Abstract
During animal development, a single fertilized egg forms a complete organism with tens to trillions of cells that encompass a large variety of cell types. Cell cycle regulation is therefore at the center of development and needs to be carried out in close coordination with cell differentiation, migration, and death, as well as tissue formation, morphogenesis, and homeostasis. The timing and frequency of cell divisions are controlled by complex combinations of external and cell-intrinsic signals that vary throughout development. Insight into how such controls determine in vivo cell division patterns has come from studies in various genetic model systems. The nematode Caenorhabditis elegans has only about 1000 somatic cells and approximately twice as many germ cells in the adult hermaphrodite. Despite the relatively small number of cells, C. elegans has diverse tissues, including intestine, nerves, striated and smooth muscle, and skin. C. elegans is unique as a model organism for studies of the cell cycle because the somatic cell lineage is invariant. Somatic cells divide at set times during development to produce daughter cells that adopt reproducible developmental fates. Studies in C. elegans have allowed the identification of conserved cell cycle regulators and provided insights into how cell cycle regulation varies between tissues. In this review, we focus on the regulation of the cell cycle in the context of C. elegans development, with reference to other systems, with the goal of better understanding how cell cycle regulation is linked to animal development in general.
Keywords: Caenorhabditis elegans, cell cycle, cell lineage, DNA replication licensing, proliferation, terminal differentiation, WormBook
IN the late 1980s, studies in yeasts, frogs, and cultured mammalian cells came together in the discovery of cyclin-dependent kinases (CDKs) as the central regulators of the eukaryotic cell division cycle (Dorée and Hunt 2002; Hartwell 2002; Nurse 2002). Although we have since learned an enormous amount, understanding the regulatory networks that link developmental and environmental signals to the cell cycle remains a major challenge. Genetic model systems offer opportunities to discover such connections. Caenorhabditis elegans has several features that make this tiny animal attractive for the analysis of cell cycle regulation in a developmental context. In particular, the ease of genetic analysis, the transparency of its body, and the reproducible pattern of its development facilitate the identification and quantitative characterization of cell cycle regulators. As a consequence, specific cell division phenotypes were described at an early stage, following screens for mutants with abnormal cell lineages (lin mutants) (Horvitz and Sulston 1980; Sulston and Horvitz 1981). For example, cells in lin-5 mutants do not complete M phase, but nevertheless continue subsequent rounds of DNA replication. Conversely, postembryonic precursor cells (“blast cells”) skip DNA replication in mcm-4 (lin-6) mutants, while initiating mitosis at the normal times. Two other mutants, cul-1 (lin-19) and lin-23, undergo supernumerary cell divisions during larval development (Kipreos et al. 1996, 2000). Subsequent molecular characterizations revealed how these genes fulfill general cell cycle functions (see below).
Homozygous cell cycle mutants are usually sterile and therefore are obtained from heterozygous mothers. In this situation, cell cycle phenotypes are generally observed during postembryonic development, as the presence of wild-type maternal product allows development through embryogenesis and masks early requirements. Since the discovery of RNA-mediated interference (RNAi) (Guo and Kemphues 1995; Fire et al. 1998), knockdown of maternal product has frequently been used to detect the requirements for cell cycle genes in the germline and during early embryogenesis. Many additional developments have facilitated progress, including the use of green fluorescent protein fusions (Chalfie et al. 1994) and recent success with clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-assisted recombineering [reviewed in: Waaijers and Boxem (2014), Dickinson and Goldstein (2016)]. An advanced molecular genetic toolkit is now available, which makes it possible to combine sophisticated genetics, cell biology, biochemistry, and genomics approaches to study cell cycle regulation at single-cell resolution in living animals.
Following pioneering studies in other systems, studies utilizing C. elegans confirmed the basic understanding of the core cell cycle machinery [reviewed in Kipreos (2005) and van den Heuvel (2005)]. The C. elegans studies also uncovered several novel cell cycle regulators. For instance, the molecular characterization of cul-1 (lin-19), which regulates cell cycle exit, revealed the evolutionarily conserved cullin family (Kipreos et al. 1996). Cullin scaffolding proteins form part of CRL (cullin-ring-ligase) E3 ubiquitin ligases, which include SCF (Skp1–cullin–F-box protein), and regulate critical cell cycle functions, among many other cellular functions. The molecular characterization of lin-5 resulted in the discovery of an evolutionarily conserved LIN-5NuMA-based protein complex (Lorson et al. 2000; Srinivasan et al. 2003). This complex is critical for the generation of microtubule pulling forces that contribute to chromosome segregation and determine the cell cleavage plane by positioning the mitotic spindle. These early examples illustrated the potential of C. elegans studies in the discovery of cell cycle control mechanisms that operate in animal development.
C. elegans is particularly attractive for discovering universal aspects of cell cycle control, and studying the integration of cell division and development. An important topic is the regulation of cell cycle entry and exit, which is regulated in substantial part during the G1 phase of the cell cycle. In this respect, it is of great importance that the critical regulators of G1 progression (described below) are evolutionarily conserved between C. elegans and more complex eukaryotes. This review will broadly cover how the cell cycle is regulated in C. elegans, including progression through variant cell cycles such as meiosis and early embryonic cell cleavages, and the integration of developmental as well as environmental signals with cell cycle entry-and-arrest decisions.
Core Regulators of the Cell Cycle
Before examining developmental control and tissue variations, this section will summarize some of the basic concepts of cell cycle regulation. Because this information is textbook level and encompasses a large number of studies, we will limit the use of references to quotations of parallels between broadly known regulators and genes with similar functions in C. elegans.
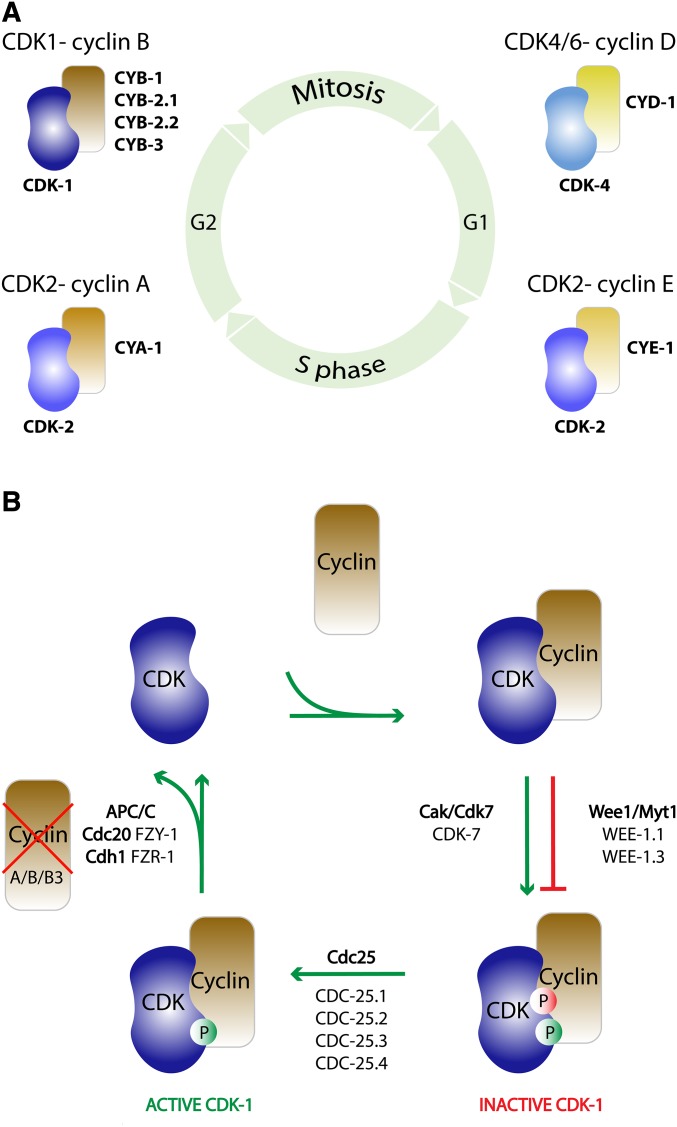
The central goals of the cell cycle include the exact duplication of genomic DNA during the S (DNA synthesis) phase and the segregation of chromosomes, as well as cleavage of the cytoplasm during the M (mitotic) phase. The S and M phases are usually separated by GAP phases in which the cells grow, repair DNA, and, when needed, arrest cell cycle progress prior to the next phase. As such, the G1 phase separates the completed M phase from S phase, and the G2 phase separates S and M phases (Figure 1A). Cells in G1 can alternatively commit to go through another cell cycle, enter a temporal cell cycle quiescent state known as G0, or permanently withdraw from the cell cycle.
Figure 1.
Key regulators of the cell division cycle. (A) Illustration of the cell division cycle and the multiple cyclin-dependent kinase (CDK) complexes that participate in the regulation of cell cycle control. The CDK catalytic subunits (blue) interact preferentially with one or more subfamilies of cyclin proteins. The circle represents a standard somatic cell cycle with sequential G1, S, G2, and M phases. CDK-cyclin complexes (mammalian names, larger font) are positioned according to their approximate time of activity in mammalian cells. The closest C. elegans homologs (names listed, smaller font) appear to share conserved functions. (B) Generic regulation of CDK activity. CDKs are positively regulated by cyclin association, activating phosphorylation (by CAK/Cdk7), and the removal of inhibitory phosphorylation (by phosphatases of the CDC25 family). CDKs are negatively regulated by inhibitory phosphorylation by Wee1/Myt1 kinases, and cyclin degradation through CRL/SCF and/or APC/C E3 ubiquitin ligases. In addition (not indicated), association with CDK-inhibitory proteins (CKIs) prevents CDK activity. This includes CKIs of the Cip/Kip family, known as CKI-1 and CKI-2 in C. elegans. See text for further details.
The transient activation of CDKs at specific cell cycle transitions drives progression through the cell cycle. Active CDKs consist of a catalytic CDK subunit and an associated cyclin protein (Figure 1A). In both budding and fission yeasts, a single CDK is required for progression through “START”, the transition in G1 at which the cell commits to go through a new division cycle, as well as for the transition from G2 into mitosis. At these different transitions, distinct cyclins (G1 or mitotic) activate the yeast cell cycle CDK (CDK1; also known as CDC2 in fission yeast and CDC28 in budding yeast). Multicellular organisms use various cyclins as well as multiple catalytic subunits to regulate cell cycle transitions [reviewed in Sherr (1996)]. Specifically, the closely related CDK4 and CDK6 kinases act in association with D-type cyclins to stimulate cell cycle entry in G1 (Figure 1A). The subsequent activation of CDK2-cyclin E kinases further promotes cell cycle commitment and progression through the G1/S transition. Next, CDK2-cyclin A becomes active during S phase and G2, and CDK1 in association with B-type cyclins controls entry into mitosis. Orthologs of CDK4/6, CDK2, and CDK1 are present as single genes in C. elegans (Boxem et al. 1999; Liu and Kipreos 2000). Moreover, cyclins of each class are expressed in C. elegans, including single D- and E-type cyclins, a single A-type cyclin, several presumed cyclin A-related pseudogenes, and multiple B-type cyclins. While the analysis currently remains incomplete, the temporal expression and cell cycle stage-specific functions of distinct CDK-cyclin combinations appear conserved between C. elegans and mammals (Figure 1A) (Kipreos 2005; van den Heuvel 2005; van der Voet et al. 2009).
Several positive and negative regulators control the activation and inactivation of CDK-cyclin complexes. This includes activating and inhibitory phosphorylations of the catalytic subunit, transcriptional regulation and protein degradation of cyclins, dephosphorylation of CDKs and their substrates, and expression as well as destruction of small CDK-inhibitory proteins (CKIs). In addition to binding cyclin, CDK activation requires the phosphorylation of a threonine residue in the kinase activation loop (T loop). This phosphorylation is carried out by a distant CDK-cyclin pair, CDK7 in association with cyclin H (Fisher and Morgan 1994) (Figure 1B). CDK7 has dual functions as a CDK-activating kinase and polymerase II C-terminal domain kinase, which appear conserved between C. elegans and mammals (Wallenfang and Seydoux 2002). In general, the regulatory phosphorylations of CDK subunits and ubiquitin-dependent degradation of cell cycle proteins involve evolutionarily conserved mechanisms, while the specific transcriptional regulators and CKIs differ substantially between single-cell eukaryotes and metazoans.
The regulation of CDKs by inhibitory phosphorylation has been best described for CDK1. Kinases of the Wee1/Myt1 family are responsible for the inhibitory CDK phosphorylation. These kinases phosphorylate a threonine–tyrosine residue pair in the ATP-binding loop, which likely interferes with CDK substrate binding and phosphate alignment (Jeffrey et al. 1995) (Figure 1B). Phosphatases of the CDC25 family remove the inhibitory phosphorylations, which is sometimes used to control the timing of CDK1 activation. At the G2/M transition, the activation of CDK1 by CDC25 initiates a positive feedback loop, in which CDK1-mediated phosphorylation promotes activation of its positive regulator CDC25 and inactivation of its negative regulator, the Wee1 kinase, to rapidly achieve complete CDK1 activation. C. elegans expresses two Wee1/Myt1-related kinases: WEE-1.1 and WEE-1.3 (wee-1.2 is a pseudogene). These kinases appear to inhibit CDK-1 in a lineage-specific manner (Wilson et al. 1999; Burrows et al. 2006). Likewise, the C. elegans genome encodes four different CDC25-family phosphatases, with some tissue specificity at least for CDC-25.1 and CDC-25.2 (Ashcroft et al. 1999; Ashcroft and Golden 2002; Kim et al. 2010; Lee et al. 2016). Developmental roles for CDC25 family members have been well documented in other animal systems, in particular Drosophila embryos [reviewed in: Yuan et al. (2016)]. Thus, regulation of CDK1 by inhibitory phosphorylation is conserved among eukaryotes, while distinct Wee1/Myt1 kinases and CDC25-related phosphatases serve developmental functions in metazoans.
The sudden activation of CDK1–mitotic cyclin complexes induces a dramatic reorganization of the cell at the onset of mitosis, leading to the formation of a bipolar spindle and chromosome alignment at the metaphase plate. Mitotic CDK phosphorylation also activates the anaphase-promoting complex/cyclosome (APC/C), a multi-subunit E3 ubiquitin ligase, in association with its coactivator CDC20 (known as Fizzy in Drosophila and FZY-1 in C. elegans; Kitagawa et al. 2002) (Figure 1B). The APC/C–CDC20 fulfills two critical functions by inducing the degradation of securin, a protein that inhibits the proteolytic enzyme separase, and mitotic cyclins (Nasmyth 2005). Securin degradation leads to the activation of separase, which then cleaves the cohesion ring complexes that hold sister chromosomes together. Through this pathway, the APC/C–CDC20 triggers sister chromosome segregation in anaphase. The degradation of mitotic cyclins induced by the APC/C–CDC20 leads to the inactivation of CDK1 and exit from mitosis. Phosphatases that dephosphorylate CDK1 substrates assist this process. Following CDK inactivation, a CDC20/Fizzy-related APC/C coactivator known as Cdh1 or Fizzy-related (FZR-1 in C. elegans) replaces CDC20/Fzy, and maintains APC/C activity and mitotic cyclin degradation during late mitosis, and into the next G1 phase (Fay et al. 2002; The et al. 2015).
Entry into the next cell cycle requires new cyclin synthesis and activation of G1 CDKs. The transcription of G1 cyclin genes is often controlled by cell external factors, while subsequent expression of G1/S, S phase, and M phase cyclins is generally under cell-intrinsic control. Heterodimeric transcription factors of the E2F/DP family (E2 promoter-binding Factor/Dimerization Partner protein) are critical in the regulation of cell cycle genes in metazoans [reviewed in van den Heuvel and Dyson (2008)]. Depending on the specific E2F subunit, the E2F/DP dimer (also named E2F) primarily acts as a transcriptional activator or a repressor. Proteins of the retinoblastoma (Rb) tumor suppressor family bind and block activating E2Fs, and act in association with repressor E2Fs to inhibit cell cycle gene expression. Upon induction of cyclin D expression by external factors, G1 CDKs (CDK4/6 associated with a D-type cyclin) become active and initiate the phosphorylation of Rb-family proteins. This is thought to reduce Rb-mediated transcriptional repression and to allow the expression of cell cycle genes that include cyclin E, cyclin A, and CDK2. CDK2-cyclin E and CDK2-cyclin A further phosphorylate and inhibit Rb. In this way, Rb/E2F and cyclin E form part of a double-negative feedback loop that controls an all-or-none decision to enter S phase. The components and mechanisms of this pathway are conserved in C. elegans, with critical roles for cyclin D (CYD-1) and CDK-4 (CDK4/6) in cell cycle entry (Park and Krause 1999; Boxem and van den Heuvel 2001).
The temporal accumulation and inactivation of CKIs provides important additional regulation of cell cycle entry. Vertebrates express two distinct classes of CKIs, the INK family members that specifically block CDK4/CDK6 kinases and the CIP/KIP class that primarily inhibits CDK2/cyclin E [reviewed in Vidal and Koff (2000)]. Induced expression of CIP/KIP inhibitors in response to external or cell intrinsic signals causes cell cycle arrest in a variety of conditions. Removal of CIP/KIP inhibitors at the G1/S transition can be triggered by phosphorylation mediated, among others, by CDK2 (Lu and Hunter 2010). Once phosphorylated, p27KIP1 is a substrate for SCF-induced ubiquitin-dependent protein degradation. SKP2, the substrate specificity factor of the SCF E3 ligase, is itself a target for the APC/C–FZR1 E3 ligase in mammals (Bashir et al. 2004; Wei et al. 2004). Promoting SKP2 degradation, and thereby p27KIP1 accumulation, is one of the mechanisms by which APC/C–FZR1 inhibits cell cycle entry.
All eukaryotes use multiple mechanisms to prevent inappropriate cell cycle entry, but the exact molecules and their relative contributions vary. Worms lack INK-type CDK inhibitors, like flies, but express two CDK inhibitors of the CIP/KIP family (Hong et al. 1998; Feng et al. 1999). CKI-1 and CKI-2 are equally similar to mammalian p21CIP1 and p27KIP1, and Drosophila dacapo. However, only CKI-1 acts as a general inhibitor of cell cycle entry, whose inactivation leads to supernumerary cell divisions (Hong et al. 1998; Fukuyama et al. 2003). By contrast, CKI-2 has only a limited contribution (Buck et al. 2009). Genetic redundancies indicate that CKIs and other negative regulators of the cell cycle (LIN-35Rb, SCF-LIN-23β-TrCP, and FZR-1FZR1/Cdh1) act in parallel (Boxem and van den Heuvel 2001; Fay et al. 2002). For instance, the G1 inhibitory role of APC/C–FZR-1 becomes apparent only when other G1/S regulators are disrupted, and probably does not result from CKI-1p27 stabilization (Fay et al. 2002; The et al. 2015).
The C. elegans CDC-14 phosphatase also promotes cell cycle quiescence, likely by antagonizing degradation of CKI-1 induced by CDK phosphorylation (Saito et al. 2004). Budding yeast Cdc14 antagonizes CDK1 phosphorylation to promote exit from mitosis by supporting the accumulation of a CKI (Sic1) and the degradation of mitotic cyclins (Stegmeier and Amon 2004). Such a role has not been reported for fission yeast or mammalian Cdc14, possibly because in these organisms the PP1 and PP2A phosphatases are more important for counteracting CDK1 phosphorylation (Grallert et al. 2015). The fact that specific regulators may be critical in some organisms while not in others may reflect different levels of redundancy, and illustrates how studying the cell cycle in multiple systems helps acquire a deeper understanding.
Several additional regulators collaborate with or antagonize CDK-cyclins to govern cell-cycle events. This includes checkpoint control pathways that, for example, arrest cell cycle progression in response to DNA damage or incomplete alignment of chromosomes at the metaphase plate. Moreover, additional kinases act in concert with CDKs to regulate DNA replication (DDK kinase) and mitosis (Plk1, Aurora A, and Aurora B). These regulators are not described in this overview section, but we have tried to include any developmental functions reported in C. elegans in the relevant sections below.
Developmental and Tissue Cell Cycle Variants
C. elegans embryonic and larval somatic cell cycles occur in the context of an almost invariant lineage. Somatic cells divide at set developmental times to produce daughter cells with reproducible cell fates (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Sulston et al. 1983). The division of somatic cells is restricted to the embryonic and larval stages of development; and adult-stage somatic cells are postmitotic. In contrast, germ cells divide throughout larval and adult stages. Below, we will briefly describe how cell cycles vary in different tissues or developmental stages. More in-depth descriptions of these differences can be found in previous reviews (Kipreos 2005; van den Heuvel 2005).
Embryonic cell cycles
“Cleavage-type” cell divisions immediately follow fertilization in many animal embryos, including C. elegans. These divisions are rapid and occur without an overall increase in size, so that daughter cells become progressively smaller with each subsequent division. In C. elegans, the first two mitotic cell cycles last only 15–20 min, with the timing lengthening progressively as cell divisions continue (Deppe et al. 1978; Sulston et al. 1983). This short cell cycle timing is likely facilitated by abundant maternal product (mRNA and protein) that is supplied to the oocyte from the hermaphrodite parent. The entire process of embryogenesis is completed in 800 min at 20° to produce 671 cells, of which 113 undergo apoptosis (Sulston et al. 1983).
The early embryonic cell cycles are streamlined to include only S phase and mitosis, as occurs in the early embryos of many other animals (Edgar and McGhee 1988). S phase is very short in the early cell cycles and only lasts ∼8.5 min in the first cell cycle (Sonneville et al. 2012). In other animals, the speed of embryonic S phase is accomplished by initiating DNA replication simultaneously at many more replication origins than in later cell cycles (Méchali 2010). Presumably, a similar strategy allows the very short S phase timing in early C. elegans embryos.
G2 phase is the first gap phase to appear during embryogenesis in C. elegans embryos. G2 phase is initially observed at the 26-cell stage in the two cells that give rise to the intestine shortly before the cells migrate into the interior of the embryo during the process of gastrulation (Edgar and McGhee 1988).
