While much is known about the enzymes that deposit and remove epigenetic modifications, less is known about the trans-acting system that determines when and where these modifications occur. By looking at four diverse...
Keywords: chromatin modification, hQTL, KRAB-ZFP, meiosis, recombination
Abstract
The epigenetic landscape varies greatly among cell types. Although a variety of writers, readers, and erasers of epigenetic features are known, we have little information about the underlying regulatory systems controlling the establishment and maintenance of these features. Here, we have explored how natural genetic variation affects the epigenome in mice. Studying levels of H3K4me3, a histone modification at sites such as promoters, enhancers, and recombination hotspots, we found tissue-specific trans-regulation of H3K4me3 levels in four highly diverse cell types: male germ cells, embryonic stem cells, hepatocytes, and cardiomyocytes. To identify the genetic loci involved, we measured H3K4me3 levels in male germ cells in a mapping population of 59 BXD recombinant inbred lines. We found extensive trans-regulation of H3K4me3 peaks, including six major histone quantitative trait loci (QTL). These chromatin regulatory loci act dominantly to suppress H3K4me3, which at hotspots reduces the likelihood of subsequent DNA double-strand breaks. QTL locations do not correspond with genes encoding enzymes known to metabolize chromatin features. Instead their locations match clusters of zinc finger genes, making these possible candidates that explain the dominant suppression of H3K4me3. Collectively, these data describe an extensive, set of chromatin regulatory loci that control the epigenetic landscape.
BOTH cellular differentiation and the functional capacities of differentiated cells are controlled by epigenetic DNA and histone modifications associated with a variety of genomic regulatory elements, including promoters, enhancers, and insulators. Although much is known about the proteins that act as writers, readers, and erasers of these marks [reviewed in Allis et al. (2015)], we know less about the underlying regulatory systems controlling their sites of action and tissue specificity. Understanding the properties of these regulatory systems is a matter of considerable significance as the majority of the known genetic variation affecting human health and disease is regulatory in nature (Maurano et al. 2012; Pickrell 2014) and chromatin changes are major features of both tumorigenesis (Verma et al. 2014; Avgustinova and Benitah 2016) and aging (Fraga et al. 2005; Martin 2005; Maegawa et al. 2014).
One avenue for exploring these questions is analyzing the genetic basis of natural variation in the epigenetic landscape (Albert and Kruglyak 2015; Lappalainen 2015; Taudt et al. 2016). Such variation can arise from mutations that act locally by altering the binding sites of transcription factors (TFs), which in turn influence levels of DNA or histone modifications (Kasowski et al. 2013; Kilpinen et al. 2013; McVicker et al. 2013). Local effects are typically cis-acting; the genetic determinant maps close to the location of the molecular phenotype it controls and only influences its own chromosome when heterozygous. The limitations of genetic mapping using human cell culture systems have largely restricted genetic studies of chromatin regulation to local cis effects (Albert and Kruglyak 2015; Pai et al. 2015; Taudt et al. 2016).
Alternatively, diffusible factors encoded by one region of the genome can act at a distance, regulating epigenetic modifications and gene expression at multiple sites in trans. The Robins laboratory (Krebs et al. 2003, 2012, 2014) have described Rsl1 (Regulator of sex-limitation 1) and Rsl2, which act in a sex- and tissue-specific manner to suppress gene expression in livers of adult male mice. Taking a functional genomics approach, one recent study using rat recombinant inbred (RI) lines found tissue-specific trans-regulation of H3K4me3 in heart but not liver by a quantitative trait locus (QTL) on chromosome 3 (Rintisch et al. 2014). Additionally, human studies are starting to identify trans-acting factors influencing DNA methylation and gene expression. One study using lymphocytes from 1748 individuals (Lemire et al. 2015) found 1657 trans-regulated sites scattered across the genome, while another study (Shi et al. 2014) examined the DNA methylome from 210 normal lung tissue samples, finding 373 QTL controlling 585 out of 33,456 CpG sites. Finally, a recent comprehensive survey of 44 human tissues found trans-regulation of gene expression in 18 tissues, with testis having the most trans-acting expression QTL (GTEx Consortium 2017).
Here, we have employed the power and precision of mouse genetics to explore natural genetic variation in trans-regulation of the mammalian epigenetic landscape in two inbred laboratory strains of Mus musculus domesticus: C57BL/6J (B6) and DBA/2J (D2), whose genomes contain over 5,000,000 single nucleotide polymorphisms (Keane et al. 2011). B6 and D2 were the founder strains for a genetic panel of well over 100 RI lines, which were created by intercrossing B6 and D2 mice followed by inbreeding the progeny to create the BXD genetic reference panel in which each line is a homozygous mosaic of the founder genomes (Taylor et al. 1973; Peirce et al. 2004). Panels of BXD lines have been used to facilitate mapping the genetic determinants of multiple phenotypes (Li et al. 2018).
As a model of epigenetic regulation, we focused on H3K4me3, a sensitive indicator of active chromatin (Santos-Rosa et al. 2002) in male germ cells. These cells provided the unique advantage that H3K4me3 sites are deposited by at least two independent enzyme systems, allowing us to distinguish between genetic variation specific to one of the enzyme systems and variation in underlying regulatory mechanisms that affect both. One system is the set of ubiquitous histone methyltransferases and demethylases that function in all cells to metabolize H3K4me3 at promoters and enhancers. The other is the meiosis-specific system placing H3K4me3 at genetic recombination hotspots. There, H3K4me3 is deposited by the histone methyltransferase PRDM9 (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010) and displaced when meiotic DNA double-strand breaks (DSBs) are subsequently repaired (Sun et al. 2015). Importantly for this study, B6 and D2 mice share the same Prdm9 allele, allowing the use of the BXD RI lines to identify genetic modifiers of hotspot H3K4me3 levels that act independently of genetic variation in PRDM9 structure.
Carrying out chromatin immunoprecipitation sequencing (ChIP-Seq) for H3K4me3 in male germ cells we found quantitative differences between B6 and D2 mice. We identified six major trans-acting histone QTL (hQTL), located on chromosomes 4, 7, 12, and 13, controlling the majority of trans-regulated H3K4me3 peaks. Additionally, by measuring H3K4me3 in embryonic stem cells (ESCs) and terminally differentiated hepatocytes and cardiomyocytes, we find that trans-regulation of the epigenome is cell-type specific. In germ cells hQTL do not coincide with any of the known writers or erasers of histone H3K4 methylation and they affect both recombination hotspots and other functional elements. In F1 hybrids, a low level of methylation is the dominant phenotype, suggesting that hQTL genes suppress methylation. The locational specificity, ability to reduce active histone modifications, and lack of overlap with known enzymes of H3K4me3 metabolism suggest that hQTL contain genes whose products recruit chromatin repressing complexes. In accordance with this hypothesis, the major hQTL regions all contain genes encoding KRAB- and BTB-zinc-finger proteins (ZFPs), which are known to promote formation of heterochromatin.
Taken together, these results indicate that a significant fraction of the epigenome is subject to trans-regulation by genes whose products regulate specific genomic locations to modulate the chromatin landscape.
Materials and Methods
Mice
C57BL/6J (stock number 000664), DBA/2J (stock number 000671), B6D2 F1/J hybrid (stock number 100006), and all BXD RI mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All animal experiments were approved by the Animal Care and Use Committee of The Jackson Laboratory (summary #04008 and #16043).
Tissue and cellular isolation
Testicular germ cell enrichment was performed on 14-day postpartum male mice as previously reported (Baker et al. 2014). This cell preparation removes somatic Sertoli and Leydig cells and results in >90% enrichment of germ cells, of which nearly 50% are spermatogonia (Ball et al. 2016).
Individual low-passage mouse ESCs were derived using protocols outlined in Czechanski et al. (2014). Briefly, 6- to 8-week-old females are mated to stud males and checked each morning for plugs. Pregnant females are euthanized on embryonic day 3.5 and the uterine horn is flushed to remove embryos. Embryos are visualized under a dissecting microscope and blastocysts are transferred to 2i (2i:CHIR99021 and PD0325901) (Ying et al. 2008) serum-free media for outgrowth of the inner cell mass. Blastocysts are allowed to hatch and attach to a mouse embryonic fibroblast (MEF) feeder layer, and the resulting outgrowth is monitored daily and fed for 8–11 days. The emergent ESCs are disaggregated and passaged onto new MEF feeders. Cultures during this time are closely monitored for unusually rapid growth (potentially indicating karyotypic instability), signs of deterioration including vacuolated cytoplasm, detachment of cells from colonies and debris, and possible signs of contamination. Successful ESC cultures were maintained on MEF feeders in serum containing 2i media supplemented with Leukemia inhibiting factor (2i/LIF) (Kiyonari et al. 2010) to maintain high levels of NANOG expression, which indicates ground-state pluripotency (Ying et al. 2008; Czechanski et al. 2014). Prior to preparing for chromatin isolation, mouse ESCs were enzymatically disassociated using trypsin and MEFs were removed by serial plating on gelatin-coated plates to which MEFs adsorb preferentially; for this, ESCs and MEFs are incubated in 2i/LIF media on fresh plates for 15 min to allow the larger MEFs to quickly attach to the plates. ESCs are aspirated and the plating procedure repeated once to further remove MEFs. ESCs were collected by centrifugation, resuspended in PBS, and cross-linked using formaldehyde.
