Abstract
The histone methyltransferase SET8 is regulated by microRNA-502 through the binding site in its 3′-untranslated region, and the rs16917496 polymorphism at the miR-502-binding site in the SET8 gene has been implicated in a number of cancer types. The rs16917496 polymorphism including CC, CT and TT genotypes was analyzed in patients with colorectal cancer; the CC genotype was identified to be independently associated with longer post-operative survival times using multivariate analysis (relative risk, 2.406; 95% confidence interval, 1.017–5.691; P=0.046). In addition, decreased SET8 expression was associated with the SET8 CC genotype and longer survival times for patients with colorectal cancer. The results of the present study indicated that miR-502 mediates SET8 expression at least partly by altering the binding affinity between miR-502 and SET8 so as to modify the colorectal cancer outcome. The results indicate that SET8 may be a novel target for colorectal cancer therapy.
Keywords: colorectal cancer, SET8, microRNA-502, polymorphism
Introduction
Colorectal cancer (CRC) is responsible for ~7×105 mortalities annually, which makes it the fourth leading cause of cancer-associated mortality for men and women worldwide (1). Its incidence has increased significantly in the majority of developing countries (2). Sporadic CRC, which usually presents as an isolated colonic or rectal lesion, is the most common type of CRC that occurs in older individuals without any family history of the disease. Genetic and epigenetic changes, dietary patterns (high fat consumption) and environmental factors are common risk factors for CRC (3,4). A number of genetic factors have been identified as predictors for the prognosis of CRC; however, the precise mechanism of CRC remains unknown (5–7).
MicroRNAs (miRNAs/miRs), RNAs with a length of ~22 nucleotides, bind to the ‘seed region’ of between 2 and 8 nucleotides at the 3′ untranslated region (UTR) to regulate targeted gene expression. The perfectly complementary base pairing between the miRNA and its target mRNA sequence may induce RNA silencing which results in decreased protein expression levels (8–11). A number of single nucleotide polymorphisms (SNPs) in the 3′ UTR of targeted genes had been identified for their association with an individual's risk of cancer by regulating targeted gene expression (12,13). As a histone H4 Lys20 monomethyltransferase implicated in normal cell cycle progression, SET8 (also known as PR-Set7 or KMT5a) is regulated by miR-502 though the binding site in the 3′ UTR of SET8 mRNA (14–16). Inappropriate SET8 expression induces S-phase defects and increased DNA damage; SET8 also interacts directly with the DNA replication factor proliferating-cell nuclear antigen and exhibits specific effects at origins of replication (17–20). During DNA double-strand break responses, SET8 activation has been identified to be essential for p53-binding protein 1 (p53BP1) recruitment (21). It has been identified that SET8 could increase the metastatic capacity of breast cancer cells by promoting epithelial-mesenchymal transition and conferring TWIST dual transcriptional activities (22).
The SNP rs16917496 was identified previously to be associated with risk of epithelial ovarian cancer and outcome of hepatocellular carcinoma, small cell lung cancer and non-Hodgkin's lymphomas (23–26). In the present study, this SNP was genotyped in patients with CRC to assess its association with cancer risk and outcome.
Materials and methods
Blood collection and DNA extraction
Genomic DNA was extracted from blood samples (0.2 ml) of 109 patients with CRC who underwent CRC resection at The Fourth Hospital of Hebei University (Shijiazhuang, China) between March 2006 and December 2008 using a Wizard Genomic DNA extraction kit (Promega Corporation, Madison, WI, USA). Blood samples were also collected from 142 age and gender matched healthy controls at the same hospital between April and December 2008. All procedures were supervised and approved by the Hospital's Human Tissue Research Committee. Written informed consent was obtained from all patients enrolled in the present study.
Polymerase chain reaction (PCR) amplification and sequence analysis
The DNA fragments flanking rs16917496 in the SET8 3′ UTR were amplified using forward primer 5′-TCACGACGGTGCTACCTAAG-3′ and reverse primer 5′-CATGCTGGTGTGACACAGTC-3′ designed according to the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/snp) using a PCR Master mix kit (Promega Corporation). The cycling conditions were one cycle of denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec and fluorescence acquisition at 72°C for 3 min. Cycle sequencing was performed using a Dye Terminator Cycle Sequencing Ready Reaction kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and analyzed using an ABI Prism Genetic Analyzer 3100 instrument (Thermo Fisher Scientific, Inc.). Polymorphisms were confirmed by repeating the analysis on the two DNA strands.