Cyclin D (CYD-1) is required for progression through G1 phase in somatic larval cells (Park and Krause 1999; Boxem and van den Heuvel 2001). Yet in the embryo, only a few, very late cell divisions require cyclin D (Boxem and van den Heuvel 2001; Yanowitz and Fire 2005). This suggests that only these late embryonic cell divisions include a G1 phase. However, it is also possible that embryonic cells have G1 phases that do not require the normal G1 regulatory program; or that cyclin D and CDK-4 are required specifically for reentering G1 phase from a quiescent (G0) state that is present in larval cell lineages but is absent from most embryonic lineages. Notably, the late-dividing embryonic cells that require CYD-1 activity have substantially longer time periods separating their previous and final divisions than the majority of embryonic cells, which may therefore include a G1 or G0 phase.
Somatic larval cell cycles
Newly hatched L1-stage larvae have 558 nuclei; some of these nuclei are in syncytial cells, and hence there are slightly less than 558 cells. Fifty-five of the cells in the newly hatched larvae will undergo cell or nuclear divisions during the larval stages. These cells are called postembryonic blast cells and include two germline precursor cells. The 53 somatic blast cells will generate an additional 403 nuclei in the hermaphrodite (Sulston and Horvitz 1977; Kimble and Hirsh 1979). The majority of the somatic postembryonic blast cells contribute to increasing the number of cells in tissues already present in the embryo, including muscle, nerves, intestine, hypodermis, and scavenger cells known as coelomocytes (Sulston and Horvitz 1977). However, two additional structures are created during the larval stages: the vulva [created by the vulval precursor cells (VPCs)] and the somatic gonad, created by the Z1 and Z4 blast cells (Kimble and Hirsh 1979).
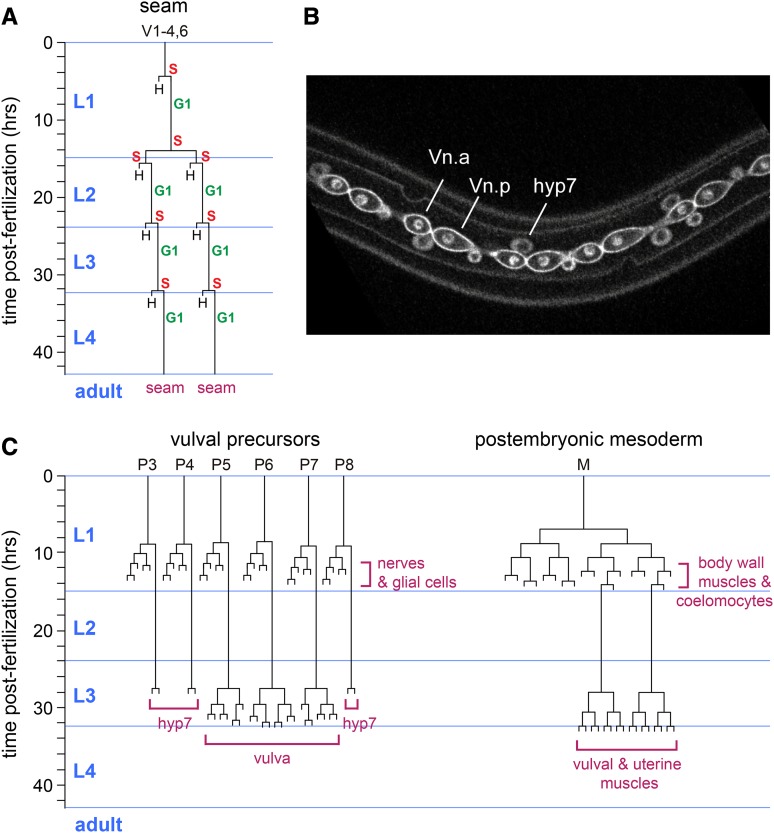
The larval-stage somatic cell divisions have full cell cycles that often include lengthy G0/G1 phases prior to initiating DNA replication, for example in the VPCs, seam cells [which produce cells for the general epidermis (hypodermis/skin)], and intestine cells (Figure 2). The length of the G2 phase varies substantially between lineages. A very short G2 phase separates S phase from mitosis in the seam cell and vulval precursor cell lineages, and a much longer G2 phase of several hours is observed in the intestine (Hedgecock and White 1985; Euling and Ambros 1996; Boxem and van den Heuvel 2001; Zhong et al. 2003).
Figure 2.
Examples of invariant postembryonic cell lineages in C. elegans. (A) Lineage of the stem-cell like epithelial seam cells. The y-axis indicates the time (hours) of development from hatching; vertical lines represent seam cells, horizontal lines are cell divisions, and an H denotes the hyp7 fusion fate; anterior is to the left. S indicates DNA synthesis and G1 temporal quiescence. Note the repeated rounds of asymmetric cell divisions, separated by a single proliferative division at the L1/L2 boundary. (B) Immunofluorescence microscopy image of a transgenic animal expressing cell membrane (GFP::PHPLC1δ) and DNA (GFP::H2B) markers in the seam cell lineage (controlled by the wrt-2 promoter). The animal just completed asymmetric seam cell divisions in the second larval stage (L2). Indicated is a seam cell that just divided, of which the anterior daughter (for simplicity named Vn.a) will fuse with the general epidermis (hyp7), while the posterior cell Vn.p will remain present as a seam cell. Also marked is the nucleus of the hyp7 syncytium. (figure courtesy of S. van der Horst). (C) Cell lineages for the vulval precursor cells that give rise to neurons and glial cells in the L1 larval stage, and the vulva in the L3 stage; and the postembryonic mesoderm lineage that gives rise to body wall muscles, coelomocytes, and sex myoblasts in the L1 stage, and vulval and uterine muscles in the L3 stage.
Endocycles
Endoreplication occurs when cells undergo rounds of DNA replication without an intervening mitosis. Two tissues in C. elegans undergo endoreplication: the hypodermis and intestine. Unlike in Drosophila, in which endocycles often replicate only the euchromatin parts of the genome (Lilly and Duronio 2005), endoreplication in the C. elegans hypodermis and intestine appears to involve complete (or largely complete) doubling of the genome in each endoreplicative cycle (Hedgecock and White 1985).
The majority of intestine cells undergo a nuclear division at the L1-to-L2 transition, producing cells with two nuclei. All intestinal nuclei then undergo endoreplication to attain a 4C DNA content. At the next three larval stage transitions, the intestinal nuclei endoreplicate to successively generate DNA contents of 8C, 16C, and 32C (Hedgecock and White 1985).
Hyp7 is a large syncytial cell that contains multiple nuclei and acts as the skin below the cuticle for the majority of the body (with the exclusion of the head and tail). Hyp7 is formed as a syncytium in embryos and the embryonically derived hyp7 nuclei remain 2C throughout development (Hedgecock and White 1985). During the larval stages, nuclei are added to the syncytium from the seam cell lineages, which produce daughter cells that fuse with hyp7 (Figure 2, A and B). The cells destined to fuse with hyp7 undergo one round of DNA replication without mitosis shortly after they are born, and then fuse with the hyp7 syncytium as 4C nuclei (Hedgecock and White 1985). In the adult stage, the hyp7 nuclei undergo additional endoreplication to attain an average DNA content of ∼10.7C (Flemming et al. 2000). The adult hyp7 nuclei do not display discrete peaks of 4C, 8C, and 16C. This suggests either that the endoreplication occurs slowly and continuously without substantial gaps between endocycles, or that the endoreplication only covers part of the genome. An analysis of copy numbers between genomic regions could distinguish these possibilities, but has not been reported. The adult endoreplication of hyp7 nuclei is positively regulated by TGF-β signaling (Nystrom et al. 2002). The extent of endoreplication of the adult hyp7 nuclei depends on the level of food intake, with diet-restricted animals exhibiting lower levels of endoreplication (Tain et al. 2008).
For both the hypodermis and intestine, a possible explanation for why the cells evolved to undergo endoreplication is to preserve the structural integrity of those tissues. Endoreplication provides increased genomic DNA content to allow cell growth without the disorganization of tissue integrity that could occur if the cells underwent mitosis (Hall and Altun 2008).
Germ cell cycles
The germline contains ∼2000 germ cells in the adult hermaphrodite, approximately twice as many cells as are present in the adult soma [reviewed in Hansen and Schedl (2013) and Kimble and Seidel (2013)]. The germline is generated from two primordial germ cells in the newly hatched L1 larvae, Z2 and Z3 (Kimble and Hirsh 1979). The terminology “germ cells” is typically used in the C. elegans field to refer to all cells derived from the Z2 and Z3 lineage, encompassing all four larval stages and adults. During the larval stages, germ cells divide continuously to increase from 2 to ∼2000 cells in early-stage adults (Kimble and Crittenden 2005). The rate of proliferation of germ stem cells (GSCs) is higher in larvae than in the adult (Roy et al. 2016). During the L3 and L4 larval stages, a subset of GSCs enter meiosis to produce sperm, and then in the adult stage, GSCs enter meiosis to produce oocytes. Germ cells in adults and later larval stages are syncytial with an opening in their plasma membrane that allows a connection to a common cytoplasm.
Adult GSCs are the only cells in C. elegans that closely match the description of an adult stem cell population that divides within a stem cell niche. The stem cell niche is located at the distal end of each gonad arm in hermaphrodites. The niche is formed by the somatic gonadal distal tip cell (DTC) that encases the distal end of the gonad, with cellular projections that extend over the region in which mitotic germ cells are present (∼20 germ cell diameters); germ cells past this point are nonmitotic and have entered meiosis. Under well-fed conditions, adult-stage GSCs divide continuously within the stem cell niche, and upon leaving the niche, they enter meiosis to produce oocytes. As discussed below, the germ cell cycle differs from somatic larval cell cycles in that the G1 phase is truncated or nonexistent in the majority of germ cells. Additionally, germ cells differ from somatic larval-stage cells in their regulation of DNA replication. These observations highlight that basic aspects of cell cycle control have evolved differently in germ cells and somatic tissues.
The Regulation of Cell Cycle Progression During Development
As described above, the pattern and timing of cell divisions in C. elegans depend on the developmental context and lineage of the cells. Cells form different lineages starting as early as the first division of the zygote (P0), when cellular components become unequally distributed among an anterior somatic blastomere (AB) and posterior germline precursor (P1). This division has been studied extensively as a model for asymmetric cell division, which create cell diversity, and often segregate the potential to proliferate and differentiate unevenly among the daughter cells. In addition to asymmetric cell division, cell–cell signaling contributes to the lineage-specific division patterns, starting as early as the four-cell stage. Moreover, environmental conditions such as the levels of nutrients and oxygen induce systemic responses that determine developmental progress. The interplay between developmental cues, environmental signals, and cell cycle regulators ultimately determines whether cells initiate or arrest cell division. In this section, we discuss how developmental and environmental processes may be integrated with the cell cycle machinery.
Oocyte meiotic arrest and maturation
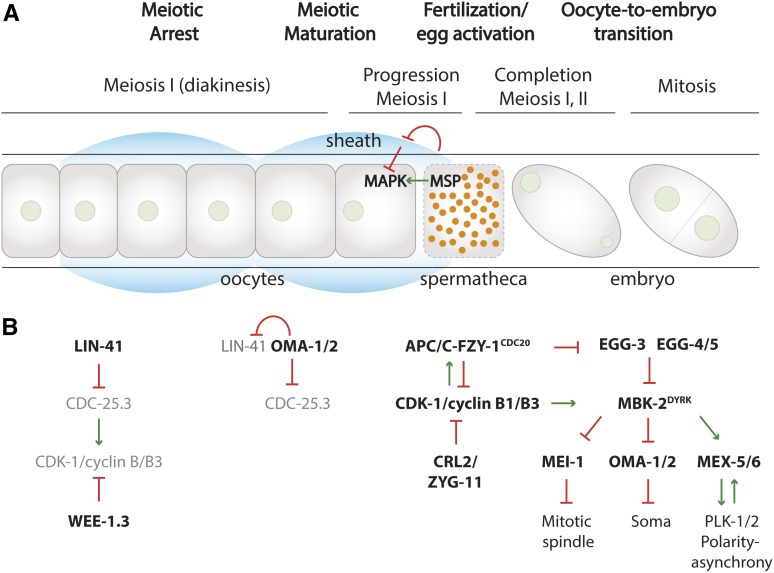
Development starts with a fertilized egg. For fertilization to be successful, oocytes first need to grow, mature, ovulate, and progress through meiosis I. As in most other animals, developing C. elegans oocytes arrest cell cycle progression in meiotic prophase I while growth continues (Figure 3). This arrest depends on low CDK-1/cyclin B activity, which is assumed to result from inhibitory phosphorylation of CDK-1 by the WEE-1.3 dual-specificity kinase (Detwiler et al. 2001; Burrows et al. 2006). The onset of maturation releases the meiotic arrest and allows oocytes to progress from diakinesis to meiotic metaphase, accompanied by nuclear envelope breakdown, meiotic spindle assembly, and cortical rearrangements [reviewed in Von Stetina and Orr-Weaver (2011) and Kim et al. (2013)]. These changes suggest that CDK-1 becomes activated during maturation, and indeed, oocyte maturation in C. elegans requires CDK-1 and mitotic cyclins (Boxem et al. 1999; Burrows et al. 2006; van der Voet et al. 2009), as well as the polo-like kinase PLK-1 (Chase et al. 2000). In this way, C. elegans oocyte maturation resembles that in other animals. In fact, studies of this process in frogs provided a breakthrough in understanding cell cycle regulation, when purified maturation-promoting factor (MPF) was found to consist of CDK1 in association with a mitotic cyclin (Masui and Markert 1971; Dunphy et al. 1988; Gautier et al. 1988).
Figure 3.
Meiotic arrest, oocyte maturation, fertilization, and the oocyte-to-embryo transition in the hermaphrodite germline. (A) Schematic illustration of the successive stages from meiosis I-arrested oocytes in the gonad arm (left) to mitotic embryos in the uterus (right). Nuclei are green, sperm are orange. (B) Diagram of various regulators that mediate the cell cycle transitions in time, from left to right. Initially, WEE-1.3 and LIN-41 are thought to antagonize CDK-1 activation. See text for further information.
What came as a surprise was the finding that C. elegans uses the sperm-specific cytoskeletal protein MSP (major sperm protein) as a hormonal trigger for maturation (Miller et al. 2001) (Figure 3A). MSPs are released from sperm in the adjacent spermathecae, and bind the Ephrin-related receptor VAB-1 on oocytes and somatic sheet cells of the gonad. This interaction overcomes VAB-1 Eph-mediated inhibition of a mitogen-activated protein kinase (MAPK) pathway in oocytes (Miller et al. 2003). In addition to MPF, meiotic progression and maturation depend on MAPK activation in C. elegans as well as vertebrate oocytes (Kim et al. 2013). Thus, in common with other animals, a hormonal signal overcomes the prophase I meiotic arrest of C. elegans oocytes by activating MAPK and MPF. However, the nature of the hormonal signal varies among animals between progesterone in frogs, luteinizing hormone (LH) in humans, and MSP in C. elegans.
Another critical step in meiotic maturation is the translational activation of maternally inherited mRNAs. In frogs, this includes the synthesis of cyclin B from mRNAs that are kept dormant in prophase-arrested oocytes. Regulators of mRNA translation are downstream targets of MSP signaling in C. elegans meiotic maturation. One of these regulators is the conserved RNA-binding protein LIN-41 TRIM-NHM (Spike et al. 2014a,b), originally discovered as a “heterochronic” gene that provides temporal control over larval development (Slack et al. 2000). LIN-41 acts as a translational repressor in the germline, both in cooperation with and antagonized by the “oocyte maturation-defective” proteins OMA-1 and OMA-2 (Tsukamoto et al. 2017) (Figure 3B). The redundant OMA-1/2 zinc finger proteins regulate multiple processes, either through binding the 3′-UTRs of specific mRNAs or protein association. Genetic and protein expression data indicate complex interactions between LIN-41 and OMA-1/2. LIN-41 is expressed throughout meiotic prophase, promotes meiotic arrest, and prevents CDK-1 activation. In contrast, OMA-1/2 is not expressed until after the pachytene stage, antagonizes LIN-41, and promotes meiotic maturation and CDK1 activation (Detwiler et al. 2001; Spike et al. 2014a,b; Tsukamoto et al. 2017). Despite their opposite functions, both LIN-41 and OMA-1/2 ribonucleoprotein (RNP) complexes associate with and repress cdc-25.3 mRNA. This probably explains the contribution of LIN-41 in meiotic arrest; however, as cdc-25.3 remains translationally repressed until early embryogenesis, derepression of cdc-25.3 mRNA translation does not trigger meiotic maturation in the wild type. Interestingly, some other mRNAs (spn-4, meg-1) are repressed by LIN-41 while being activated by OMA-1/2 (Tsukamoto et al. 2017). The translational activation of these mRNAs and the release of translation regulatory proteins from LIN-41 RNPs probably supports growth and meiotic maturation.
Completion of meiosis
As in other animals, oocyte maturation in C. elegans triggers further progression through meiosis I. RNAi experiments demonstrated that not only CDK-1 but also individual B-type cyclins exert critical roles in meiotic progression (Boxem et al. 1999; Chase et al. 2000; van der Voet et al. 2009; Deyter et al. 2010). C. elegans expresses three typical B-type cyclins (CYB-1, CYB-2.1, and CYB-2.2) and an evolutionarily conserved B3-type cyclin (CYB-3) (Kreutzer et al. 1995; van der Voet et al. 2009). B3 cyclins form a distinct subfamily of cyclins that share sequence motifs with both A- and B-type cyclins. Only simultaneous inhibition of all four B-type cyclins fully resembles cdk-1 loss and causes arrest in diakinesis of meiosis I (van der Voet et al. 2009). Nevertheless, knockdown of individual cyclins leads to distinct defects, which suggests that the B-type cyclins have overlapping as well as specific functions in substrate phosphorylation. The RNAi data indicate that CYB-1 is required specifically for the full condensation and alignment of chromosomes at the metaphase plate in meiosis, as well as mitosis. In contrast, CYB-3 is required for sister chromatid separation at the metaphase-to-anaphase transition in meiosis and mitosis (Cowan and Hyman 2006; van der Voet et al. 2009).
The data appear to fit with recent observations in Drosophila embryos and mouse oocytes, which support the idea that cyclin B3 stimulates APC/C activation and securin degradation (Yuan and O’Farrell 2015; Zhang et al. 2015). However, the mechanism by which cyclin B3 promotes APC/C activation may differ between these species. In contrast to flies and mice, C. elegans cyb-3(RNAi) embryos show increased kinetochore localization of the spindle assembly checkpoint (SAC) component MAD2 (Deyter et al. 2010). Consistently, the metaphase arrest of cyb-3(RNAi) embryos could be suppressed by co-inhibition of SAC components (Deyter et al. 2010). Thus, the absence of CYB-3 appears to trigger a strong SAC-mediated block of APC/C activation and arrest in metaphase. By contrast, cyclin B3 activation of the APC/C in fly embryos and mouse oocytes is SAC-independent (Yuan and O’Farrell 2015; Zhang et al. 2015).
While CDK-1/B-type cyclin complexes promote progression from prophase to metaphase of the meiotic divisions (van der Voet et al. 2009), the transition from metaphase to anaphase is triggered by the APC/C–FZY-1CDC20 (Golden et al. 2000; Davis et al. 2002). In agreement, forward genetics and genome-wide RNAi screens found that not only cdk-1 and cdc-25.1 are required for passage through meiosis, but also multiple mat (metaphase-to-anaphase transition defective) genes that encode APC/C components, fzy-1CDC20, and other ubiquitin-dependent proteolysis factors that include proteasome subunits (Golden et al. 2000; Davis et al. 2002; Sönnichsen et al. 2005).
An interesting question is how CDK and APC/C activities are coordinated with meiotic progression and fertilization. Interestingly, the CRL2–ZYG-11 ubiquitin ligase, containing the ZYG-11 substrate-specificity factor, is required for progression through meiosis II and cyclin B1/B3 degradation in meiosis (Liu et al. 2004; Sonneville and Gönczy 2004; Vasudevan et al. 2007). CRL2–ZYG-11 acts redundantly with the APC/C in degrading cyclin B1 and B3 (Figure 3B). CRL-2–ZYG-11 targets at the least CYB-1 directly for degradation in C. elegans, a function recently found to be conserved in human cells (Balachandran et al. 2016). The human and C. elegans CRL2–ZYG-11A/B complexes are not required for normal progression through mitosis. However, when the APC/C is kept inactive in response to SAC activation in human cells, CRL2–ZYG-11A/B-mediated degradation of cyclin B1 allows exit from mitosis, which explains a phenomenon known as “mitotic slippage” (Balachandran et al. 2016).
Similar to zyg-11 or cul-2 mutants, unfertilized oocytes fail to go through anaphase II and retain higher levels of CYB-1 than fertilized eggs (McNally and McNally 2005). It is tempting to speculate that fertilization normally triggers the activation of the CRL-2–ZYG-11 E3 ligase in meiosis II, which complements APC/C–FZY-1CDC20 in mitotic cyclin destruction. Only the combined E3 ligases would achieve the complete downregulation of CDK/cyclin activity that is needed for “licensing” of DNA replication origins during the transition from meiosis to mitotic cell division. The extensive cyclin protein degradation in meiosis and, in particular, early mitoses makes it likely that the resynthesis of cyclin B is needed for subsequent embryonic divisions.
In normal development, fertilization occurs immediately after the oocyte enters the spermatheca. Notably, when fertilization does not take place, DNA synthesis eventually reinitiates. This has been observed in multiple conditions. For instance, hermaphrodites that are exhausted of sperm at the end of the brood continue to lay mature oocytes that resume rounds of DNA replication and nuclear envelope breakdown, which is described as an endomitotic oocyte (Emo) phenotype (Ward and Carrel 1979). A similar Emo phenotype results from interfering with ovulation by ablating a proximal gonadal sheet cell (McCarter et al. 1997), and various sperm-defective (spe) or fertilization-defective (fer) mutants produce unfertilized oocytes with a single polyploid nucleus (L’Hernault et al. 1988). Oocytes in such mutants undergo maturation and ovulation, and initiate anaphase I (McNally and McNally 2005). However, the chromosomes do not become segregated into polar bodies, and meiosis II chromosome congression, spindle formation, and anaphase do not occur. Nevertheless, a female pronucleus forms without apparent delay. These data demonstrate that C. elegans oogenesis does not include a second meiotic arrest, in contrast to most other animals, in which mature oocytes remain arrested in metaphase I (Drosophila) or meiosis II (vertebrates) until fertilization occurs (Von Stetina and Orr-Weaver 2011).
