In most eukaryotes, the histone H3 variant CENP-A serves as the epigenetic mark for centromeres. CENP-A transcription is subject to cell-cycle regulation, but the molecular mechanism underlying the regulation remains elusive. Through a genetic screen...
Keywords: centromere, CENP-A, cell cycle transcriptional control, the MBF complex, Schizosaccharomyces pombe
Abstract
The centromere plays an essential role in chromosome segregation. In most eukaryotes, centromeres are epigenetically defined by the conserved histone H3 variant CENP-A. Proper centromere assembly is dependent upon the tight regulation of CENP-A level. Cell cycle regulation of CENP-A transcription appears to be a universal feature across eukaryotes, but the molecular mechanism underlying the temporal control of CENP-A transcription and how such regulation contributes to centromere function remains elusive. CENP-A in fission yeast has been shown to be transcribed before S phase. Using various synchronization methods, we confirmed that CENP-A transcription occurs at G1, leading to an almost twofold increase of the protein during S phase. Through a genetic screen, we identified the MBF (MluI box-binding factors) complex as a key regulator of temporal control of CENP-A transcription. The periodic transcription of CENP-A is lost in MBF mutants, resulting in CENP-A mislocalization and chromosome segregation defects. We identified the MCB (MluI cell cycle box) motif in the CENP-A promoter, and further showed that the MBF complex binds to the motif to restrict CENP-A transcription to G1. Mutations of the MCB motif cause constitutive CENP-A expression and deleterious effects on cell survival. Using promoters driving transcription to different cell cycle stages, we found that timing of CENP-A transcription is dispensable for its centromeric localization. Our data instead indicate that cell cycle-regulated CENP-A transcription is a key step to ensure that a proper amount of CENP-A is generated across generations. This study provides mechanistic insights into the regulation of cell cycle-dependent CENP-A transcription, as well as its importance on centromere function.
THE centromere is the special chromosomal locus where the kinetochore is assembled. The kinetochore interacts with the spindle microtubules to mediate equal segregation of sister chromatin to daughter cells during mitosis and meiosis (McKinley and Cheeseman 2016). In most eukaryotes, centromeres are epigenetically defined by a conserved histone H3 variant, CENP-A. CENP-A partially replaces the canonical histone H3 in centromeres and promotes the assembly of kinetochores (Black et al. 2007; Allshire and Karpen 2008). Proper centromere assembly is dependent upon the tight regulation of CENP-A levels. Overexpression of CENP-A in many organisms causes misincorporation of CENP-A into noncentromeric regions, leading to chromosome missegregation and growth defects (Heun et al. 2006; Olszak et al. 2011; Choi et al. 2012; Castillo et al. 2013; Gonzalez et al. 2014; Dong et al. 2016; Shrestha et al. 2017). CENP-A overexpression has been observed in a number of cancers, which might contribute to chromosome instability (Tomonaga et al. 2003; Li et al. 2007; Amato et al. 2009; Scott and Sullivan 2014; Zhang et al. 2016).
During replication, parental CENP-A appears to be partitioned equally between sister chromatids to be incorporated into two daughter centromeres (Jansen et al. 2007; Schuh et al. 2007; Black and Cleveland 2011), But the timing of the loading of newly synthesized CENP-A varies among different organisms. In plants and fission yeast Schizosaccharomyces pombe, new CENP-A is loaded into centromeres during G2 (Lermontova et al. 2006; Takayama et al. 2008; Lando et al. 2012; Gonzalez et al. 2013), whereas the loading of CENP-A in humans, Drosophila, and budding yeast occurs in G1, mitosis, and S phase, respectively (Pearson et al. 2004; Jansen et al. 2007; Mellone et al. 2011; Wisniewski et al. 2014). CENP-A transcription is also cell cycle-regulated. While CENP-A is transcribed in the G2/M window in humans, it occurs in G1/S phase in fission yeast. It appears that CENP-A transcription is generally uncoupled from canonical histone transcription (Shelby et al. 1997, 2000; Takahashi et al. 2000; Whitfield et al. 2002; Bar-Joseph et al. 2008; Rattray and Muller 2012; Grant et al. 2013). However, the molecular basis underlying cell cycle-regulated CENP-A transcription remain little known.
Interestingly, CENP-A expressed under the control of the H3 promoter fails to localize to the centromere and shows diffuse localization in the nucleus of human cells. This evidence prompted some to suggest that the timing of CENP-A expression, which is uncoupled from histone transcription during S phase, plays an important role in centromere targeting (Shelby et al. 1997). However, this hypothesis has not been formally tested. This study aims to unveil the mechanism behind CENP-A transcriptional regulation during the cell cycle, and its impact on CENP-A localization and function in fission yeast.
Here, we confirmed that CENP-A/Cnp1 in fission yeast is transcribed at G1 phase using multiple synchronization methods. We identified the MBF (MluI box-binding factors) complex as a key regulator of cell cycle-dependent CENP-A transcription. The MBF complex binds to the MCB (MluI cell cycle box) motif in the CENP-A promoter to restrict its transcription to G1 phase. However, using promoters driving transcription at different stages of the cell cycle, we found that timing of CENP-A transcription is dispensable for its centromere localization. Instead, our data indicate that cell cycle regulation of CENP-A transcription is a key step to ensure the proper of CENP-A generated across generations.
Materials and Methods
Strains, media, and genetic analysis
Fission yeast strains used in this study are listed in Supplemental Material, Table S1. Standard media and genetic analysis for fission yeast were used (Moreno et al. 1991). The mutant screen will be described in detail elsewhere (J. Yang and F. Li, unpublished data). Briefly, mutants from the Bioneer haploid deletion library were crossed with wild-type (WT) cells carrying Cnp1-GFP using a Rotor-HDA pinning robot (Singer Instruments). The resulting mutant cells carrying Cnp1-GFP were visually examined by fluorescence microscopy.
Western blot analysis
Western blot assays were performed as described with a few modifications (Huang et al. 2017). Briefly, cell extracts from exponentially growing cells were collected. Protein extracts were denatured in loading buffer (SDS 2%, glycerol 10%, Tris-Cl 60 mM, and 0.002% bromophenol blue), separated on 12% SDS-polyacrylamide gels, and transferred onto PVDF membranes. Membranes were blotted with primary anti-GFP (sc-9996; Santacruz) or α-tubulin (ab6160; Abcam) antibodies. Image J software was used for band quantification.
Cell synchronization
Centrifugal elutriations were conducted as previously described (Tormos-Pérez et al. 2016). Briefly, cells grown to the log phase at ∼23°, OD ∼1 at 600 nm, were collected and loaded into a Beckman J6 centrifuge with a JE−5.0 elutriation rotor. The smallest cells were eluted and were incubated at ∼23° for 6 hr. Samples for cell fixation (to determine septation index), RNA and protein extraction were collected every 15 min. Septation index was determined after fixing cells in 70% ethanol and staining with calcofluor (Sigma [Sigma Chemical], St. Louis, MO) and DAPI. For the cdc10 block-release experiments, cdc10-129 cells were grown at 25° to log phase and then shifted to 36° for 3.5 hr. Samples for RNA extraction were collected before and after the shift. For hydroxyurea (Sigma) treatment, exponentially growing cells were incubated in 10 mM hydroxyurea for 4 hr. Samples for RNA extraction were collected before and after hydroxyurea treatment.
Quantitative RT-PCR
RT-PCR assays were performed as previously described (He et al. 2017). Briefly, samples were homogenized using glass beads and a tube beater. Total RNA was purified using TRIzol. After treatment with RQ1 DNAse (Promega, Madison, WI), RNA was precipitated again with ethanol and used for cDNA synthesis with a PrimeScript RT Reagent Kit (Takara, Clontech). Next, 25 ng of cDNA were used for each quantitative PCR (qPCR) reaction using 2× SYBR Green qPCR Master Mix (Bimake). adh1 was used as a loading control. Fold change comparing to the corresponding experimental controls was calculated following the standard ΔΔCt method. Primers used for qPCR are listed in Table S2.
