 The aim of this study was to examine the effect of chromium(iii), iron(iii), molybdenum(iii) and nickel(ii) and their combinations on the cell cycle and mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells.
The aim of this study was to examine the effect of chromium(iii), iron(iii), molybdenum(iii) and nickel(ii) and their combinations on the cell cycle and mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells.
Abstract
The aim of this study was to examine the effect of chromium(iii), iron(iii), molybdenum(iii) and nickel(ii) and their combinations on the cell cycle and mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells. A statistically significant dose related decrease of the percentage of cells in the G0/G1 and S phases was observed. However, a statistically significant dose related increase of the percentage of cells in the G2/M phase after exposure to chromium(iii), nickel(ii) or molybdenum(iii) at 200–1000 μM concentrations in both cell lines was observed. Moreover, an increase of the percentage of cells in the subG1 phase was observed. In both cell lines a statistically significant dose related decrease of the percentage of cells in the G2/M phase after exposure to iron(iii) at 200–1000 μM concentrations was observed. However, a statistically significant dose related increase of the percentage of cells in the G0/G1 phase after exposure to iron(iii) at 200–1000 μM concentrations was observed. A concentration dependent statistically significant decrease in the level of the MTP was observed in both cell lines after exposure to chromium(iii), iron(iii), nickel(ii) and molybdenum(iii). The results obtained from both cell lines show that HepG2 cells are more sensitive when compared to BALB/3T3 cells. Additions of Cr(iii) at 200 μM plus Fe(iii) at 1000 μM showed a synergistic effect on the cell cycle and MTP. In the case of Cr(iii) at 200 μM plus Mo(iii) at 1000 μM, an antagonistic effect was observed in both analyses. In the case of Cr(iii) at 1000 μM plus Mo(iii), Ni(ii) and Fe(iii) at 200 μM, no changes in the percentage of cells in all phases were observed in both cell lines in both analyses.
Introduction
The transition elements chromium, molybdenum, iron and nickel are essential micronutrients for humans, animals and plants. Micronutrients play a crucial role in the prevention and treatment of various diseases in humans and animals. They also play an important role in the optimization of physical and mental functions. Moreover, the elements mentioned above are the components of biomaterials and dietary supplements.
The toxicity and essentiality of chromium depend on its oxidation state. There are conflicting results in the literature concerning the cytotoxicity and genotoxicity of chromium(iii). Chromium(iii) salts and chromium(iii) compounds have been shown to induce DNA damage, sister chromatid exchange, centromere positive and negative micronuclei, oxidative damage and Cr–DNA adducts. Very high chromium(iii) concentration or long exposure results in elevated intracellular concentrations and related genotoxic effects.1 Moreover, in acellular systems, chromium chloride induces genotoxic effects, including DNA strand breaks, DNA–DNA interstrand crosslinks, DNA–protein crosslinks, DNA–amino acid crosslinks, DNA–glutathione crosslinks and gene mutations. The electropositive chromium(iii) interacts with the negatively charged phosphate groups and the guanine bases forming phosphate–chromium–phosphate and guanine–chromium–guanine interstrand crosslinks. These crosslinks can result in functional disturbances in DNA and RNA polymerases.2
Iron is involved in a broad spectrum of biological functions, such as oxygen transport, electron transfer and DNA synthesis. The products of DNA damage are strand breaks, oxidatively modified bases and DNA–protein crosslinks. The damage includes decrease in membrane polyunsaturated fatty acid content, inhibition of thiol-dependent enzyme activities, lowered ATP and enhanced lysosomal fragility. Moreover, the inhibition of the activity of NADH-cytochrome c oxidoreductase (complexes I–III) and succinate dehydrogenase was observed.3 On the other hand, iron, in its free form, is a potential cytotoxic metal, especially because of its property to catalyze the formation of free radicals.4,5
Nickel exists in five valences or oxidation states of –1, +1, +2, +3, and +4, with +2 being the most common. Nickel compounds are taken up by mammalian cells, where nickel ions are released and then they can induce cytotoxicity, apoptosis, chromosomal aberrations and morphological transformation. Moreover, nickel has been found to induce chromosome deletions and genomic instability. Nickel detected in the Ames assay was also mutagenic. It has been found that nickel induces epigenetic gene silencing and histone modifications. Nickel can activate several cellular stress response signaling pathways: mitogen-activated protein kinases (MAPKs), phosphoinositide 3-kinase (PI3K), HIF-1, nuclear factor of activated T cells (NF-AT), and NF-κβ.6
Molybdenum is an essential element for humans. The most important oxidation states for the mineral are +2, +3, +4 and +6.7 Molybdenum occurs in a wide range of metalloenzymes in bacteria, fungi, algae, plants and animals where it forms a part of the active sites of these enzymes. In eukaryotic cells Mo-enzymes can be subdivided into two classes: the xanthine oxidase (XO) family that is represented by xanthine dehydrogenase, aldehyde oxidase (AO), pyridoxal oxidase and nicotinate hydroxylase, and a SO class of Mo-enzymes formed by sulfite oxidase (SO) and nitrate reductase (NR).8
Many studies have shown that the intake of the above-mentioned microelements is associated with the production of reactive oxygen species (ROS). ROS can react with proteins, DNA, and organelle lipids inside the cells. As the major site of oxidative phosphorylation in cells, mitochondria contain the richest lipid bilayer. The induction of mitochondrial malfunction leads to the activation of apoptosis.9–12 Therefore, we investigated the effects of chromium(iii), iron(iii), nickel(ii) and molybdenum(iii) on the mitochondrial transmembrane potential.