Fertilization coincides with egg activation and induces the completion of both meiotic divisions, the switch to mitotic divisions, and the formation of a protective egg shell. The mechanisms by which fertilization triggers egg activation are poorly understood, but certain spe/fer mutants are likely to miss critical steps. Specifically, the sperm-derived factor SPE-11 is essential for the completion of meiosis and polar body formation (McNally and McNally 2005). However, SPE-11 does not appear to be evolutionarily conserved and its molecular function currently remains unknown. In vertebrates, release of metaphase II arrest results from activation of the APC/C in response to a transient intracellular Ca2+ increase that follows sperm entry (Von Stetina and Orr-Weaver 2011). Ca2+ oscillations have also been described during meiotic resumption of fertilized C. elegans eggs (Singaravelu and Singson 2013). Several spe/fer mutants affect Ca2+ channels in the sperm, and the sperm-provided TRP-3 (SPE-41) Ca2+ channel is not only required for fertilization but also contributes to a local Ca2+ wave upon fertilization (Singaravelu et al. 2012). It remains to be determined whether this is critical for the fertilization signal and is linked to the cell cycle.
From meiosis to mitosis during the “oocyte-to-embryo” switch
Following meiotic progression, CDK-1 and APC/C also exert functions specific for the oocyte-to-embryo switch. This transition requires that a number of proteins present in oocytes are degraded or modified. Critical in this process is MBK-2, a member of the family of dual-specificity tyrosine phosphorylation-regulated kinases (DYRKs) (Pellettieri et al. 2003; Quintin et al. 2003; Stitzel et al. 2006). DYRKs become tyrosine autophosphorylated during their synthesis, which is critical for kinase activity. However, C. elegans MBK-2 becomes active specifically during egg activation, in part through direct phosphorylation by CDK-1 (Cheng et al. 2009) (Figure 3B). Three pseudophosphatases that are required for egg activation, EGG-3 and the closely related EGG-4/EGG-5, keep MBK-2 tethered at the cortex of oocytes in an inactive form (Maruyama et al. 2007; Stitzel et al. 2007; Cheng et al. 2009; Parry et al. 2009). Activation of the APC/C in meiotic metaphase induces the release of active MBK-2 through degradation of EGG-3 and probably EGG-4/5. Active MBK-2 phosphorylates the katanin-related protein MEI-1, which targets MEI-1 for ubiquitin-dependent degradation (Quintin et al. 2003). MEI-1 promotes the formation of compact meiotic spindles and its degradation is needed to allow the formation of extensive mitotic spindles. Further, MBK-2 phosphorylates the OMA-1/2 proteins, which induces their degradation in the somatic precursors (Shirayama et al. 2006; Stitzel et al. 2006) and redirects the function of OMA-1/2 to inhibit transcription in early germline blastomeres (Guven-Ozkan et al. 2008). Additionally, MBK-2 phosphorylates MEX-5 and MEX-6 to create a contact site for the polo-like kinase PLK-1, which is critical in the establishment of anterior–posterior polarity of cytoplasmic components (Nishi et al. 2008). Thus, parallel regulation of MBK-2 by CDK-1 and APC/C couples the oocyte-to-embryo transition to meiotic progression (Figure 3B).
Embryonic cell cycles
Cell cycle progression in the early embryo is likely driven by a single CDK (CDK-1) in association with B-type cyclins. In support of this, the alternating S and M phases of the early cleavage-type embryonic cell divisions do not require CDK-4/cyclin D or CDK-2/cyclin E kinase activity (Boxem et al. 1999; Boxem and van den Heuvel 2001). Following cye-1 knockdown, cell division continues until approximately the 100-cell stage (Fay and Han 2000). Remarkably, however, CDK-2 and CYE-1 are essential for anterior–posterior polarity in the one-cell egg (Cowan and Hyman 2006). Studies of this phenotype revealed that the CDK-2/CYE-1 kinase promotes assembly of centrosomal proteins, while the mature centrosome provides a symmetry-breaking cue during polarity establishment.
As cdk-1 knockdown results in meiotic arrest, insight into its mitotic contributions has been obtained mostly by following the inactivation of mitotic cyclins (Boxem et al. 1999; Chase et al. 2000; van der Voet et al. 2009; Deyter et al. 2010). The depletion of individual cyclins results in very similar defects in meiosis and mitosis. As described above (“completion of meiosis”), the observed phenotypes support the notion that the four B-type cyclins act partly redundantly, while CYB-1 is also specifically required for chromosome condensation and congression, and CYB-3 is essential for anaphase onset. Moreover, immunostaining of zygotes depleted of cyb-3 showed severely delayed and reduced localization of PCN-1PCNA in paternal pronuclei, probably reflecting delayed S phase progression (Michael 2016). This indicates that CYB-3 also contributes to S phase control. As such, CYB-3 could potentially complement or functionally replace cyclin E and cyclin A, which would explain why these latter cyclins are not required for DNA synthesis in early embryos (van der Voet et al. 2009).
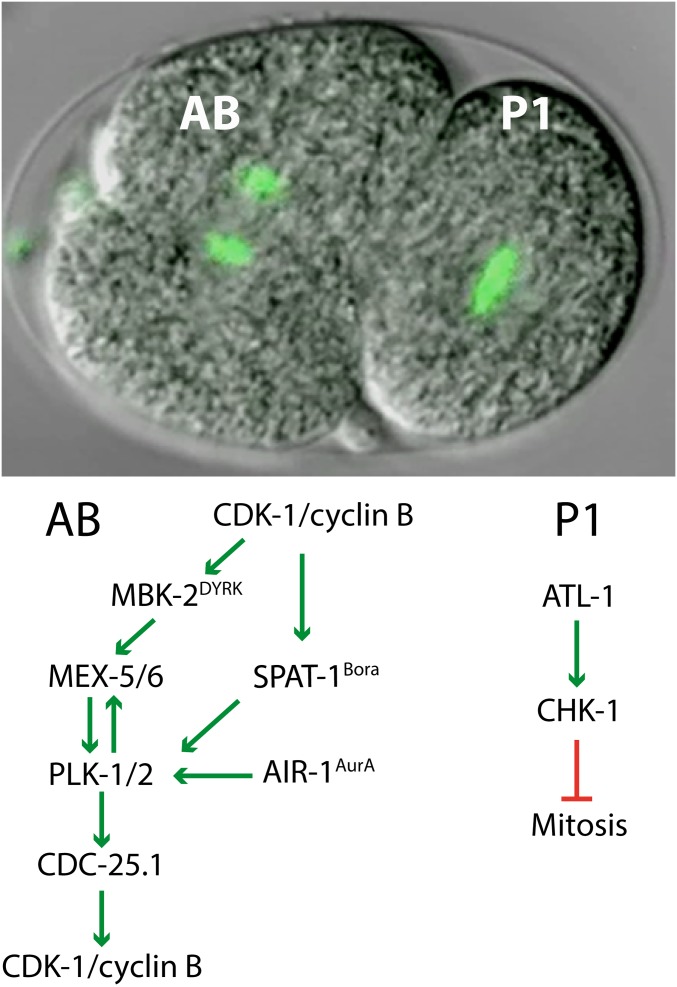
Timing asynchrony
The two daughter cells formed during the first mitotic division display asynchrony in the timing of mitotic entry, in stark contrast to the synchronous nuclear divisions in Drosophila, or cell cleavages during Xenopus and marine invertebrate early embryogenesis. The larger anterior blastomere AB enters mitosis and divides ∼2 min before the posterior blastomere P1. One mechanism underlying this asynchrony involves a checkpoint pathway that responds to DNA replication stress (Brauchle et al. 2003) (Figure 4). This pathway uses the ATR/ATM-related kinase ATL-1 and its downstream effector, the CHK-1 kinase. Inactivating this checkpoint in wild-type embryos reduced the difference in division timing between AB and P1 by ∼40% (Brauchle et al. 2003). These data suggest that a DNA replication checkpoint is normally engaged in P1 to achieve the proper timing of cell division, and possibly, through lengthening S phase, promoting the fidelity of DNA replication in the germline precursor cells.
Figure 4.
Cell division timing asynchrony in the two-cell embryo. (Top) Combined fluorescence and DIC microscopy image of a two-cell-stage embryo in which the DNA is visualized by GFP::H2B expression. The anterior blastomere (AB) initiated mitosis almost 2-min earlier than the precursor of the germline P1. (Bottom) Several cell cycle-regulatory mechanisms have been found to underlie the timing asynchrony. One is based on the anterior enrichment of the MEX-5/6 cytoplasmic determinants, which interact with the PLK-1,2 Polo kinases. The other involves preferential activation of the ATL-1ATR-CHK-1 DNA replication checkpoint pathway in P1. See text for further information.
Also contributing to timing asynchrony is the asymmetric distribution of the polo-like kinases PLK-1 and PLK-2 (Budirahardja and Gönczy 2008; Nishi et al. 2008; Rivers et al. 2008), and cyclin B3 (Michael 2016) (Figure 4). These proteins become enriched in the AB blastomere in response to anterior–posterior polarity, established through asymmetric cortical localization of PAR (partitioning-defective) proteins. The polo kinases accumulate in the anterior of the one-cell embryo by binding to the MEX-5/MEX-6 (“muscle excess”) proteins (Nishi et al. 2008). MEX-5/6 are regulators of cytoplasmic asymmetry that accumulate themselves in the anterior in response to phosphorylation by the PAR-1 kinase (Tenlen et al. 2008; Griffin et al. 2011). Additional phosphorylation by MBK-2 primes MEX-5 for interaction with and phosphorylation by PLK-1 (see above; Nishi et al. 2008). In this way, the MEX-5/6–PLK-1 interaction not only contributes to PLK-1 localization, but also promotes MEX-5/6 function in regulating the asymmetric distribution of cytoplasmic determinants in the one-cell embryo.
CDK-1 appears to act both as an upstream activator and downstream target in the PLK-1 cell cycle pathway, through mechanisms that are conserved in human cells [reviewed in Zitouni et al. (2014)]. As an activator, CDK-1 phosphorylates the PLK-1 regulator SPAT-1Bora (Tavernier et al. 2015). This promotes SPAT-1 interaction with PLK-1 and exposes the PLK-1 T loop to activating phosphorylation by Aurora A (AIR-1) (Figure 4). Active PLK-1 activates CDK-1 as a downstream target by promoting nuclear accumulation of the CDK-1-activating phosphatase CDC-25.1 (Rivers et al. 2008). Thus, the asymmetric localization of PLK-1 leads to asymmetry in nuclear CDC-25.1, which is expected to contribute to advanced mitotic entry of the anterior blastomere via more rapid activation of CDK-1/cyclin B (Figure 4). Indeed, knockdown of the wee-1.3 Myt1 kinase, which mediates inhibitory phosphorylation of CDK-1, advances the cell division timing, and reduces the asynchrony between AB and P1 (Michael 2016). This confirms that the negative phosphorylation of CDK-1 is a critical timing event in the early embryo.
Checkpoint-induced cell cycle arrest in the early embryo
In yeast, checkpoint control pathways are essential only after damage or errors occur. Yet, as described above, the C. elegans ATL-1/CHK-1 checkpoint pathway increases cell cycle length during normal embryonic development. This is reminiscent of late syncytial divisions in Drosophila embryos, in which the cell cycle lengthens during the switch to zygotic gene expression. This midblastula transition is induced by a developmentally regulated DNA replication/damage checkpoint pathway that involves the Drosophila mei-41 (ATM) and grapes (Chk1) kinases [reviewed in Yuan et al. (2016)].
Despite a constitutive role for checkpoint activation in P1, DNA damage and replication stress still induce a substantial delay in cell cycle progression of early C. elegans blastomeres. Interrupting DNA synthesis or UV irradiation generates a cell cycle delay of up to 15 min in the C. elegans zygote, with subsequent divisions showing shorter delays (Encalada et al. 2000; Brauchle et al. 2003; Stevens et al. 2016). Notably, the DNA replication checkpoint delays are substantially longer in the germline precursor cells (P0, P1, and P2) than the somatic sister cells. This increased response to DNA replication stress in germline precursors probably evolved to protect the genome of future generations.
Checkpoint genes are also used to respond to environmental conditions. For instance, a pathway related to the SAC induces an extreme form of developmental quiescence in response to severe oxygen deprivation (anoxia) (Nystul et al. 2003). This so-called “suspended animation” coincides with a cell cycle arrest of early blastomeres. A substantial percentage of the cells arrest in mitotic metaphase, dependent on the san-1MAD3/BubR1 and MDF-2MAD2 SAC pathway genes. However, anoxia also induces arrest in other phases of the cell cycle, including a prophase arrest that depends on the nucleoporin NPP-16NUP50. These arrests are critical for embryo survival and development to adulthood following reoxygenation.
Exposing adult animals to low oxygen (hypoxia) induces an “embryonic diapause” that may be a natural form of suspended animation to protect embryos in utero (Miller and Roth 2009). This response not only requires san-1, probably in the embryo, but also the hypoxia-inducible factor HIF-1 in neurons of the mother. Mitochondrial functions and reduced ATP levels are likely involved in these responses. One-cell embryos arrest prior to nuclear envelope breakdown in response to RNAi of genes encoding tricarboxylic acid cycle/Krebs cycle components (Rahman et al. 2014). These embryos arrest with CDK-1 predominantly in the inhibitory phosphorylated form. The observations point to a possible connection between the levels of ATP, or other metabolites, and cell cycle regulation via the CDC-25 phosphatase.
The SAC also shows an interesting cell size dependence. The SAC delays APC/C activation when the kinetochores of sister chromatids are not properly attached to microtubules of opposite spindle poles. Whereas early embryos of flies and frogs appear to lack this checkpoint, early C. elegans embryos display a moderate delay in anaphase onset in response to spindle defects (Encalada et al. 2005). This delay depends on conserved checkpoint proteins, including MDF-1MAD1, MDF-2MAD2, and SAN-1MAD3. Notably, C. elegans misses an MPS1 SAC kinase homolog. However, a recent study demonstrated that PLK-1Polo functionally replaces MPS1 in C. elegans and that polo kinase also participates in this checkpoint in mammals (Espeut et al. 2015; von Schubert et al. 2015). Germline precursors show a stronger SAC response than their somatic sisters, and more strikingly, the SAC-induced metaphase delay increases with each subsequent embryonic cell division (Galli and Morgan 2016). This phenomenon correlates strongly with the kinetochore-to-cytoplasm ratio, and has been explained by the larger fraction of APC/C that can be inhibited by checkpoint proteins in smaller cells. Free kinetochores generate the checkpoint signal through interaction with MAD2–CDC20. Hence, the fact that C. elegans has holocentric chromosomes and its egg has a relatively small volume may explain why C. elegans embryos display a SAC response, in contrast to the much larger embryos of frogs and flies (Galli and Morgan 2016).
Lineage-dependent introduction of GAP phases and quiescence
As described in the “Developmental and Tissue Cell Cycle Variants” section, introduction of GAP phases occurs in a lineage-dependent fashion. The first G2 phase appears at the onset of gastrulation at the 26-cell stage. The intestinal precursor cells Ea and Ep complete DNA synthesis before they migrate inward to initiate gastrulation. Mitosis is delayed until after the migration, corresponding to a G2 phase of ∼1 hr (Edgar and McGhee 1988). The precursor cell for the germline follows this inward movement of Ea/Ep, and subsequently divides one more time to form the Z2 and Z3 primordial germ cells. These germline precursor cells are the first cells to undergo a prolonged cell cycle arrest, which lasts from 140 min of embryonic development until midway through the first larval stage, a period of ∼18 hr (Sulston et al. 1983). In most embryonic lineages, the cell cycle gradually lengthens, and nearly all cells arrest division between 4 and 7 hr of embryogenesis, to either become quiescent or initiate terminal differentiation.
Several observations indicate that the extended embryonic cell cycles include a G1 or G0 state. First, a few embryonic cells depend on G1 regulators to reinitiate cell division after an extended interphase. This includes the final embryonic divisions of precursor cells in the intestinal, Q neuroblast, and coelomocyte lineages that depend on zygotic cyd-1 cyclin D expression (Boxem and van den Heuvel 2001; Yanowitz and Fire 2005; S. van den Heuvel, unpublished data). Thus, at least these specific embryonic cell cycles include a G1 phase. The blast cells that form the postembryonic lineages also appear to enter a G0 or extended G1 state, as their division during larval development requires cyclin D and CDK-4 (Park and Krause 1999; Boxem and van den Heuvel 2001). Moreover, in the absence of maternal supplies of the G1/S regulators cki-1CIP/KIP, cul-1cullin, or lin-23β-TrCP, embryos die with many additional cells (over 850 instead of 558) (Kipreos et al. 1996, 2000; Fukuyama et al. 2003). Similar to Drosophila dacapo, cki-1 loss results in a single extra division at the time of normal arrest, as observed in the intestinal and mesodermal lineages (Fukuyama et al. 2003). In contrast, the primordial germ cells normally arrest in G2/M with condensed chromosomes, and this arrest is not affected by the absence of cki-1CIP/KIP (Fukuyama et al. 2003, 2006). In short, in a few hours of embryonic development, cells change in a lineage-specific pattern from undergoing rapidly alternating S and M phases to an irreversibly arrested postmitotic state (differentiated cells), or a temporarily arrested state of quiescence (G0 or extended G1) or G2/M arrest (primordial germ cells).
Systemic regulation of cell division during larval development
The timing of the larval cell divisions depends not only on lineage-specific information, but also on environmental and systemic developmental signals. Unfavorable conditions soon after hatching trigger a developmental arrest, while such conditions experienced later in life can induce autophagy in the germline and cell cycle arrest of GSCs. These conditions require synchronization between cell cycle progression and development.
Cell cycle arrest of newly hatched larvae
The first stage larva hatches from its egg in a state without growth or active cell division. In the absence of food or nutrients, the animal remains in this arrested state, also known as L1 diapause, and can survive up to several weeks [Castro et al. 2012; reviewed in Baugh (2013)]. Uptake of food immediately triggers the larval developmental program, which includes growth and cell division. Insulin/insulin-like growth factor (IGF) signaling and several other factors have been found to control the switch between L1 arrest and development. IGF signaling has been extensively studied in C. elegans because inactivation of specific components of this pathway increases life span and allows entry into the alternative “dauer” larval state. These studies identified a single IGF receptor, DAF-2ILR, which activates the phosphoinositide 3-kinase AGE-1PI3K and downstream kinases AKT-1/2Akt/PKB, while the lipid phosphatase DAF-18PTEN acts as an antagonist [reviewed in Murphy and Hu (2013)]. A large variety of insulin-related peptides can either activate or antagonize DAF-2ILR receptor signaling. Activation of the pathway leads, through activation of AKT-1/2, to phosphorylation of the DAF-16FoxO transcription factor. This phosphorylation interferes with nuclear import of DAF-16, and thereby leads to DAF-16 inactivation. IGF signaling through DAF-16 and other downstream effectors is not only critical for the regulation of life span and dauer arrest, but also for cell cycle control during L1 development.
The initiation of L1 development in the presence of food depends on the DAF-2ILR receptor and the expression of a subset of insulin-related peptides (Baugh and Sternberg 2006; Baugh 2013). Conversely, both DAF-16FoxO and DAF-18PTEN are needed for L1 arrest in the absence of food. Arrested L1 larvae express cki-1CIP/KIP in somatic blast cells, and this expression is lost in daf-16 mutants (Hong et al. 1998). daf-16 mutants fail to properly arrest cell division and show poor survival in the absence of food. Interestingly, the Z2 and Z3 primordial germ cells undergo multiple rounds of division in daf-18 mutants (which have increased insulin signaling), independently of daf-16 and cki-1 (Fukuyama et al. 2006). This divergence from somatic cells likely reflects the unique arrest of Z2/Z3 at the G2/M transition (see above). Loss of both of the Drosophila nanos-related genes nos-1 and nos-2 also leads to Z2/Z3 divisions under starvation conditions (Subramaniam and Seydoux 1999). It is currently not known whether nos-1/nos-2 act as downstream targets of IGF-signaling to regulate the cell cycle arrest of germline precursor cells.
Several additional regulators have been found to affect L1 diapause arrest, at least in part as contributors to IGF signaling. This includes a conserved ATPase ASNA-1 and micro RNA (miRNA) miR-73, which are probably involved in the secretion of specific insulin-related peptides from sensory neurons and the intestine (Baugh 2013). The expression of another miRNA, miR-235, also contributes to L1 diapause arrest. Feeding leads to miR-235 downregulation through IGF signaling. This induces expression of the nhr-91 nuclear hormone receptor (germ cell nuclear factor), a miR-235 target that promotes L1 development (Kasuga et al. 2013). While IGF signaling and miR-235 affect L1 development in general, the DAF-16-mediated regulation of cki-1CIP/KIP is currently the only established link to the cell cycle. This connection is particularly relevant because mammalian FoxO transcription factors also have been reported to induce cell cycle arrest through p27KIP1 regulation, under the control of PI(3)K–Akt/PKB signaling (Medema et al. 2000).
Cell cycle entry during larval development
While cki-1CIP/KIP is critical for cell cycle arrest, activation of the CDK-4/cyclin D kinase likely drives the resumption of cell division during larval development. In support, precursor cells of the somatic cell lineages show transcriptional activation of the cyd-1Cyclin D and cdk-4CDK4/6 genes coincident with cell cycle entry (Park and Krause 1999; Brodigan et al. 2003). Moreover, these blast cells remain arrested in G0/G1 through all larval stages in cyd-1 and cdk-4 mutants (Park and Krause 1999; Boxem and van den Heuvel 2001). Further, ectopic expression of G1 cyclins or CDK/cyclin combinations in arrested larvae induces DNA replication and cell division (Park and Krause 1999; Korzelius et al. 2011b). Thus, although the underlying regulation currently remains unknown, the timing of cyd-1 and cdk-4 transcription and kinase activation appears to control cell cycle entry during larval development.
