Abstract
Estimating fitness differences between allelic variants is a central goal of experimental evolution. Current methods for inferring such differences from allele frequency time series typically assume that the effects of selection can be described by a fixed selection coefficient. However, fitness is an aggregate of several components including mating success, fecundity, and viability. Distinguishing between these components could be critical in many scenarios. Here, we develop a flexible maximum likelihood framework that can disentangle different components of fitness from genotype frequency data, and estimate them individually in males and females. As a proof-of-principle, we apply our method to experimentally evolved cage populations of Drosophila melanogaster, in which we tracked the relative frequencies of a loss-of-function and wild-type allele of yellow. This X-linked gene produces a recessive yellow phenotype when disrupted and is involved in male courtship ability. We find that the fitness costs of the yellow phenotype take the form of substantially reduced mating preference of wild-type females for yellow males, together with a modest reduction in the viability of yellow males and females. Our framework should be generally applicable to situations where it is important to quantify fitness components of specific genetic variants, including quantitative characterization of the population dynamics of CRISPR gene drives.
Keywords: experimental evolution, fitness, maximum likelihood, natural selection, time series data
THE concept of fitness lies at the core of Darwin’s theory of evolution by natural selection. If two allelic variants differ in fitness, the fitter allele should gradually increase in frequency at the expense of the less-fit allele. In turn, studying allele frequency changes over time can allow us to infer fitness differences (Bollback et al. 2008). This rationale has been implemented in a variety of methods for quantifying natural selection from temporal allele frequency data. Typically, such methods employ a probabilistic model for the expected allele frequency dynamics, often based on a Wright–Fisher process. By comparing the predictions of the model with empirical allele frequency measurements, one can then infer the parameters of the model. The key challenge in these approaches is to disentangle selection from stochastic processes such as random genetic drift and sampling noise, which will generate fluctuations in allele frequency estimates on top of the systematic changes due to selection. Early approaches to this end used a continuous diffusion approximation to the discrete Wright–Fisher process for calculating transition probabilities of allele frequencies (Williamson and Slatkin 1999; Bollback et al. 2008; Malaspinas et al. 2012). These probabilities were then used in a maximum likelihood (ML) framework for inferring the model parameters, including effective population size. More recently, improved analytic approximations to the likelihood function were developed using concepts such as spectral representations of the transition density (Steinrücken et al. 2014) and path augmentation (Schraiber et al. 2016). Other approaches have employed Bayesian inference methods (Foll et al. 2015; Ferrer-Admetlla et al. 2016) and likelihood ratio tests (Feder et al. 2014). Some studies have explored how selection inference can be extended to spatial populations (Mathieson and McVean 2013), fluctuating environments (Gompert 2016; Shim et al. 2016), strong selection (Lacerda and Seoighe 2014), diploid populations with dominance effects (Steinrücken et al. 2014), and scenarios of linked loci along a recombining chromosome (Illingworth et al. 2012; Illingworth and Mustonen 2013; Terhorst et al. 2015).
A commonly made assumption in these methods is that the effects of selection between two allelic variants can be described by a single selection coefficient. While this may be reasonable in many scenarios, there are also situations where it is critical to distinguish among the individual components that constitute fitness (Christiansen and Frydenberg 1973; Nadeau and Baccus 1981; Sober and Lewontin 1982). Preferential mate choice, for instance, can result in frequency-dependent dynamics that cannot be mapped onto a logistic growth model specified by a fixed selection coefficient (Barton and Servedio 2015). Individual fitness effects could also often differ between males and females, leading to systematic biases in genotype frequencies between sexes when selection is sufficiently strong.
Rigorous inference of selection can include four major selection components: zygotic selection (viability from zygote to adult), sexual selection, fecundity selection, and gametic selection that usually involves genetic elements with biased inheritance (Christiansen and Frydenberg 1973; Nadeau and Baccus 1981). Evaluation of these four selection components in a natural population requires detailed population monitoring, as observations need to be recorded at four different life cycle stages (Christiansen and Frydenberg 1973; Nadeau and Baccus 1981; Nadeau et al. 1981; Siegismund and Christiansen 1985). Typically, the analysis of selection components in the laboratory involves performing a series of isolated experiments designed to individually quantify each component (Bundgaard and Christiansen 1972). For example, progeny counts from controlled crosses and backcrosses can reveal differences in zygote-to-adult viability as measured by deviation from Mendelian proportions (Mukai et al. 1974). Egg counts from controlled crosses can reveal genotype-specific differences in fecundity. Mating arenas with either observed matings or subsequent scoring of progeny can allow for estimation of sexual selection (Dow 1976). The challenge with all of these methods is that it is extremely difficult to know if all attributes of a gene that may result in differential propagation have been considered, although it is possible to test the correspondence between individual fitness component estimates and allele frequency dynamics in cage populations (Clark and Feldman 1981; Clark et al. 1981).
In this study, we develop an ML inference framework that can disentangle sexual, fecundity, and viability selection from genotype frequency time series data. Our analytical approach employs a continuous extension of the multinomial distribution, allowing us to infer effective population size simultaneously with selection parameters. As a proof-of-principle, we apply our inference framework to empirical data obtained from cage evolution experiments of Drosophila melanogaster, in which we tracked relative genotype frequencies of a wild-type and mutant version of yellow, an X-linked gene required for black pigment formation (Drapeau et al. 2003). The mutant allele disrupts yellow, resulting in a recessive yellow body phenotype. The yellow gene is also required for the wing-extension behavior that is part of male courtship in D. melanogaster (Bastock 1956; Wilson et al. 1976). Consequently, disruption of yellow affects mating competitiveness in male flies (Heisler 1984; Drapeau et al. 2006).
We find that in cage experiments, our ML inference method can robustly distinguish between several selection models involving preferential mate choice, fecundity, and viability, while further distinguishing between male and female fitness costs. Such measurements should become particularly important in scenarios where fitness costs are large and allele frequencies are expected to change rapidly, as will likely be the case for recently proposed CRISPR gene drive approaches (Champer et al. 2016).
Methods
Plasmid construct design and generation of transgenic fly line
We designed a construct targeting the X-linked yellow gene in D. melanogaster. Disruption of this gene causes a recessive yellow phenotype, specified by a lack of dark pigment in the adult cuticle. Our construct additionally encodes a dsRed protein driven by a 3xP3 promoter, which produces an easily identifiable fluorescent phenotype. dsRed is not visible in wild-type eyes in the Canton-S background, but we observed dsRed expression in the abdomen of younger insects and in the ocelli.