All the above-mentioned microelements can interact with DNA. That is why we decided to evaluate the effect on the cell cycle. The cell cycle consists of five phases: G0, G1, S, G2 and M. Cell cycle progression depends on the activity of cyclins and cyclin-dependent kinases (cdks). The active forms of cdks occur as heterodimers that are composed of a regulatory subunit called a cyclin, and its catalytic counterpart, the cdk. The cyclin–cdk complexes are activated by phosphorylation via cyclin-activating kinases (CAKs). Progression through G1 is mediated by the expression of cyclins D, E and A, while S and M phase progression is characterized by the appearance of cyclins A and B, respectively.13
The aim of this study was to examine the effect of chromium(iii), iron(iii), molybdenum(iii) and nickel(ii) and their combinations on the cell cycle and mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells.
Materials and methods
Chemicals
Dulbecco's Modified Eagle's Medium (DMEM), Eagle's Minimum Essential Medium (EMEM), heat-inactivated calf bovine serum (CBS) and fetal bovine serum (FBS) were purchased from the American Type Culture Collection (USA). Antibiotic antimycotic solution (10 000 U mL–1 of penicillin, 10 mg mL–1 of streptomycin, and 25 μg mL–1 of amphotericin B) was purchased from Sigma Chemical Co. (St Louis, MO, USA). Cell Cycle Phase Determination Kit and JC-1 Mitochondrial Membrane Potential Assay Kit were purchased from Cayman Chemical Company (Michigan, USA). Chromium chloride (CrCl3·6H2O), iron chloride (FeCl3·6H2O), nickel chloride (NiCl2·6H2O) and molybdenum trioxide (MoO3) were purchased from Acros Organics (Belgium).
Cell culture and treatment
Mouse embryo fibroblast BALB/3T3 clone A31 cells (ATCC CCL-163) were obtained from the American Type Culture Collection. The cells were cultured as adherent monolayers in plastic tissue culture dishes in DMEM supplemented with 10% (v/v) heat-inactivated CBS and antibiotic antimycotic solution (1 mL per 100 mL of cell culture medium). The cells were maintained at 37 °C in a humidified incubator under an atmosphere of 5% CO2. The cells were subcultured three times a week. They were used for assays during the exponential phase of growth.
Liver cancer HepG2 cells (ATCC HB-8065) were obtained from the American Type Culture Collection. The cells were cultured as adherent monolayers in plastic tissue culture dishes in Eagle's Minimum Essential Medium (EMEM, ATCC) supplemented with 10% (v/v) heat-inactivated FBS and antibiotic antimycotic solution (1 mL per 100 mL of cell culture medium). The cells were maintained at 37 °C in a humidified incubator under an atmosphere of 5% CO2. The cells were subcultured three times a week. They were used for assays during the exponential phase of growth.
CrCl3·6H2O, FeCl3·6H2O, NiCl2·6H2O and MoO3 were dissolved in PBS at a concentration of 10 mM. The final concentration was obtained by dilution in complete culture medium.
In order to perform the flow cytometric cell cycle analysis and mitochondrial transmembrane potential analysis, the cells were cultured on 96-well plates (2 × 105 cells per mL) in 100 μL complete growth medium. After 24 hours, the medium was replaced with fresh media supplemented with chromium chloride or iron chloride or nickel chloride or molybdenum trioxide within the range of concentrations from 200 to 1000 μM. The cells exposed to a mixture of microelements were similarly plated at a density of 2 × 105 cells per mL and incubated for 24 hours. Next, the medium was replaced with fresh media supplemented with 200 μM of chromium chloride plus 1000 μM of iron chloride or nickel chloride or molybdenum trioxide and, in the other case, supplemented with 1000 μM of chromium chloride plus 200 μM of iron chloride or nickel chloride or molybdenum trioxide. After 24 hours of incubation, assays using Cell Cycle Phase Determination Kit and JC-1 Mitochondrial Membrane Potential Assay Kit were performed.
Flow cytometric cell cycle analysis
The cell cycle phase determination was performed according to the original manufacturer's instruction – Cell Cycle Phase Determination Kit, Cayman Chemical Company, USA. After 24 hours of incubation with chromium chloride, iron chloride, nickel chloride, molybdenum trioxide or a mixture of microelements, the cells were harvested using trypsin. The cells were resuspended to a density of 106 cells per mL in an assay buffer. One mL of the fixative was added to each sample. The cells were centrifuged at 500g for five minutes. The cell pellet was suspended in 0.5 mL of the staining solution (propidium iodide) and incubated for 30 minutes at room temperature in the dark. The samples were analyzed in an FL2 channel of an FACSCanto II flow cytometer (BD Biosciences) with a 488 nm excitation laser. The data were analyzed by using the FlowJo software (Tree Star Inc., Stanford). The percentage of cells in the subG1, G0/G1, S and G2/M phases was calculated. Six independent experiments were performed with six wells per each treatment condition.
Mitochondrial transmembrane potential (MTP)
The MTP assay was performed according to the original manufacturer's instruction – JC-1 Mitochondrial Membrane Potential Assay Kit, Cayman Chemical Company, USA.
After 24 hours of incubation with chromium chloride, iron chloride, nickel chloride, molybdenum trioxide or a mixture of microelements, 100 μL of the JC-1 staining solution per 1 mL of culture medium was added to each well. The cells were incubated at 37 °C in darkness for 20 minutes. Then the cells were harvested using trypsin. The samples were analyzed using a 448 nm band pass filter for JC-1 aggregates (red fluorescence) indicating high or normal MTP, and a 525 nm band pass filter for JC-1 monomers (green fluorescence), which represents disrupted mitochondria in apoptotic and unhealthy cells. The data were analyzed by using the FlowJo software (Tree Star Inc., Stanford). Six independent experiments were performed with six wells per each treatment condition.