To induce the G1–S phase transition, the CDK-4/Cyclin D kinase needs to overcome inhibition of cell cycle entry by lin-35Rb, cki-1CIP/KIP, and fzr-1Cdh1 (Boxem and van den Heuvel 2001; The et al. 2015). These negative regulators show substantial functional redundancy among each other and also with the SCF–LIN-23β-TrCP E3 ubiquitin ligase. Indeed, single mutation of the C. elegans Rb-related gene lin-35 or the APC/C coactivator fzr-1FZR1/Cdh1 barely affects cell division. However, combining these mutations results in substantial overproliferation, and either single mutation increases the hyperplasia associated with cki-1 or cul-1/lin-23 loss (Boxem and van den Heuvel 2001; Fay et al. 2002; Ruijtenberg and van den Heuvel 2015; The et al. 2015). While these genes all have general functions in G1/S inhibition, their relative contribution is substantially lineage-dependent. Although possibly more pronounced in C. elegans, remarkable redundancies among different regulators of the G1/S transition have also been observed in flies (Buttitta et al. 2007) and mice (Wirt et al. 2010).
The extra cell divisions in cul-1Cullin and lin-23β-TrCP mutants start during the second larval stage, at least in part because maternal product suffices at earlier developmental stages (Fukuyama et al. 2003). The overproliferation phenotype indicates that the SCF–LIN-23β-TrCP E3 ligase promotes the degradation of positive cell cycle regulators. Indeed, strong genetic evidence points to the CDC-25.1 and CDC-25.2 phosphatases as critical in vivo targets (Hebeisen and Roy 2008; Segref et al. 2010; Son et al. 2016). Substrate recognition by β-TrCP often involves interaction with a phosphorylated recognition site known as a phosphodegron. Gain-of-function mutations of CDC-25.1 and CDC-25.2 that induce extra intestinal divisions during embryogenesis appear to disrupt a β-TrCP phosphodegron. Similar to the threonine–tyrosine residues in CDK-1 and CDK-2, CDK-4 contains a conserved tyrosine residue that is probably a WEE-1/CDC-25-regulated inhibitory phosphorylation site. However, it is not known whether increased activity of CDK-4, CDK-2, or CDK-1 drives intestinal overproliferation in cdc-25 gain-of-function mutants. The fact that loss of lin-23β-TrCP or cul-1Cul1 results in much more extensive hyperplasia, compared to cdc-25.1/cdc-25.2 gain-of-function mutants, indicates that the SCF–LIN-23β-TrCP E3 ligase has critical substrates in addition to the CDC-25 phosphatase. CYD-1 remains a prime candidate, as this G1 cyclin contains a conserved β-TrCP phosphodegron, and GSK-3β phosphorylation-dependent degradation of human cyclin D1 has been reported (Diehl et al. 1998).
Candidate substrates for CDK-4/CYD-1cyclin D phosphorylation in the regulation of cell cycle entry have also been identified. One of these targets is the C. elegans Rb-related protein LIN-35, in agreement with the well-established Cdk4/6–cyclin D regulation of mammalian Rb family proteins (Leng et al. 2002; Rubin 2013). LIN-35Rb is a substrate for CDK-4/CYD-1cyclin D phosphorylation in vitro, and loss of lin-35Rb alleviates the requirement for cdk-4 and cyd-1 in vivo (Boxem and van den Heuvel 2001; The et al. 2015). Of note, CDK-4/cyclin D phosphorylates LIN-35 at residues that correspond to the CDK phosphorylation sites of Rb that disrupt E2F binding (The et al. 2015). However, most cell divisions still depend on CDK-4/cyclin D even in lin-35Rb null mutants. This demonstrates that the CDK-4/cyclin D kinase has essential functions in addition to inactivating lin-35Rb. A genetic suppressor screen identified FZR-1FZR1/Cdh1 as a second critical CDK-4/cyclin D target. The phosphorylation of the FZR-1 N-terminus by CDK-4/cyclin D resembles inhibitory phosphorylation of mammalian FZR1/Cdh1 (The et al. 2015). Substantial additional evidence supports the idea that CDK-4/cyclin D promotes cell cycle entry through the combined inhibition of LIN-35Rb-mediated transcriptional repression and APC/C–FZR-1-mediated protein degradation. These mechanisms for G1 regulation may be conserved in mammalian cells (The et al. 2015).
Progression through larval cell divisions
In addition to the CDK-4/cyclin D regulator of cell cycle entry, several additional cell cycle genes are required for the proliferation of postembryonic precursor cells. Homozygous cdk-1 null mutants complete embryogenesis due to the presence of maternal product, and their somatic blast cells go through S phase but arrest in G2 in the first stage larvae (Boxem et al. 1999). Such animals grow substantially, molt, and continue intestinal endoreplication cycles during postembryonic development. Thus, CDK-1 is specifically required for the G2/M transition. The lack of proliferation of precursor cells for the ventral nerve cord creates a typical uncoordinated (Unc) phenotype, while the absence of germline proliferation causes sterility. Several mutants with similar Sterile-Unc (Stu) phenotypes defined additional cell cycle genes, such as air-2 Aurora B and zyg-1 PLK4 (O’Connell et al. 1998; Woollard and Hodgkin 1999). The molecular genetic analysis of additional stu mutants could provide further insight in cell cycle regulation.
The timing of larval development and cell division
Genetic studies have resulted in a model in which larval stage-determining transcription factors (LIN-14 for L1 and HBL-1 Hunchback-like for L2) are downregulated by specific miRNAs (lin-4 and micro-RNAs of the let-7 family) to allow transitioning to the next larval stage. These transcription factors and miRNAs have been defined by “heterochronic” mutations that either cause reiteration or skipping of larval stages. Thus, the specific pattern of cell division is not only determined by the cell lineage, but also by the larval stage. For instance, the vulval precursor cells are formed in the L1 larval stage, and undergo a prolonged G1 arrest before entering S phase and cell division in the L3 stage (Figure 2C). In heterochronic mutants that skip the L1 or L2 stage, the VPCs enter S phase and cell division one larval stage earlier (Euling and Ambros 1996) (Figure 2C). However, it is unclear whether heterochronic transcription factors directly control cell cycle regulators. The let-7 miRNA is needed for the L4-to-adult transition and indirectly upregulates the heterochronic transcription factor LIN-29. Cells in the epidermis arrest proliferation and terminally differentiate in late L4. This coincides with high expression of CKI-1CIP/KIP and reduced expression of CDK-1, as well as other cell cycle regulators (Hong et al. 1998; Rausch et al. 2015). let-7 has been implicated in the repression of CDK-1, while upregulation of cki-1 was shown to depend on LIN-29 and its transcriptional cofactor MAB-10NAB (Harris and Horvitz 2011). However, direct transcriptional control of cki-1 by LIN-29/MAB-10 has not been demonstrated.
Dauer development
C. elegans can adjust its development and cell division pattern dependent on environmental conditions. In addition to L1 arrest in the absence of food, conditions of high population density and limited food can induce an alternative third larval stage, with animals arresting as stress-resistant and long-lived dauer larvae (Riddle and Albert 1997; Antebi 2013). Dauer development coincides with a prolonged cell cycle arrest, through mechanisms that are only partly understood. Sensory neurons in the head perceive the presence of nutrients and dauer pheromone in the milieu, and release insulin-like peptides and TGF-β-related ligands when conditions are favorable. The effects of these pathways on cell division are reasonably well understood for germ precursor cells (discussed below); however, it remains unclear how dauer-inducing systemic signals connect to the cell cycle of somatic cells. As in L1 arrest (see above), daf-16FoxO-dependent cki-1CIP/KIP expression, counteracted by IGF signaling, is probably at least partly responsible for the cell cycle arrest during dauer induction (Hong et al. 1998; Baugh and Sternberg 2006).
The Control of DNA Replication
Replication licensing in larvae
To ensure genome stability, it is essential that all genomic DNA is replicated fully, but only once per cell cycle. The replication licensing system ensures that each replication origin is activated only a single time in each S phase. Activating replication origins multiple times would result in the creation of an unstable “honeycomb” pattern of replicated DNA that has to be resolved by recombination, leading to genomic instability and gene amplification (Alexander and Orr-Weaver 2016).
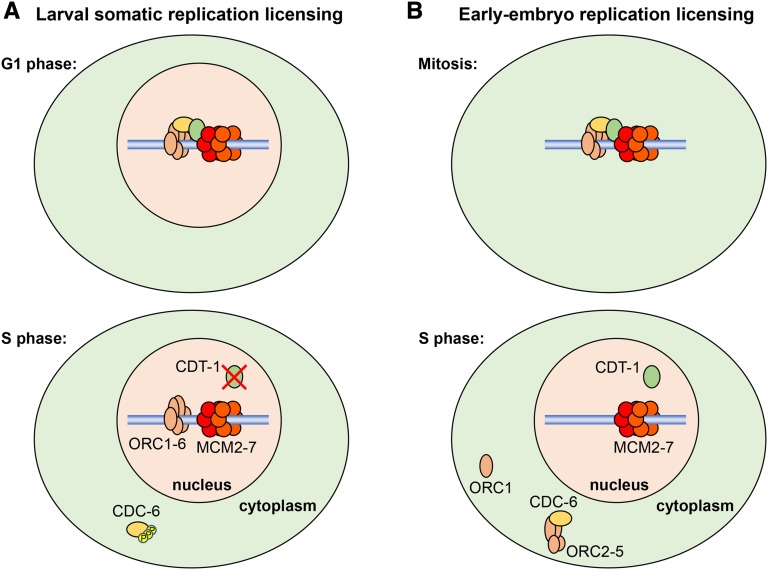
The licensing system works by restricting the licensing of DNA replication origins to late M or G1 phase. The license is equivalent to the loading of the replicative helicase onto DNA replication origins. This loading is temporally separated from the activation of the helicase in S phase. Thus, the licensing system ensures that each origin can only be “fired” once per S phase because new replicative helicases cannot be loaded onto origins in S phase to allow the refiring of origins (Tanaka and Araki 2013). In animals and yeast, the regulation of replication licensing primarily involves two replication licensing factors, Cdt1 and Cdc6, that load the MCM2-7 replicative helicase complex onto replication origins (Riera et al. 2014) (Figure 5). The replication origin is bound by the origin recognition complex (ORC). The core ORC is composed of subunits ORC2-5 (ORC-2, -3, -4, -5 in C. elegans), and it associates with ORC1 (ORC-1) and ORC6. While the core ORC complex and ORC1 are required for replication in animals, ORC6 is not required (DePamphilis et al. 2006). C. elegans ORC-1 through ORC-5 have been characterized (Sonneville et al. 2012). However, a C. elegans homolog of ORC6 has not been reported and is not readily apparent in homology searches.
Figure 5.
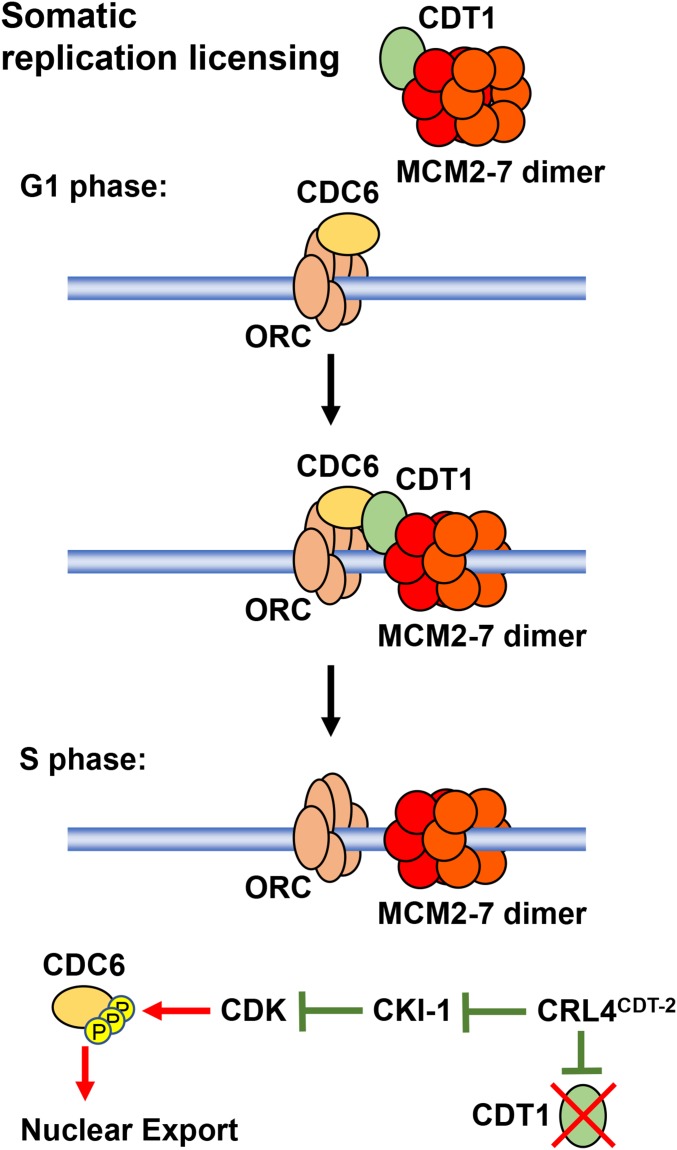
The regulation of DNA replication licensing in larval somatic cells. The prereplicative complex forms in G1 phase. In S phase, replication licensing is prevented by the nuclear export of CDC-6, which is initiated by the phosphorylation of CDC-6 on consensus CDK sites, and the ubiquitin-mediated degradation of CDT-1. Note that both processes are regulated by the ubiquitin ligase CRL4–CDT-2, which directly targets the degradation of CDT-1 and indirectly promotes CDC-6 nuclear export by targeting the degradation of the CDK-inhibitor CKI-1. See text for details.
In the process of DNA replication licensing, the licensing factor Cdc6 loads first to the ORC. Cdt1 is bound to the MCM2-7 replicative helicase and is required to load MCM2-7 onto the Cdc6–ORC complex. The ORC–Cdc6–MCM2-7 complex can recruit more replicative helicase complexes to the origin, so that each origin has several MCM2-7 complexes loaded (two of which will be utilized for bidirectional DNA replication). The loading of MCM2-7 forms the prereplicative complex (pre-RC).
The MCM2-7 helicase remains inactive during G1 phase and is activated at origins to initiate DNA replication during S phase. No further MCM2-7 complexes can be loaded onto replication origins in S phase because the licensing factors Cdt1 and Cdc6 are inactivated. In animal cells, Cdt1 is degraded and Cdc6 is exported from the nucleus. The regulation of the replication licensing system thus prevents DNA rereplication by temporally separating the loading of the replicative helicases (in late M or G1 phases) from their activation in S phase.
An additional licensing protein found in animals is the protein Geminin, which binds and inhibits Cdt1 in the S and G2 phases (Pozo and Cook 2017). In normal (full) somatic cell cycles, most Cdt1 is degraded at the initiation of S phase, and Geminin then binds to newly synthesized Cdt1 to prevent its activity in the S and G2 phases. However, other cell cycles, such as rapid embryonic cleavage divisions, may not invoke Cdt1 degradation, and so the inhibition of Cdt1 by Geminin is more important (Kermi et al. 2017). Geminin contains a destruction-box degron and its degradation is catalyzed by the APC/C ubiquitin ligase in mitosis. Geminin degradation releases Cdt1 to allow its participation in replication licensing in late M or G1 phases.
A critical regulator of replication licensing in animals and fission yeast is the CRL ubiquitin ligase CRL4–CDT-2, which contains CUL-4 as the scaffold and uses CDT-2 as the substrate receptor. The role of the CRL4 complex in this process was first identified in C. elegans. The C. elegans cul-4 mutant has a dramatic rereplication phenotype, with somatic larval cells containing up to 100C DNA content (Zhong et al. 2003). Notably, the rereplication phenotype is not observed in embryos or germ cells, suggesting that replication licensing is regulated differently in those tissues. CUL-4 was shown to be required for the degradation of the replication licensing factor CDT-1 during S phase in larvae (Zhong et al. 2003) (Figure 5). It was subsequently shown that Cdt2 is the specific CRL4 substrate receptor component for CDT-1 degradation in fission yeast and multiple animals (Havens and Walter 2011), including C. elegans (Kim and Kipreos 2007). Cdt1 degradation is restricted to S phase because CRL4–Cdt2 recognizes Cdt1 only when it is physically associated with proliferating cell nuclear antigen (PCNA), which is present in the DNA replication complex at replication forks (Havens and Walter 2011). Consistent with this mechanism, mutating C. elegans CDT-1 to remove the PCNA-binding site stabilizes the protein during S phase (Kim et al. 2007).
The CRL-4–CDT-1 complex is a master regulator that restricts the activity of multiple replication licensing factors. In addition to directly targeting CDT-1 for degradation, CUL-4 is also required for the nuclear export of the CDC-6 replication licensing factor (Kim et al. 2007). As in mammals, the nuclear localization of C. elegans CDC-6 is controlled by phosphorylation flanking its nuclear localization sequences during S phase to prevent its nuclear import (Kim et al. 2007). Because the putative nuclear export sequence is still active during S phase, CDC-6 becomes cytoplasmically localized. To promote the nuclear export of CDC-6, CRL4–CDT-2 targets the degradation of the CDK-inhibitor CKI-1. During a normal S phase, reducing CKI-1 levels allows sufficient CDK activity to phosphorylate CDC-6 on multiple CDK consensus sites clustered around the three putative nuclear localization sequences, which promotes CDC-6 nuclear export (Kim et al. 2007). If CUL-4 is inactivated, then CDC-6 does not become phosphorylated on these sites during S phase. Consequently, CDC-6 remains nuclear-localized, where it can function to reload the MCM2-7 helicase onto replication origins if CDT-1 is also present. The presence of nuclear CDT-1 and CDC-6 in cul-4 mutants during S phase allows the refiring of DNA replication origins to induce rereplication. This CRL4–CDT-2 pathway was subsequently shown to be conserved in human cells, with CRL4–CDT-2 targeting the CKI-1 ortholog p21Cip1 to regulate Cdc6 nuclear localization (Kim et al. 2008) (Figure 5).
Tissue-specific differences in regulating replication: germ cells and embryos
The observation that inactivating CRL4–CDT-2 does not result in an increase in the DNA content of germ cells or early embryonic cells, in contrast to larval somatic cells, suggests that different regulatory mechanisms control replication licensing in these tissues (Zhong et al. 2003; Kim et al. 2007). One potential difference is the contribution of the Cdt1-regulatory protein Geminin. The C. elegans Geminin homolog GMN-1 binds the CDT-1 protein, as expected, and gmn-1 RNAi reduces the numbers of germ cells (Yanagi et al. 2005). However, the level of DNA in the germ cells of gmn-1 null mutants is not increased relative to wild-type, implying that there is no rereplication (Kim and Kipreos 2007). Co-inactivating CRL4–CDT-2 and GMN-1 also does not increase DNA levels in germ cells, suggesting that there are other regulatory mechanisms to restrain replication licensing in germ cells (Kim and Kipreos 2007).
In the early embryo, which has S–M cycles that lack gap phases, replication licensing occurs during late M phase, with MCM2-7 loading onto chromosomes during metaphase and anaphase of mitosis (Korzelius et al. 2011a; Sonneville et al. 2012). This early licensing is required to allow DNA replication to commence immediately after M phase. In contrast, replication licensing occurs during G1 phase in somatic cells of larvae (Zhong et al. 2003). GMN-1Geminin depletion in the early embryo allows MCM2-7 to bind earlier in meiosis (anaphase of meiosis I rather than anaphase of meiosis II) and earlier in mitosis (prometaphase rather than metaphase and anaphase). This indicates that GMN-1 contributes to the timing of replication licensing in the early embryo. However, gmn-1 RNAi produced no phenotypes in the early embryo beyond the earlier initiation of replication licensing (Sonneville et al. 2012).
Analysis of fluorescently tagged replication components indicates that the nuclear export of multiple pre-RC components prevents origin refiring during S phase in the early embryo (Sonneville et al. 2012). During S phase, the core ORC2-5 complex and the separate ORC-1 subunit are excluded from the nucleus, and will only rebind to chromatin in early mitosis prior to the loading of the MCM2-7 complex for the subsequent round of DNA replication (Figure 6). As in somatic larval cell cycles, CDC-6 is also exported from the nucleus during S phase. Additionally, CDT-1 appears to be excluded from S phase nuclei, although this conclusion is based on relatively weak antibody staining of the endogenous protein and should be further substantiated.
Figure 6.
The regulation of DNA replication licensing differs between the early embryo and larval somatic cells. (A) The control of replication licensing in larval somatic cells (see Figure 3 for components). (B) The regulation of replication licensing in the early embryo. Replication licensing occurs in mitosis. Replication licensing is prevented from reinitiating in S phase by the export of CDC-6 and the ORC complex (ORC1 and ORC2-5 exported separately). It is not known if CDC-6 remains associated with ORC components during S phase or if CDC-6 nuclear export is regulated by CDK phosphorylation (as in larval somatic cells). See text for details.
To determine if the observed nuclear exclusion of the pre-RC components is functionally important for replication licensing, Sonneville et al. (2012) depleted the nuclear export factor Exportin-1 (XPO-1) by RNAi. In xpo-1(RNAi) embryos, there was a significant delay in the nuclear export of the ORC subunits and CDC-6. Tellingly, this led to the rereplication of genomic DNA, indicating that the nuclear export of pre-RC components forms the major control preventing replication licensing during S phase in the early embryo (Sonneville et al. 2012).
These results indicate that different tissues use different replication licensing regulations to ensure that DNA is not overreplicated. The truncated cell cycle of the early embryo, comprising S–M–S–M cycles, presumably is not compatible with the degradation of CDT-1 in every cell cycle, as occurs in larval somatic cells, which then requires the translation of new CDT-1 in the G1 phase prior to the next S phase. Why germ cells require a different DNA replication licensing control is not clear, but it may similarly result from a truncated or absent G1 phase (see below). The exact replication licensing regulatory pathway operating in germ cells remains to be determined.
Bypassing the DNA replication checkpoint
The mcm-4/lin-6 gene encodes the MCM4 component of the MCM2-7 replicative helicase. mcm-4 mutant larvae are defective in replicating their genomic DNA, as would be expected for inactivation of the replicative helicase complex (Korzelius et al. 2011a). Strikingly, despite not replicating their genomic DNA, mcm-4 mutant somatic cells nevertheless undergo mitosis at the appropriate time to create daughter cells with reduced DNA levels (Sulston and Horvitz 1981; Korzelius et al. 2011a). This phenotype implies the absence of the DNA replication checkpoint that normally would prevent mitotic entry in response to unreplicated genomic DNA. C. elegans larval somatic cells do have a functioning DNA replication checkpoint because treatment with hydroxyurea (which stalls DNA replication fork movement) inhibits entry into mitosis (Euling and Ambros 1996). Most likely, the replicative helicase cannot form in the absence of MCM-4 and therefore DNA replication never initiates, and because there is no DNA replication the DNA replication checkpoint never engages (Korzelius et al. 2011a).