ChIP-qPCR
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Kong et al. 2016). Briefly, cells were grown to log phase at 30° and cross-linked by treatment with 1% formaldehyde. Immunoprecipitation was performed with protein A agarose (KPL) and anti-GFP antibody (ab290; Abcam). Next, 2 μl of ChIP or whole-cell extract samples were analyzed by qPCR using primers targeting hht1, cdc18, and cnp1 promoter regions. act1 was used as an internal control.
In situ chromatin-binding assay
In situ chromatin-binding assays were performed as described (Yang and Li 2018). Briefly, log-phase cells were collected and incubated in ZM buffer for 30 min. Cells were then washed twice with STOP buffer. Cells were resuspended with EB (extraction buffer) buffer ± 1% Triton X-100 at room temperature for 7 min, followed by fixation with 3.7% formaldehyde and 10% methanol.
Microscopy
Live or DAPI-stained fixed cells were imaged using a Delta Vision System (Applied Precision, Issaquah, WA) coupled to an Olympus IX71 microscope. Images were taken as z-stacks of 0.2-μm increments with an oil immersion objective (×100), and deconvolved using SoftWoRX2.50 software (Applied Precision).
Thiabendazole sensitivity assay
Serial dilutions (10-fold) of log phase cells were spotted on plates with 15 μg/ml thiabendazole (TBZ) (Sigma). Cell growth was estimated by taking pictures after incubating the plates for 72 and 92 hr at 30°.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7268501.
Results
Expression level of CENP-A is cell cycle-regulated in fission yeast
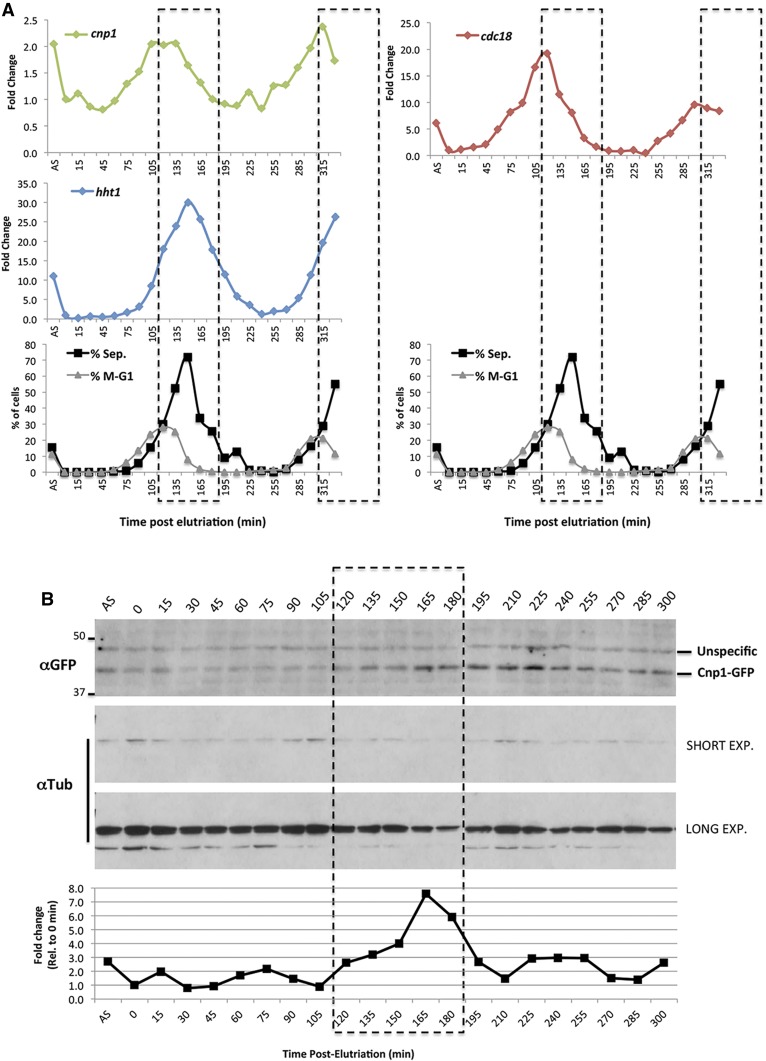
Using cells synchronized by a block-and-release approach in a cdc25-22 temperature-sensitive (ts) background, a previous report showed that the transcription of cnp1 appeared to be cell cycle-regulated in fission yeast. The mRNA is expressed around G1/S transition, peaking just before the onset of histone H3 transcription (Takahashi et al. 2000). To confirm the pattern of cnp1 expression, we synchronized WT cells at G2 phase by centrifugal elutriation and followed the expression of cnp1 mRNA from its native locus for 5.5 hr of synchronous growth using quantitative (q)RT-PCR. In fission yeast, the peak of septum-containing cells marks the time of the S phase. We found that most of the cells (∼80%) underwent the S phase 150 min after elutriation, based on the septation index. Accordingly, the peak of septation matches with the peak of expression of the histone H3 (hht1 gene) as histones are highly expressed during S phase (Figure 1A). Consistent with the previous report (Takahashi et al. 2000), cnp1 mRNA peaks before the transcription of histone H3, ∼120 min after elutriation, showing an approximately twofold increase. This pattern of expression coincides with the expression of cdc18, a well-known cell cycle-regulated gene expressed specifically in G1 phase (Figure 1A). We also observed the same transcription pattern in cells expressing Cnp1-GFP under its native promoter integrated next to the ade6 gene (Figure S1).
Figure 1.
Cell cycle-regulated expression of Cnp1. (A) Quantitative RT-PCR analysis of cnp1 expression in wild-type cells after synchronization by elutriation. Cells were synchronized in G2 by centrifugal elutriation and grown for 5 hr. Total mRNA was prepared at the indicated times postsynchronization. AS indicates a sample from the asynchronous population before elutriation. Upper panels show fold change of the indicated gene RNAs compared to a sample at 0 min. Lower panel show the percentage of septated cells and cells undergoing mitosis/G1 phase scored by DAPI and calcofluor staining at the indicated times for the same experiment. Dotted line indicates the S phase window determined by the septation index. (B) Western blot analysis of Cnp1 expression in cells after release from synchronization. Cell lysates were prepared from the same experiment described above, and processed for western blot analysis using antibodies specific for GFP and tubulin (as a loading control). Lower graph shows quantification of the Cnp1-GFP bands normalized to tubulin at each time point. Short and long time exposures (exp.) are shown for tubulin blotting.
To test whether the observed mRNA regulation has an impact on the protein level, we tagged Cnp1 with GFP under its native promoter and checked the levels of protein by western blotting using samples from the same synchronization method (Figure 1B). Cnp1-GFP protein increases approximately twofold at 165-min postelutriation. This result indicates that the mRNA increase in G1 results in augmentation of the Cnp1 protein level in S phase. Since Cnp1 in fission yeast has been suggested to be assembled into chromatin, mainly in G2 (Takayama et al. 2008; Lando et al. 2012), the timing of the increase of the Cnp1 protein level seem to be coordinated to provide the protein necessary for the next round of assembly.
We also checked the cnp1 mRNA levels in cells synchronized at G1 phase using a temperature-sensitive allele of the cdc10 gene, as well as in cells treated with hydroxyurea, which arrests cells at the beginning of S phase. cnp1 mRNA is increased in both G1 and S phase-arrested cells, whereas histone H3 transcripts are increased in S but not in G1-arrested cells (Figure S2). Other known G1 phase-regulated genes such as cdc18 and cdc22 show the same pattern as cnp1 (Figure S2). These results confirm that cnp1 is cell cycle-regulated at the mRNA and protein levels, and that its pattern of expression is uncoupled from H3 expression.