Determination of SET8 expression levels in CRC tissue
CRC tissues collected from the same 109 patients, from which blood samples were obtained, were fixed in formalin (10%) for 24 h at room temperature immediately following resection, dehydrated in absolute ethanol, embedded in paraffin and serial sections (4-µm thick). CRC tissue was immunostained using an anti-SET8 antibody (catalog no. ab3798; Abcam, Cambridge, UK) at a dilution of 1:100 at 4°C overnight, followed by incubation with a biotinylated secondary anti-mouse immunoglobulin G antibody (pre-diluted; catalog no. PV600; Zhongshan, Inc., Guangzhou, China) at room temperature for 1 h. Following incubation at room temperature for 5 min with horseradish peroxidase-conjugated streptavidin, the staining of CRC tissue was developed with 3,3-diaminobenzidine.
The stained slides were semi-quantified by two pathologists who were blinded to the sequencing data using HScore (25). Briefly, the percentage of positively stained CRC cells in each of five samples was graded (0, 1+, 2+, 3+ and 4+). The HScore was calculated as follows: HScore=(i+1)x, where i=1, 2, 3 and 4, and x varied between 0 and 100%. High expression is defined as a score of >100 and low expression is defined as a score of <100.
Statistical analysis
The distribution of expression grades for each SET8 genotype was compared using a χ2 test. Survival curves were created using the Kaplan-Meier method with a log-rank test and multivariate survival analysis was performed using a Cox proportional hazards model. Statistical analyses were performed using the SPSS software package (version 18.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
SET8 genotype is associated with CRC survival
A total of 109 patients with CRC were enrolled in the present study. The post-operative survival of these patients according to their clinical characteristics was analyzed using the Kaplan-Meier method and a log-rank test. The clinical stage and tumor length were investigated for their association with survival times of patients with CRC using univariate analysis (Table I).
Table I.
Univariate analysis of clinical characteristics associated with post-operative survival in patients with colorectal cancer.
| Characteristic | n | 5-year survival rate, % | P-value |
|---|---|---|---|
| Sex | 0.649 | ||
| Male | 61 | 55.7 | |
| Female | 48 | 58.3 | |
| Age, years | 0.664 | ||
| ≤60 | 53 | 58.5 | |
| >60 | 56 | 55.4 | |
| Tumor length, cm | 0.034 | ||
| ≤6 | 69 | 63.8 | |
| >6 | 40 | 45.0 | |
| Tumor location | 0.161 | ||
| Right colon | 57 | 50.9 | |
| Transverse colon | 7 | 100.0 | |
| Left colon | 43 | 58.1 | |
| Rectum | 2 | 50.0 | |
| Clinical stage | 0.001 | ||
| I+II | 57 | 71.9 | |
| III+IV | 52 | 40.4 |
The SNP rs16917496 of SET8 was genotyped (CC, CT and TT) in 109 patients with CRC and 142 controls; the rs16917496 distribution followed a Hardy-Weinberg equilibrium, and no difference in distribution frequency of the SET8 genotype was identified between patients with CRC and controls (data not shown). The association of SET8 genotype and post-operative survival of patients with CRC was assessed using the Kaplan-Meier method; the 5-year survival rate was 76.0% for patients with the CC genotype and 51.2% for patients with the CT + TT genotypes (Fig. 1). The patients carrying the CC genotype was associated with a significantly longer survival time compared with that of patients with the CT + TT genotypes (P=0.020). The multivariate analysis with Cox proportional hazards model was performed including all CRC survival-associated predictors (Table II). The SET8 genotype was identified as an independent predictor for the prognosis of patients with CRC (relative risk, 2.406; 95% confidence interval, 1.017–5.691; P=0.046).
Figure 1.
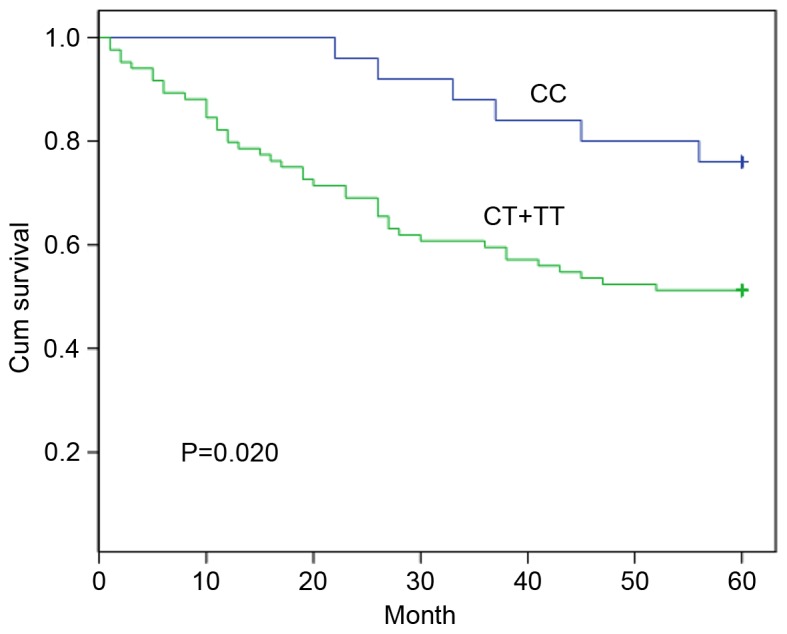
Significant difference in survival time for patients with colorectal cancer with the CC and CT + TT genotypes of rs16917496. Cum, cumulative.
