Abstract
Background
Antisense long noncoding RNAs (lncRNAs) are single‐stranded RNAs that overlapped gene‐coding regions on the opposite DNA strand and play as critical regulators in cardiovascular diseases. The high conservation and stability may be good advantages for antisense lncRNAs. However, the roles of antisense lncRNAs in cardiomyocyte proliferation and cardiac regeneration are still unknown.
Methods and Results
In this study, we found that Silent information regulator factor 2 related enzyme 1 (Sirt1) antisense lncRNA expression was significantly increased during heart development. By gain and loss function of Sirt1 antisense lncRNA using adenovirus and locked nucleic acid, respectively, we demonstrated that Sirt1 antisense lncRNA promoted cardiomyocyte proliferation in vitro and in vivo, and the suppression of Sirt1 antisense lncRNA inhibited cardiomyocyte proliferation. Moreover, overexpression of Sirt1 antisense lncRNA enhanced cardiomyocyte proliferation, attenuated cardiomyocyte apoptosis, improved cardiac function, and decreased mortality rate after myocardial infarction. Furthermore, Sirt1 antisense lncRNA can bind the Sirt1 3′‐untranslated region, enhancing the stability of Sirt1 and increasing Sirt1 abundance at both the mRNA and protein levels. Finally, we found that Sirt1 was involved in Sirt1 antisense lncRNA‐induced cardiomyocyte proliferation.
Conclusions
The present study identified Sirt1 antisense lncRNA as a novel regulator of cardiomyocyte proliferation and cardiac regeneration by interacting and stabilizing Sirt1 mRNA, which may serve as an effective gene target for preventing myocardial infarction.
Keywords: cardiac regeneration, cardiomyocyte proliferation, myocardial infarction, Sirt1, Sirt1 antisense lncRNA
Subject Categories: Myocardial Regeneration, Myocardial Infarction
Clinical Perspective
What Is New?
Sirt1 antisense long noncoding RNA promotes cardiomyocyte proliferation in vitro and in vivo.
Sirt1 antisense long noncoding RNA decreases infarct size, suppresses cardiomyocyte apoptosis, and enhances cardiac function in a mouse myocardial infarction model.
Sirt1 antisense long noncoding RNA interacts and stabilizes Sirt1 mRNA, thereby increasing Sirt1 protein expression in cardiomyocytes.
What Are the Clinical Implications?
Our results indicate that Sirt1 antisense long noncoding RNA might be a novel effective therapeutic target for treating myocardial infarction.
During the past decade, long noncoding RNAs (lncRNAs) have been identified and found to play crucial roles in cardiomyocyte proliferation,1 apoptosis,2 and so on as well as to strictly regulate several human cardiovascular diseases, such as cardiac hypertrophy,3 ventricular arrhythmias,4 and myocardial infarction (MI).5 Although lncRNAs are highly abundant in human transcripts, many lncRNAs are not conserved at the sequence level and are expressed only in primates.6 The low conservation of lncRNAs makes screening for the biological properties of only primate‐specific lncRNAs in humanized models or organoid cultures extremely challenging. Antisense lncRNAs are transcribed from the opposite direction of their specific sense protein‐coding genes or a sense strand‐derived RNA.7 Global transcriptome analyses have confirmed that several antisense lncRNAs are evolutionarily conserved in mammals.8 The conserved expression of antisense lncRNAs may be related to conserved expression of the corresponding protein‐coding genes.8 Moreover, antisense lncRNAs are more stable than the head‐to‐head bidirectional transcripts, intronic RNAs, and promote‐associated lncRNAs on the basis of genome‐wide analysis.9 The conservation and stability of antisense lncRNAs might be good indicators for their potential biological functions.
Previous studies have revealed that antisense lncRNAs are involved in various types of cell proliferation, such as Metastasis Associated In Colon Cancer 1–antisense 1 in gastric cancer cells,10 Zinc Finger E‐Box Binding Homeobox 1–antisense 1 in prostate cancer cells,11 and Hepatocyte Nuclear Factor 1‐Alpha–antisense 1 in colon cancer cells.12 Moreover, an antisense lncRNA was proved to participate in bone regeneration.13 In addition, antisense lncRNAs are potentially involved in heart development. For example, Kcnq1ot1, transcribed from the Kcnq1 locus, is imprinted in early cardiac development and experiences a loss of imprinting during cardiac development,14 which has already been associated with atherosclerotic plaques as well as arsenic trioxide–induced long‐QT syndrome.15 However, there is no direct evidence of antisense lncRNA modulation in cardiomyocyte proliferation and cardiac regeneration after MI.
Sirt1 antisense lncRNA is fully overlapping with the Sirt1 mRNA 3′ untranslated region (3′‐UTR) by base complementary pairing principle.16 Previous studies have demonstrated that Sirt1 antisense lncRNA promotes myoblast17 and endothelial progenitor cell proliferation.18 Antisense lncRNAs can increase the stability of their sense mRNAs by forming an lncRNA‐mRNA duplex and affect the sense mRNA expression.19 Sirt1 antisense lncRNA can bind to Sirt1 mRNA 3′‐UTR, subsequently increasing the stability of Sirt1 mRNA and promoting C2C12 cell proliferation.17 Recently, we successfully identified that Sirt1 promotes cardiac regeneration and prevents MI progression. On the basis of the relationship between Sirt1 antisense lncRNA and Sirt1 mRNA, we speculated that Sirt1 antisense lncRNA may induce cardiomyocyte proliferation by interacting with Sirt1 mRNA and increasing Sirt1 abundance.
In this study, we used adenovirus or adeno‐associated virus 9 (AAV9) vectors to overexpress, and locked nucleic acid (LNA) inhibitor to deplete, Sirt1 antisense lncRNA levels to elucidate the function of Sirt1 antisense lncRNA in cardiomyocyte proliferation and cardiac regeneration after MI. In addition, we proposed to clarify the molecular mechanism by which Sirt1 antisense lncRNA promotes cardiomyocyte proliferation. Our study provides a novel insight into antisense lncRNA, which could be used as an effective therapeutic approach in cardiac regeneration after MI.
Materials and Methods
The data, analytic methods, and study materials will not be made available to other researchers for purposes of reproducing the results or replicating the procedure. Other researchers can contact the corresponding authors about methodological questions.
Ventricular Cardiomyocyte Isolation
C57BL/6J mice, aged 1 and 7 days old, were purchased from the Laboratory Animal Center of Southern Medical University. All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Southern Medical University and followed the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Neonatal mice were euthanized by 2% isoflurane inhalation and cervical dislocation. Then, ventricles from neonatal mice were separated from the atria, cut into pieces, and digested in 0.25% trypsin (Gibco, CA) at 4°C overnight. Digestion was performed 2 more times using collagenase type II (Gibco) and BSA (Sigma, Darmstadt, Germany) in PBS at 37°C for 15 minutes under constant stirring. Digestion was performed at room temperature in 15‐minute steps, collecting the supernatant with fetal bovine serum (Gibco) after each step. The collected supernatant was centrifuged to harvest the cells, which were resuspended in DMEM/nutrient F‐12 Ham medium (1:1) (Hyclone, UT) supplemented with 10% fetal bovine serum, 100 U/mL penicillin (Sigma), and 100 mg/mL streptomycin (Sigma). The collected cells were seeded onto 100‐mm plastic dishes for 2 hours at 37°C in a humidified atmosphere of 5% CO2. The supernatant, composed mostly of cardiomyocytes, was then collected and pelleted. The cells were resuspended in DMEM/nutrient F‐12 Ham medium containing 10% fetal bovine serum, counted, and plated at the appropriate density. For insulin‐like growth factor 1 treatment experiments, cardiomyocytes were treated with 50 ng/mL recombinant insulin‐like growth factor 1 (Invitrogen, CA) for 48 hours.
Adenovirus and Small‐Interfering RNA Transfection
The adenovirus overexpression Sirt1 antisense lncRNA was synthesized by Vigene (Shandong, China). Sirt1 siRNA was synthesized by Ribobio (Guangzhou, China). Isolated mouse cardiomyocytes were seeded at 70% confluence and cultured for 48 hours. For adenovirus transfection, various multiplicity of infection (MOI) of adenovirus–Sirt1 antisense lncRNA (Ad‐SAS) or adenovirus‐negative control (Ad‐NC) was added to cells. For siRNA transfection, 5 μL of Lipofectamine 2000 (Invitrogen) and 50 nmol/L siRNAs were added in Opti‐MEM medium (Gibco). The mixed solution was incubated at room temperature for 20 minutes and then added to the cells. The medium containing adenovirus or siRNA was replaced after 6 hours of incubation at 37°C with an equal volume of DMEM/nutrient F‐12 Ham medium. After 48 hours, the cells were subjected to RNA or protein isolation or immunofluorescence analysis.
Injection of LNA, AAV9, and Adenovirus Vectors Into Neonatal and Adult Mice
The AAV9 overexpression Sirt1 antisense lncRNA and adenovirus depletion Sirt1 were synthesized by Vigene (Shandong, China), and the LNA GapmeR inhibition Sirt1 antisense lncRNA was synthesized by Exiqon (MA). In the experiments using LNA, neonatal C57BL/6J mice were randomly intracardially injected with the LNA‐control or LNA–Sirt1 antisense lncRNA at a dose of 5 mg/kg per animal using an insulin syringe and a 30‐gauge needle (Becton, Dickinson and Company, NJ) on postnatal days 1 and 5. The hearts of the injected mice were collected 10 days after LNA injection. For adult mouse hearts, AAV9 vectors were used for transduction in vivo. Mice were anesthetized with 2% isoflurane. Then, the AAV9 vectors (AAV9‐negative control [AAV9‐NC] or AAV9‐Sirt1 antisense lncRNA [AAV9‐SAS]) at a dose of 1×1011 viral genome particles were randomly injected into the left ventricle of each animal using an insulin syringe with a 30‐gauge needle. The AAV9 vectors were injected into the heart at 5 sites, and the total volume injected into each heart was 20 μL. After surgery, the skin was disinfected and the animals were revived by being maintained on a thermal insulation blanket. In the experiments using the adenovirus vectors, neonatal mice were anesthetized by cooling on an ice bed for 5 minutes. After the fourth intercostal was dissected, mice received random intracardiac injection with adenovirus‐control, Ad‐SAS, Ad‐SAS+Ad‐small‐interfering‐negative control (Ad‐si‐NC), or Ad‐SAS+Ad‐small‐interfering‐Sirt1 (Ad‐si‐Sirt1) at a dose of 2×1010 viral genome particles per animal using an insulin syringe with a 30‐gauge needle. The virus was injected into the neonatal heart at 3 sites, and the total volume injected into each heart was 20 μL. Then, mice were placed under a heat lamp and warmed for several minutes until recovery.