The donor plasmid BHDyR was constructed by Gibson assembly of the restriction digest of IHDyV1 (Champer et al. 2017) with StuI and XhoI, and PCR amplification of the pDsRed-attp plasmid (Gratz et al. 2014) with the oligos dsRedY_F (5′-GGGTTTTGGACACTGGGAATTCTTGCATGGCTAGACGAAGTTATCGTACGGGATCTAAT-3′) and dsRedY_R (5′-TTAGTGGTGGTATTGCCGATGCCCACGGACGCGCCGGTTAAGATACATTGATGAGTTTGG-3′) (IDT). Gibson assembly of plasmids was performed with Assembly Master Mix (New England Biolabs, Beverly, MA) and plasmids were transformed into JM109 competent cells (Zymo Research).
The transgenic line in the study was transformed at GenetiVision by injecting the BHDyR donor plasmid into Canton-S D. melanogaster embryos. Cas9 was provided by co-injection with plasmid pHsp70-Cas9 (Gratz et al. 2013) (a gift from Melissa Harrison, Kate O’Connor-Giles, and Jill Wildonger, plasmid 45945; Addgene). A guide RNA (gRNA) plasmid (BHDyg1) (Champer et al. 2017) was also included in the injection. Concentrations of donor, Cas9, and gRNA plasmids were 98, 94, and 58 ng/μl, respectively, in a 10 mM Tris-HCl and 23 μM EDTA (pH 8.1) solution. The injected plasmids were purified with ZymoPure Midiprep kit (Zymo Research). To obtain a homozygous fly line, the injected embryos were reared and crossed with wild-type Canton-S flies. The progeny with dsRed fluorescent protein in the eyes and abdomen, which indicated successful insertion of the construct, were selected and crossed with each other. The stock was considered homozygous when all male progeny had dsRed fluorescence for two consecutive generations.
Fly rearing and phenotyping
Flies were reared at 25° with a 14/10-hr day/night cycle. Bloomington Standard medium was provided as food every 2 weeks. During initial phenotyping, flies were anesthetized with and examined with a stereo dissecting microscope, and the fluorescent red phenotype was observed using the NIGHTSEA system.
For the cage studies, enclosures of internal dimensions 30 × 30 × 30 cm (BD43030D; Bugdorm) were used to house flies. At the start of an experiment, transgenic and Canton-S flies of approximately the same age were separately allowed to lay eggs in food bottles for 2 days. These food bottles (nine for cage 1, eight for cage 2, and five for cages 3–5) were then placed in cages at the desired starting ratio between transgenics and Canton-S flies. Eleven days later, bottles were replaced in the cage with fresh food (at a 1:1 ratio for cage 1 and 2:1 for other cages), while leaving adult flies in the cages. Two days later, bottles were removed again from the cages and flies retained, and fresh food bottles were added (nine for cage 1, eight for cage 2, and five for cages 3–5). One day later, all adult flies were frozen for later phenotyping and food bottles were retained for 11 days to allow the next generation to hatch. This cycle was repeated until the completion of the experiments. All frozen flies from each generation were phenotyped within 2 weeks of freezing.
Data availability
Our experimental data consists of the genotype counts in the five cage experiments and they are all reported in Figure 1. The ML inference framework was implemented in R. Supplemental material containing all the necessary scripts for reproducing the simulations and results are available at Figshare: https://doi.org/10.25386/genetics.7616171.
Figure 1.
Population sizes and genotype frequencies in our five cage experiments. Note that we define genotype frequencies relatively among males and relatively among females.
Results
Cage evolution experiments
We created a transgenic D. melanogaster line, based on the Canton-S strain, in which we inserted a construct expressing a dsRed fluorescent protein into the yellow gene. This insertion effectively disrupts the gene, producing the characteristic yellow phenotype, which is recessive. At the same time, expression of the dsRed protein produces an easily identifiable, codominant phenotype characterized by red fluorescence in the eyes, abdomen, and ocelli, though only the latter two could be observed in Canton-S flies due to eye pigmentation.
We will refer to the allele with the inserted construct as y, and we will denote the wild-type allele as . Because yellow is located on the X chromosome, males of genotype y are expected to show both the yellow (body) and red (eye) phenotypes, whereas males are expected to show neither phenotype. In females, homozygotes are expected to show both the yellow and red phenotypes, heterozygotes are expected to show only the red phenotype, and homozygotes are expected to show neither phenotype. Our system thereby allows us to unambiguously distinguish genotypes at the yellow locus from phenotypic assays of individual flies.
We evolved five laboratory cage populations, each initialized from a mixture of wild-type Canton-S flies and our transgenic yellow flies. Genotype frequencies at the yellow locus were then tracked over several generations. Our first two cages were evolved over six (cage 1) and five (cage 2) generations, starting with initial y allele frequencies of 70%. Three additional cages were each evolved for just a single generation with starting frequencies of the y allele of 20% for cage 3, 50% for cage 4, and 80% for cage 5. Census population sizes in each cage comprised between several hundred and several thousand flies per generation.
Figure 1 shows the observed genotype frequency trajectories, and changes in male and female population size, over time. Note that although we phenotyped the full adult census population, many of these flies may not have actually reproduced. The frequency of the y allele decreased systematically in all five cages, confirming that yellow disruption is likely associated with a substantial fitness cost. In cage 1, for example, the y allele decreased from 70 to 10% over just six generations, and a similar rate of decline was observed in the other cages as well.
Census population sizes fluctuated noticeably in our cages. However, we do not believe that this was due to selection at the yellow locus, but rather reflected unrelated factors, such as larval competition and variance in food preparation. We also usually observed a biased sex ratio of phenotyped flies since females had higher mortality from laying eggs in the crowded food bottles, while males approached the food bottles less often.
Evolutionary model
To estimate the fitness costs associated with yellow disruption, we will first establish an evolutionary model for allele frequency dynamics that incorporates three specific components of fitness: viability, fecundity, and mating success. We specifically consider an X-linked locus with two segregating alleles: wild-type and yellow (y). The y allele produces the yellow phenotype that has lower fitness than the wild-type phenotype. These fitness costs could be due to reduced viability, fecundity, and/or mating success in individuals with the yellow phenotype compared to wild-type individuals. In all cases, we assume that the y allele is strictly recessive, so that wild-type and heterozygous females have the same fitness. We will denote the counts of individuals observed in the present generation t, partitioned by genotype, as . Viability specifies the probability that a zygote survives to reproductive age, which we assume depends only on the phenotype and sex of the individual. Specifically, we define to be the relative viability of yellow vs. wild-type males, and to be the relative viability of yellow vs. wild-type females
| (1) |
Here, describes wild-type males, describes yellow males, describes wild-type females, and describes yellow-phenotype females.