MBF complex mutants exhibit an abnormal CENP-A distribution pattern and chromosome missegregation
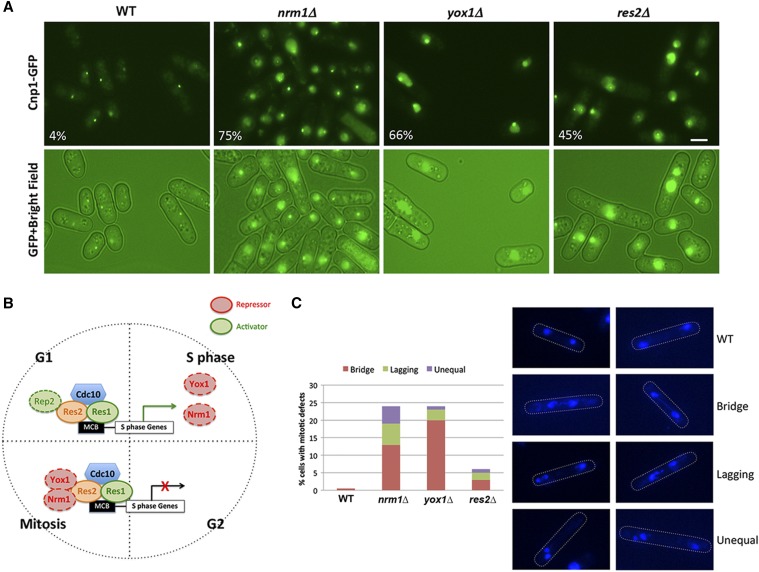
We recently initiated a visual genetic screen to identify factors involved in the regulation of CENP-A distribution using Cnp1-GFP as a visual marker. Three centromeres are clustered underneath the nuclear envelope near the spindle pole body in S. pombe WT interphase cells. Therefore, a single Cnp1-GFP focus is observed in the WT cells expressing Cnp1-GFP (Funabiki et al. 1993; He et al. 2016). Through the visual screen, we identified that three mutants, nrm1∆, yox1∆, and res2∆, showed abnormal CENP-A distribution patterns (Figure 2A). In these mutants, Cnp1-GFP at centromeres tends to be brighter and also gives a high nucleoplasmic signal (Figure 2A), suggesting an increase in Cnp1-GFP levels. Interestingly, the increased nuclear signal is only partially washed away by Triton X-100 treatment, indicating that the mislocalized Cnp1-GFP stably associates with chromatin regions other than centromeres (Figure S3). Genes corresponding to these mutants, res2, nrm1, and yox1, are all from the conserved MBF transcription factor complex. The MBF complex regulates the transcription of genes required for DNA replication during the G1/S transition of the cell cycle (Bahler 2005; Bertoli et al. 2013; Haase and Wittenberg 2014) (Figure 2B), which is the time when cnp1 mRNA levels peaks (Figure 1A). Res2, a DNA-binding protein, together with Res1 and Cdc10, forms the core of the MBF complex, and is involved in both activation and repression of the transcriptional activity (Ayté et al. 1995, 2001; Baum et al. 1997; Zhu et al. 1997; Whitehall et al. 1999; Dutta et al. 2008). Nrm1 and Yox1 are negative regulatory components of the MBF complex (Aligianni et al. 2009; Purtill et al. 2011).
Figure 2.
The MBF complex mutants show defects in CENP-A distribution and chromosome segregation. (A) Images of Cnp1-GFP localization in nrm1∆, yox1∆, and res2∆ mutants obtained by imaging live cells carrying the endogenous copy of cnp1 tagged with GFP. Bar, 2 μm. (B) Scheme showing the regulation of G1/S-expressed genes by the MBF complex. The core of the complex consists of Cdc10, Res1, and Res2, which bind to promoters containing MBF-binding motifs (MCB boxes). Rep2 normally acts as a cofactor for transcriptional activation. At the G1/S transition, the complex promotes the transcription of genes required for S phase, including Nrm1 and Yox1. The proteins coded by these two genes act as repressors of the MBF complex by binding to Res2 and limiting its transcriptional activity. This mechanism sets a negative feedback loop that restricts the activation of MBF targets during the rest of the cell cycle. (C) Overexpression of Cnp1-GFP causes chromosome segregation defects in mitotic cells. Right panel shows DAPI-staining images of nrm1∆, yox1∆, and res2∆ fixed cells. The percentage of mitotic cells showing bridge, lagging, or unequal chromosomes is indicated in the left panel. MBF, MluI box-binding factors; MCB, MluI cell cycle box; WT, wild-type.
It has been shown that increased Cnp1 levels can result in Cnp1 mislocalization, which affects chromosome segregation (Gonzalez et al. 2014). Using DAPI staining, we observed that the MBF repressor mutant cells displayed lagging chromosomes, chromosome bridges, and also unequal nuclei segregation during mitosis (Figure 2C and Figure S4). Around 25% of nrm1∆ and yox1∆ mutant cells displayed mitotic defects, while res2∆ mutants showed a relatively mild defect.
Cell cycle-regulated CENP-A transcription depends on the MBF complex
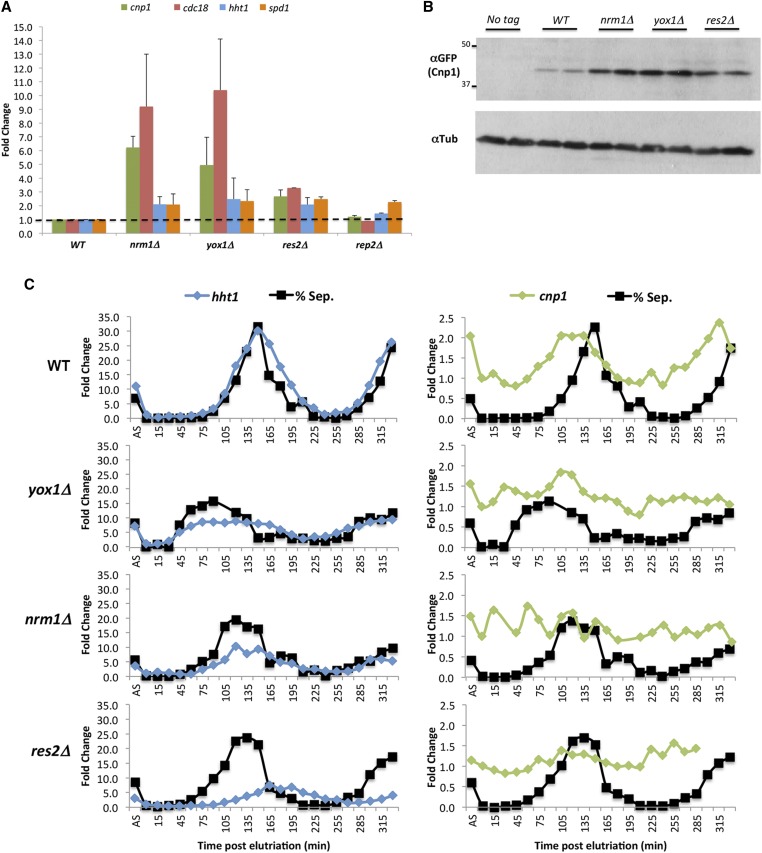
The pattern of cnp1 expression during the cell cycle and the phenotype observed in the MBF regulator mutants suggest that the MBF complex may be involved in the regulation of cnp1 expression. Accordingly, an increase in both cnp1 mRNA and protein levels was observed in asynchronous populations of nrm1, yox1, and res2 mutants (Figure 3, A and B). Deletion of nrm1 or yox1 resulted in a ∼5–6-fold increase in cnp1 mRNA levels, whereas res2 deletion led to a more modest twofold increase (Figure 3A). The magnitude of the observed defects (Figure 2, A and C) is correlated with the increase in cnp1 levels observed in these mutants (Figure 3, A and B).
Figure 3.
The MBF complex is required for cell cycle-regulated CENP-A transcription. (A) Quantitative RT-PCR analysis of cnp1 RNA expression in nrm1∆, yox1∆, res2∆, and rep2∆ cells. Total mRNA was prepared from asynchronous cells. Fold change for the indicated genes (different colored bars) was calculated compared to WT cells. (B) Western blot analysis of Cnp1-GFP levels for the MBF complex repressors mutants. Tubulin was used as a loading control. First two lines show blotting for a WT strain with no GFP tag on Cnp1 as control of antibody specificity. (C) Quantitative RT-PCR analysis of cnp1 mRNA levels throughout the cell cycle using synchronized nrm1∆, yox1∆, and res2∆ cells. Cells were synchronized by elutriation and grown for 5.5 hr. Total RNA was prepared and septation index scored every 15 min. Each graph shows the fold change for hht1 (left panels) or cnp1 (right panels) RNAs at the indicated times compared to a 0-min sample. Black curve on each graph shows the septation index of the corresponding cell population as reference. AS: asynchronous culture; MBF, MluI box-binding factors; WT, wild-type.
