Abstract
Colorectal cancer (CRC) is a multifactorial disease and one of the most malignant tumours. In addition to the sporadic form, familial occurrences, particularly hereditary non-polyposis CRC-Lynch syndrome (LS)-are often observed. LS is caused by a germline mutation in mismatch repair (MMR) genes, whose task it is to correct errors in the DNA structure that result from its replication. The aim of the present study was to stratify CRC patients using molecular diagnostics and next generation sequencing, according to the chosen criteria [positive for microsatellite instability (MSI) and negative for a BRAF mutation and MutL homolog 1 (MLH1) methylation], and subsequently to detect pathological germline mutations in MMR genes in Slovak patients. To exclude patients with MSI from further testing, the present study detected the BRAF V600E mutation and examined MLH1 methylation status. From the 300 CRC patients, 37 cases with MSI were identified. In the MSI-positive samples, 13 cases of BRAF V600E mutation were recorded. In 24 BRAF-negative patients, 11 cases of epigenetic methylation of MLH1 and 12 cases without MLH1 methylation suspected for LS were detected, and it was not possible to analyse the methylation phenotype of 1 sample. Thus, the present study reports the novel deletion of four nucleotides, 1627_1630del AAAG (Glu544Lysfs*26) in MSH6, probably associated with LS. A second case with a nonsense mutation in MSH was also detected, namely MMR_c.1030C>T (p.Q344X).
Keywords: Lynch syndrome, microsatellite instability, BRAF, methylation
Introduction
Colorectal cancer (CRC) is one of the most common malignant heterogeneous tumours in the world. Almost 70% of colorectal tumours develop from an adenocarcinoma sequence and the remaining develop via a serrated neoplasia pathway named for the pattern of crypts in the precursor polyps (1). In general, the molecular pathways that lead to colorectal carcinogenesis are distinguished by three basic pathways: Chromosomal instability (CIN), the CpG pathway of the methylation phenotype (CIMP) and microsatellite instability (MSI) (2). Hereditary non-polyposis colorectal carcinoma (HNPCC), or Lynch syndrome (LS), develops through the MSI pathway. LS is an autosomal dominant disease characterised by DNA dysfunction, namely in mismatch repair (MMR) genes. Mutations in these genes lead to errors in microsatellites. LS diagnosis was first postulated using clinical Amsterdam I and II criteria. In 2006, there was a revision of the Bethesda guidelines, which included clinical as well as morphological features of tumours. Recent studies showed that LS may be early onset or occur in people over 50 years of age. Screening criteria among patients with CRC have been expanded. Multiple guidelines recommend tumour screening through MMR protein expression or MSI. Tumour genomic profiling using next-generation sequencing (NGS) panels to define the spectrum of mutations in an individual's tumour is becoming increasingly widespread (3). Hereditary tumour groups carry germline mutations in the MMR genes: MLH1, MSH2, PMS1, PMS2, MSH3, MSH6 and EPCAM. MMR genes repair DNA damage that occurs during replication; the MMR system is responsible for replacing mismatched nucleotides (4).
MSI positivity in sporadic cancers is usually the result of epigenetic inactivation. The most common epigenetic change is CpG island hypermethylation in the MLH1 promoter region. Methylation leads to loss of function of tumour suppressor genes, MMR genes and genes responsible for regulation of cell growth and division (5). In CRC, the V600E mutation in BRAF, a gene that codes a protein in the RAS/RAF/MAPK pathway, suggests a sporadic origin for the disease. The V600E mutation is not usually found in hereditary forms of cancer and therefore allows the selection of patients diagnosed with sporadic carcinomas who are positive for MSI (6).
Screening for MSI status in CRC patient tissues, together with BRAF mutation detection and identification of hypermethylation status, represent an algorithm to stratify CRC patients. Currently, NGS is one of the most promising tools used to detect germline mutations. The Miseq system (Illumina, USA) offers an NGS workflow based on target amplification. Targeted DNA enrichment methods allow even higher genome throughput at a reduced cost per sample (7). NGS allows massive parallel sequencing that affords maximal tumour genomic assessment (8).
In the present study, we observed CRC patients with the aim of identifying suspected LS patients who were carriers of MMR mutations. For future analysis, we would like to apply molecular methods to diagnosed patients older than 50 years.
Materials and methods
Specimens
We obtained 300 DNA samples from Slovak patients with CRC in collaboration with the Department of Molecular Biology (Jessenius Faculty of Medicine in Martin), Department of Pathologic Anatomy (Jessenius Faculty of Medicine in Martin) and University Hospital in Martin. Our analyses were performed on DNA derived from microdissected formalin-fixed-paraffin-embedded (FFPE) tumour tissue, and DNA was extracted with the Black Prep FFPE DNA kit (Analytik Jena AG, Jena, Germany), according to manufacturer's protocol.
All tumours were examined with immunohistochemistry (IHC) for the expression of MMR proteins. IHC analysis was performed using 4 µm tissue sections from FFPE blocks. IHC results were diagnosed at the Department of Pathologic Anatomy (Jessenius Faculty of Medicine in Martin).
In the present study, we acquired only 5 blood samples from suspected LS patients according to our criteria. DNA was isolated from whole blood using the DNeasy Blood and Tissue kit (Qiagen GmbH, Hilden, Germany). DNA concentration was measured on a Qubit® 2.0 Fluorometer (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Samples were diluted to working solutions at a concentration of 1–2 ng/µl and stored at −20°C.