For hepatocyte isolation and purification, livers from 8-week-old female mice were perfused using a modified EGTA–collagenase perfusion protocol (Neufeld 1997). All perfusions and hepatocyte purifications were done at the same time of the day to avoid possible circadian effects on any studied parameter. EGTA buffer was used to flush the blood out of the liver and start to digest the desmosomes connecting the liver cells. After 35 ml of the 1× EGTA solution was passed through the liver, it was replaced with 7–10 ml of 1× Leffert’s buffer to flush out the EGTA, which otherwise chelates the calcium ions necessary for collagenase activity in the next step when the liver is digested by perfusion with 25–50 ml of Liberase solution (∼4.3 Wünsch units). After perfusion, the liver was removed from the abdominal cavity and passed through Nitex 80-μm nylon mesh, using extra ice-cold Leffert’s buffer with 0.02% CaCl2 and a rubber policeman. Hepatocytes were purified from the remaining cells by two consecutive centrifugations for 4 min, 50 × g each, leaving the other, smaller cell types in suspension. After each spin, the solution was decanted as waste, and the enriched cell pellet of hepatocytes was resuspended in 30 ml ice-cold Leffert’s buffer with 0.02% CaCl2. After the second centrifugation, the cell pellet contained >98.6% hepatocytes.
For cardiomyocyte isolation 8-week-old female mice were euthanized and the chest opened to expose the heart. The descending aorta and inferior vena cava were cut and an EDTA buffer was injected into the apex of the right ventricle to flush the heart. The ascending aorta was clamped and the heart transferred to a petri dish and fixed by perfusion of EDTA buffer containing 4% formaldehyde via the left ventricle. The formaldehyde was quenched by perfusing the heart with 125 mM glycine, and digested by perfusion with collagenase buffer. The ventricles were rent into smaller pieces, and triturated to complete cellular dissociation into a single-cell suspension. Cells were then filtered through a 100-μm strainer to remove tissue fragments and centrifuged at a very low speed to obtain a highly enriched fraction of fixed cardiomyocytes.
H3K4me3 chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using H3K4me3 antibody (#07–473; EMD/Millipore) on chromatin sheared enzymatically as previously reported (Baker et al. 2014). Briefly, after isolation of single-cell suspensions, cells are cross-linked with formaldehyde followed by enzymatic chromatin shearing using micrococcal nuclease (MNase). Magnetic Protein G–coated beads (Dynabead; Life Technologies) are preloaded with H3K4me3 antibody and incubated with MNase-digested nucleosomes. Beads are washed and DNA is eluted and deproteinated, and cross-links reversed.
DNA was prepared for high-throughput sequencing using either the Bioo Scientific’s NEXTflex ChIP-seq Kit without size selection for all germ cell data, or the KAPA hyper kit (Kapa Biosystems) for mESC, cardiomyocytes, and hepatocytes. Library quality and size distribution were visualized using a Bioanalyzer (Agilent). All samples are sequenced in-house using either the Illumina HiSeq 2500 or 4000 platform.
Data analysis and H3K4me3 quantification
All sequenced B6, D2, F1, and BXD H3K4me3 ChIP libraries, as well as all control input DNA samples were aligned utilizing bwa version 0.7.9a (Li and Durbin 2009). B6 parental samples were aligned to the Genome Reference Consortium Mouse Build 38 (mm10) and D2 parental samples were aligned to the de novo REL-1509 assembly, including all unplaced scaffolds, from the Mouse Genomes Project (Yalcin et al. 2012).
To ensure that H3K4me3 peaks were properly quantified across divergent genomes, we began by building a comprehensive “peakome” representing all potential H3K4me3 peaks found in the two parents. H3K4me3 peaks for B6 and D2 were called independently, utilizing alignment data from three replicate samples and one DNA input sample. Reads were filtered for a mapq alignment metric of 60 and an alignment sequence having no indels present across the entire length of the sequencing read, typically 100 bp H3K4me3 peaks were called utilizing MACS version 1.4.2 (Zhang et al. 2008) and peaks having a false discovery rate (FDR) of <1% found in two out of three replicates were accepted. Final genomic intervals for each H3K4me3 peak for each strain were derived by merging the peaks from the corresponding replicate samples using bedtools (Quinlan and Hall 2010). To link syntenic regions between B6 and D2 assemblies, which each have their own coordinate system, sequences from these genomic intervals were aligned to their alternative genome using reciprocal BLAST. In some cases, a sequence interval comprising an H3K4me3 peak in one strain aligned to multiple adjacent intervals in the alternative genome. If the sequences of these peaks in the alternate strain all fell within the boundaries of the single peak, they were merged. The boundaries of these merged peaks included the incorporated sequences from both strains. Because there are also H3K4me3 peaks that are strain specific, these peaks were accepted if, and only if, the mapped interval had a unique sequence that was found in the proper syntentic order within the alternative genome lacking that H3K4me3 peak. The final combined peakome between B6 and D2 mice was created by selecting only peaks appropriately linked across each strain, assuring that each H3K4me3 peak reciprocally aligned to only one peak in the alternative genome after merging, and that all peaks were in the same order along the chromosomes in both genomes (Supplemental Material, Table S2). Using the H3K4me3 peaks locations derived from the parental strains, final read counts for B6, D2, F1 hybrids, and BXDs were obtained by counting reads within the coordinate boundaries of the peakome intervals.
To improve mapping accuracy and utilize known sequence variation between strains, all BXDs and F1 hybrid samples were aligned separately to both the mm10 reference and the de novo D2 assembly. To reduce error in quantification of H3K4me3 levels due to genomic regions containing repetitive sequences, we removed reads with multiple alignments and retained reads with alignment metric of 60 that lacked small indels, which can often indicate misalignment. Subsequently, for each genomic interval in the peakome, final reads counts were summed for those that mapped uniquely to one of the assemblies along with those that mapped equally well to both B6 and D2 genome assemblies.
Genetic map construction
During the course of QTL mapping, we found occasional BXD lines in which there was a discrepancy between the phenotype of a given H3K4me3 peak (read abundance) and its local genotype assigned using publicly available genetic maps (Figure S1). These were particularly apparent for cis-QTL with large LOD scores where the H3K4me3 levels show distinct segregation between B6 and D2. To correct for these discrepancies and improve genetic mapping, genotype maps were derived de novo by calling genotypes of mm10 aligned reads from BXD H3K4me3 ChIP-Seq samples used in this study at known SNP positions between B6 and D2 reported in Mouse Genomes Project database REL-1505 (Keane et al. 2011; Yalcin et al. 2011). The locations of genetic crossovers were assumed to lie within the transition intervals between B6 and D2 genotypes in each BXD line. Transition intervals comprise the DNA sequence between the base-pair position of the SNP with the first detectable genotype switch and the previous SNP. In some cases, transitions could be detected within individual recombination hotspot H3K4me3 peaks (Figure S2). Overall, the sizes of the transition intervals reflected the frequency of read/SNP combination occurrences, which became sparser in regions of poorly assembled sequences in the D2 assembly. The result is a skewed distribution of sizes. The median size of transition intervals was ∼6 kb, the mean was around ∼150 kb, and the total transition interval space across the genome was found to be 362 Mb.
For our de novo genetic map marker positions were selected near the end of each transition interval. In this way, a marker represents the genotype at its location through to the next marker, with a small region of uncertainty at the end in the transition interval. Effectively each marker represents a unique genomic interval within the mapping population. In most cases, SNPs were genotyped multiple times within overlapping alignment reads. Care was taken to avoid selecting regions of rapid genotype switching due to residual heterozygosity or experimental noise. Any regions of residual heterozygosity were recognized by genotype variation among reads overlapping the same SNP and by rapid genotype switching among sequential SNPs. Rates for rapid genotype switching due to experimental noise, identified using the B6 parental strain samples, were low and avoided.
The transition intervals across all the selected BXD lines were combined to create a genome-wide map of its haplotype blocks. Using these de novo maps resolved most conflicts between genotype and phenotype (Figure S1). To estimate the functional extent of disagreement between genetic maps we characterized all H3K4me3 peaks with LOD > 25 (n = 807, mapped using the de novo genotypes). This class of H3K4me3 peaks offers the clearest test cases that allow strong separation in H3K4me3 level between B6 and D2 genotypes, driven by local variants. Of these cis-QTL, 69% (n = 560) had at least one BXD strain in which the local genotype assigned using the reported map and the de novo map disagreed, after excluding heterozygous regions. Upon visual inspection, some genotyping differences were due to one marker positioned at 5′ or 3′ to our called de novo markers, likely a result of improved resolution of our genetic maps. To exclude these cases, we next required that two markers in both directions from the QTL interval disagree between genetic maps, resulting in a broader region of discordance between maps. This resulted in 38% (n = 310, representing 198 unique genetic intervals) of cis-QTL with LOD > 25 having at least one strain with disagreement between genotypes. For 98% of these H3K4me3 peaks (304/310), the de novo genetic map led to increased LOD scores (average increase 9.1 LOD) reflecting better agreement between H3K4me3 level and local genotype (Figure S1). Two strains, BXD81 and BXD100, consistently had large regions in disagreement between the two genetic maps. Maps from all RI lines used in this study were formatted for compatibility with R/qtl2 package and are available as Table S8.
Statistical analysis and QTL mapping
All statistical analyses were performed using R (http://www.R-project.org/). Differential H3K4me3 levels between parental strains for Figure 1, and for the trans-regulation test with BXD strains, were calculated using the exact test from the edgeR package (Robinson et al. 2010). All samples were normalized using the trimmed mean of M-values method in edgeR. P-values were adjusted using the FDR method of Benjamini–Hochberg.