Statistical analysis of the data
The results were expressed as mean ± SD and the data were analyzed using one way analysis of variance (ANOVA) with Tukey's multi-comparison post-test using Statistica programme ver. 13PL. In all cases, p < 0.05 was considered significant.
Results
Flow cytometric cell cycle analysis
Flow cytometric cell cycle analysis was used to assess the effect of chromium chloride, iron chloride, nickel chloride and molybdenum trioxide, used separately and in combinations, on the distribution of the cell cycle fractions. DNA damage in both cell lines was quantified by measuring the cell cycle fractions by flow cytometry after staining with propidium iodide. Table 1 shows the analysis of fibroblasts and HepG2 cells. A statistically significant dose related decrease of the percentage of cells in the G0/G1 and S phases after Cr(iii), Ni(ii) and Mo(iii) exposure was observed. However, a statistically significant dose related increase of the percentage of cells in the G2/M and subG1 phases after exposure to chromium chloride, nickel chloride and molybdenum trioxide at concentrations of 200–1000 μM in both cell lines was observed. The results obtained from both cell lines show that HepG2 cells are more sensitive when compared to BALB/3T3 cells.
Table 1. Flow cytometric cell cycle analysis in HepG2 cells incubated with chromium chloride, iron chloride, nickel chloride, and molybdenum trioxide.
| |
BALB/3T3 clone A31 cells |
HepG2 cells |
|||||||
| Cell number [%] |
Cell number [%] |
||||||||
| subG1 | G0/G1 | S | G2/M | subG1 | G0/G1 | S | G2/M | ||
| Concentration of chromium chloride [μM] | |||||||||
| 0 | 6 ± 0.5 | 58 ± 4 | 9 ± 0.5 | 27 ± 2 | 7 ± 0.5 | 59 ± 4 | 9 ± 0.5 | 25 ± 1 | |
| 200 | 7 ± 0.4* | 55 ± 3* | 9 ± 0.6 | 29 ± 1* | 9 ± 0.7* | 54 ± 5* | 8 ± 0.3 | 29 ± 2* | |
| 400 | 10 ± 1* | 47 ± 3* | 9 ± 0.4 | 34 ± 3* | 11 ± 1* | 45 ± 3* | 8 ± 0.4 | 36 ± 2* | |
| 600 | 14 ± 1* | 33 ± 3* | 9 ± 0.7 | 44 ± 3* | 15 ± 1* | 30 ± 1* | 7 ± 0.5* | 48 ± 1* | |
| 800 | 15 ± 1.2* | 25 ± 1* | 5 ± 0.2* | 55 ± 4* | 17 ± 1* | 24 ± 2* | 3 ± 0.1* | 56 ± 4* | |
| 1000 | 16 ± 1* | 17 ± 1* | 2 ± 0.1* | 65 ± 5* | 19 ± 1* | 14 ± 1* | 1 ± 0.1* | 66 ± 4* | |
| Concentration of iron chloride [μM] | |||||||||
| 0 | 6 ± 0.5 | 58 ± 4 | 9 ± 0.5 | 27 ± 2 | 7 ± 0.5 | 59 ± 4 | 9 ± 0.5 | 25 ± 1 | |
| 200 | 7 ± 0.3 | 60 ± 3* | 9 ± 0.2 | 24 ± 1* | 8 ± 0.2 | 61 ± 3* | 9 ± 0.2 | 22 ± 1* | |
| 400 | 8 ± 0.2* | 62 ± 2* | 9 ± 0.3 | 21 ± 1* | 10 ± 1* | 64 ± 4* | 7 ± 0.3* | 19 ± 1* | |
| 600 | 9 ± 0.5* | 67 ± 5* | 8 ± 0.2* | 16 ± 1* | 12 ± 1.1* | 70 ± 5* | 6 ± 0.1* | 12 ± 0.8 | |
| 800 | 11 ± 1* | 70 ± 2* | 7 ± 0.1* | 12 ± 0.9* | 14 ± 1* | 74 ± 4* | 4 ± 0.1* | 8 ± 0.3* | |
| 1000 | 13 ± 1* | 72 ± 3* | 7 ± 0.4* | 9 ± 0.3* | 15 ± 0.8* | 75 ± 3* | 3 ± 0.1* | 7 ± 0.2* | |
| Concentration of molybdenum trioxide [μM] | |||||||||
| 0 | 6 ± 0.5 | 58 ± 4 | 9 ± 0.5 | 27 ± 2 | 7 ± 0.5 | 59 ± 4 | 9 ± 0.5 | 25 ± 1 | |
| 200 | 7 ± 0.4 | 49 ± 3* | 9 ± 0.4 | 35 ± 2* | 8 ± 0.4 | 47 ± 3* | 9 ± 0.6 | 36 ± 3* | |
| 400 | 8 ± 0.2* | 45 ± 4* | 8 ± 0.6 | 39 ± 3* | 9 ± 0.6* | 44 ± 3 | 7 ± 0.4* | 40 ± 2* | |
| 600 | 10 ± 1* | 40 ± 2* | 7 ± 0.4* | 43 ± 3* | 10 ± 1* | 38 ± 2* | 5 ± 0.1* | 47 ± 3* | |
| 800 | 12 ± 1* | 32 ± 2* | 5 ± 0.2* | 51 ± 5* | 13 ± 1* | 31 ± 1* | 4 ± 0.2* | 52 ± 4* | |