Fecundity specifies the reproductive success of a mating pair, as measured by the expected number of offspring. In contrast to viability, which is a function of an individual zygote, fecundity is a function of a mother–father pair. In our model, we assume that fecundities are defined by the following mating table, specifying the relative fecundities of matings involving parents of different phenotypes.
For our sexual selection model, we assume a scenario of female mate choice in which females will choose mates with a potential preference based on phenotype. We define the probabilities that a female of a particular phenotype chooses a male of a particular phenotype to be
| (2) |
The parameters α and β specify the relative reduction in mating success of yellow males compared to wild-type males in mating with wild-type or yellow females, respectively.
Biologically there is no reason to assume that yellow-phenotype males or females would actually have higher viability, fecundity, or mating success than their wild-type counterparts. Thus, we will assume that all selection parameters are constrained within , which will also help prevent overfitting in our ML inference approach. Finally, we assume discrete generations and random segregation of alleles within gametes due to a lack of any known mechanism for biased inheritance of y. Figure 2 illustrates the life cycle in this evolutionary model over the course of one generation, depicting where the different types of selection operate.
Figure 2.
Illustration of the different stages of the life cycle during one generation in our model.
With these definitions at hand, we can calculate the expected genotype frequencies in generation as a function of their frequencies in generation t and the selection parameters. At our X-linked locus, male zygotes always inherit their X chromosomes from their mothers, while females inherit one copy from each parent. When assuming absence of any selection , the expected genotype frequencies in the next generation will then be given by
| (3) |
Here and denote the expected relative frequencies of y and genotypes among males, and , , and denote the expected relative frequencies of , , and genotypes among females. The normalization coefficients and are defined by the conditions that gamete frequencies in males and females each have to sum to one: . Note that we do not use absolute genotype frequencies in the population, but only relative frequencies in males and females. This will later allow us to factorize likelihood calculations into male and female likelihoods.
Viability selection is straightforward to incorporate into this framework. We can simply interpret the genotype frequencies derived in (3) as the expected frequencies of zygotes, which then just need to be multiplied by the corresponding viability coefficients (and renormalized). To incorporate fecundity and mate choice, we also have to account for the probabilities that the matings that can produce the respective zygote genotypes actually occur, and then weigh them by their fecundities. Altogether, this yields
| (4) |
The coefficients and are again defined by normalization. Note that although this model is specific for an X-linked gene, it is in fact easy to extend to other inheritance patterns, for example an autosomal locus (see Supplemental Material, Supplemental Results). The general principle is always the same. To calculate the expected frequency of a given genotype, we first have to sum over all matings that can produce this genotype, weight each by its respective mating probability and expected fecundity of the mating pair, then multiply all genotypes by their respective viabilities, and finally normalize all genotype frequencies. Other selection scenarios, such as different dominance effects, could be easily implemented as well by changing the genotypes to which fitness parameters are applied.
ML inference framework
The model developed in the previous section allows us to calculate changes in genotype frequencies when stochastic fluctuations due to processes such as random genetic drift can be neglected. To extend this model to more realistic populations, we require a probabilistic model that incorporates such fluctuations. In principle, the source of this stochasticity could be explicit in our model. For example, we could describe stochasticity in the mating process by randomly drawing males with replacement. Stochasticity in fecundity could be modeled by drawing offspring numbers from some probability distribution. Finally, stochasticity at the level of viability could be implemented as a Bernoulli process.
However, such a model would require a very detailed understanding of each step of the life cycle in our cages, which may even change with the environment and population density. Since our primary goal is the inference of the selection parameters, rather than the causes of drift, we will instead use a simple Wright–Fisher-type model to describe the fluctuations in genotype frequencies, as has been employed in many previous methods (Bollback et al. 2008; Mathieson and McVean 2013; Terhorst et al. 2015; Schraiber et al. 2016). Specifically, we assume that genotypes in generation are sampled randomly from a multinomial probability distribution, specified by the expected genotype frequencies from Equation 5 and an effective population size parameter . This parameter approximates the aggregate effects of random genetic drift in the model and can be tuned such that frequency fluctuations in the model become similar in magnitude to those in the real population. Note that the parameter could, in principle, be allowed to change across generations, for instance becoming a function of census population size and/or sex ratio. We will discuss these complexities in more detail below.
There is an important complication when it comes to calculating probabilities in this model. In a Wright–Fisher population of effective size , the probability of observing a given vector of counts of all d possible genotypes is given by a multinomial distribution, which is properly defined only when . This condition will almost certainly be violated in our cages, as we expect census sizes (and thus also sample sizes, which are comprised by all adults in a cage) to be much larger than due to a variety of factors, such as a higher variance in offspring numbers in the fly populations compared to a Wright–Fisher population.
To address this problem, we will employ a continuous approximation for the multinomial distribution, where genotype counts are replaced by genotype frequencies and the discrete multinomial probabilities are replaced by a multidimensional probability density. We define this density by using the γ function as a natural continuous extension for the factorials in the multinomial distribution. Specifically, consider a diploid Wright–Fisher model with effective population size and d possible genotypes, where specifies their expected frequencies in generation . The probability density of observing a given set of genotype frequencies in generation is then defined by
| (5) |
The constant c is to ensure normalization and will be discussed below. In this framework, the probability of observing genotype frequencies that fall inside a given area of the d-dimensional frequency space is obtained by integrating the probability density over this area: .
To allow for the possibility of biased sex ratios in our model while avoiding specific assumptions about what caused these biases in our cages, we will further factorize the overall probability density into the individual probabilities of observing the male and female genotype counts independently
| (6) |
Here, and specify the observed genotype frequencies in generation in males, and , , and specify the observed frequencies in females.
The normalization coefficients and for the male and female probability densities are defined by the requirement that the integral of ρ over the d-dimensional standard simplex (defined by ) has to yield a value of 1. Unfortunately, we are not aware of any closed-form analytic expressions for this integral, but we can provide a workable approximation by discrete partitioning. For the male density, we have and can chose as the free variable, while . We can then model the area under ρ as the sum of rectangles of width and height defined by the value of ρ at the rectangles’ midpoints, , plus two rectangles of width and height, and , at the left and right boundaries of the frequency space, respectively. Setting this sum to 1 then yields
| (7) |
This approach can be directly extended to the female density, where . Here, we can chose and as the free parameters, while . The integration of ρ over the simplex can then be approximated as the sum of the volume of prisms, estimated over the triangle bounded by the lines , , and , which yields
| (8) |
Note that under these approximations, is technically no longer a proper probability density, as integration over the simplex will not always yield a value of exactly 1. However, these deviations become noticeable only for very small population sizes, where the applicability of the Wright–Fisher model becomes questionable in general. Our simulations below confirm that this discrete approximation works well in practice.