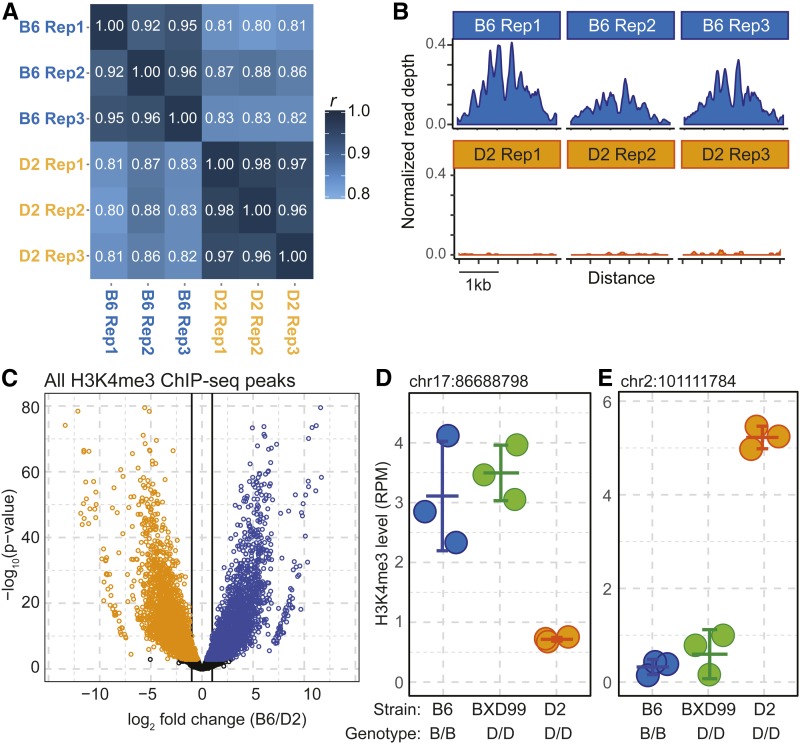
Figure 1.
Trans-regulation of H3K4me3 levels in male germ cells. (A) Heat map of the Pearson correlation coefficients of H3K4me3 level between biological replicates of male germ cells isolated from B6 and D2 mice. (B) Coverage profile for a representative locus showing reproducibility of normalized (RPM) ChIP-Seq read depth for biological replicates of a single H3K4me3 peak. (C) Volcano plot of differential H3K4me3 levels between B6 (blue) and D2 (orange) for all peaks (colored points represent peaks with significant difference after P-value adjustment, FDR < 0.05). (D) Example of an H3K4me3 peak under trans-regulation. This peak has high H3K4me3 level in B6 and low in D2; however, the BXD99 strain, which is genotypically D2 at this locus, has high levels of H3K4me3 similar to B6. Dots represent biological replicates for indicated strains. Error bars show mean and SD. The local genotype at the H3K4me3 peak is shown below the plot, the genomic position of the H3K4me3 peak is above. (E) Similar to D, showing an H3K4me3 peak with high H3K4me3 levels in the D2 background but low levels in BXD99, which is genotypically D2 at the locus.
QTL analysis was performed using R/qtl2 package (Broman et al. 2003). Prior to mapping the sample matrix was normalized using edgeR and log2 transformed. Data exploration found variation in H3K4me3 signal at recombination hotspots, even between some replicates (Figure S6A), suggesting that juvenile mice coming from different litters might be at slightly different stages of meiosis. Although each litter of mice was collected at 14 days postpartum, there might be small variation in timing of meiotic entry due to genetic or environment effects (such as litter size, time of day of birth, or mouse room). Principal component (PC) analysis found that the first PC described the variation in H3K4me3 level at hotspots (Figure S6B). While initial hierarchical clustering found that replicate samples from a few individual BXD strains did not always group together (Figure S6C), hierarchical clustering after subtracting PC1 found that replicates of the same strain clustered together (Figure S6D). Genome scans using PC1 as a phenotype did not identify any significant or suggestive QTL, further indicating that variation in the timing of recombination-specific H3K4me3 formation among these BXD lines is not a significantly variable genetic trait.
Consequently, QTL mapping was performed on PC1-subtracted data. Single-QTL scans were performed with the scan1 function of R/qtl2 using a linear mixed model and accounting for kinship. We used a permutation approach to generate genome-wide FDR thresholds. 1000 permutations were performed by randomly permuting the samples labels, and an empirical maximum LOD score distribution was computed for each H3K4me3 peak. An empirical P-value was calculated for each peak by comparing the original LOD score for each peak with the empirical LOD score distribution, and FDR was computed across the whole data set using the R package fdrtool (Strimmer 2008). For each H3K4me3 peak, only the QTL location with the highest LOD score was kept for further analysis (Table S3).
Probability of overlap between major hQTL and ZFP clusters:
There are 18 ZFP clusters in the mouse genome (Kauzlaric et al. 2017). The probability that a given hQTL contains a given cluster is (hQTL length − ZFP cluster length)/(size of genome). The average length of the 18 ZFP clusters is 2.1 Mb. Using the dimensions from the iWGCNA analysis, the average length of the six major hQTL is 7.2 Mb. If chromosome 13 hQTL is a compound of two hQTL, the average length of seven hQTL is 6.2 Mb. The probability that a single hQTL includes one ZFP cluster is (7.2 – 2.1)/2800. Since there are 18 ZFP clusters in the genome, the probability (pc) that an average major hQTL includes an average ZFP cluster is (7.2 – 2.1)18/2800 = 0.033, and for seven major hQTL is (6.2 – 2.1)18/2800 = 0.026. Using the binomial distribution, the probability that four (k) of six hQTL (n) include ZFP clusters is then [6!/(4!)(2!)] (0.033)4 (0.967)2 = 1.7 × 10−5. If we consider that the chromosome 13 hQTL is compound, with each segment containing a ZFP cluster, the probability that five of seven hQTL include ZFP clusters is 2.37 × 10−7.
Network analysis
For network analysis QTL scans were performed as described above, assuming an additive model and conditioning on the closest marker to the peak. Samples were permuted 2000 times for each H3K4me3 peak retaining LOD scores greater than the 95th percentile of the empirical maximum. Peaks found to be in linkage disequilibrium with their trans marker, or FDR > 0.05, were filtered prior to network analysis, resulting in 2689 trans-associated peaks. These were clustered into modules using a weighted gene coexpression network analysis (WGCNA) (Langfelder and Horvath 2008). To obtain tightly correlated modules, we used iterative WGCNA (Greenfest-Allen et al. 2017), with the following settings: power = 12; minKMEtoStay = 0.7; minCoreKME = 0.7, and the remaining parameters set at default values. This resulted in 1224 H3K4me3 peaks clustered into 14 tightly correlated modules (Table S4). The first PC of each module (termed an eigengene) is a representation of the summary coexpression pattern, and treated as module phenotypes for module-QTL analysis using R/qtl2.
Annotation of H3K4me3 sites
Recombination hotspot locations in both B6 and D2 mice were established by mapping the locations of the DMC1-associated single-strand DNA that arises at meiotically programmed DNA DSBs (Smagulova et al. 2011). We classified remaining sites by whether they corresponded to promoters, based on overlapping TSS, or to enhancers and CTCF sites using mouse ENCODE testis data sets (Shen et al. 2012), or to transposable elements. Homer (Heinz et al. 2010) mergePeaks function was used to compare peak sets between the four cell types.
Motif enrichment analysis
Analysis of motif enrichment (MEME Suite 4.12.0; Bailey et al. 2009) was used to identify motifs that are significantly enriched in module peaks. For controls, 10,000 sequences were randomly selected out of the total 67,110 H3K4me3 peaks. HOCOMOCO Mouse (v11 CORE) database was used to test for enrichment (Kulakovskiy et al. 2018). Motifs and their corresponding TFs, with Bonferroni-adjusted P-value < 0.01, are reported in Table S6.
Measuring DMC1-associated single-strand DNA fragments for DSB estimates
DMC1 ChIP was performed with isolated and cross-linked spermatocytes from B6 and D2 mice using an established method (Khil et al. 2012). Briefly, testes from adult B6 or D2 mice were cross-linked in 1% paraformaldehyde for 10 min, and then homogenized. Spermatocytes were washed with PBS and lysis buffers. Then, the chromatin was sheared to 500–1000 bp by sonication and incubated with DMC1 antibody (sc-8973; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°, followed by a 2 hr incubation with Protein G beads (10004D; Life Technologies). The beads were washed and the chromatin was eluted and reverse-cross-linked with 1% SDS, 0.1 M NaHCO3, and 0.2 M NaCl at 65° overnight. The libraries were constructed with TruSeq Nano DNA LD Library Prep Kit (FC-121-4001, set A; Illumina) using enzymes from New England Biolabs (Beverly, MA). DNA samples were sequenced on an Illumina platform, with 75 bp paired-end reads.
Detection of DMC1 ChIP-Seq peaks across both B6 and D2 was accomplished by aligning two replicate paired-end B6 samples and two replicate paired-end D2 samples to their associated genomes, GRCM Build 38 (mm10) and the D2 de novo REL-1504 Assembly, respectively, utilizing bwa version 0.5.10-tpx (Li and Durbin 2009). The fastq files were trimmed with Trimmomatic version 0.32 (Bolger et al. 2014) and any adapter sequences were removed.
During sample preparation, DNA single-strand fragments bound to DMC1 were enriched from background DNA by fast annealing short sequences of intramolecular microhomology, followed by exonuclease removal of 3′ overhang ends and polymerase fill-in of remaining 5′ overhang sequence (Khil et al. 2012). This generates 3′ and 5′ end homologous sequences that arise from the region of microhomology combined with filling in the 5′ overhangs. Sequencing reads were selected for the presence of these regions of 3′ and 5′ homology, and the homologous portions deleted. The trimmed reads selected in this way were then aligned to their corresponding parental genome assemblies. Because the majority of the insert sequences are shorter than the read lengths, the reads from one strand often contain a fill-in sequence. The single-strand DNA reads without fill-in were then selected by evaluation of the alignment flags and retained. This created a new single-end fastq file that was subsequently aligned for peak calling. Peaks were called using MACS version 2.1 (Zhang et al. 2008) with the extsize option set to 800 to align reads into detectable clusters. Peaks were linked between B6 and D2 similar to methods applied to H3K4me3 reads as described above.