The signal for the DNA replication checkpoint includes single-stranded DNA and stalled replication forks (Smith et al. 2010). However, if DNA replication does not initiate then there would be no stalled replication forks and the DNA replication checkpoint would not be expected to be activated. Loss of other replication components that prevent DNA replication from initiating would be expected to similarly bypass the DNA replication checkpoint. Consistently, loss of the replication licensing factors CDT-1 or CDC-6, or the helicase component MCM-5, produces a similar phenotype in embryos: continued cell division in the absence of DNA replication (Zhong et al. 2003; Kim et al. 2007; Korzelius et al. 2011a). This supports the model that the bypass of the DNA replication checkpoint results from the failure to initiate DNA replication.
While the mcm-4(e1466) homozygous mutant shows no observable DNA replication in somatic larval cells, there is a surprising level of germ cell proliferation. Germ cells still incorporate the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) in mcm-4(e1466) larval mutants, while all somatic cells fail to incorporate BrdU (Korzelius et al. 2011a). This difference between somatic and germline tissues could result from mcm-4 maternal product (mRNA or protein) in the germline that is sufficient for the continued DNA replication in the larval stages, or it may reflect a surprising lack of requirement for the full-length MCM-4 protein (as the e1466 mutation introduces a premature stop codon at amino acid 88). Overall, this provides further evidence for differences in the regulation of DNA replication between germ cells and larval somatic tissues.
Regulation of the Cell Cycle in Germ Cells
The cell cycle regulation of germ cells differs substantially from that of somatic cells. Most notably, germ cells are the only tissue that divides continuously in larval stages and adults. During the larval stages, the total number of germline cells increases rapidly, with germ cells dividing faster in larval stages than in the adult (Roy et al. 2016). Based on cell counts, the doubling time of germ cells in the L2 larval stage was reported to be ∼4 hr (Kipreos et al. 1996). Cell counts for the L3-to-adult larval period produced an average doubling time of ∼9 hr, but this includes a percentage of germ cells that enter the meiotic program and thus would no longer be actively cycling (Roy et al. 2016).
In adults, germ cell proliferation is restricted to the distal stem cell niche, which is localized within the region of the gonad encompassed by the somatic DTC (Figure 7A). The DTC forms a cap around the first 3–4 germ cell diameters, but also has extensive intercalating protrusions that extend 6–8 cell diameters from the distal end (Byrd et al. 2014). These DTC intercalations define the extent of the DTC “plexus” (Figure 7A). The DTC plexus is considered to be the true stem cell niche that encloses the GSC pool (Byrd et al. 2014); while in more proximal regions of the proliferative zone, germ cells are in various states of meiotic differentiation (Cinquin et al. 2010; Fox and Schedl 2015; Brenner and Schedl 2016). An example of more proximal cells having increased meiotic tendencies is that upon arresting cells in mitosis by inactivating the APC/C ubiquitin ligase, cells in the DTC plexus niche remain undifferentiated, while cells that are more proximal enter meiosis (Cinquin et al. 2010).
Figure 7.
The hermaphrodite gonad and the regulation of mitotic proliferation. (A) Diagram of a cross section of one arm of the hermaphrodite gonad. Germ cell nuclei are green circles. The somatic distal tip cell (DTC) at the distal end of the gonad is colored yellow. The germ cells line the side of the gonad and only one layer is shown in the cross section. The plasma membranes around the nuclei (black lines) include an opening to the central cytoplasm (rachis). (B) Simplified diagram of the interplay of several regulators of mitotic proliferation within the proliferative zone. The Delta ligands LAG-2 and APX-1 originate from the DTC, while other components are in the distal germ cells or in the transition zone as germ cells transition from mitotic proliferation to meiosis. See text for details.
Within the adult proliferative zone, all germ cells incorporate BrdU, suggesting that they are all either mitotically cycling or completing premeiotic S phase (Crittenden et al. 2006). The length of germ cell cycles has been inferred from analysis of 5-ethynyl-2′-deoxyuridine (EdU) incorporation, labeling cells in S phase, and phosphohistone H3 staining of mitotic cells. The reported doubling times of germ cells in the proliferative zone of adult hermaphrodites vary from ∼6.15 to 16–24 hr (Crittenden et al. 2006; Fox et al. 2011; Fox and Schedl 2015; Rosu and Cohen-Fix 2017). However, as the proliferative zone contains at least two distinct populations, the distal GSC niche and a more proximal premeiotic region, which regions are analyzed could have a significant impact on the cell cycle timing. Live imaging of germ cells using a photoconvertible marker showed that the region of the GSC niche, the most distal five to seven rows of germ cells, divided at the fastest rate, with on average one division every 6 hr (Rosu and Cohen-Fix 2017). The middle five to seven rows divided at an intermediate rate, and the most proximal five to seven rows showed hardly any cell division, suggesting that these cells are premeiotic. These data support the idea that, within the most distal region, all GSCs are equivalent for proliferation and stochastically enter meiosis as they are displaced proximally from the DTC plexus (Rosu and Cohen-Fix 2017). Thus, it appears that the stem cell character of GSCs is defined by their location within the niche.
GSC cell cycles differ from somatic larval-stage cell cycles in having a very short or absent G1 phase. When analyzing the entire proliferative zone, relatively few cells with 2C DNA content are observed (Fox et al. 2011; Roy et al. 2016); however, within one to five cell diameters of the distal end, a notable percentage of cells have 2C DNA content. It is possible that a G1 phase is only present in the most distal region. However, an argument against a G1 phase follows from the observation that inactivation of CDK-4 does not affect germ cell proliferation, even though it is essential for G1 phase progression in somatic cells (Park and Krause 1999; Boxem and van den Heuvel 2001). In contrast, inactivation of the G1/S cyclin CYE-1 and its binding partner CDK-2 inhibits germ cell proliferation (Fox et al. 2011). However, as we discussed previously, it has not been excluded that the CDK-4/cyclin D complex is only required when cells are transitioning into the cell cycle from the G0 phase. Under normal laboratory growth conditions, GSCs do not have a detectable G0 phase. Thus, it is possible that distal GSCs have a G1 phase despite not requiring CDK-4/cyclin D activity.
Another major difference between germ cells and other tissues is their syncytial structure, in which germ cells are in contact with a shared cytoplasm (the “rachis”) through an opening in their plasma membrane (Figure 7A). Germ cells begin to acquire their syncytial structure in the L2 larval stage and all germ cells are syncytial in adults (Amini et al. 2014). The most obvious function of the syncytial opening to the rachis is to allow maternal mRNA and protein that is created by germ cells to flow to the developing oocytes. Maternal product is observed over the entire length of the rachis (Gibert et al. 1984).
The syncytium does not appear to transmit cell cycle-state information. Active mitotic CDK1–cyclin B has been shown, when introduced into vertebrate cells, to induce the activation of CDK1–cyclin B in the recipient cell that then drives the cell into mitosis (Kishimoto 2015). However, in the C. elegans gonad, the entry of one germ cell into mitosis does not trigger mitosis in the surrounding germ cells. This suggests that active CDK1–cyclin B does not transfer from mitotic germ cells to nearby germ cells through their openings to the rachis.
Cell cycle regulators in the mitosis vs. meiosis transition
There are two major levels of control of germ cell proliferation: (1) regulation of the size of the GSC pool, which is controlled by regulating the mitosis vs. meiosis transition, and (2) regulation of the rate of germ cell proliferation. The regulation of GSC differentiation to enter meiosis has been extensively reviewed elsewhere (Hansen and Schedl 2013; Kimble and Seidel 2013). Therefore, we will focus on those aspects of the mitosis vs. meiosis decision that are known to interface with cell cycle regulators.
Notch signaling by the DTC is the major regulator of the mitosis vs. meiosis decision [reviewed in Hansen and Schedl (2013) and Kimble and Seidel (2013)]. The DTC expresses the DSL (Delta, Serrate, LAG-2) ligand proteins LAG-2 and APX-1, which are bound by the Notch receptor GLP-1 on germ cells. After GLP-1 activation, the intracellular domain of GLP-1 is cleaved to release an intracellular domain (GLP-1-ICD), which acts as a transcription factor in complex with the CSL DNA-binding protein LAG-1 and the transcriptional coactivator LAG-3/SEL-8 (Figure 7B). GLP-1 activity leads to the inhibition of GLD-1, a K homology (KH) domain RNA-binding protein that inhibits mitotic proliferation. GLP-1 also leads to the inhibition of GLD-2 and GLD-3, a cytoplasmic poly(A) polymerase and KH-domain RNA-binding protein, respectively, which promote meiosis.
The critical target genes for GLP-1-ICD/LAG-1 transcription are lst-1 and sygl-1, which are redundantly required to maintain GSCs in the mitotic state (Kershner et al. 2014) (Figure 7B). lst-1 and sygl-1 encode novel proteins, LST-1 and SYGL-1, which inhibit GLD-1 activity by acting in association with the translational repressor FBF-1 (a PUF, Pumilio and FBF, family protein) (Brenner and Schedl 2016). In addition, LST-1 also inhibits GLD-1 activity independent of FBF-1 (Figure 7B). GLP-1 signaling therefore controls the meiosis vs. mitosis cell fate decision, at least in part, by inhibiting GLD-1. The mitotic index is unchanged in glp-1 loss-of-function mutants that have reduced sizes of the proliferative zone (Fox and Schedl 2015; Roy et al. 2016). Thus, while GLP-1 signaling controls the size of the GSC niche, GLP-1 does not appear to regulate cell cycle progression directly.
A failure of germ cells to enter meiosis and instead continue mitotic division produces germline tumors, in which mitotic germ cells are present throughout the gonad. Germline tumors can arise from constitutive activation of Notch signaling, e.g., through GLP-1 gain-of-function mutations, which promotes mitotic proliferation and inhibits meiosis. Germline tumors can also arise from loss of the RNA regulators that promote meiosis and block mitotic proliferation. A complete failure to enter meiosis occurs upon loss of both the GLD-2 and GLD-3 pathway, and the GLD-1 and NOS-3 pathway (with NOS-3 a Nanos-related translational regulator). In this situation, germ cells divide mitotically even when distant from the Notch signal in the stem cell niche.
Cyclin E expression to maintain germ cells in a mitotic state:
The G1/S cyclin CYE-1 protein is present in the proliferative zone but not in the adjacent meiotic regions of the germline (Brodigan et al. 2003). Within the proliferative zone, CYE-1 expression is high in all germ cells, indicating that its expression is not cell cycle regulated (Fox et al. 2011). It is possible that the constant CYE-1 expression contributes to the apparent lack of a G1 phase in most germ cells, as other cells that have constant cyclin E expression, such as mammalian stem cells, also lack G1 phases (Orford and Scadden 2008).
CYE-1 expression in germ cells is important not only to allow proliferation within the proliferative zone but also to prevent entry into meiosis. This was shown by RNAi of cye-1 in L4 larvae, which blocked cell proliferation and induced germ cells to prematurely enter meiosis (Fox et al. 2011). In contrast, inactivation of other cell cycle regulators that are important for S and G2 phases does not induce meiotic entry (Fox et al. 2011). This suggests that either the decision to enter meiosis is directly regulated by CDK-2/CYE-1, or that it occurs early in the cell cycle (pre-S phase) and loss of CYE-1 causes arrest at that decision point.
GLD-1, which inhibits mitotic proliferation, and CDK-2/CYE-1, which promotes mitotic proliferation, mutually repress each other (Figure 7B). GLD-1 inhibits CYE-1 expression in the meiotic regions (Biedermann et al. 2009). The repression is direct and is mediated by GLD-1 binding to its target sequence in the cye-1 mRNA 3′-UTR (Biedermann et al. 2009). In gld-1 mutants, meiotic cells reenter the mitotic cell cycle, and this mitotic proliferation requires the derepression of CYE-1 that occurs in these mutants (Biedermann et al. 2009). Conversely, CYE-1 negatively represses GLD-1 protein expression in the distal region of the gonad. GLD-1 is normally absent from the most distal region of the gonad, but becomes expressed there when CYE-1 is inactivated (Jeong et al. 2011). This negative regulation of GLD-1 by CDK-2/CYE-1 also appears to be direct. CDK-2/CYE-1 can phosphorylate GLD-1 in vitro, and inactivation of CYE-1 causes the loss of GLD-1 phosphorylation in vivo (Jeong et al. 2011). Replacement of the CDK-target sites in GLD-1 with nonphosphorylatable residues extends GLD-1 protein expression to more distal regions, thereby reducing the size of the proliferative zone. Therefore, CDK-2/CYE-1 phosphorylation of GLD-1 is important for GLD-1 regulation and determining the size of the stem cell niche.
The expression of CYE-1 is also subject to negative regulation by the SCF–PROM-1 E3 ubiquitin ligase (Fox et al. 2011) (Figure 7B). Inactivation of the substrate receptor component of the E3 complex, PROM-1, allows CYE-1 expression to extend beyond its usual sharply defined region in the proliferative zone. This suggests that CYE-1 is subject to ubiquitin-mediated degradation as germ cells leave the proliferative zone, in addition to its translational regulation by GLD-1. Despite ectopic CYE-1 expression, the prom-1 mutant germ cells still cease mitotic proliferation and enter meiosis. This indicates that CYE-1 expression alone is not sufficient to maintain mitotic proliferation in germ cells. Double mutants of prom-1, with either gld-1/nos-3 or gld-2/gld-3 pathway genes, reveals that the three pathways function redundantly to promote entry into meiosis and the cessation of mitosis, with additional SCF-PROM-1 targets, beyond CYE-1, that impact this transition (Mohammad et al. 2018).
FBF represses CKI-2 expression to prevent germ cell arrest:
FBF-1 and FBF-2 are closely related PUF family RNA-binding proteins. FBF-1 and FBF-2 show the highest level of expression in the proliferative zone (Lamont et al. 2004). Together, FBF-1 and FBF-2 are required to maintain germ cells in the mitotic state (Crittenden et al. 2002). FBF represses the translation of the CDK inhibitor CKI-2 in the proliferative zone through direct interaction with the cki-2 3′-UTR (Kalchhauser et al. 2011). Inactivation of both FBF-1 and FBF-2 causes the loss of all mitotic germ cells, at least in part through the expression of CKI-2. If CKI-2 is co-inactivated with FBF-1/2 then the loss of mitotic germ cells is delayed. Thus, FBF promotes germ cell proliferation in part by repressing CKI-2 expression.
CRL2–LRR-1 promotes mitotic proliferation and prevents checkpoint activation:
Inactivation of the ubiquitin ligase CRL2–LRR-1 leads to the accumulation of CKI-1 in all germ cells (Feng et al. 1999; Starostina et al. 2010). In the absence of CRL2–LRR-1 activity, CKI-1 protein accumulates and germ cells arrest with a 2C DNA content. CRL2–LRR-1 appears to directly target CKI-1, as LRR-1 binds CKI-1 and mediates its degradation when the two proteins are coexpressed in mammalian cells (Starostina et al. 2010).
Loss of LRR-1 also activates the DNA replication checkpoint, as indicated by the presence of the phosphorylated active form of the CHK-1 kinase in embryos (Merlet et al. 2010). The lrr-1 mutant germ cell arrest can be abrogated by inactivation of CHK-1 or its upstream activator, ATL-1ATR/ATM kinase. The activation of the DNA replication checkpoint in lrr-1 mutants is associated with the accumulation of single-strand DNA-binding replication protein A (RPA-1) on chromatin, suggesting a defect in completing DNA replication (Merlet et al. 2010).
Regulation of the rate of germ cell proliferation
The germline in an adult hermaphrodite contains approximately twice as many cells (∼2000) as the entire soma (∼1000) (Kimble and Seidel 2013). A substantial percentage of total energy expenditure in C. elegans adults goes toward germ cell proliferation and the creation of oocytes that are loaded with maternal product. Given the scale of the energy consumption required for reproduction, it is not surprising that the rate of germ cell proliferation is closely linked to the level of food intake. A surprisingly large number of regulatory pathways control the rate of germ cell proliferation in the adult hermaphrodite. We will describe these pathways in this section.
TGF-β promotes the mitotic germ cell fate:
Environmental conditions can impact the rate of germ cell proliferation. These include the availability of food and signals that indicate that the food supply will soon be exhausted by the presence of many other nearby nematodes, each of which is capable of generating a steep exponential increase of 250–300 progeny approximately every 6 days. The TGF-β signaling pathway is activated in response to favorable environmental conditions, including plentiful food, and is repressed in response to unfavorable conditions that includes crowding or a lack of food (Gumienny and Savage-Dunn 2013). The chemosensory ASI neurons generate the TGF-β ligand (DAF-7) in response to low pheromone levels (signifying noncrowded conditions) and plentiful food (Dalfó et al. 2012).
The TGF-β signaling pathway is required for creating the full complement of larval germ cells (Dalfó et al. 2012). Inactivation of TGF-β signaling reduces germ cell numbers by half and produces a smaller proliferative zone (Dalfó et al. 2012). However, the mitotic index within the proliferative zone is similar to wild-type (Dalfó et al. 2012). This suggests that TGF-β signaling regulates the mitosis-to-meiosis decision to control the size of the GSC pool, rather than the rate of germ cell division.
TGF-β signaling functions in the somatic DTC to indirectly regulate germ cells (Dalfó et al. 2012). Intriguingly, the regulation of germ cells by TGF-β is independent of GLP-1 Notch signaling, as loss of TGF-β signaling still reduces germ cell numbers in a tumorous mutant that lacks the GLP-1 receptor (Dalfó et al. 2012). The TGF-β pathway functions in DTCs to increase the size of the GSC pool by inhibiting entry into meiosis. TGF-β acts through its canonical pathway to inhibit the DAF-3/DAF-5 transcriptional repressor complex (Dalfó et al. 2012), but the relevant downstream gene targets have not been reported.
Insulin/IGF-like signaling promotes germ cell proliferation:
IGF signaling cell-autonomously promotes germ cell proliferation during the larval stages (Michaelson et al. 2010). Loss of IGF signaling reduces the mitotic index, indicating that germ cell proliferation is affected rather than the mitosis-to-meiosis transition (Michaelson et al. 2010). IGF signaling affects germ cells through its canonical inhibition of the FoxO transcription factor DAF-16 (Michaelson et al. 2010). The critical transcriptional targets of DAF-16 that inhibit germ cell proliferation have not been reported.
A partial reduction-of-function allele of the daf-2 insulin receptor reduces the mitotic index of larval-stage germ cells and adult germline tumors (in which mitotic proliferation of germ cells occurs throughout the gonad as the result of particular mutations) (Pinkston et al. 2006; Michaelson et al. 2010). In contrast, the adult proliferative zone is not affected by partial daf-2 mutants. However, stronger daf-2 mutations do reduce germ cell proliferation in the adult (Narbonne et al. 2015). This suggests that IGF signaling regulates germ cell proliferation at all stages, but that larval stages and germline tumors are more sensitive to changes in IGF signaling.
There are 40 C. elegans genes that encode insulin-like peptides, INS-1 through INS-39 and DAF-28 (Baugh et al. 2011). Interestingly, germ cell proliferation appears to be solely stimulated by INS-3 and INS-33 (Michaelson et al. 2010). The expression of ins-3 is largely neuronal and ins-33 expression is largely hypodermal, suggesting that these are the sources of systemic INS-3 and INS-33 peptides (Michaelson et al. 2010). Other insulin-like peptides have been shown to control IGF signaling in other tissues, which indicates that insulin-like ligands have specialized tissue functions and that the animal can regulate germline proliferation via IGF signaling independently of other tissues (Michaelson et al. 2010).
DAF-18PTEN signaling decreases germ cell proliferation in response to unfertilized oocytes:
Germ cell proliferation decreases in hermaphrodites when their sperm are depleted and this can be reversed by additional sperm provided by mating (Narbonne et al. 2015). The inhibition of germ cell proliferation depends on the accumulation of unfertilized oocytes in the spermless gonads. The decrease in germ cell proliferation in response to unfertilized oocytes requires DAF-18PTEN but not DAF-16FoxO, suggesting that DAF-18 functions in a DAF-16-independent manner (as it does for the regulation of Z2 and Z3 in the L1 stage) (Narbonne et al. 2015).
Bacterial folates are an exogenous signal to promote germ cell proliferation:
Folates are B-complex vitamins that function to create a subset of nucleosides and amino acids in a metabolic cycle termed “one-carbon metabolism” (Selhub 2002). Animals cannot synthesize folates, in contrast to bacteria, hence C. elegans obtain folates from their diet of bacteria. Recently, it was discovered that bacterial folates act as an exogenous signal to cell-autonomously stimulate germ cell proliferation (Chaudhari et al. 2016). Bacterial folates stimulate increased DNA replication of isolated germ cells in vitro, increased mitotic index and the size of the proliferative zone in wild-type animals, and the growth of germline tumors (Chaudhari et al. 2016).
Folates are a family of related molecules that can be interconverted as part of the one-carbon metabolism cycle. Interestingly, only a subset of bacterial folates stimulates germ cell proliferation. Stimulatory folates are 10-formyl-THF-Glu(n) and 5,10-methenyl-THF-Glu(n). The latter folate converts to 10-formyl-THF-Glu(n) at neutral pHs, such as normal culture conditions; this suggests that 10-formyl-THF-Glu(n) is the only stimulatory folate. Other folates that take part in the one-carbon metabolism cycle are unable to stimulate germ cell proliferation under normal (folate replete) growth conditions (Chaudhari et al. 2016).