| 1000 | 15 ± 1* | 20 ± 1* | 5 ± 0.3* | 60 ± 4* | 17 ± 1* | 17 ± 1* | 3 ± 0.1* | 63 ± 4* | |
| Concentration of nickel chloride [μM] | |||||||||
| 0 | 6 ± 0.5 | 58 ± 4 | 9 ± 0.5 | 27 ± 2 | 7 ± 0.5 | 59 ± 4 | 9 ± 0.5 | 25 ± 1 | |
| 200 | 3 ± 0.2* | 58 ± 3* | 9 ± 0.7 | 30 ± 3* | 3 ± 0.1* | 57 ± 3 | 9 ± 0.8 | 31 ± 2* | |
| 400 | 5 ± 0.5* | 54 ± 4* | 8 ± 0.6 | 33 ± 3* | 6 ± 0.5* | 52 ± 5* | 8 ± 0.5 | 34 ± 3* | |
| 600 | 6 ± 0.4* | 50 ± 3* | 7 ± 0.5* | 37 ± 2* | 8 ± 0.6* | 46 ± 2* | 6 ± 0.2* | 40 ± 3* | |
| 800 | 11 ± 1* | 44 ± 2* | 5 ± 0.3* | 40 ± 1* | 9 ± 0.5* | 37 ± 3* | 4 ± 0.1* | 50 ± 4* | |
| 1000 | 13 ± 1* | 37 ± 2* | 1 ± 0.1* | 49 ± 4* | 14 ± 1* | 28 ± 1* | 1 ± 0.1* | 57 ± 5* | |
| Concentration (mixture of metal compounds) | |||||||||
| 200 μM | 1000 μM | ||||||||
| CrCl3·6H2O | FeCl3·6H2O | 21 ± 1*,1,2 | 77 ± 6*,1,2 | 1 ± 0.1*,1,2 | 1 ± 0.1*,1,2 | 22 ± 1*,1,2 | 76 ± 6*,1,2 | 1 ± 0.1*,1,2 | 1 ± 0.1*,1,2 |
| CrCl3·6H2O | MoO3 | 5 ± 0.2*,1,2 | 55 ± 4*,1,2 | 10 ± 1*,1,2 | 30 ± 2*,1,2 | 6 ± 0.2*,1,2 | 54 ± 4*,1,2 | 10 ± 1*,1,2 | 30 ± 2*,1,2 |
| CrCl3·6H2O | NiCl2·6H2O | 5 ± 0.1*,1,2 | 50 ± 3*,1,2 | 12 ± 1*,1,2 | 33 ± 2*,1,2 | 5 ± 0.1*,1,2 | 55 ± 2*,1,2 | 15 ± 1*,1,2 | 25 ± 1*,1,2 |
| 1000 μM | 200 μM | ||||||||
| CrCl3·6H2O | FeCl3·6H2O | 8 ± 0.2* | 50 ± 4* | 7 ± 0.2* | 35 ± 1* | 8 ± 0.2* | 40 ± 3* | 7 ± 1* | 45 ± 3* |
| CrCl3·6H2O | MoO3 | 6 ± 0.2* | 35 ± 2* | 9 ± 0.1* | 50 ± 1* | 7 ± 0.2* | 36 ± 2* | 10 ± 1* | 47 ± 2* |
| CrCl3·6H2O | NiCl2·6H2O | 5 ± 0.1* | 40 ± 1* | 10 ± 1* | 45 ± 2* | 4 ± 0.2* | 36 ± 1* | 5 ± 0.1* | 55 ± 3* |
Table 1 also shows the analysis of BALB/3T3 fibroblasts and HepG2 cells. A statistically significant dose related decrease of the percentage of cells in the S and G2/M phases after Fe(iii) exposure was observed. However, a statistically significant dose related increase of the percentage of cells in the subG1 and G0/G1 phases after exposure to iron chloride at concentrations of 200–1000 μM in both cell lines was observed as well.
Additionally Table 1 shows the changes in the cell cycle after treatment with mixtures of metals as follows: Cr(iii) at a concentration of 200 μM plus Fe(iii) or Ni(ii) or Mo(iii) at a concentration of 1000 μM. Additions of Cr(iii) at 200 μM plus Fe(iii) at 1000 μM showed the synergistic effect of an increase in the percentage of cells in the G0/G1 and subG1 phases and a decrease in the percentage of cells in the G2/M and S phases in both cell lines. In the case of Cr(iii) at 200 μM plus Mo(iii) at 1000 μM, an antagonistic effect was observed. Chromium(iii) protects against molybdenum (at 1000 μM) toxicity – a decreased percentage of cells in the subG1 phase and an increased percentage of cells in the S and G0/G1 phases in both cell lines were observed when compared to the cells incubated with molybdenum. The protective effect of Cr(iii) in decreasing the percentage of cells in the subG1 phase and increasing the percentage of cells in the S and G0/G1 phases when compared to the cells incubated with nickel was also observed in a pair of Cr(iii) at 200 μM and Ni(ii) at 1000 μM in BALB/3T3 and HepG2 cell lines.
In the case of Cr(iii) at 1000 μM plus Mo(iii), Ni(ii) and Fe(iii) at 200 μM, no changes in the percentage of cells in all phases were observed in both cell lines (Table 1).