A similar increase was observed in the cdc18 mRNA, which is a known target of the MBF complex (Kelly et al. 1993). We noticed that H3 (hht1) and spd1 genes, which are expressed in the S and G2 phases, respectively, and mediated by different transcription complexes, also showed a slight increase in MBF mutants (Figure 3A). This suggests that mutation of these genes may disturb the cell cycle program of the cells, which is not unexpected since the MBF complex regulates the G1/S transition. Interestingly, mutation of rep2, which is required for the transcriptional activation of other MBF targets (Nakashima et al. 1995), does not cause a significant change in cnp1 levels (Figure 3A).
To further investigate how the MBF mutants affect the cell cycle regulation of cnp1, we examined cnp1 mRNA levels throughout the cell cycle using synchronized nrm1∆, yox1∆, and res2∆ mutant cells. We found that, in contrast to WT cells, the mutants do not show a clear peak of cnp1 expression right before septation (Figure 3C). However, we also noticed that these mutants do not synchronize as well as WT cells, as evidenced by the percentage of cells obtained at the peak of septation (∼70% in WT vs. 35, 45, and 50% in nrm1, yox1, and res2, respectively) (Figure 3C). In addition, the fold change and timing of H3 expression also seem to be affected. Histone H3 levels are regulated by an MBF target, ams2 (Rustici et al. 2004; Takayama and Takahashi 2007), so deletion of MBF repressors is expected to indirectly impact H3 levels. MBF also regulates a set of genes required for S phase, so the overall cell cycle program is also expected to be affected. Therefore, the observed defect on cnp1 mRNA in asynchronous cells is probably resulting from a combined effect of the direct disruption of cnp1 transcription control by MBF and an indirect effect caused by a cell cycle distribution defect on the MBF mutants. Overall, these results show that mutation of MBF repressor genes leads to the deregulation and increase of cnp1 levels, and mitotic defects.
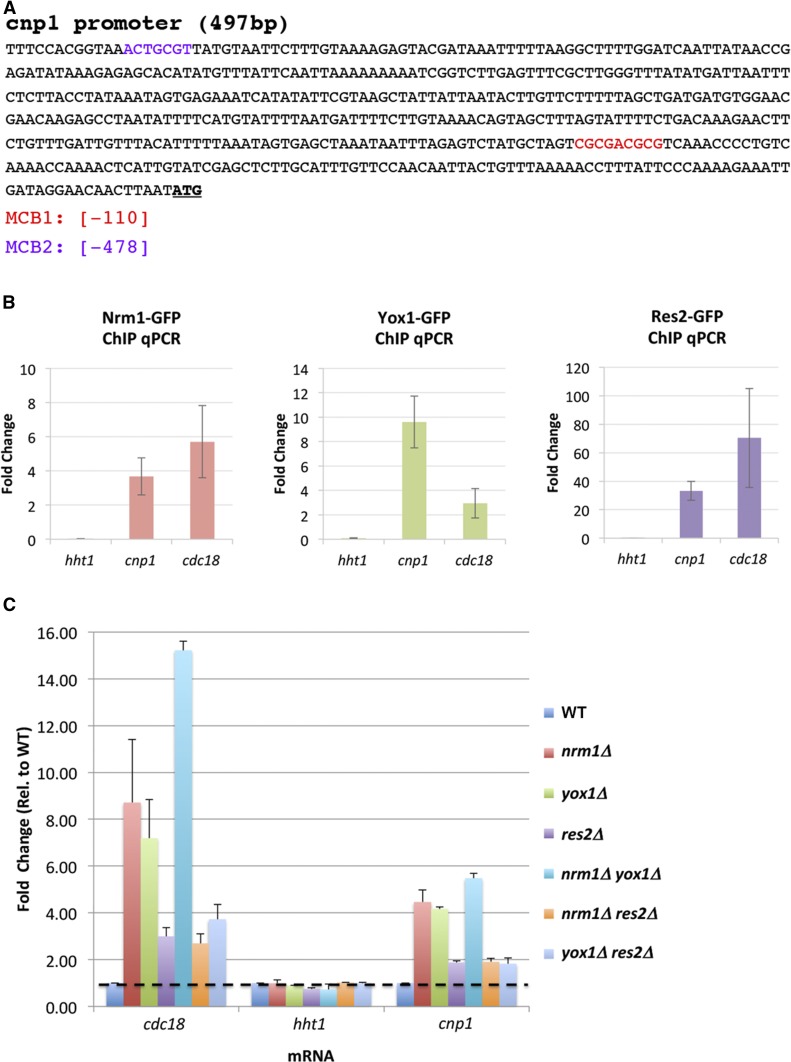
The MBF complex binds to an MCB motif in the cnp1 promoter
To investigate how the MBF complex regulates cnp1 transcription, we tested the binding of several MBF factors to the cnp1 promoter by ChIP. The MBF complex is known to target the promoters of genes such as cdc18 and cdc22. The promoter motifs bound by the MBF complex are called MCB motifs (Lowndes et al. 1992). Our in silico analysis revealed that the cnp1 promoter contains two putative MCB boxes at −110 and −478-bp upstream of the start codon, which we named MCB1 and MCB2, respectively (Figure 4A). ChIP analysis showed that Nrm1, Yox1, and Res2 bind to the cnp1 promoter (Figure 4B). Nrm1 and Yox1 are known to form a dimer that suppresses the transcriptional activity of the MBF complex outside of G1 by interacting with Res2 (de Bruin et al. 2006; Aligianni et al. 2009; Caetano et al. 2011). Consistent with these studies, we found that the nrm1∆ yox1∆ double mutants show roughly the same elevated level of cnp1 mRNA as the single mutants (Figure 4C). The observation confirmed that Nrm1 and Yox1 are functionally dependent on each other in suppressing cnp1 expression. Furthermore, the nrm1∆ res2∆ and yox1∆ res2∆ double mutants display the same cnp1 mRNA level as the res2∆ single mutant (Figure 4C), suggesting that Nrm1 and Yox1 suppress cnp1 levels by interacting with Res2.
Figure 4.
The MBF complex binds to the MCB motif on the cnp1 promoter. (A) Map of putative MCB boxes in the cnp1 promoter. MCB1 and MCB2 coordinates are indicated. ATG in bold letters marks the Cnp1 start codon. (B) Nrm1, Yox1, and Res2 bind to the promoter region of cnp1. ChIP assays were performed using WT cells carrying Nrm1-GFP, Yox1-GFP, or Res2-GFP using primers specific for the promoter region of cnp1, hht1, and cdc18. hht1 and cdc18 were used as negative and positive controls, respectively (C) Quantitative RT-PCR analysis of cnp1 mRNA levels in cells with single or double mutations of the MBF complex. Fold change compared to WT cells was calculated for cdc18, hht1, and cnp1 RNAs. Bars on different colors correspond to the fold change in the indicated mutants. ChIP, chromatin immunoprecipitation; MBF, MluI box-binding factors; MCB, MluI cell cycle box; qPCR, quantitative PCR; WT, wild-type.