RNA Isolation and Real‐Time Quantitative PCR
Total RNA from isolated cardiomyocytes or dissected ventricular heart tissue samples was extracted using the E.Z.N.A. Total RNA Kit II (Norcross, GA), according to the manufacturer's instructions. Cytoplasmic and nuclear RNAs were separated using an RNeasy Midi Kit (Qiagen, MA). The RNAs were treated with DNase I (Invitrogen) to exclude the possibility of DNA contamination. Then, cDNA was reverse transcribed from 1 μg of total RNA using the PrimeScript RT Master Mix (TaKaRa, Dalian, China). For the reverse transcription of Sirt1 antisense lncRNA, the strand‐specific primers, reverse primer of Sirt1 antisense lncRNA as the reverse transcription primers, were used. Real‐time quantitative PCR (qPCR) was performed with the SYBRs Premix Ex Taq Kit (TaKaRa) on the Lightcycler480 instrument (Roche, Basel, Switzerland). GAPDH was used as a housekeeping control to normalize gene expression using the ΔΔ cycle threshold (Ct) method. For the quantification of miRNA expression, the miRNA First Strand cDNA Synthesis (Sangon, Shanghai, China) and MicroRNAs Quantitation PCR Kit (Sangon) were used, according to the manufacturer's instructions. U6 was used as an endogenous control to normalize miRNA expression. All primers were designed by Sangon Biotech Co, Ltd (Shanghai, China). The primer sequences were as follows: Sirt1 antisense lncRNA, 5′‐AATCCAGTCATTAAACGGTCTACAA‐3′ (forward) and 5′‐AATCCAGTCATTAAACGGTCTACAA‐3′ (reverse); Sirt1, 5′‐TTGGCACC GATCCTCGAAC‐3′ (forward) and 5′‐CCCAGCTCCAGTCAGAACTAT‐3′ (reverse); Runt Related Transcription Factor 1 (RUNX1), 5′‐AACCAGGTAGCGAGATTCAACGAC‐3′ (forward) and 5′‐CAACTTGTGGCGGATTTGTAAAGA‐3′ (reverse); c‐kit, 5′‐CGGGCTAGCCAGAGACATCA‐3′ (forward) and 5′‐TCTCTGGTGCCATCCACTTCA‐3′ (reverse); Differentially‐Expressed Protein 2 (DAB2), 5′‐TAGTCCAACAGAAAGCAAAG‐3′ (forward) and 5′‐GAGGTGACTCCATTTGTTAAG‐3′ (reverse); GAPDH, 5′‐TGCTGAGTATGTCGTGGAGTCT‐3′ (forward) and 5′‐ATGCATTGCTGACAATCTTGAG‐3′ (reverse); miR‐34a, 5′‐GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAACCA‐3′ (reverse transcription primer), 5′‐CACGCATGGCAGTGTCTTAGC‐3′ (forward) and 5′‐CAGTGCAGGGTCCGAGGTA‐3′ (reverse); and U6, 5′‐GCTTCGGCAGCACATATACTAAAAT‐3′ (forward) and 5′‐CGCTTCACGAATTTGCGTGTCAT‐3′ (reverse).
Myocardial Infarction
Healthy 2‐ to 3‐week‐old male C57BL/6J mice were purchased from the Laboratory Animal Center of Southern Medical University and singly housed in a temperature‐controlled (≈22°C) environment with a 12‐hour light/dark cycle and food and water available ad libitum. The animals were habituated to the animal facilities for at least 1 week before use. Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of Southern Medical University and followed the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. MI was generated by ligating the left anterior descending (LAD) coronary artery in mice at 4 weeks. For surgical MI, mice were anesthetized with 2% isoflurane. An ALC‐V8S rodent ventilator (ALCBIO, Shanghai, China) was used to supply oxygen during the surgical procedure. The chests were opened via a horizontal incision through the muscle between the fourth and fifth intercostal space. The pericardium was then removed in mice only. The LAD coronary artery was permanently ligated with a 5‐0 silk suture (Ningbo Medical Needle Co, Ningbo, China). After surgery, the thoracic wall and skin were closed with a 5‐0 silk suture. Myocardial ischemia was confirmed by ECG ST‐segment elevation using an Animal Bio Amp (Adinstruments, NSW, Australia). After ligation, AAV9 vectors (AAV9‐NC or AAV9‐SAS) were randomly injected immediately into the myocardium bordering the infarct zone using an insulin syringe and a 30‐gauge needle. After surgery, the skin was disinfected and the animals were revived, while being maintained on a thermal insulation blanket. All animal procedures were approved by the Institutional Animal Care and Use Committee of Southern Medical University and followed the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Hearts were collected at 28 days after infarction, as described below.
Echocardiography
To evaluate cardiac function and dimensions, transthoracic 2‐dimensional electrocardiography was performed on mice anaesthetized with 2% isoflurane at 7, 14, 21, and 28 days after LAD ligation, using a Vevo 2100 Imaging System (Visual Sonics, ON, Canada) equipped with a 40‐MHz probe. M‐mode tracings in the parasternal short‐axis view were used to measure the left ventricular internal diameter at end diastole and end systole, which were used to calculate the left ventricular fractional shortening and ejection fraction.
Tissue Collection
Mice were anaesthetized with 2% isoflurane and then euthanized by an injection of 10% KCl. Their hearts and lungs were excised, briefly washed in 0.9% NaCl, weighed, and fixed in 10% formalin at room temperature. Then, the hearts were embedded in paraffin and further processed for histological or immunofluorescence analysis.
Triphenyl Tetrazolium Staining
The hearts were sliced into 4 sections and incubated in 1% triphenyl tetrazolium (Macklin, Shanghai, China) for 15 minutes at room temperature away in the dark. The staining samples were fixed in 10% neutral‐buffered formaldehyde, and slices were then photographed. The areas of infarct size were measured digitally using ImageJ software (National Institutes of Health, Bethesda, MD).
Immunofluorescence Analysis
For in vitro cultured cardiomyocytes, the culture medium was washed with PBS. Then, the cells were fixed with 4% paraformaldehyde (Leagene, Beijing, China). For terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling staining, hearts were embedded in optimal cutting temperature compound (Sakura Finetek, CA). Frozen sections of heart tissue were fixed with acetone, permeabilized with 0.2% Triton X‐100 PBS, and blocked with PBS containing 1% BSA. For tissue paraffin slices, formalin‐fixed tissue slides were deparaffinized and antigen retrieval was performed by microwaving in citrate buffer (0.1 mol/L, pH 6.0) for 14 minutes. Then, the cells or slides were incubated with primary antibodies, including cardiac troponin T (cTnT) (mouse; Abcam, Cambridge, UK), vimentin (chicken; Abcam), ki67 (rabbit; Abcam), and histone H3 phosphorylated at serine 10 (pH3) (rabbit; Abcam), for 2 hours at room temperature. This was followed by incubation with goat anti‐mouse IgG/Alexa Fluor 488, goat anti‐rabbit IgG/Alexa Fluor 594, or goat anti‐chicken IgG/Alexa Fluor 647 secondary antibodies (Biosynthesis, Beijing, China) for 1 hour at room temperature. The cells or slides were subsequently washed and incubated with Hoechst 33 342 (Bioworld, MN). Cardiomyocyte borders were defined by staining tissues with wheat germ agglutinin conjugated to Alexa Fluor 555 (Invitrogen) in PBS. Apoptotic cell death was determined by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling staining (Roche), according to the manufacturer's instructions. Image acquisition was performed with an LSM 880 confocal microscope (Zeiss, Oberkochen, Germany).
Western Blotting
Isolated cardiomyocytes or dissected mouse ventricular heart tissue samples were lysed in ice‐cold radioimmunoprecipitation assay buffer (Dingguo Changsheng, Beijing, China) with protease and phosphatase inhibitors. Protein concentrations were determined with the BCA Protein Quantitative Analysis kit (Fudebio‐tech, Hangzhou, China). Protein samples were separated by 8% to 12% SDS‐PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were incubated at room temperature for 2 hours in blocking buffer (5% BSA in Tris‐buffered saline and Tween 20 buffer). After blocking, the membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: Sirt1 (Cell Signaling Technology, MA) and β‐actin (Biosynthesis, Beijing, China). After the membranes were washed 3 times with Tris‐buffered saline and Tween 20, they were incubated with a donkey anti‐rabbit IgG horseradish peroxidase (Abcam) for 1 hour at room temperature. The membranes were developed using the enhanced chemiluminescence method, according to the manufacturer's instructions (Millipore), and detected on the chemiluminescence imager GeneGnome XRQ (Syngene, MD). To calculate the relative density, ImageJ software was used and the intensity of each protein was normalized to that of β‐actin.
RNA Fluorescence in Situ Hybridization
Isolated cardiomyocytes grown on coverslips were fixed in 4% paraformaldehyde and washed 3 times with PBS. The cells were permeabilized in 0.2% Triton X‐100 PBS, hybridized with a hybridization solution, and then incubated with a labeled Sirt1 antisense lncRNA probe overnight at 37°C. The Sirt1 antisense lncRNA probe was synthesized by Sangon. The cells were washed with 2×, 1×, and 0.5× standard saline citrate and then incubated with a mouse antidigoxin antibody conjugated with alkaline phosphatase (Sigma). Then, the cells were incubated with 4’,6‐diamidino‐2‐phenylindole (Bestbio, Shanghai, China). Image acquisition was performed with an LSM 880 confocal microscope.
Ribonuclease Protection Assay
To detect the sense‐antisense RNA duplex, ribonuclease protection assay (RPA) and real‐time qPCR analyses were performed on total RNA from isolated postnatal day 1 (P1) mouse cardiomyocytes. The sequences of oligonucleotides used for RPA were as follows: Sirt1 nonoverlapping, 5′‐CTAGGTGGTGAATATGCCAAAC‐3′ (forward) and 5′‐TCCGAAA TATGAAGAGGTGTTG‐3′ (reverse); Sirt1 overlapping, 5′‐GGTAGAGCCTGCATAGATCTT‐3′ (forward) and 5′‐ATTTATCTTATGACTTTGGGACTT‐3′ (reverse); and β‐actin, 5′‐TGCTGTCCCTGTATGCCTCTG‐3′ (forward) and 5′‐TTGATGTCACGCACGATTTCC‐3′ (reverse). Cytoplasmic RNA was orderly digested by DNaseI and RPA‐grade RNase A (Applied Biosystems, CA) to remove all genomic DNA contamination and single‐stranded RNAs. Then, cDNA synthesis and real‐time qPCR were performed, as described above.
RNA Stability Assay
To detect the stability of Sirt1 mRNA, isolated P1 mouse cardiomyocytes were treated with 2 μg/mL actinomycin D (MCE, Princeton, NJ), which suppresses transcription. The cells were harvested at 0, 2, 4, 6, 8, and 10 hours after treatment, and total RNA was extracted to detect the residual mRNAs by real‐time qPCR. GAPDH mRNA served as the internal control, because its expression is stable within 32 hours.