Significantly, the folate stimulation of germ cell proliferation occurs independently of the one-carbon metabolism cycle. Both stimulatory and nonstimulatory folates can rescue folate deficiency, indicating that the ability of stimulatory folates to increase germ cell proliferation rates under normal culture conditions is not linked to their ability to act as a vitamin. The stimulation of germ cell proliferation by folates requires the homolog of the mammalian folate receptor, FOLR-1 (Chaudhari et al. 2016). Yet, FOLR-1 is not needed to provide folates as vitamins, which instead depends on the reduced folate carrier FOLT-1, whose inactivation produces severe folate deficiency phenotypes (Balamurugan et al. 2007; Austin et al. 2010). Additionally, a folate-related molecule, dihydropteroate, which cannot take part in one-carbon metabolism, also stimulates germ cell proliferation in a FOLR-1-dependent manner (Chaudhari et al. 2016). Interestingly, the stimulatory folate, 10-formyl-THF-Glu(n), and dihydropteroate are both relatively unstable relative to other folates. It is possible that the use of these unstable folates for signaling allows a tighter linkage between the availability of high-quality bacterial food (e.g., live bacteria) and the rate of germ cell proliferation. The intracellular pathway for FOLR-1-dependent folate signaling is currently not known.
Volatile bacterial odors increase reproductive rate via neuronal signals:
Volatile odors, which can transmit through the air from specific bacteria, can affect the rate of egg production (Sowa et al. 2015). Odors from the Escherichia coli strain HB101, even when physically separated from the animals, causes hermaphrodites to lay a normal-sized brood over a shorter period of time compared to the rate with a normal diet of OP50 bacteria. The accelerated rate of egg production when grown with HB101 odors is correlated with increased proliferation of germ cells in the proliferative zone. The AWB chemosensory neurons mediate the response to the volatile odors via release of neuropeptides that are required for the stimulation of germ cell proliferation. Genetic interactions suggest that the AWB neuropeptide signaling is independent of insulin and TGF-β signaling pathways. Currently, the neuropeptides and their target tissues have not been reported.
The regulation of germ cell numbers upon starvation:
The starvation of L4-stage hermaphrodite larvae causes a dramatic reorganization of the adult germline, wherein the germ cells are dramatically reduced in number (Angelo and Van Gilst 2009). Upon starvation, mitotic germ cells in the proliferative zone undergo a G2-phase cell cycle arrest (Seidel and Kimble 2015). During starvation, adult hermaphrodites create one oocyte at a time over the course of many hours, in contrast to the situation in fed adult hermaphrodites where multiple developing oocytes are present in each gonad arm (Seidel and Kimble 2011). Embryos created during starvation either hatch in the hermaphrodite parent, leading to the death of the parent through “bagging” (eating the insides of the parent), or the embryos die in situ, presumably as a result of defects associated with the starvation (Seidel and Kimble 2011). This starvation state has been termed “adult reproductive diapause” (ARD) (Angelo and Van Gilst 2009) and “oogenic germline starvation response” (Seidel and Kimble 2011). The latter term is in reference to the fact that the shrinkage of the gonad corresponds to the creation of oocytes, presumably as gonadal cytoplasm is transferred to the developing oocytes (Seidel and Kimble 2011). A percentage of GSCs survive starvation in the DTC plexus. When starved animals are provided with food, these remaining GSCs repopulate the germline, which enlarges to the same size as the germlines of continuously fed animals (Angelo and Van Gilst 2009).
Dafachronic acid inhibits germ cell proliferation in adults and mediates the adult starvation response:
Placing young adult hermaphrodites into starvation conditions results in an increase in the bile acid-like steroid hormone dafachronic acid (DA) as well as the cytochrome P450 enzyme DAF-9, which is required to create DA (Thondamal et al. 2014). Adult hermaphrodites that are starved have reductions in the number of germ cells in the proliferative zone within 1 day. This reduction of the proliferative zone requires DA; in the absence of DA, the proliferative zone maintains the same number of germ cells over the first 1–2 days of starvation (Thondamal et al. 2014) (H. Aguilaniu, personal communication). Addition of DA inhibits germ cell proliferation in vivo in both wild-type animals and tumorous germline mutants. The inhibition depends on the canonical DA-steroid hormone receptor DAF-12 (Mukherjee et al. 2017). DA can inhibit the proliferation of isolated germ cells in vitro in a DAF-12-dependent manner, indicating a direct effect of DA on germ cells (Mukherjee et al. 2017). DAF-12 is also required in germ cells for in vivo inhibition, again suggesting that DA acts directly on germ cells. Inactivation of DAF-9 in vivo (which would block DA production) increases the rate of germ cell proliferation in wild-type and tumorous germline mutants, suggesting that DA normally functions to restrain germ cell proliferation.
DA has multiple roles in C. elegans, including inhibiting entry into the dauer pathway, regulating heterochrony, and promoting longevity in animals that lack germlines (Antebi 2013). Significantly, the larval role of DA in blocking entry into the dauer pathway has the indirect effect of increasing germ cell proliferation by preventing the dauer-stage arrest of all cell divisions. This potential functional paradox is solved by the finding that DA only inhibits germ cell proliferation in adults but not in larvae (Mukherjee et al. 2017). Thus, DA indirectly promotes the proliferation of germ cells in larvae by blocking dauer entry, and directly inhibits germ cell proliferation in adults.
Adult germ cells are more sensitive to starvation than larval germ cells, with the mitotic index in adults dropping significantly within 30 min and a complete shut down by 3 hr. In contrast, in larval stages, the germ cell mitotic index is only partially reduced after 4–5 hr of starvation, with a complete shut down only by 7–8 hr (Seidel and Kimble 2015). The adult-specific role of DA in inhibiting germ cell proliferation may contribute to the more rapid cessation of germ cell proliferation upon starvation in adults.
Further research will be required to link the distinct regulatory signals that control the rate of germ cell proliferation (described in this section) to cell division regulators that directly impact the cell cycle and, in the case of CDK-2/CYE-1, also regulate the mitosis-to-meiosis decision.
The Interplay Between Cell Proliferation, Arrest, and Differentiation
Developmentally induced signal transduction pathways and asymmetric cell divisions provide spatiotemporal control of gene expression. Transcriptional regulation of cell cycle genes is thought to form an important connection between developmental signals and the cell cycle. However, only a few molecular connections have been firmly established, and it remains poorly understood what mechanisms underlie the lineage-specific patterns of cell division and timing of terminal differentiation. In this section, we summarize insight in how transcription factors and chromatin regulators interact with the cell cycle to control the temporal pattern of cell division, and coordinate proliferation with differentiation in a lineage-specific manner.
Rb/E2F-mediated transcriptional repression
The best known transcriptional regulators of the cell cycle are the heterodimeric E2F/DP transcription factors and their binding partners of the Rb-corepressor family (van den Heuvel and Dyson 2008). Rb/E2F complexes act in concert with other transcriptional regulators and exert important functions in developmental control that go well beyond cell cycle regulation. Studies in Drosophila highlighted the contribution of repressive E2Fs in association with a fly Rb member in restricting gene expression outside the appropriate cellular context (Dimova et al. 2003). C. elegans efl-1E2F, dpl-1DP, and lin-35Rb match this paradigm, as follows from their activity as class B synthetic multivulva (synMuv) genes. Genetic analyses have shown that class B synMuv genes act redundantly with class A genes in the transcriptional repression of lin-3EGF throughout the animal (Cui et al. 2006; Saffer et al. 2011). Consequently, synMuv A,B double mutation leads to ectopic expression of lin-3EGF in hyp-7, which induces neighboring VPCs to adopt a vulval fate, and causes the formation of multiple vulvas.
Remarkably, at least seven genetically identified synMuvB genes encode proteins that form a conserved repressor complex (Harrison et al. 2006). This DRM (DP, Rb, MuvB) complex is closely related to DREAM (Drosophila RBF, E2F, and Myb), which was identified through biochemical purification of Rb/E2F complexes from Drosophila embryos (Korenjak et al. 2004). A similar mammalian DREAM complex contains repressive E2F4/E2F5 and Rb-related proteins p130 or p107, as well as homologs of five synMuvB proteins (Sadasivam and DeCaprio 2013). Interestingly, mammalian DREAM acts to repress cell cycle genes in quiescent cells. In contrast, the fly and worm complexes appear most critical for restricting tissue-specific gene expression (van den Heuvel and Dyson 2008). In addition to its contribution in lin-3 repression, DRM acts to prevent expression of germline-specific genes in somatic cells. Moreover, at least a subset of synMuv B genes, including lin-35, efl-1, and efl-2, also inhibits cell cycle entry (Boxem and van den Heuvel 2002).
Transcriptional studies confirmed the dual roles for LIN-35 and DRM in repressing developmental as well as cell cycle genes (Kirienko and Fay 2007; Goetsch et al. 2017). In addition, ectopic expression of G1 CDK-cyclins in differentiated muscle cells induced the expression of a distinct set of > 200 genes with a strong cell cycle signature and enriched for promoters with E2F-binding sites (Korzelius et al. 2011b). These data suggest that LIN-35/E2F-mediated repression normally contributes to the postmitotic state of muscle cells and that active CDKs primarily antagonize the repression of cell cycle genes. EFL-1 genetically behaves as a transcriptional repressor and its predicted amino acid sequence is most similar to repressive E2F4/5 (Ceol and Horvitz 2001).
Surprisingly, activating E2Fs have not unequivocally been identified in C. elegans. EFL-3 resembles atypical E2F7/8, which repress a subset of E2F targets in mammalian systems (van den Heuvel and Dyson 2008). EFL-2 is somewhat closer to activating E2Fs in predicted amino acid sequence, but RNAi studies have not revealed cell cycle or synMuv B functions (Ceol and Horvitz 2001; Boxem and van den Heuvel 2002). E2F (the E2F/DP heterodimer) may potentially act as a transcriptional activator in the gonad, as efl-1 and dpl-1 promote the expression of oogenesis and embryogenesis-related genes, independently of lin-35 (Chi and Reinke 2006).
The strongest indication that activating E2Fs exist in C. elegans is provided by the dual phenotype associated with loss of dpl-1DP. RNAi of dpl-1 suppresses the cell cycle arrest of cyd-1 mutants, similar to efl-1, which indicates that dpl-1 normally acts to inhibit cell cycle entry. At the same time, dpl-1 loss interferes with the expression of an S phase reporter and the proliferation of ventral cord precursor cells, which implies a cell cycle-promoting role (Ceol and Horvitz 2001; Boxem and van den Heuvel 2002). A cell cycle-promoting function of DPL-1 would be expected to depend on association with an E2F partner, but an activating partner has not been identified. Incomplete inactivation of efl-2 or redundancies among the three C. elegans E2F-related genes may have masked this role.
Lineage-dependent regulation of the cell cycle
In other model organisms, transcriptional regulation of D-type cyclins and CKIs is an important mechanism for connecting developmental and environmental signals to the cell cycle. Similarly, C. elegans cyd-1 cyclin D and cdk-4 are transcriptionally activated coincident with cell cycle entry, and in a lineage-specific manner (Park and Krause 1999). Several critical DNA sequences that are evolutionarily conserved have been identified in the enhancer/promoter regions (Brodigan et al. 2003). While these sequences are putative transcription factor-binding sites, essential trans-activating factors have not been described.
In contrast to cyd-1 and cdk-4, genetic experiments have identified transcription factors as upstream cki-1 regulators. DAF-16FoxO promotes cki-1 expression during L1 and dauer arrest, and transcription factors have been implicated in lineage-specific cell cycle arrest through cki-1 induction (described above; Baugh and Sternberg 2006). For example, the VPCs are formed during the L1 stage and remain quiescent until the induction of vulva formation in L3 larvae (Sternberg 2005) (Figure 2C). This arrest depends on cki-1, as RNAi of cki-1 causes all VPCs to undergo an extra cell division in L2 (Hong et al. 1998). A screen for extra VPC cell divisions in L2 identified the CDC-14 phosphatase, as well as the LIN-1Ets and LIN-31FoxB transcription factors, as positive regulators of cki-1 expression in the VPCs (Saito et al. 2004; Clayton et al. 2008).
The LIN-1 and LIN-31 transcription factors are downstream targets of the EGFR-RAS-MAPK-related pathway that induces vulva formation (Sternberg 2005). MAPK-mediated inhibition of LIN-1 and LIN-31 in the L3 larval stage induces one of the VPCs (P6.p) to adopt a primary vulval fate, which coincides with three full rounds of cell division (Figure 2C). Thus, the transcription factors that induce cki-1 expression in the L2 stage are downregulated at the time of cell cycle reentry. This indicates a potential direct link between EGFR-RAS-MAPK-signaling and the cell cycle (Clayton et al. 2008). Indeed, genetic experiments support the notion that LIN-31 induces cki-1 transcription, and represses cdk-4 and cye-1 transcription, until phosphorylation by MAPK inhibits LIN-31 in L3 larvae (Roiz et al. 2016). In addition, the homeodomain transcription factor LIN-39 promotes VPC proliferation by preventing their fusion with the general hypodermis, and activating the transcription of cye-1 and other cell cycle genes in the VPCs (Roiz et al. 2016).
In addition to EGFR-RAS-MAPK signaling, Notch and Wnt-signaling pathways coordinate induction of the vulval cell fates with multiple rounds of cell division. VPCs that do not adopt a vulval fate go through a single division cycle before fusing with the general hypodermis (Figure 2C; P3.p, P4.p, P8.p). Thus, the signals that trigger vulval induction appear to expand the intrinsic cell proliferation potential of the VPCs. Some components of the mediator complex, which couples sequence-specific transcription factors to the RNA Polymerase II complex, are required for the VPC quiescence in L2 and vulval induction in L3 (Sternberg 2005; Clayton et al. 2008). While the mechanism is not understood, these contributions specifically involve a CDK8 module of the mediator complex, which has been implicated in transcriptional repression.
The LIN-3EGF signal that activates the LET-23EGFR-LET-60Ras-MPK-1MAPK pathway and induces a primary vulval cell fate comes from a specific cell in the somatic gonad. This “anchor cell” (AC) also organizes the connection between the gonad and vulva, which involves breaking down the basement membranes between these organs. AC signaling and invasion depend on the nuclear hormone receptor transcription factor NHR-67 (Matus et al. 2015). Interestingly, loss of nhr-67 results in continued cell division of ACs that fail to invade, while cki-1 induction arrests cell division and restores basement membrane invasion. The data are consistent with a model in which NHR-67 induces cki-1 transcription to arrest cell division of the AC, while the arrest in G1 is critical for the fully differentiated AC state (Matus et al. 2015).
Coordinating cell cycle arrest and terminal differentiation
Cell proliferation and terminal differentiation in general appear to be mutually exclusive processes, but the underlying regulatory mechanisms are still poorly understood. The fully differentiated state is acquired through a gradual process in which sequential binary decisions progressively restrict developmental potential (Kaletta et al. 1997; Bertrand and Hobert 2010). Initially, this lineage restriction occurs coincident with proliferation and results in the formation of cell type-specific precursor cells, which eventually exit the cell cycle to acquire a permanent postmitotic state. While still poorly understood for neuronal differentiation, recent studies provide insight in the coordination between terminal differentiation and cell cycle arrest during postembryonic muscle formation in C. elegans.
Neuronal differentiation:
In the C. elegans nervous system, the generation of specific neuronal cell types follows acquisition of the postmitotic state. The “terminal selector” transcription factors that induce a specific neural identity continue to be needed to maintain this identity, but not to retain the postmitotic state (Deneris and Hobert 2014). Some of these terminal selectors are first expressed in postmitotic cells, and it is possible that the combinatorial transcription factors that induce pan-neuronal properties are responsible for cell cycle withdrawal. The redundancy among such factors, which include the HOX genes and TCF/β-catenin combinations, may explain the lack of reported mutations that allow continued cell division at the expense of neuronal differentiation. Possible exceptions are egl-44 and egl-46 mutants, in which Q neuroblast descendants undergo an extra terminal division (Feng et al. 2013). Moreover, certain mutations result in neuronal lineage reiteration. Most prominently, in unc-86POU mutants, several neuroblasts adopt a repetitive stem cell-like division pattern. This phenotype appears to reflect a defect in cell identity rather than a specific defect in cell cycle exit.
Muscle formation:
Mammalian skeletal muscle development involves a cascade of myogenic regulatory factors (MRFs) that induce muscle differentiation as well as cell cycle arrest. C. elegans and other invertebrates express a single MyoD-related MRF to induce contractile muscle formation (Krause et al. 1990). Mutation of C. elegans hlh-1MyoD does not prevent striated muscle formation, as additional factors, in particular unc-120SRF and hnd-1HAND, act partially redundantly (Fukushige et al. 2006). In addition to embryonic muscle, muscle cells are formed during larval development in the lineage of the mesoblast (M) (Figure 2C). This includes 16 striated body wall muscles, as well as 16 nonstriated muscles required for egg laying. HLH-1MyoD is present specifically in the striated muscle and appears well before the terminal division (Harfe et al. 1998). Remarkably, loss of hlh-1MyoD results in the formation of extra nonstriated uterine and vulval muscles, which normally do not express HLH-1. This discrepancy was traced to the formation of extra precursor cells of the egg-laying muscle (known as “sex myoblasts”) at the expense of differentiated body wall muscle (Harfe et al. 1998). Recent observations indicate that mesoblast-specific hlh-1 knockout interferes with the proper cell cycle arrest of body wall muscle precursor cells (M. Godfrey and S. van den Heuvel, unpublished data). Thus, HLH-1MyoD contributes to both muscle cell differentiation and cell cycle arrest.
Coordinating cell cycle exit with differentiation: SWI/SNF chromatin remodelers
Studies of the C. elegans mesoblast lineage have highlighted the role of chromatin regulators in the coordination between cell cycle arrest and differentiation. Specifically, SWI/SNF chromatin remodeling complexes were found to contribute critically to the transition from proliferating precursor cells to postmitotic muscle cells (Ruijtenberg and van den Heuvel 2015). Analyzing body wall muscle and egg-laying muscle formation in M lineage-specific mutants revealed that the cell cycle arrest during differentiation uses highly redundant controls. Double mutation of lin-35 and fzr-1, which deregulates cell divisions in some other lineages, did not alter the M lineage division pattern. A few extra M descendants were formed following RNAi of cki-1, lin-23, or cul-1, as expected, and also upon RNAi of genes encoding SWI/SNF core subunits. However, testing multiple mutant and RNAi combinations revealed that only simultaneous inhibition of SWI/SNF and G1/S inhibitor functions severely disrupts cell cycle arrest and terminal differentiation, resulting in the tumorous overproliferation of M daughter cells (Ruijtenberg and van den Heuvel 2015). These data show that canonical G1/S inhibitors and SWI/SNF chromatin remodeling complexes substantially overlap in promoting cell cycle exit and the differentiation of muscle precursor cells.
Whole-worm chromatin immunoprecipitation (ChIP) experiments have revealed thousands of genomic SWI/SNF-binding sites (Riedel et al. 2013). This includes association with the promoter/enhancer regions of nearly all negative regulators of cell division, as well as the positive regulators cdk-4 and cye-1. Similarly, analysis of ChIP data demonstrated HLH-1MYOD binding at overlapping locations in the cki-1, cdk-4, and cye-1 gene regulatory regions (Ruijtenberg and van den Heuvel 2015; ChIP data from the Krause and Snyder laboratories). Combined with single-molecule FISH studies of gene expression, the data support a model in which HLH-1MYOD acts in concert with SWI/SNF chromatin remodelers to induce expression of muscle-specific genes as well as cell cycle inhibitors. At the same time, SWI/SNF complexes directly or indirectly appear to repress cdk-4 and cye-1 (Figure 8).
Figure 8.
Model for cell cycle arrest during terminal differentiation of body wall muscle precursor cells. APC/CFZR-1- and SCFLIN-23-mediated ubiquitin-dependent protein degradation, LIN-35Rb-mediated transcriptional repression, and association of CDK-inhibitory proteins with CDK/cyclin complexes promotes cell-cycle arrest. During terminal differentiation of muscle precursor cells, SWI/SNF complexes in cooperation with lineage-specific transcription factors provide an additional level of control. The different G1/S inhibitors and SWI/SNF chromatin remodelers cooperate by providing alternative levels of control over the basic cell cycle regulators, with each level antagonizing CDK-4/CYD-1 cyclin D and/or CDK-2/CYE-1 cyclin E kinase activity. Together, these regulators provide a highly robust control network for cell cycle exit.
In Drosophila and mammals, SWI/SNF chromatin remodelers antagonize Polycomb repressor complexes (PRCs) and act as important tumor suppressor genes (Kadoch and Crabtree 2015). Interestingly, downregulation of mes-2, related to the PRC2 EZH2 methyltransferase, strongly suppressed the extra divisions of muscle precursor cells in lineage-specific swsn-1 mutants (Ruijtenberg and van den Heuvel 2015). Thus, it is conceivable that loss of SWI/SNF leads to the recruitment of PRC2-related H3K27 methylation complexes that repress the transcription of genes that are normally activated during terminal differentiation. Recent results indicate that the SWI/SNF complex is highly dosage-dependent, with complete loss-of-function preventing, rather than promoting, cell proliferation (A. van der Vaart and van den Heuvel, unpublished data). These data resemble those obtained in human cancer studies (Kadoch and Crabtree 2015), indicating that the studies of SWI/SNF remodelers in cell cycle arrest during the differentiation of C. elegans muscle cells offers insight into the frequent mutation of SWI/SNF genes in human cancers.
Combining cell proliferation and differentiation through asymmetric cell division
In C. elegans, many cell divisions create an anterior and a posterior daughter cell, which differ in nuclear levels of the POP-1TCF/LEF transcription factor and cell fate (Park and Priess 2003). Such asymmetric cell divisions may segregate the potential to proliferate and commitment to differentiate to different daughter cells. A Wnt/β-catenin asymmetry pathway controls the nuclear level of POP-1, and thereby determines whether POP-1 acts as a transcriptional repressor with UNC-37Groucho or as a transcriptional activator with SYS-1β-catenin. Thus, POP-1 is an important determinant of cell fate, which shows complex interactions with the cell cycle machinery. For instance, in the somatic gonad, Cyclin D and other G1 regulators appear to regulate axis formation and sex determination, by acting as upstream regulators of the POP-1 and FKH-6 transcription factors, respectively (Tilmann and Kimble 2005).
The Wnt/β-catenin asymmetry pathway also controls an asymmetry in CKI-1 and CYE-1 levels during specific cell divisions in the somatic gonad (Fujita et al. 2007). The second asymmetric divisions of the Z1.a and Z4.p gonadal precursor cells each form a DTC. During the asymmetric division, the DTC daughter receives a higher level of CKI-1, while the sister cell maintains more CYE-1. Reducing CYE-1 or increasing CKI-1 in the sister cell causes it to adopt a DTC fate, instead of its normal quiescence followed by continued mitotic division in the L3 stage. Based on these observations, maintenance of CDK-2/cyclin E activity at a low level has been proposed to prevent the differentiation of the sister cells (Fujita et al. 2007).