All patients were informed about the study and provided written informed consent. The present study was a part of projects that were approved by the Ethical Committee at Jessenius Faculty of Medicine in Martin.
MSI polymerase chain reaction (PCR) and fragment analyses
To determine the tumour MSI, MSI PCR was performed using the MSI Analysis System v1.2 kit (Promega Corporation, Madison, WI, USA) with a GeneAmp PCR System 9700 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. MSI analysis based on PCR is considered the gold standard approach to detect microsatellite status, and it is sensitive for identifying LS. MSI testing is a method-for-function detection which cannot be replaced under certain conditions, such as MSI tumours with intact IHC expression (non-functional protein expression). The MSI assay was performed using a pentaplex PCR with 5 quasimonomorphic mononucleotide markers (NR-27, NR-21, NR-24, BAT-25 and BAT-26) and 2 pentanucleotide markers (PENTA C and PENTA D) (9–11). The MSI Analysis System was optimised to amplify 1–2 ng/µl of genomic DNA in a 10 µl reaction volume. For a positive amplification control, we used high molecular weight K562 (Promega Corporation). After PCR, 1 µl of amplified product was added to 9.5 µl Hi-Di™ Formamide (Applied Biosystems; Thermo Fisher Scientific, Inc.) and 0.5 µl Internal Lane Standard 600 (ILS 600; Promega Corporation). Denatured samples were loaded on the ABI 3500 (Applied Biosystems; Thermo Fisher Scientific, Inc.). The results were evaluated by GeneMapper software (Applied Biosystems; Thermo Fisher Scientific, Inc.) by comparing the patient's sample with the K562 control and/or non-tumour tissue. Samples with only one altered microsatellite marker were classified as MSI-low (MSI-L). Samples with ≥2 altered markers were classified as MSI-high (MSI-H). Samples with no altered markers were classified as microsatellite stable (MSS).
Sanger sequencing for BRAF c.1799T>A (V600E)
The presence of the V600E mutation was monitored by Sanger sequencing using an ABI 3500 (Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR products for Sanger sequencing were amplified with the primers BRAF Ex15F (5′-TCATAATGCTTGCTCTGATAGGA-3′) and BRAF Ex15R (5′-GGCCAAAATTTAATCAGTGGA-3′). PCR products were evaluated by electrophoresis on a 1.75% agarose gel and purified with the NucleoSpin Gel and PCR Clean-Up kit (Machery-Nagel GmbH, Düren, Germany). Sequencing PCR was prepared using the BigDye Terminator v1.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.). PCR condition optimisation was described previously (12,13). SigmaSpin Post-Reaction Clean-Up (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used for the second purification and in this analysis according to the information provided by the manufacturer. Results were evaluated using Chromas Pro software (Technelysium Pty Ltd., South Brisbane, QLD, Australia). RKO cell lines that carried the V600E mutation were used as positive control and DNA from a healthy individual was the negative control.
Bisulfite treatment
The EpiTect Bisulfite kit (Qiagen GmbH) was used for bisulfite treatment of 1 ng/µl-2 µg/µl of genomic DNA in RNase-free water in a 20 µl volume, according to the manufacturer's instructions. The modified DNA was eluted to a final concentration of 40 ng/µl. EpiTect Controls DNAs (Qiagen GmbH), which included fully methylated or unmethylated DNA, were used as controls. The samples were stored at −20°C until further analysis.
Nested methylation-specific PCR (MS-PCR)
To detect the MLH1 methylation status, we used nested two-step accession for increased sensitivity (14), with modifications according to Lasabová et al (15). MS-PCR was performed using a GeneAmp PCR System 9700 and amplified with specific primers to distinguish methylated (M) from unmethylated (U) DNA according to Herman et al (16). The sequences of our modified external (MLH1ExF/R) and internal (MLH1UF/R and MLH1MF/R) primer sets and the size of PCR products are presented in Table I. PCR conditions for the nested PCR step with external primers were as follows: 95°C for 8 min, then 20 cycles of 95°C for 30 sec, 62°C for 30 sec and 72°C for 30 sec and finally 72°C for 8 min. The PCR products of this amplification were diluted 1:500 and we prepared the second PCR using internal primers for methylated or unmethylated DNA. Thermal cycling was identical to the above conditions, except the annealing temperature for the primers was 64°C.
Table I.