Mitochondrial transmembrane potential (MTP)
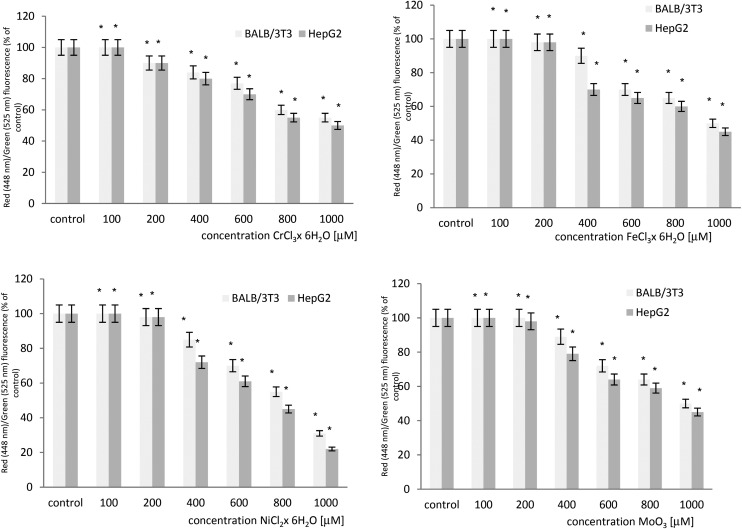
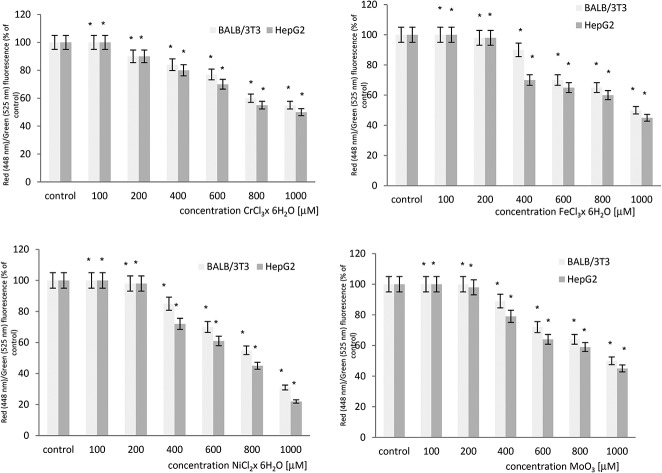
The effect of microelements on the mitochondrial transmembrane potential (MTP) was evaluated in BALB/3T3 and HepG2 cell lines. A concentration dependent statistically significant decrease in the level of the MTP was observed in both cell lines after exposure to chromium chloride, iron chloride, nickel chloride and molybdenum trioxide. The results obtained from both cell lines show that HepG2 cells are more sensitive when compared to BALB/3T3 cells. The cytometric analysis of the mitochondrial transmembrane potential in both cell lines is summarized in Fig. 1.
Fig. 1. The loss of the mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells treated with chromium chloride, iron chloride, nickel chloride or molybdenum trioxide as determined by flow cytometry using JC-1 staining. *p < 0.05, significance of difference compared to the control.
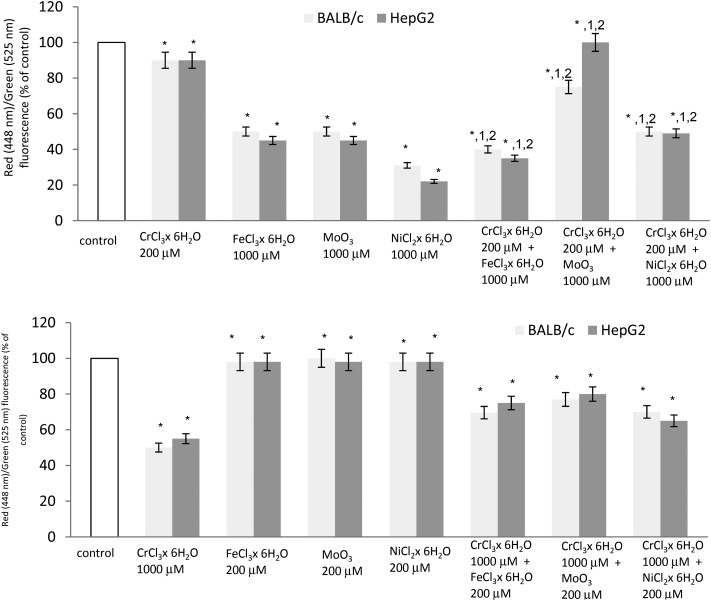
Fig. 2 shows the changes in the mitochondrial transmembrane potential by mixing metals of Cr(iii) at 200 μM concentration plus Fe(iii) or Ni(ii) or Mo(iii) at 1000 μM concentration. Additions of Cr(iii) at 200 μM plus Fe(iii) at 1000 μM showed a synergistic effect in decreasing the mitochondrial transmembrane potential in both cell lines. In the case of Cr(iii) at 200 μM plus Mo(iii) at 1000 μM, an antagonistic effect was observed. Chromium(iii) protects against molybdenum (at 1000 μM) toxicity in both cell lines. The protective effect of Cr(iii) in decreasing the mitochondrial transmembrane potential was also observed in a pair of Cr(iii) at 200 μM and Ni(ii) at 1000 μM in BALB/3T3 and HepG2 cell lines.
Fig. 2. The loss of the mitochondrial transmembrane potential (MTP) in BALB/3T3 and HepG2 cells treated with a mixture of chromium chloride with iron chloride or nickel chloride or molybdenum trioxide in BALB/3T3 and HepG2 cell lines as determined by flow cytometry using JC-1 staining. *p < 0.05, significance of difference compared to the control. 1p < 0.05, significance of difference compared to chromium chloride at a concentration of 200 μM. 2p < 0.05, significance of difference compared to iron chloride/nickel chloride/molybdenum trioxide at a concentration of 1000 μM.
In the case of Cr(iii) at 1000 μM plus Mo(iii), Ni(ii) and Fe(iii) at 200 μM, no changes in the mitochondrial transmembrane potential were observed in both cell lines (Fig. 2).
Discussion
Cell cycle analysis is a useful tool used in the analysis of the anti-proliferative activity of many substances. Moreover, the analysis of the mitochondrial potential serves to assess the pro-apoptotic activity of biologically active substances. During apoptosis several key events occur in the mitochondria, including the release of cytochrome c, changes in electron transport and loss of the mitochondrial transmembrane potential. MTP is an important parameter of mitochondrial function and has been used as an indicator of cell health.