The factorization employed in Equation 5 provides a very flexible framework for defining effective population size, which could be assumed to differ between males and females, or could vary based on the census population size. For now, we want to assume the simplest model of a constant effective population size with equal sex ratio, while more sophisticated definitions will be discussed later in the manuscript. We therefore set
| (9) |
With these definitions in place, we can formulate the log-likelihood density function for the parameter vector , given the vectors of observed genotype counts in generations t and of a population cage. Multiplying these individual log-likelihoods across all generations in an experiment (i.e., assuming independence across generations) then yields the log-likelihood density function
| (10) |
We can easily extend this approach across multiple cages by summing the log-likelihoods of the individual cages. The resulting log-likelihood density function can then be used for inference of ML estimates (MLEs) of the parameter values and their C.I.s.
Several aspects of the above approach are worth further mention. First, has now become just another parameter of the model and can thus be inferred the same way as we do with the other parameters. This way, can also take on continuous values in our model. Second, our ML inference approach is very general and could be easily applied to other evolutionary models, so long as the model provides expected values for genotype frequencies given their frequencies in the previous generation. Finally, we want to acknowledge that we have not explicitly incorporated sampling variance into our framework. This is because our genotype frequency estimates are based on counting all of the adults present in a cage. While some sampling noise due to counting, phenotyping errors, or escaped flies may of course still have occurred in any given generation, we expect that the contribution of such factors to variance in genotype frequencies should generally be negligible compared to the contribution due to drift. This is also supported by the fact that inferred effective population sizes are typically an order of magnitude smaller than the census sizes of our cages (and hence sample sizes).
Power evaluation
Even when combined over all five cages, the genotype trajectories in our experiments comprise a total of only 14 generation transitions. In light of such limited data, can we still expect our ML inference method to have sufficient power to infer the different parameters of our model? To explore this question and also provide a general proof-of-principle of our inference framework, we tested it on simulated data.
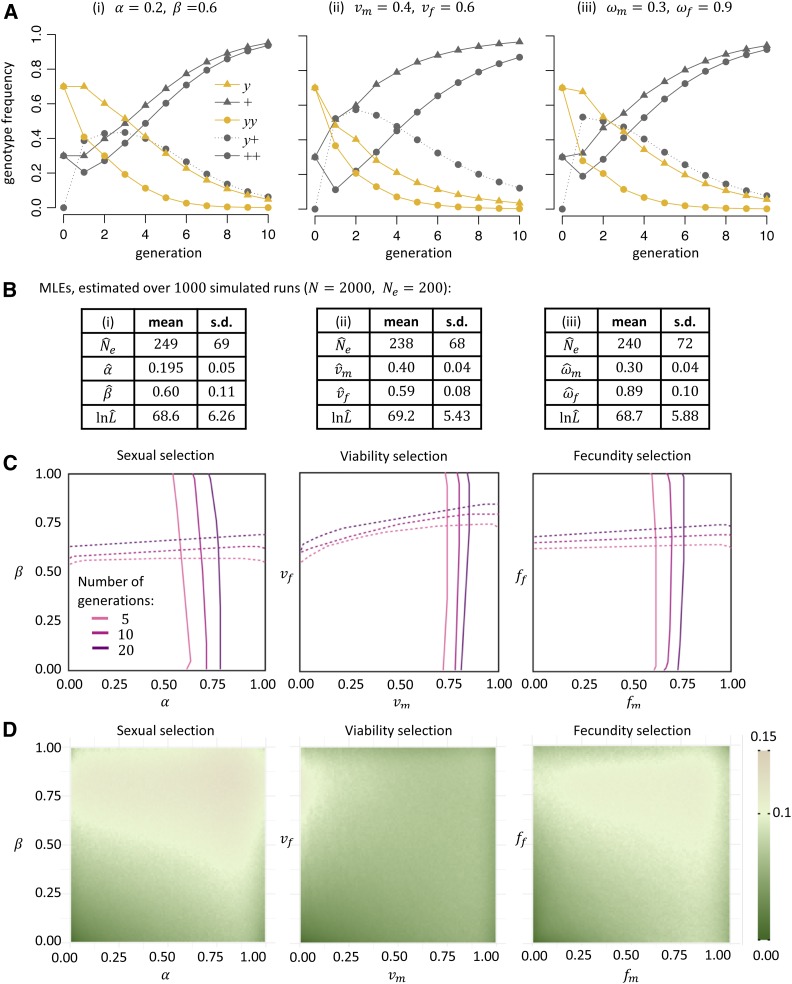
We first wanted to assess whether there are apparent differences in genotype frequency trajectories between selection scenarios where fitness costs of an X-linked allele are either due to reduced mating success, viability, or fecundity. Figure 3A shows the outcomes of three simulations in which we modeled frequency trajectories over 10 generations in a cage under these three different selection scenarios. The selection strength in each scenario was tuned to roughly conform with the observed decay rate of y allele frequencies in our experimental cages, and these simulations were run without drift (in the limit of “infinite” ). For simplicity, we further assumed that for the fecundity scenario. The simulations indeed demonstrate clear differences in the frequency trajectories between the three scenarios, suggesting that different fitness components could in principle be disentangled. While these differences are most pronounced in the early generations, where frequency changes are the largest, they also extend throughout the run, as can be seen in the different frequency ratios of y males to females.
Figure 3.
Power analysis of the maximum likelihood (ML) inference method. (A) Simulated genotype frequency trajectories under different selection scenarios in a deterministic model without drift: (i) sexual selection only (; (ii) viability selection only ; and (iii) fecundity selection only . Populations were started with a y allele frequency of 70%, equally in males and females. Only homozygous females were present in the starting generation. Selection parameters were chosen so that all three scenarios yielded similar y allele frequency at the end of the run. (B) Accuracy of our inference framework for the three selection scenarios simulated in (A). For each scenario, 1000 simulation runs were performed in populations of 2000 individuals with . In each such run, the parameter ML estimates (MLEs) were then inferred for the given model, while the other selection parameters were kept fixed at a value of 1. The table shows the mean values and SDs of these parameter MLEs for the three scenarios. (C) Selection thresholds below which the inference method can reject a value of 1 for a given selection parameter. Simulations were performed as in (B) but varying all selection parameters over a grid. For each grid point, 1000 simulations were run. Lines show the region in which the parameter can be statistically distinguished from a value of 1 with 95% confidence in 95% of simulations. Solid lines show the horizontal axis parameter and dashed lines show the vertical axis parameter. The different colors indicate different lengths of the experiment (5, 10, or 20 generations). (D) The heatmaps show the average Euclidean distance between pairs of inferred parameter MLEs and simulated values.