Data availability
All strains are available through The Jackson Laboratory Catalog. Mouse ESCs are available upon request. Supplemental files include Tables S1–S8 and Figures S1–S6. Raw sequencing data and processed files, including estimates of chromatin modification for each strain and peak location, are available at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) available under accession number GSE113192. DMC1 ChIP-Seq data for B6 is available under accession number GSE108259. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210262.
Results
Genetic variation influences histone modification levels
To obtain a genome-wide view of the similarities and differences between B6 and D2 mice in H3K4me3 sites, we performed nucleosome resolution ChIP-Seq for H3K4me3 from germ cells of juvenile B6 and D2 inbred mice. Three biological replicates were collected from each strain by pooling germ cells from three to five mice for each replicate (Table S1). Intrastrain variation between replicates was consistently low (Pearson correlation coefficient r = 0.92–0.98), while variation between strains was consistently greater (r = 0.80–0.88), suggesting genetic control of the chromatin state (Figure 1A).
In all, we identified 67,110 H3K4me3 peaks between the two strains. Among these peaks, 8319 (12%) showed significant differences between strains (FDR-adjusted P-value < 0.01), with approximately half having greater methylation in either B6 or D2. The level of differential activity varied considerably, from subtle changes in H3K4me3 level to sites that were virtually strain specific (Figure 1, B and C).
Testing for trans control of epigenetic changes
We tested for trans-regulation of chromatin features by exploiting the mosaic genetic structure of BXD lines. The location of each H3K4me3 peak identified in an RI line lies within a large, homozygous region, allowing us to identify the genetic origin of each site unambiguously. We defined an H3K4me3 peak as subject to trans control if its phenotype in an RI line differed significantly (FDR-adjusted P-value <0.01) from the phenotype of the parental strain with the same genotype at that site (Figure 1, D and E). While this test is statistically quite robust, it underestimates the number of trans-regulated sites by at least 50% because an H3K4me3 peak is only detected as subject to trans control when its putative trans-acting hQTL has the opposite genotype from the H3K4me3 site. Applying this test to three biological replicates of the RI line BXD99, we found that among the 8319 peaks that were significantly different in the parents, 1172 were trans-regulated in BXD99, indicating that at least a fourth of the differentially modified H3K4me3 sites are under trans control. For example, the H3K4me3 level of the peak at chromosome 17 86.7 Mb (Figure 1D) is low in the D2 parent and high in B6. The level of H3K4me3 at this site is also high in BXD99, even though the location of the peak in BXD99 places it in a region that is unequivocally homozygous D2. Similarly, we identified peaks with high H3K4me3 in the D2 parent but low in BXD99, even though they were genetically D2 at the H3K4me3 locus (Figure 1E).
Linkage mapping in RI lines identifies multiple trans-acting QTL regulating chromatin
We mapped the genomic locations of the loci controlling H3K4me3 levels by performing H3K4me3 ChIP-Seq in male germ cells of 59 BXD RI lines. To facilitate genetic mapping, we derived de novo genotypes using the ChIP-Seq data (Figures S1 and S2). To reduce mapping bias and improve quantification, we developed a custom alignment strategy accounting for known variation between B6 and D2 (see Materials and Methods for details).
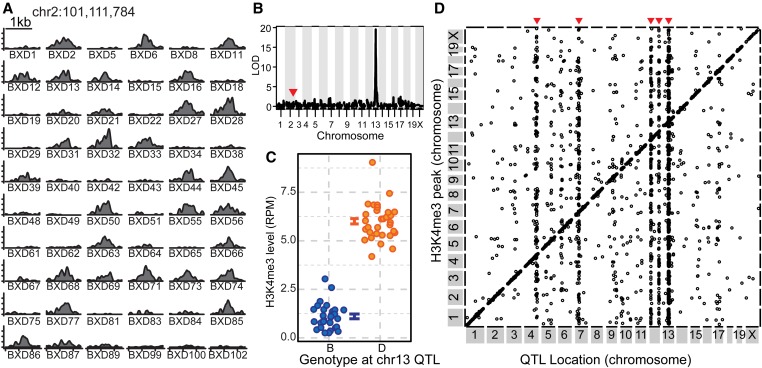
Using read depth for each peak as a quantitative phenotype, we genetically mapped the location of the hQTL controlling each H3K4me3 peak. For example, the level of H3K4me3 at the chromosome 2 10.1 Mb site, previously identified as under trans control in BXD99 (Figure 1E), segregated as two distinct phenotypes in the BXD lines (Figure 2A). Mapping the locus determining this phenotype identified a single hQTL on chromosome 13 at 64.8 Mb (Figure 2B). Having the D2 genotype at the chromosome 13 hQTL resulted in a high level of H3K4me3 at the chromosome 2 site (Figure 2C), regardless of the local genotype on chromosome 2, confirming the BXD test for trans control and identifying the location of this trans-regulator.
Figure 2.
H3K4me3 levels are extensively regulated by local and distal hQTL. (A) Coverage profile for 60 BXD lines of H3K4me3 peak from Figure 1E. (B) LOD score plotted against genetic map for H3K4me3 level shown in A. Location of the peak under regulation is indicated by a red arrowhead. (C) H3K4me3 level plotted by genotype at the trans-hQTL marker on chromosome 13 (error bars are SE). (D) Points represent maximum LOD scores from single-hQTL scans with FDR < 0.05. The location of the H3K4me3 peak is plotted on the y-axis and the location of the mapped hQTL is on the x-axis. Genetic effects acting locally (N = 8375) lie on the diagonal line. Distal acting trans-hQTL (N = 1867) lie off the diagonal; trans-hQTL affecting multiple loci appear as vertical bands (red arrows).
Overall, we identified 10,242 H3K4me3 peaks under genetic control in the BXD RI lines at a genome wide FDR < 0.05 after permutation testing (Figure 2D and Table S3). Among these, 5160 had been detected earlier as statistically different between the B6 and D2 parents. A likely explanation for the increased sensitivity of RI lines is that testing 59 RI lines, rather than only three parental replicates, greatly increased our statistical ability to detect small phenotypic differences.
Among the 10,242 H3K4me3 peaks under genetic control, 1867 were regulated in trans (FDR < 0.05) by hQTL that were on a different chromosome. This conservative definition of trans activity precluded any possibility of misidentifying cis activity at a long distance. Importantly, each trans-hQTL is highly specific, regulating a cohort of H3K4me3 sites. Among these trans-regulated H3K4me3 peaks, 1335 (72%) are collectively controlled by six major hQTL on chromosomes 4, 7, 12, and 13 (Figure 2D). The strongest hQTL, located on chromosome 13, accounts for 41% of all trans-regulated sites.
Epigenetic network analyses
We identified the same set of major hQTL by applying an iterative weighted correlation network analysis (iterativeWGCNA) (Langfelder and Horvath 2008; Greenfest-Allen et al. 2017) to maximize separation between coregulated features, a method previously used for identifying refined gene clusters (modules). We identified 1224 trans-regulated H3K4me3 peaks that grouped into 14 modules by their correlated levels of H3K4me3 modification (Table S4). We mapped the loci controlling each module by combining the data for the peaks in each module into a single eigengene. All modules mapped to loci that overlapped the location of one of the trans-acting hQTL apparent in Figure 2D, including chromosomes 4, 7, 12, and 13 (Table S4). Of the 14 modules, 9 map to chromosome 13, representing 73% (n = 897) of all module peaks, with four modules having high phenotypes when D2 at the hQTL and five having high phenotypes when B6, suggesting compound genetic regulation at this locus. Furthermore, network analysis suggested that chromosome 7 has two distinct, but closely neighboring hQTL. These data provide confirmation of the six major hQTL, and show that individual hQTL regulate distinct cohorts of H3K4me3 peaks.
Although four of the major hQTL modules (mapping to chromosomes 4, 7b, 12a, and 13) act independently, with each module controlling a unique cohort of H3K4me3 sites, the network analysis showed that module 3, which maps to both chromosomes 7a and 12b, acts jointly on a distinct set of 162 H3K4me3 sites (Table S4). Epistasis analysis did not detect interactions between 7a and 12b, suggesting that their effects are additive (data not shown). We conclude that hQTL 7a and 12b cooperate to regulate a single cohort of H3K4me3 sites.
