upregulate interleukin-6 through sponging miR-149-5p in the cytoplasm of nasopharyngeal carcinoma (NPC) cells (13). LINC00460 was distributed in the cytoplasm and nucleus in NPC cells (13). The data from the present study implied that LINC00460 distributed in the nucleus may serve its role by regulating the expression level of AR mRNA. However, the underlying mechanisms require additional investigation.
In summary, the present study demonstrated that LINC00460 has potential as a clinically promising biomarker for bladder urothelial carcinoma. LINC00460 regulated the proliferation and migration of 5637 and T24 cells, and these data may provide novel insights into molecular cancer therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Program of Translational Medicine Research on Bladder Cancer: Construction of Translational Medicine Research Center and Collaborative Network in the Area of Bladder Diseases of Liaoning Province (grant no. 2015225009).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PW, MH, MW conceived and supervised the study. LW and XZ performed the experiments. JB and LH conducted the analysis of data from The Cancer Genome Atlas. HH analyzed the experimental data. LW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.van Rhijn BW, Burger M, Lotan Y, Solsona E, Stief CG, Sylvester RJ, Witjes JA, Zlotta AR. Recurrence and progression of disease in non-muscle-invasive bladder cancer: From epidemiology to treatment strategy. Eur Urol. 2009;56:430–442. doi: 10.1016/j.eururo.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Drayton RM, Catto JW. Molecular mechanisms of cisplatin resistance in bladder cancer. Expert Rev Anticancer Ther. 2012;12:271–281. doi: 10.1586/era.11.201. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs BL, Lee CT, Montie JE. Bladder cancer in 2010: How far have we come? CA Cancer J Clin. 2010;60:244–272. doi: 10.3322/caac.20077. [DOI] [PubMed] [Google Scholar]
- 6.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivieri M, Ferro M, Terreri S, Durso M, Romanelli A, Avitabile C, De Cobelli O, Messere A, Bruzzese D, Vannini I, et al. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget. 2016;7:20636–20654. doi: 10.18632/oncotarget.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao F, Lin T, He W, Han J, Zhu D, Hu K, Li W, Zheng Z, Huang J, Xie W. Knockdown of a novel lincRNA AATBC suppresses proliferation and induces apoptosis in bladder cancer. Oncotarget. 2015;6:1064–1078. doi: 10.18632/oncotarget.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467:223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 10.He A, Liu Y, Chen Z, Li J, Chen M, Liu L, Liao X, Lv Z, Zhan Y, Zhuang C, et al. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Cancer Res. 2016;35:125. doi: 10.1186/s13046-016-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZL, Li B, Piccolo SR, Zhang XQ, Li JH, Zhou H, Yang JH, Qu LH. Integrative analysis reveals clinical phenotypes and oncogenic potentials of long non-coding RNAs across 15 cancer types. Oncotarget. 2016;7:35044–35055. doi: 10.18632/oncotarget.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ. LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene. 2018;639:77–84. doi: 10.1016/j.gene.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Tao Y, Liao Q. Long noncoding RNA: A crosslink in biological regulatory network. Brief Bioinform. 2018;19:930–945. doi: 10.1093/bib/bbx042. [DOI] [PubMed] [Google Scholar]
- 16.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;65:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 17.Hartge P, Harvey EB, Linehan WM, Silverman DT, Sullivan JW, Hoover RN, Fraumeni JF., Jr Unexplained excess risk of bladder cancer in men. J Natl Cancer Inst. 1990;82:1636–1640. doi: 10.1093/jnci/82.20.1636. [DOI] [PubMed] [Google Scholar]
- 18.Hemelt M, Yamamoto H, Cheng KK, Zeegers MP. The effect of smoking on the male excess of bladder cancer: A meta-analysis and geographicalanalyses. Int J Cancer. 2009;124:412–419. doi: 10.1002/ijc.23856. [DOI] [PubMed] [Google Scholar]