Statistical Analysis
All data are presented in graphs as the mean±SEM. Statistical analyses were performed using SPSS 20.0 software. For the statistical comparison of 2 groups, an unpaired, 2‐tailed, Student t test was used. For the comparison of ≥3 groups, a 1‐way ANOVA, followed by the least significance difference post hoc test, was used. Survival rate was determined using the Kaplan‐Meier method. The difference between survival curves was determined using the log‐rank (Mantel‐Cox) test. We calculated the significance of each treatment as a P value, and P<0.05 was considered statistically significant.
Results
Sirt1 Antisense lncRNA Promotes Neonatal Cardiomyocyte Proliferation
We first examined the expression patterns of Sirt1 antisense lncRNA in embryonic, neonatal, and adult mice hearts. Real‐time qPCR revealed a high level of Sirt1 antisense lncRNA expression in mouse embryonic day 16.5 cardiomyocytes and a sharp reduction in mouse P1 cardiomyocytes, which was further reduced in mouse postnatal day 28 cardiomyocytes (Figure 1A). Previous studies have identified that insulin‐like growth factor signaling is required for cardiomyocyte proliferation during cardiac development.20, 21, 22 We found that Sirt1 antisense lncRNA levels were increased in insulin‐like growth factor 1–treated cardiomyocytes (Figure S1), which suggested that cardiomyocyte proliferation may involve an increase in the endogenous Sirt1 antisense lncRNA expression. To elucidate a potential role of Sirt1 antisense lncRNA in cardiomyocyte proliferation, we transfected isolated cardiomyocytes with various MOIs of Ad‐SAS (Figure S2A), and the real‐time qPCR revealed that Sirt1 antisense lncRNA was overexpressed to 38.7±9.8‐fold and 117.0±10.6‐fold in the MOI=100 and MOI=200 groups, respectively (Figure S2B). Then, we used MOI=100 of Ad‐SAS to overexpress Sirt1 antisense lncRNA in the following overexpression experiments in vitro. We examined cardiomyocyte proliferation after Sirt1 antisense lncRNA overexpression using the cell cycle activity marker ki67 and the mitosis marker pH3. Sirt1 antisense lncRNA overexpression significantly increased the number of ki67‐positive cardiomyocytes (Figure 1B). We also observed an induction of cardiomyocyte proliferation in the Sirt1 antisense lncRNA overexpression cardiomyocytes, as quantified by the increase in pH3‐positive cardiomyocytes (Figure 1C and Figure S2C). In addition, the cardiomyocyte number was increased in Sirt1 antisense lncRNA overexpression cardiomyocytes (Figure 1D). We next investigated the role of Sirt1 antisense lncRNA in isolated postnatal day 7 (P7) cardiomyocytes. We successfully overexpressed Sirt1 antisense lncRNA 28.5±2.4‐fold in P7 cardiomyocytes (Figure 1E), which increased the percentage of ki67‐positive P7 cardiomyocytes to 2.38±0.58% (Figure 1F). Moreover, overexpression of Sirt1 antisense lncRNA increased the number of P7 cardiomyocytes (Figure 1G). Previous studies have revealed that proliferating cardiomyocytes in zebrafish and mouse regenerative models undergo dedifferentiation characterized by partial disassembly of sarcomeres.23, 24 Partially disassembled sarcomeric structures were observed in Sirt1 antisense lncRNA overexpression P7 cardiomyocytes (Figure 1H and 1I). It has been recently shown that the stem/progenitor markers RUNX1, c‐kit, and DAB2 identify dedifferentiated cardiomyocytes.24, 25 Real‐time qPCR results revealed that Sirt1 antisense lncRNA promotes RUNX1, c‐kit, and DAB2 expression in isolated P7 cardiomyocytes (Figure S2D). Sirt1 antisense lncRNA overexpression decreased P7 cardiomyocyte area and cardiomyocyte apoptosis (Figure 1H, 1J, and 1K). Collectively, these data suggested that Sirt1 antisense lncRNA regulates neonatal cardiomyocyte proliferation in vitro.
Figure 1.
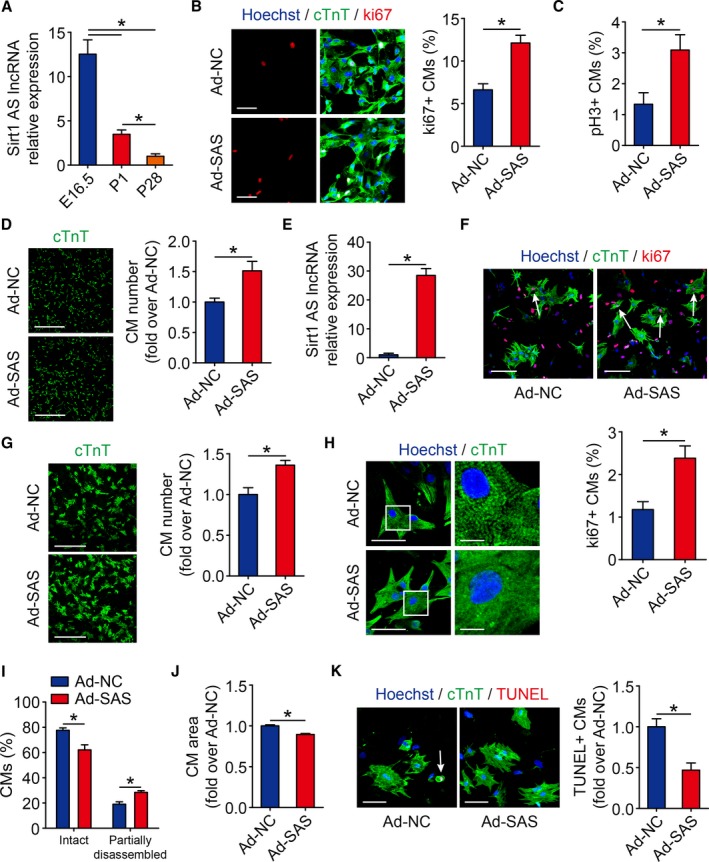
fesbSirt1 antisense (AS) long noncoding RNA (lncRNA) induces cardiomyocyte (CM) proliferation in neonatal mice. A, Real‐time quantitative PCR (qPCR) results of Sirt1 AS lncRNA between embryonic day 16.5 (E16.5), postnatal day 1 (P1), and postnatal day 18 (P28) mice hearts (n=6 mice per group). B, Isolated P1 CMs transfected with adenovirus‐control (Ad‐NC) or Ad–Sirt1 AS lncRNA (SAS) were immunostained for ki67 and quantification of ki67‐positive CMs (652 CMs from 4 mice in Ad‐NC group, 857 CMs from 4 mice in Ad‐SAS group). Bar=50 μm. C, Quantification of pH3‐positive CMs in isolated P1 CMs transfected with Ad‐NC or Ad‐SAS (440 CMs from 4 mice in Ad‐NC group, 678 CMs from 4 mice in Ad‐SAS group). Bar=30 μm. D, Immunofluorescence of cTnT and quantification of the counts of P1 CMs transfected with Ad‐NC or Ad‐SAS (n=3 mice per group). Bar=500 μm. E, Real‐time qPCR results of Sirt1 AS lncRNA level in isolated P7 CMs transfected with Ad‐NC and Ad‐SAS (n=3 mice per group). F, P7 CMs transfected with Ad‐NC or Ad‐SAS were immunostained for ki67 and quantification of ki67‐positive CMs (419 CMs from 4 mice in Ad‐NC group, 421 CMs from 4 mice in Ad‐SAS group). Ki67‐positive CMs were indicated by arrows. Bar=100 μm. G, Immunofluorescence of cTnT and quantification of the counts of P7 CMs transfected with Ad‐NC or Ad‐SAS (n=3 mice per group). Bar=500 μm. H, Immunofluorescence of cTnT and Hoechst 33 342 and quantification of the CM area and CM sarcomeric status in isolated P7 CMs transfected with Ad‐NC or Ad‐SAS. Bars: 50 μm (left panels); 10 μm (right panels). I, Quantification of CM sarcomeric status in isolated P7 CMs transfected with Ad‐NC or Ad‐SAS (713 CMs from 3 mice in Ad‐NC group, 752 CMs from 3 mice in Ad‐SAS group). J, Quantification of CM area in isolated P7 CMs transfected with Ad‐NC or Ad‐SAS (90 CMs from 3 mice in Ad‐NC group, 103 CMs from 3 mice in Ad‐SAS group). K, Cell apoptosis was detected by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) staining in Ad‐NC– and Ad‐SAS–transfected isolated P1 CMs and quantitative analysis of TUNEL‐positive CMs (727 CMs from 3 mice in Ad‐NC group, 764 CMs from 3 mice in Ad‐SAS group). TUNEL‐positive CMs were indicated by arrows. Bar=50 μm. Statistical significance was calculated using 1‐way ANOVA, followed by least significant difference post hoc test in A and 2‐tailed unpaired Student t test in B through G and I through K. Data represent mean±SEM. *P<0.05.
Sirt1 Antisense lncRNA Is Required for Neonatal Cardiomyocyte Proliferation
To further elucidate the role of Sirt1 antisense lncRNA in cardiomyocyte proliferation, we knocked down Sirt1 antisense lncRNA using LNA in P1 mice cardiomyocytes in vitro. Transfecting P1 cardiomyocytes with LNA targeting Sirt1 antisense lncRNA decreased Sirt1 antisense lncRNA expression to 42.3±10.9% (Figure 2A). The depletion of Sirt1 antisense lncRNA significantly inhibited the number of ki67‐positive P1 cardiomyocytes (Figure 2B). In addition, knockdown of Sirt1 antisense lncRNA using LNA in P1 mice decreased the number of P1 cardiomyocytes (Figure 2C). Sirt1 antisense lncRNA depletion increased cell area and apoptosis in isolated P1 cardiomyocytes (Figure 2D and 2E). Next, we depleted Sirt1 antisense lncRNA to 53.7±16.2% in isolated P7 cardiomyocytes using LNA (Figure 2F). The inhibition of Sirt1 antisense lncRNA levels also decreased the cardiomyocyte cell cycle activity, determined by assaying ki67 immunofluorescence in P7 cardiomyocytes (Figure 2G). The cardiomyocyte number was decreased in Sirt1 antisense lncRNA depletion P7 cardiomyocytes (Figure 2H). These results indicated that Sirt1 antisense lncRNA is an endogenous regulator of neonatal cardiomyocyte proliferation in vitro.
Figure 2.