Table II.
Multivariate analysis of prognostic factors associated with post-operative survival in patients with colorectal cancer with Cox hazard model.
| Factor | RR | 95% CI | P-value |
|---|---|---|---|
| rs16917496 | 2.406 | 1.017–5.691 | 0.046 |
| Tumor length | 1.425 | 0.788–2.576 | 0.241 |
| Clinical stage | 2.406 | 1.289–4.491 | 0.006 |
RR, relative risk; CI, confidence interval.
SET8 expression mediated by miR-502 is associated with CRC outcome
The association between the miR-502-binding site SNP rs16917496 and SET8 expression was investigated in CRC tissues (Fig. 2). The HScore of SET8 was calculated and the SNP rs16917496-based SET8 expression in patients with CRC is presented in Table III. The SET8 CC genotype was associated with lower levels of SET8 expression compared with that of the TT genotype by χ2 test (P<0.001). Survival analysis referring to SET8 expression was performed, the patients with low SET8 expression had a longer lifespan compared with that of patients with high SET8 expression (5-year survival rate, 75.7 vs. 47.2%; P=0.005).
Figure 2.
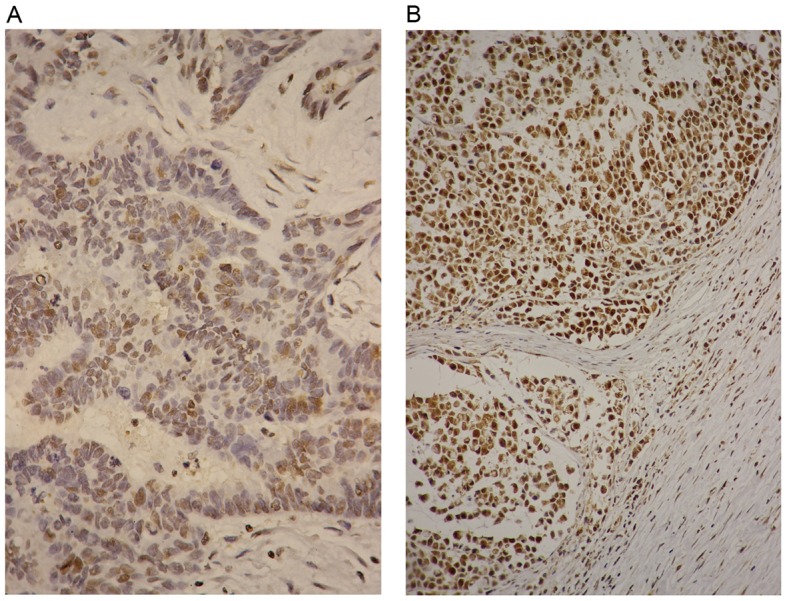
SET8 expression in patients with CRC. (A) Low SET8 expression with an HScore of 40 for the SET8 CC genotype in CRC tissues. (B) High SET8 expression with an HScore of 300 for the SET8 TT genotype in CRC tissues. Original magnification, ×400. CRC, colorectal cancer.
Table III.
Association of SET8 expression with SET8 genotype.
| Genotype | Low expression | High expression | P-value |
|---|---|---|---|
| CC | 17 | 8 | <0.001 |
| CT+TT | 20 | 64 |
Discussion
The methyltransferase SET8 has been implicated in a number of cancer processes (23–28). The results of the present study indicated that the SNP rs16917496 in the miR-502-binding site of the SET8 3′ UTR was associated with the survival time of patients with CRC. In addition, this SNP may eliminate the binding affinity between miR-502 and SET8 so as to modulate SET8 expression, with altered SET8 expression contributing to the progression of CRC.
The association of the miRNA-binding site SNP with cancer risk and outcome has been well-studied (25,29–32). Consistent with previous studies that identified the prognostic value of the rs16917496 on the outcome of patients with small cell lung cancer, hepatocellular carcinoma and non-Hodgkin's lymphomas with the CC genotype being associated with longer survival (23–26), the results of the present study indicate that the CC genotype tends to lead to a long lifespan in patients with CRC. The convergence of the SET8 CC genotype and low SET8 expression which was identified in patients with breast cancer and hepatocellular carcinoma was also confirmed in CRC tissue (25,33).