Trans-regulation of H3K4me3 is cell-type specific
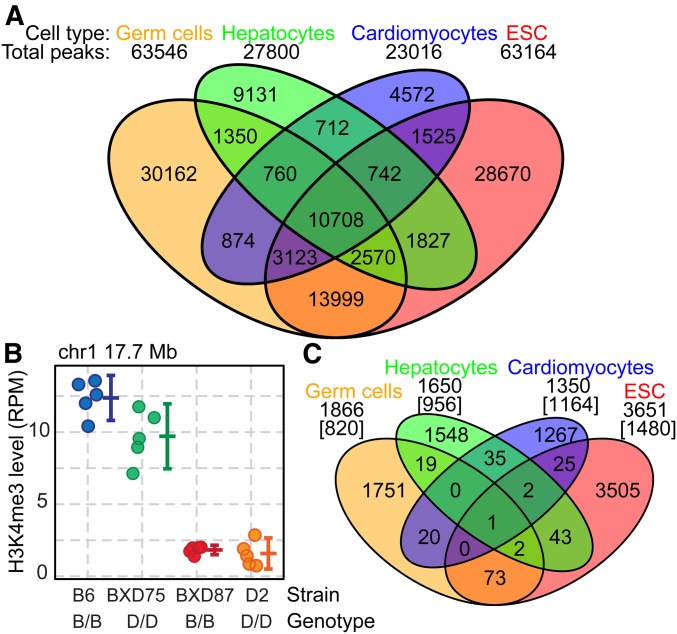
Germ cells preparations used in the above study represent a mixture of cell types including both spermatogonial stem cells and differentiating meiotic cells (Ball et al. 2016). To reduce cellular complexity and test for cell-type specificity in regulation of H3K4me3, we applied the RI line test for trans-regulation of H3K4me3 to three highly enriched cell types: ESCs, hepatocytes, and cardiomyocytes. For each cell type, we performed H3K4me3 ChIP in a minimum of three biological replicates from B6 and D2 mice as well as from two RI lines, BXD75 and BXD87 (Figure S3). Each cell type contains both overlapping and cell-specific H3K4me3 peaks (Figure 3A). ESCs and germ cells both contain the largest number of H3K4me3 peaks (>63,000 peaks each), with many sites overlapping (n = 30,400). Applying our BXD test for trans-regulation outlined above, these data predicted hundreds of peaks under trans-regulation in each cell type (Table S5). Using two different BXD lines provided internal controls for both reproducibility and specificity. For example, the H3K4me3 peak located on chromosome 1 17.7 Mb in ESCs, which has high methylation in B6 and low methylation in D2, was identified in both BXD75 and BXD87 as under trans-regulation (Figure 3B). Additionally, we found the expected ∼50% overlap between the two BXD lines when we considered all predicted trans-regulated peaks (Table S5). Very importantly, while all four cell types shared a considerable percentage of their total H3K4me3 sites, virtually all trans-regulated peaks were tissue specific (Figure 3C). Even though many H3K4me3 sites are found in multiple tissues (Figure 3C, brackets), trans-regulation of those peaks only occurred in a single cell type. Combining the results from germ cells, ESCs, hepatocytes, and cardiomyocytes, we conclude that an extensive chromatin regulatory system operates in a cell-type-specific manner to modulate H3K4me3 in trans.
Figure 3.
Trans-regulation of H3K4me3 is cell-type specific. (A) Venn diagram showing the number and relationship of all observed H3K4me3 sites between germ cells, hepatocytes, cardiomyocytes, and embryonic stem cells (ESCs) from B6 and D2 parents. (B) Example H3K4me3 site predicted to be under trans-regulation in ESCs. Both the strain and genotype at the H3K4me3 site is indicated under the graph (error bars represent SD). (C) Venn diagram of the number and relationship of either mapped H3K4me3 peaks regulated by trans-QTL (germ cells) or predicted trans-regulated H3K4me3 sites (ESCs, hepatocytes, and cardiomyocytes). Total trans-regulated peaks are indicated under the cell type, numbers in brackets indicate peaks that, while being uniquely subject to trans-regulation within the indicated cell type, are present in at least one of the other cell types.
Germ cell hQTL dominantly suppress H3K4me3
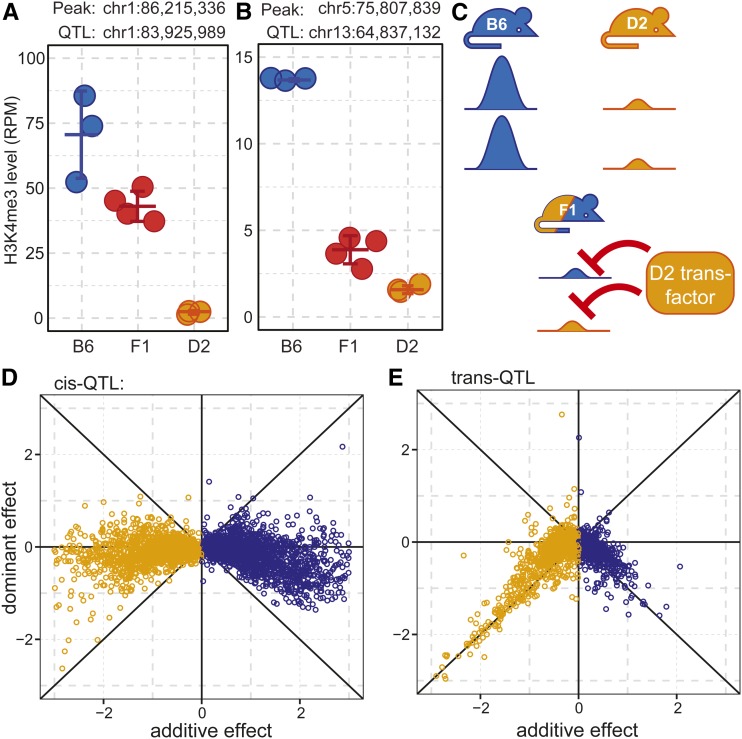
To determine whether hQTL alleles act additively or dominantly in germ cells, we compared H3K4me3 levels from germ cells in heterozygous (B6×D2) F1 hybrids with those in the two parental strains. A cis-acting local variant is expected to control each chromosome independently in an F1 heterozygote, resulting in additive inheritance, with the phenotype of the F1 intermediate between B6 and D2 (Figure 4A). In contrast, a trans-acting hQTL that either induces or suppresses H3K4me3 levels would likely show a dominant phenotype in the F1 that would be closer to one of the parents. For example, the D2 allele of the chromosome 13 hQTL dominantly suppresses H3K4 trimethylation at site on chromosome 5 in trans (Figure 4, B and C).
Figure 4.
Trans-hQTL suppress H3K4me3 levels in F1 hybrids. (A) Example of a local-hQTL with additive inheritance in F1 hybrids. (B) Example of a distal-hQTL showing dominant repression of H3K4me3 activity in F1 hybrids, similar to the D2 parent. (C) Model illustrating the dominant effect in F1 hybrids. A D2-derived trans-acting factor results in reduced H3K4me3 on both chromosomes in the heterozygous state. (D) All cis-hQTL with LOD >20 showing most loci exhibit additive inheritance. (E) All trans-hQTL showing the general dominant repression of H3K4me3 levels.
To generalize these observations, we adopted the method of Tian et al. (2016) in which the measure of dominance (d), defined as the F1 minus the parental average, is plotted against the size of the additive effect (a), which is defined as half the distance between the mean of the two parents. In this formulation d is 0 when there is no dominance, i.e., additive inheritance; when high methylation is fully dominant d is positive on the 45° line, and when low methylation is fully dominant d is negative on the 45° line. As expected, additive inheritance is generally seen at cis-QTL (LOD score > 20, Figure 4D). When plotting all trans-acting hQTL together, regardless of whether the H3K4me3 site is high in B6 or D2, we observe the trend expected for fully dominant reduction in methylation (Figure 4E). We conclude that the trans-hQTL we observe in germ cells act by suppressing H3K4me3.
hQTL loci can be genetically compound
Many of the major hQTL are defined by H3K4me3 peaks whose QTL map to adjacent genetic intervals along the chromosome. This, and the fact that multiple network modules can be assigned to the same regions, suggest the possibility that some hQTL are compound, containing more than one causal gene. To refine the location of candidate regions we took advantage of the homozygous genetic structure of the BXD genomes, focusing on the RI lines with crossovers in the QTL regions and the H3K4me3 peaks that showed strong difference in phenotype between B6 and D2 (e.g., Figure 2, A–C). When individually mapped, 613 peaks (FDR < 0.05) regulated by chromosome 13 are associated with six consecutive intervals across an ∼8 Mb region (Table S3). The RI lines that are most informative about a causal gene’s exact location are the lines with crossovers that divide the QTL region in question. Out of the 59 strains used in this study, seven have recombination breakpoints across this region (Figure S4). Plotting the H3K4me3 level for several informative peaks based on the genotype at each of the six intervals shows a clear split in trans-regulation across this region, defined by a crossover in BXD71, and suggests multiple closely spaced hQTL. In the case of chromosome 13, this analysis found hQTL in at least two genetically distinct regions in close proximity: Mb 60,418,152–65,890,145 and 65,890,145–69,026,189 (Figure S4). Repeating this analysis on other hQTL regions, resulted in a suggestive split at the chromosome 4 hQTL (Mb 143,302,894–144,911,401 and 144,911,401–148,865,285) determined by a crossover in BXD42.