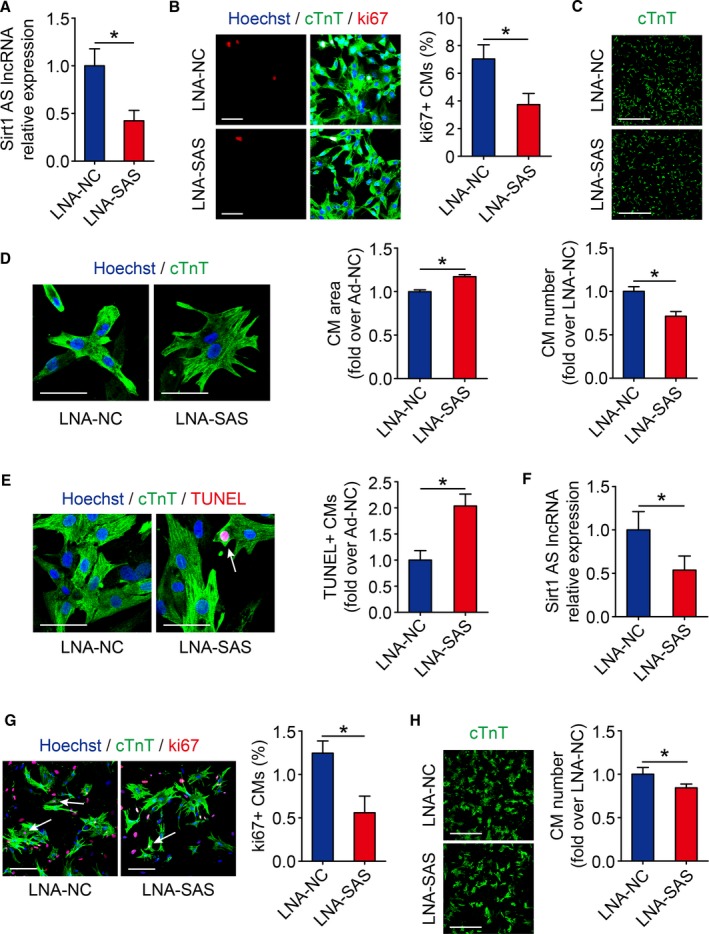
Sirt1 antisense (AS) long noncoding RNA (lncRNA) is required for cardiomyocyte (CM) proliferation in neonatal mice. A, The mRNA levels of Sirt1 AS lncRNA in isolated postnatal day 1 (P1) CMs transfected with si‐NC or si‐SAS (n=3 mice per group). B, Ki67 immunofluorescence staining in isolated P1 CMs transfected with si‐NC or si‐SAS and quantification of ki67‐positive CMs (738 CMs from 4 mice in locked nucleic acid–control [LNA‐NC] group, 699 CMs from 4 mice in LNA–Sirt1 AS lncRNA [SAS] group). Bar=50 μm. C, Immunofluorescence of cTnT and quantification of the counts of P1 CMs transfected with si‐NC or si‐SAS (n=3 mice per group). Bar=500 μm. D, Immunofluorescence of cTnT and Hoechst 33 342 and quantification of the CM area in isolated P1 CMs transfected with si‐NC or si‐SAS (105 CMs from 3 mice in LNA‐NC group, 82 CMs from 3 mice in LNA‐SAS group). Bar=50 μm. E, Cell apoptosis was detected by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) staining in si‐NC– and si‐SAS–transfected isolated P1 CMs and quantitative analysis of TUNEL‐positive CMs (771 CMs from 3 mice in LNA‐NC group, 772 CMs from 3 mice in LNA‐SAS group). TUNEL‐positive CMs were indicated by arrows. Bar=50 μm. F, Real‐time quantitative PCR results of Sirt1 AS lncRNA level in isolated postnatal day 7 (P7) CMs transfected with si‐NC and si‐SAS (n=3 mice per group). G, P7 CMs transfected with si‐NC or si‐SAS were immunostained for ki67 and quantification of ki67‐positive CMs (556 CMs from 4 mice in LNA‐NC group, 562 CMs from 4 mice in LNA‐SAS group). Ki67‐positive CMs were indicated by arrows. Bar=100 μm. H, Immunofluorescence of cTnT and quantification of the counts of P7 CMs transfected with si‐NC or si‐SAS (n=3 mice per group). Bar=500 μm. Statistical significance was calculated using 2‐tailed unpaired Student t test in A through H. Data represent mean±SEM. Ad‐NC indicates adenovirus‐control. *P<0.05.
Depletion of Sirt1 Antisense lncRNA Inhibits Cardiomyocyte Proliferation in Vivo
Next, we detected whether Sirt1 antisense lncRNA is required for cardiomyocyte proliferation in vivo. At 3 days after the intracardial injection of LNA targeting Sirt1 antisense lncRNA, Sirt1 antisense lncRNA expression was decreased to 51.6±12.4% in neonatal mouse hearts (Figure 3A), which had no significant effect on heart/body weight ratio (Figure 3B). The cardiomyocyte size was increased in Sirt1 antisense lncRNA depletion hearts (Figure 3C), and the inhibition of Sirt1 antisense lncRNA levels also decreased the cardiomyocyte cell cycle activity, as determined by assaying ki67 and pH3 immunofluorescence in vivo (Figure 3D and 3E). These results indicated that Sirt1 antisense lncRNA is required for neonatal cardiomyocyte proliferation in vivo.
Figure 3.
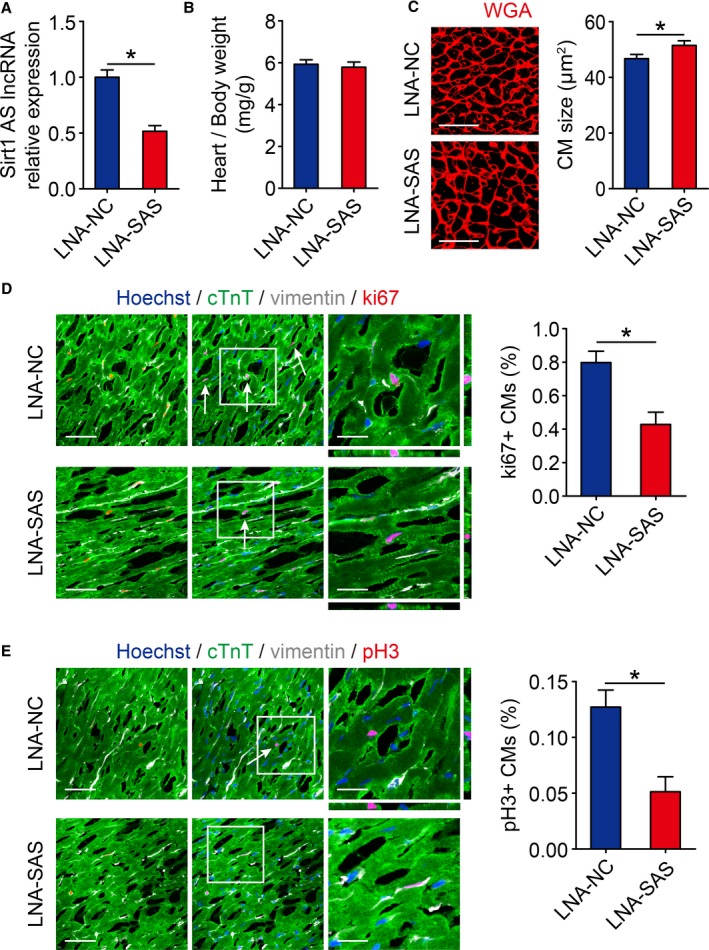
Inhibition of Sirt1 antisense (AS) long noncoding RNA (lncRNA) suppresses cardiomyocyte (CM) proliferation in vivo. A, Real‐time quantitative PCR analyses of Sirt1 AS lncRNA levels in locked nucleic acid–control (LNA‐NC) or LNA–Sirt1 AS lncRNA (SAS) injected neonatal mouse hearts 10 days after injection (n=6 mice per group). B, Ratios of heart weight/body weight in neonatal mouse hearts injected with LNA‐NC or LNA‐SAS (n=5 mice per group). C, Wheat germ agglutinin (WGA) staining of heart sagittal sections for LNA‐NC and LNA‐SAS adult mice hearts and quantification of CM size (167 CMs from 5 mice in LNA‐NC group, 167 CMs from 5 mice in LNA‐SAS group). Bar=25 μm. D, Immunofluorescence of ki67 in neonatal hearts injected with LNA‐NC or LNA‐SAS and quantification of ki67‐positive CMs (1627 CMs from 5 mice in LNA‐NC group, 1671 CMs from 5 mice in LNA‐SAS group). Ki67‐positive CMs were indicated by arrows. Bars: 50 μm (left panels); 20 μm (right panels). E, Immunofluorescence of pH3 in neonatal hearts injected with LNA‐NC or LNA‐SAS and quantification of pH3‐positive CMs (6244 CMs from 5 mice in LNA‐NC group, 7416 CMs from 5 mice in LNA‐SAS group). pH3‐positive CMs were indicated by arrows. Bars: 50 μm (left panels); 20 μm (right panels). Statistical significance was calculated using 2‐tailed unpaired Student t test in A through E. Data represent mean±SEM. *P<0.05.
Sirt1 Antisense lncRNA Induces Cardiomyocyte Proliferation in Adult Mouse Hearts
To investigate whether Sirt1 antisense lncRNA promotes cardiomyocyte proliferation in adult mice, we induced Sirt1 antisense lncRNA overexpression in adult mouse hearts by the intracardial injection of AAV9 vectors (Figure 4A). The heart/body weight ratios of the Sirt1 antisense lncRNA overexpression mice were not significantly different from those of the AAV‐NC group hearts (Figure 4B). The cardiomyocyte size was not altered in Sirt1 antisense lncRNA overexpression hearts (Figure 4C). Increased cell proliferation in Sirt1 antisense lncRNA overexpression hearts was further confirmed by ki67 immunofluorescence and quantification (Figure 4D). The mitosis marker pH3 was also enhanced in Sirt1 antisense lncRNA overexpression hearts (Figure 4E). These data showed that Sirt1 antisense lncRNA significantly increases cardiomyocyte proliferation in adult mice.
Figure 4.