The concentrations of chromium chloride, iron chloride, molybdenum trioxide and nickel chloride were chosen for these studies on the basis of our earlier investigations. At 200 μM concentration in all used microelements, no changes in cell viability were observed. Moreover, in some cases the cell viability was slightly stimulated. However, at a concentration of 1000 μM and above that, a decrease of cell viability and apoptosis was observed.14
Table 1 presents the normal distribution of DNA in the whole cell cycle in normal and cancer cells. It has been noticed that the population is maximal in the G0/G1 phase, which is the state of active growth before DNA replication. The DNA content in the S phase is smaller when compared to the cells in the G2/M phase of the cell cycle.
In our study DNA decreases in the G0/G1 phase and in the S phase, while in the S phase it practically disappears in the cells treated with chromium chloride in both cell lines. Moreover, subG1 peak (aneuploidy peak) was observed. This peak may represent the cells that escaped mitotic arrest and replicate as multinuclear cells without dividing. Our earliest investigations confirm these observations. In the cells treated with chromium chloride, giant and multinuclear cells were observed.14 Some authors suggest that Cr(iii) ions may interact with nucleus causing changes in chromatin structure.15 Similar results were obtained by Shivastava et al.16 It can be concluded that trivalent chromium arrests the cell cycle in the G2/M phase. The appearance of the aneuploidy peak is a major characteristic of apoptotic cells – the ploid DNA loses its normal ploidy due to the degradation of DNA.16 Moreover, Dai et al.17 show that trivalent chromium increases the frequency of mutations: substitutions, transversions, deletions and insertions. The Cr(iii) ion strongly inhibits the activity and fidelity of the DNA polymerase-mediating in vitro replication. The G2/M phase plays an important role in mitosis. The G2/M DNA damage check-point serves to prevent the cells from entering mitosis with genomic DNA damage.18 It has been proved that chromium induces ROS production. ROS are reactive chemical species in biological systems; when present in excess, they can cause not only oxidative DNA damage including DNA base modifications and DNA strand breaks, but they can also damage lipids, proteins, and mitochondria. In this study a decrease of the mitochondrial transmembrane potential was observed. This supports the observation that ROS are responsible for chromium(iii)-induced DNA and mitochondria damage.
Iron is an essential element for cell metabolism. In most eukaryotic cells, iron is necessary to facilitate the assembly of functional Fe–S cluster proteins, heme-binding proteins, and ribonucleotide reductases. The iron-binding proteins are abundantly present in the mitochondria, cytosol, and nucleus. These proteins are involved in electron transfer, ribosome maturation, DNA replication and repair, and cell cycle control.19 However, cellular oxidative stress induced by iron can cause alterations on membrane lipids, proteins and DNA; it promotes functional organelle impairment and genetic mutations, such as mutation in the p53 gene, a key gene of cell cycle control.20
The relationship between iron and the cell cycle has mainly been studied during iron depletion. Iron chelators induce cell cycle blockage during the G1, S, or G2 phases.20–22 This blockage may be related to the deficiency of iron incorporation in ribonucleotide reductase, consequently inhibiting DNA synthesis. Moreover, it has been described that cyclin D-, cyclin E-, and cyclin A-associated kinase activity is inhibited and the cyclin D1 protein level is decreased after iron chelator exposure. Iron exposure induces cell cycle disturbances. Iron-induced cyclin D overexpression could play a role in this priming. It has been shown that cyclin overexpression was sufficient to induce G1/S transition.20 In our study a decrease of the percentage of cells in the G2/M phase was observed, while an increase in the G0/G1 and subG1 phases was observed in both cell lines.
Incubation of both cell lines with iron chloride resulted in a dose dependent loss of the mitochondrial transmembrane potential. A decrease in the loss of the MTP can be caused by ROS production after iron(iii) treatment. Iron induces the production of superoxides and hydroxyl radicals via the Haber–Weiss and Fenton reactions.23 These radicals can interact with membrane lipids leading to the loss of the MTP. Moreover, ROS are involved in the destruction of DNA and the inhibition of DNA replication.
Nickel compounds are recognized as carcinogens. The possible mechanisms of nickel-induced carcinogenesis include the generation of DNA strand breaks, DNA protein crosslinks, and induction of reactive oxygen species which interact with DNA.24 Moreover, nickel can inhibit mitochondrial succinate dehydrogenase activity leading to the loss of the mitochondrial membrane potential (MTP) and downregulation of the expression of Bcl-2.25 In our experiments the incubation of both cell lines with nickel chloride caused an increase of the percentage of cells in the G2/M phase, while the percentage of cells in the S and G0/G1 phases decreased. Lee et al.26 have shown that nickel(ii) has a strong potential to cause apoptosis in RPMI-2650 cells. With increasing nickel(ii) concentration, the percentage of cells in the G1 and S phases tends to decrease, while it increases in the G2/M phase, indicative of an arrest. Accumulation of cells in the G2/M phase was observed in kidney cells and Chinese hamster ovary cells after nickel(ii) treatment.27,28 The investigations conducted by Lee et al.26 suggest that nickel(ii) induces G2/M cell accumulation by inhibiting the activity of cdk1 through directly stimulating the expression of p53 and p21WAF1/CIP1.
Incubation of both cell lines with nickel chloride resulted in a dose dependent loss of the mitochondrial transmembrane potential. In investigations by Lee et al.26 a decrease in the loss of the MTP and an increase of dose dependent ROS generation were observed. Similar results were obtained by Wang et al.25 This is in agreement with our investigations.