Trans-regulation of chromatin involves an array of genomic regulatory features
The chromatin regulatory genes within the hQTL differ from traditional TFs in that they are capable of affecting all classes of H3K4me3 sites in male germ cells: promoters, enhancers, insulators, transposons, and recombination hotspots, as well as sites of unknown function (Figure S5). Overall, trans-QTL regulate all classes of functional elements, although individual modules differ in the proportions of functional elements they influence (Figure S5). In particular, the H3K4me3 peaks regulated by hQTL on chromosome 7b and chromosome 12a (represented by modules 2 and 5) are enriched for hotspots, while hQTL 7a and 12b (module 3), are virtually devoid of hotspots. Of the nine modules that map to chromosome 13, most are depleted for hotspots, with the exception module 8. While many individual modules are depleted for promoters and genic regions, module 1 on chromosome 4 is enriched. Our male germ cell preparations include both spermatogonial stem cells and spermatocytes; given the evidence of cell-type specificity in trans-regulation of H3K4me3 outlined in the previous section, one explanation for the differences among enrichments of functional elements is that the hQTL may act at different stages of meiosis.
hQTL influence cellular function
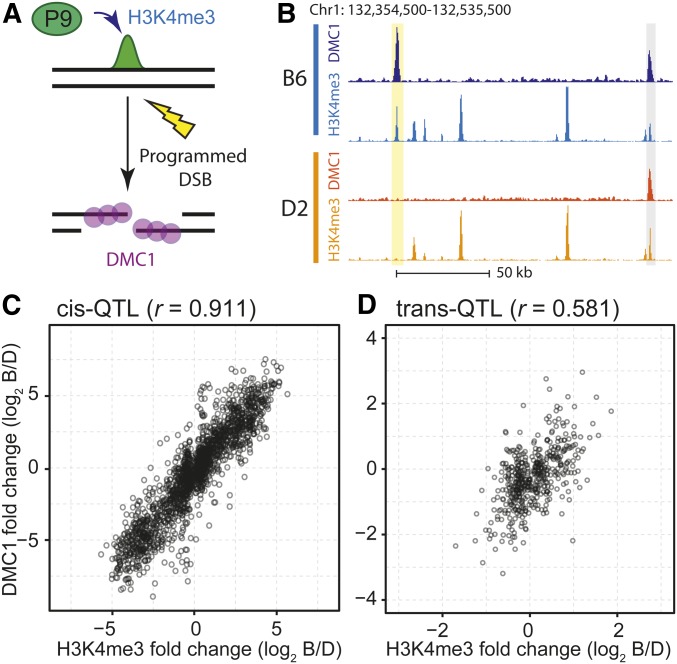
A unique property of meiotic cells offered a direct test of whether the hQTL have functional consequences. The meiotic DSBs that give rise to genetic recombination occur following activation of hotspots by PRDM9, in part through local deposition of H3K4me3 (Smagulova et al. 2011; Baker et al. 2014; Lange et al. 2016). Accordingly, we tested whether hQTL regulation of H3K4me3 levels at recombination hotspots influences the subsequent likelihood of DSB formation by comparing H3K4me3 levels in BXD mice to DMC1 levels in the founders (Figure 5, A and B).
Figure 5.
hQTL control DSB frequency at recombination hotspots. (A) Illustration of events leading up to meiotic DSBs. PRDM9 (P9) binds DNA and locally trimethylates H3K4, leading to targeting of meiotic DSB machinery. The DSB is resected and single-strand DNA ends are coated with the meiosis-specific recombinase DMC1. (B) Coverage profile showing H3K4me3 and DMC1 single-strand DNA levels in parental B6 and D2 mice; yellow indicates an example recombination hotspot that is active in B6 but not D2; gray indicates an example hotspot found in both strains. (C) Scatterplot of the log2 ratio of B6/D2 H3K4me3 levels found in BXD strains vs. parental B6/D2 DMC1 levels at hotspots for all cis-hQTL, showing Pearson correlation (r). (D) Similar to C, showing results for all hotspots under regulation by trans-hQTL.
Doing so, we found a clear correlation between hQTL determined changes in H3K4me3 levels and the consequent changes in DSB frequency at both cis- and trans-hQTL (Figure 5, C and D). For cis-acting hQTL, the differences in H3K4me3 levels account for nearly 83% of the variance in DSB frequency; the fraction of the variance was less for trans-acting QTL, which are likely subject to additional genetic variation. We conclude that hQTL modulation of PRDM9-dependent H3K4me3 influences the subsequent likelihood of DSB formation at hotspots. In effect, chromatin regulatory loci influence the recombination landscape and thus inheritance patterns between generations.
Additional evidence that hQTL are functionally distinct is supported by the observation that the H3K4me3 peaks found in several modules contain distinct sets of TF binding sites (Table S6). For example, module 3 (chromosomes 7a and 12b) is highly enriched for the YY1 motif (P-value < 8.98 × 10−29) and Yy1 is located on chromosome 12, 2 Mb from the chromosome 12b critical region. YY1 is an architectural protein that bridges the physical interactions between enhancers and promoters in a cell-type-specific manner (Beagan et al. 2017; Weintraub et al. 2017), suggesting that the H3K4me3 peaks regulated by module 3 could be meiosis-specific functional elements. Additionally, several modules are enriched for TF motifs that are predicted to form regulatory networks in meiotic cells (Ball et al. 2016). For example, module nine is enriched for the retinoic acid receptor Rarg (P-value < 2.19 × 10−5), which is upregulated in preleptotene spermatocytes, a meiotic substage present in male germ cell preparations. Module 1 is enriched for the Zbtb33 motif (P-value < 5.73 × 10−3), a TF expressed later in meiosis, and is also enriched for H3K4me3 peaks that overlap promoters and known testis enhancers. Finally, modules 3 and 11 are enriched for Zfp42 motifs to different extents (P-values < 1.46 × 10−20 and 6.93 × 10−3, respectively). Zfp42 is expressed exclusively in pluripotent ESCs and male germ cells (Rogers et al. 1991); it is a known marker for pluripotency and functions to repress transposable elements (Schoorlemmer et al. 2014). Together, these data show that different modules are enriched for different TF binding motifs, suggesting these modules may represent functionally related sets of H3K4me3 peaks that interact with known meiotic TFs.
Cis regulation of H3K4me3 level
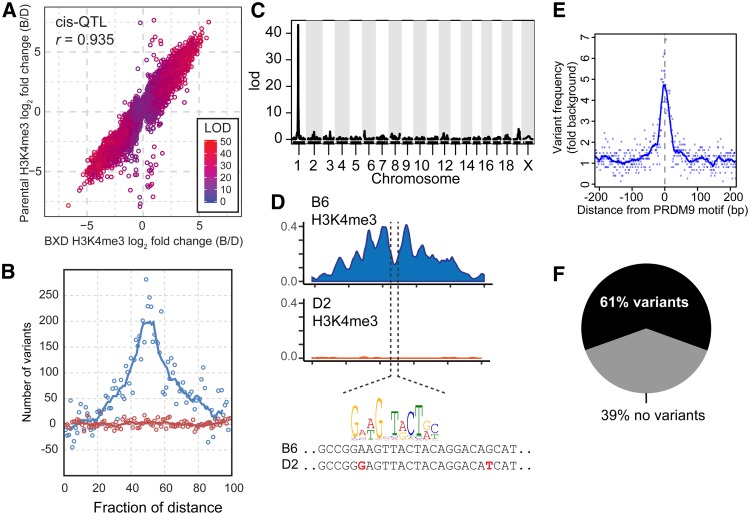
In addition to trans-acting hQTL, genetic variation of 8375 H3K4me3 peaks map back to their genomic locations. At each of these peaks, the difference in H3K4me3 level between BXD lines whose genotype at the peak is B6 vs. those with a D2 genotype are highly correlated with the differences in H3K4me3 level between the B6 and D2 parents (Figure 6A). Defining cis-regulated H3K4me3 sites as those whose hQTL was within 10 Mb, we found an increased frequency of sequence variants concentrated near the center of H3K4me3 peaks compared to trans-regulated sites (Figure 6B). Because sequence variants are shared among all cells, we next determined whether cis-acting variation in histone modification is shared between different cell types. As a conservative estimate, we selected H3K4me3 sites regulated by cis-hQTL in germ cells with LOD scores >10 (n = 4358), and compared their H3K4me3 levels to the differences in H3K4me3 levels between parental strains in ESCs, hepatocytes and cardiomyocytes (Table S7). Of the H3K4me3 peaks that overlap, only a few were significantly different in parental strains (FDR < 0.05) and had the same direction of effect (i.e., had high H3K4me3 levels when B6). This suggests that even though different cells will share the same genetic variants, the trans-acting factors that determine H3K4me3 level at these sites are cell-type specific.
Figure 6.
Local cis-hQTL are enriched for genetic variants within the H3K4me3 peak. (A) Correlation of the log fold change (B6/D2) between the founders and BXD lines for H3K4me3 levels at local-hQTL (points shaded by LOD score). (B) Number of background-subtracted genomic variants between B6 and D2 (SNPs and short indels) as a fraction of the distance across H3K4me3 peaks (blue is local-QTL, red is distal-QTL, solid lines indicate 5 bp moving average). (C) LOD plot for local-QTL for an example recombination hotspot on distal chromosome 1. (D) Top: coverage profile of H3K4me3 level (loess-smoothed RPM) for hotspot shown in C for both genetic backgrounds. Bottom: DNA sequence from nucleosome-depleted center of recombination hotspot showing variants between strains in red. Prdm9Dom2 motif is indicated above best-matching DNA sequence. (E) Enrichment of genomic variants between B6 and D2 plotted against distance from Prdm9 motif for all local-QTL at recombination hotspots (solid line indicates 10 bp moving average). (F) Percent of recombination hotspot local-QTL that have known genetic variants within the Prdm9 binding site.
Recombination hotspots undergo selection for motif-disrupting mutations in their PRDM9 binding sites (Myers et al. 2010; Baker et al. 2015). Cis-hQTL were significantly enriched for recombination hotspots compared to all peaks (Figure S5). An example is the locally controlled PRDM9-dependent H3K4me3 peak shown in Figure 1B. The nucleosome depleted region where PRDM9 binds in this H3K4me3 peak (Baker et al. 2014) contains a match to the PRDM9 consensus sequence in the B6 genome, but there are two potentially disruptive SNPs near the PRDM9 binding site in the D2 genome (Figure 6, C and D). Extending this observation, we found an increase in single nucleotide variants at PRDM9 binding motifs (Figure 6E). Out of the 1516 recombination hotspots with a cis-hQTL, 61% (n = 929) have a variant in proximity to the PRDM9 binding site (Figure 6F, 36 bp motif ± 4 bp) (Walker et al. 2015). That 39% of cis-regulated hotspots lack variants near their PRDM9 binding motif indicates the existence of additional mechanisms for cis control of hotspots. An appealing possibility is that these hotspots are in the vicinity of genetically variable DNA binding sites for other trans-regulatory proteins that do not differ genetically between B6 and D2.