The MBF complex restricts CENP-A transcription to G1 phase
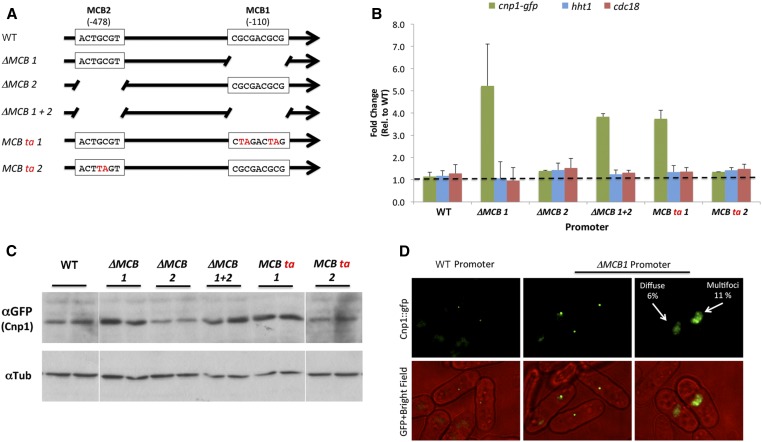
To probe the functionality of the two putative MCB boxes in the cnp1 promoter, we made single (∆MCB1 and ∆MCB2) and double deletions of the boxes (∆MCB 1+2) (Figure 5A). We also mutagenized the core CG nucleotides in each MCB box into TA (MCB ta 1 and MCB ta 2) as an alternative way to disturb the putative binding of MBF proteins (Figure 5A). The cnp1 gene containing these mutations was tagged with GFP and introduced at the ade6 site by homologous recombination. Our qRT-PCR analysis showed that mutation or deletion of the MCB1 box (located −110 bp from the start codon) results in a four-to-fivefold increase in the cnp1 mRNA level (Figure 5B). Mutation of the MCB2 box (−478 bp from the start codon) does not have any detectable effect on the mRNA levels of cnp1 and therefore this box was considered to not be functional. The increase in mRNA levels in the MCB1 mutant also results in an increase in protein levels (Figure 5C). Interestingly, mutation of the MCB1 mimics the increase in cnp1 levels observed in the MBF repressor mutants (nrm1, yox1, and res2). Even though most of the MCB1 mutant cells show a single Cnp1-GFP focus corresponding to its normal centromeric localization, we observed several differences when comparing with cnp1-gfp under the control of the WT promoter. The MCB1 mutant cells display brighter centromeric foci with an average increase of twofold in signal intensity, suggesting an increase in the amount of Cnp1 assembled at the centromere (Figure 5D and Figure S5). Additionally, we observed that some of these mutant cells show either diffuse or multifoci Cnp1-GFP localization (Figure 5D). These results suggest that mutation of the MCB1 box leads to CENP-A overexpression and defective localization.
Figure 5.
Cell cycle-regulated expression of Cnp1 is abnormal when the MCB1 box is mutated. (A) Schematic representation of the cnp1 promoter mutant constructs. Predicted MCB box 1 or 2 was disrupted by either deletion or substitution of the core GC bases to TA. (B) Quantitative RT-PCR analysis of cnp1-gfp, hht1, and cdc18 RNAs in cells carrying cnp1-gfp under control of a promoter with the indicated mutations in the MCB boxes. hht1 and cdc18 were used as negative controls since their expression should not be affected by mutations in the cnp1 promoter (C) Western blot analysis of Cnp1-GFP level from the same experiment shown in (B). Tubulin was used as a loading control. (D) Images of Cnp1-GFP in live cells carrying the WT cnp1 promoter or ∆MCB1 promoter. Percentages of cells showing the indicated abnormal Cnp1 localization are indicated. MCB, MluI cell cycle box; WT, wild-type.
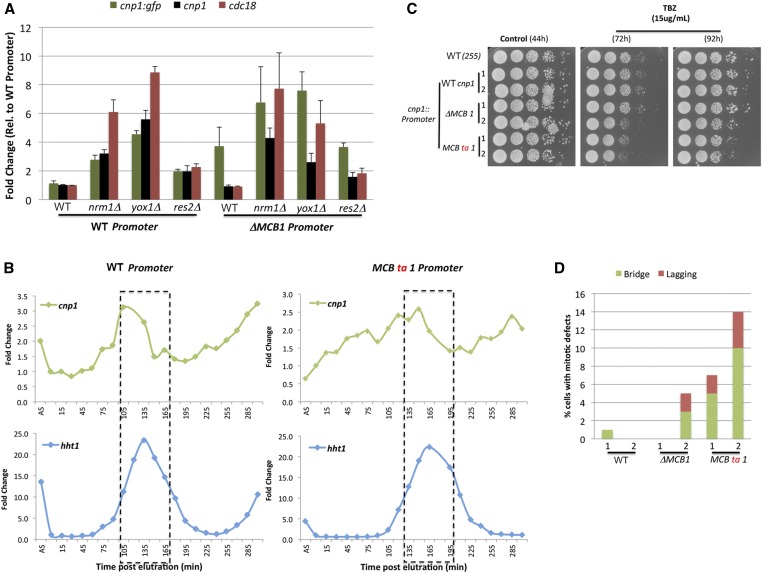
Based on these observations, we hypothesized that an intact MCB1 box is required for the binding of the MBF repressors to the cnp1 promoter to restrict the expression of cnp1. To test this idea, we checked the expression of the cnp1-gfp reporter gene (extra copy, inserted at the ade6 site) with a WT or MCB1 mutant promoter in the background of the MBF repressor mutants (Figure 6A). Mutation of the MBF repressors causes an increase in the expression of the cnp1-gfp reporter gene with the WT promoter (three-, five-, and twofold for nrm1Δ, yox1Δ, and res2Δ, respectively). A similar increase was observed for the cnp1 endogenous copy. However, in cells carrying cnp1-gfp controlled by the promoter with MCB1 deletion, mutation of nrm1 and yox1 caused just a modest increase of 1.5- and 1.7-fold change, respectively, and no increase was observed in res2 mutants. This indicates that the MBF repressors require the MCB1 box to fully repress cnp1 levels.
Figure 6.
The MBF complex restricts CENP-A transcription to G1 phase. (A) Quantitative RT-PCR analysis of cnp1 RNA levels in cells with a WT or ∆MCB1 cnp1 promoter in combination with mutation of MBF genes. Cells carrying an extra copy of cnp1 tagged with GFP under control of the WT or ∆MCB1 cnp1 promoter were crossed with mutants for nrm1, yox1, or res2 genes. Graph shows RNA fold changes for the cnp1-gfp transgene, endogenous cnp1 and cdc18 genes for the indicated genotypes. Fold change was calculated normalizing to levels in WT cells with the cnp1-gfp under the WT cnp1 promoter. (B) cnp1 and hht1 mRNA levels measured by quantitative RT-PCR in synchronized cells with a WT or MCB1 box mutant promoter on cnp1. Dotted line indicates the time window when most of the cells are undergoing S phase, measured by the septation index. (C) TBZ sensitivity assays for Cnp1 promoter mutants. Serial dilutions of indicated strain cultures were plated onto YES medium supplemented with 15 μg/ml TBZ or control YES medium without TBZ. Pictures of colony growth were taken 72 or 96 hr postplating. (D) Overexpression of Cnp1-GFP causes chromosome segregation defects in MCB1 mutant cells. Cells were fixed and stained with DAPI staining. The percentage of mitotic cells showing bridged, and lagging chromosomes was quantified as in Figure 2C. MBF, MluI box-binding factors; MCB, MluI cell cycle box; TBZ, thiabendazole; WT, wild-type; YES, yeast extract with supplements.
To further test the functional consequences of disrupting the MCB1 box, we mutated this box in the cnp1 endogenous gene. This caused disturbed expression of cnp1 during the cell cycle in a synchronized cell population (Figure 6B), with cnp1 RNA levels being induced well in advance of the G1/S transition. This suggests that the MCB1 box is required to restrict cnp1 expression to the G1 phase. We also noticed that cells carrying the MCB1 deletion showed an ∼30-min delay in the cell cycle with most of the cells undergoing S phase (hht1 RNA peak) ∼165 min postelutriation, compared to 135 min in WT cells. As observed with the reporter gene, mutation or deletion of the endogenous MCB1 box resulted in an increase in the levels of cnp1 (Figure S6). The magnitude of this increase was variable among different clones containing the same MCB1 mutation. When we subjected these cells to mitotic stress by treatment with the microtubule polymerization inhibitor TBZ, we observed a slight but consistent decrease in growth of the cells (Figure 6C). This was correlated with the presence of bridged and lagging chromosomes in these mutants (Figure 6D). The cells showing slower growth on TBZ and more mitotic errors were the ones expressing higher levels of cnp1 mRNA (Figure S6), suggesting that tight regulation of cnp1 levels is required to avoid mitotic defects. Together, these results demonstrate that the MBF repressor genes bind to the cnp1 promoter via the MCB1 box to restrict its expression, and avoid cnp1 overexpression and chromosome missegregation.