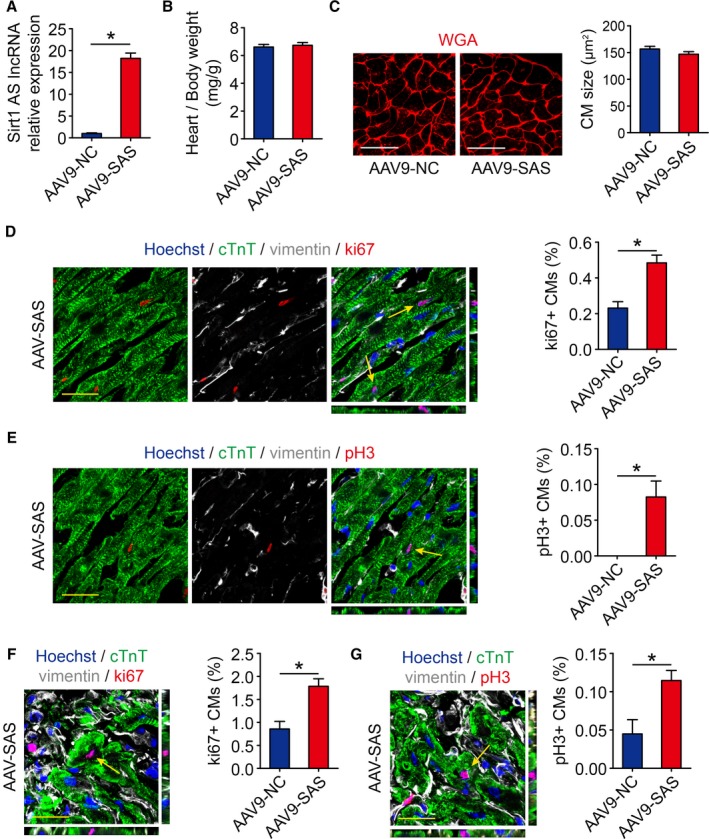
Sirt1 antisense (AS) long noncoding RNA (lncRNA) drives cardiomyocyte (CM) proliferation in adults. A, Real‐time quantitative PCR analyses of Sirt1 AS lncRNA levels in adeno‐associated virus 9–negative control (AAV9‐NC) or AAV9–Sirt1 AS lncRNA (SAS) injected adult mice hearts 28 days after injection (n=6 mice per group). B, Ratios of heart weight/body weight in neonatal mouse hearts injected with AAV9‐NC or AAV9‐SAS (n=5 mice per group). C, Wheat germ agglutinin (WGA) staining of heart sagittal sections for AAV9‐NC and AAV9‐SAS adult mice hearts and quantification of CM size (175 CMs from 5 mice in AAV9‐NC group, 175 CMs from 5 mice in AAV9‐SAS group). Bar=25 μm. D, Ki67 immunofluorescence staining in AAV9‐NC and AAV9‐SAS adult mice hearts and quantification of ki67‐positive CMs (2591 CMs from 5 mice in AAV9‐NC group, 2680 CMs from 5 mice in AAV9‐SAS group). Ki67‐positive CMs were indicated by arrows. Bar=20 μm. E, pH3 immunofluorescence staining in AAV9‐NC and AAV9‐SAS adult mice hearts and quantification of pH3‐positive CMs (5714 CMs from 5 mice in AAV9‐NC group, 5870 CMs from 5 mice in AAV9‐SAS group). pH3‐positive CMs were indicated by arrows. Bar=20 μm. F, Ki67 immunofluorescence staining in AAV9‐NC and AAV9‐SAS adult mice hearts 28 days after myocardial infarction (MI) and quantification of ki67‐positive CMs (2690 CMs from 5 mice in AAV9‐NC group, 2695 CMs from 5 mice in AAV9‐SAS group). Ki67‐positive CMs were indicated by arrows. Bar=20 μm. G, pH3 immunofluorescence staining in AAV9‐NC and AAV9‐SAS adult mice hearts 28 days after MI and quantification of pH3‐positive CMs (6778 CMs from 5 mice in AAV9‐NC group, 6858 CMs from 5 mice in AAV9‐SAS group). pH3‐positive CMs were indicated by arrows. Bar=20 μm. Statistical significance was calculated using 2‐tailed unpaired Student t test in A through G. Data represent mean±SEM. *P<0.05.
Overexpression of Sirt1 Antisense lncRNA Improves Cardiac Function in Adult Mouse Hearts After MI
To test whether Sirt1 antisense lncRNA is involved in the regulation of cardiac repair in response to ischemic injury, an MI model was established. We performed LAD permanent ligation on 4‐week‐old mice (Figure S3A), and ST‐segment elevation was observed via ECG when LAD was ligated (Figure S3B). We then injected AAV9 vectors expressing Sirt1 antisense lncRNA or a control vector into the peri‐infarcted area. At 4 weeks after MI, we noted a significantly higher cumulative survival rate in the AAV9‐SAS group than in the AAV9‐NC group (P=0.0334, Figure 5A). A ki67 immunofluorescence assay showed that the ki67 signal in cardiomyocytes of the border zones of Sirt1 antisense lncRNA overexpression hearts was markedly increased (Figure 4F). A significant number of pH3‐positive cardiomyocytes were detected in the infarct border zone of Sirt1 antisense lncRNA overexpression hearts (Figure 4G). To evaluate left ventricular function, transthoracic 2‐dimensional echocardiography was performed on mice at 7, 14, 21, and 28 days after MI (Figure 5A and Figure S3A). As evaluated by echocardiography, significantly less ventricular chamber dilation, both in diastole and systole, was observed in Sirt1 antisense lncRNA overexpression hearts (Figure 5B and 5C). The preserved chamber dimensions of Sirt1 antisense lncRNA overexpression hearts were associated with significantly better left ventricular function, as reflected by preserved fractional shortening and ejection fractions (Figure 5D and 5E). Analysis of triphenyl tetrazolium staining and Masson's trichrome staining clearly showed that the infarct size was significantly reduced in Sirt1 antisense lncRNA overexpression hearts (Figure 5F). Overexpression of Sirt1 antisense lncRNA inhibited cardiomyocyte apoptosis after MI, as determined by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling assays (Figure 5G). Sirt1 antisense lncRNA also decreased the cardiomyocyte size after MI (Figure 5H). These results suggested that Sirt1 antisense lncRNA upregulation contributes to endogenous regeneration in adult hearts after ischemic injury.
Figure 5.
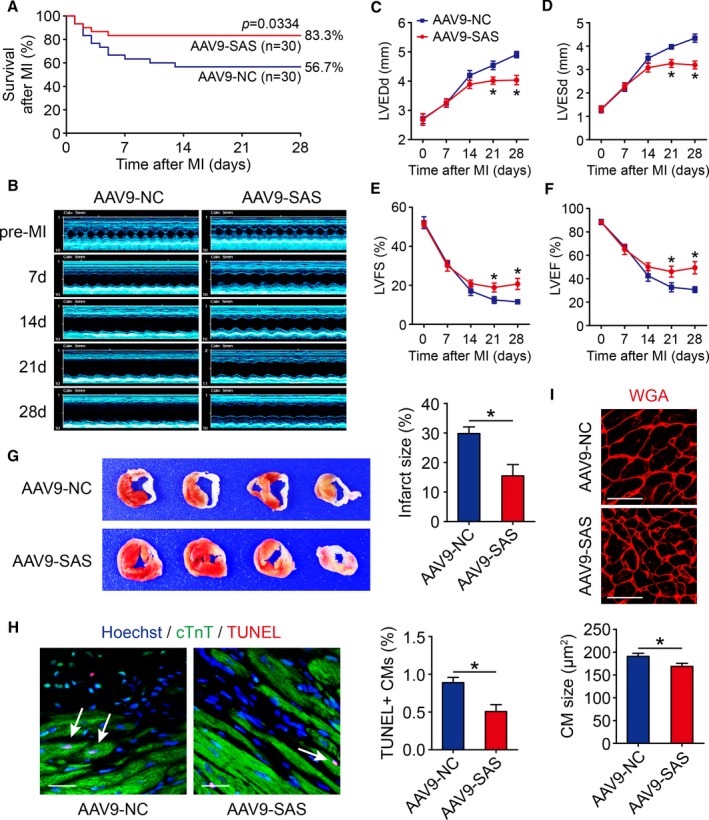
Sirt1 antisense (AS) long noncoding RNA (lncRNA) induces cardiac regeneration after myocardial infarction (MI) in adult mice. A, Kaplan‐Meier survival curves in adeno‐associated virus 9–negative control (AAV9‐NC) and AAV9–Sirt1 AS lncRNA (SAS) adult mice after MI (n=30 mice per group). B through F, The M‐mode ultrasonic cardiography change of mice hearts detected by echocardiography in AAV9‐NC and AAV9‐SAS adult mice at pre‐MI and 7, 14, 21, and 28 days after MI and quantitative analyses of left ventricular internal diameter at end diastole (LVEDd; C), left ventricular internal diameter at end systole (LVESd; D), left ventricular fractional shortening (LVFS; E), and left ventricular ejection fraction (LVEF; F) (n=5 mice per group). G, Triphenyl tetrazolium (TTC) staining of mice ventricular cross sections in AAV9‐NC and AAV9‐SAS adult mice 28 days after MI and quantitative analyses of TTC staining infarct size (n=5 mice per group). H, Cell apoptosis was detected by terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) staining in AAV9‐NC and AAV9‐SAS adult mice 28 days after MI and quantitative analyses of TUNEL‐positive cardiomyocytes (CMs; 1343 CMs from 5 mice in AAV9‐NC group, 1387 CMs from 5 mice in AAV9‐SAS group). TUNEL‐positive CMs were indicated by arrows. Bar=20 μm. I, Wheat germ agglutinin (WGA) staining in AAV9‐NC and AAV9‐SAS adult mice 28 days after MI and quantitative analyses of CM size (139 CMs from 5 mice in AAV9‐NC group, 139 CMs from 5 mice in AAV9‐SAS group). Bar=25 μm. Statistical significance was calculated using log‐rank (Mantel‐Cox) test in A, 1‐way ANOVA followed by least significant difference post hoc test in C through F, and 2‐tailed unpaired Student t test in G through I. Data represent mean±SEM. *P<0.05.
Sirt1 Antisense lncRNA Increases the Stability of Sirt1 mRNA
Because Sirt1 antisense lncRNA transcribed from and overlapped with the Sirt1 mRNA 3′‐UTR, we next investigated the possible relationship between Sirt1 antisense lncRNA and Sirt1 in cardiomyocytes. The expression of Sirt1 mRNA was significantly decreased during heart development and was consistent with the expression patterns of Sirt1 antisense lncRNA (Figure 6A). Sirt1 antisense lncRNA knockdown decreased the Sirt1 mRNA and protein levels (Figure 6B and 6C). We previously demonstrated that diverse Sirt1 overexpression could have different effects on cardiomyocyte proliferation; thus, we detected how Sirt1 changes when we overexpressed Sirt1 antisense lncRNA. We found that Sirt1 protein level was significantly increased when MOI=50 of Ad‐SAS, and maintained 2.81‐fold to ≈3.25‐fold when MOI of Ad‐SAS increased to 100 to 300 (Figure 6D and 6E). Moreover, Sirt1 antisense lncRNA promoted Sirt1 mRNA and protein levels in MI mouse hearts (Figure S4A and Figure 6F). To further elucidate the functional mechanisms, we examined the cellular location of Sirt1 antisense lncRNA in cardiomyocytes. Real‐time qPCR revealed that Sirt1 antisense lncRNA was mainly located in the cytoplasm of cardiomyocytes (Figure 6G). Moreover, Sirt1 antisense lncRNA was shown to locate in the cytoplasm of cardiomyocytes using the fluorescence in situ hybridization assay (Figure 6H). We used an RPA on RNA from isolated cardiomyocytes to test the possibility of RNA duplex formation. Real‐time qPCR data showed that the overlapping part of both transcripts was protected from degradation (Figure 6I and 6J), indicating that Sirt1 antisense lncRNA and Sirt1 mRNA form an RNA duplex. Then, isolated cardiomyocytes were treated with actinomycin D to suppress transcription, and the loss of Sirt1 mRNA expression was measured. Sirt1 antisense lncRNA elongated the half‐life of Sirt1 mRNA (Figure 6K), suggesting that Sirt1 antisense lncRNA is required for the regulation of Sirt1 mRNA stability. These results demonstrated that Sirt1 antisense lncRNA increases the stability of Sirt1 mRNA and upregulates Sirt1 expression.