Discussion
The totality of our data implies the existence of an extensive set of regulatory genes controlling chromatin modifications that are likely to be distinct from the enzymes that establish or remove methylation at H3K4me3. These genes, represented by trans-acting hQTL, control specific cohorts of chromatin sites, and act dominantly to suppress trimethylation of H3K4. Testing four diverse cell populations, we find cell-type-specific trans-regulation of H3K4me3. Several of the hQTL are genetically compound, suggesting they contain multiple regulatory genes. Two independent lines of evidence suggest that these chromatin regulatory genes have functional significance. They affect the likelihood that recombination hotspots can acquire the DSBs that give rise to genetic crossing over, and cohorts of H3K4me3 sites controlled by individual hQTL contain distinct sets of TF binding sites.
The broad extent of epigenomic regulation makes it likely that the activities of many trans-acting chromatin regulatory factors are themselves influenced by other chromatin regulatory factors, including both those that act positively, such as TFs, and those that act negatively, like the QTL described here. This creates a series of regulatory loops that interact to determine the overall chromatin state of a cell. In this case, the process of differentiation can be viewed as a progression of epigenomic states determined by feedback driven, consequential changes in expression of chromatin regulatory genes, until a final, stable homeostatic state is reached. In essence, this is simply putting a molecular face to Waddington’s original seminal concept of epigenetic canalization (Waddington 1957).
Distinctions among hQTL
Male germ cell hQTL modules differ among themselves in several respects. Module 3 is unique in that both loci act on the same cohort of H3K4me3 peaks, it does not modulate recombination hotspots at all, and is unidirectional in the sense that nearly all H3K4me3 peaks under regulation are high when the same allele of the hQTL is present. In contrast bidirectional hQTL are mixtures of high and low H3K4me3 peaks when the B6 (or D2) allele is present. In contrast to module 3, the other major hQTL modules are bidirectional, affect hotspots, and each controls a unique cohort of H3K4me3 peaks. Bidirectionality can be explained if the alleles of a chromatin regulatory gene differ in their DNA binding specificity, act at different stages of meiosis, or if the QTL is genetically compound. Cohort sharing and lack of influence on hotspots by module 3 can be explained if the gene products function together in a pathway that is only expressed at a stage of meiosis when recombination hotspots are not present, for example in spermatogonia.
Potential candidate genes
It is unlikely that the genes underlying the hQTL are enzymes that write or erase H3K4me3. First, many of the hQTL, including hQTL 4, 7b, 12a, and 13, regulate both recombination hotspots and conventional promoter and enhancer sites, locations where H3K4me3 is metabolized by independent enzyme systems. Second, all of the hQTL show great locational specificity, but the known writers or erasers of H3K4me3 are not known to possess DNA binding activity on their own. And third, the locations of our major hQTL did not coincide with the locations of any of the genes encoding these enzymes. H3K4 trimethylation is catalyzed by six SET domain proteins, SET1A, SET1B, and MLL1-4, which nucleate the COMPASS complexes containing ASH2L, RBBP5, WDR5, and DPY30 plus two to four additional proteins chosen from among a battery of possible accessory proteins (CFP1, WDR82, BOD1/1L, HCF1/2, NCOA6, MENIN, KDM6A, PSIP1, PAXIP1, and PA1) (Gu and Lee 2013). H3K4me3 at transcriptional regulatory sites can also be formed by the action of SMYD1, 2, and 3, ASH1L, SET7/9, and SETMAR. In addition to these writers, there are several known H3K4 demethylases of the LSD and JARID families (Gu and Lee 2013). Importantly, none of the genes encoding these proteins coincide with the locations of the six major QTL regions.
While we cannot rule out that the hQTL may modulate the abundance or activity of one of the above genes, this type of regulation does not provide an explanation for the observed locational specificities of our hQTL. Instead the properties of the identified hQTL, ability to suppress H3K4 methylation, and the physical location of the hQTL all make ZFPs, particularly KRAB-ZFPs, attractive candidates. In germ cells, there is a considerable coincidence in location between the observed hQTL and ZFPs. Most of the hundreds of ZFP genes occur in a small number of tightly linked clusters (Walker et al. 2015; Kauzlaric et al. 2017), and the major hQTL on chromosomes 4, 7b, 12a, and 13 all overlap such clusters (probability of overlap P = 1.7 × 10−5, binomial test), while the hQTL at chromosomes 7a and 12b each overlap a single zinc finger gene. Moreover, in the case of the chromosome 13 QTL, which is compound, each of the subregions overlap a different ZFP cluster. The presence of multiple ZFPs within an hQTL could help to explain bidirectionality as different ZFPs would affect different sites in the B6 and D2 genomes. Finally, the Rsl1 and Rsl2 genes provide precedents. The KRAB-ZFPs encoded by these genes repress expression of a variety of genes in female liver, but have no effect in the kidney (Krebs et al. 2003). At one target gene, Slp (Sex limited protein), RSL1 binds at an LTR-based enhancer upstream of the Slp promoter, where suppression correlates with methylation of CpG dinucleotides (Krebs et al. 2012). Incidentally, Rsl1 and Rsl2 are located in the distal chromosome 13 hQTL ZFP cluster, although we do not yet have sufficient evidence to rule these genes in or out as candidates in germ cells.
ZFPs make up the single largest class of genes in mammalian genomes (Ecco et al. 2017), and many possess secondary domains that nucleate chromatin-modifying complexes capable of closing chromatin. This is particularly true of KRAB-ZFPs, of which there are several hundred in both mice and humans (Ecco et al. 2017), and BTB-ZFPs, with ∼50 identified in both species (Stogios et al. 2005). Their zinc fingers provide DNA binding specificity, accounting for the locational specificity of H3K4me3 peaks regulated by hQTL, and their KRAB and BTB domains generally recruit chromatin-repressing complexes to these specific genomic locations, accounting for their dominant suppression of H3K4me3. KRAB domains frequently bind TRIM28/KAP1 (Friedman et al. 1996), leading to deacetylation of H3K27 and trimethylation of H3K9 (Ecco et al. 2017), two reactions that serve to repress or close chromatin. BTB domains can close chromatin by associating with histone deacetylase complexes including N-CoR and SMRT (Huynh and Bardwell 1998). Together these chromatin modifying features of ZFPs combine to establish cell-type specific gene regulatory networks (Krebs et al. 2012; Ecco et al. 2017; Yang et al. 2017). Furthermore, a recent report found a correlation between reduced levels of meiotic recombination and proximity to KRAB-ZFP binding sequences (Altemose et al. 2017). Combined, these properties of our identified hQTL suggest a model in which DNA-binding factors guide chromatin repressing complexes to specific genomic locations in a tissue-specific manner, thereby establishing major features of the epigenome.
Testing for trans-regulation of genomic features
Classically, testing for trans-regulation relies on asking whether chromosomal sites that are regulated differently in two parents behave the same in the heterozygous background of an F1 hybrid (Wittkopp et al. 2008). In attempting to apply this test to B6 and D2 mice, we found it limited by the paucity of informative SNPs between the two haplotypes. Instead, using the homozygous genome of an RI line, we can reliably conclude that a chromosomal site is subject to trans-regulation if its molecular phenotype does not correspond to the genetic origin of the DNA at that site. If an H3K4me3 peak in a BXD line is genetically B6 in origin but no longer has the phenotype seen in B6 mice, we conclude that it is under trans control by one or more distal hQTL, and similarly for sites of D2 origin. In practice this advantage more than compensated for the 50% efficiency resulting from the fact that the test is only positive when the distal QTL has the opposite genotype of the regulated H3K4me3 site.
Our hQTL mapping provided an opportunity to confirm the utility and accuracy of this test. hQTL mapping in 59 RI lines identified 4429 cis-hQTL that were also significantly different in the parental lines. Testing a single RI line, BXD99, over 95% (4226) of H3K4me3 sites showed similar phenotypes to their parents of origin and were correctly predicted to be under local regulation. Conversely, we identified 731 trans-hQTL in RI lines that showed significant parental differences and 43% of these were predicted as trans in BXD99, a value similar to the estimated 50% efficiency. Additionally, in the other three cell types tested, we see nearly 50% overlap in the identity of the H3K4me3 peaks predicted to be under trans-regulation between different BXD lines.
In summary, we have observed trans-regulation of the epigenome in four functionally very diverse cell types. ESCs are pluripotent, hepatocytes are capable of tissue regeneration and are endodermal in origin, cardiomyocytes are not capable of regeneration and are mesodermal in origin, and our germ cell preparations include a mixture of meiotic cell stages progressing from spermatogonial stem cells to early meiosis. Extrapolating these results, it seems likely that trans-regulation of the epigenome occurs in many mammalian cells, with significant implications for human health. An estimated 93% of mapped genetic variation affecting human health lies outside the coding regions of genes in functional elements (Maurano et al. 2012; Albert and Kruglyak 2015). Further, a recent survey of TF DNA binding domains in human populations identified extensive functional variation (Barrera et al. 2016), likely resulting in the type of variation in the epigenome we see between mouse strains. In this way chromatin regulatory loci, such as those identified as hQTL here, have the potential to subtly shape the activity of functional elements in trans and therefore influence the manifestation of complex traits.
Acknowledgments
This manuscript is dedicated to the memory of Pavlina Petkova, a wonderful scientist, colleague, and wonderful friend. We thank members of the Baker, Paigen, Petkov, and Carter laboratories for their discussion of the data and manuscript. This work was assisted by The Jackson Laboratory scientific services, which are supported through National Institutes of Health Cancer Core grant CA34196. BXD ESC lines were kindly provided by Anne Czechanski and Laura Reinholdt, funded by the Special Mouse Strain Resource OD011102-18. Funding for the work was provided by NIGMS F32-GM101736 and The Jackson Laboratory start-up funds supporting C.L.B., and P01-GM099640 to K.P. and G.W.C.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210262.