Primer sets for nested methylation-specific polymerase chain reaction.
| Primer | Sequence (5′-3′) | Size (bp) |
|---|---|---|
| MLH1UF | GTAGATGTTTTATTAGGGTTGT | 113 |
| MLH1UR | CACCTCATCATAACTACCCACA | |
| MLH1MF | GTAGACGTTTTATTAGGGTCGC | 113 |
| MLH1MR | CCTCATCGTAACTACCCGCG | |
| MLH1ExF | GAGTAGTTTTTTTTTTAGGAGTGA | 192 |
| MLH1ExR | ATAAAACCCTATACCTAATCTATC |
F, forward; R, reverse; MLH1, MutL homolog 1; Ex, modified external; U/M, modified internal
NGS analysis
For NGS analysis, we examined 5 blood samples from suspected LS patients. NGS analysis was performed on the MiSeq sequencer (Illumina, Inc., San Diego, CA, USA), according to the manufacturer's protocol, using the HNPCC MASTR PlusKit (Agilent Technologies, Inc., Santa Clara, CA, USA). The kit was used with the complementary MASTR Plus product, the molecular identifier (MID) for Illumina Miseq kit [1-48] (Agilent Technologies, Inc.). HNPCC MASTR Plus identifies single nucleotide variants and copy number variation in MLH1, MSH2, MSH6, PMS2 and the 3′ untranslated region (UTR) of EPCAM, genes associated with HNPCC. In the first step of library preparation, we amplified all targeted regions in separate multiplex PCR amplifications for each sample. We used 50 ng of genomic DNA per reaction. The multiplex PCR products for each sample were then combined together into one tube and this amplicon library was purified from small residual DNA fragments with Agencourt AM Pure XP beads (Beckman Coulter, Inc., Brea, CA, USA). PCR products were diluted 1:1,000. In the second step, universal PCR amplification tagged amplicons with MID adaptors. For each sample, we prepared a universal PCR with a unique combination of MID p5-p7 primer mix sequences. The average amplicon size after universal PCR was 469 base pairs (bp). The number of cycle reads was 2×251 bp. After this PCR, we performed electrophoresis on a chip with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) for quality control. We then purified the tagged amplicon library again with Agencourt AM Pure XP beads, determined the library concentrations and pooled equal volumes of the libraries into the final library. Concentrations of the obtained tagged amplicons were measured with the Qubit™ dsDNA HS Assay kit (Thermo Fisher Scientific, Inc.). The library pool was processed using kits with all necessary consumables and a reagent cartridge for sequencing. For sequencing, we used Miseq Reagent Nano kit v.2 (Illumina, Inc.) with a 500 megabase (Mb) capacity. The obtained data were evaluated by the Variant Effect Predictor software (https://www.ensembl.org). Only mutations reported in Polyphen, SIFT and Cosmic were taken into account, and silent or intronic mutations were not reported.
Results
Specimens and MSI PCR analysis
From a total of 300 patients identified with CRC, 103 (34%) were women, 163 (54%) were men and the gender was unknown for 34 (11.3%) cases (Table II). Patient age ranged from 28 to 94 years (average 60.5). Histopathological parameters of the cases, including histological grade, morphological subtype and localisation, were known. Most carcinomas were classified as adenocarcinomas except: 1 (0.3%) synchronous carcinoma, 1 (0.3%) undifferentiated carcinoma, 2 (0.6%) medullar carcinomas, 4 (1.3%) signet ring cell carcinomas and 36 (12%) mucinous carcinomas (Table II). Fifty-three (18%) cases were grade 3, 136 (45%) were grade 2 and 39 (13%) were grade 1 (Table II).
Table II.
Clinicopathological features of cohort.
| Clinicopathological features | |
|---|---|
| Gender | N |
| Female | 103 |
| Male | 163 |
| Not determined | 34 |
| Grade | |
| Grade 1 | 39 |
| Grade 2 | 136 |
| Grade 3 | 53 |
| Not determined | 77 |
| Localization | |
| Colon ascendens/descendens | 155 |
| Rectum | 59 |
| Caecum | 39 |
| Rectosigmoideum | 13 |
| Not determined | 34 |
| Morphological subtype | |
| Synchronous carcinoma | 1 |
| Medullar carcinoma | 2 |
| Signet ring cell | 4 |
| Mucinous carcinoma | 36 |
| Undifferentiated carcinoma | 1 |
| Adenocarcinoma NOS | 222 |
| Not determined | 34 |
The present cohort consisted of 300 patients (163 men, 103 women and 34 not determined). NOS, not other specified adenocarcinoma
From our cohort (n=300), we captured 33 (11%) MSI-H and 4 (1.3%) MSI-L cases. In 263 (88%) samples, we did not detect MSI. These samples were evaluated as MSS. The results of MMR IHC and MSI testing were shown to be largely concordant. Genetic examination of MSI was confirmed in 33 cases based on IHC examination. This examination included 4 MSI-L cases grouped as MSS carcinomas, but according to genetic analysis they belonged to the MSI-positive tumour group. Although we stratified more patients suspected for LS, we were only able to obtain 5 blood samples. The blood samples, which were from suspected LS patients based on immunohistochemical and molecular analyses, were submitted for genetic counselling and NGS analysis.
Sanger sequencing for BRAF and nested methylation PCR
In 37 (12.3%) samples with a positive MSI status, we subsequently detected the BRAF V600E mutation. There was no significant association between BRAF-and MSI-positive statuses. We focused on detection of the BRAF c.1799T>Amutation in the samples. Twenty-four (64.9%) samples did not have the c.1799T>A mutation in BRAF exon 15. To further stratify patients, we continued the analysis of the 24 samples negative for the BRAF mutation (Fig. 1). Since the presence of the above mutation generally indicates sporadic CRC, only negative samples were subjected to the more detailed analysis. Due to the low DNA concentration (<50 ng/µl) required for bisulfite treatment, it was not possible to analyse the presence of MLH1 methylation for 1 sample. Eleven (45.8%) samples were positive for MLH1 methylation while 12 (50%) samples were without methylation. We also determined MLH1 methylation status in 13 BRAF-mutation-positive samples, which were excluded from additional testing for LS. From these 13 samples, 12 cases (92%) were also positive for MLH1 methylation (Table III).