We next applied our ML inference method to these three selection scenarios, but this time simulating a Wight–Fisher model with drift . Figure 3B demonstrates that our approach can successfully infer the individual model parameters from a single 10-generation run in these simulated experiments. When averaged over many simulation runs, MLEs for the selection parameters converge to the true values, with SD among runs being on the order of of the parameter values. However, the estimates inferred by our approach were consistently higher than the true values. This is likely due to the fact that, in any single experiment, drift alone should already result in an upward or downward shift in the y allele frequency that our method presumably mistakes for “additional” selection, thereby underestimating the true amount of drift. Consider a completely neutral scenario, for instance. In this case, our method would still presumably infer some positive or negative selection in any given run that ended at an allele frequency different from the starting frequency. This would lead to a lower inferred level of drift, even though the direction of selection would be random and average out over many runs.
The simulations shown in Figure 3, A and B all involved rather strong fitness costs. To test how inference power depends on the strength of selection, we systematically varied the two selection parameters in each of the three scenarios, as well as the duration of the experiment. We first wanted to know the threshold for each parameter where our method can reject a value of 1 with 95% confidence in 95% of simulations. Figure 3C shows that even for a short experiment of only five generations, our approach can reliably detect selection as long as individual fitness parameters are ∼0.6 or smaller (i.e., fitness costs are 40% or larger for the particular parameter). Increasing the duration of the experiment to 10 or 20 generations raises these thresholds.
We next studied the accuracy of the parameter MLEs across the parameter space in the three selection models. Figure 3D shows that relative errors are typically smaller than 15% in the test scenarios when run for 10 generations. Note that although each selection scenario has a male and female selection parameter, inference power is asymmetrical for them. For the fecundity and viability scenarios, this is due to the fact that we are considering an X-linked locus with a recessive phenotype. For the sexual selection scenario, there are additional systematic differences due to the fact that females are the choosy sex in our model.
So far, we have only tested our inference method under the assumption that we already know which type of selection has acted (fecundity, viability, or sexual selection), validating that the method can reliably infer the respective selection parameters in this case. But can it also distinguish the different types of selection from each other? To test this, we applied our method to each of the three selection scenarios, using either the correct inference model or models in which a different type of selection was inferred. This can be easily implemented in our framework by fixing specific selection parameters. For example, to devise a sexual-selection-only inference model, the α and β parameters would be allowed to vary, while all other selection parameters would be set to a value of 1.
Figure 4 shows the power of our ML inference method to identify the correct selection type among the three selection scenarios. Specifically, for each parameter setting, we measured the fraction of simulations in which the selection inference models with the correct type yielded a higher maximum log-likelihood value than either of the incorrect types. Figure S1 further shows how these results subdivide into individual two-way comparisons between the correct model and only one of the incorrect models. These results demonstrate that our ML inference method has surprisingly good power in identifying the correct selection type, with power generally improving as selection strength increases. However, we also observe distinct patterns for the three selection scenarios in which it becomes difficult to distinguish the correct model. In a sexual selection simulation where α and β have similar values, we have little power to distinguish the sexual selection model from the fecundity selection model with (Figure S1). This is because when , the sexual selection model is mathematically identical to the fecundity selection model with . Similarly, we observe that in a fecundity selection simulation where , the power to infer the correct selection type is actually inferior to a random guess (Figure S1), presumably due to a better capacity of the sexual selection inference model to account for stochastic variation in this situation. Finally, we note that viability selection can almost always be correctly distinguished from the two other types in our test scenarios unless selection is very weak.
Figure 4.
Power of the inference method to distinguish the three different types of selection. Simulations were run as in Figure 3, C and D. Parameter maximum likelihood estimates were then obtained for each run using the three individual selection inference models. For each grid point, the heatmaps show the fraction of simulation runs in which the correct inference model yielded the highest maximum log-likelihood value among the three models.
Our evolution experiments tracked an X-linked locus with a recessive phenotype. Consequently, the inference method we developed focuses on such a scenario. However, there are some peculiarities that distinguish evolutionary dynamics at an X-linked locus from the more conventional case of an autosomal locus. For example, at an X-linked locus, it will take longer for the population to approach Hardy–Weinberg equilibrium when it started from a state of disequilibrium, as was the case in our experiments. This raises the question of whether the power of our method to distinguish selection types and infer selection parameters may in part be due to such peculiarities. To explore this question, we extended our inference method to an autosomal locus and verified that it still retains power in this more general case, although distinguishing between male and female selection parameters becomes more difficult. A detailed study of the autosomal model is provided in the Supplemental Results.
Parameter estimation in cage experiments and model comparison
We applied our ML inference method to the combined data from our five cage populations by adding the individual log-likelihoods of each of the 14 generational transitions observed in our experiments. Table 1 shows the resulting parameter MLEs and maximum log-likelihood values for several different inference models.
Table 1. Parameter MLEs and model comparison.
| Model description | P | AICc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Full model | 165 | 0.84 | 0.75 | 0.92 | 0.95 | 1.00 | 0.07 | 1.00 | 79.4 | 8 | −114 |
| No sexual sel. | 103 | 0.98 | 1.00 | 0.10 | 0.40 | 0.29 | 1* | 1* | 69.6 | 6 | −115 |
| No viability sel. | 154 | 1* | 1* | 0.77 | 0.86 | 0.73 | 0.09 | 0.98 | 77.9 | 6 | −132 |
| No fecundity sel. | 165 | 0.83 | 0.77 | 1* | 1* | 1* | 0.06 | 1.00 | 79.4 | 5 | −141 |
| Sexual sel. only | 134 | 1* | 1* | 1* | 1* | 1* | 0.03 | 0.69 | 74.9 | 3 | −141 |
| Viability sel. only | 16 | 0.79 | 0.56 | 1* | 1* | 1* | 1* | 1* | 32.0 | 3 | −56 |
| Fecundity sel. only | 103 | 1* | 1* | 0.10 | 0.39 | 0.29 | 1* | 1* | 69.6 | 4 | −127 |
| Simplistic model | 32 | 1* | 1* | 0.49 | 1* | 1* | 44.0 | 2 | −83 | ||
| Minimal model 1 | 164 | 0.81 | 1* | 1* | 1* | 0.06 | 1* | 79.2 | 3 | −150 | |
| Minimal model 2 | 155 | 1* | 1* | 0.86 | 0.09 | 1* | 77.9 | 3 | −147 |
Each row shows the parameter maximum likelihood estimates and associated maximum log-likelihood values when estimated from the combined data across all cages (1–5). An entry of 1* indicates that the parameter was fixed at a value of 1 in the particular inference model. “=” indicates that the value of a parameter is fixed in relation to another. P denotes the number of free parameters of the model, and AICc specifies its corrected Akaike information criterion value, AICc where is the sample size. Smaller AICc values indicate better support for the model. sel., selection.