Stem cell-like seam cells in the epidermis undergo asymmetric cell divisions during each larval stage (Figure 2, A and B). These asymmetric divisions create a new seam cell, and a cell that either fuses with the general epidermis (hyp7) or forms neurons (Sulston and Horvitz 1977). Around the L1/L2 molt, six of the seam cells (V1-4p-V6p and Tp) also undergo a symmetric cell division that expands the seam cell number (Figure 2A). The extra divisions substantially depend on a Runx family transcription factor, RNT-1, in association with the cofactors BRO-1CBFβ and UNC-37Groucho (Nimmo et al. 2005; Kagoshima et al. 2007; Xia et al. 2007). Based on genetic interactions, this repressor complex has been proposed to antagonize the cki-1, lin-35, and fzr-1 cell cycle inhibitor genes. This regulation, in combination with Wnt/β catenin signaling and the heterochronic network, establishes the proper cell division–differentiation patterns in the seam cell lineages.
Perspectives
The use of C. elegans as a model system has yielded considerable insight into how this simple animal regulates its cell divisions in the context of its physiology and development. The analysis of C. elegans has broken new ground in areas that include the regulation of stem cell divisions in the hermaphrodite germline and asymmetric cell divisions in the early embryo. Cell cycle regulatory functions have been described for many different CRL/SCF ubiquitin ligases, and links have been established between transcriptional regulators and cell cycle genes in the decision between proliferation and terminal differentiation. Moreover, the studies of cell cycle regulation in C. elegans have uncovered differences between tissues and developmental stages in the regulation of DNA replication and cell division.
Much research has been focused on how external conditions are associated with systemic factors, such as insulin or other hormonal signals, in the regulation of cell division. External signals that come from the environment impact how C. elegans responds in its boom and bust life cycle. In response to replete food conditions without crowding, the animal rapidly develops and reproduces. In the absence of food, animals undergo regulated quiescence in the L1 and dauer larval stages, and as adults reorganize the germline to conserve GSCs while shrinking the remainder of the germline to generate limited numbers of oocytes. Thus, cell cycle regulation in C. elegans is more tightly linked to its surroundings than, for example, the typical cell within a mammalian tissue.
Studies of the early embryo have yielded many insights into how the cell cycle is coupled to the regulation of cell polarity and positioning of the mitotic spindle, the regulation of spindle forces, and cell division asymmetry. The early embryo has also provided extensive insight into the control of centrosome duplication. The biology of the spindle and centrosomes were not covered extensively in this review, and are instead extensively described in another WormBook chapter on cell division and other reviews (Rose and Gönczy 2014).
When considering the field as a whole, what remains less well understood is the question how cell proliferation is regulated to produce the invariant cell lineage. Cell cycle regulators that are required to allow the proper cell lineage have been identified, and substantial redundancies among such regulators have been detected. The inactivation of specific negative regulators interferes with the normal program of cell cycle exit resulting in supernumerary cell divisions, and the inactivation of positive regulators prevents normal cell divisions. However, what is still largely missing is an understanding of how these cell cycle regulators are controlled within normal development to produce the stereotypical cell division pattern where, for example, a vulval lineage will undergo three rounds of cell division and then exit the cell cycle. Global regulators of these cell division patterns, such as heterochronic genes that control the timing of larval stage programs, have not yet been linked at the molecular level to the control of specific cell cycle regulators. Therefore, the promise of using C. elegans to understand how an invariant cell lineage is generated still remains largely an outstanding question.
In addition to the number and timing of cell divisions, the coordination between cell cycle arrest and terminal differentiation remains an important topic for future studies. Such studies will continue to benefit from the transparency of C. elegans, by allowing detailed observations in intact living animals. Many recent technical developments will help future analyses through advanced genome engineering, time-lapse fluorescence microscopy, and single-cell analysis methods. The possibilities for reverse genetics continue to expand, for instance with the addition of lineage-specific gene knockout and protein degradation strategies, and whole-genome sequencing has significantly accelerated conventional mutational analysis. Individual cells, proteins, and protein complexes can be followed in vivo, and can be purified efficiently from stage-synchronized animals. Many insights are expected to be obtained from genomic, transcriptomic, and proteomic analyses of individual cells, or pools of cells, extracted at well-defined developmental times. With the current knowledge of the central cell cycle regulators, the stage is now set for the connection of a large variety of cell cycle phenotypes to molecular pathways and the acquisition of a more complete understanding of the regulatory networks that control the cell cycle in animal development.
Acknowledgments
We thank all of our colleagues for their support and inspiration over the years, and apologize for all of the good research that could not be included. S.v.d.H. thanks J. Teapal for help with the figures. This work was financed by a grant from the National Institutes of Health, National Institute of General Medical Sciences (R01 GM 074212) to E.T.K. and by the Netherlands Organization for Scientific Research (Nederlandse Organisatie voor Wetenschappelijk Onderzoek), in part through a ZonMW TOP project (91216058) to S.v.d.H.
Footnotes
Communicating editor: G. Seydoux
Literature Cited
- Alexander J. L., Orr-Weaver T. L., 2016. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 30: 2241–2252. 10.1101/gad.288142.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini R., Goupil E., Labella S., Zetka M., Maddox A. S., et al. , 2014. C. elegans anillin proteins regulate intercellular bridge stability and germline syncytial organization. J. Cell Biol. 206: 129–143 [corrigenda: J. Cell Biol. 209: 467 (2015)] 10.1083/jcb.201310117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo G., Van Gilst M. R., 2009. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 326: 954–958. 10.1126/science.1178343 [DOI] [PubMed] [Google Scholar]
- Antebi A., 2013. Steroid regulation of C. elegans diapause, developmental timing, and longevity. Curr. Top. Dev. Biol. 105: 181–212. 10.1016/B978-0-12-396968-2.00007-5 [DOI] [PubMed] [Google Scholar]
- Ashcroft N., Golden A., 2002. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis 33: 1–7. 10.1002/gene.10083 [DOI] [PubMed] [Google Scholar]
- Ashcroft N. R., Srayko M., Kosinski M. E., Mains P. E., Golden A., 1999. RNA-mediated interference of a cdc25 homolog in Caenorhabditis elegans results in defects in the embryonic cortical membrane, meiosis, and mitosis. Dev. Biol. 206: 15–32. 10.1006/dbio.1998.9135 [DOI] [PubMed] [Google Scholar]
- Austin M. U., Liau W. S., Balamurugan K., Ashokkumar B., Said H. M., et al. , 2010. Knockout of the folate transporter folt-1 causes germline and somatic defects in C. elegans. BMC Dev. Biol. 10: 46 10.1186/1471-213X-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran R. S., Heighington C. S., Starostina N. G., Anderson J. W., Owen D. L., et al. , 2016. The ubiquitin ligase CRL2ZYG11 targets cyclin B1 for degradation in a conserved pathway that facilitates mitotic slippage. J. Cell Biol. 215: 151–166. 10.1083/jcb.201601083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan K., Ashokkumar B., Moussaif M., Sze J. Y., Said H. M., 2007. Cloning and functional characterization of a folate transporter from the nematode Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 293: C670–C681. 10.1152/ajpcell.00516.2006 [DOI] [PubMed] [Google Scholar]
- Bashir T., Dorrello N. V., Amador V., Guardavaccaro D., Pagano M., 2004. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature 428: 190–193. 10.1038/nature02330 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16: 780–785. 10.1016/j.cub.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., Kurhanewicz N., Sternberg P. W., 2011. Sensitive and precise quantification of insulin-like mRNA expression in Caenorhabditis elegans. PLoS One 6: e18086 10.1371/journal.pone.0018086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V., Hobert O., 2010. Lineage programming: navigating through transient regulatory states via binary decisions. Curr. Opin. Genet. Dev. 20: 362–368. 10.1016/j.gde.2010.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke A., Fielenbach N., Wang Z., Mangelsdorf D. J., Antebi A., 2009. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324: 95–98. 10.1126/science.1164899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., et al. , 2009. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17: 355–364. 10.1016/j.devcel.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Boxem M., van den Heuvel S., 2001. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128: 4349–4359. [DOI] [PubMed] [Google Scholar]
- Boxem M., van den Heuvel S., 2002. C. elegans class B synthetic multivulva genes act in G1 regulation. Curr. Biol. 12: 906–911. 10.1016/S0960-9822(02)00844-8 [DOI] [PubMed] [Google Scholar]
- Boxem M., Srinivasan D. G., van den Heuvel S., 1999. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126: 2227–2239. [DOI] [PubMed] [Google Scholar]
- Brauchle M., Baumer K., Gönczy P., 2003. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13: 819–827. 10.1016/S0960-9822(03)00295-1 [DOI] [PubMed] [Google Scholar]
- Brenner J. L., Schedl T., 2016. Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics 202: 1085–1103. 10.1534/genetics.115.185678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodigan T. M., Liu J., Park M., Kipreos E. T., Krause M., 2003. Cyclin E expression during development in Caenorhabditis elegans. Dev. Biol. 254: 102–115. 10.1016/S0012-1606(02)00032-5 [DOI] [PubMed] [Google Scholar]
- Buck S. H., Chiu D., Saito R. M., 2009. The cyclin-dependent kinase inhibitors, cki-1 and cki-2, act in overlapping but distinct pathways to control cell cycle quiescence during C. elegans development. Cell Cycle 8: 2613–2620. 10.4161/cc.8.16.9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budirahardja Y., Gönczy P., 2008. PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development 135: 1303–1313. 10.1242/dev.019075 [DOI] [PubMed] [Google Scholar]
- Burrows A. E., Sceurman B. K., Kosinski M. E., Richie C. T., Sadler P. L., et al. , 2006. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development 133: 697–709. 10.1242/dev.02241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L. A., Katzaroff A. J., Perez C. L., de la Cruz A., Edgar B. A., 2007. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 12: 631–643. 10.1016/j.devcel.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Byrd D. T., Knobel K., Affeldt K., Crittenden S. L., Kimble J., 2014. A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS One 9: e88372 10.1371/journal.pone.0088372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro P. V., Khare S., Young B. D., Clarke S. G., 2012. Caenorhabditis elegans battling starvation stress: low levels of ethanol prolong lifespan in L1 larvae. PLoS One 7: e29984 10.1371/journal.pone.0029984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol C. J., Horvitz H. R., 2001. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7: 461–473. 10.1016/S1097-2765(01)00194-0 [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C., 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. 10.1126/science.8303295 [DOI] [PubMed] [Google Scholar]
- Chase D., Serafinas C., Ashcroft N., Kosinski M., Longo D., et al. , 2000. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26: 26–41. [DOI] [PubMed] [Google Scholar]
- Chaudhari S. N., Mukherjee M., Vagasi A. S., Bi G., Rahman M. M., et al. , 2016. Bacterial folates provide an exogenous signal for C. elegans germline stem cell proliferation. Dev. Cell 38: 33–46. 10.1016/j.devcel.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C.-C., Klancer R., Singson A., Seydoux G., 2009. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139: 560–572. 10.1016/j.cell.2009.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Reinke V., 2006. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development 133: 3147–3157. 10.1242/dev.02490 [DOI] [PubMed] [Google Scholar]
- Cinquin O., Crittenden S. L., Morgan D. E., Kimble J., 2010. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. USA 107: 2048–2053. 10.1073/pnas.0912704107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. E., van den Heuvel S. J. L., Saito R. M., 2008. Transcriptional control of cell-cycle quiescence during C. elegans development. Dev. Biol. 313: 603–613. 10.1016/j.ydbio.2007.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A., 2006. Cyclin E-Cdk2 temporally regulates centrosome assembly and establishment of polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8: 1441–1447. 10.1038/ncb1511 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., et al. , 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417: 660–663. 10.1038/nature754 [DOI] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T., Kimble J., 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17: 3051–3061. 10.1091/mbc.e06-03-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Chen J., Myers T. R., Hwang B. J., Sternberg P. W., et al. , 2006. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10: 667–672. 10.1016/j.devcel.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Dalfó D., Michaelson D., Hubbard E. J., 2012. Sensory regulation of the C. elegans germline through TGF-β-dependent signaling in the niche. Curr. Biol. 22: 712–719. 10.1016/j.cub.2012.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. S., Wille L., Chestnut B. A., Sadler P. L., Shakes D. C., et al. , 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneris E. S., Hobert O., 2014. Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17: 899–907. 10.1038/nn.3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Blow J. J., Ghosh S., Saha T., Noguchi K., et al. , 2006. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 18: 231–239. 10.1016/j.ceb.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Deppe U., Schierenberg E., Cole T., Krieg C., Schmitt D., et al. , 1978. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 75: 376–380. 10.1073/pnas.75.1.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R., 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1: 187–199. 10.1016/S1534-5807(01)00026-0 [DOI] [PubMed] [Google Scholar]
- Deyter G. M. R., Furuta T., Kurasawa Y., Schumacher J. M., 2010. Caenorhabditis elegans cyclin B3 is required for multiple mitotic processes including alleviation of a spindle checkpoint-dependent block in anaphase chromosome segregation. PLoS Genet. 6: e1001218 10.1371/journal.pgen.1001218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J. A., Cheng M., Roussel M. F., Sherr C. J., 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12: 3499–3511. 10.1101/gad.12.22.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D. K., Stevaux O., Frolov M. V., Dyson N. J., 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17: 2308–2320. 10.1101/gad.1116703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorée M., Hunt T., 2002. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J. Cell Sci. 115: 2461–2464. [DOI] [PubMed] [Google Scholar]
- Dunphy W. G., Brizuela L., Beach D., Newport J., 1988. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 54: 423–431. 10.1016/0092-8674(88)90205-X [DOI] [PubMed] [Google Scholar]
- Edgar L. G., McGhee J. D., 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599. 10.1016/0092-8674(88)90575-2 [DOI] [PubMed] [Google Scholar]
- Encalada S. E., Martin P. R., Phillips J. B., Lyczak R., Hamill D. R., et al. , 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. 10.1006/dbio.2000.9965 [DOI] [PubMed] [Google Scholar]
- Encalada S. E., Willis J., Lyczak R., Bowerman B., 2005. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 16: 1056–1070. 10.1091/mbc.e04-08-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Lara-Gonzalez P., Sassine M., Shiau A. K., Desai A., et al. , 2015. Natural loss of Mps1 kinase in nematodes uncovers a role for Polo-like Kinase 1 in spindle checkpoint initiation. Cell Rep. 12: 58–65. 10.1016/j.celrep.2015.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euling S., Ambros V., 1996. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell 84: 667–676. 10.1016/S0092-8674(00)81045-4 [DOI] [PubMed] [Google Scholar]
- Fay D. S., Han M., 2000. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127: 4049–4060. [DOI] [PubMed] [Google Scholar]
- Fay D. S., Keenan S., Han M., 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517. 10.1101/gad.952302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G., Yi P., Yang Y., Chai Y., Tian D., et al. , 2013. Developmental stage-dependent transcriptional regulatory pathways control neuroblast lineage progression. Development 140: 3838–3847. 10.1242/dev.098723 [DOI] [PubMed] [Google Scholar]
- Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., et al. , 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1: 486–492. 10.1038/70272 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Morgan D. O., 1994. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell 78: 713–724. 10.1016/0092-8674(94)90535-5 [DOI] [PubMed] [Google Scholar]
- Flemming A. J., Shen Z. Z., Cunha A., Emmons S. W., Leroi A. M., 2000. Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc. Natl. Acad. Sci. USA 97: 5285–5290. 10.1073/pnas.97.10.5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Schedl T., 2015. Analysis of germline stem cell differentiation following loss of GLP-1 Notch activity in Caenorhabditis elegans. Genetics 201: 167–184. 10.1534/genetics.115.178061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. M., Vought V. E., Hanazawa M., Lee M. H., Maine E. M., et al. , 2011. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138: 2223–2234. 10.1242/dev.059535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Takeshita H., Sawa H., 2007. Cyclin E and CDK2 repress the terminal differentiation of quiescent cells after asymmetric division in C. elegans. PLoS One 2: e407 10.1371/journal.pone.0000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Brodigan T. M., Schriefer L. A., Waterston R. H., Krause M., 2006. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 20: 3395–3406. 10.1101/gad.1481706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Gendreau S. B., Derry W. B., Rothman J. H., 2003. Essential embryonic roles of the CKI-1 cyclin-dependent kinase inhibitor in cell-cycle exit and morphogenesis in C elegans. Dev. Biol. 260: 273–286. 10.1016/S0012-1606(03)00239-2 [DOI] [PubMed] [Google Scholar]
- Fukuyama M., Rougvie A. E., Rothman J. H., 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16: 773–779. 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- Galli M., Morgan D. O., 2016. Cell size determines the strength of the spindle assembly checkpoint during embryonic development. Dev. Cell 36: 344–352. 10.1016/j.devcel.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Norbury C., Lohka M., Nurse P., Maller J., 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54: 433–439. 10.1016/0092-8674(88)90206-1 [DOI] [PubMed] [Google Scholar]
- Gibert M. A., Starck J., Beguet B., 1984. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50: 77–85. 10.1111/j.1768-322X.1984.tb00254.x [DOI] [PubMed] [Google Scholar]
- Goetsch P. D., Garrigues J. M., Strome S., 2017. Loss of the Caenorhabditis elegans pocket protein LIN-35 reveals MuvB’s innate function as the repressor of DREAM target genes. PLoS Genet. 13: e1007088 10.1371/journal.pgen.1007088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. 10.1083/jcb.151.7.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A., Boke E., Hagting A., Hodgson B., Connolly Y., et al. , 2015. A PP1–PP2A phosphatase relay controls mitotic progression. Nature 517: 94–98. 10.1038/nature14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. E., Odde D. J., Seydoux G., 2011. Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell 146: 955–968. 10.1016/j.cell.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny, T. L., and C. Savage-Dunn, 2013 TGF-β signaling in C. elegans (July 10, 2013), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.22.2, http://www.wormbook.org. 10.1895/wormbook.1.22.2 [DOI] [PMC free article] [PubMed]
- Guo S., Kemphues K. J., 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T., Nishi Y., Robertson S. M., Lin R., 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 135: 149–160. 10.1016/j.cell.2008.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Altun Z. F., 2008. C. elegans Atlas. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hansen D., Schedl T., 2013. Stem cell proliferation vs. meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 71–99. 10.1007/978-1-4614-4015-4_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Branda C. S., Krause M., Stern M. J., Fire A., 1998. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development 125: 2479–2488. [DOI] [PubMed] [Google Scholar]
- Harris D. T., Horvitz H. R., 2011. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138: 4051–4062. 10.1242/dev.065417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Ceol C. J., Lu X., Horvitz H. R., 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA 103: 16782–16787. 10.1073/pnas.0608461103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., 2002. Nobel Lecture. Yeast and cancer. Biosci. Rep. 22: 373–394. 10.1023/A:1020918107706 [DOI] [PubMed] [Google Scholar]
- Havens C. G., Walter J. C., 2011. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25: 1568–1582. 10.1101/gad.2068611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeisen M., Roy R., 2008. CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans. Development 135: 1259–1269. 10.1242/dev.014969 [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., White J. G., 1985. Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 107: 128–133. 10.1016/0012-1606(85)90381-1 [DOI] [PubMed] [Google Scholar]
- Hong Y., Roy R., Ambros V., 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey P. D., Russo A. A., Polyak K., Gibbs E., Hurwitz J., et al. , 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313–320. 10.1038/376313a0 [DOI] [PubMed] [Google Scholar]
- Jeong J., Verheyden J. M., Kimble J., 2011. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 7: e1001348 10.1371/journal.pgen.1001348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Crabtree G. R., 2015. Mammalian SWI/SNF chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. Sci. Adv. 1: e1500447 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H., Nimmo R., Saad N., Tanaka J., Miwa Y., et al. , 2007. The C. elegans CBFβ homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development 134: 3905–3915. 10.1242/dev.008276 [DOI] [PubMed] [Google Scholar]
- Kalchhauser I., Farley B. M., Pauli S., Ryder S. P., Ciosk R., 2011. FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. EMBO J. 30: 3823–3829. 10.1038/emboj.2011.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T., Schnabel H., Schnabel R., 1997. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature 390: 294–298. 10.1038/36869 [DOI] [PubMed] [Google Scholar]
- Kasuga H., Fukuyama M., Kitazawa A., Kontani K., Katada T., 2013. The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497: 503–506. 10.1038/nature12117 [DOI] [PubMed] [Google Scholar]
- Kermi C., Lo Furno E., Maiorano D., 2017. Regulation of DNA replication in early embryonic cleavages. Genes (Basel) 8: 42 10.3390/genes8010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A. M., Shin H., Hansen T. J., Kimble J., 2014. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 111: 3739–3744. 10.1073/pnas.1401861111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Feng H., Kipreos E. T., 2007. C. elegans CUL-4 prevents rereplication by promoting the nuclear export of CDC-6 via a CKI-1-dependent pathway. Curr. Biol. 17: 966–972. 10.1016/j.cub.2007.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kawasaki I., Shim Y.-H., 2010. cdc-25.2, a C. elegans ortholog of cdc25, is required to promote oocyte maturation. J. Cell Sci. 123: 993–1000. 10.1242/jcs.060442 [DOI] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. 10.1007/978-1-4614-4015-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kipreos E. T., 2007. The Caenorhabditis elegans replication licensing factor CDT-1 is targeted for degradation by the CUL-4/DDB-1 complex. Mol. Cell. Biol. 27: 1394–1406. 10.1128/MCB.00736-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Starostina N. G., Kipreos E. T., 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22: 2507–2519. 10.1101/gad.1703708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J., and S. L. Crittenden, 2005 Germline proliferation and its control (August 15, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.13.1, http://www.wormbook.org. 10.1895/wormbook.1.13.1 [DOI] [PMC free article] [PubMed]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. 10.1016/0012-1606(79)90035-6 [DOI] [PubMed] [Google Scholar]