Communicating editor: J. Schimenti
Literature Cited
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Allis C. D., Caparros M. L., Jenuwin T., Reinberg D. (Editors), 2015. Epigenetics, Ed. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Altemose N., Noor N., Bitoun E., Tumian A., Imbeault M., et al. , 2017. A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis. eLife 6: e28383. 10.7554/eLife.28383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgustinova A., Benitah S. A., 2016. The epigenetics of tumour initiation: cancer stem cells and their chromatin. Curr. Opin. Genet. Dev. 36: 8–15. 10.1016/j.gde.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Petkov P. M., Paigen K., 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24: 724–732. 10.1101/gr.170167.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Kajita S., Walker M., Saxl R. L., Raghupathy N., et al. , 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11: e1004916 10.1371/journal.pgen.1004916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L., Fujiwara Y., Sun F., Hu J., Hibbs M. A., et al. , 2016. Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes. BMC Genomics 17: 628 10.1186/s12864-016-2865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera L. A., Vedenko A., Kurland J. V., Rogers J. M., Gisselbrecht S. S., et al. , 2016. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science 351: 1450–1454. 10.1126/science.aad2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 (erratum: Science 328: 690) 10.1126/science.1183439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan J. A., Duong M. T., Titus K. R., Zhou L., Cao Z., et al. , 2017. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27: 1139–1152. 10.1101/gr.215160.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. 10.1093/bioinformatics/btg112 [DOI] [PubMed] [Google Scholar]
- Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L. R., et al. , 2014. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 9: 559–574. 10.1038/nprot.2014.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G., Imbeault M., Trono D., 2017. KRAB zinc finger proteins. Development 144: 2719–2729. 10.1242/dev.132605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M. F., Ballestar E., Paz M. F., Ropero S., Setien F., et al. , 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 102: 10604–10609. 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., et al. , 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10: 2067–2078. 10.1101/gad.10.16.2067 [DOI] [PubMed] [Google Scholar]
- Greenfest-Allen E., Cartailler J.-P., Magnuson M. A., Stoeckert C. J., Jr., 2017. iterativeWGCNA: iterative refinement to improve module detection from WGCNA co-expression networks. bioRxiv 234062. DOI: 10.1101/234062. [Google Scholar]
- GTEx Consortium , 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213 (erratum: Nature 553: 530). 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Lee M. G., 2013. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 3: 39 10.1186/2045-3701-3-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K. D., Bardwell V. J., 1998. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17: 2473–2484. 10.1038/sj.onc.1202197 [DOI] [PubMed] [Google Scholar]
- Kasowski M., Kyriazopoulou-Panagiotopoulou S., Grubert F., Zaugg J. B., Kundaje A., et al. , 2013. Extensive variation in chromatin states across humans. Science 342: 750–752. 10.1126/science.1242510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauzlaric A., Ecco G., Cassano M., Duc J., Imbeault M., et al. , 2017. The mouse genome displays highly dynamic populations of KRAB-zinc finger protein genes and related genetic units. PLoS One 12: e0173746 10.1371/journal.pone.0173746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil P. P., Smagulova F., Brick K. M., Camerini-Otero R. D., Petukhova G. V., 2012. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 22: 957–965. 10.1101/gr.130583.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Waszak S. M., Gschwind A. R., Raghav S. K., Witwicki R. M., et al. , 2013. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 342: 744–747. 10.1126/science.1242463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyonari H., Kaneko M., Abe S., Aizawa S., 2010. Three inhibitors of FGF receptor, ERK, and GSK3 establishes germline-competent embryonic stem cells of C57BL/6N mouse strain with high efficiency and stability. Genesis 48: 317–327. [DOI] [PubMed] [Google Scholar]
- Krebs C. J., Larkins L. K., Price R., Tullis K. M., Miller R. D., et al. , 2003. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes Dev. 17: 2664–2674. 10.1101/gad.1135703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C. J., Schultz D. C., Robins D. M., 2012. The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration. Mol. Cell. Biol. 32: 3732–3742. 10.1128/MCB.00615-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C. J., Zhang D., Yin L., Robins D. M., 2014. The KRAB zinc finger protein RSL1 modulates sex-biased gene expression in liver and adipose tissue to maintain metabolic homeostasis. Mol. Cell. Biol. 34: 221–232. 10.1128/MCB.00875-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy I. V., Vorontsov I. E., Yevshin I. S., Sharipov R. N., Fedorova A. D., et al. , 2018. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46: D252–D259. 10.1093/nar/gkx1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J., Yamada S., Tischfield S. E., Pan J., Kim S., et al. , 2016. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167: 695–708.e16. 10.1016/j.cell.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T., 2015. Functional genomics bridges the gap between quantitative genetics and molecular biology. Genome Res. 25: 1427–1431. 10.1101/gr.190983.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire M., Zaidi S. H., Ban M., Ge B., Aissi D., et al. , 2015. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat. Commun. 6: 6326 10.1038/ncomms7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang X., Rukina D., Huang Q., Lin T., et al. , 2018. An integrated systems genetics and omics toolkit to probe gene function. Cell Syst. 6: 90–102.e4. 10.1016/j.cels.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Maegawa S., Gough S. M., Watanabe-Okochi N., Lu Y., Zhang N., et al. , 2014. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 24: 580–591. 10.1101/gr.157529.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. M., 2005. Epigenetic drift in aging identical twins. Proc. Natl. Acad. Sci. USA 102: 10413–10414. 10.1073/pnas.0504743102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., van de Geijn B., Degner J. F., Cain C. E., Banovich N. E., et al. , 2013. Identification of genetic variants that affect histone modifications in human cells. Science 342: 747–749. 10.1126/science.1242429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. 10.1126/science.1182363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld D. S., 1997. Isolation of rat liver hepatocytes. Methods Mol. Biol. 75: 145–151. [DOI] [PubMed] [Google Scholar]
- Pai A. A., Pritchard J. K., Gilad Y., 2015. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 11: e1004857 10.1371/journal.pgen.1004857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835 10.1126/science.1181495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce J. L., Lu L., Gu J., Silver L. M., Williams R. W., 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5: 7 10.1186/1471-2156-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., 2014. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94: 559–573 (erratum: Am. J. Hum. Genet. 95: 126) 10.1016/j.ajhg.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintisch C., Heinig M., Bauerfeind A., Schafer S., Mieth C., et al. , 2014. Natural variation of histone modification and its impact on gene expression in the rat genome. Genome Res. 24: 942–953. 10.1101/gr.169029.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. B., Hosler B. A., Gudas L. J., 1991. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113: 815–824. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., et al. , 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. 10.1038/nature01080 [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J., Perez-Palacios R., Climent M., Guallar D., Muniesa P., 2014. Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front. Oncol. 4: 14 10.3389/fonc.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., et al. , 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Marconett C. N., Duan J., Hyland P. L., Li P., et al. , 2014. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat. Commun. 5: 3365 10.1038/ncomms4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I. V., Brick K., Khil P., Camerini-Otero R. D., et al. , 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. 10.1038/nature09869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Prive G. G., 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6: R82 10.1186/gb-2005-6-10-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K., 2008. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24: 1461–1462. 10.1093/bioinformatics/btn209 [DOI] [PubMed] [Google Scholar]
- Sun F., Fujiwara Y., Reinholdt L. G., Hu J., Saxl R. L., et al. , 2015. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma 124: 397–415. 10.1007/s00412-015-0511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudt A., Colome-Tatche M., Johannes F., 2016. Genetic sources of population epigenomic variation. Nat. Rev. Genet. 17: 319–332. 10.1038/nrg.2016.45 [DOI] [PubMed] [Google Scholar]
- Taylor B. A., Heiniger H. J., Meier H., 1973. Genetic analysis of resistance to cadmium-induced testicular damage in mice. Proc. Soc. Exp. Biol. Med. 143: 629–633. 10.3181/00379727-143-37380 [DOI] [PubMed] [Google Scholar]
- Tian J., Keller M. P., Broman A. T., Kendziorski C., Yandell B. S., et al. , 2016. The dissection of expression quantitative trait locus hotspots. Genetics 202: 1563–1574. 10.1534/genetics.115.183624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M., Rogers S., Divi R. L., Schully S. D., Nelson S., et al. , 2014. Epigenetic research in cancer epidemiology: trends, opportunities, and challenges. Cancer Epidemiol. Biomarkers Prev. 23: 223–233. 10.1158/1055-9965.EPI-13-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H., 1957. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. G. Allen and Unwin, London. [Google Scholar]
- Walker M., Billings T., Baker C. L., Powers N., Tian H., et al. , 2015. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8: 31 10.1186/s13072-015-0024-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub A. S., Li C. H., Zamudio A. V., Sigova A. A., Hannett N. M., et al. , 2017. YY1 is a structural regulator of enhancer-promoter loops. Cell 171: 1573–1588.e28. 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 40: 346–350. 10.1038/ng.77 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329. 10.1038/nature10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Adams D. J., Flint J., Keane T. M., 2012. Next-generation sequencing of experimental mouse strains. Mamm. Genome 23: 490–498. 10.1007/s00335-012-9402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Wang Y., Macfarlan T. S., 2017. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 33: 871–881. 10.1016/j.tig.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., et al. , 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available through The Jackson Laboratory Catalog. Mouse ESCs are available upon request. Supplemental files include Tables S1–S8 and Figures S1–S6. Raw sequencing data and processed files, including estimates of chromatin modification for each strain and peak location, are available at the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) available under accession number GSE113192. DMC1 ChIP-Seq data for B6 is available under accession number GSE108259. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210262.