We first tested our full inference model with all three types of selection possible (eight free parameters total, including ). The resulting parameter MLEs for this model suggest that all three types of selection were indeed acting in the cages, as indicated by the fact that at least one parameter for each type of selection was inferred to be different from one. However, we suspected that there could be substantial overfitting in this model, given the rather large number of parameters for the amount of data available. Furthermore, various parameters could be intertwined with each other and difficult to disentangle in practice. For example, it should generally be possible to approximate many scenarios of preferential mate choice by appropriately tuning a model with only fecundity selection, especially since the latter has more free parameters when we drop the assumption that , but let be another free parameter.
To assess which fitness components are indeed most essential to incorporate to accurately describe the genotype dynamics observed in our cages, we tested several inference models, systematically reducing model complexity. The quality of these models was then compared by calculating the corrected Akaike information criterion (AICc) values (Hurvich et al. 1989; Akaike 1998), which provide an estimator for the goodness-of-fit of a given model, while also penalizing an increase in the number of model parameters. Although the correction term is strictly accurate only for linear models, it serves as an adequate approximation for more complex scenarios in most cases (Hurvich et al. 1989). Note that in contrast to log-likelihood values where higher values indicate a better-fitting model, a lower value indicates a better-fitting model for AICc values.
We first evaluated inference models in which one type of selection was completely eliminated (by setting all parameters describing that component to a value of 1). In this case, the largest reduction in maximum log-likelihood was observed when sexual selection was excluded. All of these models yielded better AICc values than the full model because of their reduced number of parameters.
We next tested models with only one type of selection acting. Consistent with the above result, we found that a model with only sexual selection achieved a higher maximum log-likelihood compared to models with only viability or only fecundity selection, and the AICc value of this model was again better than the full model. The model with only viability selection was the least-supported model we tested overall.
A simplistic model with only a single fecundity parameter, , assumed to be equal in males and females, and multiplicative , also produced a very poor fit to the observed data, emphasizing the need for more complex models for understanding the fitness costs of the yellow phenotype.
Finally, we sought to identify a minimal model of low complexity, based on a limited number of biological assumptions, which can still describe the observed dynamics reasonably well. Motivated by the observation that the β parameter was often estimated to be one in the previously studied inference models, we assumed for this minimal model that sexual selection acts only by lowering the mating success of yellow-phenotype males when wild-type females choose their mate. In addition, we assumed that yellow-phenotype individuals have either reduced viability (minimal model 1) or reduced fecundity (minimal model 2), with equal costs between the sexes and multiplicative costs in the case of fecundity. Both of these minimal models have only three free parameters (including ), yet yielded better AICc values than any of the other models. In fact, our minimal model 1 yielded the best AICc value overall, suggesting that the data can be explained well with this simple model, in which wild-type females show severe mating bias against males of yellow phenotype and both sexes experience a modest reduction in viability when having a yellow phenotype . However, note that the inclusion of preferential mate choice is crucial in this model, given that the model with only viability selection produced the worst fit among all models tested.
One key advantage of ML-based approaches is that confidence intervals of parameter estimates can be easily calculated by likelihood ratio tests. Figure 5A demonstrates this for the effective population size parameter in our full model, which we inferred to be with a confidence intervals of ∼, or ∼5–12% of the census population size. This estimate is consistent with previous cage evolution experiments in D. melanogaster, where values were observed ranging from 4–25% of population census sizes (Malpica and Briscoe 1981; Mueller et al. 2013). Importantly, in our framework, the inference parameter will also depend on the overall goodness-of-fit of the inference model. A misspecified model would be expected to result in lower estimates, because the ML approach would have to assume higher amounts of drift to explain the larger deviations between the predicted and observed frequency trajectories.
Figure 5.
Log-likelihood surfaces and confidence intervals (A) The log-likelihood curve of our full model varying , while keeping other parameters fixed at their maximum likelihood estimate. The vertical dashed lines indicate the maximum likelihood estimate and the corresponding C.I., estimated from a likelihood ratio test with one d.f. (B) The log-likelihood surface for minimal model 1 with the mating success parameter α, viability parameter , and fixed . (C) The log-likelihood surface for minimal model 2 with the mating success parameter α, fecundity parameter , , and fixed . Darker colors correspond to higher likelihoods. The three contour lines specify C.I.s according to a likelihood ratio test with two d.f.
Figure 5, B and C show the likelihood surface plots and confidence intervals for the selection parameters in each of our two minimal models, demonstrating that our ML approach has good power in inferring these parameters. The diagonal elongation of the contour areas in minimal model 2 suggest that the two parameters are in fact partially dependent on one another in this model.
Discussion
In this study, we developed an ML inference method for estimating selection parameters from genotype frequency time series. The key advancement of our approach over existing methods is the ability to explicitly distinguish among the different components that constitute fitness, including mating success, fecundity, and viability. Our analysis indicates that these different types of selection can indeed result in distinct genotype frequency trajectories, which can be reliably distinguished from each other using our inference method.
As a proof-of-principle, we applied our method to study the fitness costs associated with a disrupted yellow allele in D. melanogaster, using data from cage evolution experiments. Consistent with previous studies (Heisler 1984; Drapeau et al. 2006), we found that yellow-phenotype males indeed experience a large fitness cost compared to their wild-type counterparts. Our method allowed us to further show that this primarily results from wild-type females strongly preferring to mate with wild-type males over yellow males. Additionally, yellow-phenotype males and females may both experience a moderate reduction in viability according to our minimal model 1, which had the highest statistical support (Table 1).
Figure 6 shows that dissecting individual fitness components is crucial for an accurate description of the genotype frequency dynamics at the yellow locus. A simplistic model that seeks to approximate the action of selection with a single selection parameter would predict significantly different dynamics from those observed in the cages, whereas our minimal model captures the true dynamics with substantially higher accuracy.
Figure 6.
Comparison of the observed genotype trajectories in our population cages (averaged over cages 1 and 2) with the predicted trajectories of the simplistic one-parameter fecundity-selection-only model , , and our minimal model 1 (, ), using the parameter maximum likelihood estimates inferred in Table 1. Simulations were run without drift.