- Kimble, J., and H. Seidel, 2014 C. elegans germline stem cells and their niche. StemBook [Internet]. Stem Book. Available at http//dx..org/10.3824/stembook.1.95.1. [Google Scholar]
- Kipreos E. T., 2005. C. elegans cell cycles: invariance and stem cell divisions. Nat. Rev. Mol. Cell Biol. 6: 766–776. 10.1038/nrm1738 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Lander L. E., Wing J. P., He W. W., Hedgecock E. M., 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85: 829–839. 10.1016/S0092-8674(00)81267-2 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Gohel S. P., Hedgecock E. M., 2000. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development 127: 5071–5082. [DOI] [PubMed] [Google Scholar]
- Kirienko N. V., Fay D. S., 2007. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev. Biol. 305: 674–684. 10.1016/j.ydbio.2007.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T., 2015. Entry into mitosis: a solution to the decades-long enigma of MPF. Chromosoma 124: 417–428. 10.1007/s00412-015-0508-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R., Law E., Tang L., Rose A. M., 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12: 2118–2123. 10.1016/S0960-9822(02)01392-1 [DOI] [PubMed] [Google Scholar]
- Korenjak M., Taylor-Harding B., Binné U. K., Satterlee J. S., Stevaux O., et al. , 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193. 10.1016/j.cell.2004.09.034 [DOI] [PubMed] [Google Scholar]
- Korzelius J., The I., Ruijtenberg S., Portegijs V., Xu H., et al. , 2011a. C. elegans MCM-4 is a general DNA replication and checkpoint component with an epidermis-specific requirement for growth and viability. Dev. Biol. 350: 358–369. 10.1016/j.ydbio.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J., The I., Ruijtenberg S., Prinsen M. B. W., Portegijs V., et al. , 2011b. Caenorhabditis elegans cyclin D/CDK4 and cyclin E/CDK2 induce distinct cell cycle re-entry programs in differentiated muscle cells. PLoS Genet. 7: e1002362 10.1371/journal.pgen.1002362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Fire A., Harrison S. W., Priess J., Weintraub H., 1990. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell 63: 907–919. 10.1016/0092-8674(90)90494-Y [DOI] [PubMed] [Google Scholar]
- Kreutzer M. A., Richards J. P., De Silva-Udawatta M. N., Temenak J. J., Knoblich J. A., et al. , 1995. Caenorhabditis elegans cyclin A- and B-type genes: a cyclin A multigene family, an ancestral cyclin B3 and differential germline expression. J. Cell Sci. 108: 2415–2424. [DOI] [PubMed] [Google Scholar]
- Lamont L. B., Crittenden S. L., Bernstein D., Wickens M., Kimble J., 2004. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev. Cell 7: 697–707. 10.1016/j.devcel.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Lee Y. U., Son M., Kim J., Shim Y. H., Kawasaki I., 2016. CDC-25.2, a C. elegans ortholog of cdc25, is essential for the progression of intestinal divisions. Cell Cycle 15: 654–666. 10.1080/15384101.2016.1146839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X., Noble M., Adams P. D., Qin J., Harper J. W., 2002. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol. Cell. Biol. 22: 2242–2254. 10.1128/MCB.22.7.2242-2254.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault S. W., Shakes D. C., Ward S., 1988. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics 120: 435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M. A., Duronio R. J., 2005. New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775. 10.1038/sj.onc.1208610 [DOI] [PubMed] [Google Scholar]
- Liu J., Kipreos E. T., 2000. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 17: 1061–1074. 10.1093/oxfordjournals.molbev.a026387 [DOI] [PubMed] [Google Scholar]
- Liu J., Vasudevan S., Kipreos E. T., 2004. CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior-posterior axis in C. elegans. Development 131: 3513–3525. 10.1242/dev.01245 [DOI] [PubMed] [Google Scholar]
- Lorson M. A., Horvitz H. R., van den Heuvel S., 2000. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J. Cell Biol. 148: 73–86. 10.1083/jcb.148.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Hunter T., 2010. Ubiquitylation and proteasomal degradation of the p21Cip1, p27Kip1 and p57Kip2 CDK inhibitors. Cell Cycle 9: 2342–2352. 10.4161/cc.9.12.11988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R., Velarde N. V., Klancer R., Gordon S., Kadandale P., et al. , 2007. EGG-3 regulates cell-surface and cortex rearrangements during egg activation in Caenorhabditis elegans. Curr. Biol. 17: 1555–1560. 10.1016/j.cub.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Masui Y., Markert C. L., 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J. Exp. Zool. 177: 129–145. 10.1002/jez.1401770202 [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Lohmer L. L., Kelley L. C., Schindler A. J., Kohrman A. Q., et al. , 2015. Invasive cell fate requires G1 cell-cycle arrest and histone deacetylase-mediated changes in gene expression. Dev. Cell 35: 162–174. 10.1016/j.devcel.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T., 1997. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181: 121–143. 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- McNally K. L., McNally F. J., 2005. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev. Biol. 282: 218–230. 10.1016/j.ydbio.2005.03.009 [DOI] [PubMed] [Google Scholar]
- Méchali M., 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11: 728–738. 10.1038/nrm2976 [DOI] [PubMed] [Google Scholar]
- Medema R. H., Kops G. J., Bos J. L., Burgering B. M., 2000. AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404: 782–787. 10.1038/35008115 [DOI] [PubMed] [Google Scholar]
- Merlet J., Burger J., Tavernier N., Richaudeau B., Gomes J. E., et al. , 2010. The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137: 3857–3866. 10.1242/dev.054866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., 2016. Cyclin CYB-3 controls both S-phase and mitosis and is asymmetrically distributed in the early C. elegans embryo. Development 143: 3119–3127. 10.1242/dev.141226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y., Hubbard E. J., 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. 10.1242/dev.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. L., Roth M. B., 2009. C. elegans are protected from lethal hypoxia by an embryonic diapause. Curr. Biol. 19: 1233–1237. 10.1016/j.cub.2009.05.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., et al. , 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291: 2144–2147. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Ruest P. J., Kosinski M., Hanks S. K., Greenstein D., 2003. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 17: 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A., Broek K. V., Wang C., Daryabeigi A., Jantsch V., et al. , 2018. Initiation of meiotic development is controlled by three posttranscriptional pathways in Caenorhabditis elegans. Genetics 209: 1197–1224. 10.1534/genetics.118.300985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M., Chaudhari S. N., Balachandran R. S., Vagasi A. S., Kipreos E. T., 2017. Dafachronic acid inhibits C. elegans germ cell proliferation in a DAF-12-dependent manner. Dev. Biol. 432: 215–221. 10.1016/j.ydbio.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T., and P. J. Hu, 2013 Insulin/insulin-like growth factor signaling in C. elegans (December 26, 2013), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.164.1, http://www.wormbook.org. 10.1895/wormbook.1.164.1 [DOI] [PMC free article] [PubMed]
- Narbonne P., Maddox P. S., Labbe J. C., 2015. DAF-18/PTEN locally antagonizes insulin signalling to couple germline stem cell proliferation to oocyte needs in C. elegans. Development 142: 4230–4241. 10.1242/dev.130252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., 2005. How do so few control so many? Cell 120: 739–746. 10.1016/j.cell.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Nimmo R., Antebi A., Woollard A., 2005. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development 132: 5043–5054. 10.1242/dev.02102 [DOI] [PubMed] [Google Scholar]
- Nishi Y., Rogers E., Robertson S. M., Lin R., 2008. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development 135: 687–697. 10.1242/dev.013425 [DOI] [PubMed] [Google Scholar]
- Nurse P., 2002. Cyclin dependent kinases and cell cycle control (Nobel Lecture). ChemBioChem 3: 596–603. [DOI] [PubMed] [Google Scholar]
- Nystrom J., Shen Z. Z., Aili M., Flemming A. J., Leroi A., et al. , 2002. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics 161: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystul T. G., Goldmark J. P., Padilla P. A., Roth M. B., 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science 302: 1038–1041. 10.1126/science.1089705 [DOI] [PubMed] [Google Scholar]
- O’Connell K. F., Leys C. M., White J. G., 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149: 1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K. W., Scadden D. T., 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9: 115–128. 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Pacquelet A., 2017. Asymmetric cell division in the one-cell C. elegans embryo: multiple steps to generate cell size asymmetry. Results Probl. Cell Differ. 61: 115–140. 10.1007/978-3-319-53150-2_5 [DOI] [PubMed] [Google Scholar]
- Park F. D., Priess J. R., 2003. Establishment of POP-1 asymmetry in early C. elegans embryos. Development 130: 3547–3556. 10.1242/dev.00563 [DOI] [PubMed] [Google Scholar]
- Park M., Krause M. W., 1999. Regulation of postembryonic G1 cell cycle progression in Caenorhabditis elegans by a cyclin D/CDK-like complex. Development 126: 4849–4860. [DOI] [PubMed] [Google Scholar]
- Parry J. M., Velarde N. V., Lefkovith A. J., Zegarek M. H., Hang J. S., et al. , 2009. EGG-4 and EGG-5 link events of the oocyte-to-embryo transition with meiotic progression in C. elegans. Curr. Biol. 19: 1752–1757. 10.1016/j.cub.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J., Reinke V., Kim S. K., Seydoux G., 2003. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev. Cell 5: 451–462. 10.1016/S1534-5807(03)00231-4 [DOI] [PubMed] [Google Scholar]
- Pinkston J. M., Garigan D., Hansen M., Kenyon C., 2006. Mutations that increase the life span of C. elegans inhibit tumor growth. Science 313: 971–975. 10.1126/science.1121908 [DOI] [PubMed] [Google Scholar]
- Pozo P. N., Cook J. G., 2017. Regulation and function of Cdt1; a key factor in cell proliferation and genome stability. Genes (Basel) 8: 2 10.3390/genes8010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S., Mains P. E., Zinke A., Hyman A. A., 2003. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 4: 1175–1181. 10.1038/sj.embor.7400029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M., Rosu S., Joseph-Strauss D., Cohen-Fix O., 2014. Down-regulation of tricarboxylic acid (TCA) cycle genes blocks progression through the first mitotic division in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 111: 2602–2607. 10.1073/pnas.1311635111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch M., Ecsedi M., Bartake H., Müllner A., Grosshans H., 2015. A genetic interactome of the let-7 microRNA in C. elegans. Dev. Biol. 401: 276–286. 10.1016/j.ydbio.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Albert P. S., 1997. Genetic and environmental regulation of dauer larva development in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Riedel C. G., Dowen R. H., Lourenco G. F., Kirienko N. V., Heimbucher T., et al. , 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15: 491–501. 10.1038/ncb2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera A., Tognetti S., Speck C., 2014. Helicase loading: how to build a MCM2–7 double-hexamer. Semin. Cell Dev. Biol. 30: 104–109. 10.1016/j.semcdb.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Rivers D. M., Moreno S., Abraham M., Ahringer J., 2008. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J. Cell Biol. 180: 877–885. 10.1083/jcb.200710018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiz D., Escobar-Restrepo J. M., Leu P., Hajnal A., 2016. The C. elegans hox gene lin-39 controls cell cycle progression during vulval development. Dev. Biol. 418: 124–134. 10.1016/j.ydbio.2016.07.018 [DOI] [PubMed] [Google Scholar]
- Rose, L., and P. Gönczy, 2014 Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos (December 30, 2014), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.30.2, http://www.wormbook.org. 10.1895/wormbook.1.30.2 [DOI] [PubMed]
- Rosu S., Cohen-Fix O., 2017. Live-imaging analysis of germ cell proliferation in the C. elegans adult supports a stochastic model for stem cell proliferation. Dev. Biol. 423: 93–100. 10.1016/j.ydbio.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Michaelson D., Hochman T., Santella A., Bao Z., et al. , 2016. Cell cycle features of C. elegans germline stem/progenitor cells vary temporally and spatially. Dev. Biol. 409: 261–271. 10.1016/j.ydbio.2015.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin S. M., 2013. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 38: 12–19. 10.1016/j.tibs.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S., 2015. G1/S Inhibitors and the SWI/SNF complex control cell-cycle exit during muscle differentiation. Cell 162: 300–313. 10.1016/j.cell.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Sadasivam S., DeCaprio J. A., 2013. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 13: 585–595 (erratum: Nat. Rev. Cancer 13: 752) 10.1038/nrc3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer A. M., Kim D. H., van Oudenaarden A., Horvitz H. R., 2011. The Caenorhabditis elegans synthetic multivulva genes prevent Ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF. PLoS Genet. 7: e1002418 10.1371/journal.pgen.1002418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R. M., Perreault A., Peach B., Satterlee J. S., van den Heuvel S., 2004. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol. 6: 777–783. 10.1038/ncb1154 [DOI] [PubMed] [Google Scholar]
- Segref A., Cabello J., Clucas C., Schnabel R., Johnstone I. L., 2010. Fate specification and tissue-specific cell cycle control of the Caenorhabditis elegans intestine. Mol. Biol. Cell 21: 725–738. 10.1091/mbc.e09-04-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Kimble J., 2011. The oogenic germline starvation response in C. elegans. PLoS One 6: e28074 10.1371/journal.pone.0028074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Kimble J., 2015. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. Elife 4: e10832 10.7554/eLife.10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selhub J., 2002. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J. Nutr. Health Aging 6: 39–42. [PubMed] [Google Scholar]
- Sherr C. J., 1996. Cancer cell cycles. Science 274: 1672–1677. 10.1126/science.274.5293.1672 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Soto M. C., Ishidate T., Kim S., Nakamura K., et al. , 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16: 47–55. 10.1016/j.cub.2005.11.070 [DOI] [PubMed] [Google Scholar]
- Singaravelu G., Singson A., 2013. Calcium signaling surrounding fertilization in the nematode Caenorhabditis elegans. Cell Calcium 53: 2–9. 10.1016/j.ceca.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu G., Chatterjee I., Rahimi S., Druzhinina M. K., Kang L., et al. , 2012. The sperm surface localization of the TRP-3/SPE-41 Ca2+ -permeable channel depends on SPE-38 function in Caenorhabditis elegans. Dev. Biol. 365: 376–383. 10.1016/j.ydbio.2012.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Smith J., Tho L. M., Xu N., Gillespie D. A., 2010. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108: 73–112. 10.1016/B978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- Son M., Kawasaki I., Oh B.-K., Shim Y.-H., 2016. LIN-23, an E3 ubiquitin ligase component, is required for the repression of CDC-25.2 activity during intestinal development in Caenorhabditis elegans. Mol. Cells 39: 834–840. 10.14348/molcells.2016.0238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Gönczy P., 2004. zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131: 3527–3543. 10.1242/dev.01244 [DOI] [PubMed] [Google Scholar]
- Sonneville R., Querenet M., Craig A., Gartner A., Blow J. J., 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196: 233–246. 10.1083/jcb.201110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., et al. , 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469. 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Sowa J. N., Mutlu A. S., Xia F., Wang M. C., 2015. Olfaction modulates reproductive plasticity through neuroendocrine signaling in Caenorhabditis elegans. Curr. Biol. 25: 2284–2289. 10.1016/j.cub.2015.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D., et al. , 2014a. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics 198: 1535–1558. 10.1534/genetics.114.168831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Nishi Y., Guven-Ozkan T., Oldenbroek M., et al. , 2014b. Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics 198: 1513–1533. 10.1534/genetics.114.168823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D. G., Fisk R. M., Xu H., van den Heuvel S., 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 17: 1225–1239. 10.1101/gad.1081203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina N. G., Simpliciano J. M., McGuirk M. A., Kipreos E. T., 2010. CRL2(LRR-1) targets a CDK inhibitor for cell cycle control in C. elegans and actin-based motility regulation in human cells. Dev. Cell 19: 753–764. 10.1016/j.devcel.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A., 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38: 203–232. 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., 2005 Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.6.1, http://www. wormbook.org. 10.1895/wormbook.1.6.1 [DOI] [PMC free article] [PubMed]
- Stevens H., Williams A. B., Michael W. M., 2016. Cell-type specific responses to DNA replication stress in early C. elegans embryos. PLoS One 11: e0164601 10.1371/journal.pone.0164601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel M. L., Pellettieri J., Seydoux G., 2006. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr. Biol. 16: 56–62. 10.1016/j.cub.2005.11.063 [DOI] [PubMed] [Google Scholar]
- Stitzel M. L., Cheng K. C.-C., Seydoux G., 2007. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr. Biol. 17: 1545–1554. 10.1016/j.cub.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G., 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82: 41–55. 10.1016/0012-1606(81)90427-9 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Tain L. S., Lozano E., Saez A. G., Leroi A. M., 2008. Dietary regulation of hypodermal polyploidization in C. elegans. BMC Dev. Biol. 8: 28 10.1186/1471-213X-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Araki H., 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 5: a010371 10.1101/cshperspect.a010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernier N., Noatynska A., Panbianco C., Martino L., Van Hove L., et al. , 2015. Cdk1 phosphorylates SPAT-1/Bora to trigger PLK-1 activation and drive mitotic entry in C. elegans embryos. J. Cell Biol. 208: 661–669. 10.1083/jcb.201408064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenlen J. R., Molk J. N., London N., Page B. D., Priess J. R., 2008. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development 135: 3665–3675. 10.1242/dev.027060 [DOI] [PubMed] [Google Scholar]
- The I., Ruijtenberg S., Bouchet B. P., Cristobal A., Prinsen M. B. W., et al. , 2015. Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells. Nat. Commun. 6: 5906 10.1038/ncomms6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thondamal M., Witting M., Schmitt-Kopplin P., Aguilaniu H., 2014. Steroid hormone signalling links reproduction to lifespan in dietary-restricted Caenorhabditis elegans. Nat. Commun. 5: 4879 10.1038/ncomms5879 [DOI] [PubMed] [Google Scholar]
- Tilmann C., Kimble J., 2005. Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev. Cell 9: 489–499. 10.1016/j.devcel.2005.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Gearhart M. D., Spike C. A., Huelgas-Morales G., Mews M., et al. , 2017. LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 206: 2007–2039. 10.1534/genetics.117.203174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, S., 2005 Cell-cycle regulation (September 21, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.28.1, http://www.wormbook.org. 10.1895/wormbook.1.28.1 [DOI] [PMC free article] [PubMed]
- van den Heuvel S., Dyson N. J., 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724. 10.1038/nrm2469 [DOI] [PubMed] [Google Scholar]
- van der Voet M., Lorson M. A., Srinivasan D. G., Bennett K. L., van den Heuvel S., 2009. C. elegans mitotic cyclins have distinct as well as overlapping functions in chromosome segregation. Cell Cycle 8: 4091–4102. 10.4161/cc.8.24.10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Starostina N. G., Kipreos E. T., 2007. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8: 279–286. 10.1038/sj.embor.7400895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A., Koff A., 2000. Cell-cycle inhibitors: three families united by a common cause. Gene 247: 1–15. [DOI] [PubMed] [Google Scholar]
- von Schubert C., Cubizolles F., Bracher J. M., Sliedrecht T., Kops G., et al. , 2015. Plk1 and Mps1 cooperatively regulate the spindle assembly checkpoint in human cells. Cell Rep. 12: 66–78. 10.1016/j.celrep.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Von Stetina J. R., Orr-Weaver T. L., 2011. Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol. 3: a005553 10.1101/cshperspect.a005553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Boxem M., 2014. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods 68: 381–388. [DOI] [PubMed] [Google Scholar]
- Wallenfang M. R., Seydoux G., 2002. cdk-7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99: 5527–5532. 10.1073/pnas.082618399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Carrel J. S., 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73: 304–321. 10.1016/0012-1606(79)90069-1 [DOI] [PubMed] [Google Scholar]
- Wei W., Ayad N. G., Wan Y., Zhang G. J., Kirschner M. W., et al. , 2004. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428: 194–198. 10.1038/nature02381 [DOI] [PubMed] [Google Scholar]
- Wilson M. A., Hoch R. V., Ashcroft N. R., Kosinski M. E., Golden A., 1999. A Caenorhabditis elegans wee1 homolog is expressed in a temporally and spatially restricted pattern during embryonic development. Biochim. Biophys. Acta 1445: 99–109. 10.1016/S0167-4781(99)00027-5 [DOI] [PubMed] [Google Scholar]
- Wirt S. E., Adler A. S., Gebala V., Weimann J. M., Schaffer B. E., et al. , 2010. G1 arrest and differentiation can occur independently of Rb family function. J. Cell Biol. 191: 809–825. 10.1083/jcb.201003048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A., Hodgkin J., 1999. stu-7/air-2 is a C. elegans aurora homologue essential for chromosome segregation during embryonic and post-embryonic development. Mech. Dev. 82: 95–108. 10.1016/S0925-4773(99)00020-9 [DOI] [PubMed] [Google Scholar]
- Xia D., Zhang Y., Huang X., Sun Y., Zhang H., 2007. The C. elegans CBFβ homolog, BRO-1, regulates the proliferation, differentiation and specification of the stem cell-like seam cell lineages. Dev. Biol. 309: 259–272. 10.1016/j.ydbio.2007.07.020 [DOI] [PubMed] [Google Scholar]
- Yanagi K., Mizuno T., Tsuyama T., Tada S., Iida Y., et al. , 2005. Caenorhabditis elegans geminin homologue participates in cell cycle regulation and germ line development. J. Biol. Chem. 280: 19689–19694. 10.1074/jbc.C500070200 [DOI] [PubMed] [Google Scholar]
- Yanowitz J., Fire A., 2005. Cyclin D involvement demarcates a late transition in C. elegans embryogenesis. Dev. Biol. 279: 244–251. 10.1016/j.ydbio.2004.12.022 [DOI] [PubMed] [Google Scholar]
- Yuan K., O’Farrell P. H., 2015. Cyclin B3 is a mitotic cyclin that promotes the metaphase-anaphase transition. Curr. Biol. 25: 811–816. 10.1016/j.cub.2015.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Seller C. A., Shermoen A. W., O’Farrell P. H., 2016. Timing the Drosophila mid-blastula transition: a cell cycle-centered view. Trends Genet. 32: 496–507. 10.1016/j.tig.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Qi S.-T., Huang L., Ma X.-S., Ouyang Y.-C., et al. , 2015. Cyclin B3 controls anaphase onset independent of spindle assembly checkpoint in meiotic oocytes. Cell Cycle 14: 2648–2654. 10.1080/15384101.2015.1064567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Feng H., Santiago F. E., Kipreos E. T., 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889. 10.1038/nature01747 [DOI] [PubMed] [Google Scholar]
- Zitouni S., Nabais C., Jana S. C., Guerrero A., Bettencourt-Dias M., 2014. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15: 433–452. 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]