The present study shows that DNA decreases in the G0/G1 phase, while it increases in the G2/M phase after incubation with molybdenum(iii). This suggests that molybdenum activates the G2/M cell cycle checkpoint. Molybdenum trioxide inhibits cell proliferation by inducting G2/M arrest in both cell lines. The G2/M DNA damage checkpoint serves to prevent cells from entering mitosis with genomic DNA damage. Similar results were obtained by Siddiqui et al.18
In this study we have noticed a decrease in the mitochondrial transmembrane potential (MTP) in both cell lines exposed to molybdenum trioxide. Similar results were obtained by Siddiqui et al.18They found that molybdenum nanoparticles decrease the MTP in L929 cell lines. Moreover, the same investigation found that molybdenum induced intracellular ROS generation in a concentration dependent manner. This oxygen radical production during mitochondrial respiration suggests possible correlation between oxidative stress and mitochondrial activity and DNA damage.
The mixture of Cr(iii) plus Mo(iii) or Cr(iii) plus Ni(ii) shows an antagonistic effect – chromium(iii) protects against nickel(ii) or molybdenum(iii) toxicity. In cultures simultaneously treated with 200 μM of chromium chloride and 1000 μM of nickel chloride or molybdenum trioxide, a decreased percentage of cells in the subG1 phase was observed when compared to the cells incubated with nickel chloride or molybdenum trioxide at a concentration of 1000 μM. Moreover, the percentage of cells in the G0/G1, S and G2/M phases is comparable to the control cells. Investigations performed by Snow and Xu29 show that chromium chloride at low concentrations enhanced nucleotide incorporation during the replication of single-stranded DNA. What is more, the processivity of DNA polymerase increases in low concentrations of chromium chloride.29 The same mechanism was observed in the mitochondrial transmembrane potential assay – chromium(iii) protects against nickel(ii) or molybdenum(iii) toxicity. The MTP increases when compared to the control cells and the cells incubated with nickel chloride or molybdenum trioxide at a concentration of 1000 μM. These two results support the observations confirming the participation of ROS in this interaction. ROS levels are regulated by pro-oxidant and anti-oxidant systems and the correct balance between these two mechanisms is essential for cellular health.30 It has been proved that chromium(iii) at low concentrations statistically significantly increases superoxide dismutase and catalase activity, which are the most important enzymes involved in antioxidant activity.31,32 This suggests that chromium(iii) may activate antioxidant enzymes and protect against the toxic action of other metals. The catalase activity decreases at higher concentrations of Cr(iii), which is probably caused by the destruction of an active site of catalase due to excess Cr(iii).32 For this reason in the cells incubated with chromium chloride at a concentration of 1000 μM with nickel chloride or molybdenum trioxide at a concentration of 200 μM, the protective effect of chromium(iii) was not observed.
Cr(iii) and Fe(iii) show synergistic effects in both assays. In cultures simultaneously treated with 200 μM of chromium chloride and 1000 μM of iron chloride, an increase in the percentage of cells in the subG1 and G0/G1 phases was observed when compared to the control cells and the cells incubated with chromium chloride and iron chloride. Moreover, the same mechanism was observed in the mitochondrial transmembrane potential assay – chromium(iii) potentiates the decrease of the mitochondrial membrane potential. It has been reported that both of them can generate reactive oxygen species. In this case chromium(iii) does not protect against iron toxicity, mainly because other products are synthesized after iron(iii) incubation, i.e. lipid hydroxyl-peroxide: ROOH, lipid peroxidative products such as malondialdehyde and 4-hydroxy-2-nonenal are increased, and they form the radicals: ROO-(alkyl oxyradical) and RO-(alkoxy radical). These radicals cannot be neutralized by catalase and SOD. The lipid-based radicals possess longer half-lives than hydroxyl radicals, and they also have a stronger effect on chronic cell toxicity and DNA damage. They attack biomolecules and organelles: membrane phospholipids, DNA and mitochondria.33 Additionally, Cr(iii) interacts with both the base and phosphate groups of DNA, and Fe(iii) interacts with DNA bases.34 These independent mechanisms can cause a synergistic effect.
These initial findings indicate the need to carry out further investigations to identify the different mechanisms of interaction between microelements. They could be useful to define the influence of these elements on human and animal health.
Conflicts of interest
There are no conflicts of interest to declare.