- 19.Overdevest JB, Knubel KH, Duex JE, Thomas S, Nitz MD, Harding MA, Smith SC, Frierson HF, Conaway M, Theodorescu D. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci USA. 2012;109:E3588–E3596. doi: 10.1073/pnas.1113960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawahara T, Shareef HK, Aljarah AK, Ide H, Li Y, Kashiwagi E, Netto GJ, Zheng Y, Miyamoto H. ELK1 is up-regulated by androgen in bladder cancer cells and promotes tumor progression. Oncotarget. 2015;6:29860–29876. doi: 10.18632/oncotarget.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding G, Yu S, Cheng S, Li G, Yu Y. Androgen receptor (AR) promotes male bladder cancer cell proliferation and migration via regulating CD24 and VEGF. Am J Transl Res. 2016;8:578–587. [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, Burke M, Füllgrabe A, Fuentes AM, George N, et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46(D1):D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Pasin E, Josephson DY, Mitra AP, Cote RJ, Stein JP. Superficial bladder cancer: An update on etiology, molecular development, classification, and natural history. Rev Urol. 2008;10:31–43. [PMC free article] [PubMed] [Google Scholar]
- 26.Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M, et al. Radical cystectomy in the treatment of invasive bladder cancer: Long-term results in 1,054 patients. J Clin Oncol. 2001;19:666–675. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- 27.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et al. SEER cancer statistics review, 1975–2012. Natl Cancer Inst. 2015 [Google Scholar]
- 28.Sternberg CN, de Mulder P, Schornagel JH, Theodore C, Fossa SD, van Oosterom AT, Witjes JA, Spina M, van Groeningen CJ, Duclos B, et al. Seven year update of an EORTC phase III trial of high-dose intensity M-VAC chemotherapy and G-CSF versus classic M-VAC in advanced urothelial tract tumours. Eur J Cancer. 2006;42:50–54. doi: 10.1016/j.ejca.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 29.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–4608. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 30.Saxman SB, Propert KJ, Einhorn LH, Crawford ED, Tannock I, Raghavan D, Loehrer PJ, Sr, Trump D. Long-term follow-up of a phase III intergroup study of cisplatin alone or in combination with methotrexate, vinblastine, and doxorubicin in patients with metastatic urothelial carcinoma: A cooperative group study. J Clin Oncol. 1997;15:2564–2569. doi: 10.1200/JCO.1997.15.7.2564. [DOI] [PubMed] [Google Scholar]
- 31.Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, Fucile C, Richheimer S, Beckham CJ. Expression of the long non-coding RNA HOTAIR Correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One. 2016;11:e0147236. doi: 10.1371/journal.pone.0147236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 33.Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 2016;107:18–27. doi: 10.1111/cas.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JR, Sun CH, Li W, Chao RF, Huang CC, Zhou XJ, Liu CC. Cancer RNA-Seq Nexus: A database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016;44:D944–D951. doi: 10.1093/nar/gkv1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atienza JM, Roth RB, Rosette C, Smylie KJ, Kammerer S, Rehbock J, Ekblom J, Denissenko MF. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther. 2005;4:361–368. doi: 10.1158/1535-7163.MCT-04-0241. [DOI] [PubMed] [Google Scholar]
- 36.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Zheng Y, Izumi K, Ishiguro H, Ye B, Li F, Miyamoto H. Androgen activates β-catenin signaling in bladder cancer cells. Endocr Relat Cancer. 2013;20:293–304. doi: 10.1530/ERC-12-0328. [DOI] [PubMed] [Google Scholar]
- 38.Shiota M, Takeuchi A, Yokomizo A, Kashiwagi E, Tatsugami K, Kuroiwa K, Naito S. Androgen receptor signaling regulates cell growth and vulnerability to doxorubicin in bladder cancer. J Urol. 2012;188:276–286. doi: 10.1016/j.juro.2012.02.2554. [DOI] [PubMed] [Google Scholar]
- 39.Wu JT, Han BM, Yu SQ, Wang HP, Xia SJ. Androgen receptor is a potential therapeutic target for bladder cancer. Urology. 2010;75:820–827. doi: 10.1016/j.urology.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 40.Zheng Y, Izumi K, Yao JL, Miyamoto H. Dihydrotestosterone upregulates the expression of epidermal growth factor receptor and ERBB2 in androgen receptor-positive bladder cancer cells. Endocr Relat Cancer. 2011;18:451–464. doi: 10.1530/ERC-11-0010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.