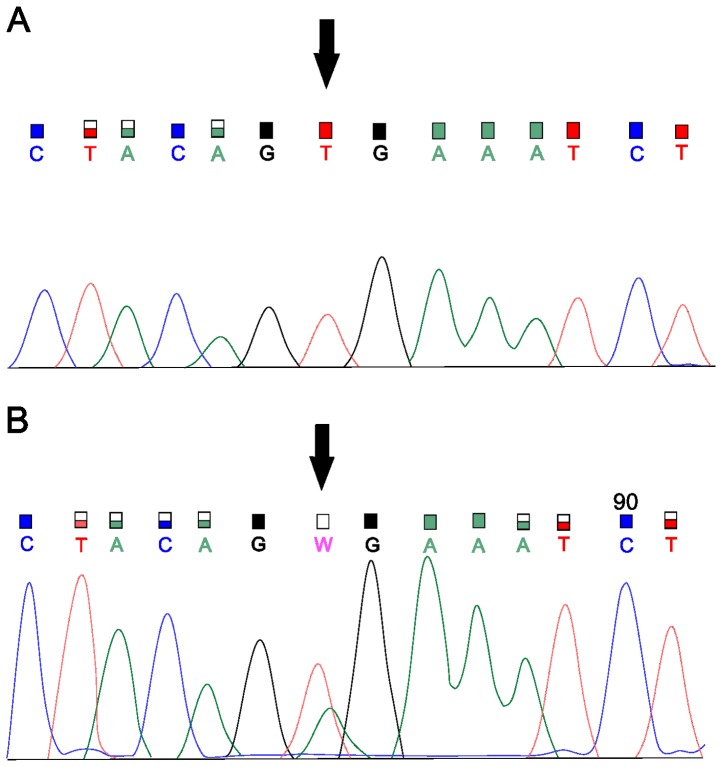
Figure 1.
Sanger sequencing for the detection of V600E. (A) Wild-type allele T in exon 15 of the BRAF gene. (B) Mutation V600E (c.1799T>A) in BRAF.
Table III.
Table of MSI cases with MSI polymerase chain analysis and BRAF mutation analysis.
| Case | MSI | BRAF | Methylation |
|---|---|---|---|
| 1 | MSI-H | − | + |
| 2 | MSI-H | + | + |
| 3 | MSI-H | − | + |
| 4 | MSI-H | + | + |
| 5 | MSI-H | − | − |
| 6 | MSI-H | − | + |
| 7 | MSI-H | − | − |
| 8 | MSI-H | + | + |
| 9 | MSI-L | − | − |
| 10 | MSI-H | − | − |
| 11 | MSI-H | + | Not determined |
| 12 | MSI-H | − | Not determined |
| 13 | MSI-H | − | − |
| 14 | MSI-H | + | + |
| 15 | MSI-H | − | + |
| 16 | MSI-H | + | + |
| 17 | MSI-H | + | + |
| 18 | MSI-H | − | + |
| 19 | MSI-H | + | + |
| 20 | MSI-H | − | − |
| 21 | MSI-H | − | − |
| 22 | MSI-H | − | − |
| 23 | MSI-H | + | + |
| 24 | MSI-H | − | − |
| 25 | MSI-H | − | − |
| 26 | MSI-H | − | − |
| 27 | MSI-H | − | + |
| 28 | MSI-H | − | + |
| 29 | MSI-H | − | + |
| 30 | MSI-H | + | + |
| 31 | MSI-H | + | + |
| 32 | MSI-L | − | + |
| 33 | MSI-L | − | + |
| 34 | MSI-H | + | + |
| 35 | MSI-H | − | + |
| 36 | MSI-H | + | + |
| 37 | MSI-L | − | − |
A total of 37 MSI cases were selected from the present cohort. Twenty-four (64.9%) samples did not have the c.1799T>A mutation in BRAF exon 15. MLH1 methylation was determined in 23 cases. For NGS analysis 12 suspected samples were selected (BRAF and MLH1 negative). +, positive status; -, negative status; MSI, microsatellite instability; -L, low; -H, high
NGS analysis to detect germline mutations
To confirm LS, we utilised NGS analysis with a Miseq system. From the available blood samples (n=5), we detected 2 samples with suspected pathogenic germline mutations. In the first sample, we captured the previously unpublished deletion of the four nucleotides 1627_1630del AAAG (Glu544Lysfs*26) in MSH6, a mutation with a high impact on the gene (based on Assembly GRCh37). According to IHC, we assumed there was some variant in MSH6, because the protein was not detected. This patient was positive for MSI and negative for the BRAF mutation and MLH1 methylation. This frameshift variant was found in MSH6 exon 4. We verified the deletion by Sanger sequencing (Fig. 2). In the sample with the newly detected MSH6 deletion, we detected 42 additional nonpathogenic variants, most of which were missense (66%), synonymous (8%) or frameshift (6%) variants with benign or uncertain significance. Our results were verified by a commercial laboratory. In another sample, we confirmed the pathogenic stop-gain variant MMR_c.1030C>T (Gln344Ter) for MSH2 according to a five-tiered classification from the International Agency for Research on Cancer (based on Assembly GRCh37). This variant leads to the stop codon p.Q344X. We were not able to verify this mutation by Sanger sequencing. In this case, there were also mainly missense variants (81%), followed by synonymous (11%) and stop-gain variants (8%). These variants were classified as benign or of uncertain significance. We also recommended verification by a commercial laboratory, but that result is unknown to us. In the 3 other blood samples, we identified several variants with uncertain significance.