Despite its generality, there are a number of important limitations to our method. In contrast to many existing methods that are able to analyze sparse allele frequency data obtained from distant time points, our method requires frequency estimates over discrete, consecutive generations. This requirement allowed us to establish a fully analytical model based on the Wright–Fisher process, which does not rely on simulations or numerical approximations of the transition probabilities across multiple generations, as are typically invoked by other methods. However, it does likely limit the applicability of our approach to experimental studies where discrete generations can be enforced and data can be obtained for each generation.
There are also potential issues regarding model complexity and overfitting in our inference method. With its large number of parameters, certain combinations of values can create very similar outcomes, and these scenarios may only be distinguishable if there are major sex-based differences between the parameters. This became apparent when examining the power to distinguish sexual vs. fecundity selection. Similarly, even though both our minimal models suggested strong sexual selection against yellow males, the first model also suggested a viability cost for yellow-phenotype individuals while the second suggested a fecundity cost. These results may have been influenced by small sample sizes, so it remains unclear whether or how the yellow phenotype induces a modest reduction in viability, fecundity, or perhaps both. In more complex models, such as when parameters are allowed to vary with time, it will be even more difficult to distinguish between different models that could produce similar genotype trajectories. Finally, it is not always clear how well the evolutionary model actually resembles the mechanisms at play in the real population. For example, in our implementation of a mate choice model, we assumed that females are the choosy sex, yet males could potentially be choosy too in some situations.
To calculate the probabilities of specific allele frequency changes between consecutive generations, it was necessary to incorporate an effective population size in our model. We defined this parameter to be equal for males and females, and constant across generations, yet other valid possibilities exist. For example, the effective population size for each generational transition could be defined as a fixed percentage of the census population size, using either the size in the previous generation, the current generation, or some weighted combination of both. This issue illustrates the shortcomings of the Wright-Fisher model, where it is the population size in the current generation that determines the amount of drift, whereas in real populations we would often assume that the size in the previous generation should be more relevant because it determines the number of mating pairs. Another possibility would be to treat male and female effective population sizes separately, which would also allow for a lower effective proportion of males compared to females. In some scenarios, these considerations may yield a model that is more representative of the true level of stochasticity in each generational transition. However, in exploring these possibilities, we found that for our experiments, none of the more sophisticated definitions of we tested yielded significantly better likelihoods than the simple model of a single parameter that is equal in males and females, and constant over time.
Disrupted yellow alleles are classic genetic markers in Drosophila genetics and have long been known to affect the mating success of male flies due to less effective courtship displays. In particular, wing-extension display is known to be defective in yellow males, resulting in their securing only ∼10% of matings with a wild-type female when in one-to-one competitions with wild-type males (Drapeau et al. 2006), which is broadly consistent with our results. Interestingly, in long-term yellow-phenotype stocks, yellow females were thought to have evolved to be more receptive to their male counterparts (Bastock 1956), but it was later determined that females with the yellow phenotype were themselves more receptive to yellow males, irrespective of their genetic background (Dow 1976). Our study corroborates this result. While it is possible that yellow females do show a modest preference for wild-type males over yellow males, our study lacked the statistical power to determine this.
Many evolutionary scenarios will still be well served by classical models invoking only a single, fixed selection coefficient. However, certain scenarios will likely require more detailed fitness models such as the one we have presented here. Perhaps the primary application for such models involves the spread of selfish genetic elements. The potential use of engineered gene drives (Unckless et al. 2015; Champer et al. 2016) is one such example, as it demands accurate and detailed models for their performance in natural populations, which will likely have to be based on cage trials. For instance, while an existing D. melanogaster gene drive (Champer et al. 2017) that targeted the yellow gene would not be able to spread, a multiplexed gRNA drive (Champer et al. 2018) at this locus could be expected to spread in a frequency-dependent fashion in either of our minimal models involving sexual selection, a feature not commonly associated with such homing-type gene drives. Thus, determination of individual fitness components is not only essential for understanding the spread of gene drives, but could potentially inform their design as well.
Acknowledgments
We thank Yassi Hafezi for helpful advice on the experiments, and Joshua Schraiber and two anonymous reviewers for their excellent comments and suggestion that helped improve the manuscript. This work was supported by funds from the College of Agriculture and Life Sciences at Cornell University to P.W.M., and the National Institutes of Health (award R01 GM-127418 to P.W.M.; award R21 AI-130635 to J.C., A.G.C., and P.W.M.; and award F32 AI-138476 to J.C.). A.M.L. was supported by a Ph.D. fellowship of the Vetmeduni Vienna, the Austrian Science Funds, and the Austrian Marshall Plan Foundation.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7616171.
Communicating editor: G. Coop
Literature Cited
- Akaike H., 1998. Information theory and an extension of the maximum likelihood principle, pp. 199–213 in 2nd Int. Symp. Inf. Theory, edited by Petrov B., Csaki F. Akadémiai Kiadó, Budapest, Hungary: 10.1007/978-1-4612-1694-0_15 [DOI] [Google Scholar]
- Barton N. H., Servedio M. R., 2015. The interpretation of selection coefficients. Evolution 69: 1101–1112. 10.1111/evo.12641 [DOI] [PubMed] [Google Scholar]
- Bastock M., 1956. A gene mutation which changes a behavior pattern. Evolution 10: 421–439. 10.1111/j.1558-5646.1956.tb02868.x [DOI] [Google Scholar]
- Bollback J. P., York T. L., Nielsen R., 2008. Estimation of . Genetics 179: 497–502. 10.1534/genetics.107.085019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundgaard J., Christiansen F. B., 1972. Dynamics of polymorphisms. I. Selection components in an experimental population of Drosophila melanogaster. Genetics 71: 439–460. [DOI] [PubMed] [Google Scholar]
- Champer J., Buchman A., Akbari O. S., 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17: 146–159. 10.1038/nrg.2015.34 [DOI] [PubMed] [Google Scholar]