The timing of CENP-A expression is dispensable for its targeting to centromeres
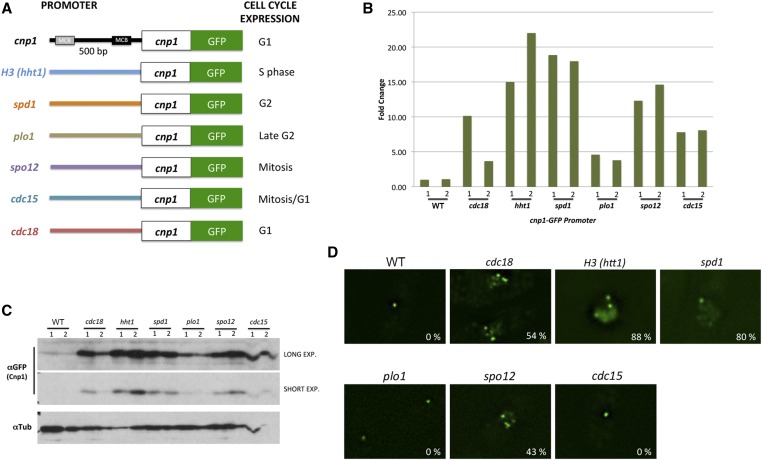
We next tested the previously suggested hypothesis that the timing of CENP-A transcription contributes to the proper targeting of cnp1 (Shelby et al. 1997). This hypothesis was proposed based on the observation that forcing expression of CENP-A in S phase using the histone H3 promoter leads to the mislocalization of the protein in human cells (Shelby et al. 1997). To determine whether the timing of CENP-A expression is important for its targeting to centromeres, we expressed a cnp1-gfp reporter gene under the control of a variety of promoters that drive expression at different stages of the cell cycle (Figure 7A). They include promoters from histone H3 (hht1), spd1, plo1, spo12, cdc15, and cdc18, which direct transcription at S, G2, late G2, mitosis, mitosis/G1, and G1 phase, respectively. cnp1-gfp mRNA and protein levels produced by the different promoters were analyzed (Figure 7, B and C). We also analyzed the localization of Cnp1-GFP expressed from different promoters (Figure 7D). In agreement with the previous report in human cells (Shelby et al. 1997), expression of cnp1-gfp under control of the histone H3 promoter led to multiple foci of Cnp1-GFP with strong signal in nuclei, suggesting ectopic localization of the protein. Multiple Cnp1-GFP foci were also found in cells expressing Cnp1-GFP under the spd1, spo12, and cdc18 promoters. However, most cells expressing Cnp1-GFP driven from the plo1 and cdc15 promoter contain a single Cnp1-GFP focus, though the cdc15 promoter results in a brighter Cnp1-GFP spot. Accordingly, we found that Cnp1-GFP is highly expressed in cells carrying Cnp1-GFP under the hht1, spd1, and spo12 promoter (Figure 7, B and C). Expression driven by these promoters is higher compared to the plo1 and cdc15 promoters. These results indicate that the distribution pattern of CENP-A correlates with the strength of the promoters, rather than the timing of CENP-A expression for its association with centromeres. Consistent with this, a strain with cnp1-gfp under control of the cdc18 promoter, which generates an ∼10-fold increase in cnp1 mRNA (Figure 7B), exhibited ectopic localization of Cnp1-GFP. However, a second strain generated with the cdc18 promoter showed only a fourfold increase in cnp1 mRNA and displayed WT-like single foci Cnp1-GFP localization (data not shown). This result is especially interesting since the cdc18 promoter drives expression at G1, the same timing as the cnp1 promoter. The previously reported mislocalization of CENP-A driven by the histone H3 promoter is likely due to overexpression rather than misregulation of the timing of CENP-A expression. This result suggests that tight cell cycle regulation of CENP-A transcription ensures that the proper level of CENP-A is generated.
Figure 7.
The timing of CENP-A expression is dispensable for its centromeric targeting. (A) Schematic diagram of the cnp1-gfp constructs under promoters that drive expression at different cell cycle stages. (B) Quantitative RT-PCR analysis of RNA levels of cnp1-gfp driven by different promoters. Graphic shows fold change compared to the WT cnp1 promoter. (C) Western blotting analysis of Cnp1-GFP protein levels expressed by different promoters. Tubulin was used as a loading control. Short and long exposures are shown for anti-GFP blotting. (D) Distribution patterns of Cnp1-GFP expressed from different promoters. Percentage of cells showing multifoci phenotype is indicated for each sample. Exp., exposure; WT, wild-type.
Discussion
Cell cycle regulation of CENP-A transcription appears to be a universal feature across eukaryotes, but how transcription of CENP-A is regulated remains elusive. Here, we confirmed that CENP-A transcription in fission yeast peaks during G1 phase. We further identified components of the MBF complex, including Nrm1, Yox1, and Res2, that are required for cell cycle-dependent CENP-A transcription regulation. The MBF complex binds to the MCB motif in the CENP-A promoter to restrict its transcription to G1 phase (Figure 8). However, CENP-A can still target to centromeres when forced to express at non-G1 phase, indicating that timing of CENP-A transcription is not essential for its centromere localization. We conclude that cell cycle regulation of CENP-A transcription is a key control mechanism to ensure that the proper amount of CENP-A is generated across generations.
Figure 8.
Model for cell cycle-regulated cnp1 transcription by the MBF complex. During the G1/S transition, the MBF core complex (Cdc10, Res1, and Res2) binds to the MCB motif in the cnp1 promoter to activate cnp1 transcription. The MBF complex also induces the transcription of the yox1 and nrm1 genes, as well as other genes required for DNA replication. Yox1 and Nrm1 form a dimer and bind to the MBF core complex through interacting with Res2. The binding of Nrm1 and Yox1 inhibits the transcriptional induction activity of the complex, and establishes a negative feedback loop preventing the constitutive activation of cnp1 for the rest of the cell cycle. In the absence of Nrm1 and Yox1, the MBF activator core complex remains active throughout the cell cycle, leading to an aberrant accumulation of cnp1 transcripts. If the level of Cnp1 surpasses a certain threshold, the cell starts showing mitotic defects as a consequence of Cnp1 mislocalization. MBF, MluI box-binding factors; MCB, MluI cell cycle box.
Using a visual genetic screen, we identified nrm1∆, yox1∆, and res2∆ mutants that exhibited excessive CENP-A-GFP and chromosome segregation defects. Nrm1, Yox1, and Res2 are components of the MBF complex, which regulates transcription during the G1/S transition of the cell cycle. We showed that CENP-A mRNA is significantly upregulated in nrm1∆ and yox1∆, consistent with its role as a negative regulator of the MBF complex. Deletion of res2 has a weaker effect on CENP-A transcription and chromosome segregation, likely resulting from the fact that Res2 has both negative and positive roles in MBF-mediated transcription. Cdc10, another component of the MBF complex, also has a dual role in regulating gene expression during G1/S (McInerny et al. 1995). Accordingly, we found that CENP-A transcription is abnormal in cdc10 mutant cells (Figure S2). Rep2 is considered to be the transcriptional activator of other MBF targets (Nakashima et al. 1995), but disruption of Rep2 does not have a strong effect on cnp1 transcription. This suggests that Cnp1 regulation by the MBF proteins does not totally fit with the canonical regulation of the complex.