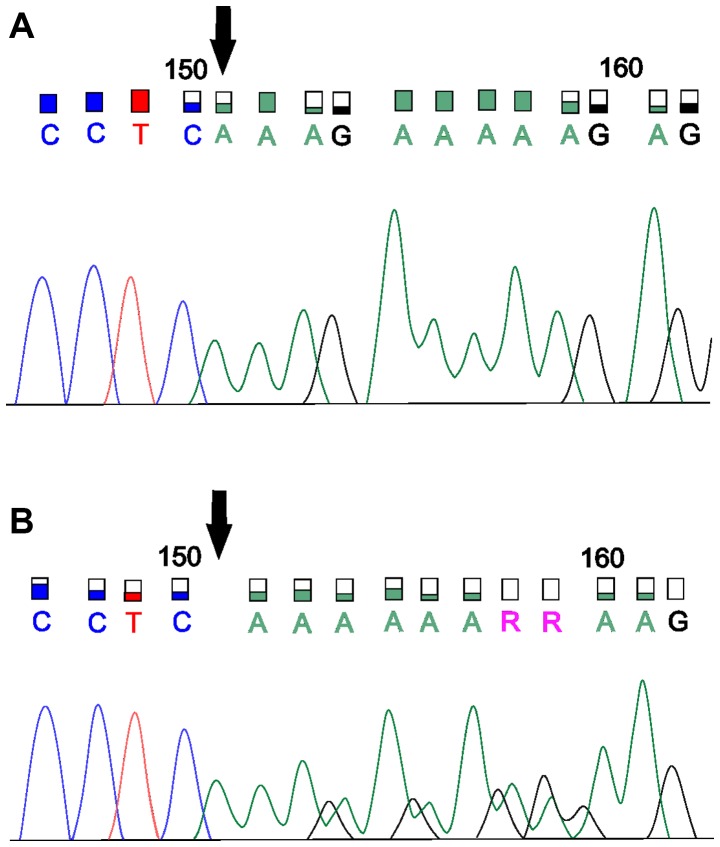
Figure 2.
Sanger sequencing for the verification of the novel deletion in the MSH6 gene. (A) Electropherogram without deletion and (B) an electropherogram presenting the novel deletion in the MSH6 gene. MSH6, MutS homolog 6.
Discussion
The MSI pathway affects CRC, particularly in terms of genetic instability such as oncogene activation or tumour suppressor gene inactivation. The aim of our study was to improve a stratification procedure for Slovak CRC patients and to determine the significance of the utilised procedures. We focused on the possibilities of stratification, which is important for rapid selection of appropriate prevention and treatment especially for patients with hereditary forms of the disease. Research shows that patients with LS have earlier disease onset compared to sporadic CRC patients (17). A useful screening tool for capturing LS has been MSI detection (7) since suspected LS cases usually exhibit MSI. MSI testing with MMR deficiency has 93% sensitivity, but it does not predict which MMR gene(s) is/are altered (18). Nevertheless, MSI testing is effective for screening but there is significant uncertainty surrounding what balance of sensitivity and specificity will be achieved in clinical practice (17).
According to some authors, Bethesda criteria are sensitive but not very specific for MSI status. Hereditary forms of the disease occur in patients with a family history of the same affinity or adenoma (19). Screening MMR genes is a gold standard for LS diagnosis, and MSI has a role in CRC patient stratification. It is a part of targeted screening for LS patients, and MSI status is recommended as the first step (20) because screening all MMR genes is more expensive, demanding and time consuming (21). Determining MSI status can also be used as a predictor for treatment (22). Recent studies showed that patients with MMR-deficient tumours are more responsive to programmed cell death-1 (PD-1) blockade than MMR-competent tumours, so patients with MMR deficiency may benefit from anti-PD-1 therapy (23). Differentiation between hereditary and sporadic MSI CRC is currently the basic diagnostic step for patient stratification (8). Carcinomas with MSI represent approximately 15% of all CRC cases, LS accounts for 3% and sporadic MSI carcinomas represent 12% (24). From our specimens of the 300 CRC cases, 37 (12.3%) were MSI positive. To exclude sporadic MSI carcinomas from further testing, we chose to detect the BRAF V600E mutation by Sanger sequencing. According to Snowsill et al (25), cascade testing is used in every strategy for LS patients. Additionally, strategies that utilise MSI and BRAF testing are the most cost effective. This cascade strategy was also used by Buchanan et al (26) to identify LS patients, and it is recommended in study of Cohen et al (27) together with MLH1 methylation testing. BRAF mutations were not detected in cases with a germline mutation in MLH1, MSH2 or MSH6 (28). The BRAF V600E mutation and MLH1 methylation are associated with sporadic MSI carcinomas, but they are not associated with MMR deficiency in LS (29). Adar et al (30) stated that the presence of the V600E mutation in tumours had a positive predictive value for sporadic cancer, but negative predictive value for MLH1 methylation; the presence of the BRAF V600E mutation excludes LS diagnosis. Seppälä et al (31) declared that MSI carcinomas had better prognosis compared to MSS, and BRAF-positive tumours were associated with poor prognosis.