Figure 6.
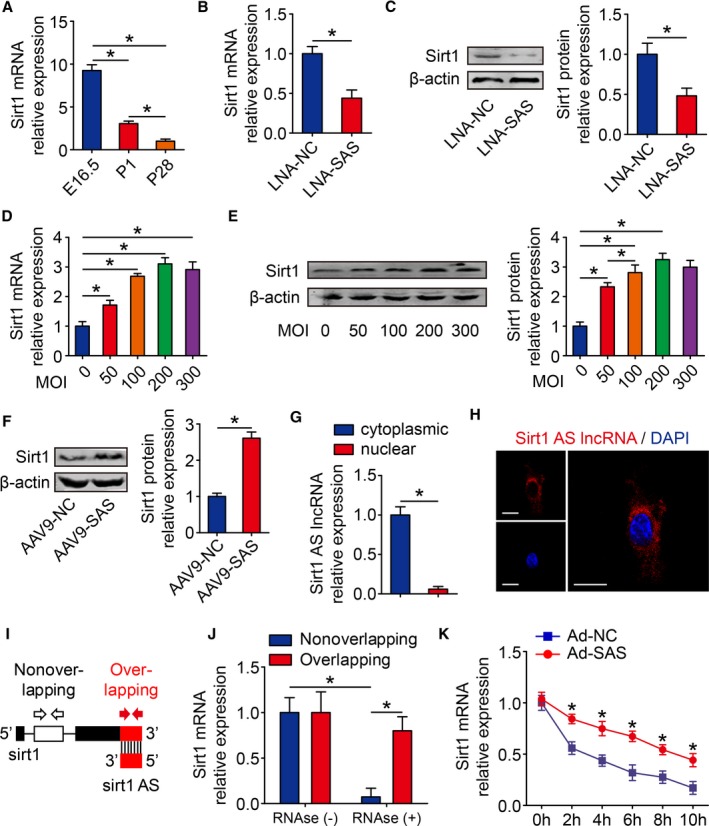
Sirt1 antisense (AS) long noncoding RNA (lncRNA) increases stability of Sirt1 mRNA. A, Real‐time quantitative PCR (qPCR) analysis of Sirt1 mRNA levels in embryonic day 16.5 (E16.5), postnatal day 1 (P1), and postnatal day 28 (P28) mouse hearts (n=6 mice per group). B, Real‐time qPCR analysis of Sirt1 mRNA levels in isolated cardiomyocytes (CMs) transfected with locked nucleic acid–control (LNA‐NC) or LNA–Sirt1 AS lncRNA (SAS) (n=3 mice per group). C, Western blotting analysis and quantitative analyses of Sirt1 protein levels in isolated CMs transfected with LNA‐NC or LNA‐SAS. β‐Actin was used as a loading control (n=3 mice per group). D, Real‐time qPCR analysis of Sirt1 mRNA levels in isolated CMs transfected with various multiplicities of infection (MOIs) of adenovirus (Ad)–SAS (n=3 mice per group). E, Western blotting analysis and quantitative analyses of Sirt1 protein levels in isolated CMs transfected with various MOIs of Ad‐SAS. β‐Actin was used as a loading control (n=3 mice per group). F, Western blotting analysis and quantitative analyses of Sirt1 protein levels in adeno‐associated virus 9 (AAV9)–NC– or AAV9‐SAS–injected adult mice hearts 28 days after injection. β‐Actin was used as a loading control (n=6 mice per group). G, Real‐time qPCR of cytoplasmic and nuclear fractions to detect Sirt1 AS lncRNA expression in isolated CMs (n=3 mice per group). H, RNA fluorescence in situ hybridization assay of Sirt1 AS lncRNA immunofluorescence detection in isolated CMs. Bar=20 μm. I, Nonoverlapping and overlapping primer positions for ribonuclease protection assay (RPA). J, RPA performed on RNA samples from isolated CMs (n=3 mice per group). K, RNA stability assay of Sirt1 mRNA expression in isolated CMs transfected with Ad‐NC or Ad‐SAS (n=3 mice per group). Statistical significance was calculated using 1‐way ANOVA followed by least significant difference post hoc test in A, D, E, J, and K, and 2‐tailed unpaired Student t test in B, C, F, and G. Data represent mean±SEM. DAPI indicates 4’,6‐diamidino‐2‐phenylindole. *P<0.05.
Sirt1 Is Involved in Sirt1 Antisense lncRNA‐Induced Cardiomyocyte Proliferation
A previous study revealed that Sirt1 promotes cardiomyocyte proliferation. We also detected whether the depletion of Sirt1 in isolated cardiomyocytes inhibits their proliferation. An siRNA targeting Sirt1 was transfected to isolated cardiomyocytes and inhibited ki67‐positive cardiomyocytes to 3.26±0.79% compared with cardiomyocytes transfected with small‐interfering‐negative control (si‐NC) (Figure 7A). To explore whether Sirt1 antisense lncRNA promotes cardiomyocyte proliferation via inducing Sirt1 expression, we cotransfected Sirt1 antisense lncRNA and small‐interfering‐Sirt1 (si‐Sirt1) in isolated cardiomyocytes. Western blot analysis results showed that Sirt1 protein expression was increased 3.06±0.38‐fold in Sirt1 antisense lncRNA overexpression cardiomyocytes and decreased to 1.24±0.22‐fold in cardiomyocytes cotransfected with Sirt1 antisense lncRNA and si‐Sirt1 (Figure 7B). We also used the ki67 immunofluorescence to verify the coeffects of Sirt1 antisense lncRNA and si‐Sirt1 on cardiomyocyte proliferation. The overexpression of Sirt1 antisense lncRNA increased the number of ki67‐positive cardiomyocytes, and this effect was decreased in cardiomyocytes cotransfected with Sirt1 antisense lncRNA and si‐Sirt1 (Figure 7C). To further clarify whether Sirt1 antisense lncRNA promotes cardiomyocyte proliferation through Sirt1 in vivo, we injected the adenovirus mixture of Sirt1 antisense lncRNA and si‐Sirt1 into neonatal mouse hearts. Overexpression of Sirt1 antisense lncRNA significantly promoted Sirt1 protein levels, and this effect was significantly attenuated by transfecting with Ad‐SAS and Ad‐si‐Sirt1 (Figure 7D). The ki67‐positive cardiomyocytes were increased to 1.62±0.17% in the Ad‐SAS group, and this number was decreased to 0.78±0.18% in the Ad‐SAS+Ad‐si‐Sirt1 group (Figure 7E). These results showed that Sirt1 is involved in Sirt1 antisense lncRNA‐induced cardiomyocyte proliferation.
Figure 7.
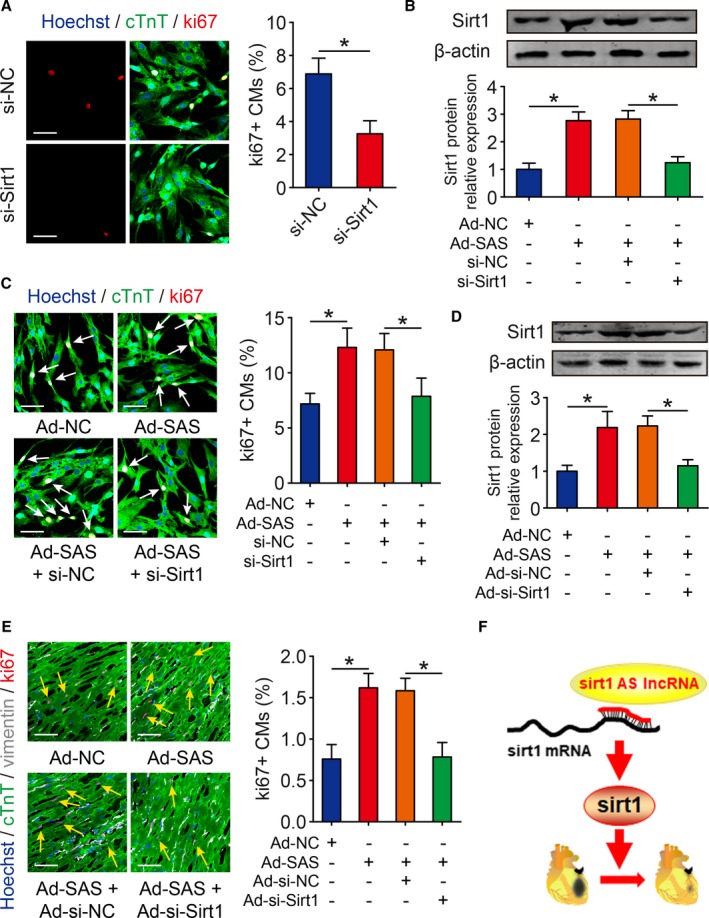
Sirt1 is involved in Sirt1 antisense (AS) long noncoding RNA (lncRNA)–regulated cardiomyocyte (CM) proliferation. A, Ki67 immunofluorescence staining in isolated postnatal day 1 CMs transfected with si‐NC or si‐Sirt1 and quantification of ki67‐positive CMs (349 CMs from 4 mice in si‐NC group, 308 CMs from 4 mice in si‐Sirt1 group). Bar=50 μm. B, The protein levels of Sirt1 in isolated CMs transfected with adenovirus‐control (Ad‐NC), Ad–Sirt1 AS lncRNA (SAS), Ad‐SAS+si‐NC, or Ad‐SAS+si‐Sirt1 and quantitative analyses of protein intensity. β‐Actin was used as a loading control (n=3 mice per group). C, Ki67 immunofluorescence staining in CMs transfected with Ad‐NC, Ad‐SAS, Ad‐SAS+si‐NC, or Ad‐SAS+si‐Sirt1 and quantification of ki67‐positive CMs (391 CMs from 4 mice in Ad‐NC group, 399 CMs from 4 mice in Ad‐SAS group, 413 CMs from 4 mice in Ad‐SAS+si‐NC group, 347 CMs from 4 mice in Ad‐SAS+si‐Sirt1 group). Ki67‐positive CMs were indicated by arrows. Bar=50 μm. D, Western blotting analysis and quantitative analysis of Sirt1 protein levels in neonatal hearts injected with Ad‐NC, Ad‐SAS, Ad‐SAS+Ad‐si‐NC, or Ad‐SAS+Ad‐si‐Sirt1. β‐Actin was used as a loading control (n=5 mice per group). E, Immunofluorescence of ki67 in neonatal hearts injected with Ad‐NC, Ad‐SAS, Ad‐SAS+Ad‐si‐NC, or Ad‐SAS+Ad‐si‐Sirt1 and quantification of ki67‐positive CMs (1722 CMs from 5 mice in Ad‐NC group, 1733 CMs from 5 mice in Ad‐SAS group, 1769 CMs from 5 mice in Ad‐SAS+Ad‐si‐NC group, 1666 CMs from 5 mice in Ad‐SAS+Ad‐si‐Sirt1 group). Ki67‐positive CMs were indicated by arrows. Bar=50 μm. F, The schematic model of Sirt1 AS lncRNA interacted and stabilized Sirt1, thereby regulating CM proliferation and cardiac regeneration after myocardial infarction. Statistical significance was calculated using 2‐tailed unpaired Student t test in A and 1‐way ANOVA followed by least significant difference post hoc test in B through E. Data represent mean±SEM. *P<0.05.