References
- Novotnik B., Scancar J., Milacic R., Filipic M., Zegura B. Chemosphere. 2016;154:124–131. doi: 10.1016/j.chemosphere.2016.03.118. [DOI] [PubMed] [Google Scholar]
- Santonen T., Inorganic Chromium(III) Compounds, World Health Organization, 2009. [Google Scholar]
- Ponka P., Tenenbein M. and Eaton J. W., in Handbook on the Toxicology of Metal, ed. G. F. Nordberg and B. A. Fowler, Elsevier, 4th edn, 2015, ch. 41 – Iron, pp. 879–902. [Google Scholar]
- Hirsh M., Konijn A. M., Iancu T. C. J. Hepatol. 2002;36:30–38. doi: 10.1016/s0168-8278(01)00221-5. [DOI] [PubMed] [Google Scholar]
- Viatte L., Vaulont S. Biochimie. 2009;91:1223–1228. doi: 10.1016/j.biochi.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Klein C. and Costa M., in Handbook on the Toxicology of Metals, ed. G. F. Nordberg and B. A. Fowler, Elsevier, 4th edn, 2015, ch. 48 – Nickel, pp. 1091–1111. [Google Scholar]
- Tallkvist J. and Oskarsson A. G., in Handbook on the Toxicology of Metals, ed. G. F. Nordberg and B. A. Fowler, Elsevier, 4th edn, 2015, ch. 47 – Molybdenum, pp. 1077–1089. [Google Scholar]
- Mendel R. R., Bittner F. Biochim. Biophys. Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Liu P., Wang L., Luo J., Zhang C., Guo X., Hi G., Cao H. Biol. Trace Elem. Res. 2016;170:106–114. doi: 10.1007/s12011-015-0450-0. [DOI] [PubMed] [Google Scholar]
- Sharma B., Singh S., Siddiqi N. J. BioMed Res. Int. 2014;2014:26. doi: 10.1155/2014/640754. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Singh N., Gupta V. K., Kumar A., Sharma B. Front. Chem. 2017;5:70. doi: 10.3389/fchem.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Singh N., Pandey R., Kumar V. G. and Sharma B. in Biomedical Applications of Metals, ed. M. Rai, et al., Springer International Publishing AG, part of Springer Nature 2018, Synergistic Effects of Heavy Metals and Pesticides in Living Systems, 2018, pp. 297–319. [Google Scholar]
- Le N. T. V., Richardson D. R. Biochim. Biophys. Acta. 2002;1603:31–46. doi: 10.1016/s0304-419x(02)00068-9. [DOI] [PubMed] [Google Scholar]
- Terpiłowska S., Siwicki A. K. Chemosphere. 2018;201:780–789. doi: 10.1016/j.chemosphere.2018.03.062. [DOI] [PubMed] [Google Scholar]
- Rudolf E., Červinka M. Acta Med. (Hradec Králové) 2003;46(4):139–146. [PubMed] [Google Scholar]
- Shrivastava H. Y., Ravikumar T., Shanmugasundaram N., Babu M., Nair B. U. Free Radicals Biol. Med. 2005;38:58–69. doi: 10.1016/j.freeradbiomed.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Dai H., Liu J., Malkas L. H., Catalano J., Alagharu S., Hickey R. J. Toxicol. Appl. Pharmacol. 2009;236:154–165. doi: 10.1016/j.taap.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui M. A., Saquiba Q., Ahamed M., Farshorid N. N., Ahmad J., Wahab R., Khan S. T., Alhadlaq H. A., Musarrat J., Abdulaziz Al-Khedhairy A., Pant A. B. Colloids Surf., B. 2015;125:73–81. doi: 10.1016/j.colsurfb.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Zhang C. Protein Cell. 2014;5(10):750–760. doi: 10.1007/s13238-014-0083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troadec M.-B., Courselaud B., Detivaud L., Haziza-Pigeon Ch., Leroyer P., Brissot P., Loreal O. J. Hepatol. 2006;44:391–399. doi: 10.1016/j.jhep.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Chenoufi N., Drenou B., Loréal O., Pigeon C., Brissot P., Lescoat G. Biochem. Pharmacol. 1998;56:431–437. doi: 10.1016/s0006-2952(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Rakba N., Loyer P., Gilot D., Delcros J. G., Glaise D., Baret P. Carcinogenesis. 2000;21:943–951. doi: 10.1093/carcin/21.5.943. [DOI] [PubMed] [Google Scholar]
- Chan S., Chen M., Cao J., Chan G. C. F., Cheung Y. Acta Haematol. 2014;132:200–210. doi: 10.1159/000356808. [DOI] [PubMed] [Google Scholar]
- Błasiak J., Arabski M., Petryński T., Woźniak K., Drzewoski J. Cell Biol. Toxicol. 2002;18:279–288. doi: 10.1023/a:1016059112829. [DOI] [PubMed] [Google Scholar]
- Wang Y.-F., Shyu H.-W., Chang Y.-Ch., Tseng W.-Ch., Huang Y.-L., Lin K.-H., Chou M.-Ch., Liu H.-L., Chen Ch.-Y. Toxicol. Appl. Pharmacol. 2012;259(2):177–186. doi: 10.1016/j.taap.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Lee Y.-J., Lim S.-S., Baek B. J., An J.-M., Nam H.-S., Woo K.-M., Cho M.-K., Kim S.-H., Lee S.-H. Environ. Toxicol. Pharmacol. 2016;42:76–84. doi: 10.1016/j.etap.2016.01.005. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Lin T. K., Chang Y. C., Wang Y. F., Shyu H. W., Lin K. H., Chou M. C. J, Toxicol. Environ. Health., Part A. 2010;73:529–539. doi: 10.1080/15287390903421250. [DOI] [PubMed] [Google Scholar]
- Shiao Y. H., Lee S. H., Kasprzak K. S. Carcinogenesis. 1998;19:1203–1207. doi: 10.1093/carcin/19.7.1203. [DOI] [PubMed] [Google Scholar]
- Snow E. T., Xu L.-S. Biol. Trace Elem. Res. 1989;21:61–71. doi: 10.1007/BF02917237. [DOI] [PubMed] [Google Scholar]
- Pilo F., Angelucci E. Blood Rev. 2018;32:29–35. doi: 10.1016/j.blre.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Długosz A., Rembacz K. P., Pruss A., Durlak M., Lembas-Bogaczyk J. Pol. J. Environ. Stud. 2012;21(2):331–335. [Google Scholar]
- Chen L., Zhang J., Zhu Y., Zhang Y. Food Chem. 2018;244:378–385. doi: 10.1016/j.foodchem.2017.10.062. [DOI] [PubMed] [Google Scholar]
- Kohgo Y., Ikuta K., Ohtake T., Torimoto Y., Kato J. Int. J. Hematol. 2008;88:7–15. doi: 10.1007/s12185-008-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki H., Osborne M., Phillips D. H. Toxicol. in Vitro. 2008;22(1):36–44. doi: 10.1016/j.tiv.2007.07.011. [DOI] [PubMed] [Google Scholar]