The third stratification step in the MSI-positive group without the BRAF V600E mutation was detection of methylation in the most commonly methylated MMR gene, namely MLH1. Previous studies indicated the presence of alterations in MMR genes, which led to the loss of gene product expression and development of malignancy. The role of epigenetic changes, especially aberrant DNA methylation, not only in polyposis sporadic carcinomas but also in hereditary tumours, is not yet accurately known (3). Approximately one third of carcinomas are thought to occur due to epigenetics (32). In the process of patient stratification, detection of MLH1 promoter region methylation serves to exclude sporadic forms of carcinoma from further testing to allow the selection of suspected hereditary tumours. The primary target sequence for DNA methylation is 5′-CpG-3′ dinucleotides. Promoter CpG-islands are normally protected from methylation, while CpG dinucleotides that are not associated with CpG-islands are heavily methylated (10). However, in rare cases of LS, MLH1 hypermethylation can serve as a second hit (33). Based on these findings, we introduced an NGS analysis that clearly confirmed the presence of pathological changes in DNA and provided a comprehensive view of genetic information.
Heterozygous germline variants in MMR genes are responsible for LS. Patients with LS have one functional allele of the MMR gene and the second allele carries the germline mutation. Malignant tumours result from the second (defective) allelic copy, which becomes nonfunctional because of other somatic mutations or methylation (34). Only a few LS cases with homozygous MMR mutations were described previously; these patients exhibited early cancer onset (33). The majority of mutations are located in MSH2 and MLH1, followed by MSH6 (35,36). Indeed, almost 90% of LS cases have a germline mutation in MLH1 or MSH2 (37). The hot spot areas for the appearance of MMR gene variants are MSH6 exon 4 and MSH2 exon 3 or 12. MSH6 and PMS2 mutations also prevail in the Icelandic population (38), but genetic drift can influence the appearance of some mutations. In a study from 2007, missense MMR gene mutations were common in LS (39). Usually, we identify chromosomal mutations at position 3p21 of MLH1, 2p16 of MSH2, 7p22 of PMS2 and 2p16 of MSH6 (34). In a 2017 study, the authors declared that MSH6 mutations accounted for approximately 18% of LS cases (40). In our study of Slovak CRC patients, we stratified the subjects, and in suspected LS cases we tried to find germline mutations. In the first suspected case, we identified a novel missense mutation, a deletion in MSH6 exon 6. This mutation, c.1627_1630AAAG (p.Glu544Lysfs), should be included in future studies. In the second suspected LS case, we detected a previously reported nonsense mutation (c.1030C>T (p.Q344X)) in MSH2 (41,42). While we identified this pathogenic variant by NGS, we were unable to verify it by the Sanger sequencing. Nevertheless, all tested patients were recommended for genetic counselling and diagnosis from a certified commercial diagnostic laboratory.
Undetermined clinical information, limited sample size for NGS analysis and NGS method were limitations of this study. We were unable to get more information about patients and about their family history. However, our findings provide stratification for routine diagnostics. NGS analysis in our study had a few limitations. For this method only 5 blood samples from 12 suspected LS cases were available. For next studies is necessary to get more blood samples. Although, we identified a novel deletion probably associated with LS and detected known mutation in MSH2 gene, our findings are insufficient to prove the association to LS. These limitations demonstrated that other information about patients are important, especially family history and results of IHC. However, based on our findings we recommend testing for all CRC patients, also without unrecorded family history.
Known mutations in hereditary CRC occur with a low frequency and thus it is difficult to predict penetrance. There is also heterogeneity in phenotypic expression. We found some new variants of uncertain clinical significance, and so it was difficult to predict the variant impacts on the genes (38). Our findings provide a few useful and fast stratification steps for routine diagnostics. Indeed, this study was conducted to address the need for clearer patient stratification for suspected LS cases and for clearer information about diagnostic results based on MSI testing. A targeted NGS panel for LS evaluated the ability to detect germline mutation. Our results directly demonstrated the greatly contribute to rapid patient diagnosis and optimal treatment settings. We recommend a cascade strategy (MSI for MMR genes, BRAF and MLH1 testing) for determination of sporadic MSI-positive cancer and LS-like cancer. We recommend this strategy despite of study limitation-NGS analysis. We did not have MSI-H, MSI-L, MSS categories of patients in NGS analysis necessary for LS validation. On the other hand, detection of a new deletion was confirmed by the commercial laboratory.
A cascade screening strategy is suitable for patients who meet the Amsterdam criteria or revised Bethesda guidelines, and so we recommend use of this screening for stratification of all patients with CRC in addition to cases with a negative or unrecorded family history. In the future, we would like to continue with our study. Experiment is a suggestion for future studies in area of LS. Also new detected deletions should be as a suggestion for further studies. We need more knowledges about this variant because it was previously not described in ClinVar, Human Gene Mutation Database, Ensembl and InSight databases. Based on our findings, new screening strategies are necessary and LS needs to be further studied. In future we would like to study new insights into patient testing.