SET8 methylates Lys382 of p53 to modulate p53 activity, which is implicated in cell death and cell cycle arrest following DNA damage; furthermore, SET8 depletion could abrogate the accumulation of 53BP1 in DNA double-strand breaks to render cells sensitive to apoptosis (21,27). SET8 could methylate the promoters of the TWIST target genes of breast cancer cell to promote epithelial-mesenchymal transition and enhance the invasive potential (22). Although SET8 would be a novel therapy target for CRC treatment, the underlying molecular mechanism of miR-502-mediated SET8 expression and whether SET8 modifies the development of CRC via its methyltransferase requires further investigation.
Acknowledgements
The authors would like to thank Professor Xiaoling Wang (Department of Pathology, The Fourth Hospital of Hebei Medical University) for their assistance with immuohistochemistry.
Funding
The present study was supported by Key Basic Research Program of Hebei (grant no. 14967713D).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SL collected PCR and sequence data, and wrote the manuscript. HD collected the immunostaining data. JW contributed to data collection and writing of the manuscript. CW contributed to project design, data collection and manuscript writing. All authors read and approved the final manuscript. The authors agreed to be accountable for all aspects of the study, ensuring that questions related to the accuracy or integrity of any part of the study are appropriately investigated and resolved.
Ethics approval and consent to participate
All procedures were supervised and approved by the Human Tissue Research Committee of The Fourth Hospital of Hebei University (Shijiazhuang, China). Written informed consent was obtained from all patients enrolled in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F, editors. Globocan 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012 v1.0. IARC CancerBase. http://globocan.iarcfr 2014;(11) IARC, Lyon.
- 2.Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. CancerEpidemiol Biomarkers Prev. 2009;18:1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 3.World Cancer Research Fund, American Institute for Cancer Research Imperial College, London, corp-author. WCRF/AICR Systematic Literature Review Continuous Update Project Report: The Associations between Food, Nutrition and Physical Activity and the Risk of Colorectal Cancer. https://www.wcrf.org/sites/default/files/SLR_colorectal_cancer_2010.pdf. [Oct;2010 ];
- 4.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, Berndt SI, Bézieau S, Brenner H, Butterbach K, et al. Colon cancer family registry and the genetics and epidemiology of colorectal cancer consortium: Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology. 2013;144:799–807. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/S1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 12.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, Hemminki K, Lenner P, Försti A. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–1399. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 14.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/S0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 15.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/S1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dulev S, Tkach J, Lin S, Batada NN. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep. 2014;15:1163–1174. doi: 10.15252/embr.201439434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Guo Z, Wu C, Li Y, Kang S. A polymorphism at the miR-502 binding site in the 3′ untranslated region of the SET8 gene is associated with the risk of epithelial ovarian cancer. Cancer Genet. 2012;205:373–376. doi: 10.1016/j.cancergen.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Ding C, Li R, Peng J, Li S, Guo Z. A polymorphism at the miR-502 binding site in the 3′ untranslated region of the SET8 gene is associated with the outcome of small-cell lung cancer. Exp Ther Med. 2012;3:689–692. doi: 10.3892/etm.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Z, Wu C, Wang X, Wang C, Zhang R, Shan B. A polymorphism at the miR-502 binding site in the 3′-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int J Cancer. 2012;131:1318–1322. doi: 10.1002/ijc.27352. [DOI] [PubMed] [Google Scholar]
- 26.Diao L, Su H, Wei G, Li T, Gao Y, Zhao G, Guo Z. Prognostic value of microRNA 502 binding site SNP in the 3′-untranslated region of the SET8 gene in patients with non-Hodgkin's lymphoma. Tumori. 2014;100:553–558. doi: 10.1177/1660.18180. [DOI] [PubMed] [Google Scholar]
- 27.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhami GK, Liu H, Galka M, Voss C, Wei R, Muranko K, Kaneko T, Cregan SP, Li L, Li SS. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol Cell. 2013;50:565–576. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 29.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, Novotny J, Forsti A, Hemminki K, Canzian F, Landi S. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 30.Gao Y, He Y, Ding J, Wu K, Hu B, Liu Y, Wu Y, Guo B, Shen Y, Landi D, et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30:2064–2069. doi: 10.1093/carcin/bgp283. [DOI] [PubMed] [Google Scholar]
- 31.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song F, Zheng H, Liu B, Wei S, Dai H, Zhang L, Calin GA, Hao X, Wei Q, Zhang W, Chen K. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;19:6292–6300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