- Champer J., Reeves R., Oh S. Y., Liu C., Liu J., et al. , 2017. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13: e1006796 10.1371/journal.pgen.1006796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J., Liu J., Oh S. Y., Reeves R., Luthra A., et al. , 2018. Reducing resistance allele formation in CRISPR gene drive. Proc. Natl. Acad. Sci. USA 115: 5522–5527. 10.1073/pnas.1720354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen F. B., Frydenberg O., 1973. Selection component analysis of natural polymorphisms using population samples including mother-offspring combinations. Theor. Popul. Biol. 4: 425–445. 10.1016/0040-5809(73)90019-1 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Feldman M. W., 1981. The estimation of epistasis in components of fitness in experimental populations of drosophila melanogaster II. Assessment of meiotic drive, viability, fecundity and sexual selection. Heredity 46: 347–377. 10.1038/hdy.1981.45 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Feldman M. W., Christiansen F. B., 1981. The estimation of epistasis in components of fitness in experimental populations of Drosophila melanogaster I. A two-stage maximum likelihood model. Heredity 46: 321–346. 10.1038/hdy.1981.44 [DOI] [PubMed] [Google Scholar]
- Dow M. A., 1976. The genetic basis of receptivity of yellow mutant Drosophila melanogaster females. Behav. Genet. 6: 141–143. 10.1007/BF01067144 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Radovic A., Wittkopp P. J., Long A. D., 2003. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 55: 53–72. 10.1002/neu.10196 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Cyran S. A., Viering M. M., Geyer P. K., Long A. D., 2006. A cis-regulatory sequence within the yellow locus of Drosophila melanogaster required for normal male mating success. Genetics 172: 1009–1030. 10.1534/genetics.105.045666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder A. F., Kryazhimskiy S., Plotkin J. B., 2014. Identifying signatures of selection in genetic time series. Genetics 196: 509–522 [corrigenda: Genetics 210: 1559 (2018)] 10.1534/genetics.113.158220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., Leuenberger C., Jensen J. D., Wegmann D., 2016. An approximate Markov model for the Wright-Fisher diffusion and its application to time series data. Genetics 203: 831–846. 10.1534/genetics.115.184598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Shim H., Jensen J. D., 2015. WFABC: a Wright-Fisher ABC-based approach for inferring effective population sizes and selection coefficients from time-sampled data. Mol. Ecol. Resour. 15: 87–98. 10.1111/1755-0998.12280 [DOI] [PubMed] [Google Scholar]
- Gompert Z., 2016. Bayesian inference of selection in a heterogeneous environment from genetic time-series data. Mol. Ecol. 25: 121–134. 10.1111/mec.13323 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler I. L., 1984. Inheritance of female mating propensities for yellow locus genotypes in Drosophila melanogaster. Genet. Res. 44: 133–149. 10.1017/S0016672300026343 [DOI] [Google Scholar]
- Hurvich C. M., Tsai C.-L., Jun N., 1989. Regression and time series model selection in small samples. Biometrika 76: 297–307. 10.1093/biomet/76.2.297 [DOI] [Google Scholar]
- Illingworth C. J., Parts L., Schiffels S., Liti G., Mustonen V., 2012. Quantifying selection acting on a complex trait using allele frequency time series data. Mol. Biol. Evol. 29: 1187–1197. 10.1093/molbev/msr289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C. J. R., Mustonen V., 2013. Quantifying selection in evolving populations using time-resolved genetic data. J. Stat. Mech. Theory Exp. P01004 Available at: https://iopscience.iop.org/article/10.1088/1742-5468/2013/01/P01004/meta. [Google Scholar]
- Lacerda M., Seoighe C., 2014. Population genetics inference for longitudinally-sampled mutants under strong selection. Genetics 198: 1237–1250. 10.1534/genetics.114.167957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas A. S., Malaspinas O., Evans S. N., Slatkin M., 2012. Estimating allele age and selection coefficient from time-serial data. Genetics 192: 599–607. 10.1534/genetics.112.140939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica J. M., Briscoe D. A., 1981. Effective population number estimates of laboratory populations of Drosophila melanogaster. Experientia 37: 947–948. 10.1007/BF01971769 [DOI] [Google Scholar]
- Mathieson I., McVean G., 2013. Estimating selection coefficients in spatially structured populations from time series data of allele frequencies. Genetics 193: 973–984. 10.1534/genetics.112.147611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L. D., Joshi A., Santos M., Rose M. R., 2013. Effective population size and evolutionary dynamics in outbred laboratory populations of Drosophila. J. Genet. 92: 349–361. 10.1007/s12041-013-0296-1 [DOI] [PubMed] [Google Scholar]
- Mukai T., Cardellino R. A., Watanabe T. K., Crow J. F., 1974. The genetic variance for viability and its components in a local population of Drosophila melanogaster. Genetics 78: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau H. J., Baccus R., 1981. Selection components of four allozymes in natural populations of Peromyscus maniculatus. Evolution 35: 11–20. 10.1111/j.1558-5646.1981.tb04854.x [DOI] [PubMed] [Google Scholar]
- Nadeau J. H., Dietz K., Tamarin R. H., 1981. Gametic selection and the selection component analysis. Genet. Res. 37: 275–284. 10.1017/S0016672300020279 [DOI] [Google Scholar]
- Schraiber J. G., Evans S. N., Slatkin M., 2016. Bayesian inference of natural selection from allele frequency time series. Genetics 203: 493–511. 10.1534/genetics.116.187278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Laurent S., Matuszewski S., Foll M., Jensen J. D., 2016. Detecting and quantifying changing selection intensities from time-sampled polymorphism data. G3 (Bethesda) 6: 893–904. 10.1534/g3.115.023200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegismund H. R., Christiansen H. B., 1985. Selection component analysis of natural polymorphisms using population samples including mother-offspring combinations, III. Theor. Popul. Biol. 27: 268–297. 10.1016/0040-5809(85)90002-4 [DOI] [PubMed] [Google Scholar]
- Sober E., Lewontin R. C., 1982. Artifact, cause and genic selection. Philos. Sci. 49: 157–180. 10.1086/289047 [DOI] [Google Scholar]
- Steinrücken M., Bhaskar A., Song Y. S., 2014. A novel spectral method for inferring general diploid selection from time series genetic data. Ann. Appl. Stat. 8: 2203–2222. 10.1214/14-AOAS764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst J., Schlötterer C., Song Y. S., 2015. Multi-locus analysis of genomic time series data from experimental evolution. PLoS Genet. 11: e1005069 10.1371/journal.pgen.1005069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless R. L., Messer P. W., Connallon T., Clark A. G., 2015. Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201: 425–431. 10.1534/genetics.115.177592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E. G., Slatkin M., 1999. Using maximum likelihood to estimate population size from temporal changes in allele frequencies. Genetics 152: 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Burnet B., Eastwood L., Connolly K., 1976. Behavioural pleiotropy of the yellow gene in Drosophila melanogaster. Genet. Res. 28: 75–88. 10.1017/S0016672300016748 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Our experimental data consists of the genotype counts in the five cage experiments and they are all reported in Figure 1. The ML inference framework was implemented in R. Supplemental material containing all the necessary scripts for reproducing the simulations and results are available at Figshare: https://doi.org/10.25386/genetics.7616171.
Figure 1.
Population sizes and genotype frequencies in our five cage experiments. Note that we define genotype frequencies relatively among males and relatively among females.