Acknowledgements
The authors would like to thank Dr Jana Kršiaková (M-Genetik, Martin, Slovakia) for assisting with patient recruitment.
Funding
The present study was supported by the Biomedical Center Martin Grant (grant no. ITMS 26220220187), which was co-financed by EU sources. The study was also supported by the project ‘Genomic Profile And Transcriptional Signature Of Colorectal Cancer’ (grant no. APVV-16-0066) and the project ‘Pilot study of utility of DNA mutational analysis of tissue and so-called liquid biopsy in the diagnosis and prediction of therapeutic response in two malignancies’ (grant no. APVV-14-0273).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
IK designed the study and wrote the manuscript. IK, MK, KJ, TB, BM, AV and SM conducted the experiments and analyzed the data. ZL and LP designed the study, and compiled and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients were informed about the present study and provided written informed consent. This research was a part of projects that were approved by the Ethical Committee at Jessenius Faculty of Medicine in Martin (Martin, Slovakia).
Patient consent for publication
The patients provided written informed consent for the publication of the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: Current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 2.Worthley DL, Leggett BA. Colorectal cancer: Molecular features and clinical opportunities. Clin Biochem Rev. 2010;31:31–38. [PMC free article] [PubMed] [Google Scholar]
- 3.Stadler ZK, Battaglin F, Middha S, Hechtman JF, Tran C, Cercek A, Yaeger R, Segal NH, Varghese AM, Reidy-Lagunes DL, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34:2141–2147. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llinàs-Arias P, Esteller M. Epigenetic inactivation of tumour suppressor coding and non-coding genes in human cancer: An update. Open Biol. 2017;7(170152) doi: 10.1098/rsob.170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27:1423–1431. doi: 10.1111/j.1440-1746.2012.07200.x. [DOI] [PubMed] [Google Scholar]
- 6.Sharma SG, Gulley ML. BRAF mutation testing in colorectal cancer. Arch Pathol Lab Med. 2010;134:1225–1228. doi: 10.5858/2009-0232-RS.1. [DOI] [PubMed] [Google Scholar]
- 7.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: A Practical perspective. Clin Biochem Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 8.Serrati S, De Summas S, Pilato B, Petriella D, Lacalamita R, Tommasi S, Pinto R. Next-generation sequencing: Advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355–7365. doi: 10.2147/OTT.S99807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF, Duval A, Hamelin R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol. 2006;24:241–251. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- 10.Søreide K. High-fidelity of five quasimonomorphic mononucleotide repeats to high-frequency microsatellite instability distribution in early-stage adenocarcinoma of the colon. Anticancer Res. 2011;31:967–971. [PubMed] [Google Scholar]
- 11.Benlloch S, Payá A, Alenda C, Bessa X, Andreu M, Jover R, Castells A, Llor X, Aranda FI, Massutí B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jasek K, Buzalkova V, Minarik G, Stanclova A, Szepe P, Plank L, Lasabova Z. Detection of mutations in the BRAF gene in patients with KIT and PDGFRA wild-type gastrointestinal stromal tumors. Virchows Arch. 2017;470:29–36. doi: 10.1007/s00428-016-2044-4. [DOI] [PubMed] [Google Scholar]
- 13.Minárik G, Plank L, Lasabová Z, Szemes T, Burjanivová T, Szépe P, Buzalková V, Porubský D, Sufliarsky J. Spectrum of mutations in gastrointestinal stromal tumor patients-a population based study from Slovakia. APMIS. 2013;121:539–548. doi: 10.1111/apm.12019. [DOI] [PubMed] [Google Scholar]
- 14.House MG, Guo M, Iacobuzio-Donahue C, Herman JG. Molecular progression of promoter methylation in intraductal mucinous neoplasm (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–198. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 15.Lasabova Z, Tilandyova P, Kajo K, Zubor P, Burjanivova T, Danko J, Plank L. Hypermethylation of the GSTP1 promoter region in breast cancer is associated with prognostic clinicopathological parameters. Neoplasma. 2010;57:35–40. doi: 10.4149/neo_2010_01_035. [DOI] [PubMed] [Google Scholar]
- 16.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JPJ, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coelho H, Jones-Hughes T, Snowsill T, Briscoe S, Huxley N, Frayling IM, Hyde C. A systematic review of test accuracy studies evaluating molecular micro-satellite instability testing for the detection of individuals with lynch syndrome. BMC Cancer. 2017;17:836. doi: 10.1186/s12885-017-3820-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendriks YM, de Jong AE, Morreau H, Tops CM, Vasen HF, Wijnen JT, Breuning MH, Bröcker-Vriends AH. Diagnostic approach and management of lynch syndrome (hereditary nonpolyposis colorectal carcinoma): A guide for clinicians. CA Cancer J Clin. 2006;56:213–225. doi: 10.3322/canjclin.56.4.213. [DOI] [PubMed] [Google Scholar]