As expected, we found that the MBF complex binds to the promoter region of CENP-A. Mutations in the putative MBF-binding MCB motif −110-bp upstream of the start codon abolished the cell cycle transcription pattern of CENP-A, and resulted in its deregulated expression and a higher level of CENP-A transcripts, demonstrating that it is a bona fide MCB box. Disruption of the MCB box at its endogenous site gave rise to a cell cycle delay, TBZ sensitivity, and mitotic defects, indicative of chromosome missegregation. Our data suggest that the MBF repressors bind to the MCB box at the CENP-A promoter to restrict its expression to G1 phase, which prevents cnp1 overexpression. The second putative MCB motif, localized at −475-bp upstream of the start codon appears not to be functional.
The level of CENP-A is tightly controlled. CENP-A overexpression leads to erroneous deposition of CENP-A to noncentromeric regions. This results in chromosome missegregation and growth defects. Ubiquitin-dependent proteolysis has been shown to be a conserved post-translational mechanism to degrade excessive CENP-A (Collins et al. 2004; Moreno-Moreno et al. 2006; Hewawasam et al. 2010; Ranjitkar et al. 2010; Au et al. 2013; Gonzalez et al. 2014; Yang et al. 2018). This study demonstrates that transcriptional regulation of CENP-A is another important control mechanism to ensure that the proper amount of CENP-A is expressed. Disruption of the CENP-A transcription machinery also generates mistargeted CENP-A and chromosome segregation defects. Nevertheless, these mutants also show cell cycle deregulation of multiple genes required for S phase progression (Caetano et al. 2014). We cannot exclude the possibility that the observed mitotic phenotypes in the mutants could be, at least partially, due to replicative stress.
Interestingly, CENP-A transcription and the loading of CENP-A usually occur in different stages of the cell cycle. In fission yeast, Cnp1 is transcribed at G1, resulting in the protein pool increasing by approximately twofold during S phase. The increase in Cnp1 probably occurs in preparation for the next round of Cnp1 assembly that occurs in G2. The timing of CENP-A transcription is also often different from canonical histone transcription. It has been proposed that the restriction of histone gene transcription to S phase may prevent regular histones from competing with histone variants for incorporation into the chromosomes of nonreplicating cells (Bahler 2005). CENP-A in fission yeast is expressed at G1/S before histone transcription. How CENP-A is prevented from assembling into noncentromeric nucleosomes during replication is unclear. In addition to ubiquitin-dependent proteolysis, the nucleosome assembly protein (NAP) domain-containing Ccp1 and the chromatin remodeling complexes, such as facilitates chromatin transcription (FACT) and human histone cell cycle regulator (HIRA), also play important roles in removing mistargeted CENP-A (Choi et al. 2012; Deyter and Biggins 2014; Dong et al. 2016; Ciftci-Yilmaz et al. 2018). Perhaps these factors maintain the integrity of chromosome arms by excluding ectopic CENP-A during replication.
It has been shown that CENP-A expressed under the promoter of histone H3 in human cells is distributed throughout the nucleus. The observation led to the hypothesis that the timing of CENP-A expression acts as an important component of the CENP-A-targeting mechanism (Shelby et al. 1997, 2000). Our study also showed that cells expressing Cnp1 under the histone H3 promoter display excessive Cnp1-GFP foci with diffuse nuclear signal in fission yeast. However, Cnp1 driven by the plo1 and cdc15 promoters, which induce transcription at late G2 and mitosis/G1, respectively, can properly target to centromeres, indicating that the timing of CENP-A transcription is not absolutely required for its centromere localization. Consistent with this, deletion of the MCB1 box results in constitutive overexpression of CENP-A, but the protein is still assembled mostly at centromeres. Careful examination reveals that the distribution pattern of CENP-A driven by different promoters correlates with its expression level. Strong promoters, such as that of histone H3, lead to an abnormal distribution of CENP-A. On the other hand, CENP-A expressed from weaker promoters, including these of plo1 and cdc15, still targets to centromeres. We noted that the expression levels expressed from the plo1 and cdc15 promoters are modestly higher than the level expressed by the native CENP-A promoter. These data are consistent with previous studies showing that when weakly overexpressed, CENP-A in fission yeast still associates specifically with centromeres (Choi et al. 2012; Gonzalez et al. 2014). Together, our results support our model that precise control of the timing of CENP-A transcription ensures that the proper level of CENP-A is expressed, rather than its targeting to centromeres.
Interestingly, we often observed significant variation in the level of Cnp1 expression in different clones carrying the same mutations of the MCB1 box in the endogenous cnp1 promoter. Clones with higher Cnp1 overexpression exhibited increased TBZ sensitivity and mitotic defects. This result suggests that there may be a threshold of tolerance for Cnp1 overexpression, above which cells are strongly selected against. This may lead to clonal variation by selecting cells that activate mechanisms preventing excessive Cnp1.
The MBF complex is functionally analogous to the E2F complex in plants and metazoans, which has been shown to control the G1/S and also G2/M transitions (Zhu et al. 2004; Bertoli et al. 2013). In Arabidopsis, E2fa binds to the promoter of CENP-A in vivo, and mutating the E2F-binding sites at the promoter results in increased reporter gene activity (Heckmann et al. 2011). Putative E2F-binding sites were also found in the human CENP-A promoter by an in silico study (Amato et al. 2009). Downregulation of E2F/RBR in human cells leads to increased RNA and protein levels of CENP-A (Sullivan et al. 2011). Thus, the mechanism underlying MBF-mediated transcriptional regulation of CENP-A expression may be conserved in plants and humans.
Acknowledgments
We thank Q. Dong for critical reading of the manuscript, the Japan Yeast Genetic Resource Center of Japan for kindly providing the strains used in this study, and J. Ayté for discussion and input in some results, as well as providing some strains. F.L. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. This project was supported by National Institutes of Health grant R01 GM-106037 (to F.L.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7268501.
Communicating editor: M. Freitag
Literature Cited
- Aligianni S., Lackner D. H., Klier S., Rustici G., Wilhelm B. T., et al. , 2009. The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Genet. 5: e1000626 10.1371/journal.pgen.1000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Karpen G. H., 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9: 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A., Schillaci T., Lentini L., Di Leonardo A., 2009. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., et al. , 2013. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayté J., Leis J. F., Herrera A., Tang E., Yang H., et al. , 1995. The Schizosaccharomyces pombe MBF complex requires heterodimerization for entry into S phase. Mol. Cell. Biol. 15: 2589–2599. 10.1128/MCB.15.5.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayté J., Schweitzer C., Zarzov P., Nurse P., DeCaprio J. A., 2001. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat. Cell Biol. 3: 1043–1050. [DOI] [PubMed] [Google Scholar]
- Bahler J., 2005. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 39: 69–94. [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z., Siegfried Z., Brandeis M., Brors B., Lu Y., et al. , 2008. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc. Natl. Acad. Sci. USA 105: 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Wuarin J., Nurse P., 1997. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 16: 4676–4688. 10.1093/emboj/16.15.4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J. M., de Bruin R. A., 2013. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14: 518–528. 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Cleveland D. W., 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. E., Jansen L. E., Maddox P. S., Foltz D. R., Desai A. B., et al. , 2007. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25: 309–322. 10.1016/j.molcel.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Caetano C., Klier S., de Bruin R. A., 2011. Phosphorylation of the MBF repressor Yox1p by the DNA replication checkpoint keeps the G1/S cell-cycle transcriptional program active. PLoS One 6: e17211 10.1371/journal.pone.0017211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano C., Limbo O., Farmer S., Klier S., Dovey C., et al. , 2014. Tolerance of deregulated G1/S transcription depends on critical G1/S regulon genes to prevent catastrophic genome instability. Cell Rep. 9: 2279–2289. 10.1016/j.celrep.2014.11.039 [DOI] [PubMed] [Google Scholar]
- Castillo A. G., Pidoux A. L., Catania S., Durand-Dubief M., Choi E. S., et al. , 2013. Telomeric repeats facilitate CENP-A(Cnp1) incorporation via telomere binding proteins. PLoS One 8: e69673 (erratum: Cell 8). 10.1371/journal.pone.0069673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. S., Stralfors A., Catania S., Castillo A. G., Svensson J. P., et al. , 2012. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast. PLoS Genet. 8: e1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S., Au W. C., Mishra P. K., Eisenstatt J. R., Chang J., et al. , 2018. A genome-wide screen reveals a role for the HIR histone chaperone complex in preventing mislocalization of budding yeast CENP-A. Genetics 210: 203–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14: 1968–1972. 10.1016/j.cub.2004.10.024 [DOI] [PubMed] [Google Scholar]