Discussion
In the present study, we demonstrated that Sirt1 antisense lncRNA drives cardiomyocyte proliferation and cardiac regeneration after MI. Sirt1 antisense lncRNA was a positive regulator of cardiomyocyte proliferation and contributed to protecting cardiac function, attenuating cardiac remodeling after MI. Our data further showed that Sirt1 antisense lncRNA directly bound Sirt1 mRNA 3′‐UTR, increased the stability of Sirt1 mRNA, and upregulated Sirt1 mRNA and protein expression; Sirt1 mediated cardiomyocyte proliferation induced by Sirt1 antisense lncRNA (Figure 7F). More important, Sirt1 antisense lncRNA is potentially more conducive to the precise regulation of Sirt1, consequently overcoming the barrier of the bidirectional effect of Sirt1 on the regulation of cardiomyocyte proliferation, which might provide a potential target for treating MI.
Previous studies have revealed that antisense lncRNAs play as critical regulators in cardiovascular diseases, including heart development,14 cardiac hypertrophy,26 and coronary atherosclerosis.27 Herein, we extended the role of antisense lncRNA into cardiomyocyte proliferation by reporting that Sirt1 antisense lncRNA can positively regulate cardiomyocyte proliferation in vitro and in vivo. We examined cardiomyocyte proliferation using ki67 and pH3 immunofluorescence, which are classic markers of cell cycle activity and mitosis, respectively, used in previous studies.24, 28 More important, we used diverse primary antibodies to distinguish cardiomyocytes and cardiac fibroblasts in our in vivo data, performed high magnification (×100) using confocal microscopy, and provided high‐magnification z‐stack images, which place more emphasis on clearly demonstrating that the induction of cardiomyocyte proliferation in vivo is true. Our data suggested that Sirt1 antisense lncRNA overexpression increased the number of ki67‐ and pH3‐positive cardiomyocytes, and led to cardiomyocyte dedifferentiation characterized by sarcomere disassembly and increased expression of cardiomyocyte dedifferentiation markers. In contrast, suppression of Sirt1 antisense lncRNA inhibited cardiomyocyte proliferation. Moreover, Sirt1 antisense lncRNA overexpression significantly regenerated cardiomyocytes, reduced the fibrotic area, inhibited cardiomyocyte apoptosis, improved left ventricular function, and decreased mortality rate after MI. Previous studies have shown that Sirt1 antisense lncRNA can promote cell proliferation in myoblasts17 and endothelial progenitor cells18 and upregulate the expression of cell cycle genes, such as CCNB, CCND, and CCNE, in C2C12 cells.17 Herein, we confirmed that the induction of Sirt1 antisense lncRNA expression accounts for cardiomyocyte proliferation and protecting hearts from MI injury.
We subsequently showed that Sirt1 antisense lncRNA bound to Sirt1 mRNA 3′‐UTR and formed the antisense lncRNA/mRNA duplex, thereby increasing Sirt1 mRNA and protein levels. Sirt1 antisense lncRNA was located more in the cytoplasm than in the nucleus, suggesting that Sirt1 antisense lncRNA is mostly involved in mRNA stability at the epigenetic level. RPA assays further confirmed that the Sirt1 antisense‐sense pair prevented Sirt1 mRNA from RNase degradation. In addition, the RNA stability assay indicated that Sirt1 antisense lncRNA extended the Sirt1 mRNA half‐life. Actually, antisense lncRNA and its sense mRNA transcripts can hybridize and form an lncRNA/mRNA duplex because of their ability to base pair, and this hybrid complex in the cytoplasm can further alter mRNA stability and modulate translation.19, 29 Consistent with our results, previous researchers have also shown that antisense lncRNAs, such as Beta‐Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1)‐antisense, Type II Mesothelial Keratin K7 (KRT7)‐antisense, and Fibroblast Growth Factor Receptor 3 (FGFR3)–antisense 1, interact with the sense target genes BACE1, KRT7, and FGFR3, respectively, to exert their functions.30, 31, 32 In this study, we identified that Sirt1 antisense lncRNA stabilized Sirt1 to increase its expression at both the mRNA and protein levels in cardiomyocytes. Furthermore, the antisense lncRNA/mRNA duplex may also cover microRNA‐binding sites, thus prohibiting the inhibitory role of microRNAs.19, 33 Previous studies have found that the miR‐34a target sequence on the Sirt1 mRNA 3′‐UTR overlaps with the corresponding region of Sirt1 antisense lncRNA on the Sirt1 mRNA 3′‐UTR, and Sirt1 antisense lncRNA increases the stability of Sirt1 mRNA by competing with miR‐34a.17 Actually, miR‐34a was confirmed to regulate cardiomyocyte proliferation and cardiac regeneration after MI.34 Thus, we suspected that Sirt1 antisense lncRNA may also block the miR‐34–binding site on Sirt1 mRNA 3′‐UTR in cardiomyocytes, despite the fact that Sirt1 antisense lncRNA may have no effect on miR‐34a expression in cardiomyocytes (Figure S4B). Collectively, our mechanistic findings indicated that Sirt1 antisense lncRNA stabilizes Sirt1 mRNA by forming an lncRNA/mRNA duplex, which subsequently upregulates Sirt1 overexpression in cardiomyocytes.
Interestingly, Sirt1 antisense lncRNA was found to overcome the Sirt1 bidirectional regulation problem in cardiomyocyte proliferation. In a previous study, low (2.5‐fold) to moderate (7.5‐fold) overexpression of Sirt1 in the heart could protect against oxidative stress, attenuate cardiomyocyte apoptosis, and attenuate the appearance of age‐related cardiomyopathy involving hypertrophy. In contrast, high levels (12.5‐fold) of Sirt1 increased oxidative stress and induced cardiomyopathy in the heart.35 We previously reported that low Sirt1 overexpression (2.1‐fold to ~2.8‐fold) may promote cardiomyocyte proliferation. On the other hand, high Sirt1 overexpression (~25‐fold) may have no proliferative effect in cardiomyocytes. The reason may be that low overexpression of Sirt1 reduces reactive oxygen species production and then promotes cardiomyocyte proliferation, whereas high overexpression of Sirt1 decreases nicotinamide‐adenine dinucleotide, oxidized form, levels and induces reactive oxygen species production in cardiomyocytes. In the present study, we overexpressed Sirt1 antisense lncRNA using various MOIs of adenovirus in cardiomyocytes. We found that, although Sirt1 antisense lncRNA was overexpressed up to 160‐fold, Sirt1 protein levels were increased barely no more than 3.2‐fold in cardiomyocytes. In a previous study, Sirt1 antisense lncRNA was overexpressed ≈200‐fold, and the Sirt1 protein levels were increased 1.5‐fold in myoblast cells.17 These could be explained by another regulator, such as miR‐34a or miR‐449a, that could also regulate Sirt1 expression in cardiomyocytes. Considering the interesting data, Sirt1 antisense lncRNA could potentially constitute a therapeutic target that is well suited to modulate Sirt1 expression, thus facilitating cardiomyocyte proliferation.
Our experimental study did have the following limitations. First, the intramyocardial injection of virus vectors was validated as an efficient gene delivery tool in vivo.36, 37 Of course, gene editing, like CRISP/Cas9, and transgenic technology may also be the good methods. Second, although we identified that Sirt1 antisense lncRNA plays a role in cardiomyocyte proliferation by regulating the stability of Sirt1, 2 questions remain: (1) Is Sirt1 involved in other Sirt1 antisense lncRNA‐regulated physiological and pathological cardiomyocyte processes, such as cardiomyocyte apoptosis and cardiomyocyte hypertrophy? (2) Do other regulators, such as splicing factors, transcription factors, and microRNAs, participate in the binding process between Sirt1 antisense lncRNA and Sirt1 mRNA? These questions need to be addressed in future studies.
In conclusion, our findings characterized a long noncoding antisense transcript for Sirt1, Sirt1 antisense lncRNA, which functions as a regulator of Sirt1 gene expression and further affects cardiomyocyte proliferation. Specifically, Sirt1 antisense lncRNA exerts these functions by forming an lncRNA‐mRNA duplex with Sirt1 mRNA, enhancing the stability of Sirt1 mRNA and increasing Sirt1 protein levels. Our findings suggested that Sirt1 antisense lncRNA may be an effective gene target for treating MI.
Sources of Funding
This work was supported by grants to Bin from the National Natural Science Foundation of China (No. 81771857, No. 81571698, and No. 81271640).
Disclosures
None.
Supporting information
Figure S1. Expression of Sirt1 AS lncRNA in PBS‐ or IGF‐1‐treated CMs. Real‐time qRCR analyses of Sirt1 AS lncRNA levels in isolated P1 CMs treated with PBS or IGF‐1 (n=3 mice per group). Statistical significance was calculated using two‐tailed unpaired Student t test. *P<0.05; error bars represent mean±SEM.