- 19.Kheirelseid EAH, Miller N, Kerin MJ. Molecular biology of colorectal cancer: Review of the literature. Am J Mol Biol. 2013;3:72–80. doi: 10.4236/ajmb.2013.32010. [DOI] [Google Scholar]
- 20.Schofield L, Watson N, Grieu F, Li WQ, Zeps N, Harvey J, Stewart C, Abdo M, Goldblatt J, Iacopetta B. Population-based detection of Lynch syndrome in young colorectal cancer patients using microsatellite instability as the initial test. Int J Cancer. 2009;124:1097–1102. doi: 10.1002/ijc.23863. [DOI] [PubMed] [Google Scholar]
- 21.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 22.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatalica Z, Vranic S, Xiu J, Swensen J, Reddy S. High microsatellite instability (MSI-H) colorectal carcinoma: A brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer. 2016;15:405–412. doi: 10.1007/s10689-016-9884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snowsill T, Huxley N, Hoyle M, Jones-Hughes T, Coelho H, Cooper C, Frayling I, Hyde C. A systematic review and economic evaluation of diagnostic strategies for Lynch syndrome. Health Technol Assess. 2014;18:1–406. doi: 10.3310/hta18580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchanan DD, Clendenning M, Rosty C, Eriksen SV, Walsh MD, Walters RJ, Thibodeau SN, Stewart J, Preston S, Win AK, et al. Tumor testing to identify lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol. 2017;32:427–438. doi: 10.1111/jgh.13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen R, Buhard O, Cervera P, Hain E, Dumont S, Bardier A, Bachet JB, Gornet JM, Lopez-Trabada D, Dumont S, et al. Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. Eur J Cancer. 2017;86:266–274. doi: 10.1016/j.ejca.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191–195. doi: 10.1158/1078-0432.CCR-1118-3. [DOI] [PubMed] [Google Scholar]
- 29.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 30.Adar T, Rodgers LH, Shannon KM, Yoshida M, Ma T, Mattia A, Lauwers GY, Iafrate AJ, Chung DC. A tailored approach to BRAF and MLH1 methylation testing in a universal screening program for Lynch syndrome. Mod Pathol. 2017;30:440–447. doi: 10.1038/modpathol.2016.211. [DOI] [PubMed] [Google Scholar]
- 31.Seppälä TT, Böhm JP, Friman M, Lahtinen L, Väyrynen VM, Liipo TK, Ristimäki AP, Kairaluoma MV, Kellokumpu IH, Kuopio TH, Mecklin JP. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1–10. doi: 10.1016/j.humpath.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Wolf AI, Buchanan AH, Farkas LM. Historical review of Lynch syndrome. J Coloproctol (Rio J) 2013;33 [Google Scholar]
- 34.Le S, Ansari U, Mumtaz A, Malik K, Patel P, Doyle A, Khachemoune A. Lynch syndrome and muir-torre syndrome: An update and review on the genetics, epidemiology, and management of two related disorders. Derma Online J. 2017;23:2. [PubMed] [Google Scholar]
- 35.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition-Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg Y, Kedar I, Kariiv R, Halpern N, Plesser M, Hubert A, Kaduri L, Sagi M, Lerer I, Abeliovich D, et al. Lynch syndrome in high risk Ashkenazi Jews in Israel. Fam Cancer. 2014;13:65–73. doi: 10.1007/s10689-013-9675-2. [DOI] [PubMed] [Google Scholar]
- 37.Cravo M, Afonso AJ, Lage P, Albuquerque C, Maia L, Lacerda C, Fidalgo P, Chaves P, Cruz C, Nobre-Leitão C. Pathogenicity of missense and splice site mutations in hMSH2 and hMLH1 mismatch repair genes: Implications for genetic testing. Gut. 2002;50:405–412. doi: 10.1136/gut.50.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haraldsdottir S, Rafnar T, Frankel WL, Einarsdottir S, Sigurdsson A, Hampel H, Snaebjornsson P, Masson G, Weng D, Arngrimsson R, et al. Comprehensive population-wide analysis of Lynch syndrome in Iceland reveals founder mutations in MSH6 and PMS2. Nat Commun. 2017;8:14755. doi: 10.1038/ncomms14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagerstedt Robinson K, Liu T, Vandrovcova J, Halvarsson B, Clendenning M, Frebourg T, Papadopoulos N, Kinzler KW, Vogelstein B, Peltomäki P, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 40.Houlleberghs H, Goverde A, Lusseveld J, Dekker M, Bruno MJ, Menko FH, Mensenkamp AR, Spaander MCW, Wagner A, Hofstra RMW, Te Riele H. Suspected Lynch syndrome associated MSH6 variants: A functional assay to determine their pathogenicity. PLoS Genet. 2017;13:e1006765. doi: 10.1371/journal.pgen.1006765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartosova Z, Fridrichova I, Bujalkova M, Wolf B, Ilencikova D, Krizan P, Hlavcak P, Palaj J, Lukac L, Lukacova M, et al. Novel MLH1 and MSH2 germline mutations in the first HNPCC families identified in Slovakia. Hum Mutat. 2003;21:449. doi: 10.1002/humu.9127. [DOI] [PubMed] [Google Scholar]
- 42.Rey JM, Noruzinia M, Brouillet JP, Sarda P, Maudelonde T, Pujol P. Six novel heterozygous MLH1, MSH2, and MSH6 and one homozygous MLH1 germline mutations in hereditary nonpolyposis colorectal cancer. Cancer Genet Cytogenet. 2004;155:149–151. doi: 10.1016/j.cancergencyto.2004.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.