- de Bruin R. A., Kalashnikova T. I., Chahwan C., McDonald W. H., Wohlschlegel J., et al. , 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23: 483–496. [DOI] [PubMed] [Google Scholar]
- Deyter G. M., Biggins S., 2014. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 28: 1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Yin F. X., Gao F., Shen Y., Zhang F., et al. , 2016. Ccp1 homodimer mediates chromatin integrity by antagonizing CENP-A loading. Mol. Cell 64: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C., Patel P. K., Rosebrock A., Oliva A., Leatherwood J., et al. , 2008. The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol. Cell. Biol. 28: 5977–5985. 10.1128/MCB.00596-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M., 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121: 961–976. 10.1083/jcb.121.5.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., He H., Sun S., Li C., Li F., 2013. Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proc. Natl. Acad. Sci. USA 110: 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., He H., Dong Q., Sun S., Li F., 2014. Ectopic centromere nucleation by CENP-A in fission yeast. Genetics 198: 1433–1446. 10.1534/genetics.114.171173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G. D., Brooks L., III, Zhang X., Mahoney J. M., Martyanov V., et al. , 2013. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell 24: 3634–3650. 10.1091/mbc.e13-05-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase S. B., Wittenberg C., 2014. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 196: 65–90. 10.1534/genetics.113.152595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Zhang S., Wang D., Hochwagen A., Li F., 2016. Condensin promotes position effects within tandem DNA repeats via the RITS complex. Cell Rep. 14: 1018–1024. 10.1016/j.celrep.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Li Y., Dong Q., Chang A. Y., Gao F., et al. , 2017. Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex. Proc. Natl. Acad. Sci. USA 114: 12524–12529. 10.1073/pnas.1712961114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann S., Lermontova I., Berckmans B., De Veylder L., Baumlein H., et al. , 2011. The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J. 68: 646–656. 10.1111/j.1365-313X.2011.04715.x [DOI] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M. D., Weiss S., Skora A. D., et al. , 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10: 303–315. 10.1016/j.devcel.2006.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40: 444–454. 10.1016/j.molcel.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Zhang P., Li W., Zhao T., Zhang Z., et al. , 2017. G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 8: e2726 10.1038/cddis.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L. E., Black B. E., Foltz D. R., Cleveland D. W., 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176: 795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., et al. , 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74: 371–382. 10.1016/0092-8674(93)90427-R [DOI] [PubMed] [Google Scholar]
- Kong L., Zhang P., Li W., Yang Y., Tian Y., et al. , 2016. KDM1A promotes tumor cell invasion by silencing TIMP3 in non-small cell lung cancer cells. Oncotarget 7: 27959–27974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Endesfelder U., Berger H., Subramanian L., Dunne P. D., et al. , 2012. Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol. 2: 120078 10.1098/rsob.120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Fuchs J., Klatte S., Macas J., et al. , 2006. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451. 10.1105/tpc.106.043174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. M., Liu X. H., Cao X. Z., Wang L., Zhu M. H., 2007. Expression of centromere protein A in hepatocellular carcinoma. Zhonghua Bing Li Xue Za Zhi 36: 175–178. [PubMed] [Google Scholar]
- Lowndes N. F., McInerny C. J., Johnson A. L., Fantes P. A., Johnston L. H., 1992. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature 355: 449–453. 10.1038/355449a0 [DOI] [PubMed] [Google Scholar]
- McInerny C. J., Kersey P. J., Creanor J., Fantes P. A., 1995. Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res. 23: 4761–4768. 10.1093/nar/23.23.4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K. L., Cheeseman I. M., 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17: 16–29. 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B. G., Grive K. J., Shteyn V., Bowers S. R., Oderberg I., et al. , 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 7: e1002068 10.1371/journal.pgen.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., Azorin F., 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34: 6247–6255. 10.1093/nar/gkl902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N., Tanaka K., Sturm S., Okayama H., 1995. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 14: 4794–4802. 10.1002/j.1460-2075.1995.tb00161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak A. M., van Essen D., Pereira A. J., Diehl S., Manke T., et al. , 2011. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat. Cell Biol. 13: 799–808. 10.1038/ncb2272 [DOI] [PubMed] [Google Scholar]
- Pearson C. G., Yeh E., Gardner M., Odde D., Salmon E. D., et al. , 2004. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14: 1962–1967. 10.1016/j.cub.2004.09.086 [DOI] [PubMed] [Google Scholar]
- Purtill F. S., Whitehall S. K., Williams E. S., McInerny C. J., Sharrocks A. D., et al. , 2011. A homeodomain transcription factor regulates the DNA replication checkpoint in yeast. Cell Cycle 10: 664–670. 10.4161/cc.10.4.14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40: 455–464. 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. M., Muller B., 2012. The control of histone gene expression. Biochem. Soc. Trans. 40: 880–885. 10.1042/BST20120065 [DOI] [PubMed] [Google Scholar]
- Rustici G., Mata J., Kivinen K., Lio P., Penkett C. J., et al. , 2004. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 36: 809–817. 10.1038/ng1377 [DOI] [PubMed] [Google Scholar]
- Schuh M., Lehner C. F., Heidmann S., 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17: 237–243. 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Scott K. C., Sullivan B. A., 2014. Neocentromeres: a place for everything and everything in its place. Trends Genet. 30: 66–74. 10.1016/j.tig.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R. D., Vafa O., Sullivan K. F., 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136: 501–513. 10.1083/jcb.136.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R. D., Monier K., Sullivan K. F., 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151: 1113–1118. 10.1083/jcb.151.5.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R. L., Ahn G. S., Staples M. I., Sathyan K. M., Karpova T. S., et al. , 2017. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8: 46781–46800. 10.18632/oncotarget.18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L. L., Boivin C. D., Mravinac B., Song I. Y., Sullivan B. A., 2011. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 19: 457–470. 10.1007/s10577-011-9208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E. S., Yanagida M., 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. 10.1126/science.288.5474.2215 [DOI] [PubMed] [Google Scholar]
- Takayama Y., Takahashi K., 2007. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 35: 3223–3237. 10.1093/nar/gkm213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y., Sato H., Saitoh S., Ogiyama Y., Masuda F., et al. , 2008. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell 19: 682–690. 10.1091/mbc.e07-05-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., et al. , 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63: 3511–3516. [PubMed] [Google Scholar]
- Tormos-Pérez M., Pérez-Hidalgo L., Moreno S., 2016. Fission yeast cell cycle synchronization methods. Methods Mol. Biol. 1369: 293–308. 10.1007/978-1-4939-3145-3_20 [DOI] [PubMed] [Google Scholar]
- Whitehall S., Stacey P., Dawson K., Jones N., 1999. Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol. Biol. Cell 10: 3705–3715. 10.1091/mbc.10.11.3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., et al. , 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13: 1977–2000. 10.1091/mbc.02-02-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J., Hajj B., Chen J., Mizuguchi G., Xiao H., et al. , 2014. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. Elife 3: e02203 10.7554/eLife.02203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li F., 2018. In situ chromatin-binding assay using epifluorescent microscopy in S. pombe. Methods Mol. Biol. 1721: 155–165. 10.1007/978-1-4939-7546-4_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Sun S., Zhang S., Gonzalez M., Dong Q., et al. , 2018. Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation. PLoS Genet. 14: e1007572 10.1371/journal.pgen.1007572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Mao J. H., Zhu W., Jain A. K., Liu K., et al. , 2016. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 7: 12619 10.1038/ncomms12619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Giangrande P. H., Nevins J. R., 2004. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23: 4615–4626. 10.1038/sj.emboj.7600459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Takeda T., Whitehall S., Peat N., Jones N., 1997. Functional characterization of the fission yeast Start-specific transcription factor Res2. EMBO J. 16: 1023–1034. 10.1093/emboj/16.5.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7268501.