Figure S2. Sirt1 AS lncRNA promotes CM proliferation and dedifferentiation in vitro. A, Infection efficiency of various MOI of Ad‐SAS transfected into isolated P1 CMs. Scale bar, 500 μm. B, Real‐time qPCR analysis of Sirt1 AS lncRNA levels in isolated P1 CMs transfected with various MOI of Ad‐SAS (n=3 mice per group). C, CMs in prophase, prometaphase, metaphase, anaphase and cytokinesis in the Sirt1 AS lncRNA overexpression group immunostained for pH3. Scale bar, 30 μm, (D) Isolated P7 CMs were transfected with Ad‐NC or Ad‐SAS. mRNA expression levels of CM dedifferentiated markers (RUNX1, c‐kit, DAB2) were detected by Real‐time qPCR (n=3 mice per group). Statistical significance was calculated using one‐way ANOVA followed by LSD post hoc test in B, and two‐tailed unpaired Student t test in D. *P<0.05, error bars represent mean±SEM.
Figure S3. Schematic of the MI experiments in adult mice. A, Schematic of in vivo experiments in adult mice with MI surgery. Echo, echocardiography, IF, immunofluorescence. B, Elevation of ST‐segment from ECG in MI model.
Figure S4. Sirt1 AS lncRNA induces Sirt1 mRNA levels in MI hearts and has no effect on miR‐34a expression in CMs. A, Real‐time qRCR analyses of Sirt1 mRNA levels in AAV9‐NC‐ or AAV9‐SAS‐injected adult mice hearts 28 days after injection (n=6 mice per group). B, Real‐time qRCR analyses of miR‐34a levels in isolated P1 CMs transfected with Ad‐NC or Ad‐SAS (n=3 mice per group). Statistical significance was calculated using 2‐tailed unpaired Student t test in (A and B). *P<0.05; error bars represent mean±SEM.
(J Am Heart Assoc. 2018;7:e009700 DOI: 10.1161/JAHA.118.009700.)
Contributor Information
Zhonghua Teng, Email: tengzhh@126.com.
Jianping Bin, Email: jianpingbin@126.com, Email: jianpingbin@hotmail.com.
References
- 1. Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, Hua B, Xu J, Li J, Bi C, Guo S, Yang F, Han Z, Li Y, Yan G, Yu Y, Bao Z, Yu M, Li F, Tian Y, Pan Z, Yang B. The long noncoding rna carel controls cardiac regeneration. J Am Coll Cardiol. 2018;72:534‐550. [DOI] [PubMed] [Google Scholar]
- 2. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia‐induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR‐539‐dependent PHB2 downregulation. Nat Commun. 2014;5:3596. [DOI] [PubMed] [Google Scholar]
- 3. Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR‐489. Circ Res. 2014;114:1377–1388. [DOI] [PubMed] [Google Scholar]
- 4. Long QQ, Wang H, Gao W, Fan Y, Li YF, Ma Y, Yang Y, Shi HJ, ChenW BR, Meng HY, Wang QM, Wang F, Wang ZM, Wang LS. Long noncoding RNA Kcna2 antisense RNA contributes to ventricular arrhythmias via silencing Kcna2 in rats with congestive heart failure. J Am Heart Assoc. 2017;6:e005965 DOI: 10.1161/JAHA.117.005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y, Jiao L, Sun L, Li Y, Gao Y, Xu C, Shao Y, Li M, Li C, Lu Y, Pan Z, Xuan L, Zhang Y, Li Q, Yang R, Zhuang Y, Zhang Y, Yang B. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca(2+) overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res. 2018;122:1354–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui‐Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. [DOI] [PubMed] [Google Scholar]
- 8. Ling MH, Ban Y, Wen H, Wang SM, Ge SX. Conserved expression of natural antisense transcripts in mammals. BMC Genom. 2013;14:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark MB, Johnston RL, Inostroza‐Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome‐wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, Dong S, Wen Z, Rao J, Liao W, Shi M. The lncRNA MACC1‐AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mrna stability of MACC1. Mol Cancer. 2018;17:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Su W. Long noncoding RNA ZEB1‐AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Cell Death Dis. 2017;16:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, Wu X, Yan D. Long non‐coding RNA HNF1A‐AS1 mediated repression of miR‐34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. [DOI] [PubMed] [Google Scholar]
- 13. Babajko S, Petit S, Fernandes I, Meary F, LeBihan J, Pibouin L, Berdal A. Msx1 expression regulation by its own antisense RNA: consequence on tooth development and bone regeneration. Cells Tissues Organs. 2009;189:115–121. [DOI] [PubMed] [Google Scholar]
- 14. Korostowski L, Sedlak N, Engel N. The Kcnq1ot1 long non‐coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8:e1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang Y, Du W, Chu Q, Qin Y, Tuguzbaeva G, Wang H, Li A, Li G, Li Y, Chai L, Yue E, Sun X, Wang Z, Pavlov V, Yang B, Bai Y. Downregulation of long non‐coding RNA Kcnq1ot1: an important mechanism of arsenic trioxide‐induced long QT syndrome. Cell Physiol Biochem. 2018;45:192–202. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Pang WJ, Wei N, Xiong Y, Wu WJ, Zhao CZ, Shen QW, Yang GS. Identification, stability and expression of Sirt1 antisense long non‐coding RNA. Gene. 2014;539:117–124. [DOI] [PubMed] [Google Scholar]
- 17. Wang GQ, Wang Y, Xiong Y, Chen XC, Ma ML, Cai R, Gao Y, Sun YM, Yang GS, Pang WJ. Sirt1 as lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR‐34a. Sci Rep. 2016;6:21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ming GF, Wu K, Hu K, Chen Y, Xiao J. NAMPT regulates senescence, proliferation, and migration of endothelial progenitor cells through the SIRT1 AS lncRNA/miR‐22/SIRT1 pathway. Biochem Biophys Res Commun. 2016;478:1382–1388. [DOI] [PubMed] [Google Scholar]
- 19. Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kajstura J, Cheng W, Reiss K, Anversa P. The IGF‐1‐IGF‐1 receptor system modulates myocyte proliferation but not myocyte cellular hypertrophy in vitro. Exp Cell Res. 1994;215:273–283. [DOI] [PubMed] [Google Scholar]
- 21. Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, Lien CL. IGF signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS ONE. 2013;8:e67266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reiss K, Cheng W, Pierzchalski P, Kodali S, Li B, Wang S, Liu Y, Anversa P. Insulin‐like growth factor‐1 receptor and its ligand regulate the reentry of adult ventricular myocytes into the cell cycle. Exp Cell Res. 1997;235:198–209. [DOI] [PubMed] [Google Scholar]
- 23. Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. D'Uva G, Aharonov A, Lauriola M, Kain D, Yahalom‐Ronen Y, Carvalho S, Weisinger K, Bassat E, Rajchman D, Yifa O, Lysenko M, Konfino T, Hegesh J, Brenner O, Neeman M, Yarden Y, Leor J, Sarig R, Harvey RP, Tzahor E. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. 2015;17:627–638. [DOI] [PubMed] [Google Scholar]
- 25. Kubin T, Poling J, Kostin S, Gajawada P, Hein S, Rees W, Wietelmann A, Tanaka M, Lorchner H, Schimanski S, Szibor M, Warnecke H, Braun T. Oncostatin m is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell. 2011;9:420–432. [DOI] [PubMed] [Google Scholar]
- 26. Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z. Long non‐coding RNA ANRIL regulates inflammatory responses as a novel component of NF‐kappaB pathway. RNA Biol. 2016;13:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Bostrom P, Che L, Zhang C, Spiegelman BM, Rosenzweig A. miR‐222 is necessary for exercise‐induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non‐coding RNAs. Int J Mol Sci. 2015;16:3251–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med. 2008;14:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z, Ke X, Meltzer SJ. Long non‐coding antisense RNA KRT7‐AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun J, Wang X, Fu C, Wang X, Zou J, Hua H, Bi Z. Long noncoding RNA FGFR3‐AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol Biol Rep. 2016;43:427–436. [DOI] [PubMed] [Google Scholar]
- 33. Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA‐PXN‐AS1. Nat Cell Biol. 2017;19:820–832. [DOI] [PubMed] [Google Scholar]
- 34. Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI, Liao R. MicroRNA‐34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res. 2015;117:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. SIRT1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. [DOI] [PubMed] [Google Scholar]
- 36. Chu D, Thistlethwaite PA, Sullivan CC, Grifman MS, Weitzman MD. Gene delivery to the mammalian heart using AAV vectors. Methods Mol Biol. 2004;246:213–224. [DOI] [PubMed] [Google Scholar]
- 37. Mathison M, Gersch RP, Nasser A, Lilo S, Korman M, Fourman M, Hackett N, Shroyer K, Yang J, Ma Y, Crystal RG, Rosengart TK. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc. 2012;1:e005652 DOI: 10.1161/JAHA.112.005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression of Sirt1 AS lncRNA in PBS‐ or IGF‐1‐treated CMs. Real‐time qRCR analyses of Sirt1 AS lncRNA levels in isolated P1 CMs treated with PBS or IGF‐1 (n=3 mice per group). Statistical significance was calculated using two‐tailed unpaired Student t test. *P<0.05; error bars represent mean±SEM.
Figure S2. Sirt1 AS lncRNA promotes CM proliferation and dedifferentiation in vitro. A, Infection efficiency of various MOI of Ad‐SAS transfected into isolated P1 CMs. Scale bar, 500 μm. B, Real‐time qPCR analysis of Sirt1 AS lncRNA levels in isolated P1 CMs transfected with various MOI of Ad‐SAS (n=3 mice per group). C, CMs in prophase, prometaphase, metaphase, anaphase and cytokinesis in the Sirt1 AS lncRNA overexpression group immunostained for pH3. Scale bar, 30 μm, (D) Isolated P7 CMs were transfected with Ad‐NC or Ad‐SAS. mRNA expression levels of CM dedifferentiated markers (RUNX1, c‐kit, DAB2) were detected by Real‐time qPCR (n=3 mice per group). Statistical significance was calculated using one‐way ANOVA followed by LSD post hoc test in B, and two‐tailed unpaired Student t test in D. *P<0.05, error bars represent mean±SEM.
Figure S3. Schematic of the MI experiments in adult mice. A, Schematic of in vivo experiments in adult mice with MI surgery. Echo, echocardiography, IF, immunofluorescence. B, Elevation of ST‐segment from ECG in MI model.
Figure S4. Sirt1 AS lncRNA induces Sirt1 mRNA levels in MI hearts and has no effect on miR‐34a expression in CMs. A, Real‐time qRCR analyses of Sirt1 mRNA levels in AAV9‐NC‐ or AAV9‐SAS‐injected adult mice hearts 28 days after injection (n=6 mice per group). B, Real‐time qRCR analyses of miR‐34a levels in isolated P1 CMs transfected with Ad‐NC or Ad‐SAS (n=3 mice per group). Statistical significance was calculated using 2‐tailed unpaired Student t test in (A and B). *P<0.05; error bars represent mean±SEM.


